Abstract
Myxobacteria are social microbial predators that use cell–cell contacts to identify bacterial or fungal prey and to differentiate kin relatives to initiate cellular responses. For prey killing, they assemble Tad-like and type III-like secretion systems at contact sites. For kin discrimination (KD), they assemble outer membrane exchange complexes composed of the TraA and TraB receptors at contacts sites. A type VI secretion system and Rhs proteins also mediate KD. Following cellular recognition, these systems deliver appropriate effectors into target cells. For prey, this leads to cell death and lysis for nutrient consumption by myxobacteria. In KD, a panel of effectors are delivered, and if adjacent cells are clonal cells, resistance ensues because they express a cognate panel of immunity factors; while nonkin lack complete immunity and are intoxicated. This review compares and contrasts recent findings from these systems in myxobacteria.
Keywords: Myxococcus xanthus, outer member exchange, rearrangement hotspot, tight adherence transport system, type III secretion system, type VI secretion system
Introduction
Conflict systems are widespread among bacteria and often result in a cell death response [1, 2]. These systems serve different roles, including nutrient acquisition, self-defence, and ensuring populations are homogenous and cooperative. In some cases, conflict systems behave altruistically, such as when a phage attack triggers a programmed cell death response, which prevents phage propagation and sibling infection. Other conflict systems kill competitors to promote individual fitness. In this review, we focus on a subset of systems used by myxobacteria for predation and kin discrimination (KD).
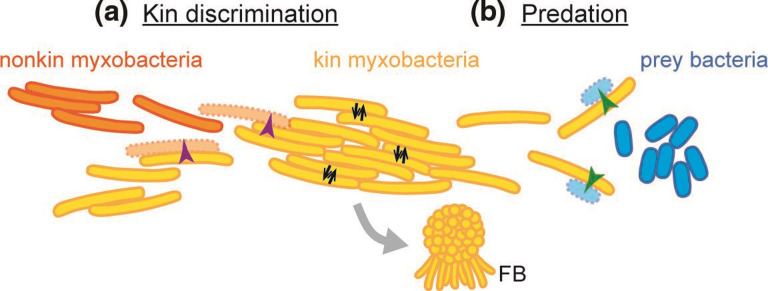
Myxobacteria are ubiquitous soil-dwelling microbes that combine both single-cell and multicellular behaviours, enabling them to maintain a complex social lifestyle (Fig. 1) [3]. Single cells glide on surfaces with the aid of lateral motors, but they also frequently assemble into multicellular swarms that move coordinately, primarily driven by type IV pili-dependent social motility. Multicellular swarms are believed to be involved in social feeding because myxobacteria secrete costly hydrolytic enzymes to externally digest biopolymers; and in groups, cells share this metabolic burden by providing a division of public good production [4, 5]. Rippling is another motile social behaviour in which waves of cells move rhythmically back and forth, apparently to improve predation efficiency [6]. However, the best-described social behaviour of myxobacteria is arguably fruiting body development. In response to starvation, thousands of cells aggregate from their surroundings to construct cooperative multicellular fruiting bodies, wherein cells differentiate into different cell types including environmentally resistant spores [3]. By making this transition, myxobacteria benefit from both single-cell and multicellular lifestyles, as the latter provides erect macroscopic structures that are thought to facilitate spore dispersion and division of labour by differentiation into different cell types. Many myxobacteria species use specific conflict systems for nutrient acquisition by predation or for KD, the latter of which allows congruent assembly of multicellular populations to ensure spores belong to the same social group. As outlined here, these conflict systems use different delivery mechanisms to trigger cell death in target cells.
Fig. 1.
Conflict systems in the myxobacterial life cycle. (a) KD allows myxobacteria to engage in costly social behaviours only with their closest kin (yellow), which ensures congruent vegetative populations and homogenous developmental fruiting bodies (FB). Nonkin myxobacteria (orange) are recognized and eliminated (purple arrows). Within a kin group, OME (black arrows) can benefit the fitness of a population. (b) Predation serves the acquisition of nutrients, as myxobacteria specifically kill nonkin cells of other species (blue) and feed on the released biomass. Part of the multilayered predation strategy is cell-contact-dependent prey killing, where a single myxobacterium kills and lyses a prey bacterium in a cell-contact-dependent manner (green arrows).
Predation: killing others for nutrients
Predatory bacteria of different phyla live in terrestrial and aquatic habitats and have developed various strategies to kill, disintegrate and consume cells of other species [7, 8]. Myxobacteria, and especially understudied families such as the Haliangiaceae , constitute a dominant fraction of micropredators in soil environments and might strongly affect the composition of soil microbiomes [9]. For this reason, they were recently proposed to be classified as ‘keystone taxa.’ Notably, predation behaviour is facultative and myxobacteria also obtain nutrients saprophytically from decaying organic material. So far, killing and consumption of micro-organisms has been observed for the majority of described myxobacteria species throughout all suborders, with only few non-predatory exceptions [4, 10].
Generally, myxobacteria are epibiotic predators, as they kill and lyse prey cells from the outside. The biomass released from micro-organisms is rich in amino acids and lipids, which are their primary sources of carbon and energy. Furthermore, purines and pyrimidines released from prey nucleic acids are incorporated via salvage pathways [11, 12]. Predatory myxobacteria are considered generalists, as they kill and lyse various Gram-positive and Gram-negative bacteria, including plant and human pathogens, but they also feed on eukaryotic fungi [5, 13–15]. This broad spectrum of structurally different prey is made possible by a multilayered predation strategy with different predation mechanisms that vary in their efficacy and specificity, and that are predominant in single cells or in multicellular swarms, respectively.
Cell-contact-dependent prey killing
Single myxobacteria kill bacterial prey cells one-on-one in a contact-dependent manner: when, for example, a single Myxococcus xanthus cell approaches a prey cell using gliding motility, it stops when in direct cell contact and induces prey cell death within 5 to 10 min [16–18]. In most cases, the M. xanthus cell then moves on to kill the next prey cell, leaving behind the prey biomass for consumption by fellow members of its population [18]. While this behaviour was observed decades ago [19], the molecular mechanisms were revealed only recently. Notably, contact-dependent killing involves the combined action of two protein secretion systems, which are specialized for their function in predation: a Tad-like secretion apparatus and a type III-like system (T3SS*) [16, 17].
The M. xanthus Tad apparatus, which was also termed the ‘Kil complex,’ is instrumental in inducing prey cell death, presumably by attaching to prey and/or delivering toxic effectors; however, the molecular details of these processes and how they recognize/attach to prey cells are not known (Fig. 2) [16, 17]. Kil proteins are encoded in two separate gene clusters, MXAN_3102–3108 and MXAN_4648–4661. Similar to Tad systems of other species, the putative Kil complex spans the M. xanthus cell envelope with an inner membrane platform (KilG and KilH) and an outer membrane secretin (KilC), associated with KilB. Additional components are major and minor pilins (KilK, KilL, KilM), a pre-pilin peptidase (KilA) and a cytoplasmic ATPase (KilF). The functions of all remaining proteins encoded within the Kil gene clusters are unknown but, notably, several genes encode fork-head associated domains (FHA), which often mediate signalling processes. KilD, one of the FHA-domain proteins, is essential for prey killing [16].
Fig. 2.
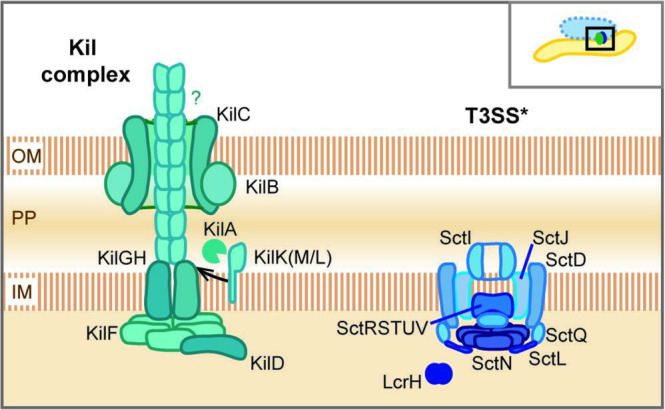
Protein secretion systems that act during predation. The Kil complex is an envelope-spanning protein complex and similar to systems of the Tad (tight adherence) family. Kil is required to induce cell death of prey bacteria, although the putative toxic effectors are unknown. T3SS* is an atypical type III secretion system, as it lacks any OM components and a needle structure. T3SS* is required to initiate prey cell lysis and functionally interacts with the Kil complex in an unknown manner. Both systems accumulate at the predator–prey contact site. See the main text for details on the individual components. PP, Periplasm; IM, inner membrane.
Tad (tight adherence) secretion machineries are mechanistically related to type II secretion systems, and in many bacteria they produce type IVc pili for surface attachment and biofilm stabilization [20, 21]. In several Bdellovibrio species, which are endobiotic predators that invade the periplasm of Gram-negative bacteria, the tad locus and the pilin PilA were shown to be involved in prey cell attachment [22–24]. It is, therefore, tempting to speculate that M. xanthus attaches to prey cells via a Kil-dependent filamentous structure, but the formation of such a filament has not yet been experimentally verified, and while polar type IV pili are essential for social motility, they are not required for cell-contact-dependent prey killing [16]. Whether Kil functions in the secretion of toxic effectors, either directly or in conjunction with T3SS*, remains to be investigated.
The second protein secretion system involved in cell-contact-dependent prey killing is an incomplete type III secretion system (T3SS*), which shows important differences to the canonical T3SS/injectisome of pathogenic bacteria [17, 25, 26]. The respective gene cluster (MXAN_2434–2464) contains protein components to form an inner membrane platform (SctD, SctJ, SctI) and C-ring (SctQ), the export apparatus (SctRSTUV) and ATPase complex (SctN and SctL), as well as several homologues to the LcrH chaperone (Fig. 2). Interestingly, the M. xanthus T3SS* gene cluster lacks a secretin that would span the outer membrane (OM), and it does not contain homologues to a T3SS needle or a translocon structure, raising the question how putative protein substrates are translocated by this system. From an evolutionary point of view, the needle-less T3SS* complex possibly represents an intermediate between the flagellar T3SS and the injectisome and is conserved within the myxobacteria [26, 27]. This argues for a functional, rather than a degenerate system, which has been specifically adapted to act during bacterial predation.
T3SS* mutants are still able to induce prey cell death; however, on a slower time scale than wild-type M. xanthus . Notably, Escherichia coli cells killed by T3SS* mutants do not lyse, but appear to retain their overall structure, suggesting that T3SS* initiates the disintegration of prey cells [17]. While putative T3SS* effectors are unknown at this point, this hypothesis is corroborated by the fact that M. xanthus populations of T3SS* mutants cannot efficiently utilize prey biomass to maintain swarm expansion.
Microscopy analysis revealed that components of the Kil and T3SS* complexes specifically accumulate at the M. xanthus /prey contact site shortly before the prey cell dies [16, 17]. This hints at a functional interplay of the Kil and T3SS* secretion systems during prey cell killing, but its nature and molecular details need further investigation. Possibilities include the sequential delivery of killing and lytic effectors via a mixed Kil/T3SS* secretion complex, or separate but overlapping functions in prey attachment and secretion of killing factors.
Interestingly, contact-dependent killing mechanisms discriminate between kin and prey cells: only contact with a prey cell, but not with other M. xanthus cells, causes accumulation of the Kil/T3SS* machinery and cell death [16, 17, C. Kaimer, unpublished observations]. This implies that the Kil/T3SS* machinery, directly or indirectly, distinguishes self from non-self-interacting cells by a currently unknown mechanism, which may or may not overlap with the KD processes described below. Both Kil and T3SS* were shown to act on different bacterial species, although killing and lysis of fungi or other eukaryotes has not yet been tested [16, 17]. However, while Kil is equally required for predation of Gram-negative and Gram-positive bacteria, T3SS* seems to be expendable when preying on Gram-positives [17]. It is likely that here the lytic contribution of T3SS* can be compensated by other systems that independently secrete a bacteriolytic enzyme cocktail, as discussed below.
Mutation of either the Kil or T3SS* system leaves residual predation activity, but in the absence of both systems M. xanthus is unable to move into and lyse bacterial prey colonies [17]. This observation emphasizes the critical role of cell–cell contact by single predator cells, which presumably start killing individual prey cells and make way for multicellular swarms, also described as ‘wolf packs’ (see below) to enter and release high concentrations of bacteriolytic proteins and antibiotics.
Secreted predation factors
The described Kil/T3SS* machinery presumably translocates toxic effectors at contact sites onto or into prey cells and acts in response to an unknown, cell-contact-dependent recognition mechanism. However, myxobacteria also constitutively secrete a multitude of hydrolytic enzymes in a non-targeted manner and independently of Kil/T3SS*, including proteases, lipidases, nucleases, amidases and glycoside hydrolases, which externally degrade biopolymers [3, 10, 17]. During predation, the main function of these secreted enzymes presumably lies in processing biomass from dead prey, and thereby overlaps with a saprophytic lifestyle. Nevertheless, killing and lysis of live prey by secreted proteins isolated from M. xanthus can be observed at high concentrations, and Gram-positive bacteria and fungi with an exposed cell wall are lysed more efficiently than Gram-negative bacteria with a protective OM [28]. A considerable fraction of bacteriolytic enzymes and antibiotics are secreted as cargo of lipid vesicles that emerge from the M. xanthus OM [29–31]. After being released from the M. xanthus cell, outer membrane vesicles (OMVs) are thought to fuse with the membranes of target cells to deliver their toxic contents [30, 32]. Both bacteriolytic and growth-inhibiting activities have been reported for OMVs isolated from M. xanthus , but their efficacy appears to be low compared to other killing mechanisms and might be limited to a few prey species [28, 30, 31].
The bacteriolytic activity of myxobacteria culture supernatants was demonstrated years ago [3, 33]; however, few individual enzymes have since been isolated and characterized. The ones that are include a family 19 glucoside hydrolase, LlpM, which is secreted by M. xanthus and targets the bacterial peptidoglycan cell wall similar to lysozyme. It is, however, not strictly required for growth on prey bacteria, and its activity against fungi has not been tested [28]. MepA, a M63 metalloprotease, is enriched in OMVs of M. xanthus and contributes to extracellular proteolytic activity [29]. Enzymes with glucanase activity play an important role, especially in fungi and oomycetes predation, with the membrane-associated 1,6-β-glucanase GluM from Corallococcus sp. and secreted 1,3-β-glucanase from Archangium sp. as recently characterized examples [15, 34].
Secondary metabolites with fungicidal, bactericidal or “toxic” activity on ciliates are produced by myxobacteria in great variety [35–37], but relatively few compounds have been investigated specifically in the context of predation. Here, it is difficult to differentiate whether the predominant function of an antibiotic lies in killing prey for consumption, or in inhibiting the growth of competitors, a strategy also used by non-predatory bacteria. Myxovirescin A (also known as TA) inhibits lipoprotein maturation and causes the formation of lethal cross-links between the membrane and cell wall. Due to this mode of action, the bactericidal activity of myxovirescin A is limited to growing bacteria, suggesting its main function lies in minimizing prey growth in the presence of M. xanthus [38, 39]. Some prey species, such as Bacillus licheniformis , survive M. xanthus encounters specifically by modifying myxovirescin A [40]. Myxoprincomide, a peptide antibiotic with an unknown mode of action, is also involved in predation, but acts solely on Gram-positive bacteria [41, 42].
Benefits of social predation
Above, we discussed the importance of single, scouting cells for predation. However, the majority of a preying population is engaged in social interactions within a swarm. In an early study by Rosenberg et al., it was shown that saprophytic growth of M. xanthus on casein in liquid culture requires a high cell density, which suggested that high concentrations of secreted digestive enzymes are a prerequisite for nutrient acquisition [4]. Also during predatory growth, secreted enzymes, antibiotics and OMVs can accumulate to high concentrations, which theoretically increases the predation efficiency of myxobacteria swarms over that of single cells. This hypothesis has prompted the comparison of preying myxobacteria to wolf packs, where many individuals cooperate and benefit from a shared pool of public goods, which in the case of myxobacteria consists of secreted predation factors; thus, making local prey biomass more accessible for consumption [3, 8, 43]. In support of this, enhanced predation was found in strain mixing experiments. Here, three isogenic M. xanthus strains that secreted different 1,3-β-glucanases, produced a cooperative cocktail of enzymes for prey consumption [44]. Nevertheless, the wolf pack idea requires further experimentation to understand how social behaviours during predation affect the fitness of single cells and to determine whether predation is a cooperative behaviour or reflects the synergy of individual cells acting side by side [45].
Another striking group behaviour is rippling. This behaviour is seen as macroscopic wave patterns, arising from a swarm of cells that have coordinated cell reversals, and presumably increases the contact time of M. xanthus with its prey [6, 44, 46]. Rippling also occurs in response to certain biomolecules, such as peptidoglycan from lysed bacteria, and requires the regulatory activity of a chemosensory two-component system, the Frz pathway [6, 46, 47]. However, it is unknown how a signal is perceived and propagated among single cells to coordinate motility. Rippling can also be observed during development in some strains, presumably caused by autolysis of a subpopulation of M. xanthus cells [6].
Regulation of predation and establishing a reciprocal predator–prey relationship
Predation-specific mechanisms, such as the ATP-dependent protein translocation by the Kil/T3SS* complexes or production of digestive enzymes, are energy intensive, raising the question to what extent are these processes regulated in response to prey. Transcriptome analysis indicates an induction of Kil gene expression upon starvation, but no further increase in the presence of prey [16, 48]. However, expression of several hydrolytic enzymes appears to be upregulated by prey cells, although further studies are needed [49, 50]. Interestingly, M. xanthus engaging in predation appear to adapt their lipid metabolism by using alternative pathways for fatty acid and lipid biosynthesis, which might culminate in an altered membrane composition [49]. Upregulation of the mevalonate pathway for isoprenoid synthesis, for example, appears to be a common and preferred pathway, as suggested by comparative genome analysis of predatory and non-predatory bacteria [51]. Transcriptional changes in myxobacteria were also observed in response to isolated acylhomoserine lactones (AHLs), which are released by other bacteria as quorum sensing signals [52, 53]. However, there is no direct link of AHL-regulated genes to known prey killing mechanisms, and it remains to be investigated how this potential eavesdropping by the predator might benefit the predation process.
The interaction of myxobacteria with prey goes beyond killing and consumption, and involves the bilateral competition for scarce resources. This is reflected by transcriptional changes in iron uptake, such as the synthesis of siderophores, which were observed for both predator and prey, e.g. co-cultures of M. xanthus with Sinorhizobium meliloti or Streptomyces coelicolor [49, 50, 54]. Moreover, the production of defensive antibiotics is induced in prey in response to predation, e.g. actinorhodin in Streptomyces coelicolor [55] or bacillaene in Bacillus subtilis [56]. However, biosynthesis of antibiotics myxovirescin and myxoprincomide in M. xanthus is not transcriptionally upregulated by prey or in response to AHLs [49, 52]. Additionally, long-term co-culture experiments of M. xanthus with E. coli revealed co-evolution by both parties [57]. Here, the genes eatB of M. xanthus , encoding a membrane protein of unknown function, and ompT of E. coli , encoding an OM protease, were identified as selection hotspots, although their putative role in predation is currently unknown.
Kin discrimination
KD in myxobacteria refers to their ability to distinguish between close relatives, particularly clonemates, from more distant myxobacteria [58]. Discrimination plays a central role in their transitions from single cell to multicellularity. However, their transitions face a steep challenge: intrusion of nonkin into vegetative swarms or aggregative multicellular fruiting bodies that disrupt cooperative kin interactions. Such exploitation is likely because soil microbial diversity is high, often consisting of tens of thousands of species per gram [59], as well as dozens of disruptive conspecific isolates [60, 61]. During vegetative predation and growth, cheater cells can scavenge nutrients without necessarily contributing to the shared pool of secreted digestive enzymes and secondary metabolites [34], broadly called public goods. During development, exploitation occurs when a subset of genotypes differentiates into mature spores, while the majority of other cells lyse, in perhaps altruistic acts, to provide nutrients for the metabolically costly programme that takes days to unfold [62].
To prevent exploitation and to ensure social groups remain homogenous, myxobacteria use elaborate KD mechanisms to distinguish self from nonself, and to eliminate nonkin (Fig. 1). The effectiveness of these systems is evident when analysing fruiting body assemblies from natural sources, where cells within fruiting bodies are nearly all clonal [63], despite being derived from environments rich in diversity. Importantly, KD often includes intraspecific discrimination; thus, preventing the formation of mosaic fruiting bodies consisting of different genotypes.
KD systems typically work by delivering polymorphic toxins, which leads to cell death of distant relatives, while protecting clonemates because they express cognate immunity factors. These toxin-immunity cassettes are often associated with mobile genetic elements (MGEs) that promote horizontal transfer [64], where the tight linkage between these genes is critical to ensure the new host simultaneously acquires immunity. To provide exquisite levels of discriminatory specificity against related genotypes, myxobacteria contain dozens of toxin-immunity loci that fluidly move within populations and form unique ‘self-identity barcodes’ in different isolates [65].
Myxobacteria contain three known KD systems – outer membrane exchange (OME), the type VI secretion system (T6SS) and the rearrangement hotspot (Rhs) – all of which deliver toxins that act on fellow myxobacteria and, to date, there are no reports these systems act on other bacterial species [66]. While the specificity of KD varies among these systems, the outcomes are the same: cell death. Myxobacteria likely also contain undiscovered KD systems, some of which may work by nonlethal mechanisms [67].
Outer membrane exchange
OME is a dual functioning platform that mediates KD as well as kin cooperation [68]. It does so through the function of a polymorphic cell surface receptor called TraA and its partner receptor TraB, whose genes overlap in an operon [69]. Self-recognition is mediated by homotypic binding between identical or nearly identical TraA receptors between neighbouring cells; thus, providing highly selective partner recognition (Fig. 3a, b) [68, 70]. TraAB function in cell adhesion, and when overexpressed, cells adhere in end-to-end chains and side-by-side rafts, which alters their motility and social behaviours [71, 72]. In contrast, when adjacent cells express different or incompatible TraA receptors, there is no cell binding nor OME. By using fluorescent fusion reporters, TraAB proteins are found to coalesce into foci upon cell–cell contact and recognition [73], analogous to the Kil and T3SS* complexes described above. Following engagement, OM proteins and lipids are bidirectionally and robustly exchanged between collaborating cells [69, 74]. Although no cytoplasmic proteins nor DNA are exchanged, transferred cargo nevertheless includes hundreds of different OM proteins. Since lipophilic fluorescent dyes and lipopolysaccharides are also transferred [69, 75], the mechanism of exchange is likely mediated by OM fusion where TraAB serve as fusogens.
Fig. 3.
Kin recognition and OME by TraA. (a) AlphaFold2 structure prediction of TraA [101], excluding the N-terminal and C-terminal signal peptide and sorting tag, respectively. The variable domain provides specificity for homotypic TraA–TraA binding between cells. Full length proteins contain ~80 cysteine residues where the mature C-terminal residue shown is a cysteine that is predicted to be lipidated, which thus anchors TraA on the cell surface [102]. TraB is an OM β-barrel protein that interacts with TraA and for simplicity is not shown. (b) Cells undergoing OME and subsequent consequences. The top centre cell (blue glow) is the effector and when nonkin cells (red) express a TraA compatible receptor, cell poisoning ensues following OME, because they lack immunity to the suite of transferred SitA lipoprotein toxins. Transferred toxins initially reside in the OM of the recipient, where they can subsequently be serially transferred and poison other cells following OME (left red cell). Toxins are eventually transported into the cytoplasm where they act as nucleases in the absence of cognate immunity factors [80]. OME between kin (blue cells) results in no KD. The inset shows an AlphaFold2 prediction of TraA–TraA binding between cells.
The outcomes of OME vary widely depending on the nature of partnering cells [68]. As alluded, OME can facilitate cooperation. For example, heterogeneous populations that share and mix cell envelope components via OME transition toward homogeneity, which presumably helps synchronize cooperative behaviours and transition populations toward a tissue-like collective [68, 73]. In other cases, healthy donors transfer and replenish defective components to low fitness siblings resulting in their revitalization [69, 75]. In turn, the healthy cells also benefit by increasing the population size of fit cells to threshold levels for multicellular functions, including development [76].
In cases where cells express compatible TraA receptors, but are not siblings, antagonism ensues by the transfer of a suite of lipoprotein toxins (Fig. 3b) [77]. These effectors are named SitA, for swarm inhibition toxins, because in mixed colony populations between nonkin, outward swarm expansion is blocked by OME-mediated cell poisoning [78, 79]. SitA effectors belong to six distinct families (SitA1/2–SitA7) defined by their unique N-terminal ‘escort domains’ [65]. At the C-terminus, each effector contains a toxin domain that belongs to one of >30 different families, many of which are found in other conflict systems. Interestingly, all the C-terminal domains that are characterized or have predicted functions are nucleases [65, 77]. Consequently, once delivered to the inner leaflet of target cells by OME, the toxin module must traverse the cell envelope to reach the cytoplasm (Fig. 3b). This migration involves a secondary step to OME and is mediated by their escort domains, which facilitate transport across the cytoplasmic membrane [80]. This secondary step of toxin migration is not instant and, consequently, cells become ‘infected’ and can transfer toxins to their siblings by OME (Fig. 3b) [77]. As a result, this KD system is extremely potent where one inhibitor cell can infect >1000 target cells by serial toxin transfer among cells. In contrast, if the recipient cell is clonal, it will contain the cognate suite of SitI immunity proteins and is not harmed.
The number of sitAI loci in myxobacterial genomes is large, ranging from 15 to >80, where a single difference can result in lethal OME [65]. However, toxic encounters only occur when strains express compatible TraA receptors, which again are highly polymorphic; thus, guarding against unintentional lethal exchanges with distant relatives. Nevertheless, since the number of functional TraA polymorphisms is finite, lethal encounters occur. Moreover, unique sitAI loci are rapidly disseminated through populations by their association with MGEs, resulting in KD between recent siblings. With this knowledge, comparative genomics serves as a viable approach to predict OME social compatibilities among natural isolates based on their sitAI and traA allele composition [65, 66]. Finally, because OME is a mutual decision, where both cells must express compatible TraA receptors to share their private cellular goods, we hypothesize OME originally evolved as a cooperative mechanism that subsequently was co-opted for KD [68].
Genomic analysis of sitAI loci frequently reveals they reside on a prophage, plasmid or transposons [65, 66, 77]. These and other findings led to our conclusion that MGEs exploited OME as a means for their expansion and retention in populations. For instance, when a cell acquires a selfish element with a unique sitAI locus, that transformed cell can now kill and outcompete its siblings by cell–cell intoxication [77, 80]. Additionally, if the element is lost by excision or deletion, then that cell is susceptible to poisoning by neighbouring siblings. As revealed by genomics, the benefits of sitAI loci outweighs their loss, explaining their large numbers in genomes, even though they represent a significant genomic burden (e.g. 100 kb of DNA). In turn, along with the immediate host benefiting from acquired selfish elements, populations at large may also benefit because new sitAI loci provide renewed policing mechanisms to guard against exploitation of their cooperative interactions [64]. Finally, we propose the evolutionary forces that drive traA diversification and maintenance of its polymorphism were done so to select against lethal OME encounters between nonkin. In addition, since OME plays dual roles in both cooperation and KD, this explanation provides a solution to Crozier’s paradox, namely how cooperative genes can be polymorphic [77, 81].
T6SS
The T6SS is a multicomponent protein secretion system that is widely distributed amongst Gram-negative bacteria and is thought to be evolutionarily related to contractile phage tails [82]. It punctures target cell envelopes and delivers effectors, which are often associated with the VgrG or PAAR spike proteins or the Hcp tube. T6SS effectors act on the cell wall, membrane, nucleic acids or other essential components [83]. T6SS is frequently used to attack other bacteria and in some cases eukaryotic cells. In M. xanthus , there is one T6SS gene cluster composed of 14 genes that encode the apparatus. In contrast, effector-immunity loci are scattered around the chromosome. Similar to sitAI loci, many T6SS effector-immunity cassettes are found on prophage, particularly a family called Mx-alpha. In turn, these MGEs enable rapid ecological dissemination that results in social diversification of populations [64, 66]. Cryo-electron microscopy revealed expanded and contracted forms of the T6SS apparatus in M. xanthus cells, as well as associated cell surface filaments or ‘antenna’, which may be involved in target cell recognition and, if so, work independently of TraA recognition [84].
Because of their predatory behaviours and the widespread use of T6SS as microbial weapons, it was reasoned myxobacteria use them in bacteria predation. However, studies by different labs found no evidence for this role [16, 17, 41]. Instead, T6SS is primarily involved in social policing or KD. This finding was first made by Troselj et al., who found that phenotypically less fit cells were killed by more fit siblings [85]. Experimentally, this was shown with isogenic strains where prototrophs killed their auxotroph siblings via the T6SS. However, antagonism only occurred when an essential metabolite, e.g. histidine for auxotrophs, was absent from media, thus blocking their growth, while prototrophs grew in mixed colonies. Interestingly, the responsible T6SS effector-immunity locus, tsxEI, is conserved within the genus Myxococcus , unlike other T6SS polymorphic effectors, suggesting a conserved mechanism for social policing against less fit siblings. Although the auxotroph strains contain the identical tsxEI locus, their sensitivity to intoxication occurs because during metabolite starvation the level of the TsxI immunity protein decreases with concurrent loss of protection to TsxE.
In separate work, Li and colleagues used a genetic screen for swarm incompatibility to identify M. xanthus mutants that exhibited KD by the parent strain [86]. They identified mutations in six gene clusters, encoding T6SS toxin-immunity cassettes, which resulted in cell death when mixed with the parent strain [87]. One of the toxins encodes a nuclease that when expressed from a plasmid kills the E. coli host, while co-expression of the downstream immunity gene neutralizes toxicity [88].
In another study, involving 22 sympatric M. xanthus soil isolates, the effect of T6SS and OME on KD was assessed. Here, for strains containing compatible TraA receptors, it was necessary to create double mutants to relieve antagonism; thus, OME and T6SS act redundantly in KD. However, for two isolates antagonism remained and involved a third system (see below) [66]. Strikingly, the discriminatory T6SS effectors were located on prophage Mx-alpha, as well as many of the OME effectors. Consistent with these findings, T6SS are also involved in KD in other genera, such as Vibrio [89, 90] and Proteus [91, 92]. In summary, T6SS provides a broad-spectrum KD mechanism in myxobacteria, while OME provides a narrow-spectrum KD mechanism based on TraA compatibility.
Finally, in a recent study, a specific T6SS toxin showed anti-fungal activity, but was not active in KD when a partner cell lacked the cognate immunity gene [87]. To date, this is the only known case in M. xanthus where T6SS is involved in inter-species competition or predation. This protein, MXAN_1813, contains a PAAR domain, such domains are frequently associated with T6SS toxins, and a C-terminal domain of unknown function that is only found in myxobacteria. Because of this finding, it is possible other T6SS effectors may function, in a limited sense, in predation.
Rhs proteins
Rhs proteins are widely found in bacteria, contain YDxxGRL(I/T) repeat sequences related to YD-repeats and C-terminal polymorphic toxin domains [93]. Rhs proteins can be divided into two broad classifications, where one group are very large proteins, usually >1500 amino acids, contain signal peptides and likely form filamentous cell surface appendages. In contrast, the second group, typically found in Gram-negative bacteria, are much smaller proteins, e.g. several hundred amino acids, lack signal peptides and are frequently delivered by T6SS. Many of the characterized Rhs proteins function in intra- and inter-species antagonism [94, 95], such as WapA protein of B. subtilis . Additionally, some Rhs proteins are virulence factors in pathogenic organisms [96].
In the above study of 22 M. xanthus soil isolates, nine large rhs genes were identified from genomic sequences. Seven of these genes were conserved among clade members, while two rhs genes (rhs4 and 5), each encoding ~4000 amino acids, were unique and shown to be involved in KD [60, 66]. Their role in KD was discovered after the OME and T6S systems were inactivated and antagonism persisted. Thus, antagonism was relieved only after knocking out these rhs genes, as well as OME and T6SS [66]. Strikingly, in contrast to their other rhs genes, rhs4 and 5 are located on different Mx-alpha prophage, as are unique OME and T6SS effectors. Moreover, since antagonism remained in T6SS mutants, the delivery of Rhs4 and Rhs5 is not T6SS dependent. These Rhs proteins contain signal sequences and likely reside on the cell surface where they bind and deliver C-terminal toxins to neighbouring cells [66], presumably analogous to contact-dependent growth inhibition (CDI) toxin delivery [2]. In support of this, an Rhs protein from Pseudomonas aeruginosa localizes on the cell surface where it is involved in virulence [96]. Whether M. xanthus Rhs proteins function in interspecies competition remains an open question.
While there is no evidence that myxobacteria use Rhs proteins for predation, one study found they play an inhibitory role. Here, a transposon insertion in the rhs gene (MXAN_6679) resulted in a gain-of-function or hyper-predation phenotype on B. subtilis [41]. In a separate genetic screen, a MXAN_6679 insertion mutant resulted in a motility defect [97]. Clearly, further studies are needed to understand the roles Rhs proteins play in myxobacterial social interactions.
Concluding remarks
Predation and KD serve central but different roles in the lifecycles of myxobacteria (Fig. 1). However, both functions rely on conflict systems that recognize target cells to initiate effector transfer resulting in target cell death (Fig. 4). Recent efforts by different groups have revealed important insights of the molecular basis of these systems; but one remaining question is, to what extent predation and KD might overlap. Theoretically, the TraA receptor or a similar polymorphic marker could also define the target spectrum for predation, e.g. by blocking the assembly of the Kil/T3SS* machinery between kin via a, yet to be identified, signalling mechanism. Given the very broad prey spectrum of myxobacteria, such a protective or exclusion mechanism might be more likely than the active recognition of different structures on prey cells. Additionally, the killing factors released by the Kil/T3SS* machinery are unknown, but might be similar or possibly identical to the toxin-immunity pairs used during KD, and/or involve separate effector classes, such as digestive enzymes. Finally, the ability to use one or more of the described KD systems to kill siblings might lead to cannibalistic behaviours, such as those reported in B. subtilis and suggested in other species [98–100]. Although killing by related myxobacterial occurs, to date there are no reports that the killer acquires nutrients that supports measurable growth as found against distant prey species. It is clear at this point, however, that with their conflict systems the social, yet predatory, myxobacteria have established an efficient way to manage the manifold interactions with other bacteria to their advantage.
Fig. 4.
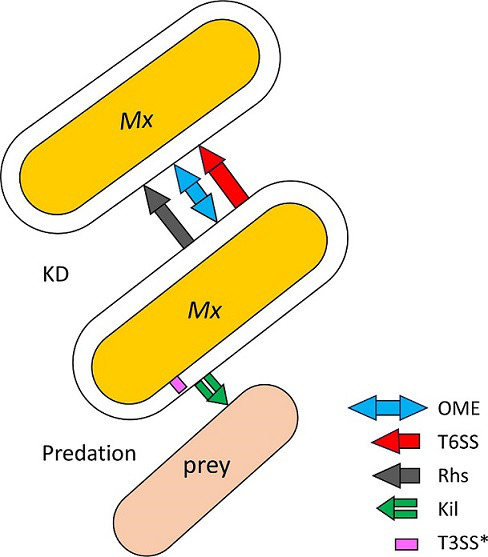
Conflict systems mediated by cell–cell contact in M. xanthus (Mx) and their biological roles.
Funding information
The work of C.K. was supported by Deutsche Forschungsgemeinschaft grant Ka3361/3-1. The work of D.W. was supported by National Institutes of Health grant GM140886.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AHL, acylhomoserine lactone; KD, kin discrimination; MGE, mobile genetic element; OM, outer membrane; OME, outer membrane exchange; T6SS, type VI secretion system.
References
- 1.Aravind L, Iyer LM, Burroughs AM. Discovering biological conflict systems through genome analysis: evolutionary principles and biochemical novelty. Annu Rev Biomed Data Sci. 2022;5:367–391. doi: 10.1146/annurev-biodatasci-122220-101119. [DOI] [PubMed] [Google Scholar]
- 2.Ruhe ZC, Low DA, Hayes CS. Polymorphic toxins and their immunity proteins: diversity, evolution, and mechanisms of delivery. Annu Rev Microbiol. 2020;74:497–520. doi: 10.1146/annurev-micro-020518-115638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muñoz-Dorado J, Marcos-Torres FJ, García-Bravo E, Moraleda-Muñoz A, Pérez J. Myxobacteria: moving, killing, feeding, and surviving together. Front Microbiol. 2016;7:781. doi: 10.3389/fmicb.2016.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg E, Keller KH, Dworkin M. Cell density-dependent growth of Myxococcus xanthus on casein. J Bacteriol. 1977;129:770–777. doi: 10.1128/jb.129.2.770-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiery S, Kaimer C. The predation strategy of Myxococcus xanthus . Front Microbiol. 2020;11:2. doi: 10.3389/fmicb.2020.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berleman JE, Chumley T, Cheung P, Kirby JR. Rippling is a predatory behavior in Myxococcus xanthus . J Bacteriol. 2006;188:5888–5895. doi: 10.1128/JB.00559-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jurkevitch E. Predatory Prokaryotes: Biology, Ecology and Evolution. Berlin, Heidelberg: Springer; 2006. [DOI] [Google Scholar]
- 8.Pérez J, Moraleda-Muñoz A, Marcos-Torres FJ, Muñoz-Dorado J. Bacterial predation: 75 years and counting! Environ Microbiol. 2016;18:766–779. doi: 10.1111/1462-2920.13171. [DOI] [PubMed] [Google Scholar]
- 9.Petters S, Groß V, Söllinger A, Pichler M, Reinhard A, et al. The soil microbial food web revisited: predatory myxobacteria as keystone taxa? ISME J. 2021;15:2665–2675. doi: 10.1038/s41396-021-00958-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimkets LJ, Dworkin M, Reichenbach H. The Prokaryotes - A Handbook Onthe Biology of Bacteria. Springer; 2006. [DOI] [Google Scholar]
- 11.Curtis PD, Shimkets LJ. In: Myxobacteria: Multicellularity and Differentiation. Whitworth DE, editor. Washington, DC: American Society for Microbiology; 2007. Metabolic pathways relevant to predation, signaling, and development; pp. 241–258. [DOI] [Google Scholar]
- 12.Bretscher AP, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978;133:763–768. doi: 10.1128/jb.133.2.763-768.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan AD, MacLean RC, Hillesland KL, Velicer GJ. Comparative analysis of Myxococcus predation on soil bacteria. Appl Environ Microbiol. 2010;76:6920–6927. doi: 10.1128/AEM.00414-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livingstone PG, Morphew RM, Whitworth DE. Myxobacteria are able to prey broadly upon clinically-relevant pathogens, exhibiting a prey range which cannot be explained by phylogeny. Front Microbiol. 2017;8:1593. doi: 10.3389/fmicb.2017.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Ye X, Liu M, Xia C, Zhang L, et al. A novel outer membrane β-1,6-glucanase is deployed in the predation of fungi by myxobacteria. ISME J. 2019;13:2223–2235. doi: 10.1038/s41396-019-0424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seef S, Herrou J, de Boissier P, My L, Brasseur G, et al. A Tad-like apparatus is required for contact-dependent prey killing in predatory social bacteria. Elife. 2021;10:e72409. doi: 10.7554/eLife.72409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiery S, Turowski P, Berleman JE, Kaimer C. The predatory soil bacterium Myxococcus xanthus combines a Tad- and an atypical type 3-like protein secretion system to kill bacterial cells. Cell Rep. 2022;40:111340. doi: 10.1016/j.celrep.2022.111340. [DOI] [PubMed] [Google Scholar]
- 18.Zhang W, Wang Y, Lu H, Liu Q, Wang C, et al. Dynamics of solitary predation by Myxococcus xanthus on Escherichia coli observed at the single-cell level. Appl Environ Microbiol. 2020;86:e02286-19. doi: 10.1128/AEM.02286-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride MJ, Zusman DR. Behavioral analysis of single cells of Myxococcus xanthus in response to prey cells of Escherichia coli . FEMS Microbiol Lett. 1996;137:227–231. doi: 10.1111/j.1574-6968.1996.tb08110.x. [DOI] [PubMed] [Google Scholar]
- 20.Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 21.Ellison CK, Whitfield GB, Brun YV. Type IV pili: dynamic bacterial nanomachines. FEMS Microbiol Rev. 2022;46:fuab053. doi: 10.1093/femsre/fuab053. [DOI] [PubMed] [Google Scholar]
- 22.Chanyi RM, Koval SF. Role of type IV pili in predation by Bdellovibrio bacteriovorus . PLoS One. 2014;9:e113404. doi: 10.1371/journal.pone.0113404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avidan O, Petrenko M, Becker R, Beck S, Linscheid M, et al. Identification and characterization of differentially-regulated type IVb pilin genes necessary for predation in obligate bacterial predators. Sci Rep. 2017;7:1013. doi: 10.1038/s41598-017-00951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capeness MJ, Lambert C, Lovering AL, Till R, Uchida K, et al. Activity of Bdellovibrio hit locus proteins, Bd0108 and Bd0109, links type IVa pilus extrusion/retraction status to prey-independent growth signalling. PLoS One. 2013;8:e79759. doi: 10.1371/journal.pone.0079759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konovalova A, Petters T, Søgaard-Andersen L. Extracellular biology of Myxococcus xanthus . FEMS Microbiol Rev. 2010;34:89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- 26.Abby SS, Rocha EPC. The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet. 2012;8:e1002983. doi: 10.1371/journal.pgen.1002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milne-Davies B, Wimmi S, Diepold A. Adaptivity and dynamics in type III secretion systems. Mol Microbiol. 2021;115:395–411. doi: 10.1111/mmi.14658. [DOI] [PubMed] [Google Scholar]
- 28.Arend KI, Schmidt JJ, Bentler T, Lüchtefeld C, Eggerichs D, et al. Myxococcus xanthus predation of Gram-positive or Gram-negative bacteria is mediated by different bacteriolytic mechanisms. Appl Environ Microbiol. 2021;87:e02382-20. doi: 10.1128/AEM.02382-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berleman JE, Allen S, Danielewicz MA, Remis JP, Gorur A, et al. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol. 2014;5:474. doi: 10.3389/fmicb.2014.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans AGL, Davey HM, Cookson A, Currinn H, Cooke-Fox G, et al. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology. 2012;158:2742–2752. doi: 10.1099/mic.0.060343-0. [DOI] [PubMed] [Google Scholar]
- 31.Remis JP, Wei D, Gorur A, Zemla M, Haraga J, et al. Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ Microbiol. 2014;16:598–610. doi: 10.1111/1462-2920.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitworth DE. Myxobacterial vesicles death at a distance? Adv Appl Microbiol. 2011;75:1–31. doi: 10.1016/B978-0-12-387046-9.00001-3. [DOI] [PubMed] [Google Scholar]
- 33.Sudo S, Dworkin M. Bacteriolytic enzymes produced by Myxococcus xanthus . J Bacteriol. 1972;110:236–245. doi: 10.1128/jb.110.1.236-245.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Dong C, Wang J, Liu M, Wang J, et al. Predation of oomycetes by myxobacteria via a specialized CAZyme system arising from adaptive evolution. ISME J. 2023;17:1089–1103. doi: 10.1038/s41396-023-01423-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korp J, Vela Gurovic MS, Nett M. Antibiotics from predatory bacteria. Beilstein J Org Chem. 2016;12:594–607. doi: 10.3762/bjoc.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weissman KJ, Müller R. Myxobacterial secondary metabolites: bioactivities and modes-of-action. Nat Prod Rep. 2010;27:1276–1295. doi: 10.1039/c001260m. [DOI] [PubMed] [Google Scholar]
- 37.Yu U, Kim J, Park S, Cho K. Tubulysins are essential for the preying of ciliates by myxobacteria. J Microbiol. 2023;61:627–632. doi: 10.1007/s12275-023-00056-2. [DOI] [PubMed] [Google Scholar]
- 38.Xiao Y, Wei X, Ebright R, Wall D. Antibiotic production by myxobacteria plays a role in predation. J Bacteriol. 2011;193:4626–4633. doi: 10.1128/JB.05052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao Y, Gerth K, Müller R, Wall D. Myxobacterium-produced antibiotic TA (myxovirescin) inhibits type II signal peptidase. Antimicrob Agents Chemother. 2012;56:2014–2021. doi: 10.1128/AAC.06148-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Liu X, Zhang P, Wang Y, Li Z, et al. Bacillus licheniformis escapes from Myxococcus xanthus predation by deactivating myxovirescin A through enzymatic glucosylation. Environ Microbiol. 2019;21:4755–4772. doi: 10.1111/1462-2920.14817. [DOI] [PubMed] [Google Scholar]
- 41.Müller S, Strack SN, Ryan SE, Shawgo M, Walling A, et al. Identification of functions affecting predator-prey interactions between Myxococcus xanthus and Bacillus subtilis . J Bacteriol. 2016;198:3335–3344. doi: 10.1128/JB.00575-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortina NS, Krug D, Plaza A, Revermann O, Müller R. Myxoprincomide: a natural product from Myxococcus xanthus discovered by comprehensive analysis of the secondary metabolome. Angew Chem Int Ed Engl. 2012;51:811–816. doi: 10.1002/anie.201106305. [DOI] [PubMed] [Google Scholar]
- 43.Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev. 2009;33:942–957. doi: 10.1111/j.1574-6976.2009.00185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Vaksman Z, Litwin DB, Shi P, Kaplan HB, et al. The mechanistic basis of Myxococcus xanthus rippling behavior and its physiological role during predation. PLoS Comput Biol. 2012;8:e1002715. doi: 10.1371/journal.pcbi.1002715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall RC, Whitworth DE. Is “wolf-pack” predation by antimicrobial bacteria cooperative? Cell behaviour and predatory mechanisms indicate profound selfishness, even when working alongside kin. Bioessays. 2019;41:e1800247. doi: 10.1002/bies.201800247. [DOI] [PubMed] [Google Scholar]
- 46.Berleman JE, Scott J, Chumley T, Kirby JR. Predataxis behavior in Myxococcus xanthus . Proc Natl Acad Sci. 2008;105:17127–17132. doi: 10.1073/pnas.0804387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimkets LJ, Kaiser D. Induction of coordinated movement of Myxococcus xanthus cells. J Bacteriol. 1982;152:451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livingstone PG, Millard AD, Swain MT, Whitworth DE. Transcriptional changes when Myxococcus xanthus preys on Escherichia coli suggest myxobacterial predators are constitutively toxic but regulate their feeding. Microb Genom. 2018;4:e000152. doi: 10.1099/mgen.0.000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pérez J, Contreras-Moreno FJ, Muñoz-Dorado J, Moraleda-Muñoz A. Development versus predation: transcriptomic changes during the lifecycle of Myxococcus xanthus . Front Microbiol. 2022;13:1004476. doi: 10.3389/fmicb.2022.1004476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C, Xiao Y, Wang Y, Liu Y, Yao Q, et al. Comparative genomics and transcriptomics insight into myxobacterial metabolism potentials and multiple predatory strategies. Front Microbiol. 2023;14:1146523. doi: 10.3389/fmicb.2023.1146523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasternak Z, Pietrokovski S, Rotem O, Gophna U, Lurie-Weinberger MN, et al. By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J. 2013;7:756–769. doi: 10.1038/ismej.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Akbar S, Phillips KE, Misra SK, Sharp JS, Stevens DC. Differential response to prey quorum signals indicates predatory specialization of myxobacteria and ability to predate Pseudomonas aeruginosa . Environ Microbiol. 2022;24:1263–1278. doi: 10.1111/1462-2920.15812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lloyd DG, Whitworth DE. The myxobacterium Myxococcus xanthus can sense and respond to the quorum signals secreted by potential prey organisms. Front Microbiol. 2017;8:439. doi: 10.3389/fmicb.2017.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee N, Kim W, Chung J, Lee Y, Cho S, et al. Iron competition triggers antibiotic biosynthesis in Streptomyces coelicolor during coculture with Myxococcus xanthus . ISME J. 2020;14:1111–1124. doi: 10.1038/s41396-020-0594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pérez J, Jiménez-Zurdo JI, Martínez-Abarca F, Millán V, Shimkets LJ, et al. Rhizobial galactoglucan determines the predatory pattern of Myxococcus xanthus and protects Sinorhizobium meliloti from predation. Environ Microbiol. 2014;16:2341–2350. doi: 10.1111/1462-2920.12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Müller S, Strack SN, Hoefler BC, Straight PD, Kearns DB, et al. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus . Appl Environ Microbiol. 2014;80:5603–5610. doi: 10.1128/AEM.01621-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nair RR, Vasse M, Wielgoss S, Sun L, Yu Y-TN, et al. Bacterial predator-prey coevolution accelerates genome evolution and selects on virulence-associated prey defences. Nat Commun. 2019;10:4301. doi: 10.1038/s41467-019-12140-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wall D. Kin recognition in bacteria. Annu Rev Microbiol. 2016;70:143–160. doi: 10.1146/annurev-micro-102215-095325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bickel S, Or D. Soil bacterial diversity mediated by microscale aqueous-phase processes across biomes. Nat Commun. 2020;11:116. doi: 10.1038/s41467-019-13966-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wielgoss S, Didelot X, Chaudhuri RR, Liu X, Weedall GD, et al. A barrier to homologous recombination between sympatric strains of the cooperative soil bacterium Myxococcus xanthus . ISME J. 2016;10:2468–2477. doi: 10.1038/ismej.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vos M, Velicer GJ. Social conflict in centimeter-and global-scale populations of the bacterium Myxococcus xanthus . Curr Biol. 2009;19:1763–1767. doi: 10.1016/j.cub.2009.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee B, Holkenbrink C, Treuner-Lange A, Higgs PI. Myxococcus xanthus developmental cell fate production: heterogeneous accumulation of developmental regulatory proteins and reexamination of the role of MazF in developmental lysis. J Bacteriol. 2012;194:3058–3068. doi: 10.1128/JB.06756-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wielgoss S, Wolfensberger R, Sun L, Fiegna F, Velicer GJ. Social genes are selection hotspots in kin groups of a soil microbe. Science. 2019;363:1342–1345. doi: 10.1126/science.aar4416. [DOI] [PubMed] [Google Scholar]
- 64.Weltzer ML, Wall D. Social diversification driven by mobile genetic elements. Genes. 2023;14:648. doi: 10.3390/genes14030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vassallo CN, Wall D. Self-identity barcodes encoded by six expansive polymorphic toxin families discriminate kin in myxobacteria. Proc Natl Acad Sci. 2019;116:24808–24818. doi: 10.1073/pnas.1912556116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vassallo CN, Troselj V, Weltzer ML, Wall D. Rapid diversification of wild social groups driven by toxin-immunity loci on mobile genetic elements. ISME J. 2020;14:2474–2487. doi: 10.1038/s41396-020-0699-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rendueles O, Zee PC, Dinkelacker I, Amherd M, Wielgoss S, et al. Rapid and widespread de novo evolution of kin discrimination. Proc Natl Acad Sci. 2015;112:9076–9081. doi: 10.1073/pnas.1502251112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sah GP, Wall D. Kin recognition and outer membrane exchange (OME) in myxobacteria. Curr Opin Microbiol. 2020;56:81–88. doi: 10.1016/j.mib.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, et al. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS Genet. 2012;8:e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao P, Wei X, Awal RP, Müller R, Wall D. A highly polymorphic receptor governs many distinct self-recognition types within the Myxococcales order. mBio. 2019;10:e02751-18. doi: 10.1128/mBio.02751-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cao P, Wall D. Self-identity reprogrammed by a single residue switch in a cell surface receptor of a social bacterium. Proc Natl Acad Sci. 2017;114:3732–3737. doi: 10.1073/pnas.1700315114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Balagam R, Cao P, Sah GP, Zhang Z, Subedi K, et al. Emergent myxobacterial behaviors arise from reversal suppression induced by kin contacts. mSystems. 2021;6:e0072021. doi: 10.1128/mSystems.00720-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao PB, Wall D. Direct visualization of a molecular handshake that governs kin recognition and tissue formation in myxobacteria. Nat Commun. 2019;10:3073. doi: 10.1038/s41467-019-11108-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol. 2011;81:315–326. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- 75.Vassallo C, Pathak DT, Cao P, Zuckerman DM, Hoiczyk E, et al. Cell rejuvenation and social behaviors promoted by LPS exchange in myxobacteria. Proc Natl Acad Sci. 2015;112:E2939–E2946. doi: 10.1073/pnas.1503553112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vassallo CN, Wall D. Tissue repair in myxobacteria: a cooperative strategy to heal cellular damage. BioEssays. 2016;38:306–315. doi: 10.1002/bies.201500132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vassallo CN, Cao P, Conklin A, Finkelstein H, Hayes CS, et al. Infectious polymorphic toxins delivered by outer membrane exchange discriminate kin in myxobacteria. Elife. 2017;6:e29397. doi: 10.7554/eLife.29397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dey A, Wall D. A genetic screen in Myxococcus xanthus identifies mutants that uncouple outer membrane exchange from a downstream cellular response. J Bacteriol. 2014;196:4324–4332. doi: 10.1128/JB.02217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dey A, Vassallo CN, Conklin AC, Pathak DT, Troselj V, et al. Sibling rivalry in Myxococcus xanthus is mediated by kin recognition and a polyploid prophage. J Bacteriol. 2016;198:994–1004. doi: 10.1128/JB.00964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vassallo CN, Sah GP, Weltzer ML, Wall D. Modular lipoprotein toxins transferred by outer membrane exchange target discrete cell entry pathways. mBio. 2021;12:e0238821. doi: 10.1128/mBio.02388-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Strassmann JE, Gilbert OM, Queller DC. Kin discrimination and cooperation in microbes. Annu Rev Microbiol. 2011;65:349–367. doi: 10.1146/annurev.micro.112408.134109. [DOI] [PubMed] [Google Scholar]
- 82.Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci. 2009;106:4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jurėnas D, Journet L. Activity, delivery, and diversity of type VI secretion effectors. Mol Microbiol. 2021;115:383–394. doi: 10.1111/mmi.14648. [DOI] [PubMed] [Google Scholar]
- 84.Chang Y-W, Rettberg LA, Ortega DR, Jensen GJ. In vivo structures of an intact type VI secretion system revealed by electron cryotomography. EMBO Rep. 2017;18:1090–1099. doi: 10.15252/embr.201744072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Troselj V, Treuner-Lange A, Søgaard-Andersen L, Wall D. Physiological heterogeneity triggers sibling conflict mediated by the type VI secretion system in an aggregative multicellular bacterium. mBio. 2018;9:e01645-17. doi: 10.1128/mBio.01645-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gong Y, Zhang Z, Zhou X-W, Anwar MN, Hu X-Z, et al. Competitive interactions between incompatible mutants of the social bacterium Myxococcus xanthus DK1622. Front Microbiol. 2018;9:1200. doi: 10.3389/fmicb.2018.01200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, Wang J, Zhang Z, Wang F, Gong Y, et al. Two PAAR proteins with different C-terminal extended domains have distinct ecological functions in Myxococcus xanthus . Appl Environ Microbiol. 2021;87:e00080-21. doi: 10.1128/AEM.00080-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gong Y, Zhang Z, Liu Y, Zhou X-W, Anwar MN, et al. A nuclease-toxin and immunity system for kin discrimination in Myxococcus xanthus . Environ Microbiol. 2018;20:2552–2567. doi: 10.1111/1462-2920.14282. [DOI] [PubMed] [Google Scholar]
- 89.Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, et al. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci. 2018;115:E8528–E8537. doi: 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Speare L, Woo M, Dunn AK, Septer AN. A putative lipoprotein mediates cell-cell contact for type VI secretion system-dependent killing of specific competitors. mBio. 2022;13:e0308521. doi: 10.1128/mbio.03085-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saak CC, Gibbs KA. The self-identity protein IdsD is communicated between cells in swarming Proteus mirabilis colonies. J Bacteriol. 2016;198:3278–3286. doi: 10.1128/JB.00402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tipping MJ, Gibbs KA. Peer pressure from a Proteus mirabilis self-recognition system controls participation in cooperative swarm motility. PLoS Pathog. 2019;15:e1007885. doi: 10.1371/journal.ppat.1007885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jamet A, Nassif X. New players in the toxin field: polymorphic toxin systems in bacteria. mBio. 2015;6:e00285-15. doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t’Kint de Roodenbeke C, Kaplan MD, et al. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci. 2013;110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alcoforado Diniz J, Coulthurst SJ. Intraspecies competition in Serratia marcescens is mediated by type VI-secreted Rhs effectors and a conserved effector-associated accessory protein. J Bacteriol. 2015;197:2350–2360. doi: 10.1128/JB.00199-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kung VL, Khare S, Stehlik C, Bacon EM, Hughes AJ, et al. An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proc Natl Acad Sci. 2012;109:1275–1280. doi: 10.1073/pnas.1109285109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Youderian P, Hartzell PL. Triple mutants uncover three new genes required for social motility in Myxococcus xanthus . Genetics. 2007;177:557–566. doi: 10.1534/genetics.107.076182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.López D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis . Mol Microbiol. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.González-Pastor JE. Cannibalism: a social behavior in sporulating Bacillus subtilis . FEMS Microbiol Rev. 2011;35:415–424. doi: 10.1111/j.1574-6976.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- 100.Gophna U. Et tu, Vibrio cholerae? Kin-cannibalism and a bacterial secretion system. Cell. 2022;185:4039–4040. doi: 10.1016/j.cell.2022.09.040. [DOI] [PubMed] [Google Scholar]
- 101.Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, et al. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sah GP, Cao P, Wall D. MYXO-CTERM sorting tag directs proteins to the cell surface via the type II secretion system. Mol Microbiol. 2020;113:1038–1051. doi: 10.1111/mmi.14473. [DOI] [PMC free article] [PubMed] [Google Scholar]