Abstract
Influenza A viruses are zoonotic pathogens that continuously circulate and change in several animal hosts, including birds, pigs, horses and humans. The emergence of novel virus strains that are capable of causing human epidemics or pandemics is a serious possibility. Here, we discuss the value of surveillance and characterization of naturally occurring influenza viruses, and review the impact that new developments in the laboratory have had on our understanding of the host tropism and virulence of viruses. We also revise the lessons that have been learnt from the pandemic viruses of the past 100 years.
Influenza A viruses constantly circulate in many animal hosts, such as humans, birds, horses, dogs and pigs. Seasonal influenza virus infections in humans cause annual epidemics that result in millions of human infections worldwide and have significant health and economic burdens1; influenza pandemics can also have devastating effects globally, resulting in millions of deaths2. Influenza A virus has a segmented genome of eight single-stranded negative-sense RNA molecules that typically encode 11 or 12 viral proteins3, including N40, a newly identified protein that is expressed from the PB1 segment4 (FIG. 1 a). It is well known that simultaneous infection of a single cell by two distinct influenza A viruses can lead to gene mixing, or reassortment, which can result in the generation of a novel influenza virus strain, and it is believed that most human pandemic viruses arose in this manner.
Figure 1 ∣. Replication and antigenic classification of influenza A viruses.
a ∣ The influenza A virus genome consists of eight single-stranded RNAs that encode 11 or 12 proteins. These are nuclear export protein (NEP; also known as NS2) and the host antiviral response antagonist non-structural protein 1 (NS1), which are encoded by the NS segment; the matrix protein M1 and the ion channel M2, which are encoded by the M segment; the receptor-binding protein haemagglutinin (HA), the sialic acid-destroying enzyme neuraminidase (NA), nucleoprotein (NP), and the components of the RNA-dependent RNA polymerase complex (PB1, PB2 and PA), all expressed from their respective genome segments; and the newly identified N40 protein, which is expressed from the PB1 segment and has an unknown function4. In addition, some viruses express the pro-apoptotic protein PB1-F2, which is encoded by a second ORF in the PB1 segment. Within the virion, each of the eight viral segments forms a viral ribonucleoprotein (RNP) complex: viral RNA is wrapped around NP, and this structure is then bound to the viral polymerase complex. b ∣ The antigenic properties of HA allow the classification of influenza A viruses into two major groups, 1 and 2, which are further classified into five clades and 16 subtypes. c ∣ In the initial stages of influenza A virus replication, the viral HA attaches to host cell receptors that contain terminal α-2,6-linked or α-2-3-linked sialic acid (α-2,6-SA or α-2,3-SA) moieties, and the virus enters the cell by receptor-mediated endocytosis. Cleavage of HA by cellular proteases is required to expose the HA peptide that is responsible for the fusion between the viral envelope and the endosomal membrane (see below). Acidification of the endocytic vesicle opens the M2 ion channel, resulting in acidification of the inside of the virion, a process that is required for proper uncoating of the RNP complexes that contain the viral genome. Acidification of the endosome also triggers the pH-dependent fusion step that is mediated by HA and results in the cytoplasmic release of the RNP complexes. These translocate to the nucleus, where the RNA-dependent RNA polymerase transcribes and replicates the negative-sense viral RNA ((−)vRNA), giving rise to three types of RNA molecules: the complementary positive-sense RNA ((+)cRNA), which it uses as a template to generate more vRNA; negative-sense small viral RNAs (svRNAs), which are thought to regulate the switch from transcription to replication153,154; and the viral mRNAs, which are exported to the cytoplasm for translation. Viral proteins that are needed in replication and transcription are translocated back to the nucleus, and progeny RNPs are then exported to the cytoplasm for packaging, assisted by M1 and NEP. Viral HA, NA and M2 are transported by the trans-Golgi secretory pathway, and the mature proteins arrive at the plasma membrane, where M1 assists in the formation of virus particles. Budding then occurs, and release from the host cells is mediated by the neuraminidase activity of NA, which destroys the SA of the cellular and viral glycoproteins that would otherwise retain the new virions at the cell surface.
Influenza A viruses can be subtyped according to the antigenic properties of their haemagglutinin (HA) and neuraminidase (NA) glycoproteins. HA has an important role in determining host tropism, as it binds to host cell receptors that contain terminal α-2,6-linked or α-2,3-linked sialic acid (α-2,6-SA or α-2,3-SA) moieties. It also contains a cleavage site that must be cleaved by host cell proteases. The amino acid sequence of this cleavage site modulates tissue tropism and systemic spread, affecting disease severity (discussed below). The neuraminidase activity of NA is crucial for destroying the SA-containing receptors of the host and viral membranes, a process that is required for proper budding and release of progeny virions from the host cell surface. Currently, there are virus strains from 16 HA subtypes and nine NA subtypes circulating in birds, and strains from two virus subtypes circulating in humans: H1N1 and H3N2 (H2N2 strains were also circulating in humans from 1957 to 1968). Overall, the HA subtypes are classified into two groups (or lineages) based on their antigenic properties and their major structural features5-8 (FIG. 1b). Group 1 encompasses the H1a, H1b and H9 clades, which include the H1 subtype that contains both the 1918 and 2009 pandemic H1N1 strains and the human seasonal H1N1 strains, and the H5 HA subtype that includes the highly pathogenic avian influenza (HPAI) H5N1 strains. Group 2 consists of the H3 and H7 clades, which contain the human H3N2 strains and the HPAI H7N7 strains, respectively (FIG. 1b). Antigenic evolution of human seasonal influenza A viruses occurs through antigenic drift and is characterized by the seasonal selection of new strains containing amino acid changes in HA and NA. These changes partially overcome pre-existing immunity in humans, and these new strains are largely responsible for seasonal influenza epidemics. More dramatic changes in HA subtype resulting from antigenic shift have traditionally been associated with the emergence of pandemic viruses9, although this notion has been challenged by the 2009 H1N1 pandemic (discussed below). Therefore, HA not only has a crucial role in the influenza virus life cycle (FIG. 1c) but also, through variations in its genotype, is a determinant of host susceptibility and pathogenesis.
The zoonotic origin of human influenza pandemics is well described9 and is highlighted by the increasing number of lethal human infections with HPAI H5N1 viruses that have spread in domestic birds throughout parts of East and Southeast Asia, the Middle East, Africa and Europe. Fortunately, these antigenically novel viruses have yet to sustain human–human transmission and have therefore failed to generate a potentially devastating human pandemic. By contrast, and as a surprise to the influenza virus research community, a novel H1N1 swine origin influenza virus (SOIV) emerged in 2009 to produce the first human influenza pandemic of the twenty-first century. Within 1 year, this virus spread to 214 countries and caused >18,000 confirmed deaths worldwide10. It is estimated that, by April 2010, 43 million to 89 million people had been infected with this virus in the United States alone11.
As the 2009 pandemic H1N1 SOIV spread, modern-day surveillance systems, as well as recently established experimental tools and animal models, were rapidly deployed to characterize its genomic sequence and pathogenicity and to investigate its transmission competence, antigenic characteristics and antiviral susceptibility. Outstanding collaborative work from many laboratories worldwide ensured that the pandemic was dealt with as swiftly as possible. During this period, it was apparent that progress in several areas of current influenza virus research greatly enhanced our ability to react quickly. However, it was also apparent that the pandemic caught the world off guard and that specific aspects of the pandemic-preparedness plans are still in need of improvement. This Review discusses the recent advances and future needs for comprehensive and systematic animal and human surveillance, improved assessment of the virulence of novel strains in humans and a better understanding of viral tropism and of the underlying mechanisms of pathogenesis.
Influenza A strains of the twenty-first century
The pandemic potential of the HPAI H5N1 virus, as well as the numerous outbreaks in wild birds with viruses of the H5, H7 and H9 subtypes in the past decade12,13, has prompted the surveillance of influenza viruses in avian species in several regions around the world14,15. However, the emergence of the 2009 H1N1 pandemic from pigs revealed the lack of systematic surveillance of other susceptible hosts. Large-scale surveillance efforts have been aided by the development of several key technologies, including high-throughput and deep-sequencing techniques (which have been used to obtain full viral genome sequences from field and clinical isolates), dedicated sequence databases and sophisticated phylogenetic and coalescent analysis tools. These tools are allowing faster, more comprehensive epidemiological studies of influenza viruses in human and natural reservoirs.
Surveillance of avian viruses.
HPAI H5N1 viruses probably arose from mutations in the HA cleavage site through the introduction of a low-pathogenic avian H5N1 virus from wild birds into domestic birds16. Several major outbreaks of HPAI H5N1 viruses in domestic birds have occurred, and the first human case of HPAI H5N1 infection was documented in 1997 (REF 17). Overall, 562 human cases from 15 countries, with a fatality rate of ~59% (329 deaths), have been reported to the WHO18, and so far most cases have been associated with direct human contact with infected avian species. Surveillance of wild birds is key to understanding the origin, pathogenesis, evolution and global spread of these viruses. Since 2002, genotype Z (which contains small deletions in the genes encoding the NA and NS1 proteins) has been the predominant H5N1 genotype in southern China19. Nonetheless, ten main distinct clades of the HPAI H5N1 virus have been identified to date20,21, demonstrating the complex and dynamic evolution of these viruses in nature. Comprehensive genomic studies of a diverse collection of avian influenza viruses (AIVs) demonstrated that the greatest variability lies in the HA, NA and NS1 proteins15, and led to the discovery of a putative PDZ domain ligand at the carboxyl terminus of NS1 (REF 15) that might be involved in virulence22. Surveillance of North American wild birds indicated that Eurasian and American strains rarely mix and detected no evidence of HPAI H5N1 viruses14. However, AIV isolates from the United States demonstrated a high rate of reassortment rates among circulating strains, although no clear association patterns among RNA segments were found (that is, there seems to be no requirement for a pair or group of segments to be reassorted together). This is suggestive of a constant mixing of AIV genomes rather than the spreading of a restricted and stable set of segments that characterizes mammalian-adapted influenza A viruses23.
In 2005, an HPAI H5N1 virus was responsible for an outbreak in waterfowl in Qinghai Lake, western China, resulting in high bird mortality and raising concerns of the potential for sustained transmission of HPAI H5N1 viruses among migratory birds24. Subsequently, genetically and antigenically distinct HPAI H5N1 clades have emerged throughout a broad geographical range in Southeast Asia and have become endemic in domestic poultry25,26. More recently, HPAI H5N1 viral lineages have emerged through reassortment with the endemic viruses that are present in local aquatic poultry, leading to the selection of viruses that are capable of infecting multiple avian hosts and allowing transmission to other geographical regions16. Both transport of poultry and bird migration appear to have important roles in the spread of HPAI H5N1 viruses over long distances, and possibly explain the outbreaks in Europe, the Middle East and Africa. Nonetheless, the lack of further reassortment of these viruses after they have been exported out of China indicates that different factors affecting the epidemiology of AIVs may exist in other areas of the world16.
In addition to outbreaks in wild birds and human disease caused by HPAI H5N1 viruses, the past two decades have seen outbreaks in poultry12,13 and also zoonotic infection of humans with viruses of the H7 (REFS 27,28) and H9 (REFS 29,30) subtypes in Europe, Asia and the Americas31,32. These outbreaks have motivated enhanced surveillance efforts to better understand the ecology, genomic characteristics and global circulation of these and other AIVs12,13. A large-scale phylogenetic analysis of the H9N2 viruses revealed marked geographical and host-specific patterns that reflect the complex evolution of these viruses33. In southern China, the long-term establishment of multiple AIV lineages, such as the H9N2 and H5N1 lineages, is thought to contribute to the high level of reassortment that is seen in this region, thus giving rise to the great genetic diversity of these viruses34. Interestingly, the genes encoding the internal proteins (PB2, PB1, PA, NP, M and NS) of H9N2 viruses have been found to be similar to those of the AIV and HPAI H5N1 viruses that were isolated from humans during 1997 (REFS 30,33), whereas the HA protein of the H9N2 viruses that are endemic to China, and were surveyed throughout 2007, appear to have undergone positive selection for about 13 years rather than undergoing reassortment35. Extensive segment reassortments have also been observed within the H7 viruses, which have multiple NA subtypes that are associated and maintained with specific H7 HA proteins throughout different geographical regions (for example, in Australia H7 viruses form a monophyletic clade based on their HA protein, which can combine with N2, N3, N4, N6 and N7 subtypes)36. Recent H7 viruses that were isolated through surveillance of wild birds in Europe were closely related to those H7 viruses that caused outbreaks in poultry in Italy (1999–2000) and the Netherlands (2003)37, highlighting the value of systematic surveillance efforts for detecting avian viruses that are potential threats to humans.
Molecular epidemiology of human seasonal viruses.
The seasonal H1N1 and H3N2 viruses have been cocirculating in humans since 1977. Although both H1N1 and H3N2 subtypes have distinct evolutionary dynamics, with the H1N1 subtype drifting at a slower rate38, they each undergo frequent reassortment among the lineages within their subtype39-41. Thus, multiple lineages co-circulate, and random intra-lineage reassortment contributes to the overall viral genetic pool that is present in a season40. Large-scale analysis of H1N1 and H3N2 virus sequences led to the ‘sink–source’ model to explain the origin of annual seasonal strains. This model proposes that a population in the tropics undergoes strong antigenic selection and serves as the ‘source’ for influenza virus epidemics, such that viruses are exported linearly from this source to ‘sink’ populations in the Northern and Southern Hemispheres38. The strongly unidirectional nature of global epidemics was also shown by genetic and antigenic analysis of the 2002–2007 seasonal H3N2 viruses42. This study showed that overlapping epidemics in East and Southeast Asia generate a continuous circulation of H3N2 influenza viruses within this region, from which viruses are then seeded (through travel and trade) to Oceania, North America, Europe and, subsequently, South America. Hence, close surveillance of influenza viruses in East and Southeast Asia may help to determine the antigenic characteristics of the viruses that might circulate later in other parts of the world42. In agreement with these findings, the emergence of influenza viruses that cause seasonal epidemics is largely influenced by the global migration of viruses and is not a result of latent influenza viruses within the host being reactivated during winter43. Dry, cold, wintery conditions appear to contribute to the efficient transmission and spread of influenza viruses44. However, the 2009 pandemic H1N1 virus emerged during spring in the Northern Hemisphere, spread during summer and produced a larger second wave of infections that peaked in early autumn45, indicating that although climate conditions influence the epidemiology of influenza A virus, its transmission and spread can occur efficiently in naive populations regardless of seasonality, and that this spread can be modulated by other factors such as close gathering of the susceptible population46,47.
Emergence of the 2009 pandemic H1N1 influenza virus.
Despite an increased focus on surveillance of HPAI H5N1 viruses in Southeast Asia, the emergence of an H1N1 pandemic SOIV in April 2009 was largely unexpected. Fortunately, several years of coordinated international efforts to avoid a potential H5N1 pandemic allowed the prompt detection and continuous surveillance of the novel pandemic H1N1 strain as it spread worldwide (BOX 1). Unprecedented efforts using modern epidemiological and molecular tools allowed the rapid characterization of human transmission rates48 and the determination of the pathogenic potential48 and the sequence and origin49,50 of the novel H1N1 virus. However, the accuracy and value of evaluating pathogenesis early during a pandemic remains a controversial issue. For instance, the limited epidemiological data that were available at the beginning of the outbreak in Mexico led to an overestimation of the severity of the novel pandemic virus48. Nonetheless, early genetic and evolutionary analyses revealed that this virus contains a complex set of genes which originate from the viruses that infect birds, humans and pigs49,50 (FIG. 2), and that it was likely to be derived from viruses which had been circulating in swine populations, undetected, for approximately one decade50. The emergence of the 2009 pandemic H1N1 virus thus underscored the importance of animal and human surveillance in understanding and responding to emerging and re-emerging zoonotic pathogens.
Box 1 ∣. Molecular epidemiology of the 2009 H1N1 influenza A virus pandemic.
The 2009 H1N1 influenza A virus possibly emerged in April 2009 through a single introduction into humans49 in North America (most likely in Mexico)147. Its genome has none of the previously known markers of human adaptation, and its predecessors possibly circulated undetected in pigs for some time, suggesting that reassortment of the precursor swine lineages probably occurred years before the detection of the virus in humans49,50. Early epidemiological data indicated that the basic reproduction number (R0; a measure of human–human transmission) was approximately 1.2, similar to the lower-end R0 estimates of previous influenza pandemics, and in children <15 years old the clinical attack rate was double that in adults48. Antigenic analysis confirmed the similarity of this virus to swine viruses circulating in North America and its difference to human seasonal H1N1 viruses49. Shortly after worldwide outbreaks began, the dynamic spread of the virus was rapidly tracked and mapped147-149. Outbreaks were characterized by multiple regional introductions of the novel pandemic strain150. The global geographical spread occurred in three major stages: first, the spread from Mexico to the United States; second, sustained transmission in North America and an initial spread to other parts of the world; and third, continuous global spread and secondary outbreaks outside the Americas147,149. Coalescent-based studies demonstrated that the virus circulated in the human population for a period of up to 3 months before it was first identified (the date of the original human infection (that is, the date of the most recent ancestor) was estimated to have been between 29 December 2008 and 22 February 2009)147,148, with its spread resembling the epidemiological dynamics of seasonal influenza viruses38. Of note, during the 2009 pandemic multiple introductions of the virus into domestic pigs were detected in several parts of the world, and a study in Hong Kong reported the presence of a swine reassortant isolate containing an NA protein similar to that of the human pandemic H1N1 virus151. A comprehensive longitudinal study conducted in southern China showed that extensive reassortment among circulating swine and human-avian lineages (such as those shown in FIG. 1b) led to the emergence of antigenically and genetically diverse swine influenza viruses in around 2007 (REF. 152). The potential for further reassortment of the 2009 pandemic H1N1 virus warrants further systematic and comprehensive surveillance of pigs worldwide to characterize and detect circulating viral strains with a potential risk for humans.
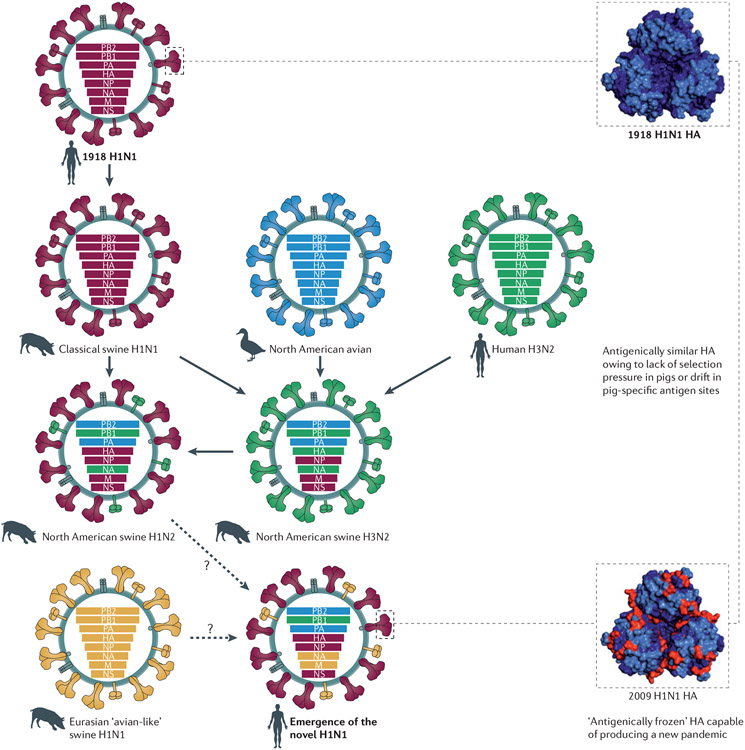
Figure 2 ∣. Emergence of an ‘antigenically frozen’ 2009 pandemic H1N1 virus.
Influenza viruses similar to the 1918 pandemic H1N1 virus became established in domestic pigs between 1918 and 1920; this lineage is referred to as the classical swine lineage. In 1979, a distinct Eurasian ‘avian-like’ H1N1 virus emerged in European pigs and has since co-circulated with the classical swine H1N1 viruses. Triple-reassortant swine origin influenza virus (SOIV) H1 viruses of different strains and subtypes (for example, H3N2 and H1N2) emerged and became predominant among North American pig herds in the 1990s. All of these viruses provided the genetic pool for the genesis of the 2009 pandemic H1N1 SOIV, possibly owing to further reassortment in pigs. Thus, the 2009 pandemic H1N1 virus is composed of PB2 and PA segments from North American avian viruses, the PB1 segment of the human H3N2 viruses, haemagglutinin (HA; of the H1 subtype), nucleoprotein (NP) and NS segments derived from classical swine H1N1 viruses, and the neuraminidase (NA; of the N1 subtype) and M segments of Eurasian ‘avian-like’ swine viruses. Sequence and antigenic analyses of the 2009 pandemic H1N1 virus show that there are similarities between the HA of this virus and that of the 1918 and human H1N1 viruses that circulated sometime between 1918 and the 1950s. The antigenic similarities between the 1918 and 2009 pandemic H1N1 viruses are represented in the crystal structure models of the trimeric configuration of the HA protein globular head, as seen from a top view. The antigenic sites of the HA proteins are shown in light blue, non-antigenic sites are shown in dark blue. The sites that differ between the 1918 and 2009 HA proteins are depicted in red.
Novel concepts in host tropism
Most influenza virus subtypes are restricted to specific hosts, but some seem to be more promiscuous and circulate in several species (for example, H1N1 and H3N2 viruses are endemic in humans, birds and pigs). Since 1918, the H1N1, H2N2 and H3N2 subtypes have initiated influenza pandemics in humans9. The fact that only sporadic infections have occurred (in humans who were in direct contact with avian species infected with HPAI H5N1 viruses) emphasizes the notion that host factors restrict influenza virus infection in new species (reviewed in REF. 51). Nonetheless, the 2009 H1N1 pandemic and previous pandemics are reminders that certain viruses can readily bypass host restriction barriers.
Tissue tropism and receptor specificity.
The HA proteins of the human seasonal H1 and H3 virus subtypes mainly recognize receptors with terminal α-2,6-SA moieties, which are found on bronchial epithelial cells of the human upper respiratory tract (URT)52,53. By contrast, AIVs bind predominantly to galactose linked to α-2,3-SA54, which is found abundantly on epithelial cells in the intestine of birds and in the lower respiratory tract (LRT) of humans55,56 (FIG. 3). Pigs have receptors containing both α-2,3-SA and α-2,6-SA in their trachea and have therefore been proposed as a ‘mixing vessel’ (REF. 57) for the reassortment of human and avian viruses, leading to the potential generation of pandemic viruses58. Similarly, pheasants, turkeys, quail and guinea fowl contain both receptor types in their respiratory tract and intestine59,60, so may also serve as mixing vessels. Interestingly, the H1N1 SOIV responsible for the 2009 pandemic has been reported to bind to α-2,6-SA and, to a limited extent, to α-2,3-SA61-63, and can infect cells of the URT and LRT64,65. Binding to the LRT is thought to induce the viral pneumonia that is seen in individuals infected with HPAI viruses and occurred in some severe cases from the 2009 pandemic. However, HPAI H5N1 viruses can also infect and replicate in cells of the nasopharyngeal and oropharyngeal epithelia, and thus might use other receptors to infect cells of the URT66. The specificity of avian viruses for α-2,3-SA-containing receptors, which in humans are mainly present in the LRT, probably contributes to the limited avian–human viral transmission, although exchange of the HPAI H5N1 virus surface glycoproteins with those of a URT transmission-competent seasonal virus did not confer transmissibility67. Hence, in addition to HA–receptor specificity, other viral, host and environmental factors44 probably influence the fitness and transmission of influenza viruses in different hosts.
Figure 3 ∣. Influenza A virus tropism.
The anatomical expression patterns of the viral receptors in different hosts restricts infection with and replication of influenza A viruses. The swine trachea contains receptors with α-2,3-linked and α-2,6-linked sialic acid (α-2,3-SA and α-2,6-SA) moieties that allow for binding of both avian and human viruses, leading to the idea that pigs can serve as the ‘mixing vessel’ (REF 57) in which reassortment of human and avian viruses can occur. Avian viruses bind preferentially to α-2,3-SA, which is found on receptors in the gut and respiratory tract of birds. By contrast, human-adapted viruses (for example, seasonal H1N1, H3N2 and 2009 pandemic H1N1 viruses) have a higher affinity for α-2,6-SAs, which are expressed in the upper respiratory tract of humans. Human infection with a non-human-adapted virus is rare and is usually a result of a direct spillover transmission event. Viral proteins and their specific residues that affect receptor binding and have been established as adaptations to the human host are listed; H1, H3 and H5 are variations of the haemagglutinin (HA) protein, and PB2 is an RNA-dependent RNA polymerase component.
Nevertheless, the specificity and affinity of the viral HA for its receptor is one of the crucial determinants of host tropism and transmission (FIG. 3). For example, the airborne transmission of the 1918 pandemic H1N1 influenza virus in a ferret model is modulated by amino acids at positions 190 and 225 in HA (H3 numbering is used for standardization). D225G variants have decreased α-2,6-SA-binding affinity52,68, resulting in reduced attachment to goblet cells of the human trachea (which express α-2,6-SA-containing receptors)52,68 and decreased transmission in ferrets69. The D225G variant in combination with the D190E change, which matches the consensus amino acids that are found in avian H1N1 strains, results in a binding preference for receptors containing α-2,3-SA52. Although this virus can replicate efficiently in the ferret URT, its transmissibility is abolished69. Efficient human–human transmission of HPAI H5N1 viruses has not occurred efficiently in nature or in experimental mammalian models of transmission67,70. Therefore, specific adaptations might be needed for efficient infection and transmission of HPAI viruses in humans. Mutations G225D and E190D reduced the avidity of an HPAI H5N1 virus to α-2,3-SA, but α-2,6-SA specificity was not favoured71, indicating that different residues are responsible for receptor-binding specificity in H5N1 viruses72,73 (FIG. 3; TABLE 1). Similarly, different residues located around the receptor-binding site have been implicated in the receptor-binding specificities of other avian71,74 and pandemic viruses75,76 (TABLE 1), indicating that receptor specificity is differentially modulated in diverse HA subtypes. Importantly, isolates of H1N1 SOIV viruses from the 2009 pandemic that were found to contain a D225G mutation have been associated with severe human disease and death77,78. These isolates show increased α-2,3-SA binding, so this mutation confers dual receptor specificity79,80. The ability of the H1N1 SOIV responsible for the 2009 pandemic to partially bind α-2,3-SA61-63 contrasts with the seasonal human H1N1 viruses that emerged previous to 2009, which bind predominantly to α-2,6-SA and possibly reflect years of human adaptation. Interestingly, these human viruses have higher affinity for long than for short sugars containing α-2,6-SA53, suggesting that glycan topology may also modulate the binding affinity of HA and viral receptor adaptations. The slight differences in receptor-binding specificity of the 2009 pandemic strain might partly explain the increased replication, transmission and pathogenesis that were observed in animal models for this virus compared with seasonal viruses64,79,81,82. Of note, the severity of the disease induced by the 2009 pandemic H1N1 virus in the general human population was not drastically different to that seen with seasonal influenza, suggesting that additional factors, such as pre-existing immunity and host adaptations (discussed below), modulate the pathogenic potential of influenza A viruses in humans.
Table 1 ∣.
Molecular virulence markers and pathogenic determinants of influenza A viruses
| Virulence marker and pathogenic determinant |
Pandemic virus | HPAI virus | Contemporary seasonal virus |
|||||
|---|---|---|---|---|---|---|---|---|
| 1918 H1N1 | 1957 H2N2 | 1968 H3N2 | 2009 H1N1 | H5N1 | H7N7 | H1N1 | H3N2 | |
| HA (binding and fusing with the host cell; antigenic determinant) | ||||||||
| Sialic acid linkage specificity | α-2,6* | α-2,6‡ | α-2,6‡ | α-2,6 and α-2,3§ | α-2,3 | α-2,3∥ | α-2,6 | α-2,6 |
| Residues involved in binding specificity¶ | D190 and D225 | Q226 and N186, or L226 and S228 | L226 and S228 | D190 and D225§; K133# K145#and K222# | G143, T160, N186, Q196, Q226 and G228 | Q226** | D190 and D225 | L226‡‡ and S228 |
| Multibasic cleavage site | No | No | No | No | Yes | Yes | No | No |
| PB1-F2 (induction of apoptosis; promotion of secondary bacterial infection) | ||||||||
| S66 (associated with increased virulence), N66 or truncation | S66 | N66 | N66 | Truncation | S66 or N66§§ | N66 | Truncation | N66 |
| PB2 (temperature-dependent replication competence) | ||||||||
| Adaptation to mammalian hosts | K627 | K627 | K627 | R591 | K627 or N701∥∥ | K627 or N701∥∥ | K627 | K627 |
| NS1 (host antiviral response antagonist) | ||||||||
| PDZ domain-binding motif¶¶ | KSEV | RSKV | RSKV | Truncated | ESEV or EPEV; ~3.3% of H5N1 isolates in 2003 were truncated | ESEV | RSEV | RSKV |
| CPSF30 binding | Yes | Yes## | Yes## | No | Yes## | Yes*** | Yes | Yes |
| E91 associated with virulence | No | No | No | No | Yes‡‡‡ | No | No | No |
| M2 (ion channel) | ||||||||
| Resistant to adamantanes (presence of N31) | No | No | No | Yes | Yes and no§§§ | No | No | Yes |
| NA (receptor-destroying enzyme) | ||||||||
| Resistant to oseltamavir (presence of Y275 and S294) | No | No | No | No∥∥∥ | No∥∥∥ | No | Yes | No |
CPSF30, cleavage and polyadenylation specificity factor 30 kDa subunit; HA, haemagglutinin; HPAI, highly pathogenic avian influenza; NA, neuraminidase; NS1, non-structural protein 1. *Some 1918 virus variants have differential receptor binding, allowing dual α-2,6-linked sialic acid (α-2,6-SA) and α-2,3-SA specificity (see main text for details). ‡Some early human virus isolates contained the α-2,3-SA-binding residues Q226 and Q228. §Residues D190 and D225 are needed for α-2,6-SA binding. Binding to α-2,3-SA is limited; however, the D225G amino acid mutation that is found in a small subset of 2009 pandemic H1N1 isolates from some severe cases confers an increased α-2,3-SA-binding specificity. ∥H7 viruses also show moderate binding to α-2,6-SA owing to a K193 residue. ¶For the HA protein, amino acid positions are according to the H3 numbering. #Part of a positively charged ‘lysine fence’ at the base of the receptor-binding site (identified through structural prediction models) that allows binding to α-2,3-SA and compensates for the lack of the avian E190. **Predicted from amino acid sequence analysis and from a crystal structure of an H7N3 HA in complex with receptor analogues. ‡‡Recent viruses contain either a V (from years 1996–2002) or I (from years 2003–2011) at position 226. §§Natural virus variants can contain either S or N. ∥∥Found in a mouse-adapted H7N7 virus and in some avian H7N7 isolates, and in some avian and human H5N1 isolates. ¶¶Binding to the PDZ domain, a common structural domain of 80–90 amino-acids that is found in the signalling proteins of diverse organisms, has been implicated in virulence: (K/E)(S/P)EV is a strong binding motif, RS(E/K)V displays weak or no binding. ##Some variants have weaker binding. ***Predicted binding; not confirmed experimentally. ‡‡‡Found in viruses that were isolated during the 1997 H5N1 outbreak. §§§Avian and human H5N1 isolates containing the resistant N31 residue or the sensitive S31 residue have been isolated and exist in nature. ∥∥∥ Resistant viruses may occasionally emerge in patients undergoing treatment or prophylaxis.
Replication competence.
Receptor affinity alone does not guarantee successful infection and replication in the host83, as overall viral fitness is crucial for influenza virus growth. The viral polymerase confers host-specific adaptations that enhance replication efficiency. The K627 residue of the RNA-dependent RNA polymerase protein PB2 has long been recognized as a host range determinant that confers the ability to infect humans84 and is present in most human H1N1 and H3N2 viruses but not in many avian viruses. Viruses containing the K627 residue can grow at 33 °C and replicate efficiently in the URT of mice85, indicating that they can readily replicate in the URT of humans (FIG. 3). K627 has been correlated with enhanced virulence of human HPAI H5N1 isolates and was found in a fatal human case of infection with HPAI H7N7 virus during an outbreak in the Netherlands in 2003 (REF. 86). Viruses that were isolated from birds during the 2005 Qinghai Lake outbreak in China also possessed a K627 substitution24, indicating that this residue can evolve in nature without previous selection in humans. A D701N mutation in PB2 has also been implicated in the adaptation of AIVs to growth in mammalian cells87,88 and has been shown to modulate transmission in the guinea pig70,73 and ferret89 models. Surprisingly, the 2009 pandemic H1N1 strain can efficiently replicate and transmit, and can even outcompete the seasonal human-adapted strains90, despite its PB2 having neither the K627 nor the N701 adaptations. It was recently established that an R591 residue, which is present in the 2009 pandemic strain, can confer efficient replication in mammals and compensates for the lack of the K627 and N701 adaptations91,92. Other recent studies have shown that HA, NA, PB2 and PA (another RNA-dependent RNA polymerase protein) contribute to the replication competence of H7N7 viruses in human cells86. Similarly, human-adapted PB2 and HA proteins were found to confer replication competence and transmissibility to some AIVs in the ferret model89. Thus, adaptations in the polymerase proteins and in HA allow successful viral binding to, entry into and replication in the relevant human cells of the respiratory tract.
Host factors that influence influenza virus infection.
Recent genome-wide approaches have identified various host factors that are required for efficient influenza virus replication93-97 (reviewed in REF. 98). These include host factors that are involved in viral fusion and uncoating; transport of viral RNP complexes into the nucleus; replication, transcription and translation of the viral genome; export of viral RNP complexes from the nucleus; and viral assembly and budding. Knockdown of several of the required host factors substantially reduced viral infection rates. For example, the vacuolar ATPase ATP6V0D1, which is involved in the endocytosis pathway, is required for influenza virus entry, and small interfering RNA-mediated depletion of ATP6V0D1 mRNA in human HEK-293 cells specifically decreased the replication of H1N1 and H5N1 viruses93. Other proteins that were found to be required for optimal influenza A virus replication were: CAMK2B, a calcium sensor that is expressed ubiquitously and implicated in the regulation of several cell processes; CDC-like kinase 1 (CLK1), which is involved in regulation of alternative splicing in mammalian cells; and p27 (also known as CDKN1B), a cell cycle regulator94,96. A different study identified several physical and regulatory interactions between viral proteins and host factors that map to novel cellular pathways and that affect influenza virus replication. For instance, deletion of WNT pathway components increased viral replication and reduced interferon (IFN) production, indicating that this pathway regulates influenza virus infection through as-yet-unknown mechanisms95. Furthermore, two independent studies reported that IFN-induced transmembrane protein 3 (IFITM3) restricts an early step of influenza virus replication97,99. Additional studies to establish the strain-specific effects of these host factors will not only aid our understanding of the influenza virus–host interactions, but also possibly lead to the development of novel broad-spectrum antiviral approaches. The inhibition of certain host factors (without affecting host-specific functions) could be used successfully as a pharmacological intervention to combat influenza viruses and could minimize the development of resistant viral strains.
Novel concepts about influenza virus pathogenesis
Genetic manipulation of influenza viruses has allowed the identification of several viral markers that are associated with virulence and pathogenesis (TABLE 1). Most notably, the reconstruction of the H1N1 influenza virus responsible for the 1918 Spanish influenza pandemic100 and studies of HPAI H5N1 influenza viruses have clearly established the complexity and multigenic characteristics of influenza virus pathogenesis. In addition, animal model systems have been developed and used extensively to elucidate many of the virus–host interactions and host factors that contribute to pathogenesis (FIG. 4). Similarly, the 2009 H1N1 pandemic has expanded our understanding of the underlying risk factors and the molecular mechanisms that lead to severe disease in humans.
Figure 4 ∣. Summary of viral and host factors that influence the pathogenesis of influenza A virus.
Virus–host interactions affect viral replication. Variation in many of the influenza A virus proteins contributes to pathogenesis, and host factors can also influence susceptibility to infection and disease progression. The risk factors obesity and diabetes were identified during the 2009 H1N1 pandemic. HA, haemagglutinin; NA, neuraminidase; NP, nucleoprotein; NS1, non-structural protein 1.
Pre-existing immunity and pathogenesis.
As discussed above, the antigenic properties of influenza HA are a major determinant of pathogenesis. The main concern about antigenic shift is that it could generate a human pandemic virus belonging to an HA subtype to which the human population is naive. However, a novel concept from the 2009 H1N1 pandemic is that a change of HA subtype is not required for the generation of a human pandemic virus, as younger members of the population are immunologically naive and thus susceptible to strains that the older generation has been exposed to and is protected against (FIG. 2). During the 1918 H1N1 pandemic, the H1N1 virus established independent lineages in both humans and pigs. Constant antigenic drift of the H1N1 viruses in humans resulted in the modern seasonal H1N1 viruses, which are notably different, antigenically, from the original parental 1918 virus. However, the swine H1N1 virus that was established during the 1918 pandemic, from which the HA of the new 2009 pandemic H1N1 virus is derived, did not undergo extensive drift and was thus was maintained ‘antigenically frozen’ in pigs. This could be due to the short life span of domestic pigs, which are not exposed to multiple influenza virus infections in their lifetime and are therefore under no major humoral selection pressure to stimulate antigenic drift. Alternatively, host-specific immune selection pressure might drive antigenic drift in pigs at sites other than those that are important for antigenicity in humans. Both processes could lead to an ‘antigenically frozen’ HA. Thus, given the right conditions (for example, zoonotic transmission within a large immunologically naive human population), a spillover event of a virus that has been circulating for some time in pigs can result in the emergence of a novel human pandemic virus containing an HA from a previously circulating subtype.
Early epidemiological data gathered during the 2009 pandemic indicated that the young (<35 years of age) were more vulnerable to infection than the old (>65 year of age)101. The sera of elderly individuals were found to contain cross-reactive antibodies to the novel strain81,102, suggestive of previous exposure to antigenically similar viruses. Remarkably, immunization of mice with 1918 H1N1 virus-like particles, human H1N1 viruses that circulated before 1947 or classical swine H1N1 viruses elicited cross-reactive antibodies and conferred protection to a 2009 H1N1 virus challenge82,103,104, demonstrating the close antigenic similarity of these viruses. The highest antigenic similarity is between the new 2009 and the old 1918 pandemic viruses, which have high similarity at the antigenic sites (named Sa, Sb, Ca1, Ca2 and Cb) of the globular head of the HA protein, particularly at site Sa82. Vaccination of humans with the novel 2009 H1N1 vaccine strain elicits significant levels of cross-reactive antibodies against the 1918 H1N1 virus, confirming their antigenic similarity, and treatment of mice with human sera that were positive for these antibodies protected the mice against a lethal challenge with 1918 H1N1 virus105. The vast global spread of the 2009 H1N1 virus, which resulted in millions of natural infections, and the widespread immunization with the 2009 H1N1 vaccine strain suggest that a large proportion of the global population now has antibodies that are cross-reactive to the 1918 virus, an unanticipated benefit of the current vaccine strain. By contrast, the vaccines against seasonal H1N1 viruses have a low cross reactivity to the 1918 virus. This is due to the accumulation of mutations in the antigenic sites of HA during antigenic drift, and to the acquisition of glycosylation sites in the globular head of the protein, which can shield antigenically relevant regions82,106-108; overall, these changes result in drastic antigenic differences between the seasonal strains and the 1918 and 2009 pandemic viruses. Thus, young adults who have been exposed to only modern seasonal H1N1 viruses are immunologically naive to the 2009 pandemic H1N1 virus, partially explaining their higher incidence of severe disease caused by infection with the 2009 pandemic virus101.
Viral determinants of pathogenesis.
The sequence of the HA cleavage site is a major determinant of disease severity. The HA cleavage site of most low-pathogenic influenza virus strains contains a single arginine amino acid that is recognized by specific extracellular trypsin-like proteases (for example, serine proteases such as human transmembrane protease serine 2 (TMPRSS2) and trypsin-like proteases of the human airway epithelium) that are present only in the intestinal and respiratory mucosal surfaces of the host. However, the HA proteins of H5 and H7 HPAI viruses are characterized by the presence of a multibasic cleavage site, which is cleaved by intracellular ubiquitous proteases that are found in multiple organs, such as the subtilisin-like proteases furin and proprotein convertase subtilisin-kexin type 6. This can lead to systemic infection and increased virulence, especially in birds and small mammals109. In humans, infections with the HPAI H5N1 virus are also characterized by the presence of viral RNA outside the respiratory tissue and by increased lethality as a result of multiple organ failure88,109. However, despite the 1918 pandemic H1N1 virus lacking an HA multibasic cleavage site, infection with this virus in animal models results in high levels of viral replication and induction of pro-inflammatory cytokines, leading to severe disease110. Recombinant viruses expressing only the nucleoprotein (NP) and RNA-dependent RNA polymerase proteins PB1, PB2 and PA from the 1918 virus, along with a seasonal HA, showed efficient replication in the URT and LRT of ferrets111; furthermore, the HA, NA and PB1 proteins from the 1918 virus are also critical determinants of virulence in mice112. A newly identified virulence factor is the protein PB1-F2, which localizes to mitochondria in infected cells, inducing dissipation of the mitochondrial membrane and leading to apoptosis113-115. The 1918 pandemic virus is unusual in that it contains a S66 residue in PB1-F2 that is only present in various isolates of the HPAI H5N1 viruses. The most common residue found at this position in PB1-F2 of other strains is N, and a S66N substitution dramatically decreases the virulence of the 1918 virus and of other viruses containing a HPAI PB1 segment116. PB1-F2 also promotes and increases pathogenesis of secondary pneumonia infection117. More recently, it was found that PB1-F2 cooperates with the viral NS1 protein to inhibit the host IFN response118. However, in some viruses, particularly swine and modern human H1N1 isolates (including the 2009 pandemic H1N1 strain), stop codons in PB1-F2 lead to the expression of a truncated protein, but restoring the PB1-F2-coding capacity of the 2009 virus had only a minimal impact on virulence119, probably owing to additional sequence requirements in PB1-F2 (REF. 120). Another virulence determinant of influenza virus is the multifunctional protein NS1 (reviewed in REF. 121), which is an antagonist of the IFN-mediated antiviral host response122 and a modulator of adaptive immune responses123, and can inhibit global host gene expression through its interaction with the nuclear protein cleavage and polyadenylation specificity factor 30 kDa subunit (CPSF30; also known as CPSF4)124. NS1 probably contributes to the cytokine dysregulation that is seen in macaques infected with the 1918 virus110 and in humans that succumb to HPAI H5N1 virus infection125. The E92 residue of the HPAI H5N1 virus NS1 has also been implicated in virulence126. The NS1 protein from the 2009 pandemic H1N1 strain contains both mutations that abolish its CPSF30-binding properties and an 11 amino acid deletion at its carboxyl terminus, and it therefore lacks the PDZ-binding domain that was previously implicated in virulence22. Unexpectedly, when these functions were reintroduced they had no major effects on the replication, pathogenesis or transmission127,128 of the 2009 pandemic virus, suggesting that certain NS1 functions are not needed for successful human infection.
Host determinants of pathogenesis.
In humans, HPAI viruses can induce high plasma levels of 10 kDa IFNγ-induced protein (IP10; also known as CXCL10), monocyte chemoattractant protein 1 (MCP1; also known as CCL2), interleukin-8 (IL-8), IL-6, IL-10 and other pro-inflammatory cytokines that correlate with an elevated pharyngeal virus load and increased frequency of death caused by viral pneumonia88 (reviewed in REF. 125). Mammalian models of infection with 1918 H1N1 and HPAI H5N1 viruses show an increased and rapid activation of host immune response genes. Infection with the 1918 H1N1 virus is associated with severe pulmonary pathology as compared to infection with the contemporary H1N1 strain129. These infection models also show induction of macrophage and neutrophil infiltration for both strains (HPAI H5N1 and 1918 H1N1 viruses), resulting in acute lung inflammation130 and in the deregulation of antiviral responses that are insufficient for protection and possibly contribute to pathogenesis110. The severity of the disease resulting from infection with the 1918 virus correlates with a sustained pathology resulting from upregulation of inflammatory and cell death genes131. Similarly, increased and prolonged transcription of type I IFN, inflammation and innate immune induction are seen during infection with the HPAI H5N1 virus, resulting in apoptosis of dendritic cells and severe lung pathology132. Furthermore, influenza virus infection of mice that are deficient in IL-15 (REF 133), IL-17 (REF. 134) or CC-chemokine receptor 2 (CCR2)135 signalling and of mice that lack both tumour necrosis factor (TNF) and IL-1 receptors136 results in increased survival and/or delayed mortality compared with those seen on infection of wild-type mice, owing to a reduced level of leukocyte133, neutrophil134,136 or macrophage135,136 infiltration. Infections of IL-10-knockout mice cause increased production of antibodies in the lungs compared with the levels in infected wild-type mice, resulting in lower viral titres and increased survival137. However, a different study demonstrated that CD8+ effector T cells, by producing IL-10, could control the lung inflammation and injury that is induced during acute influenza virus infection138. Thus, further investigation is needed to define the role of IL-10, particularly in the context of highly virulent influenza virus strains. Moreover, mice that are deficient in TNF, IL-6 or CC-chemokine ligand 2 (CCL2) succumb to infection with HPAI H5N1 virus similarly to wild-type mice139, indicating that although acute host immune responses may contribute to pathogenesis, the elimination of some pro-inflammmatory signals does not prevent severe disease. The development of replication-competent fluorescence-labelled influenza A viruses provides a novel tool aimed at elucidating the kinetics and cell tropism of influenza virus infection in vivo. This tool has revealed that natural killer cells, B cells and antigen-presenting cells are also susceptible to infection, in addition to epithelial cells140. Such model systems will allow further investigation of the specific host cells that contribute to pathogenesis and to recovery from disease.
During the 2009 H1N1 pandemic, a subset of individuals developed rapid and severe viral pneumonia that was often associated with failure of other organs and resulted in considerable deterioration of underlying asthma or chronic obstructive airway disease141. The highest risks of severe or fatal illness were found in pregnant women (especially during the third trimester of pregnancy), children younger than 2 years of age and people with chronic lung diseases, including asthma141. Obesity142 and diabetes143,144 were newly identified risk factors for severe infection with the 2009 pandemic H1N1 virus. A severe infection with this virus was also observed in some healthy individuals. However, the underlying cause of severity is still unknown, and a close observation of the clinical course and the rapid management of individuals with severe disease therefore remain the main approaches for mitigating lethality. Interestingly, a recent study reported a high correlation for immune complex-mediated disease and severe symptoms in adults after infection with the 2009 H1N1 virus. Severe disease was associated with high levels of low-avidity, non-protective antibodies and immune complex-mediated complement activation in the respiratory tract of infected adults, uncovering a novel mechanism which might explain the increased disease severity that was observed in middle-aged individuals during the 2009 H1N1 pandemic145. Additional studies expanding these observations to animal models would be an important step towards understanding the severity of the disease caused by pandemic influenza A viruses.
What does the future hold?
The 2009 H1N1 strain has drastically changed the circulation dynamics of the previous seasonal influenza virus strains (since the emergence of the pandemic, almost no seasonal H1N1 has been detected worldwide). Immune selection pressure will probably cause the 2009 H1N1 virus to undergo changes in its antigenic characteristics (that is, to undergo antigenic drift). Exposure to contemporary seasonal viruses, immunization with the current vaccine strains and pre-existing immunity to older H1N1 viruses together result in a complex spectrum of immunity in the population, and this will probably select for previously unseen drift variants, reflecting the plasticity of antigenic evolution of the influenza virus HA protein. High-throughput whole-genome sequencing from original clinical isolates will allow a systematic surveillance of human influenza viruses in real time to establish drift variants and genetic changes that might be of epidemiological relevance.
Additional emphasis should also be placed on surveying influenza viruses from humans and from wild and domestic animal hosts in geographical regions that are currently poorly represented or not represented in the genomic databases (for example, Latin America and Africa). Continuous and enhanced surveillance of the wild hosts that harbour influenza viruses will not only allow the detection of novel strains, but also contribute to our understanding of influenza virus ecology (by establishing a map of influenza A virus hot spots worldwide), of the species that harbour these viruses and of the extent to which natural avian migration and/or localized avian communities modulate the generation of reassortant viruses. Additional viral sequences will also aid in the refinements of current models of influenza virus evolution, improve our ability to assess the underlying molecular basis of virulence and help in the design of more efficient long-term, worldwide surveillance efforts.
Two families of antiviral drugs are currently used to treat human influenza virus infections. Oseltamivir (Tamiflu; Roche) and zanamivir (Relenza; GlaxoSmithKline) inhibit the neuraminidase activity of the NA protein, thus blocking release of the newly formed virions from infected cells. The adamantanes (amantadine and rimantadine) inhibit the ability of the viral ion channel protein M2 to exchange H+ in order to lower the pH inside the virus, a step that is needed for viral uncoating during entry into the host cell. Although these antiviral therapies are efficacious against the current influenza virus strains, their use can result in the selection of resistant viruses146, and resistant strains do occur in nature (TABLE 1). This emphasizes the great need for additional broad-spectrum therapeutic approaches. Drugs aimed at diverse viral or host targets and at distinct stages of influenza virus replication would represent formidable tools to combat infection by multiple approaches and would minimize the development of resistant viruses.
The complex dynamics of influenza viruses continuously challenges host species barriers, and the emergence of novel virulent virus strains in humans is therefore a constant possibility. To gain insights into the mechanism by which reassorted viruses arise, further studies are necessary to elucidate the specific receptor requirements and the specific cell type, or types, and site (organ) where such an event might take place. Furthermore, elucidating the molecular requirements of reassortment at the viral factor and host factor levels will greatly help our understanding of how these viruses arise in nature and should therefore lead to additional and enhanced antiviral interventions. Moreover, in this complex interplay between host and influenza virus, host factors play an important part in disease severity and outcome, and studies focusing on identifying the human susceptibility factors are therefore key for understanding the host determinants of influenza virus pathogenesis. Current animal model systems only partially recapitulate human influenza, so complementary, carefully designed clinical studies are needed to validate such models and to find the genetic susceptibility factors and polymorphisms, as well as the specific underlying conditions, that modulate disease outcome in humans.
Acknowledgements
We thank B. Hale for comments on the manuscript and E. Nistal-Villan for help with the crystal structure models in figure 2. Work in the A.G.-S. laboratory is supported by US National Institute of Allergy and Infectious Disease (NIAID) grants R01AI046954, U01AI070469, U19AI083025, U54AI057158 and P01AI058113; by the Center for Research of Influenza Pathogenesis (CRIP) — an NIAID-funded Center of Excellence for Influenza Research and Surveillance — (HHSN266200700010C); and by the W. M. Keck Foundation.
Glossary
- Reassortment
The exchange of segments of the viral genome between two distinct virus strains
- Clades
Groups of biological taxa or species, the members of which share homologous features that were inherited from a common ancestor
- Antigenic drift
A gradual change in genotype that is due to antibody-mediated immune selection pressure driving the accumulation of mutations
- Antigenic shift
The reassortment of viral genomes, leading to the generation of a new subtype with a dramatic change in antigenic potential
- Genotype
The classification of an influenza A virus subtype based on the genetic characteristics of the eight gene segments
- Zoonotic
Pertaining to an infectious disease: originating in non-human animals, both wild and domestic, and able to be transmitted from those animals to humans
- Coalescent analysis
A retrospective study of a genetic population (in this case, the influenza A virus genomes or segments), allowing all the alleles of each gene in question to be traced to a single ancestral gene
- Nasopharyngeal
Pertaining to the area near the nasopharynx, which is the area of the upper throat that lies behind the nose
- Oropharyngeal
Pertaining to the area near the oropharynx, which is the area of the upper throat that lies behind the mouth
- Spillover
Infection of a distinct host by an infected natural host reservoir
- Immune complex
A cluster of antibodies bound to a soluble antigen. These can cause disease if they are deposited in organs where they lead to tissue damage
Footnotes
Competing interests statement
The authors declare competing financial interests: see Web version for details.
References
- 1.Molinari NA et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25, 5086–5096 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Johnson NP & Mueller J Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med 76, 105–115 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Palese P & Shaw ML in Fields Virology 5th edn (eds Knipe DM et al. ) 1647–1689 (Lippincott Williams & Wilkins, Philadelphia, USA, 2007). [Google Scholar]
- 4.Wise HM et al. A complicated message: identification of a novel PB1-related protein translated from influenza A virus segment 2 mRNA. J. Virol 83, 8021–8031 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouchier RA et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol 79, 2814–2822 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell RJ et al. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325, 287–296 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Nobusawa E. et al. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182, 475–485 (1991). [DOI] [PubMed] [Google Scholar]
- 8. Air GM Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc. Natl Acad. Sci. USA 78, 7639–7643 (1981). A seminal study that has led to the current group and clade classification of influenza virus subtypes.
- 9.Wright PF, Neumann G, & Kawaoka Y in Fields Virology 5th edn (eds Knipe DM et al. ) 1691–1740 (Lippincott Williams & Wilkins, Philadelphia, USA, 2007). [Google Scholar]
- 10.WHO. Pandemic (H1N1) 2009 - update 109. WHO Global Alert and Response; [online], http://www.who.int/csr/don/2010_07_16/en/index.html (2009). [Google Scholar]
- 11.CDC. Updated CDC estimates of 2009 H1N1 influenza cases, hospitalizations and deaths in the United States, April 2009 - April 10, 2010. CDC; [online], http://www.cdc.gov/h1n1flu/pdf/CDC_2009_H1N1_Est_PDF_May_4_10_fulltext.pdf (2010). [Google Scholar]
- 12.Brown IH Summary of avian influenza activity in Europe, Asia, and Africa, 2006–2009. Avian Dis. 54, 187–193 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Senne DA Avian influenza in North and South America, the Caribbean, and Australia, 2006–2008. Avian Dis. 54, 179–186 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Krauss S. et al. Influenza in migratory birds and evidence of limited intercontinental virus exchange. PLoS Pathog. 3, e167 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Obenauer JC et al. Large-scale sequence analysis of avian influenza isolates. Science 311, 1576–1580 (2006). The first comprehensive sequencing and genomic analysis of AIVs, leading to the identification of the NS1 PDZ domain-binding motif.
- 16.Vijaykrishna D. et al. Evolutionary dynamics and emergence of panzootic H5N1 influenza viruses. PLoS Pathog. 4, e1000161 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Claas EC et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351, 472–477 (1998). A clinical study describing one of the first H5N1 influenza virus infections to be suggestive of a direct avian–human transmission.
- 18.WHO. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO. WHO Global Alert and Response; [online], http://www.who.int/csr/disease/avian_influenza/country/cases_table_2011_06_22/en/index.html (2011). [Google Scholar]
- 19.Li KS et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430, 209–213 (2004). [DOI] [PubMed] [Google Scholar]
- 20.WHO. Continuing progress towards a unified nomenclature system for the highly pathogenic H5N1 avian influenza viruses. WHO; [online], http://www.who.int/csr/disease/avian_influenza/guidelines/nomenclature/en/index.html (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.WHO/OIE/FAO H5N1 Evolution Working Group. Toward a unified nomenclature system for highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis 14, e1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson D, Hossain MJ, Hickman D, Perez DR & Lamb RA A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl Acad. Sci. USA 105, 4381–4386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dugan VG et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 4, e1000076 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H. et al. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436, 191–192 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Chen H. et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl Acad. Sci. USA 103, 2845–2850 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith GJ et al. Emergence and predominance of an H5N1 influenza variant in China. Proc. Natl Acad. Sci. USA 103, 16936–16941 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fouchier RA et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl Acad. Sci. USA 101, 1356–1361 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Koopmans M. et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363, 587–593 (2004). The original account of a highly pathogenic H7N7 influenza virus outbreak in humans that was produced by direct avian-to-human transmission.
- 29.Peiris M. et al. Human infection with influenza H9N2. Lancet 354, 916–917 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Lin YP et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl Acad. Sci. USA 97, 9654–9658 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik Peiris JS Avian influenza viruses in humans. Rev. Sci. Tech 28, 161–173 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Kalthoff D, Globig A & Beer M (Highly pathogenic) avian influenza as a zoonotic agent. Vet. Microbiol 140, 237–245 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Dong G. et al. Phylogenetic diversity and genotypical complexity of H9N2 influenza A viruses revealed by genomic sequence analysis. PLoS ONE 6, e17212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu KM et al. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J. Virol 81, 10389–10401 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song XF, Han P & Chen YP Genetic variation of the hemagglutinin of avian influenza virus H9N2. J. Med. Virol 83, 838–846 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Bulach D. et al. Molecular analysis of H7 avian influenza viruses from Australia and New Zealand: genetic diversity and relationships from 1976 to 2007. J. Virol 84, 9957–9966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munster VJ et al. Mallards and highly pathogenic avian influenza ancestral viruses, northern Europe. Emerg. Infect. Dis 11, 1545–1551 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rambaut A. et al. The genomic and epidemiological dynamics of human influenza A virus. Nature 453, 615–619 (2008). A comprehensive phylogeographical analysis that led to the proposal of the ‘sink–source’ model of influenza virus epidemics.
- 39.Nelson MI et al. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 4, e1000012 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes EC et al. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 3, e300 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson MI et al. Stochastic processes are key determinants of short-term evolution in influenza a virus. PLoS Pathog. 2, e125 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Russell CA et al. The global circulation of seasonal influenza A (H3N2) viruses. Science 320, 340–346 (2008). A genetic and antigenic analysis of human seasonal H3N2 viruses, demonstrating the dominant seeding characteristics of these viruses that originated from local epidemics in West and Southeast Asia.
- 43.Nelson MI, Simonsen L, Viboud C, Miller MA & Holmes EC Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. 3, 1220–1228 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowen AC, Mubareka S, Steel J & Palese P Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 3, 1470–1476 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.CDC. Update: influenza activity - United States, 2009–2010 season. MMWR Morb. Mortal. Wkly Rep 59, 901–908 (2010). [PubMed] [Google Scholar]
- 46.Zepeda-Lopez HM et al. Inside the outbreak of the 2009 influenza A (H1N1)v virus in Mexico. PLoS ONE 5, e13256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chao DL, Halloran ME & Longini IM Jr. School opening dates predict pandemic influenza A(H1N1) outbreaks in the United States. J. Infect. Dis 202, 877–880 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraser C. et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324, 1557–1561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garten RJ et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325, 197–201 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith GJ et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122–1125 (2009). References 49 and 50 present two detailed genetic, phylogenetic and coalescent-based analyses demonstrating the complex reassortment history and genesis of the 2009 pandemic H1N1 virus.
- 51. Kuiken T. et al. Host species barriers to influenza virus infections. Science 312, 394–397 (2006). An excellent review on the host, viral and environmental factors that affect and contribute to host tropism.
- 52.Stevens J. et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol 355, 1143–1155 (2006). [DOI] [PubMed] [Google Scholar]
- 53. Chandrasekaran A. et al. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nature Biotech. 26, 107–113 (2008). The first demonstration that the size and shape of SA-containing receptors can modulate the receptor-binding properties of influenza A viruses.
- 54.Matrosovich M, Zhou N, Kawaoka Y & Webster R The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol 73, 1146–1155 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shinya K. et al. Avian flu: influenza virus receptors in the human airway. Nature 440, 435–436 (2006). [DOI] [PubMed] [Google Scholar]
- 56.van Riel D. et al. H5N1 virus attachment to lower respiratory tract. Science 312, 399 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Ito T et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol 72, 7367–7373 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith GJ et al. Dating the emergence of pandemic influenza viruses. Proc. Natl Acad. Sci. USA 106, 11709–11712 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimble B, Nieto GR & Perez DR Characterization of influenza virus sialic acid receptors in minor poultry species. Virol. J 7, 365 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan H & Perez DR Quail carry sialic acid receptors compatible with binding of avian and human influenza viruses. Virology 346, 278–286 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soundararajan V. et al. Extrapolating from sequence—the 2009 H1N1 ‘swine’ influenza virus. Nature Biotech. 27, 510–513 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Chen LM et al. Receptor specificity of subtype H1 influenza A viruses isolated from swine and humans in the United States. Virology 412, 401–410 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Childs RA et al. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nature Biotech. 27, 797–799 (2009). A study showing the binding activity of the 2009 pandemic H1N1 virus with respect to SA specificity.
- 64.Munster VJ et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325, 481–483 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maines TR et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325, 484–487 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nicholls JM et al. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nature Med. 13, 147–149 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Maines TR et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc. Natl Acad. Sci. USA 103, 12121–12126 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Srinivasan A. et al. Quantitative biochemical rationale for differences in transmissibility of 1918 pandemic influenza A viruses. Proc. Natl Acad. Sci. USA 105, 2800–2805 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tumpey TM et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315, 655–659 (2007). The demonstration that the transmission of influenza A virus can be modulated by receptor-binding specificity.
- 70.Steel J, Lowen AC, Mubareka S & Palese P Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 5, e1000252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Stevens J. et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–410 (2006). The first crystal structure of the HA of a HPAI virus.
- 72. Yamada S et al. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature 444, 378–382 (2006). A study that elucidates the specific residues needed for the avian H5N1 influenza viruses to switch from avian to human SA specificity.
- 73.Gao Y. et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 5, e1000709 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Russell RJ, Stevens DJ, Haire LF, Gamblin SJ & Skehel JJ Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconj. J 23, 85–92 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Xu R, McBride R, Paulson JC, Basler CF & Wilson IA Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J. Virol 84, 1715–1721 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu J. et al. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc. Natl Acad. Sci. USA 106, 17175–17180 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan PK et al. Clinical and virological course of infection with haemagglutinin D222G mutant strain of 2009 pandemic influenza A (H1N1) virus. J. Clin. Virol 50, 320–324 (2011). [DOI] [PubMed] [Google Scholar]
- 78.Kilander A, Rykkvin R, Dudman SG & Hungnes O Observed association between the HA1 mutation D222G in the 2009 pandemic influenza A(H1N1) virus and severe clinical outcome, Norway 2009–2010. Euro Surveill. 15, 19498 (2010). [DOI] [PubMed] [Google Scholar]
- 79.Chutinimitkul S. et al. Virulence-associated substitution D222G in the hemagglutinin of 2009 pandemic influenza A(H1N1) virus affects receptor binding. J. Virol 84, 11802–11813 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y. et al. Altered receptor specificity and cell tropism of D222G hemagglutinin mutants isolated from fatal cases of pandemic A(H1N1) 2009 influenza virus. J. Virol 84, 12069–12074 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Itoh Y. et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460, 1021–1025 (2009). A report describing comprehensive in vitro and in vivo studies of the 2009 pandemic H1N1 virus strain, characterizing its pathogenicity, drug susceptibility and human antibody cross reactivity.
- 82. Manicassamy B. et al. Protection of mice against lethal challenge with 2009 H1N1 influenza a virus by 1918-like and classical swine H1N1 based vaccines. PLoS Pathog. 6, e1000745 (2010). The first study to demonstrate the close antigenic similarities between the 2009 and 1918 pandemic H1N1 viruses.
- 83.Salomon R. et al. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med 203, 689–697 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Subbarao EK, London W & Murphy BR A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol 67, 1761–1764 (1993). A seminal work which demonstrates that residue K627 of PB2 is an adaptation to the human host.
- 85.Hatta M. et al. Growth of H5N1 influenza A viruses in the upper respiratory tracts of mice. PLoS Pathog. 3, 1374–1379 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Wit E. et al. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J. Virol 84, 1597–1606 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gabriel G. et al. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl Acad. Sci. USA 102, 18590–18595 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. de Jong MD et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nature Med. 12, 1203–1207 (2006). A study showing a correlation between increased viral titres and potent immune responses (in the form of a cytokine storm) in fatal cases of human infection with HPAI H5N1 virus.
- 89.Van Hoeven N. et al. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc. Natl Acad. Sci. USA 106, 3366–3371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Perez DR et al. Fitness of pandemic H1N1 and seasonal influenza A viruses during co-infection: evidence of competitive advantage of pandemic H1N1 influenza versus seasonal influenza. PLoS Curr. 1, RRN1011 (2009). A timely study that demonstrates the in vivo infection dynamics of the 2009 pandemic H1N1 virus as compared with the dynamics of sesonal H1N1 and the H3N2 viruses.
- 91.Mehle A & Doudna JA Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl Acad. Sci. USA 106, 21312–21316 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamada S. et al. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 6 e1001034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hao L. et al. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454, 890–893 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Konig R. et al. Human host factors required for influenza virus replication. Nature 463, 813–817 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shapira SD et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139, 1255–1267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karlas A. et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463, 818–822 (2010). [DOI] [PubMed] [Google Scholar]
- 97.Brass AL et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139, 1243–1254 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Watanabe T, Watanabe S & Kawaoka Y Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7, 427–439 (2010). A comprehensive review summarizing the key findings of five large genome-wide studies (references 92-96) that identified host factors which are required at different stages of influenza virus replication.
- 99.Yount JS et al. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nature Chem. Biol 6, 610–614 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tumpey TM et al. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310, 77–80 (2005). The first study to describe the increased pathogenesis of the extinct 1918 pandemic H1N1 virus (reconstructed from plasmid DNA).
- 101. Dawood FS et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med 360, 2605–2615 (2009). The original article from the US CDC reporting the detection of the novel swine origin 2009 H1N1 influenza virus that resulted in the first influenza pandemic of the twenty-first century.
- 102. Hancock K. et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med 361, 1945–1952 (2009). A seroprevalence study which shows that individuals who were>65 years of age in 2009 had a considerable level of cross-reactive antibodies against the 2009 H1N1 virus, indicative of previous exposure to an older, antigenically similar virus.
- 103.Skountzou I. et al. Immunity to pre-1950 H1N1 influenza viruses confers cross-protection against the pandemic swine-origin 2009 A (H1N1) influenza virus. J. Immunol 185, 1642–1649 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kash JC et al. Prior infection with classical swine H1N1 influenza viruses is associated with protective immunity to the 2009 pandemic H1N1 virus. Influenza Other Respi. Viruses 4, 121–127 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Medina RA et al. Pandemic 2009 H1N1 vaccine protects against 1918 Spanish influenza virus. Nature Commun. 1, 28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu R. et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328, 357–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wei CJ et al. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci. Transl. Med 2, 24ra21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang CC et al. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl Acad. Sci. USA 106, 18137–18142 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jang H. et al. Highly pathogenic H5N1 influenza virus can enter the central nervous system and induce neuroinflammation and neurodegeneration. Proc. Natl Acad. Sci. USA 106, 14063–14068 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Kobasa D. et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445, 319–323 (2007). This work shows that increased immune responses exacerbate and contribute to the pathogenesis of the 1918 H1N1 virus in non-human primates.
- 111.Watanabe T. et al. Viral RNA polymerase complex promotes optimal growth of 1918 virus in the lower respiratory tract of ferrets. Proc. Natl Acad. Sci. USA 106, 588–592 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pappas C. et al. Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc. Natl Acad. Sci. USA 105, 3064–3069 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gibbs JS, Malide D, Hornung F, Bennink JR & Yewdell JW The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J. Virol 77, 7214–7224 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chen W. et al. A novel influenza A virus mitochondrial protein that induces cell death. Nature Med. 7, 1306–1312 (2001). The first identification of the PB1-F2 protein, which is encoded by a second ORF of the PB1 segment in influenza A viruses.
- 115.Zamarin D, Garcia-Sastre A, Xiao X, Wang R & Palese P Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 1, e4 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Conenello GM, Zamarin D, Perrone LA, Tumpey T & Palese P A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 3, 1414–1421 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. McAuley JL et al. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2, 240–249 (2007). A study showing that PB1-F2 promotes secondary bacterial infection, thus increasing influenza A virus pathogenesis.
- 118.Varga ZT et al. The influenza virus protein PB1-F2 inhibits the induction of type I interferon at the level of the MAVS adaptor protein. PLoS Pathog. 7, e1002067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hai R. et al. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J. Virol 84, 4442–4450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McAuley JL et al. PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLoS Pathog. 6, e1001014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Hale BG, Randall RE, Ortin J & Jackson D The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol 89, 2359–2376 (2008). A thorough review discussing the many functions and properties of the NS1 protein of influenza viruses.
- 122.Gack MU et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5, 439–449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fernandez-Sesma A. et al. Influenza virus evades innate and adaptive immunity via the NS1 protein. J. Virol 80, 6295–6304 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nemeroff ME, Barabino SM, Li Y, Keller W & Krug RM Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3’end formation of cellular pre-mRNAs. Mol. Cell 1, 991–1000 (1998). [DOI] [PubMed] [Google Scholar]
- 125.Beigel JH et al. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med 353, 1374–1385 (2005). [DOI] [PubMed] [Google Scholar]
- 126.Seo SH, Hoffmann E & Webster RG Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nature Med. 8, 950–954 (2002). [DOI] [PubMed] [Google Scholar]
- 127.Hale BG et al. Mutations in the NS1 C-terminal tail do not enhance replication or virulence of the 2009 pandemic H1N1 influenza A virus. J. Gen. Virol 91, 1737–1742 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hale BG et al. Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. J. Virol 84, 6909–6922 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kash JC et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443, 578–581 (2006). The first global analysis of the host response to an in vivo infection (in mice) with the reconstructed 1918 pandemic H1N1 virus.
- 130.Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM & Tumpey TM H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4, e1000115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cilloniz C. et al. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog. 5, e1000604 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Baskin CR et al. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc. Natl Acad. Sci. USA 106, 3455–3460 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nakamura R. et al. Interleukin-15 is critical in the pathogenesis of influenza a virus-induced acute lung injury. J. Virol 84, 5574–5582 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Crowe CR et al. Critical role of IL-17RA in immunopathology of influenza infection. J. Immunol 183, 5301–5310 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Herold S. et al. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J. Exp. Med 205, 3065–3077 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Perrone LA, Szretter KJ, Katz JM, Mizgerd JP & Tumpey TM Mice lacking both TNF and IL-1 receptors exhibit reduced lung inflammation and delay in onset of death following infection with a highly virulent H5N1 virus. J. Infect. Dis 202, 1161–1170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sun K, Torres L & Metzger DW A detrimental effect of interleukin-10 on protective pulmonary humoral immunity during primary influenza A virus infection. J. Virol 84, 5007–5014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sun J, Madan R, Karp CL & Braciale TJ Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nature Med. 15, 277–284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Salomon R, Hoffmann E & Webster RG Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection. Proc. Natl Acad. Sci. USA 104, 12479–12481 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Manicassamy B. et al. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl Acad. Sci. USA 107, 11531–11536 (2010). The demonstration that natural killer cells, B cells and antigen-presenting cells, in addition to epithelial cells, are susceptible to influenza A virus infection in vivo.
- 141.WHO. Clinical features of severe cases of pandemic influenza. Pandemic (H1N1) 2009 briefing note 13. WHO Global Alert and Response; [online], http://www.who.int/csr/disease/swineflu/notes/h1n1_clinical_features_20091016/en/index.html (2009). [Google Scholar]
- 142.Rothberg MB & Haessler SD Complications of seasonal and pandemic influenza. Crit. Care Med 38, e91–e97 (2010). [DOI] [PubMed] [Google Scholar]
- 143.Carcione D. et al. Comparison of pandemic (H1N1) 2009 and seasonal influenza, Western Australia, 2009. Emerg. Infect. Dis 16, 1388–1395 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Peiris JS, Poon LL & Guan Y Emergence of a novel swine-origin influenza A virus (S-OIV) H1N1 virus in humans. J. Clin. Virol 45, 169–173 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Monsalvo AC et al. Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nature Med. 17, 195–199 (2010). An interesting study that correlates the severity of disease seen in some middle-aged individuals infected with the 2009 pandemic H1N1 virus and the presence of immune complexes in their lungs.
- 146.Le QM et al. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437, 1108 (2005). [DOI] [PubMed] [Google Scholar]
- 147. Lemey P, Suchard M & Rambaut A Reconstructing the initial global spread of a human influenza pandemic: a Bayesian spatial-temporal model for the global spread of H1N1pdm. PLoS Curr. 1, RRN1031 (2009). An original study showing the worldwide geographical spread of the 2009 H1N1 virus during the first 4 months of the pandemic outbreak.
- 148.Rambaut A & Holmes E The early molecular epidemiology of the swine-origin A/H1N1 human influenza pandemic. PLoS Curr. 1, RRN1003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jombart T, Eggo RM, Dodd P & Balloux F Spatiotemporal dynamics in the early stages of the 2009 A/H1N1 influenza pandemic. PLoS Curr. 1, RRN1026 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nelson M. et al. The early diversification of influenza A/H1N1pdm. PLoS Curr. 1, RRN1126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Vijaykrishna D. et al. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328, 1529 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Vijaykrishna D. et al. Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473, 519–522 (2011). The first large-scale, systematic longitudinal surveillance study of swine influenza A viruses, which shows that reassortment contributes to the emergence of antigenic-drift lineages.
- 153. Perez JT et al. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl Acad. Sci. USA 107, 11525–11530 (2010). A study showing the generation and accumulation of a small viral RNA that seems to regulate the switch from transcription to replication in influenza viruses during infection.
- 154.Umbach JL, Yen HL, Poon LL & Cullen BR Influenza a virus expresses high levels of an unusual class of small viral leader RNAs in infected cells. mBio 1, e00204–e00210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]