Abstract
Tripterygium glycosides (TG) have been reported to ameliorate Alzheimer's disease (AD), although the mechanism involved remains to be determined. In the present study, the lncRNA and circRNA expression profiles of an AD mouse model treated with TG were assessed using microarrays. lncRNAs, mRNAs, and circRNAs in the hippocampi of 3 AD+normal saline (NS) mice and 3 AD+TG mice were detected using microarrays. The most differentially expressed lncRNAs, mRNAs, and circRNAs were screened between the AD+NS and AD+TG groups. The differentially expressed lncRNAs and circRNAs were analyzed using GO enrichment and KEGG analyses. Co-expression analysis of lncRNAs, circRNAs, and mRNAs was performed by calculating the correlation coefficients. Protein-protein interaction (PPI) network analysis was performed on mRNAs using STRING. The lncRNA-target-transcription factor (TF) network was analyzed using the Network software. In total, 661 lncRNAs, 64 circRNAs, and 503 mRNAs were found to be differentially expressed in AD mice treated with TG. Pou4f1, Egr2, Mag, and Nr4a1 were the hub genes in the PPI network. The KEGG results showed that the mRNAs that were co-expressed with lncRNAs were enriched in the TNF, PI3K-Akt, and Wnt signaling pathways. LncRNA-target-TF network analysis indicated that TFs, including Cebpa, Zic2, and Rxra, were the most likely to regulate the detected lncRNAs. The circRNA-miRNA interaction network indicated that 275 miRNAs may bind to the 64 circRNAs. In conclusion, these findings provide a novel perspective on AD pathogenesis, and the detected lncRNAs, mRNAs, and circRNAs may serve as novel therapeutic targets for the management of AD.
Keywords: lncRNA, circRNA, Alzheimer's disease, Tripterygium glycoside
Introduction
Alzheimer's disease (AD) is a progressive, persistent, and degenerative disease of the central nervous system (CNS), with clinical manifestations of cognitive impairment and memory impairment (1,2). The pathogenesis of AD is not clear, and there is a lack of effective treatments. Currently, cholinesterase inhibitors are commonly used in the clinical treatment of AD, but these drugs can only improve and relieve symptoms. Natural medicines, including polysaccharides (3,4), phenylpropanoids (5,6), flavonoids (7,8), alkaloids (9), saponins (10,11), and polyphenols (12,13), have also been considered for the treatment of AD.
Tripterygium glycoside (TG), also known as Tripterygium, is the total glycoside extracted from the peeled roots of Tripterygium wilfordii Hook.f. It has been used in the treatment of rheumatoid arthritis (RA) (14), lupus nephritis (LN) (15), diabetes mellitus (DM) (16), and Guillain-Barre syndrome (GBS) (17). Animal experiments have shown that TG has protective effects on the CNS. It has been suggested that TG can significantly improve the inflammatory damage to astrocytes induced by lipopolysaccharide (LPS) by decreasing the expression of TNF-α, iNOS, and IL-6(18). In our previous study, TG suppressed the release of inflammatory factors and inhibited the phosphorylation of IκBα and p38 MAPK in Aβ25-35-induced AD mice (19). However, the mechanism of action of TG in AD remains to be determined.
Noncoding RNAs, including long noncoding RNAs (lncRNAs), circular RNAs (circRNAs), and microRNAs (miRNAs), have been studied due to the extent of their expression and their involvement in several biological processes (20). LncRNAs, such as BACE1- AS (21), 17A (22), NDM29(23), 51A (24), BC200(25), and NAT-RAD18(26), have been reported to be involved in the formation of senile plaques, DNA repair or synaptic formation, as well as in the pathogenesis of AD. CircRNAs are derived from mRNA precursors, which may affect normal cell differentiation, maintain tissue homeostasis, and influence the progression of various diseases (27). Additionally, circRNAs have been shown to play an important role in the occurrence and development of AD by influencing neuronal genesis and injury, Aβ deposition, neuroinflammation, autophagy, and synaptic function through the function of microRNA (miRNA/miR) sponges (28-30). Thus, lncRNAs and circRNAs may be considered risk factors, progression biomarkers, and therapeutic targets for AD.
However, the lncRNAs and circRNAs regulated by TG in AD treatment have not been determined. In the present study, the expression profiles of the lncRNAs and circRNAs of an AD mouse model treated with TG were determined using microarrays.
Materials and methods
Animal model of AD
The study was performed in accordance with the ARRIVE guidelines (https://arriveguidelines.org/), and the protocols followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals (31). The research procedures were approved by the Ethics Committee of Changsha Medical University (approval no. EC20190114). A total of Twenty-four C57BL/6 J mice (male, 28±5 g, 6 months old) were purchased from Silaike Jingda Biotechnology Co., Ltd. (12th Sep 2021; Changsha, China). The mice were fed standard chow, and housed under controlled conditions (temperature, 23±0.6˚C; relative humidity, 55±8%; 12 h light: dark cycle). Animal health and behavior of all mice were monitored weekly. The neuroprotective effects of TG on AD mouse models were identified in our previous study (19). A normal control group was excluded from the present study. Thus, the mice were divided into two groups: AD+TG group and AD+normal saline (NS) group. The AD model was constructed as described in our previous study (19). Except for 8 mice that died, a total of 16 AD mice models were included in further study. Mice in the AD+TG group (n=8) were treated with TG (0.25 mg/10 g.d, 1 mg/ml). The dosing and duration of TG followed the study conducted by Wang et al (18). Mice in the AD+NS group (N=8) were treated with NS (0.9%) 0.5 ml/d. The treatments were administered by intraperitoneal injection once per day for 4 weeks. At the end of treatment, a total of 6 and 7 mice were obtained in the AD+TG group and AD+NS group, respectively. From each group, 3 samples were selected randomly for lncRNA and mRNA microarray analysis. Any surviving animals (n=3 in the AD+TG group and n=4 in the AD+NS group) at the end of the experiment were euthanized by exposure to carbon dioxide (CO2) overdose with the CO2 displacement rate of 30-70% of the cage volume per min. Death was confirmed based on a lack of heartbeat and brain death (no environmental response, pupil reflex to light or spontaneous breathing).
Tissue collection and RNA extraction
General anesthesia was performed by intraperitoneal injection of 0.2% sodium pentobarbital (40 mg/kg). Hippocampal tissues were isolated from mouse brains and stored at -80˚C until required for further analysis. TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract total RNA from hippocampal tissues. Then, DNAse I was used to remove DNA contamination. The quality of RNA was quantified using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc.).
Microarray analysis
PCR amplification and fluorescence labeling of total RNA from 6 samples [AD+NS (n=3) and AD+TG (n=3)] were performed using an Agilent expression spectrum chip kit (Agilent, California, USA). The labeled cRNA was purified with an RNA extraction and purification kit (Sigma-Aldrich, St. Louis, USA). A total of 600 µg cRNA was hybridized using an Agilent-085631 microarray (Agilent Technologies, Inc.) with the following conditions: 65˚C for 17 h at 3.354 x g. Feature Extraction Software version 12.0 (Agilent Technologies, Inc.) was used to read the data. The Limma package (release 3.16; https://www.bioconductor.org/packages/release/bioc/html/limma.html) (32) in R-4.2.3 software (https://cran.r-project.org/bin/windows/base/) (33,34) was used for normalization. The differentially expressed lncRNAs, circRNAs, and mRNAs with a fold change ≥2 and P<0.05 were selected and identified. Clustering analyses were performed with hierarchical and average linkage algorithms using Mev version 4.9.0(35).
Reverse transcription-quantitative (RT-q)PCR
Four lncRNAs, mRNAs, and circRNAs were randomly selected and identified by qPCR. Trans-script II First-strand cDNA Synthesis SuperMix Kit (Sigma-Aldrich, St. Louis, USA) was used for reverse transcription according to the manufacturer's protocol. The TB Green Premix ExTaq™ kit (Takara Bio, Inc.) was used for qPCR. The reaction conditions were as follows: 95˚C for 5 min, followed by 38 cycles of 94˚C for 30 sec, 58˚C for 30 sec and 72˚C for 60 sec. The relative expression levels of lncRNAs and mRNAs were analyzed using the 2-ΔΔCq method (36) and normalized to GAPDH. Amplification was performed on an ABI7500 quantitative PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.). The sequences of the primers are listed in Table I.
Table I.
Sequences of the primers used for quantitative PCR detection of the selected lncRNAs, circRNAs, and mRNAs.
| Gene names | Forward, 5'-3' | Reverse, 5'-3' |
|---|---|---|
| NONMMUG042458.2 | GACACTGGCGTGAAAAAGGAAC | GACAATCAAGGCAGGAGGAG |
| NONMMUG004800.3 | CCTCCACCCCTACAGGAAGAG | GACAGGAGCAAAAGTGACTT |
| NONMMUG007415.2 | GAAATGGCAACTCAGCGGAGAG | GAAATTATCCAGGATGAGAAAG |
| NONMMUT141368.1 | GTCTCGGTGGGCGGGCATG | GTGTCTGAGACAAAAGGGAG |
| Skint8 | ACCACTCCCA CAAGACACCT | GAAGGAGGCCATTGGAGAAG |
| Lce1b | AGATGTCCTG CCAGCAGAAC | GGTGGTTGCTGCAGTTCTGG |
| Padi4 | GGTGAAAGCAGCCAGCAGCAG | GAATGGACTTTGAGGATGAC |
| Slc4a9 | CTTCATTCAACTAAATGAGC TG | AGCCCAGAACTGAGAGGACA GCT |
| MMU_CIRCpedia_20654 | ACATGAGCCTTCAGAGATAC | CAGAGGCAACAACTACCC |
| mmu_circ_0010693 | GGAACATTTCCATCAACATT | CTCTGAATTACTGC |
| MMU_CIRCpedia_214399 | TGATGTCATCCTGATAGTTG | GGTTGACATCGACCAA |
| mmu_circ_0010830 | AGGATATTCACAGACATGC | GAACATTGAGCCTACTCAAG |
Co-expression network analysis
Pearson's correlation analysis was used to calculate the correlation coefficients (r) and P-values. The screening criteria were r>|0.85| and P<0.05. The co-expression network was generated using Circos software (version 0.69-6; http://circos.ca/software/download/) (37).
PPI network of the differentially expressed mRNAs
The PPI network of differentially expressed mRNAs was generated using STRING (version 11.5) (38) (https://cn.string-db.org/) with a threshold score >0.4.
Functional classification and pathway analysis
Gene Ontology (GO) analysis (39,40) (http://geneontology.org/page/go-database) from three aspects [biological process (BP), cellular component (CC), and molecular function (MF)], and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.kegg.jp/) (41) were used to determine the roles of the differentially expressed mRNAs. P<0.01 was used as the selection criterion to analyze the difference in mRNAs involved in the pathways.
Cis/trans target gene prediction of lncRNA
All coding genes within 100k bp upstream and downstream of differentially expressed lncRNAs were selected as the target gene for cis-regulation. Cis-regulation was assessed using the FEELNC software version 0.2.1(42). Trans-regulation was predicted using RIsearch version 2.0 (Center for non-coding RNA in Technology and Health, Department of Veterinary and Animal Sciences, Faculty for Health and Medical Sciences, University of Copenhagen Frederiksberg, Denmark) (43,44) (https://rth.dk/resources/risearch/) with the following parameters: Number of interacting bases between lncRNA and gene, <10; binding free energy, <-100.
lncRNA-target-transcription factor (TF) network analysis
Potential lncRNA binding TFs were predicted based on the JASPAR database (http://jaspar.genereg.net/). The gene-TF pairs provided by the GTRD database (http://gtrd.biouml.org/) and the co-expression relationship of lncRNA-mRNA pairs were used to construct the 3-element regulatory network of the lncRNA-TF-mRNA. The top 500 relationship pairs were extracted, and the regulatory network diagram of lncRNA-TF-mRNA was drawn using the network software.
Prediction of circRNA-miRNA interactions
The interaction between circRNA and related miRNAs was predicted using miRDB database (http://www.mirdb.org/). The miRNA response element (MRE) of different circRNAs was studied. Target miRNAs were selected based on complementary pairing sequences. Cytoscape software (version.3.9.1) (45) (http://circos.ca/software/download/) was used to construct the circRNA-miRNA network.
Statistical analysis
Data were analyzed using SPSS 22.0 (IBM Corp.). Data are presented as the mean ± SD, and a Student's t-test was used to compare differences between the two groups. All tests were two-sided, and P<0.05 was considered to indicate a statistically significant difference.
Results
Differentially expressed lncRNAs, mRNAs, and circRNAs in the Aβ25-35-induced AD mouse model treated with TG
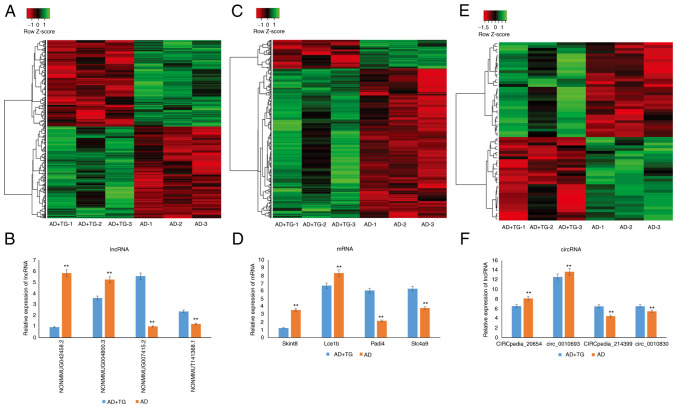
In total, 108,510 lncRNAs, 15,321 circRNAs, and 43,093 mRNAs were obtained by high-throughput sequencing. Compared with the control group, 661 lncRNAs, 64 circRNAs, and 503 mRNAs were significantly differentially expressed following TG treatment, including 422, 341, and 34 upregulated, as well as 81, 320, and 30 downregulated mRNAs, lncRNAs, and circRNAs, respectively. Amongst them, NONMMUG090228.1 (FC=268.27) and NONMMUG011123.2 (FC=-36.51) were the most upregulated and downregulated lncRNAs, respectively, Pou4f1 (FC=26.01) and Gm14781 (FC=-10.75) were the most upregulated and downregulated mRNAs, respectively, and MMU_CIRCpedia_35174 (FC=5.47) and mmu_circ_0007187 (FC=-4.04) were the most upregulated and downregulated circRNAs, respectively. The lncRNA, mRNA, and circRNA expression patterns in different samples are shown in Fig. 1A-C.
Figure 1.
The hierarchical clustering of the DE lncRNAs, mRNAs, and circRNAs in AD (n=3/group) and AD treated with TG (n=3/group). Clustering analysis of the (A) DElncRNAs, (C) DEmRNAs and (E) DEcircRNAs. qPCR validation of 4 randomly selected (B) DElncRNAs, (D) DEmRNAs, and (F) DEcircRNAs. The qPCR results were consistent with the microarray data. **P<0.05. DE, differentially expressed; AD, Alzheimer's disease.
To verify the microarray results, the expression of 4 randomly selected lncRNAs (2 upregulated and 2 downregulated), 4 mRNAs (2 upregulated and 2 downregulated), and 4 circRNAs (2 upregulated and 2 downregulated) using qPCR. The results showed that the expression trend of the selected genes was consistent with the microarray analysis (Fig. 1D-F).
Co-expression of lncRNAs with mRNAs, and circRNAs with mRNAs
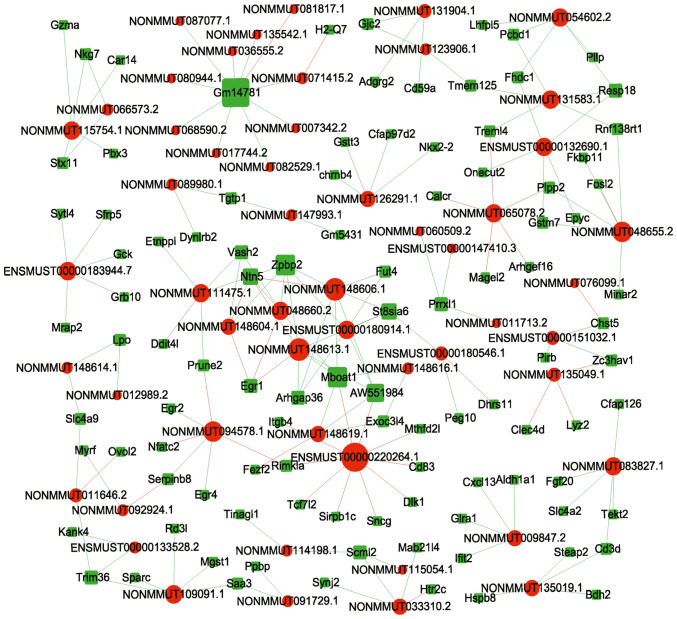
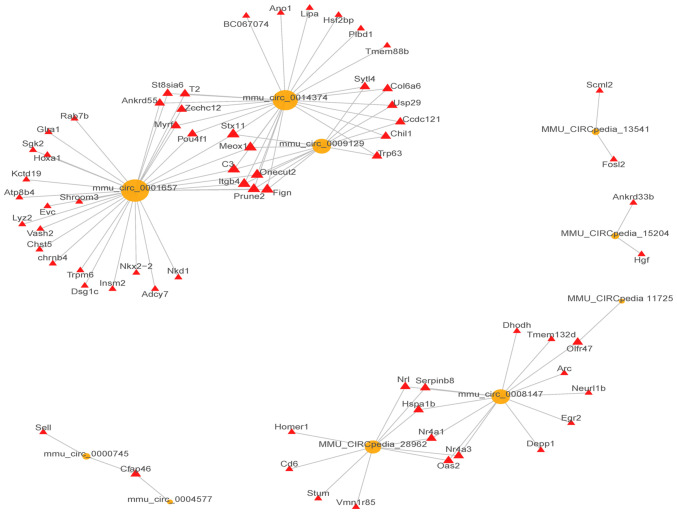
Overall, 648 network nodes and 152,453 connections (64,141 negative and 88,312 positive interactions) were identified in the lncRNA-mRNA co-expression network. The correlations between the top 20 (10 up and 10 downregulated) dysregulated lncRNAs and mRNAs are shown in Fig. 2. In total, 19,553 pairs of mRNA-circRNA interactions were identified (8,871 negative and 10,682 positive interactions) in the circRNA-mRNA co-expression network. The correlations between the top 10 (5 up and 5 downregulated) dysregulated lncRNAs and mRNAs are shown in Fig. 3.
Figure 2.
The top 20 lncRNA-mRNA interactions based on the network analysis. Red circles represent dysregulated lncRNAs, green squares represent dysregulated mRNAs.
Figure 3.
The top 10 circRNA-mRNA interactions based on the network analysis. Yellow circles represent dysregulated circRNAs, red triangles represent dysregulated mRNAs.
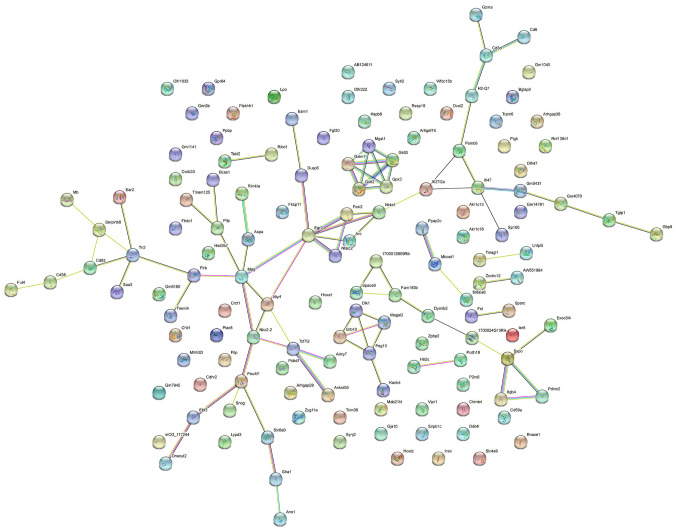
PPI network of differentially expressed mRNAs
The PPI network analysis conducted on the top 200 differentially expressed mRNAs consisted of 139 nodes and 88 edges, with an average node degree of 1.27 (Fig. 4; Table SI). Amongst the mRNAs, Pou4f1, Egr2, Mag, and Nr4a1 were the hub genes in the PPI network. The protein-encoding genes in the PPI network were primarily enriched in ‘Oligodendrocyte development’ (GO: 0014003), ‘Myelination’ (GO: 0042552), and ‘Glial cell development’ (GO: 0021782), and were involved in the ‘Glutathone metabolism’ pathway (mmu00480).
Figure 4.
Protein-protein interaction network based on the top 200 differentially expressed mRNAs.
GO and KEGG analysis of lncRNAs co-expressed with mRNAs
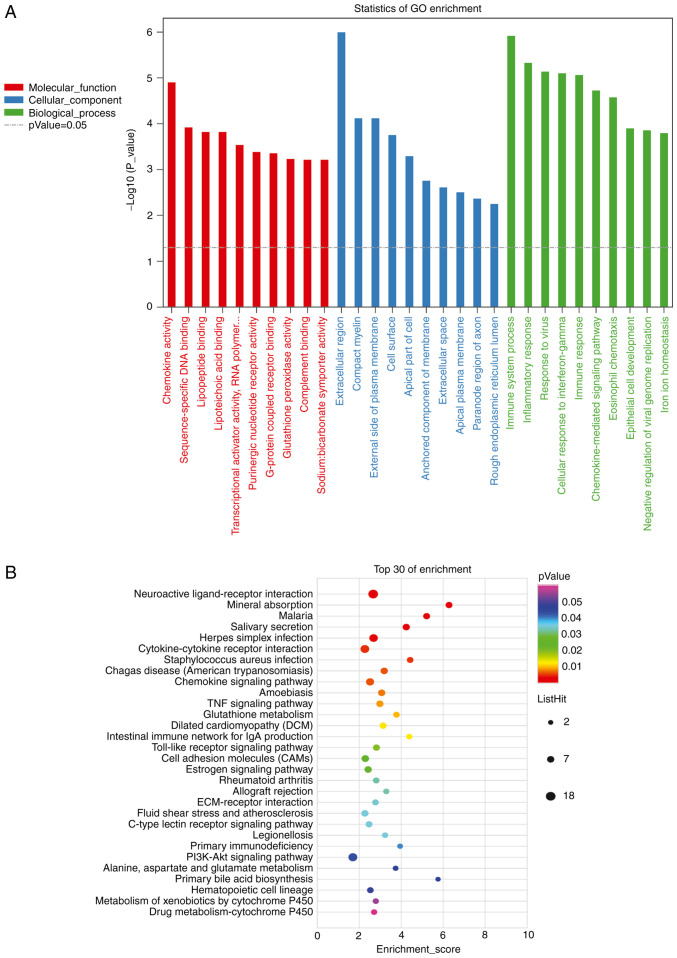
Overall, 499 genes were identified, including 396 genes annotated by BP, 402 genes annotated by CC, and 398 genes annotated by MF. There were 284 terms with P-values ≤0.05. ‘Extracellular region’ (GO:0005576, P=9.9x10-7), ‘immune system process’ (GO:0002376, P=1.2x10-6), and ‘inflammatory response’ (GO:0006954, P=4.6x10-6) had the lowest P-values (Fig. 5A; Table SII).
Figure 5.
KEGG pathway and GO enrichment analysis of differentially expressed lncRNAs. The top 30 most enriched GO categories and pathways were calculated and plotted. (A) GO enrichment analysis and (B) KEGG pathway analysis. KEGG, Kyoto encyclopedia of genes and genomes.
In total, 171 genes were annotated using KEGG pathway analysis, with 28 pathways having P-values ≤0.05. The top three pathways with minimum P-values were ‘Neuroactive ligand-receptor interaction’ (path: MMU04080, P=0.00013), ‘Mineral absorption’ (path: mmu04978, P=0.00034), and ‘Malaria’ (path: mmu05144, P=0.00095). KEGG pathway analysis suggested that lncRNAs were also involved in the ‘TNF signaling pathway’, ‘PI3K-Akt signaling pathway’, and ‘Wnt signaling pathway’ (Fig. 5B; Table SIII).
Cis/trans target genes of lncRNAs
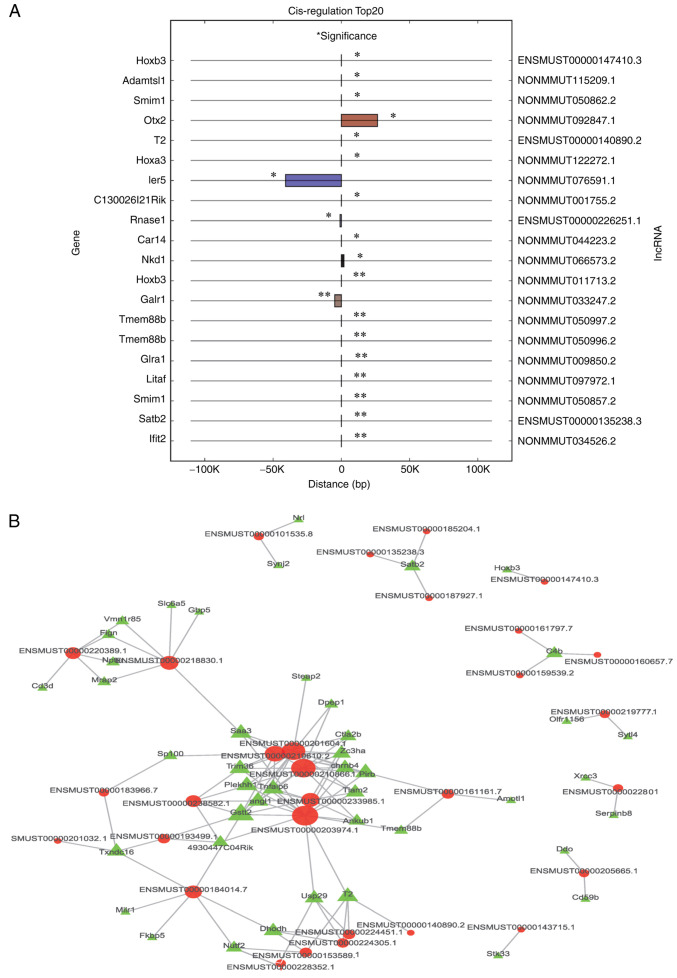
The top 20 cis-regulatory results are shown in Fig. 6A, and 46 cis-regulated pairs were identified, most of which were positive regulation pairs (91.3%). Additionally, the top 500 trans-regulated results are shown in Fig. 6B, where 748 trans-regulation pairs were identified.
Figure 6.
The trans- and cis- regulation of lncRNAs and genes. (A) the top 20 cis regulated lncRNAs and genes. mRNAs and lncRNAs are shown on the left and right Y-axis respectively. The X-axis shows the distance between the mRNA and lncRNA in the genome; negative values represent an upstream position and positive value represent a downstream position. Same colored bars represent the same lncRNAs. (B) The top 500 trans regulated lncRNAs and genes. The red node represents lncRNAs, the green node represents genes, and the node size represents connectivity (the number of connections of a node with other nodes). The larger the node size, the greater its connectivity. *P<0.05, **P<0.01.
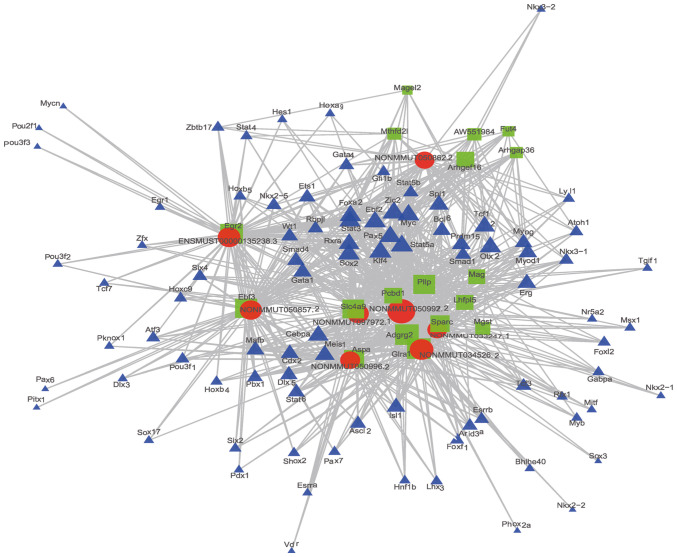
LncRNA-Target-TF network
In total, 98 TFs and 75 mRNAs were predicted to regulate or be the target of the top 20 cis-lncRNAs. Among the TFs, Cebpa (n=170), Zic2 (n=166), and Rxra (n=159) were predicted to regulate most of the lncRNAs. The 2 most related lncRNA-mRNA and lncRNA-TF pairs according to the P-value were used to construct the lncRNA-target-TF network (Fig. 7).
Figure 7.
lncRNA-target-TF network of the 2 most differently expressed lncRNAs. Red circles, lncRNAs; green squares, target mRNAs; purple triangles, TFs. TFs, transcription factors.
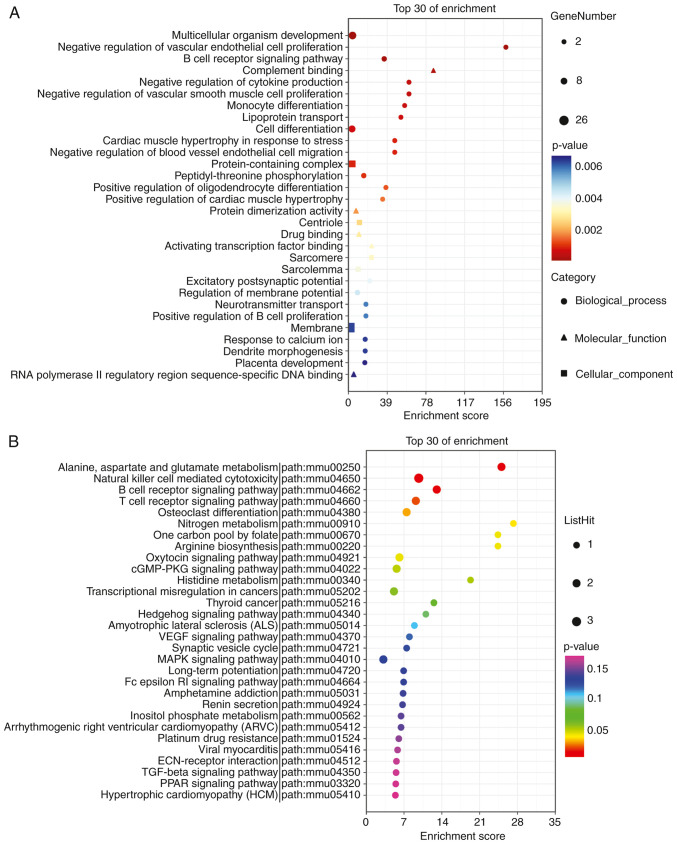
Enrichment analysis of circRNA host genes
Overall, 61 circRNA host genes were analyzed. Of these, 51 were annotated by BP, 53 genes were annotated by CC, and 52 genes were annotated for MF. ‘Multicellular organism development’ (GO: 0007275, P=2.1x10-5), ‘negative regulation of vascular endothelial cell proliferation’ (GO: 1905563, P=6.2x10-5), and ‘B-cell receptor signaling pathway’ (GO: 0050853, P=7.8x10-5) had the smallest P-values (Fig. 8A). In addition, 61 genes were identified in this study, and 17 genes were annotated as KEGG pathways. The top three pathways with minimum P-values were ‘Alanine, aspartate, and glutamate metabolism’ (path: mmu00250, P=0.0028), ‘Natural killer cell-mediated cytotoxicity’ (path: mmu04650, P=0.0033), and ‘B-cell receptor signaling pathway’ (path: mmu04662, P=0.01, Fig. 8B).
Figure 8.
KEGG pathway and GO enrichment analysis of differently expressed circRNA host genes. The top 30 most enriched GO categories and pathways were calculated and plotted. (A) GO and (B) KEGG pathway enrichment analyses. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
CircRNA-miRNA interactions
Overall, 275 miRNAs that bind to the 64 circRNAs were predicted. The top 4 differentially expressed circRNAs (2 upregulated and 2 downregulated) were mmu_circ_0007187, mmu_circpedia_35174, mmu_circpedia_35014, and mmu_circpedia_15204, and they were shown to bind to 5 miRNAs. The top 2 circRNA-miRNA interaction pairs are shown in Table II.
Table II.
The circRNA-miRNA interaction pairs of mmu_circ_0007187, mmu _circpedia_35174, mmu_circpedia_35014, and mmu_circpedia_15204.
| miRNA | circRNA | Total score | Total energy | Max score | Max energy | miRNA length | circRNA length | Positions |
|---|---|---|---|---|---|---|---|---|
| mmu_circ_0007187 | mmu-miR-92a-2-5p | 881 | -144.01 | 165 | -27.79 | 22 | 1,816 | 461, 1,337, 76, 1,301 1,373, 1,310 |
| mmu-miR-7006-5p | 753 | -129.77 | 167 | -30.12 | 25 | 1,816 | 1,329, 1,127, 1,409, 1,295, 1,300 | |
| mmu-miR-7092-3p | 706 | -84.66 | 145 | -22.43 | 24 | 1,816 | 618, 269, 207, 1,470, 1,786 | |
| mmu-miR-6931-5p | 620 | -123.24 | 169 | -36.94 | 21 | 1,816 | 1,343, 1,303, 464, 1,330 | |
| mmu-miR-6914-5p | 615 | -146.74 | 167 | -40.26 | 24 | 1,816 | 1,281, 1,415, 781, 947 | |
| MMU_CIRCpedia_35314 | mmu-miR-669h-5p | 326 | -35.45 | 165 | -19.12 | 25 | 378 | 273, 93 |
| mmu-miR-7048-3p | 314 | -39.08 | 168 | -23.15 | 24 | 378 | 274, 96 | |
| mmu-miR-1966-5p | 302 | -31.78 | 159 | -17.38 | 22 | 378 | 1, 73 | |
| mmu-miR-1a-2-5p | 295 | -44.5 | 154 | -22.58 | 22 | 378 | 209, 63 | |
| mmu-miR-1a-1-5p | 292 | -43.52 | 148 | -23.17 | 22 | 378 | 130, 74 | |
| MMU_CIRCpedia_35174 | mmu-miR-7680-5p | 288 | -30.18 | 144 | -16.93 | 20 | 393 | 86, 310 |
| mmu-miR-205-5p | 180 | -29.63 | 180 | -29.63 | 22 | 393 | 63 | |
| mmu-miR-669h-3p | 168 | -15.87 | 168 | -15.87 | 22 | 393 | 170 | |
| mmu-miR-8118 | 166 | -18.86 | 166 | -18.86 | 22 | 393 | 1 | |
| mmu-miR-669i | 163 | -14.74 | 163 | -14.74 | 21 | 393 | 169 | |
| MMU_CIRCpedia_35014 | mmu-miR-500-5p | 326 | -35.45 | 165 | -19.12 | 25 | 378 | 273, 93 |
| mmu-miR-362-5p | 314 | -39.08 | 168 | -23.15 | 24 | 378 | 274, 96 | |
| mmu-miR-7028-3p | 302 | -31.78 | 159 | -17.38 | 22 | 378 | 1, 73 | |
| mmu-miR-3074-5p | 295 | -44.5 | 154 | -22.58 | 22 | 378 | 209, 63 | |
| mmu-miR-1903 | 292 | -43.52 | 148 | -23.17 | 22 | 378 | 130, 74 | |
| MMU_CIRCpedia_15204 | mmu-miR-203-5p | 448 | -54.12 | 163 | -27.81 | 22 | 1,332 | 361, 515, 185 |
| mmu-miR-7028-5p | 438 | -70.06 | 153 | -25.95 | 23 | 1,332 | 211, 594, 1303 | |
| mmu-miR-432 | 437 | -57.75 | 151 | -20.8 | 23 | 1,332 | 599, 314, 953 | |
| mmu-miR-1291 | 333 | -38.53 | 180 | -21.55 | 26 | 1,332 | 534, 358 | |
| mmu-miR-761 | 319 | -45.01 | 168 | -23.52 | 22 | 1,332 | 431, 1,124 |
Discussion
In the present study, hippocampal tissues of AD+NS mice and AD+TG mice were used for microarray analysis. In total, 661 differentially expressed lncRNAs, 503 differentially expressed mRNAs, and 64 differentially expressed circRNAs were screened using bioinformatics analysis. A total of 12 RNAs (four lncRNAs, four mRNAs and four circRNAs) were randomly selected and analyzed using RT-qPCR, and the results were consistent with the microarray analyses.
There were 661 differentially expressed lncRNAs (341 upregulated and 320 downregulated) identified between the AD+NS mouse group and the AD mouse model treated with TG. Abnormal regulation of lncRNAs may cause cancer, epilepsy, heart disease, and neurodegenerative diseases (46-48). In addition, an increasing number of lncRNAs have been associated with the pathogenesis of AD. The innate immune system and inflammatory signaling are critical for homeostasis, repair, and neuroprotection. Excess oxygen free radicals and proinflammatory cytokines trigger an inflammatory cascade that ultimately leads to neurodegeneration (49). LncRNA-cox-2 is located downstream of cyclo-oxygenase 2 (COX2) and was reported to activate and inhibit the expression of various immune genes in macrophages and regulate NF-κB, which in turn affects aging and age-related diseases, including AD (50). In addition, the accumulation of mitochondrial superoxide free radicals and transformation to hydrogen peroxide can cause oxidative stress, the release of cytochrome C and apoptosis, which are also important mechanisms of AD (51). Several mitochondrial lncRNAs, including LNCND5, LNCND6, and LNCCYTB, may be involved in the regulation of mitochondrial genes and in maintaining normal mitochondrial function, which has been shown to be associated with neurodegenerative diseases (52). In our previous study, multiple lncRNAs associated with the pathogenesis of AD induced by LPS were detected. NONMMUT034127.2 and NONMMUT079254.1 were the most differentially expressed lncRNAs in the LPS-induced AD mouse model (53). In the present study, it was found that NONMMUG090228.1 and NONMMUG011123.2 were the most differentially expressed lncRNAs in an AD mouse model treated with TG, which indicated that these two lncRNAs may participate in the development of AD. However, to confirm this hypothesis, functional identification of the two lncRNAs in AD is necessary.
Four hub genes, Pou4f1, Egr2, Mag, and Nr4a1, were identified in the PPI network. Notably, these genes were shown to be involved in the formation, differentiation, maturation, apoptosis, and autophagy of neurons. POU4F1 (POU Class 4 Homeobox 1) is an important molecule in the POU TF family and is expressed in central neural precursor cells in the early stage of embryonic development (54). The TF POU4F1 can bind to the promoter region of the Bcl-2 gene to regulate its expression and interact with the P53 protein to regulate its transcriptional activity (55). Thus, the TF POU4F1 can promote the differentiation of the nervous system and inhibit the apoptosis of nerve cells. In the present study, Pou4f1 expression was significantly increased in the AD+TG group, which indicated that TG may exert a neuroprotective effect by inhibiting neuronal apoptosis via the regulation of Pou4f1.
The NR4A1 nuclear receptor belongs to the orphan nuclear receptor subfamily NR4As. Studies have shown that NR4A1 is involved in long-term memory formation (56). In the central nervous system, NR4A1 expression in the hippocampus increases after learning tasks related to hippocampal memory (57). The NR4A1 receptor is related to a variety of signaling molecules involved in memory formation, such as ERK, CREB, and BDNF (58). NR4A1 deficiency leads to late-phase long-term potentiation (L-LTP) and impaired long-term memory formation (59). In the present study, Nr4a1 expression was significantly increased in the AD+TG group, which indicated that TG may also improve memory in AD by regulating Nr4a1.
EGR2 plays an important role in peripheral nerve myelination, T-cell maturation, posterior brain segmentation, and lipid biosynthesis (60), whereas MAG plays an important role in maintaining myelin integrity and inhibiting axon regeneration in the central nervous system (61). Therefore, the results indicate that TG may ameliorate AD by regulating Pou4f1, Egr2, Mag, and Nr4a1 expression.
Furthermore, 64 differentially expressed circRNAs (34 upregulated and 30 downregulated) were identified between the AD+NS and AD+TG groups. circRNAs tend to accumulate in the aging brain, as well as in cells with low proliferation rates (such as neurons), especially neurons with synaptic development and differentiation (30,62). Zhang et al (63) found that there were several differentially expressed circRNAs in the brains of patients with AD. Knockdown/overexpression of some of these differentially expressed circRNAs was also assessed in AD cells and animal models to reduce AD-like pathological manifestations, suggesting that circRNAs may be involved in the regulation of AD pathology (64). Neuronal injury and apoptosis are the most intuitive pathological manifestations of neurodegenerative diseases, such as AD (65). Abnormal deposition of Aβ, neuroinflammation, oxidative stress injury, abnormal autophagy levels, and other factors can lead to neuronal injury and apoptosis (66,67). circRNAs are hypothesized to be involved in the above pathogenesis, as well as in AD. In previous studies, circHIPK2(68), circ-0002468(69), circ HDAC9(70), and circ-0000950(71) were shown to be involved in neuronal injury and to affect the pathogenesis and pathological process of AD. Circ-7(72), hsa_circ RNA-405619, and hsa_circ RNA-000843(73) were shown to participate in Aβ metabolism. In addition, circNF1-419(74) and circHECTD1(75) were shown to participate in autophagy and to affect the occurrence and development of AD. In the present study, 64 differentially expressed circRNAs were identified between the hippocampus of AD+NS mice and AD mice treated with TG. mmu_CIRCpedia_35174 and mmu_circ_0007187 were the most upregulated and downregulated circRNAs, respectively, which indicated that TG may treat AD by regulating the expression of these two circRNAs. However, the relationships of mmu_CIRCpedia_35174 and mmu_circ_0007187 with AD remain to be determined. Therefore, it is necessary to investigate the functions of these two circRNAs in AD treated with TG in the future.
In summary, several dysregulated lncRNAs, mRNAs, and circRNAs that may serve as potential biomarkers or targets in AD treated with TG were identified. Future studies should elucidate the detailed mechanisms underlying the regulation of the identified differentially expressed lncRNAs, mRNAs, and circRNAs.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: This work was supported by the National Natural Science Foundation of China (grant no. 81873780), The Changsha Outstanding Innovative Young People Training Scheme (grant nos. kq2206058 and kq2206056), The Foundation of Project of Hunan Health and Family Planning Commission (grant no. 202202082739), The Foundation of the Education Department of Hunan Province (grant nos. 21A0586 and 22A0662), The Foundation of the Education Department of Guangxi Province (grant no. 2021KY1959); The Hunan Key Laboratory Cultivation Base of the Research and Development of Novel Pharmaceutical Preparations (grant no. 2016TP1029), and the Application Characteristic Discipline of Hunan Province.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request. The relevant data can also be accessed via a public repository (GEO series accession no. GSE204817; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE204817).
Author's contributions
LT and JML designed the study, collected the data, performed the initial analysis, and drafted the manuscript. YW and QX aided in data acquisition, data analysis, and statistical analysis. JX, YZ and DWY performed the literature search, assisted with data acquisition, and edited the manuscript. YZ, JX and JML reviewed the manuscript. QX, DWY and YZ confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The research procedures were approved by the Ethics Committee of the Changsha Medical University, China (approval no. EC20190114).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mucke L. Alzheimer's disease. Nature. 2009;461:895–897. doi: 10.1038/461895a. [DOI] [PubMed] [Google Scholar]
- 2.Wenk GL. Neuropathologic changes in Alzheimer's disease. J Clin Psychiatry. 2003;64:7–10. [PubMed] [Google Scholar]
- 3.Du Q, Zhu X, Si J. Angelica polysaccharide ameliorates memory impairment in Alzheimer's disease rat through activating BDNF/TrkB/CREB pathway. Exp Biol Med (Maywood) 2020;245:1–10. doi: 10.1177/1535370219894558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou B, Tan J, Zhang C, Wu Y. Neuroprotective effect of polysaccharides from Gastrodia elata blume against corticosterone-induced apoptosis in PC12 cells via inhibition of the endoplasmic reticulum stress-mediated pathway. Mol Med Rep. 2018;17:1182–1190. doi: 10.3892/mmr.2017.7948. [DOI] [PubMed] [Google Scholar]
- 5.Wei M, Liu Y, Pi Z, Li S, Hu M, He Y, Yue K, Liu T, Liu Z, Song F, Liu Z. Systematically characterize the anti-Alzheimer's disease mechanism of lignans from S. chinensis based on in-vivo ingredient analysis and target-network pharmacology strategy by UHPLC-Q-TOF-MS. Molecules. 2019;24(1203) doi: 10.3390/molecules24071203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo X, Ye YJ, Song K, An LP, Sheng Y. Mechanism of schisandrae chinensis fructus lignans in alleviating learning and memory ability in D-galactose aging mice. Chin J Exp Trad Med Formulae. 2020;26:85–91. [Google Scholar]
- 7.Wu J, Qu JQ, Zhou YJ, Zhou YJ, Li YY, Huang NQ, Deng CM, Luo Y. Icariin improves cognitive deficits by reducing the deposition of β-amyloid peptide and inhibition of neurons apoptosis in SAMP8 mice. Neuroreport. 2020;31:663–671. doi: 10.1097/WNR.0000000000001466. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Zhang XB, Gu JH, Zeng YQ, Li JT. Breviscapine exerts neuroprotective effects through multiple mechanisms in APP/PS1 transgenic mice. Mol Cell Biochem. 2020;468:1–11. doi: 10.1007/s11010-020-03698-7. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Chen Y, Liang Y, Chen H, Ji X, Huang M. Berberine mitigates cognitive decline in an Alzheimer's disease mouse model by targeting both tau hyperphosphorylation and autophagic clearance. Biomed Pharmacother. 2020;121(109670) doi: 10.1016/j.biopha.2019.109670. [DOI] [PubMed] [Google Scholar]
- 10.Quan QK, Li X, Feng JJ, Hou JX, Li M, Zhang BW. Ginsenoside Rg1 reduces β-amyloid levels by inhibiting CDΚ5-induced PPARγ phosphorylation in a neuron model of Alzheimer's disease. Mol Med Rep. 2020;22:3277–3288. doi: 10.3892/mmr.2020.11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B, Li Q, Chu X, Sun S, Chen S. Salidroside reduces tau hyperphosphorylation via up-regulating GSK-3β phosphorylation in a tau transgenic Drosophila model of Alzheimer's disease. Transl Neurodegener. 2016;5:1–6. doi: 10.1186/s40035-016-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao J, Liu W, Zhou HY, Gui YR, Yan YH, Wu MJ, Xiao YF, Shang JT, Long GF, Shu XJ. Epigallocatechin-3-gallate alleviates cognitive deficits in APP/PS1 mice. Curr Med Sci. 2020;40:18–27. doi: 10.1007/s11596-020-2142-z. [DOI] [PubMed] [Google Scholar]
- 13.Hase T, Shishido S, Yamamoto S, Yamashita R, Nukima H, Taira S, Toyoda T, Abe K, Hamaguchi T, Ono K, et al. Rosmarinic acid suppresses Alzheimer's disease development by reducing amyloid β aggregation by increasing monoamine secretion. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-45168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo J, Song W, Xu Y, Chen GY, Hu Q, Tao QW. Benefits and safety of tripterygium glycosides and total glucosides of paeony for rheumatoid arthritis: An overview of systematic reviews. Chin J Integr Med. 2019;25:696–703. doi: 10.1007/s11655-019-3221-5. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Huang Y, Zhang Y, He C, Zhao Y, Wang L, Gao J. Efficacy of tripterygium glycosides combined with ARB on diabetic nephropathy: A meta-analysis. Biosci Rep. 2020;40(BSR20202391) doi: 10.1042/BSR20202391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li T, Zhou HC, Chen J. Effects of tripterygium wilfordii polyglycosides on the proliferation, apoptosis and PI3K/AKT signaling pathway of human oral cancer KB cells. J Guangdong Coll Pharm. 2019;35:653–657. (In Chinese) [Google Scholar]
- 17.Ma CL, Zhang BL, Liu XM. Effects of tripterygium glycosides on migration and angiogenesis of lung adenocarcinoma cell by inhibiting PI3K/AKT signaling. Anhui Med Pharm J. 2022;26:235–238. (In Chinese) [Google Scholar]
- 18.Wang M, Chen TG, Yang XL, Zhang DL, Zhou KS, Nan W, Zhang HH. Effect of tripterygium glycosides on inflammatory factors induced by lipopolysaccharide in rat astrocytes. Chin J Clin Pharmacol. 2019;35:154–158. (In Chinese) [Google Scholar]
- 19.Tang L, Xiang Q, Xiang J, Zhang Y, Li J. Tripterygium glycoside ameliorates neuroinflammation in a mouse model of Aβ25-35-induced Alzheimer's disease by inhibiting the phosphorylation of IκBα and p38. Bioengineered. 2021;12:8540–8554. doi: 10.1080/21655979.2021.1987082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun X, Malhotra A. Noncoding RNAs (ncRNA) in hepato cancer: A review. J Environ Pathol Toxicol Oncol. 2018;37:15–25. doi: 10.1615/JEnvironPatholToxicolOncol.2018025223. [DOI] [PubMed] [Google Scholar]
- 21.Faghihi M, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, Laurent GS III, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massone S, Vassallo I, Fiorino G, Castelnuovo M, Barbieri F, Borghi R, Tabaton M, Robello M, Gatta E, Russo C, et al. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol Dis. 2011;41:308–317. doi: 10.1016/j.nbd.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Massone S, Ciarlo E, Vella S, Nizzari M, Florio T, Russo C, Cancedda R, Pagano A. NDM29, a RNA polymerase III-dependent non coding RNA, promotes amyloidogenic processing of APP and amyloid β secretion. Biochim Biophys Acta. 2012;1823:1170–1177. doi: 10.1016/j.bbamcr.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Ciarlo E, Massone S, Penna I, Nizzari M, Gigoni A, Dieci G, Russo C, Florio T, Cancedda R, Pagano A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer's disease brain samples. Dis Model Mech. 2013;6:424–433. doi: 10.1242/dmm.009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mus E, Hof PR, Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc Natl Acad Sci USA. 2007;104:10679–10684. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parenti R, Paratore S, Torrisi A, Cavallaro S. A natural antisense transcript against Rad18, specifically expressed in neurons and upregulated during beta-amyloid-induced apoptosis. Eur J Neurosci. 2007;26:2444–2457. doi: 10.1111/j.1460-9568.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- 27.Verduci L, Tarcitano E, Strano S, Yarden Y, Blandino G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021;12:1–12. doi: 10.1038/s41419-021-03743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, Ibanez L, Fernandez MV, Farias F, Norton J, et al. An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci. 2019;22:1903–1912. doi: 10.1038/s41593-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma N, Pan J, Ye X, Yu B, Zhang W, Wan J. Whole-transcriptome analysis of APP/PS1 mouse brain and identification of circRNA-miRNA-mRNA networks to investigate AD pathogenesis. Mol Ther Nucleic Acids. 2019;18:1049–1062. doi: 10.1016/j.omtn.2019.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gruner H, Cortes-Lopez M, Cooper DA, Bauer M, Miura P. CircRNA accumulation in the aging mouse brain. Sci Rep. 2016;6(38907) doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. National Research Council (US) Institute for Laboratory Animal Research. Guide for the Care and Use of Laboratory Animals. National Academies Press, Washington, DC, 1996. [PubMed] [Google Scholar]
- 32.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(e47) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. R Core Team: R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, 2012. http://www.R-project.org/. [Google Scholar]
- 34. RStudio Team: RStudio, Integrated Development for R. RStudio, Inc., Boston MA, 2015. http://www.rstudio.com/. [Google Scholar]
- 35.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. The Gene Ontology Consortium. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M. Post-genome Informatics. Oxford University press Inc., New York, NY, 2000. [Google Scholar]
- 42.Wucher V, Legeai F, Hedan B, Rizk G, Lagoutte L, Leeb T, Jagannathan V, Cadieu E, David A, Lohi H, et al. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017;45(e57) doi: 10.1093/nar/gkw1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alkan F, Wenzel A, Palasca O, Kerpedjiev P, Rudebeck AF, Stadler PF, Hofacker IL, Gorodkin J. RIsearch2: Suffix array-based large-scale prediction of RNA-RNA interactions and siRNA off-targets. Nucleic Acids Res. 2017;45(e60) doi: 10.1093/nar/gkw1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wenzel A, Akbasli E, Gorodkin J. RIsearch: Fast RNA-RNA interaction search using a simplified nearest-neighbor energy model. Bioinformatics. 2012;28:2738–2746. doi: 10.1093/bioinformatics/bts519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen Y, Peng X, Shen C. Identification and validation of immune-related lncRNA prognostic signature for breast cancer. Genomics. 2020;112:2640–2646. doi: 10.1016/j.ygeno.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 47.Luo ZH, Walid AA, Xie Y, Long H, Xiao W, Xu L, Fu Y, Feng L, Xiao B. Construction and analysis of a dysregulated lncRNA-associated ceRNA network in a rat model of temporal lobe epilepsy. Seizure. 2019;69:105–114. doi: 10.1016/j.seizure.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 48.Wei CW, Luo T, Zou SS, Wu AS. The role of long noncoding RNAs in central nervous system and neurodegenerative diseases. Front Behav Neurosci. 2018;12(175) doi: 10.3389/fnbeh.2018.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh A, Kukreti R, Saso L, Kukreti S. Oxidative stress: A key modulator in neurodegenerative diseases. Molecules. 2019;24(1583) doi: 10.3390/molecules24081583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Z, Jiang S, Shang J, Jiang Y, Dai Y, Xu B, Yu Y, Liang Z, Yang Y. LncRNA: Shedding light on mechanisms and opportunities in fibrosis and aging. Ageing Res Rev. 2019;52:17–31. doi: 10.1016/j.arr.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Swerdlow RH. Mitochondria and mitochondrial cascades in Alzheimer's disease. J Alzheimers Dis. 2018;62:1403–1416. doi: 10.3233/JAD-170585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hezroni H, Perry RBT, Ulitsky I. Long noncoding RNAs in development and regeneration of the neural lineage. Cold Spring Harb Symp Quant Biol. 2019;84:165–177. doi: 10.1101/sqb.2019.84.039347. [DOI] [PubMed] [Google Scholar]
- 53.Tang L, Liu L, Li G, Jiang P, Wang Y, Li J. Expression profiles of long noncoding RNAs in intranasal LPS-mediated Alzheimer's disease model in mice. BioMed Res Int. 2019;2019(9642589) doi: 10.1155/2019/9642589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng M, Yang H, Xie X, Liang G, Gan L. Comparative expression analysis of POU4F1, POU4F2 and ISL1 in developing mouse cochleovestibular ganglion neurons. Gene Expr Patterns. 2014;15:31–37. doi: 10.1016/j.gep.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yadav R, Srivastava P. Establishment of resveratrol and its derivatives as neuroprotectant against monocrotophos-induced alteration in NIPBL and POU4F1 protein through molecular docking studies. Environ Sci Pollut Res Int. 2020;27:291–304. doi: 10.1007/s11356-019-06806-3. [DOI] [PubMed] [Google Scholar]
- 56.Jeanneteau F, Barrère C, Vos M, De Vries CJM, Rouillard C, Levesque D, Dromard Y, Moisan MP, Duric V, Franklin TC, et al. The stress-induced transcription factor NR4A1 adjusts mitochondrial function and synapse number in prefrontal cortex. J Neurosci. 2018;38:1335–1350. doi: 10.1523/JNEUROSCI.2793-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McNulty SE, Barrett RM, Vogel-Ciernia A, Malvaez M, Hernandez N, Davatolhagh MF, Matheos DP, Schiffman A, Wood MA. Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long-term memory. Learn Mem. 2012;19:588–592. doi: 10.1101/lm.026385.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z, Yu J. NR4A1 promotes cerebral ischemia reperfusion injury by repressing Mfn2-mediated mitophagy and inactivating the MAPK-ERK-CREB signaling pathway. Neurochem Res. 2018;43:1963–1977. doi: 10.1007/s11064-018-2618-4. [DOI] [PubMed] [Google Scholar]
- 59.Chen YL, Wang Y, Erturk A, Kallop D, Jiang Z, Weimer RM, Kaminker J, Sheng M. Activity-induced Nr4a1 regulates spine density and distribution pattern of excitatory synapses in pyramidal neurons. Neuron. 2014;83:431–443. doi: 10.1016/j.neuron.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 60.LeBlanc SE, Srinivasan R, Ferri C, Mager GM, Gillian-Daniel AL, Wrabetz L, Svaren J. Regulation of cholesterol/lipid biosynthetic genes by Egr2/Krox20 during peripheral nerve myelination. J Neurochem. 2005;93:737–748. doi: 10.1111/j.1471-4159.2005.03056.x. [DOI] [PubMed] [Google Scholar]
- 61.Llorens F, Gil V, del Río JA. Emerging functions of myelin-associated proteins during development, neuronal plasticity, and neurodegeneration. FASEB J. 2011;25:463–475. doi: 10.1096/fj.10-162792. [DOI] [PubMed] [Google Scholar]
- 62.Wang Z, Xu P, Chen B, Zhang Z, Zhang C, Zhan Q, Huang S, Xia ZA, Peng W. Identifying circRNA-associated-ceRNA networks in the hippocampus of Aβ1-42-induced Alzheimer's disease-like rats using microarray analysis. Aging (Albany NY) 2018;10:775–788. doi: 10.18632/aging.101427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Yu F, Bao S, Sun J. Systematic characterization of circular RNA-associated CeRNA network identified novel circRNA biomarkers in Alzheimer's disease. Front Bioeng Biotechnol. 2019;7(222) doi: 10.3389/fbioe.2019.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castillo E, Leon J, Mazzei G, Abolhassani N, Haruyama N, Saito T, Saido T, Hokama M, Iwaki T, Ohara T, et al. Comparative profiling of cortical gene expression in Alzheimer's disease patients and mouse models demonstrates a link between amyloidosis and neuroinflammation. Sci Rep. 2017;7(17762) doi: 10.1038/s41598-017-17999-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fielder E, Von Zglinicki T, Jurk D. The DNA damage response in neurons: Die by apoptosis or survive in a senescence-like state? J Alzheimer's Dis. 2017;60:S107–S131. doi: 10.3233/JAD-161221. [DOI] [PubMed] [Google Scholar]
- 66.Coimbra-Costa D, Alva N, Duran M, Carbonel T, Rama R. Oxidative stress and apoptosis after acute respiratory hypoxia and reoxygenation in rat brain. Redox Biol. 2017;12:216–225. doi: 10.1016/j.redox.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Galluzzi L, Pedro JMBS, Blomgren K, Kroemer G. Autophagy in acute brain injury. Nat Rev Neurosci. 2016;17:467–484. doi: 10.1038/nrn.2016.51. [DOI] [PubMed] [Google Scholar]
- 68.Wang G, Han B, Shen L, Wu S, Yang L, Liao J, Wu F, Li M, Leng S, Zang F, et al. Silencing of circular RNA HIPK2 in neural stem cells enhances functional recovery following ischaemic stroke. EBioMedicine. 2020;52(102660) doi: 10.1016/j.ebiom.2020.102660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang M, Xiang G, Yu D, Yang G, He W, Yang S, Zhou G, Liu A. Hsa_circ_0002468 regulates the neuronal differentiation of SH-SY5Y cells by modulating the MiR-561/E2F8 axis. Med Sci Monit. 2019;25:2511–2519. doi: 10.12659/MSM.915518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang N, Gao Y, Yu S, Sun XH, Shen K. Berberine attenuates Aβ42-induced neuronal damage through regulating circHDAC9/miR-142-5p axis in human neuronal cells. Life Sci. 2020;252(117637) doi: 10.1016/j.lfs.2020.117637. [DOI] [PubMed] [Google Scholar]
- 71.Yang H, Wang H, Shang H, Chen X, Yang S, Qu Y, Ding J, Li X. Circular RNA circ_0000950 promotes neuron apoptosis, suppresses neurite outgrowth and elevates inflammatory cytokines levels via directly sponging miR-103 in Alzheimer's disease. Cell Cycle. 2019;18:2197–2214. doi: 10.1080/15384101.2019.1629773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shi Z, Chen T, Yao Q, Zheng L, Zhang Z, Wang J, Hu Z, Cui H, Han Y, Han X, et al. The circular RNA cirs-7 promotes APP and BACE 1 degradation in an NF-κB-dependent manner. FEBS J. 2017;284:1096–1109. doi: 10.1111/febs.14045. [DOI] [PubMed] [Google Scholar]
- 73.Li Y, Lv Z, Zhang J, Ma Q, Li Q, Song L, Gong L, Zhu Y, Li X, Hao Y, Yang Y. Profiling of differentially expressed circular RNAs in peripheral blood mononuclear cells from Alzheimer's disease patients. Metab Brain Dis. 2020;35:201–213. doi: 10.1007/s11011-019-00497-y. [DOI] [PubMed] [Google Scholar]
- 74.Chen D, Guo Y, Qi L, Tang X, Liu Y, Yang X, Hu GY, Shuai Q, Yong Y, Wang D, et al. Circular RNA NF1-419 enhances autophagy to ameliorate senile dementia by binding Dynamin-1 and adaptor protein 2 B1 in AD-like mice. Aging (Albany NY) 2019;11:12002–12031. doi: 10.18632/aging.102529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han B, Zhang Y, Zhang Y, Bai Y, Chen XF, Huang R, Wu FF, Shou L, Chao J, Zhang J, et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: Implications for cerebral ischemic stroke. Autophagy. 2018;14:1164–1184. doi: 10.1080/15548627.2018.1458173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request. The relevant data can also be accessed via a public repository (GEO series accession no. GSE204817; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE204817).