Abstract
Grainyhead-like 2 (GRHL2) is a transcription factor that suppresses epithelial-to-mesenchymal transition (EMT). It has been previously shown that GRHL2 can confer both oncogenic and tumor-suppressive roles in human cancers, including breast, pancreatic and colorectal cancers. However, its role in lung cancer remains elusive. In the present study, a meta-analysis of multiple gene expression datasets with clinical data revealed that GRHL2 expression was increased in lung cancer compared with that in the normal tissues. Copy number analysis of GRHL2, performed using datasets of whole exome sequencing involving 151 lung cancer cell lines, revealed frequent amplifications, suggesting that the increased GRHL2 expression may have resulted from gene amplification. A survival meta-analysis of GRHL2 using The Cancer Genome Atlas (TCGA) dataset showed no association of GRHL2 expression with overall survival. GRHL2 expression was found to be associated with EMT status in lung cancer in TCGA dataset and lung cancer cell lines. GRHL2 knockdown induced partial EMT in the hTERT/Cdk4-immortalized normal lung epithelial cell line HBEC4KT without affecting proliferation measured by CCK-8 assays. In addition, GRHL2 silencing caused three lung cancer cell lines, H1975, H2009 and H441, to undergo partial EMT. However, the proliferative effects differed significantly. GRHL2 silencing promoted proliferation but not colony formation in H1975 cells whilst suppressing colony formation without affecting proliferation in H2009 cells, but it did not affect proliferation in H441 cells. These results suggest cell type-dependent effects of GRHL2 knockdown. Downstream, GRHL2 silencing enhanced the phosphorylation of AKT and ERK, assessed by western blotting with phospho-specific antibodies, in HBEC4KT, H1975 and H2009 cell lines but not in the H441 cell line. By contrast, transient GRHL2 overexpression did not affect A549 cell proliferation, which lack detectable endogenous expression of the GRHL2 protein. However, GRHL2 overexpression did suppress E-cadherin expression in A549 cells. These results suggested that GRHL2 does not only function as a tumor suppressor of EMT but can also behave as an oncogene depending on the lung cancer cell-type context.
Keywords: epithelial-to-mesenchymal transition, human bronchial epithelial cell line, The Cancer Genome Atlas
Introduction
During epithelial-mesenchymal transition (EMT), which was first discovered in embryonic developmental studies, epithelial cells undergo notable changes in morphology, as characterized by polarized and tightly connected to adjacent cells to being less polarized and loosely connected, with spindle-like morphology and acquire functional properties of mesenchymal cells, such as a motile and invasive phenotype (1–3). Cancer cells acquires invasiveness and metastatic potential, by exploiting the EMT machinery that endows the cells with a motile and invasive phenotype (4).
EMT is regulated by several EMT-inducing transcription factors that serve as master regulators of EMT (3,4). By contrast, a number of EMT-repressing transcription factors have also been discovered (3). Among them, Grainyhead-like 2 (GRHL2) encodes a transcription factor that has been reported to regulate wound healing, tubulogenesis and cancer (5,6). However, GRHL2 can exert both tumor-promoting and -suppressive functions depending on the human cancer involved (7–21). In pancreatic cancer, GRHL2 functions as a tumor-promoting gene by conferring metastatic potential to cancer cells, by retaining epithelial but acquiring stem cell properties (11). By contrast, in breast cancer GRHL2 inhibits anchorage-independent growth, where the loss of GRHL2 expression has been found to be associated with higher tumor stages (14). This suggests the tumor-suppressive role of GRHL2 in breast cancer (14). In addition, another study has previously reported tumor-suppressive roles of GRHL2 in breast cancer, by demonstrating that suppression of EMT by GRHL2 increased sensitivity to anoikis (10).
To the best of our knowledge, few reports have investigated the roles of GRHL2 in lung cancer (12). GRHL2 is overexpressed in lung cancer compared with that in normal cells, where it was associated with inferior prognosis. GRHL2 silencing was reported to suppress proliferation and colony formation whilst enhancing migration and invasion. These data are difficult to interpret, because the results of proliferation and colony formation, together with increased GRHL2 expression, suggested that GRHL2 serves oncogenic roles, but those from migration and invasion assays suggested a tumor-suppressive role. Furthermore, GRHL2 has been demonstrated to exert oncogenic roles by stabilizing a ‘hybrid epithelial/mesenchymal phenotype’ in H1975 lung cancer cells (22). In that particular study, this ‘hybrid epithelial/mesenchymal phenotype’ was considered to be a promoter of cancer metastasis. These findings are contradictory to some extent and suggest a highly complex role of GRHL2 in lung cancer. This justifies further investigations into the roles of GRHL2 in lung cancer pathophysiology.
Therefore, in the present study, the role of GRHL2 in lung cancer was investigated using the online bioinformatic tool, Lung Cancer Explorer (LCE) and preclinical models, including hTERT/Cdk4-immortalized normal lung epithelial cells and lung cancer cell lines (23–26).
Materials and methods
Cell culture
All 10 of lung cancer cell lines (HCC827, HCC4006, HCC4017, A549, H460, H441, H661, H1792, H1975 and H2009) used in the present study and the hTERT/Cdk4-immortalized normal human bronchial epithelial cell lines HBEC3KT and HBEC4KT were purchased from American Type Culture Collection and were derived from the Hamon Center Collection University of Texas Southwestern Medical Center (Dallas, USA) (27,28). The lung cancer cell lines were cultured in RPMI-1640 (cat. no. 189-02025; FUJIFILM Wako Pure Chemical Corporation) supplemented with 10% FBS (cat. no 35-079-CV; Corning, Inc.). HBEC3KT and HBEC4KT cells were cultured in keratinocyte-serum free medium (cat. no. 10724-011; Thermo Fisher Scientific, Inc.) supplemented with 5 ng/ml epidermal growth factor (cat. no. PHG0314; Thermo Fisher Scientific, Inc.) and 50 ng/ml bovine pituitary extract (cat. no. 13028-014; Thermo Fisher Scientific, Inc.). All cells were cultured at 37°C in a humidified 5% CO2 incubator. The provenance of H1975, H2009 and H441 was confirmed by DNA fingerprinting short tandem repeat analysis. Negativity for mycoplasma contamination in the cultured cell lines was confirmed using the VenorGeM Onestep kit (cat. no. 11-8050) according to the manufacturer's protocol (Minerva Biolabs GmbH) or by mycoplasma group-specific PCR (29).
Small-interfering RNA (siRNA) transfection
A total of 3×105 cells (HBEC4KT, H1975, H2009 and H441) were transfected with predesigned siRNA (Silencer Select RNAi; cat. no. 4427037, Thermo Fisher Scientific, Inc.) to a final concentration of 6.25 nM using Lipofectamine® RNAiMAX (cat. no. 13778-150; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Knockdown of GRHL2 was performed using two siRNA oligos, si-GRHL2#1 (cat. no. 4427037; ID, s36755; Thermo Fisher Scientific, Inc.) and si-GRHL2#2 (cat. no. 4427037, ID, s36756; Thermo Fisher Scientific, Inc.). The negative control used was Silencer™ Select Negative Control No. 1 siRNA (siNC; cat. no. 4390844; Thermo Fisher Scientific, Inc.). All transfections were performed at 37°C in a humidified 5% CO2 incubator. Durations of transfection were two days for cell proliferation and colony formation assays and four days for harvesting cells for RNA and protein extractions. Subsequent experimental procedures were performed immediately after completion of the transfections.
GRHL2 overexpression
A custom-ordered GRHL2 expression vector, pLenti6/V5-GW/GRHL2, was constructed by replacing the coding sequence of LacZ in a pLenti6/V5-GW/lacZ vector (cat. no. K4955-10; Thermo Fisher Scientific, Inc.) with the GRHL2 reference sequence (accession no. NM_024915.4), purchased from GenScript. 2×105 of A549 cells plated in 6 cm dishes were transiently transfected with 1.5 ug of either the pLenti6/V5-GW/GRHL2 or the pLenti6/V5-GW/lacZ vector used as a control using Fugene®4K transfection reagent (cat. no. E5911; Promega Corporation) as aforementioned. Subsequently, 2 days of culture at 37°C in a humidified 5% CO2 incubator after the transfection, cells were harvested for protein or RNA extraction, or they were re-plated for proliferation or colony formation assays. Transfection efficiency was evaluated in 1×105 A549 cells plated in 6-well plates by co-transfecting with 0.32 µg EGFP-expressing vector pEGFP-N3 (cat. no. 6080; Clontech.) at the ratio of 1:1 to the pLenti6/V5-GW/GRHL2 or pLenti6/V5-GW/lacZ vector (cat. no. K4955-10, Thermo Fisher Scientific, Inc.).
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Four days after transfection, total RNA was extracted from the cells (HBEC4KT, H1975, H2009 and H441) using the RNeasy mini kit (cat. no. 74106; Qiagen GmbH) following the manufacturer's protocol. Reverse transcription was completed using the SuperScript IV First-Strand Synthesis System with a random primer system (cat. no. 18091050; Thermo Fisher Scientific, Inc.) Using the PowerUp™ SYBR™ Green Master Mix (cat. no. A25742; Thermo Fisher Scientific, Inc.), qPCR was performed for E-cadherin, VIMENTIN, TWIST1, Zinc finger E-box-binding homeobox 1 (ZEB1), snail homolog (SNAI)1 and SNAI2. The thermocycling conditions were follows: 95°C for 20 sec, 40 cycles of 95°C for 3 sec and 60°C for 30 sec. The melt condition was follows; 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. GAPDH was used as an internal control, and the relative expression level was calculated by the ΔΔCq method (30). The primer sequences are shown in Table SI.
Cell proliferation assays
In total, 2 days after the transfection of siRNA, the cells (HBEC4KT, H1975, H2009, or H441) were plated into 96-well plates at 5,000 cells/well. Subsequently, 2 days later, a colorimetric proliferation assay was performed using the Cell Counting Kit-8 (CCK-8: cat. no. CK04; Dojindo Molecular Technologies, Inc.) according to the manufacturer's protocol. Cells were incubated at 37°C, 5% CO2 for 2 h, measured absorbance at 450 nm and at 600 nm as a reference wavelength.
Colony formation assay
In total, 2 days after siRNA transfection, the cells (H441, H1975, H2009, and HBEC4KT) were replated into six-well plates at 1,000 cells/well for GRHL2 silencing experiments. After 10 days of culture at 37°C, 5% CO2, the colonies were stained with a solution containing 0.5% methylene blue tetrahydrate (cat. no. 137-06982; FUJIFILM Wako Pure Chemical Corporation) and 50% ethanol for fixation) at room temperature for 30 min. Colonies >1 mm in diameter were counted manually.
Western blot analysis
Western blotting was conducted as described previously using whole cell lysates (31). Lysate of transfected cells was collected 4 days after siRNA transfection and 2 days after GRHL2 overexpression. Cell lysate of untransfected normal and lung cancer cell lines was collected at ~80% confluency. The primary antibodies used were rabbit anti-GRHL2 (cat. no. HPA062839; Merck KGaA; 1:500), mouse anti-E-cadherin (cat. no. 610181; BD Biosciences; 1:1,000), mouse anti-vimentin (cat. no. HPA001762; Merck KGaA; 1:500), rabbit anti-AKT (pan; cat. no. 4691; Cell Signaling Technology, Inc.; 1:1,000), rabbit anti-phosphorylated (p-)-AKT (ser473; cat. no. 4060; Cell Signaling Technology, Inc.; 1:1,000), rabbit anti-p44/42MAPK (Erk1/2; cat. no. 9102; Cell Signaling Technology, Inc.; 1:1,000), rabbit anti-p-p44/42MAPK (Erk1/2; cat. no. 4370; Cell Signaling Technology, Inc.; 1:1,000) and mouse anti-β-actin (cat. no. A2228; Merck KGaA; 1:2,000). The β-actin protein level was used as a control for protein loading. The primary antibodies were incubated at 4°C overnight. The secondary antibodies conjugated with horseradish peroxidase (HRP) were anti-rabbit (cat. no. NA934-1ML; Cytiva) and anti-mouse antibodies (cat. no. NA931-1ML; Cytiva) at a 1:2,000 dilution and incubated at room temperature for 1 h. Primary and secondary antibodies of GRHL2 were diluted by Can Get Signal™ (cat. no. NKB-101; Toyobo Life Science). The intensities of the bands were quantified with Fiji (v.2.9.0/1.53t) (https://imagej.net/software/fiji/) (32) or ImageJ (v.1.53k) (https://imagej.nih.gov/ij/index.html) (33), where values of intensities normalized to β-actin, AKT or ERK were shown below the corresponding band images in the figures.
DNA copy number analysis
To perform a DNA copy number analysis, whole-exome sequencing (dbGaP Study Accession: phs001823.v1.p1; http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001823.v1.p1) (34). A total of 68 lung adenocarcinoma, 20 lung squamous cell carcinoma and 63 non-small cell lung cancer (NSCLC)-histology not otherwise specified lung cancer cell lines were used (Table SII). Copy number variation (CNV) was determined using the R package ‘DNAcopy’ (R version 3.6, r-project.org/; DNAcopy version 1.36, https://bioconductor.org/packages/release/bioc/html/DNAcopy.html) with several modifications.
Choice of diploid controls
Because of differences in target capture enrichment during whole-exome sequencing, diploid controls were selected from the same batch as tumor samples. These batches were determined computationally by estimating the amount of noise (deviation from copy number segments) for each pair of tumor and control samples, followed by the hierarchical clustering of these noise values. Typically, tumor-control pairs from the same batch will have noticeably lower noise values. Next diploid controls were compared to other controls (from the same batch) to generate CNV segmentation plots. This allowed a further selection of controls with no or few CNV.
Recalibration
Because tumor samples are not diploid and frequently do not have an equal amount of copy number gains and losses, copy numbers were recalibrated by the visual inspection of each plot. In general, the lowest segments were set to one copy [log2(tumor/control)=−1], which shifted the subsequent segments so that log2(tumor/control) can be 0 (2 copies), 0.58 (3 copies), 1 (4 copies), and so on.
Statistical analysis
The Lung Cancer Explorer (LCE; http://lce.biohpc.swmed.edu/lungcancer/) was used to analyze the data from The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov/) and to perform meta-analyses on expression levels of GRHL2 in tumor vs. normal tissues and impact of GRHL2 expression on survival in patients with lung cancer. Univariate and multivariate analyses were performed using EZR (v. 1.55; jichi.ac.jp/saitama-sct/SaitamaHP.files/statmedEN.html) (35). Pearson's correlation coefficients with associated P-values were calculated for correlation analyses by using EZR (v. 1.55). Statistically significant differences between two comparisons (control and one of the investigated groups) or >2 groups (control and multiple of the investigated groups) were analyzed by unpaired t-test or one-way ANOVA with Dunnett's post hoc test respectively, using the SPSS software (v.28; IBM Corp.). P<0.05 were considered to be statistically significant difference.
Results
GRHL2 expression is upregulated in lung cancer but is not associated with patient survival
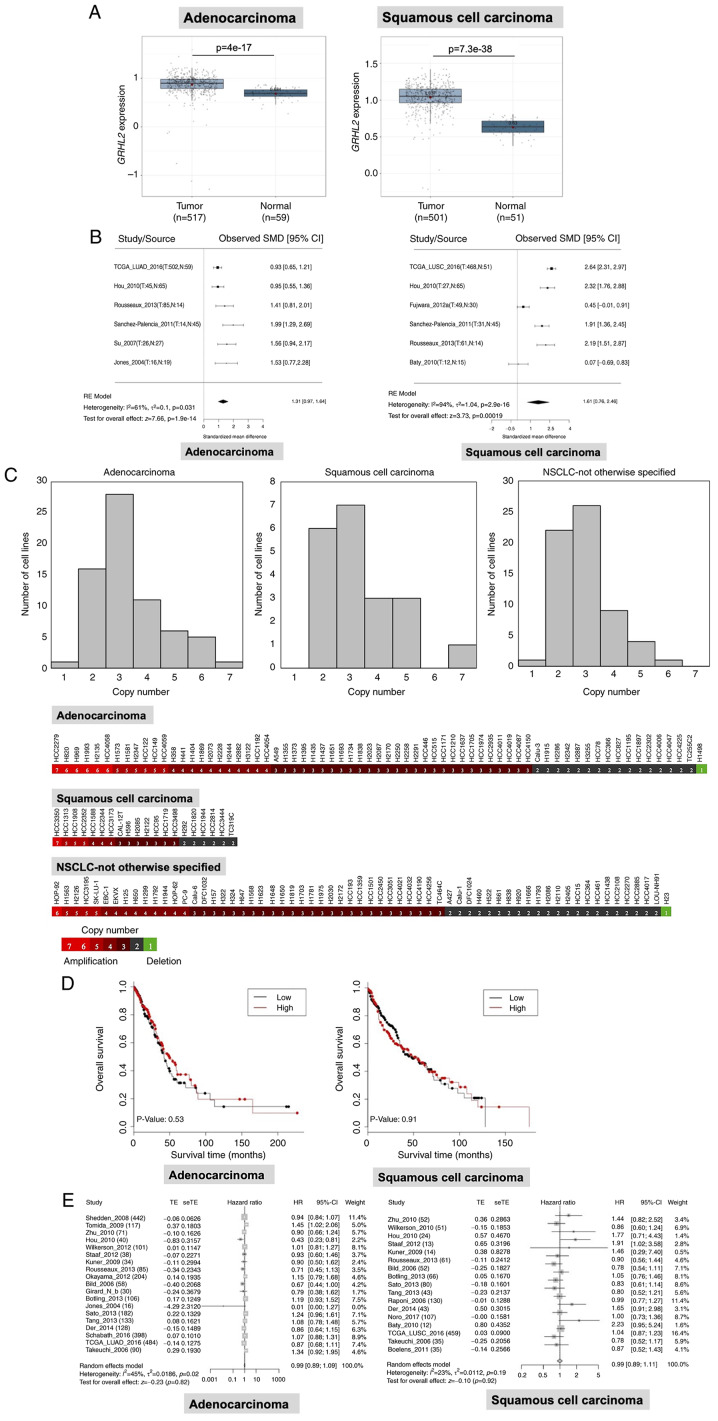
To investigate the GRHL2 expression profile in lung cancer and its impact on patient survival, statistical analyses were performed using LCE (23), with focus on a dataset of TCGA (cancer. gov/ccg/research/genome-sequencing/tcga). GRHL2 expression was revealed to be significantly upregulated in both lung adenocarcinoma and squamous cell carcinoma tissues compared with that in normal lung tissues (Fig. 1A) in TCGA dataset. This result was confirmed by a meta-analysis of six datasets, including TCGA. GRHL2 expression in adenocarcinoma and squamous cell carcinoma tissues was 1.31 and 1.61-fold higher compared with that in normal tissue, respectively (Fig. 1B). In addition, copy number analysis by whole-exome sequencing, carried out as part of a different study (34), revealed increases in GRHL2 copy numbers. Specifically, 51/68 (75.0%) adenocarcinoma, 14/20 (70.0%) squamous cell carcinoma and 40/63 (63.5%) NSCLC-not otherwise specified cell lines had > three GRHL2 copies (mean ± SD; 3.35±1.25 in adenocarcinoma, 3.35±1.31 in squamous cell carcinoma and 2.94±0.97 in NSCLC-otherwise specified cell lines; Fig. 1C). These data suggest that the increased GRHL2 expression may have resulted from gene amplification. Subsequently, it was investigated whether GRHL2 expression was associated with patient survival in patients with adenocarcinoma or squamous cell carcinoma. Kaplan-Meier analysis in TCGA dataset revealed that overall survival was not significantly different in the GRHL2 high- and low-expression adenocarcinoma and squamous cell carcinoma groups, following division using the median expression value (Fig. 1D). Univariate and multivariate Cox regression analysis in TCGA dataset showed that GRHL2 expression was not associated with survival in patients with either adenocarcinoma (Table I) or squamous cell carcinoma (Table II). In addition, the absence of correlation between GRHL2 expression between patient survival in lung cancer was also confirmed by a metanalysis using LCE (Fig. 1E). These data suggest that the expression of GRHL2 was upregulated in both adenocarcinoma and squamous cell carcinoma, but this phenomenon was not associated with patient survival.
Figure 1.
GRHL2 is expressed at high levels in lung cancer. (A) Expression analysis of GRHL2 in lung adenocarcinoma and squamous cell carcinoma in TCGA dataset. (B) Meta-analysis of GRHL2 expression in lung adenocarcinoma and squamous cell carcinoma from six studies. T, tumor, N, normal, RE, random effects. (C) DNA copy number analysis of GRHL2 showing frequent increases in the copy number of GRHL2 in NSCLC cell lines. (D) Kaplan-Meier survival curves for patients with high or low GRHL2 expression (the cut-off value is the median expression level). (E) Survival meta-analysis for GRHL2 in adenocarcinoma (left) and squamous cell carcinoma (right). In each forest plot, the name of each study is followed by the number of total tumor samples. GRHL2, Grainyhead-like 2; SMD, standardized mean difference; TE, estimated treatment effect; seTE, standard error of treatment effect; HR, hazard ratio; CI, confidence interval, TCGA, The Cancer Genome Atlas; NSCLC, non-small cell lung cancer.
Table I.
Univariate and multivariate analyses of cox multivariate regression analysis in adenocarcinoma in TCGA cohort.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Sex | ||||||
| Female | Reference | Reference | ||||
| Male | 0.9058 | 0.632–1.298 | 0.5903 | 0.8083 | 0.5362–1.219 | 0.3097 |
| Age, years | ||||||
| >65 | Reference | Reference | ||||
| ≤65 | 0.7696 | 0.5353–1.107 | 0.1575 | 0.6787 | 0.4589–1.004 | 0.0522 |
| Smoking status | ||||||
| Never | Reference | Reference | ||||
| Former | 0.9214 | 0.5095–1.666 | 0.7864 | 1.2300 | 0.6490–2.332 | 0.5255 |
| Current | 0.7176 | 0.3668–1.404 | 0.3324 | 0.7643 | 0.3741–1.561 | 0.4609 |
| Stage | ||||||
| I | Reference | Reference | ||||
| II | 2.516 | 1.582–4.002 | <0.0001a | 3.0790 | 1.8620–5.091 | <0.0001a |
| III | 4.426 | 2.774–7.062 | <0.0001a | 4.8250 | 2.9390–7.922 | <0.0001a |
| IV | 3.266 | 1.665–6.407 | 0.0006a | 4.4210 | 2.1770–8.981 | <0.0001a |
| GRHL2 mRNA | ||||||
| Low | Reference | Reference | ||||
| High | 0.9162 | 0.6413–1.309 | 0.1575 | 0.9025 | 0.6140–1.327 | 0.6018 |
P<0.05. GRHL2 expression levels were classified as high and low with median as the cut-off value. HR, hazard ratio; GRHL2, grainyhead-like 2; CI, confidence interval.
Table II.
Univariate and multivariate analyses of cox multivariate regression analysis in squamous cell carcinoma in TCGA cohort.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Sex | ||||||
| Female | Reference | Reference | ||||
| Male | 1.038 | 0.7188–1.498 | 0.8434 | 1.0120 | 0.6924–1.478 | 0.9528 |
| Age, years | ||||||
| >65 years | Reference | Reference | ||||
| ≤65 years | 0.767 | 0.5447–1.080 | 0.1286 | 0.6727 | 0.4708–0.9611 | 0.0294 |
| Smoking status | ||||||
| Never | Reference | Reference | ||||
| Former | 0.4628 | 0.1695–1.264 | 0.1327 | 0.4100 | 0.1462–1.150 | 0.0903 |
| Current | 0.5929 | 0.2114–1.663 | 0.3205 | 0.5453 | 0.1894–1.570 | 0.2611 |
| Stage | ||||||
| I | Reference | Reference | ||||
| II | 1.202 | 0.8161–1.771 | 0.3514 | 1.1420 | 0.7654–1.704 | 0.5150 |
| III | 1.499 | 0.9979–2.251 | 0.0512 | 1.6030 | 1.060–2.424 | 0.0254 |
| IV | 2.042 | 0.6373–6.542 | 0.2295 | 1.8550 | 0.5745–5.992 | 0.3014 |
| GRHL2 mRNA | ||||||
| Low | Reference | Reference | ||||
| High | 1.008 | 0.7317–1.387 | 0.963 | 0.9540 | 0.6845–1.330 | 0.7812 |
GRHL2 expression levels were classified as high and low with median as the cut-off value. HR, hazard ratio; CI, confidence interval; GRHL2, grainyhead-like 2.
GRHL2 expression is associated with the epithelial phenotype in lung cancer
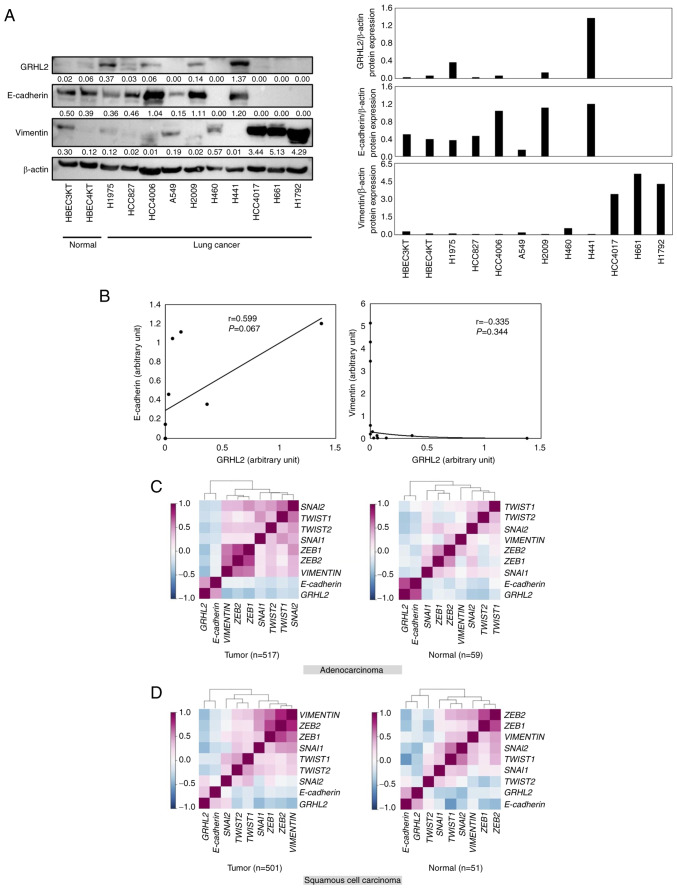
The association between GRHL2 protein expression and the EMT status in lung cancer was next investigated by evaluating the expression levels of GRHL2, E-cadherin (an epithelial marker) and vimentin (a mesenchymal marker) in a panel of lung cancer cell lines (HCC827, HCC4006, HCC4017, A549, H460, H441, H661, H1792, H1975, and H2009). As shown in Fig. 2A and B, expression levels of GRHL2 were not significantly positively correlated with E-cadherin (Pearson's correlation coefficient=0.059; P=0.067) or negatively correlated with Vimentin (Pearson's correlation coefficient=0.335; P=0.344). The correlation among the expression levels of GRHL2, E-cadherin, vimentin and master EMT transcription factors ZEB1, ZEB2, SNAI1, SNAI2, TWIST1 and TWIST2, was then investigated using LCE. It was shown that GRHL2 expression was positively and negatively correlated with E-cadherin and vimentin, respectively, in both patients with adenocarcinoma and patients with squamous cell carcinoma (Fig. 2C and D; Table SIII). This suggests that GRHL2 expression is associated with the epithelial phenotype in lung cancer. In addition, GRHL2 expression was found to be negatively correlated with five of the six (except SNAI2) of the master EMT transcription factors (Fig. 2C and D; Table SIII).
Figure 2.
GRHL2 is associated with an epithelial phenotype in lung cancer. (A) Western blotting of GRHL2, E-cadherin and Vimentin in a panel of lung cancer and immortalized bronchial epithelial cell lines. Values below bands indicate the semi-quantitation of band intensities normalized to β-actin. (B) Correlations between bands intensities of GRHL2 and E-cadherin (left graph) or Vimentin (right graph) in (A). Correlation analysis between the expression of GRHL2, E-cadherin, VIMENTIN and six master epithelial-to-mesenchymal transition regulator genes in (C) adenocarcinomas and (D) squamous cell carcinomas in The Cancer Genome Atlas dataset. The degrees of positive or negative correlation, revealed by Pearson's correlation coefficients, are colored by the darkness of purple or blue, respectively. GRHL2, Grainyhead-like 2; ZEB, Zinc finger E-box-binding homeobox; SNAI, snail homolog.
GRHL2 silencing induces a partial EMT in an hTERT/Cdk4-immortalized normal lung epithelial cell line without affecting growth
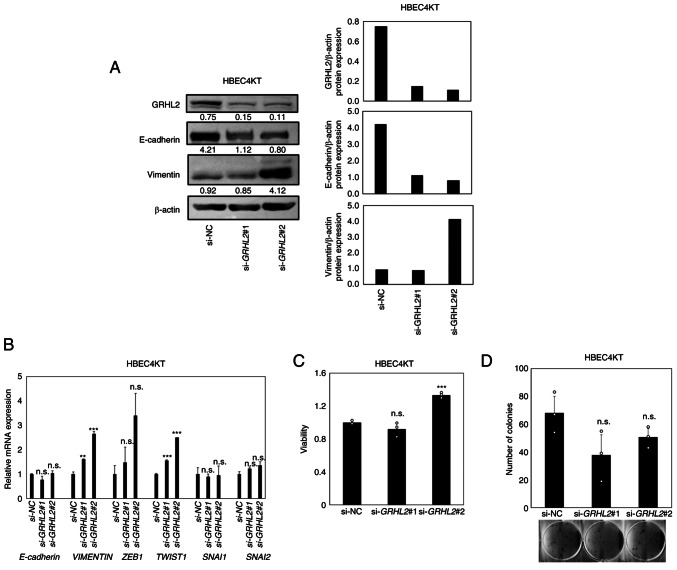
Next, to evaluate the impact of GRHL2 silencing on EMT in normal lung epithelial cells, knockdown experiments were performed using the hTERT/Cdk4 immortalized normal lung epithelial cell line HBEC4KT (28). This cell line does not harbor genetic alterations that may affect EMT phenotypes (28). GRHL2 silencing was found to decrease E-cadherin whilst increasing vimentin expression, with upregulation in expression also observed in master EMT transcription factors TWSIT1 (statistically significant) and ZEB1 (not statistically significant; Fig. 3A and B). These suggest that partial EMT, characterized by decreased E-cadherin and increased Vimentin, albeit without completely switching their expression, has occurred (36). To evaluate the effects of this partial EMT on cell proliferation and colony formation, CCK-8 and colony formation assays were performed but no significant differences could be identified except for increased viability in si-GRHL2#2-treated cells (Fig. 3C and D). Therefore, these data suggest that GRHL2 silencing promoted partial EMT in this cell line, but it did not significantly affect cell proliferation.
Figure 3.
GRHL2 silencing induces a partial EMT in hTERT/Cdk4-immortalized normal lung epithelial cell line (HBEC4KT) without affecting its growth. (A) Western blotting of GRHL2, E-cadherin and Vimentin in the immortalized normal bronchial epithelial cell line HBEC4KT transfected with either control or GRHL2 siRNA. Values below bands indicate quantitation of band intensities normalized to β-actin. Quantification of band intensities is shown in the right graph. (B) Reverse transcription-quantitative PCR of E-cadherin, VIMENTIN and four master EMT regulators in HBEC4KT cells, transfected with either control (si-NC) or GRHL2 siRNA. (C) Proliferation and (D) colony formation assays for HBEC4KT cells transfected with either control or GRHL2 siRNA. Results are shown as the mean ± SD (n=3), where comparisons were performed with one-way ANOVA followed by the Dunnett test. **P<0.01 and ***P<0.001 vs. si-NC. GRHL2, Grainyhead-like 2, siRNA, small-interfering RNA; NC, negative control; n.s., non-significant; EMT, epithelial-to-mesenchymal transition; ZEB, Zinc finger E-box-binding homeobox; SNAI, snail homolog.
GRHL2 silencing induces partial EMT in lung cancer with cell type-dependent effects on growth
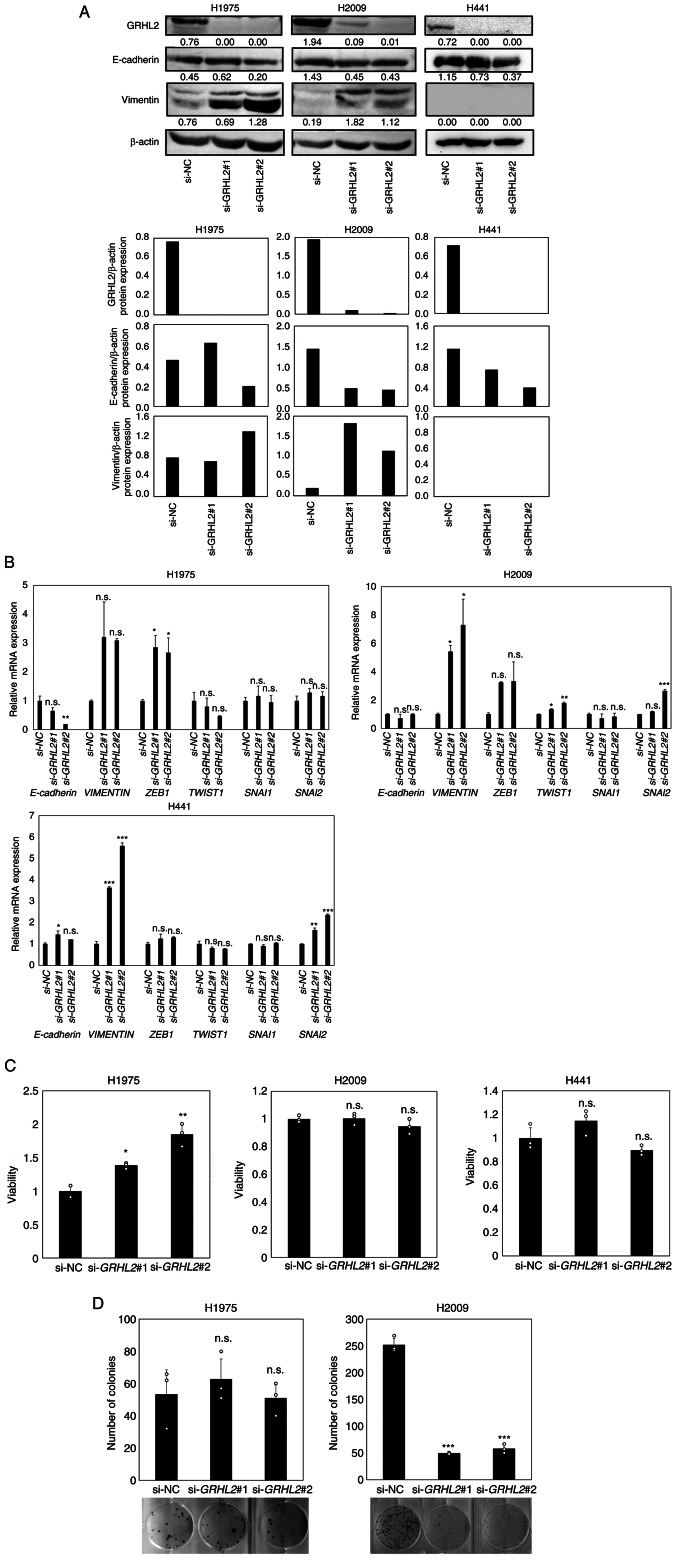
Next, three lung adenocarcinoma cancer cell lines H1975, H2009 and H441 were selected for evaluating the effects of GRHL2 silencing on EMT status and growth. H1975 [TP53 (Arg273His) and EGFR (Thr790Met, Leu858Arg) mutant) and H2009 (TP53 (Arg273Leu) and KRAS (Gly12Ala) mutant] were selected due to the expression of significant levels of GRHL2 protein (Fig. 2A) and due to the possession of two major representative lung adenocarcinoma driver mutations (37,38). In addition, H441 [TP53 (Arg158Leu) and KRAS (Gly12Val) mutant] was selected for experimentation because it expressed the highest levels of GRHL2 protein among the lung cancer cell line panel tested (Fig. 2A). GRHL2 silencing did not alter E-cadherin protein or mRNA expression in any of the three aforementioned cell lines (Fig. 4A). By contrast, increased vimentin protein and mRNA expression levels in H1975 and H2009 cells were observed. However, GRHL2 silencing significantly increased vimentin mRNA expression but did not affect protein expression in H441 cells (Fig. 4A and B). GRHL2 silencing resulted in the upregulation of ZEB1 expression in H1975, TWIST1 expression in H2009 and SNAI2 in H2009 and H441, suggesting a partial EMT and variable response to this silencing (Fig. 4A and B). GRHL2 silencing also increased cell viability in H1975 cells but not in either H2009 or H441 cells (Fig. 4C). The colony formation assay revealed a significant decrease in colony formation by H2009 cells, but no change was observed in H1975 cells (Fig. 4D). H441 cells did not form colonies and therefore could not be tested (Fig. 4D). These results suggest that GRHL2 silencing induced EMT in the lung cancer cell lines, but its effects on proliferation and colony formation were cell type-dependent and varied greatly among the lung adenocarcinoma cell lines.
Figure 4.
GRHL2 silencing induces a partial EMT in lung cancer but its effects on growth are cell type-dependent. (A) Western blotting of GRHL2, E-cadherin and vimentin in the lung cancer cell lines H1975 and H2009 transfected with control or GRHL2 siRNA. Values below bands indicate quantitation of band intensities normalized to β-actin. Quantification of band intensities is shown in the bottom graph. (B) Reverse transcription-quantitative PCR of E-cadherin, vimentin and four master EMT regulator genes in H1975 and H2009 cells transfected with control (si-NC) or GRHL2 siRNA. (C) Proliferation and (D) colony formation assays of H1975 and H2009 cells transfected with either control or GRHL2 siRNA. Results are shown as the mean ± SD, assessed by one-way ANOVA followed by the Dunnett test. *P<0.05, **P<0.01 and ***P<0.001 vs. si-NC. GRHL2, Grainyhead-like 2; siRNA, small-interfering RNA; NC, negative control; n.s., non-significant; EMT, epithelial-to-mesenchymal transition; ZEB, Zinc finger E-box-binding homeobox; SNAI, snail homolog.
GRHL2 silencing promotes the phosphorylation of AKT and ERK
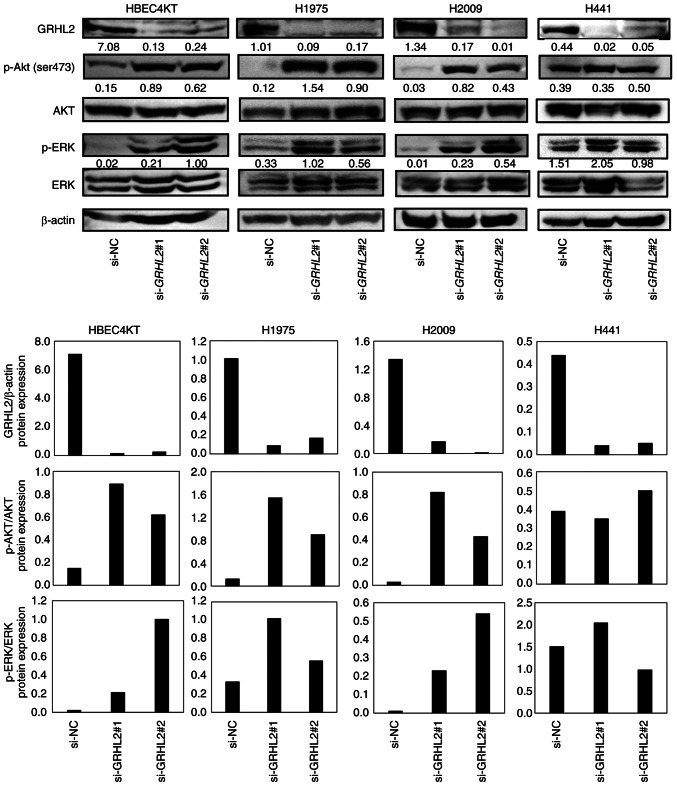
Compared with the present study, numerous studies have reported that EMT can promote cancer cell proliferation (39–41). Additionally, several studies have found oncogenic roles in GRHL2 by showing growth suppression in several cancer cell types, including breast, pancreatic and colorectal cancers, after GRHL2 silencing (5,6,11,16,18,20). Growth suppression induced by GRHL2 silencing occurred through inhibition of the AKT or ERK pathways (18,20). This finding that GRHL2 silencing causes the inhibition of AKT and ERK is inconsistent with the function of GRHL2 as an EMT suppressor because the AKT and ERK pathways are associated with EMT in various types of canceer including lung and colorectal cancers (42–44). It was therefore hypothesized that the differential effects of GRHL2 silencing on growth may be due to whether the AKT or ERK pathway was induced or suppressed by GRHL2 silencing. Therefore, the effect of GRHL2 silencing on the phosphorylation of AKT and ERK was next assessed in HBEC4KT, H1975, H2009 and H441 cell lines by western blotting. GRHL2 silencing enhanced the phosphorylation of both AKT and ERK in HBEC4KT, H1975 and H2009 cell lines, but it did not affect either AKT or ERK in H441 (Fig. 5). The finding that both p-AKT and p-ERK were equally upregulated in HBEC4KT, H1975 and H2009 cells was unexpected, because the effects of GRHL2 silencing in these cells further downstream differed substantially. This suggests that the differential effects of GRHL2 silencing on cell proliferation among normal bronchial epithelial and lung cancer cells do not occur through the differences in the phosphorylation of the AKT or ERK pathways.
Figure 5.
GRHL2 knockdown promotes the phosphorylation of AKT and ERK in HBEC4KT and lung cancer cell lines. Western blotting of GRHL2, p- or total AKT and p- or total ERK in HBEC4KT, H1975 and H2009 cells transfected with control or GRHL2 siRNA. Values below bands indicate the semi-quantitation of band intensities normalized to AKT for p-AKT (ser473), ERK for p-ERK or β-actin for GRHL2. GRHL2, Grainyhead-like 2; siRNA, small-interfering RNA; NC, negative control; p-, phosphorylated.
GRHL2 overexpression does not affect the proliferation in A549 cells
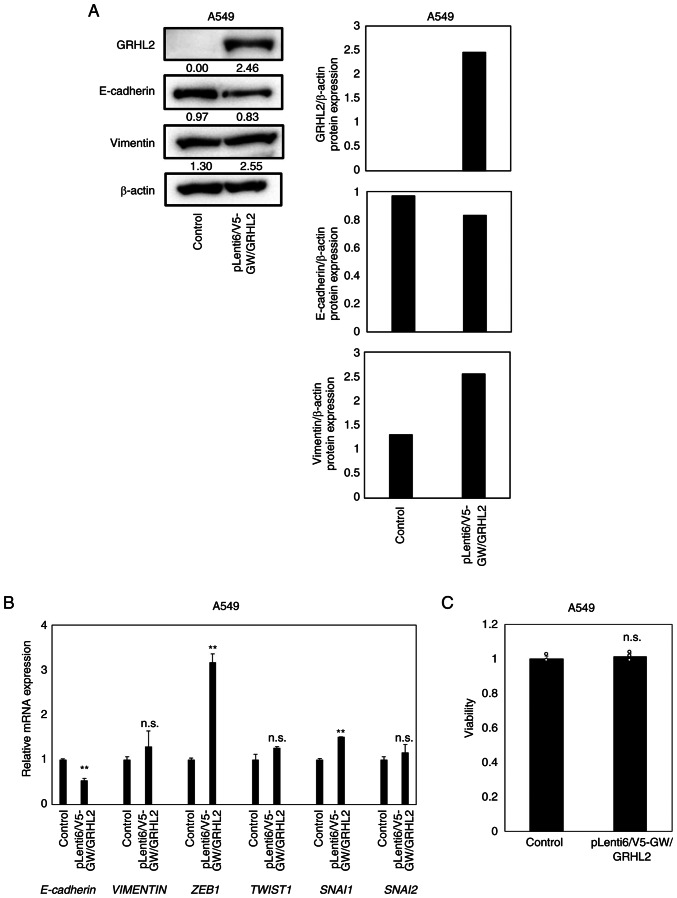
To evaluate the effects of GRHL2 overexpression in lung cancer cells, GRHL2 overexpression experiments were performed using A549 cells [TP53 wild-type, KRAS(Gly12Ser) and liver kinase B1 (Gln37Ter) mutant], which lacked detectable expression levels of the GRHL2 protein (Fig. 2A). In total, ~50% transfection efficiency was confirmed by co-transfection with the EGFP-expressing vector pEGFP-N3 (Fig. S1). Overexpression of the GRHL2 protein was achieved in the A549 cells transfected with the GRHL2-expressing vector (Fig. 6A). GRHL2 overexpression reduced the protein expression of E-cadherin, whilst the protein expression of vimentin remained unaltered (Fig. 6A). GRHL2 overexpression decreased the mRNA expression of E-cadherin and the increased mRNA expression of ZEB1 and SNAI1 without affecting TWIST1 or SNAI2 expression (Fig. 6B). However, GRHL2 overexpression did not affect A549 proliferation (Fig. 6C).
Figure 6.
GRHL2 overexpression does not affect proliferation in A549 cells. (A) Western blotting of GRHL2, E-cadherin and Vimentin in the lung cancer cell line A549 transiently transfected with GRHL2- or LacZ-expressing vectors. Quantification of band intensities is shown in the right graph. (B) Reverse transcription-quantitative PCR of E-cadherin, vimentin and four master epithelial-to-mesenchymal regulator genes in A549 transiently transfected with GRHL2- or LacZ-expressing (control) vectors. **P<0.01 vs. control. (C) Proliferation assay for A549 transiently transfected with GRHL2- or LacZ-expressing vectors. Results are shown as the means ± SD (n=3), where comparisons with controls were performed by the unpaired t-test. n.s., indicates ‘no significant difference’. GRHL2, Grainyhead-like 2; ZEB, Zinc finger E-box-binding homeobox; n.s., not significant; SNAI, snail homolog.
Discussion
Both oncogenic and tumor suppressive roles of GRHL2 have been reported in various malignancies (7–21). A study previously showed that GRHL2 is overexpressed in lung cancer, which is in turn associated with poor prognosis. In addition, silencing GRHL2 expression was found to suppress proliferation and colony formation, but it also promoted migration and invasion in lung cancer cell lines (12). Based on the finding that GRHL2 silencing promoted migration and invasion in a lung cancer cell line H1299, this previous study concluded that GRHL2 primarily functions as a tumor suppressor by suppressing metastasis (12). However, this previous study also presented data suggesting tumor-promoting roles of GRHL2, including that higher expression levels of GRHL2 in lung cancer samples are associated with poorer patient prognosis. In addition, suppressed proliferation and colony formation resulted from GRHL2 silencing (12). Therefore, the roles of GRHL2 in lung cancer remain unclear. In the present study, increased expression of GRHL2 was observed in lung cancer tissues without association with patient survival. Expression data from a public database together with the experimental results of the present study indicated that GRHL2 likely serves an important role in maintaining an epithelial phenotype in lung cancer. Nevertheless, despite its EMT-suppressing function in lung cancer, it was shown that GRHL2 silencing resulted in either growth-promoting or -suppressing effects in lung cancer cell lines.
Consistent with a previous study (12), it was shown that GRHL2 is overexpressed in lung cancer in a meta-analysis of gene expression datasets. In addition, this previous study also showed that patients with lung cancer and higher protein expression of GRHL2 had significantly shorter survival (12). However, the meta-analysis in the present study revealed the absence of association between GRHL2 expression and patient survival in lung cancer, suggesting that GRHL2 expression is not associated with survival in patients with lung cancer. This was unexpected because of the apparent EMT-suppressing function of GRHL2. Bi-directional effects of GRHL2 on the growth of lung cancer cells were observed, which likely explains the reason underlying this ambiguous result.
GRHL2 is one of the several transcription factors that can suppress EMT (3,6). However, the predominant roles that GRHL2 plays in maintaining the epithelial phenotypes in lung cancer remain to be clarified. The present study suggested an association between GRHL2 expression and epithelial phenotypes was identified in a panel of lung cancer cell lines and clinical tumors tissues. Additionally, GRHL2 silencing induced partial EMT in lung cancer cell lines and an immortalized normal bronchial epithelial cell line. These results suggest an important role of GRHL2 in maintaining an epithelial phenotype in lung cancer.
Despite the partial EMT induced by GRHL2 silencing in the lung cancer cell lines, its impact on growth differed substantially among the cell lines tested, as shown by enhanced viability in H1975 and inhibited clonogenic growth in H2009. To gain insights into the interpretation of these results, the phosphorylation of AKT and ERK was investigated because these growth-promoting proteins were previously found to be suppressed by GRHL2 silencing (18,20). GRHL2 silencing resulted in increased levels of p-AKT and p-ERK in all cell lines studied, suggesting that the differential activation of either AKT or ERK may not be attributable to the observed differences in the effects of GRHL2 silencing on cell proliferation. Notably, one previous study (20) reported that GRHL2 silencing suppressed the proliferation of two colorectal cancer cell lines through decreased AKT phosphorylation. However, in the present study, it was shown that GRHL2 silencing resulted in the increased activation of AKT even in the H2009 lung cancer cell line, which in turn inhibited colony formation (Fig. 4D). These findings suggested that the effects of GRHL2 on the AKT or ERK pathways markedly vary among cancer types. Therefore, the effects of GRHL2 silencing on the phosphorylation status of AKT and ERK should be evaluated in a variety of human cancer cell lines, such as breast and pancreatic cancer. ERK can serve a pro-apoptotic function when it is translocated into the mitochondria instead of the nucleus (45). Therefore, it remains possible that upregulated ERK activity may not promote, but rather suppress, cell proliferation in the experiments performed in the present study.
A discrepancy was observed in the results of cell viability and colony formation assays in H2009 cells, whereby GRHL2 silencing did not affect proliferation but inhibited colony formation. It was hypothesized that these contradictory results could be explained by the differences in the properties of cancer cells these two assays evaluate. The viability assay evaluates cell proliferation, whereas colony formation assays measure the ability of individual cells to survive and propagate over long periods of time, typically over 10–14 days (46,47). Notably, a previous study (46) described that colony formation assays can be used to evaluate self-renewing capacity in vitro. Relevant roles of GRHL2 in self-renewal have been reported (48,49). The growth suppression induced by GRHL2 silencing in the present study was only observed in colony formation but not in viability assays in H2009, suggesting the involvement of GRHL2 in cell stemness. Collectively, results of the present study in H2009 cells suggest that GRHL2 may confer anti-apoptotic and/or self-renewing abilities. Therefore, this hypothesis warrants further investigation in future studies.
GRHL2 overexpression did not affect proliferation in A549 cells, which lacked detectable expression levels of the endogenous GRHL2 protein. Both growth-promoting and -suppressive effects of GRHL2 overexpression have been reported in numerous types of different cell models (8,10,13–16,20). To further evaluate the effects of GRHL2 overexpression on proliferation and clonogenic growth in lung cells, GRHL2 overexpression experiments using additional lung cancer cell lines and immortalized normal lung epithelial cells are planned as future experiments. In addition, it was an unexpected observation that GRHL2 overexpression decreased the expression of E-cadherin on both mRNA and protein levels because of its function of positive transcription factor for E-cadherin (3). One possible explanation for this result is the increased ZEB1 expression in A549 cell, which was also induced in response to GRHL2 overexpression. It was possible that ZEB1 counteracted the function of GRHL2 as a positive regulator of E-cadherin expression, leading to decreased levels of E-cadherin expression. To test this hypothesis, luciferase reporter assay is required for evaluating transcriptional activity of E-cadherin in GRHL2-overexpressing cells with or without ZEB1 silencing.
Altogether, the present study revealed that GRHL2 expression was associated with an epithelial phenotype, but its expression was not associated with prognosis in lung cancer. Gene silencing experiments suggested that GRHL2 is not likely to be a definitive tumor suppressor or an oncogene, instead acting as either of them depending on the cell context.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported, in part, by the Grant-in-Aid for Exploratory Research (grant no. 19K22617), Grant-in-Aid for Scientific Research (grant no. 21H02924), the 45th (2020) Aichi Cancer Research Foundation to M. Sato, Nagoya University CIBoG program from MEXT WISE program and National Cancer Institute, SPORE: Developing New Rationale, Personalized Medicine for Lung Cancer (grant no.,P50 CA070907).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
NK and MS conceived and designed the study. NK, KM, KK, NM, MT, YT, YI, MY, NS and MS acquired data. NK, KM, LG, LC, YX, IT, MM, LG, JDM and MS analyzed and interpreted the data. LG and JDM provided technical or material support. NK, NS, LG, JDM and MS wrote, reviewed and revised the manuscript. All authors have read and approved the final version of the manuscript. NK and MS confirm the authenticity of all the raw data.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
JM receives royalties from the NIH and University of Texas Southwestern for distribution of human cell lines. The remaining authors declare that they have no competing interests.
References
- 1.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 2.Lim J, Thiery JP. Epithelial-mesenchymal transitions: Insights from development. Development. 2012;139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 3.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 5.Frisch SM, Farris JC, Pifer PM. Roles of grainyhead-like transcription factors in cancer. Oncogene. 2017;36:6067–6073. doi: 10.1038/onc.2017.178. [DOI] [PubMed] [Google Scholar]
- 6.He J, Feng C, Zhu H, Wu S, Jin P, Xu T. Grainyhead-like 2 as a double-edged sword in development and cancer. Am J Transl Res. 2020;12:310–331. [PMC free article] [PubMed] [Google Scholar]
- 7.Chung VY, Tan TZ, Tan M, Wong MK, Kuay KT, Yang Z, Ye J, Muller J, Koh CM, Guccione E, et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci Rep. 2016;6:19943. doi: 10.1038/srep19943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and grainyhead-like-2. Cancer Res. 2013;73:6299–6309. doi: 10.1158/1538-7445.FBCR13-A15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cieply B, Riley P IV, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM. Suppression of the epithelial-mesenchymal transition by grainyhead-like-2. Cancer Res. 2012;72:2440–2453. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farris JC, Pifer PM, Zheng L, Gottlieb E, Denvir J, Frisch SM. Grainyhead-like 2 reverses the metabolic changes induced by the oncogenic epithelial-mesenchymal transition: Effects on anoikis. Mol Cancer Res. 2016;14:528–538. doi: 10.1158/1541-7786.MCR-16-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishino H, Takano S, Yoshitomi H, Suzuki K, Kagawa S, Shimazaki R, Shimizu H, Furukawa K, Miyazaki M, Ohtsuka M. Grainyhead-like 2 (GRHL2) regulates epithelial plasticity in pancreatic cancer progression. Cancer Med. 2017;6:2686–2696. doi: 10.1002/cam4.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan X, Zhang R, Xie C, Gan M, Yao S, Yao Y, Jin J, Han T, Huang Y, Gong Y, et al. GRHL2 suppresses tumor metastasis via regulation of transcriptional activity of RhoG in non-small cell lung cancer. Am J Transl Res. 2017;9:4217–4226. [PMC free article] [PubMed] [Google Scholar]
- 13.Shen J, Lv X, Zhang L. GRHL2 acts as an anti-oncogene in bladder cancer by regulating ZEB1 in epithelial-mesenchymal transition (EMT) process. Onco Targets Ther. 2020;13:2511–2522. doi: 10.2147/OTT.S239120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner S, Frey S, Riethdorf S, Schulze C, Alawi M, Kling L, Vafaizadeh V, Sauter G, Terracciano L, Schumacher U, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem. 2013;288:22993–23008. doi: 10.1074/jbc.M113.456293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang J, Fu X, Ran W, Wang Z. Grhl2 reduces invasion and migration through inhibition of TGFβ-induced EMT in gastric cancer. Oncogenesis. 2017;6:e284. doi: 10.1038/oncsis.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang X, Deng Z, Zhuang X, Ju S, Mu J, Jiang H, Zhang L, Yan J, Miller D, Zhang HG. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One. 2012;7:e50781. doi: 10.1371/journal.pone.0050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, Wu D, Chen Y, Min Z, Quan Y. GRHL2 inhibits colorectal cancer progression and metastasis via oppressing epithelial-mesenchymal transition. Cancer Biol Ther. 2019;20:1195–1205. doi: 10.1080/15384047.2019.1599664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Kang KL, Alshaikh A, Varma S, Lin YL, Shin KH, Kim R, Wang CY, Park NH, Walentin K, et al. Grainyhead-like 2 (GRHL2) knockout abolishes oral cancer development through reciprocal regulation of the MAP kinase and TGF-β signaling pathways. Oncogenesis. 2018;7:38. doi: 10.1038/s41389-018-0047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faddaoui A, Sheta R, Bachvarova M, Plante M, Gregoire J, Renaud MC, Sebastianelli A, Gobeil S, Morin C, Ghani K, Bachvarov D. Suppression of the grainyhead transcription factor 2 gene (GRHL2) inhibits the proliferation, migration, invasion and mediates cell cycle arrest of ovarian cancer cells. Cell Cycle. 2017;16:693–706. doi: 10.1080/15384101.2017.1295181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu F, He Z, Sun C, Rong D. Knockdown of GRHL2 inhibited proliferation and induced apoptosis of colorectal cancer by suppressing the PI3K/Akt pathway. Gene. 2019;700:96–104. doi: 10.1016/j.gene.2019.03.051. [DOI] [PubMed] [Google Scholar]
- 21.Quan Y, Jin R, Huang A, Zhao H, Feng B, Zang L, Zheng M. Downregulation of GRHL2 inhibits the proliferation of colorectal cancer cells by targeting ZEB1. Cancer Biol Ther. 2014;15:878–887. doi: 10.4161/cbt.28877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jolly MK, Tripathi SC, Jia D, Mooney SM, Celiktas M, Hanash SM, Mani SA, Pienta KJ, Ben-Jacob E, Levine H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7:27067–27084. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai L, Lin S, Girard L, Zhou Y, Yang L, Ci B, Zhou Q, Luo D, Yao B, Tang H, et al. LCE: An open web portal to explore gene expression and clinical associations in lung cancer. Oncogene. 2019;38:2551–2564. doi: 10.1038/s41388-018-0588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato M, Shames DS, Hasegawa Y. Emerging evidence of epithelial-to-mesenchymal transition in lung carcinogenesis. Respirology. 2012;17:1048–1059. doi: 10.1111/j.1440-1843.2012.02173.x. [DOI] [PubMed] [Google Scholar]
- 25.Sato M, Shay JW, Minna JD. Immortalized normal human lung epithelial cell models for studying lung cancer biology. Respir Investig. 2020;58:344–354. doi: 10.1016/j.resinv.2020.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Sato M, Vaughan MB, Girard L, Peyton M, Lee W, Shames DS, Ramirez RD, Sunaga N, Gazdar AF, Shay JW, Minna JD. Multiple oncogenic changes (K-RAS(V12), p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 2006;66:2116–2128. doi: 10.1158/0008-5472.CAN-05-2521. [DOI] [PubMed] [Google Scholar]
- 27.Phelps RM, Johnson BE, Ihde DC, Gazdar AF, Carbone DP, McClintock PR, Linnoila RI, Matthews MJ, Bunn PA, Jr, Carney D, et al. NCI-navy medical oncology branch cell line data base. J Cell Biochem Suppl. 1996;24:32–91. doi: 10.1002/jcb.240630505. [DOI] [PubMed] [Google Scholar]
- 28.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 29.van Kuppeveld FJ, Johansson KE, Galama JM, Kissing J, Bölske G, van der Logt JT, Melchers WJ. Detection of mycoplasma contamination in cell cultures by a mycoplasma group-specific PCR. Appl Environ Microbiol. 1994;60:149–152. doi: 10.1128/aem.60.1.149-152.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Sato M, Girard L, Sekine I, Sunaga N, Ramirez RD, Kamibayashi C, Minna JD. Increased expression and no mutation of the Flap endonuclease (FEN1) gene in human lung cancer. Oncogene. 2003;22:7243–7246. doi: 10.1038/sj.onc.1206977. [DOI] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. NIH image to imageJ: 25 Years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMillan EA, Ryu MJ, Diep CH, Mendiratta S, Clemenceau JR, Vaden RM, Kim JH, Motoyaji T, Covington KR, Peyton M, et al. Chemistry-first approach for nomination of personalized treatment in lung cancer. Cell. 2018;173:864–878.e29. doi: 10.1016/j.cell.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saitoh M. Involvement of partial EMT in cancer progression. J Biochem. 2018;164:257–264. doi: 10.1093/jb/mvy047. [DOI] [PubMed] [Google Scholar]
- 37.Das AK, Sato M, Story MD, Peyton M, Graves R, Redpath S, Girard L, Gazdar AF, Shay JW, Minna JD, Nirodi CS. Non-small-cell lung cancers with kinase domain mutations in the epidermal growth factor receptor are sensitive to ionizing radiation. Cancer Res. 2006;66:9601–9608. doi: 10.1158/0008-5472.CAN-06-2627. [DOI] [PubMed] [Google Scholar]
- 38.Sunaga N, Imai H, Shimizu K, Shames DS, Kakegawa S, Girard L, Sato M, Kaira K, Ishizuka T, Gazdar AF, et al. Oncogenic KRAS-induced interleukin-8 overexpression promotes cell growth and migration and contributes to aggressive phenotypes of non-small cell lung cancer. Int J Cancer. 2012;130:1733–1744. doi: 10.1002/ijc.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Liu H, Ji Y, Ma Z, Shen K, Shangguan X, Qian H, Zhao Y, Pan CW, Xue W. Downregulation of SHMT2 promotes the prostate cancer proliferation and metastasis by inducing epithelial-mesenchymal transition. Exp Cell Res. 2022;415:113138. doi: 10.1016/j.yexcr.2022.113138. [DOI] [PubMed] [Google Scholar]
- 40.He F, Feng G, Ma N, Midorikawa K, Oikawa S, Kobayashi H, Zhang Z, Huang G, Takeuchi K, Murata M. GDF10 inhibits cell proliferation and epithelial-mesenchymal transition in nasopharyngeal carcinoma by the transforming growth factor-β/Smad and NF-κB pathways. Carcinogenesis. 2022;43:94–103. doi: 10.1093/carcin/bgab122. [DOI] [PubMed] [Google Scholar]
- 41.Huang W, Huang H, Xiao Y, Wang L, Zhang T, Fang X, Xia X. UBE2T is upregulated, predicts poor prognosis, and promotes cell proliferation and invasion by promoting epithelial-mesenchymal transition via inhibiting autophagy in an AKT/mTOR dependent manner in ovarian cancer. Cell Cycle. 2022;21:780–791. doi: 10.1080/15384101.2022.2031426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding C, Luo J, Li L, Li S, Yang L, Pan H, Liu Q, Qin H, Chen C, Feng J. Gab2 facilitates epithelial-to-mesenchymal transition via the MEK/ERK/MMP signaling in colorectal cancer. J Exp Clin Cancer Res. 2016;35:5. doi: 10.1186/s13046-015-0280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karimi Roshan M, Soltani A, Soleimani A, Rezaie Kahkhaie K, Afshari AR, Soukhtanloo M. Role of AKT and mTOR signaling pathways in the induction of epithelial-mesenchymal transition (EMT) process. Biochimie. 2019;165:229–234. doi: 10.1016/j.biochi.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 44.Xu M, Cao FL, Li N, Gao X, Su X, Jiang X. Leptin induces epithelial-to-mesenchymal transition via activation of the ERK signaling pathway in lung cancer cells. Oncol Lett. 2018;16:4782–4788. doi: 10.3892/ol.2018.9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cook SJ, Stuart K, Gilley R, Sale MJ. Control of cell death and mitochondrial fission by ERK1/2 MAP kinase signalling. FEBS J. 2017;284:4177–4195. doi: 10.1111/febs.14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brix N, Samaga D, Belka C, Zitzelsberger H, Lauber K. Analysis of clonogenic growth in vitro. Nat Protoc. 2021;16:4963–4991. doi: 10.1038/s41596-021-00615-0. [DOI] [PubMed] [Google Scholar]
- 47.Franken NAP, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 48.Chen AF, Liu AJ, Krishnakumar R, Freimer JW, DeVeale B, Blelloch R. GRHL2-Dependent enhancer switching maintains a pluripotent stem cell transcriptional subnetwork after exit from naive pluripotency. Cell Stem Cell. 2018;23:226–238.e4. doi: 10.1016/j.stem.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao X, Bali AS, Randell SH, Hogan BL. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J Cell Biol. 2015;211:669–682. doi: 10.1083/jcb.201506014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.