Significance
Brown adipocytes are specialized thermogenic cells able to dissipate nutritional energy in the form of heat, which is conducive to metabolic homeostasis. Obesity limits thermogenesis due to unknown inhibitory signals for brown fat. Here, we identify CLCF1 signaling as a brake signal on thermogenesis, which is activated in obesity and inhibited during thermogenesis. CLCF1 activates CNTFR-STAT3 signaling pathway; thereafter, STAT3 transcriptionally inhibits PGC-1α and PGC-1β expression, thus restraining mitochondrial biogenesis in brown adipocytes. Consequently, CLCF1 transgenic mice exhibit impaired energy expenditure and thermogenic capacity and spontaneously develop metabolic abnormalities without external metabolic stress. Our finding defines CLCF1 signaling as an explanation for obesity-related brown adipocyte degeneration, which may be a target for intervention in metabolic disorders.
Keywords: brown adipose tissue, thermogenesis, metabolism, cardiotrophin-like cytokine factor 1
Abstract
Great progress has been made in identifying positive regulators that activate adipocyte thermogenesis, but negative regulatory signaling of thermogenesis remains poorly understood. Here, we found that cardiotrophin-like cytokine factor 1 (CLCF1) signaling led to loss of brown fat identity, which impaired thermogenic capacity. CLCF1 levels decreased during thermogenic stimulation but were considerably increased in obesity. Adipocyte-specific CLCF1 transgenic (CLCF1-ATG) mice showed impaired energy expenditure and severe cold intolerance. Elevated CLCF1 triggered whitening of brown adipose tissue by suppressing mitochondrial biogenesis. Mechanistically, CLCF1 bound and activated ciliary neurotrophic factor receptor (CNTFR) and augmented signal transducer and activator of transcription 3 (STAT3) signaling. STAT3 transcriptionally inhibited both peroxisome proliferator-activated receptor-γ coactivator (PGC) 1α and 1β, which thereafter restrained mitochondrial biogenesis in adipocytes. Inhibition of CNTFR or STAT3 could diminish the inhibitory effects of CLCF1 on mitochondrial biogenesis and thermogenesis. As a result, CLCF1-TG mice were predisposed to develop metabolic dysfunction even without external metabolic stress. Our findings revealed a brake signal on nonshivering thermogenesis and suggested that targeting this pathway could be used to restore brown fat activity and systemic metabolic homeostasis in obesity.
In mammals, white adipocytes are specialized for lipid storage and release, while classical brown adipocytes are specialized thermogenic cells able to dissipate nutritional energy in the form of heat (1, 2). Beige adipocytes are inducible brown adipocytes residing in white adipose tissue (WAT) that acquire thermogenic properties following external stimulation such as cold exposure, adrenergic stimuli, or exercise (3–5). Brown and beige adipocyte activation is considered a promising strategy for increasing systemic energy consumption and counteracting metabolic disorders (6). Studies in humans have demonstrated that the presence of brown adipose tissue (BAT) was independently correlated with reduced incidence of type 2 diabetes, dyslipidemia, and coronary artery disease (7). In addition to energy storage and expenditure, adipose tissue is also an essential endocrine organ that secretes adipokines such as adiponectin, leptin, and neuregulin-4 to maintain metabolic homeostasis in adipose tissue and distant organs (8, 9). Thus, dysregulation either in adipose tissue energy balance or endocrine function is strongly associated with the global increase in metabolic disorders.
Mitochondria are central organelles in thermogenic adipocytes. In mitochondria of brown or beige adipocytes, uncoupling protein 1 (UCP1) mediates proton leakage across the mitochondrial inner membrane and uncouples oxidative respiration from ATP synthesis to generate heat (3). In addition to the UCP1-dependent mechanism, mitochondria also drive UCP1-independent thermogenesis such as creatine futile cycling (3, 10). We and others previously showed that genes involved in mitochondrial biogenesis and function are significantly up-regulated during thermogenic activation (11, 12). For example, we found that coiled-coil-helix-coiled-coil-helix domain containing 10 (CHCHD10) protein is responsible for the assembly of mitochondrial cristae in adipocytes under thermogenic stimulation, deficiency of which causes obvious impairment in thermogenesis (11). Indeed, mitochondrial defects can result in profoundly impaired energy balance. A recent study showed that adipose mitochondrial levels strongly predicted metabolic syndrome traits in both the mouse and the human, and this was negatively correlated with both body weight and homeostatic model assessment for insulin resistance (13).
Members of the peroxisome proliferator-activated receptor-γ coactivator (PGC) family (PGC-1α and PGC-1β) are master regulators of mitochondrial biogenesis in thermogenic adipocytes. Simultaneous loss of PGC-1α and PGC-1β dramatically reduces mitochondrial density and respiration, thereby leading to functional defects in brown adipocytes and thermogenesis (14, 15). PGC-1α links external stimuli to the regulation of mitochondrial content and function. For example, cold activates β-adrenergic receptors in adipocytes, induces the elevation of intracellular cAMP, and subsequently activates cAMP-responsive element binding protein, thereby promoting the expression of PGC-1α, which turns on adaptive thermogenic programs including mitochondrial biogenesis, fatty acid oxidation, uncoupled respiration, and increased thermogenic gene expression (16–18). Although many physiological stimuli such as cold exposure, adrenergic stimulation, and exercise can increase mitochondrial function, the regulatory signals involved in obesity-associated disturbance of mitochondrial biogenesis remain poorly understood. These inhibitory signals might cause thermogenesis impairment in obesity, which largely limits the application of thermogenic fat in obesity intervention.
We previously conducted transcriptomics analyses of cold-induced thermogenic fat to identify thermogenic regulators (11), and up-regulated genes were highly enriched in mitochondrial function, lipolysis, and cellular respiration (11, 19). To explore the extracellular repressive signals of thermogenesis, we focused on genes encoding cytokines that were decreased during thermogenic activation and found that cardiotrophin-like cytokine factor 1 (CLCF1) was significantly down-regulated. CLCF1 was initially identified as a cytokine of the interleukin (IL)-6 family in T cell lymphomas, with relatively higher expression in the immune system (20, 21). CLCF1 performs vital functions in organ development, tumorigenesis, and motor neuron survival (22–24). Mice lacking CLCF1 exhibit decreased facial motility and are therefore unable to suckle, primarily due to motor neuron deficits in the facial nucleus (23). These mice die from starvation shortly after birth (23). Mutations in the CLCF1 gene can cause Crisponi/cold-induced sweating syndrome (CS/CISS) in humans, characterized by noninfectious hyperthermia, difficulties during feeding and respiration in the neonatal period, and cold-induced (<20 °C) sweating in adolescence (25, 26). However, the roles of CLCF1 and its downstream signals in adipocyte function are unknown. Here, we found that CLCF1 expression was negatively correlated with thermogenic capacity, and we identified a CLCF1-mediated autocrine signaling pathway that negatively regulates mitochondrial biogenesis and thermogenesis in adipocytes.
Results
CLCF1 Levels in Adipocytes Are Negatively Correlated with Thermogenic Capacity.
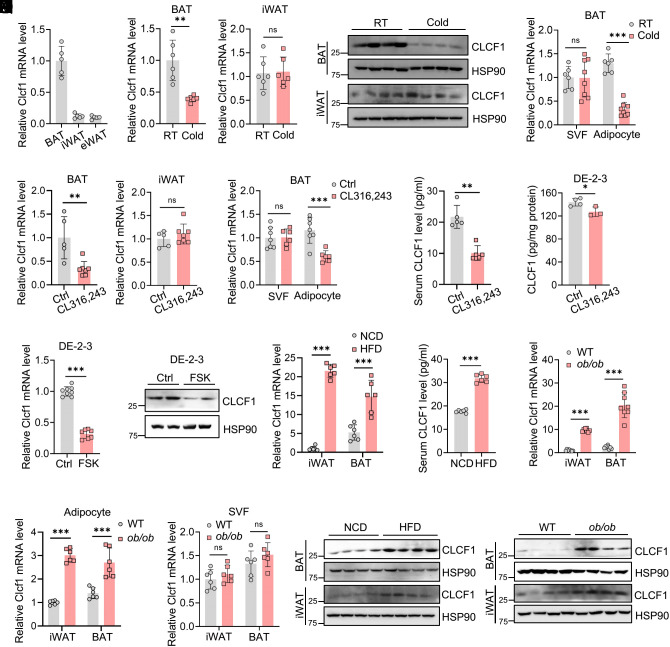
We first measured CLCF1 levels in adipose tissues and observed that Clcf1 mRNA expression in BAT was higher than in inguinal WAT (iWAT) and epididymal WAT (eWAT) (Fig. 1A and SI Appendix, Fig. S1A). Under cold exposure, CLCF1 expression at both mRNA and protein levels was significantly reduced in BAT of mice and remained unchanged in iWAT (Fig. 1 B and C). Clcf1 expression in mature adipocytes of BAT was decreased in response to cold exposure, whereas its expression in stromal vascular fraction (SVF) was not changed (Fig. 1D). We then fractionated BAT into mature adipocytes, CD45+ SVF cells, and CD45- SVF cells and found that Clcf1 was expressed in all three fractions and that CD45+ fraction had relatively higher expression of Clcf1 than mature adipocytes and CD45- cells (SI Appendix, Fig. S1B), which was consistent with the WAT single-cell data (27). Notably, Clcf1 levels were specifically reduced in mature adipocytes upon cold stimulation, while remained unchanged in CD45+ and CD45- SVF cells (SI Appendix, Fig. S1 C–E). Consistently, Clcf1 expression in BAT, but not in iWAT, was decreased following treatment with β3-adrenoreceptor agonist CL316, 243 (Fig. 1E). Clcf1 expression in adipocytes was decreased under CL316,243 treatment, while its expression in SVF remained unchanged (Fig. 1F). Serum CLCF1 levels were also significantly decreased in CL316,243-treated mice (Fig. 1G).
Fig. 1.
CLCF1 levels are decreased by thermogenic stimulation and increased in obesity. (A) The mRNA levels of Clcf1 in BAT, iWAT, and eWAT of male 8-wk-old mice (n = 5). (B and C) CLCF1 mRNA (n = 6) and protein levels in BAT and iWAT of male 8-wk-old mice exposed to 4 °C for 72 h. Data were analyzed by a two-tailed unpaired Student’s t test (iWAT) or with Welch’s correction (BAT). (D) Clcf1 mRNA levels in adipocytes and SVF in the BAT of male 8-wk-old mice exposed to 4 °C for 72 h (n = 6 to 8). Data were analyzed by a two-tailed unpaired Student’s t test. (E) Clcf1 mRNA levels in BAT and iWAT of male 8-wk-old mice injected with CL316,243 (1 mg/kg/d) for 7 d (n = 5 to 7). Data were analyzed by the Mann–Whitney test (BAT) or a two-tailed unpaired Student’s t test (iWAT). (F) Clcf1 mRNA levels in adipocytes and SVF in the BAT of CL316,243-treated male mice (n = 6 to 7). Data were analyzed by a two-tailed unpaired Student’s t test. (G) Serum CLCF1 levels in CL316,243-treated male mice (n = 5). Data were analyzed by the Mann–Whitney test. (H) CLCF1 level in the conditional medium from DE-2-3 mature adipocyte treated with CL316,243 (n = 3 to 4). Data were analyzed by a two-tailed unpaired Student’s t test. (I and J) The mRNA (n = 7 to 8) and protein levels of CLCF1 in DE-2-3 treated with FSK for 48 h. Data in I were analyzed with a two-tailed unpaired Student’s t test. (K and L) Clcf1 mRNA levels in iWAT and BAT (n = 6) and serum levels of CLCF1 of male mice fed with NCD and HFD for 16 wk. Data were analyzed by a two-tailed unpaired Student’s t test or with Welch’s correction. (M) Clcf1 mRNA levels in iWAT and BAT of male 8-wk-old WT and ob/ob mice (n = 8). Data were analyzed by a two-tailed unpaired Student’s t test (iWAT) or with Welch’s correction (BAT). (N and O) Clcf1 mRNA levels in the adipocytes or SVF of iWAT and BAT in WT and ob/ob mice. Data of adipocytes in the iWAT were analyzed by a two-tailed unpaired Student’s t test with Welch’s correction; other data in N and O by a two-tailed unpaired Student’s t test. (P and Q) CLCF1 protein levels in BAT of HFD-fed and ob/ob mice. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
CLCF1 was readily detectable in medium from immortalized DE-2-3 brown adipocytes and its levels in medium were significantly decreased by CL316,243 treatment (Fig. 1H). We mimicked thermogenic activation by treating immortalized brown adipocytes with forskolin (FSK), a specific stimulator of adenylyl cyclase. FSK treatment significantly reduced CLCF1 mRNA and protein levels (Fig. 1 I and J). We then sought to identify the transcription factors mediating CLCF1 downregulation. We searched the Cistromic (ChIP-Seq) regulation report from The Signaling Pathways Project (SPP) in the GeneCards database (https://www.genecards.org/) and predicted that PR domain containing 16 (PRDM16) might bind to the Clcf1 promoter, which is a master regulator of brown fat identity maintenance and thermogenesis. For validation, we conducted ChIP-qPCR and confirmed that PRDM16 was enriched at the Clcf1 promoter relative to nonspecific control sites (SI Appendix, Fig. S2A). Luciferase reporter assays indicated that PRDM16 inhibited Clcf1 promoter activity (SI Appendix, Fig. S2B). Ablation of PRDM16 promoted Clcf1 expression (SI Appendix, Fig. S2C), as well as the amount of CLCF1 secreted into the medium (SI Appendix, Fig. S2D). Consistent with that PRDM16 inhibited CLCF1 expression, expression of PRDM16 in BAT was increased by CL316,243 treatment (SI Appendix, Fig. S2E), contrary to the trend in CLCF1 expression.
Next, we evaluated CLCF1 levels in obesity, under which condition thermogenesis in adipocytes was impaired. Clcf1 mRNA expression was significantly elevated in both iWAT and BAT of high-fat-diet (HFD)-fed and ob/ob mice, and serum CLCF1 was also increased in HFD mice (Fig. 1 K–M). Clcf1 expression in adipocytes from iWAT and BAT was increased in ob/ob mice (Fig. 1N), whereas Clcf1 in SVF remained unchanged in ob/ob mice (Fig. 1O). Consistently, CLCF1 protein levels were increased in iWAT and BAT of HFD-fed and ob/ob mice (Fig. 1 P and Q).
Transgenic Expression of CLCF1 Impairs Thermogenesis and Energy Expenditure by Promoting Whitening of BAT.
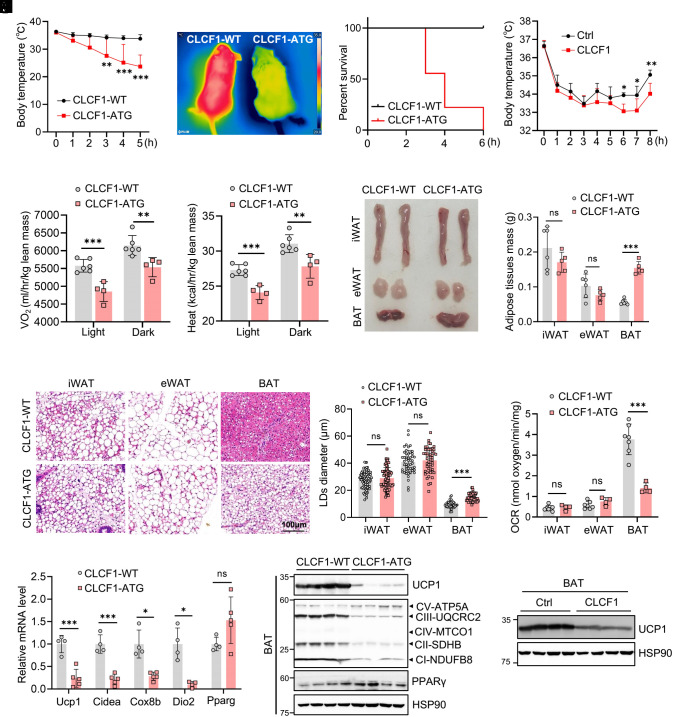
Having observed that CLCF1 levels in mature adipocytes were decreased during thermogenic activation and increased in obesity, we examined the role of CLCF1 in thermogenic adipocytes by generating adipocyte-specific CLCF1 transgenic (CLCF1-ATG) mice using adiponectin-promoter-triggered Cre to specifically drive CLCF1-3×Flag expression in adipose tissues (SI Appendix, Fig. S3A). Clcf1 was specifically overexpressed in adipose tissues, with no expression changes in other tissues (SI Appendix, Fig. S3B). CLCF1 levels in circulation were increased in CLCF1-ATG mice (SI Appendix, Fig. S3C). CLCF1-ATG mice had body weights similar to WT littermates when fed a normal chow diet (NCD; SI Appendix, Fig. S3D). To determine the role of CLCF1 in adaptive thermogenesis, we performed cold tolerance tests (4 °C) and found that CLCF1-ATG mice presented significantly lower rectal and surface temperatures than WT (Fig. 2 A and B); strikingly, all CLCF1-ATG mice died from hypothermia after 6 h of cold exposure (Fig. 2C). Consistently, mice with acute injection of recombinant CLCF1 protein also exhibited cold intolerance (Fig. 2D), but they could survive under cold exposure, possibly because they were transiently stimulated by CLCF1.
Fig. 2.
CLCF1-ATG impairs energy expenditure and thermogenesis. (A) Rectal temperature of male 8-wk-old CLCF1-WT and CLCF1-ATG mice exposed to 4 °C (n = 6/group). Data were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. (B) Representative dorsal images of infrared thermography under exposure to 4 °C for 3 h. (C) Kaplan–Meier survival curve of CLCF1-WT (n = 10) and CLCF1-ATG (n = 9) mice during cold exposure. (D) Rectal temperature of male 8-wk-old mice injected with CLCF1 recombinant protein after exposure to 4 °C (n = 5/group). Data were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. (E and F) Oxygen consumption (VO2) and heat production of male 8-wk-old CLCF1-WT (n = 6) and CLCF1-ATG mice (n = 4) at room temperature. (G) Representative images of adipose tissues from CLCF1-WT and CLCF1-ATG mice. (H) Weights of fat pads of male 8-wk-old CLCF1-WT (n = 6) and CLCF1-ATG mice (n = 5). Data in E–H were analyzed by a two-tailed unpaired Student’s t test. (I) Representative images of H&E staining of iWAT, eWAT, and BAT from male CLCF1-WT and CLCF1-ATG mice (Scale bar, 100 μm.) (J) Quantification of lipid droplet diameter in H&E staining using the Image J software. Each dot represents a lipid droplet. Data were analyzed by a two-tailed unpaired Student’s t test (eWAT and BAT) or with Welch’s correction (iWAT). (K) Oxygen consumption of iWAT, eWAT, and BAT from male CLCF1-WT (n = 7) and CLCF1-ATG mice (n = 4). Data were analyzed by a two-tailed unpaired Student’s t test. (L) qPCR analysis of thermogenic genes in BAT of CLCF1-WT and CLCF1-ATG mice (n = 4 to 5). Data were analyzed using a two-tailed unpaired Student’s t test or with Welch’s correction. (M) Western blot for UCP1 and the indicated mitochondrial complex components in BAT of CLCF1-WT and CLCF1-ATG mice. (N) Western blot for UCP1 in BAT of CLCF1-injected mice. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
The indirect calorimetry study showed that CLCF1-ATG mice had significantly lower rates of O2 consumption and CO2 production, and decreased heat production relative to WT mice, but no significant difference in respiratory exchange ratio (RER) or food intake was detected (Fig. 2 E and F and SI Appendix, Fig. S3 E–G). The morphology of BAT from CLCF1-ATG mice appeared abnormal, enlarged, and “whitened” compared with WT, but with no obvious changes in iWAT or eWAT (Fig. 2G). Weights of BAT in CLCF1-ATG mice were consistently increased (Fig. 2H). H&E staining revealed increased lipid deposition and a switch from small to large lipid droplets in BAT of CLCF1-ATG, morphologically similar to WAT, indicative of hypertrophy in BAT (Fig. 2 I and J). The oxygen consumption rate of BAT in CLCF1-ATG was dramatically reduced compared with WT (Fig. 2K). In contrast, the masses of iWAT and eWAT in CLCF1-ATG mice, as well as oxygen consumption of WAT, were comparable to WT mice (Fig. 2 H–K). Thermogenic-related genes such as Ucp1, Cidea, and Cox8b were markedly decreased in BAT of CLCF1-ATG mice, with no changes in general markers for adipogenesis such as Pparg (Fig. 2L). Remarkedly, UCP1 protein levels in BAT of CLCF1-ATG mice were too low to be easily detected (Fig. 2M). Mitochondrial oxidative phosphorylation (OXPHOS) complex component levels were dramatically reduced in BAT of CLCF1-ATG (Fig. 2M). Because all CLCF1-ATG mice died shortly (within 6 h) following cold exposure (Fig. 2C), UCP1 in iWAT of WT mice was not strongly activated at that time; hence, only a mild decrease in UCP1 was observed in iWAT of CLCF1-ATG mice (SI Appendix, Fig. S3H). Moreover, acute injection of recombinant CLCF1 protein also inhibited UCP1 levels in BAT under cold stimulation (Fig. 2N). These results collectively provided strong evidence that CLCF1-ATG mice had defective adaptive thermogenesis.
CLCF1 Inhibits Mitochondrial Biogenesis in BAT.
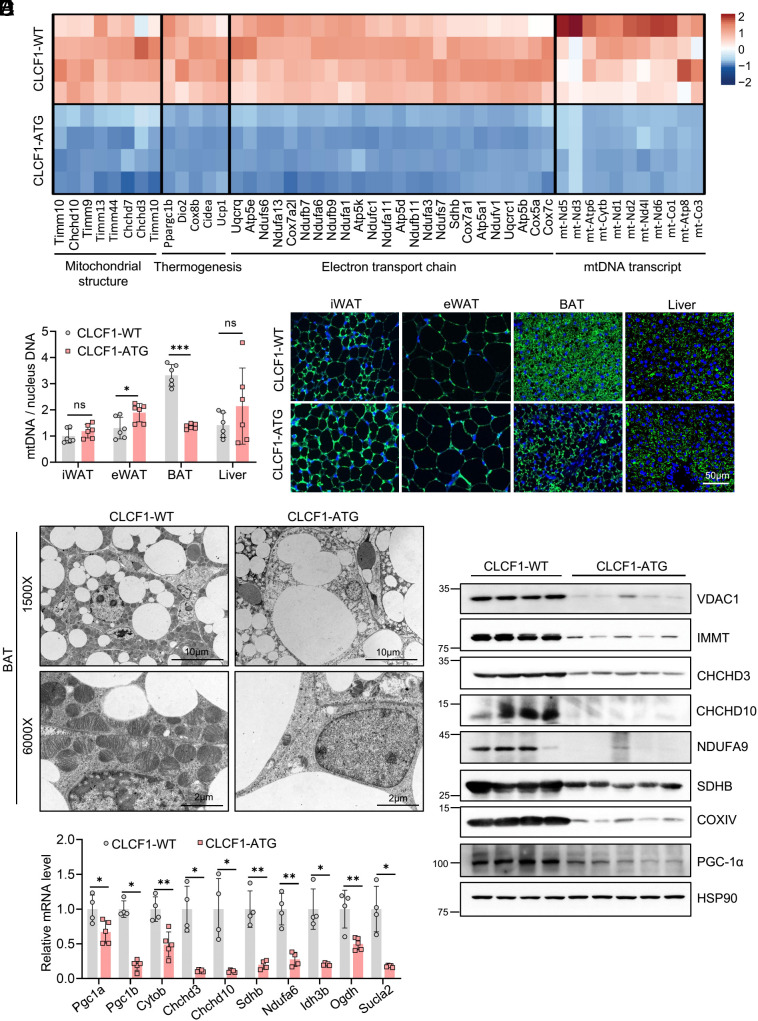
To understand the mechanism of CLCF1-mediated BAT dysfunction, we performed RNA sequencing (RNA-seq) on BAT from WT and CLCF1-ATG mice (SI Appendix, Fig. S4A). Gene annotation enrichment analysis found that most down-regulated genes were strongly related to mitochondrial components and functions, including the mitochondrial respiratory chain and oxidative phosphorylation (SI Appendix, Fig. S4B). Heat map analysis showed that genes involved in mitochondrial structure, the electron transport chain, mitochondrial DNA (mtDNA) transcription and thermogenesis were dramatically reduced in BAT of CLCF1-ATG mice (Fig. 3A). The mtDNA copy number, which reflects the number of mitochondria, was significantly decreased (Fig. 3B); meanwhile, mtDNA levels in iWAT and liver remained unchanged, and the level in eWAT was even slightly increased (Fig. 3B). Immunofluorescence analysis using TOM20 antibody, a mitochondrial marker, revealed that mitochondrial content in BAT was obviously decreased in CLCF1-ATG mice (Fig. 3C). Consistently, transmission electron microscopy showed that mitochondrial density in BAT of CLCF1-ATG mice was dramatically reduced (Fig. 3D). The qPCR confirmed the RNA-seq results and further indicated that expression of mitochondrial-related genes was significantly decreased in BAT of CLCF1-ATG mice, including mitochondrial biogenesis-related genes such as Pgc1a and Pgc1b, mtDNA transcripts such as Cytob, genes involved in mitochondrial crista assembly such as Chchd3 and Chchd10, and electron transport chain-related genes including Sdhb and Ndufa6 (Fig. 3E). Levels of multiple mitochondrial biogenesis and structural proteins, as well as OXPHOS complex components, were robustly reduced (Fig. 3F). As master regulators of mitochondrial biogenesis, decreases in PGC-1α and PGC-1β might be the cause of mitochondrial depletion. Together, these results strongly indicated that CLCF1 overexpression led to a sharp decline in the number of mitochondria in BAT.
Fig. 3.
CLCF1-ATG dramatically suppresses mitochondrial biogenesis in BAT. (A) Heat map showing part of the down-regulated genes in the BAT of male CLCF1-ATG mice compared with CLCF1-WT (n = 4). (B) mtDNA copy number of iWAT, eWAT, BAT, and liver in male 8-wk-old CLCF1-WT and CLCF1-ATG mice (n = 6). Data were analyzed by the Mann–Whitney test (iWAT), two-tailed unpaired Student’s t test (eWAT), or with Welch’s correction (BAT and liver). (C) Immunofluorescent TOM20 staining of indicated tissues. (D) Transmission electron microscopy showing mitochondrial morphology of BAT. (E and F) The mRNA (n = 4 to 5) and protein levels of mitochondrial-related genes in BAT of CLCF1-WT and CLCF1-ATG mice. Pgc1a, Cytob, Ndufa6, and Ogdh were analyzed using a two-tailed unpaired Student’s t test; Chchd3, Chchd10, Sdhb, Idh3b, and Sucla2 using a two-tailed unpaired Student’s t test with Welch’s correction; and Pgc1b using the Mann–Whitney test. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
To further verify the above observation, we generated brown adipocyte-specific CLCF1 transgenic mice (CLCF1-BTG) using Ucp1-Cre to trigger CLCF1-3×Flag expression in brown adipocytes (SI Appendix, Fig. S5A). The qPCR confirmed that Clcf1 was specifically overexpressed in BAT (SI Appendix, Fig. S5B). Like CLCF1-ATG mice, CLCF1-BTG mice showed severe cold intolerance (SI Appendix, Fig. S5 C and D) and BAT dysfunction, as indicated by enlarged and hypertrophic BAT with increased lipid accumulation (SI Appendix, Fig. S5 E–G), but there was no change in morphology or the masses of WAT (SI Appendix, Fig. S5 E–G). The number of mitochondria was largely reduced in BAT of CLCF1-BTG mice (SI Appendix, Fig. S5H). Thermogenic genes and mitochondrial-related genes expression were significantly decreased in BAT of CLCF1-BTG mice (SI Appendix, Fig. S5 I and J) but showed minimal changes in iWAT (SI Appendix, Fig. S5K). Additionally, acute overexpression of CLCF1 in BAT following injection of CLCF1 adenovirus also yielded a phenotype with increased lipid deposition and reduced UCP1 levels in BAT (SI Appendix, Fig. S5L). Together, these results demonstrated that increased CLCF1 resulted in a loss of brown adipocyte identity.
CNTFR Is Required for CLCF1-Induced Mitochondrial Defects and Thermogenic Impairment.
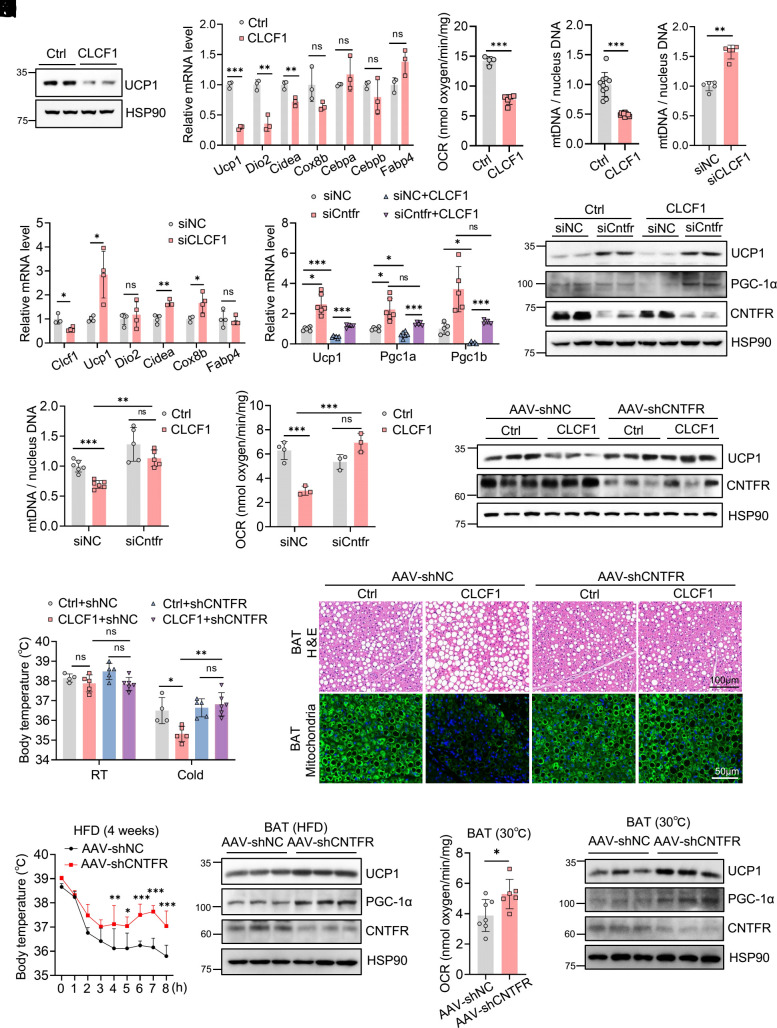
To determine whether CLCF1 regulates energy expenditure and mitochondrial density in a cell-autonomous manner, we performed in vitro studies. We induced differentiation of immortal brown preadipocytes to mature adipocytes and treated them with CLCF1 recombinant protein or siCLCF1 to disrupt CLCF1 expression. General adipogenesis was not affected (SI Appendix, Fig. S6A). CLCF1 inhibited expression of thermogenic genes including Ucp1, Dio2, and Cidea but had little effect on expression of adipogenic genes such as Cebpa, Cebpb, and Fabp4 (Fig. 4 A and B). Remarkedly, CLCF1 treatment blunted oxygen consumption in adipocytes and potently reduced mitochondrial content (Fig. 4 C and D). Knock-down of CLCF1 in brown adipocytes significantly increased mtDNA levels and up-regulated expression of thermogenic genes (Fig. 4 E and F). These in vitro results suggested that CLCF1 directly targeted mature adipocytes to attenuate thermogenic capacity.
Fig. 4.
CNTFR mediates the inhibitory effect of CLCF1 on thermogenic capacity. (A and B) Thermogenic gene expression in immortalized brown adipocytes DE-2-3 treated with 50 ng/mL CLCF1 (n = 3). Data of Ucp1, Dio2, Cidea, Cox8b, Cebpb, and Fabp4 were analyzed by a two-tailed unpaired Student’s t test; Cebpa was analyzed by a two-tailed unpaired Student’s t test with Welch’s correction. (C and D) OCR (n = 4 to 5) and mtDNA copy number (n = 10) of immortalized brown adipocytes treated with 50 ng/mL CLCF1. Data were analyzed by a two-tailed unpaired Student’s t test (C) or with Welch’s correction (D). (E) mtDNA copy number in immortalized brown adipocytes treated with siNC or siCLCF1 (n = 5). Data were analyzed by the Mann–Whitney test. (F) qPCR analysis of thermogenic genes (n = 3 to 4). Clcf1, Cidea, and Cox8b were analyzed using a two-tailed unpaired Student’s t test; Ucp1 using a two-tailed unpaired Student’s t test with Welch’s correction; and Dio2 and Fabp4 using the Mann–Whitney test. (G–J). Immortalized brown adipocytes were transfected with siCntfr and treated with CLCF1 as indicated, then subjected to following detection. (G) qPCR analysis of Ucp1, Pgc1a, and Pgc1b (n = 4 to 6). (H) Protein levels of UCP1 and PGC-1α. (I) mtDNA copy number (n = 5 to 6). (J) Oxygen consumption of indicated adipocytes (n = 3 to 4). Data in G and I were analyzed by Brown–Forsythe and Welch ANOVA tests followed by Tamhane’s T2 multiple comparisons test; data in J were analyzed by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test. (K–M) Adipoq-promoter-driven AAV-shNC and AAV-shCNTFR were locally injected into BAT, followed by injection of adenovirus for CLCF1. The indicated mice fed with NCD were subjected to the following test. (K) Rectal temperature of indicated mice at RT and 4 °C for 3 h (n = 4 to 6). Data were analyzed by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test. (L) Protein levels of UCP1 in BAT after cold stimulation. (M) H&E staining and immunofluorescent TOM20 staining of BAT in indicated mice after cold stimulation. (N and O) AAV-shNC and AAV-shCNTFR were locally injected into BAT, and mice were fed with an HFD for 4 wk. (N) Rectal temperature of mice exposed to 4 °C for indicated times (n = 5/group). Data were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. (O) Protein levels of UCP1 and PGC-1α in the BAT. (P and Q) Three wk after injection of AAV-shCNTFR, mice with a normal diet were housed at 30 °C for 5 d and subjected to the following measurements. (P) OCR of BAT (n = 6 to 7). Data were analyzed by a two-tailed unpaired Student’s t test. (Q) Protein levels of UCP1 and PGC-1α. Data are presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001.
We then investigated the receptor and intracellular signaling in adipocytes responsible for the effect of CLCF1. The receptor of CLCF1 is the ciliary neurotrophic factor receptor (CNTFR), a member of the IL-6 receptor family that forms a trimeric complex with leukemia inhibitory factor receptor (LIFR) and glycoprotein 130 (Gp130) (28). CLCF1 activates the CNTFR-LIFR-Gp130 complex, leading to phosphorylation of LIFR and Gp130, which induces downstream signals (28). We found that Cntfr had higher expression in BAT than iWAT (SI Appendix, Fig. S6B), whereas both Lifr and Gp130 had identical expression levels in BAT and iWAT (SI Appendix, Fig. S6B). In BAT, Cntfr was highly expressed in mature adipocytes compared with SVF (SI Appendix, Fig. S6C). Notably, CNTFR was significantly reduced in BAT under cold exposure and CL316,243 treatment (SI Appendix, Fig. S6 D–F) and obviously increased in adipose tissues under thermoneutrality and in obesity (SI Appendix, Fig. S6 G and H), exhibiting a negative correlation with thermogenic capacity, similar to CLCF1. Coimmunoprecipitation assays showed that CLCF1 could interact with CNTFR (SI Appendix, Fig. S6I). To test whether CLCF1 functioned via its canonical receptor, we first knocked down CNTFR in cultured brown adipocytes and evaluated the phenotype. Ablation of CNTFR significantly rescued CLCF1-mediated decreases in thermogenic genes (Fig. 4 G and H). Loss of CNTFR boosted mtDNA levels and consequently abolished the inhibitory effect of CLCF1 on mitochondrial content and oxygen consumption in adipocytes (Fig. 4 I and J). We then assessed the role of CNTFR in vivo by injecting adiponectin promoter-driven AAV-shCNTFR in BAT depots. Cold tolerance tests showed that depletion of CNTFR in brown adipocytes increased the body temperature (SI Appendix, Fig. S6J) and restored the impaired thermogenesis capacity caused by CLCF1 (Fig. 4K). Deficiency of CNTFR totally reversed downregulation of UCP1 expression (Fig. 4L) and strongly blocked lipid deposition and deceased mitochondrial density in BAT induced by CLCF1 (Fig. 4M).
We noticed that while knockdown of CNTFR reversed inhibitory effect of CLCF1 on thermogenesis, inactivation of CNTFR had a weak effect on thermogenesis in WT mice. The possible reason is that BAT thermogenesis is highly active under cold stimulation, effects that enhanced the thermogenesis were not easily to be observed. Thus, we disrupted CNTFR by AAV in the BAT and challenged the mice with an HFD, under which condition basal BAT thermogenesis was attenuated. There was no difference in body weights between AAV-shNC and AAV-shCNTFR groups after HFD for 4 wk (SI Appendix, Fig. S6K). CNTFR depletion in brown adipocytes led to an increase in cold tolerance under HFD (Fig. 4N) and resulted in enhanced expression of UCP1 and PGC-1α in BAT (Fig. 4O). Additionally, we also housed AAV-shCNTFR-treated mice at 30 °C with a normal diet. Under the thermoneutrality condition, knockdown of CNTFR promoted oxygen consumption in BAT (Fig. 4P), potently enhanced thermogenic genes expression (Fig. 4Q and SI Appendix, Fig. S6L), increased mitochondrial content, and inhibited lipid accumulation in BAT (SI Appendix, Fig. S6M). These data together suggested that CNTFR deficiency promoted BAT thermogenic capacity.
STAT3 Signaling Activated by CLCF1 Represses Mitochondrial Biogenesis and Thermogenesis.
Having confirmed that CLCF1 induced mitochondrial defects in BAT via its receptor CNTFR, thereby modulating thermogenesis, we then sought to identify the downstream signal and transcriptional factors that mediate the effect of CLCF1-CNTFR. The Janus kinase (JAK)-STAT pathway is reported to be one of downstream signals of CNTFR (28). For validation, we treated adipocytes with CLCF1 and found that CLCF1 robustly stimulated phosphorylation of STAT3, indicative of STAT3 activation induced by CLCF1 (SI Appendix, Fig. S7A). Phosphorylated STAT3 (p-STAT3) was also potently augmented in adipose tissues of CLCF1-TG mice (SI Appendix, Fig. S7B). CNTFR was indispensable for CLCF1-induced activation of STAT3 (SI Appendix, Fig. S7C). Additionally, STAT3 was inhibited in the BAT by cold and activated in BAT of ob/ob mice or under the thermoneutrality condition, coinciding with the negative correlation of CLCF1 with thermogenesis (SI Appendix, Fig. S7 D–F).
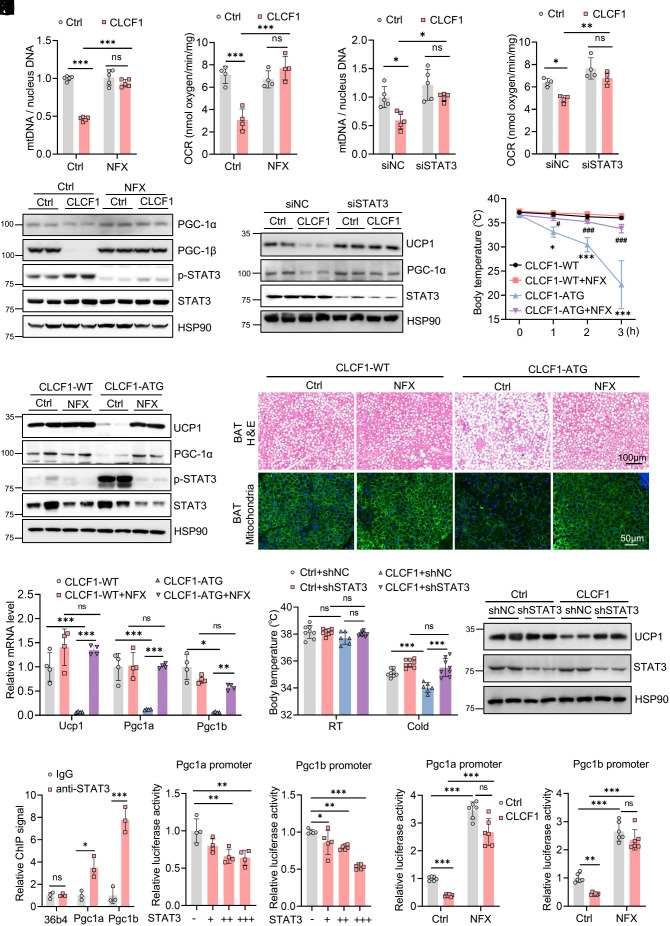
Next, we set out to elucidate whether STAT3 was involved in the inhibitory effect of CLCF1 on thermogenesis. We first treated adipocytes in vitro with nifuroxazide (NFX), a potent inhibitor of STAT3 that suppresses STAT3 phosphorylation (29). We found that inhibition of STAT3 by NFX recovered the reduced mitochondrial content in CLCF1-treated cells (Fig. 5A) and completely restored the decreased oxygen consumption induced by CLCF1 (Fig. 5B). Moreover, NFX reversed the decreased expression of thermogenic genes and mitochondrial-related genes caused by CLCF1 (Fig. 5C). Consistently, knock-down of STAT3 also rescued the decreased mtDNA content and oxygen consumption, and the reduced thermogenesis-related proteins induced by CLCF1 (Fig. 5 D–F). We then evaluated the effects of STAT3 in vivo by intraperitoneal injection of NFX in WT and CLCF1-ATG mice. Cold tolerance tests showed that inhibition of STAT3 could improve hypothermia induced by CLCF1 (Fig. 5G). NFX completely recovered the decreased expression of Ucp1, Pgc1a, and Pgc1b in BAT of CLCF1-ATG mice (Fig. 5 H and I). Moreover, NFX potently inhibited lipid deposition in BAT caused by CLCF1-ATG (Fig. 5J) and abolished the reduced mitochondrial content (Fig. 5J). We then disrupted STAT3 expression in brown fat depots by injecting AAV-shSTAT3. Deficiency of STAT3 totally reversed the impaired cold tolerance and the downregulation of UCP1 expression (Fig. 5 K and L) and strongly blocked lipid deposition in BAT induced by CLCF1 (SI Appendix, Fig. S7G). These results showed that CLCF1 failed to attenuate thermogenic function when STAT3 was inhibited.
Fig. 5.
STAT3 signal is necessary for CLCF1 to inhibit the thermogenic function. (A–C) Immortalized brown adipocytes DE-2-3 were treated with 5 μM NFX or 50 ng/mL CLCF1 for 72 h and subjected to the following detection: mtDNA (A, n = 5), OCR (B, n = 4), and western blot analysis of PGC-1α and PGC-1β (C). Data in A and B were analyzed using ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test. (D–F) Immortalized brown adipocytes with knockdown of STAT3 were treated with CLCF1, and subjected to mtDNA (D, n = 5) and OCR measurement (E, n = 4) and detection of UCP1 and PGC-1α (F). Data in D and E were analyzed by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test. (G–J) Male 16-wk-old CLCF1-WT and CLCF1-ATG mice were daily intraperitoneally injected with NFX (10 mg/kg) for 10 d and then subjected to the following measurement. (G) Rectal temperature of indicated mice exposed to 4 °C (n = 3 to 6/group). Data were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. (H) Western blot analysis of UCP1 and PGC-1α in the BAT of indicated mice. (I) qPCR analysis in the BAT (n = 4/group). Ucp1 and Pgc1a were analyzed using ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test; Pgc1b was analyzed by Brown–Forsythe and Welch ANOVA tests followed by Tamhane’s T2 multiple comparisons test. (J) H&E staining and immunofluorescent TOM20 staining of BAT. (K and L) AAV-shNC and AAV-shSTAT3 were locally injected into BAT, followed by injection of adenovirus for CLCF1. (K) Rectal temperature of indicated mice at RT and 4 °C for 5 h (n = 6 to 8). Data were analyzed by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test. (L) Protein levels of UCP1 in the BAT. (M) ChIP analysis of occupation of STAT3 on Pgc1a, Pgc1b, and 36b4 gene (n = 3), with 36b4 as a nonspecific control. Pgc1a and Pgc1b data were analyzed by a two-tailed unpaired Student’s t test; 36b4 by the Mann–Whitney test. (N) Luciferase reporter assay of Pgc1a or Pgc1b promoter with different dosages of STAT3 (n = 4 to 6). Data were analyzed by ordinary one-way ANOVA followed by Bonferroni’s multiple comparisons test. (O) Luciferase reporter assay of Pgc1a or Pgc1b promoter treated with CLCF1 and NFX as indicated (n = 6). Pgc1a data using Brown–Forsythe and Welch ANOVA tests followed by Tamhane’s T2 multiple comparisons test; Pgc1b using ordinary one-way ANOVA followed by Bonferroni's multiple comparisons test. *P < 0.05; **P < 0.01; ***P < 0.001. #P < 0.05; ##P < 0.01; ###P < 0.001 compared with CLCF1-ATG group in G.
We also treated HFD-fed mice with NFX (SI Appendix, Fig. S7H). Treatment of NFX for 1 wk had no effect on body weights (SI Appendix, Fig. S7I). Cold tolerance tests showed that NFX potently improved cold tolerance (SI Appendix, Fig. S7J). Additionally, we treated mice under NCD with NFX by daily intraperitoneal injection and housed them at 30 °C for 5 d. Inhibition of STAT3 by NFX reduced lipid deposition in BAT, increased mitochondrial content, and enhanced protein levels of thermogenic genes in BAT (SI Appendix, Fig. S7 K and L).
PGC-1α and PGC-1β, master regulators of mitochondrial biogenesis and thermogenesis, were down-regulated by CLCF1. Thus, we wondered whether STAT3 could transcriptionally regulate Pgc1 expression. We used the JASPAR database (https://jaspar.genereg.net/) and identified multiple potential binding sites for STAT3 on the promoters of Pgc1a and Pgc1b. ChIP-qPCR assays validated the prediction and showed that STAT3 was specifically enriched at the promoter of both Pgc1a and Pgc1b (Fig. 5M). STAT3 dose dependently inhibited the promoter activity of both Pgc1a and Pgc1b (Fig. 5N). CLCF1 treatment also reduced Pgc1a and Pgc1b promoter activity, while inhibition of STAT3 by NFX totally reversed this (Fig. 5O). These results suggested that activation of STAT3 by CLCF1 transcriptionally inhibited expression of Pgc1a and Pgc1b.
CLCF1-TG Mice Are Prone to Developing Metabolic Disorders Even under Normal Diet.
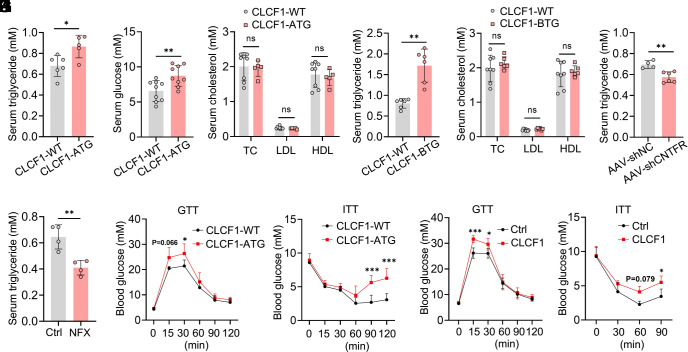
The above results indicated that CLCF1-CNTFR-STAT3 signaling inhibited mitochondrial biogenesis, thereby disrupting thermogenic capacity in brown adipocytes. Because brown adipocytes are strongly related to metabolic homeostasis, we therefore explored the metabolic phenotype of CLCF1-TG mice. Upon an NCD, there was no significant difference in body weight between WT and CLCF1-ATG mice (SI Appendix, Fig. S3D). Nevertheless, we found that CLCF1-ATG mice exhibited increased serum triglyceride and glucose levels compared with WT mice (Fig. 6 A and B), with no effect on serum total cholesterol (TC), LDL, or HDL levels (Fig. 6C). Consistently, CLCF1-BTG mice also exhibited elevated serum triglyceride levels and identical cholesterol levels compared with WT mice (Fig. 6 D and E). To complement the gain-of-function model, we blocked CLCF1 signaling in BAT via AAV-mediated disruption of CNTFR. Deficiency of CNTFR down-regulated triglyceride levels (Fig. 6F), consistent with observations in CLCF1-TG mice. Inhibition of STAT3 by NFX also down-regulated serum triglyceride levels (Fig. 6G). Moreover, CLCF1-ATG mice displayed glucose intolerance and insulin resistance compared with WT mice (Fig. 6 H and I). Acute administration of recombinant CLCF1 by intraperitoneal injection analogously induced impaired glucose tolerance and insulin sensitivity (Fig. 6 J and K). These results suggested that CLCF1-TG mice spontaneously developed metabolic abnormalities without external metabolic stress.
Fig. 6.
Elevated CLCF1 can predispose to metabolic dysfunction. (A–C) Serum triglyceride (n = 5 to 6), glucose (n = 9), cholesterol, LDL, and HDL (n = 5 to 8) levels of male 8-wk-old CLCF1-WT and CLCF1-ATG mice. Triglyceride, glucose, LDL, and HDL levels were analyzed by a two-tailed unpaired Student’s t test; the cholesterol level was analyzed by the Mann–Whitney test. (D and E) Serum triglyceride (n = 6), cholesterol, LDL, and HDL (n = 6 to 8) levels of male 8-wk-old CLCF1-WT and CLCF1-BTG mice. Data were analyzed by a two-tailed unpaired Student’s t test (cholesterol, LDL, and HDL) or with Welch’s correction (triglyceride). (F and G) Serum triglyceride (n = 4 to 6) of mice with CNTFR-deficiency in brown adipocytes or treated with NFX. Data were analyzed by a two-tailed unpaired Student’s t test. (H and I) GTT (n = 4 to 5) and ITT (n = 4) of 8-wk-old CLCF1-WT and CLCF1-ATG mice. (J and K) GTT (n = 4 to 5) and ITT (n = 4) of 8-wk-old mice treated with CLCF1 recombinant protein. Data in H–K were analyzed by two-way ANOVA followed by Bonferroni’s multiple comparisons test. *P < 0.05; **P < 0.01; ***P < 0.001.
We also challenged CLCF1-WT and CLCF1-ATG mice with an HFD. No significant body changes have been observed between two groups (SI Appendix, Fig. S8A). CLCF1-ATG mice had significantly lower rates of O2 consumption and CO2 production and decreased heat production relative to WT mice (SI Appendix, Fig. S8 B–D). CLCF1-ATG mice exerted glucose intolerance and remarkedly increased serum triglyceride and glucose levels, with no changes in serum total cholesterol (SI Appendix, Fig. S8 E–H). CLCF1-ATG mice were cold intolerant (SI Appendix, Fig. S8I). The expressions of thermogenic and mitochondrial-related genes were dramatically decreased in the BAT of CLCF1-ATG mice (SI Appendix, Fig. S8J). These data together suggested that CLCF1-ATG mice exhibited a greater susceptibility to diet-induced metabolic disorders, independent of changes in body weight.
Discussion
The thermogenic functions of brown fat are highly responsive to external stimuli and systemic metabolic status. Catecholamines, thyroid hormones, BMPs, and FGF21 are among a growing group of cytokines known to drive activation of thermogenic programs (30–33). Although numerous inducers have been identified, application of thermogenic adipocytes continues to prove challenging, especially for obesity and aging populations, due to unknown negative regulatory factors. Here, we revealed a mechanism in which the obesity-triggered CLCF1 signal inhibited mitochondrial biogenesis and thermogenesis. CLCF1-expressed adipocytes may serve as a type of thermogenesis-inhibitory cells. Under physiological conditions, such inhibitory signaling likely protects against energy overexpenditure, which can be blocked under thermogenic stimulation. In obesity, such inhibitory signals are activated and stimulate resistance to thermogenesis. Similarly, a recent study uncovered an acetate-mediated paracrine signaling pathway that represses thermogenesis in BAT (34). The authors identified a subset of thermogenesis-inhibitory adipocytes in mouse and human BAT that restrains the thermogenic capacity of brown adipocytes via local production of acetate. These inhibitory adipocytes are enriched in BAT under thermoneutral conditions. Another recent study identified two distinct populations of thermogenic cells in mouse BAT, classical brown adipocytes and low-thermogenic brown adipocytes (35). Cold exposure activates low thermogenic cells to become highly thermogenic. Mounting evidence indicates that BAT is heterogeneous, and BAT function is regulated by the coordinated activity of distinct adipocyte subpopulations. It would be interesting to investigate CLCF1 signals in these cells with differences in thermogenic capacity. Notably, inhibition of these negative regulators might improve obesity-associated disturbances and facilitate thermogenesis. Kim et al. recently reported a high-affinity soluble receptor (eCNTFR-Fc) that effectively sequesters CLCF1, thereby inhibiting CLCF1 oncogenic effects (24). It would be interesting to test the thermogenic phenotype following administration of this type of decoy receptor or neutralizing antibody for CLCF1 in obesity.
Our finding demonstrated an autocrine CLCF1 signaling pathway in brown fat that restrains thermogenic capacity through activation of CNTFR-LIFR-Gp130 tripartite receptor complex and downstream STAT3 pathway. In agreement with our observations, it was recently reported that adipocyte-specific knockout of Gp130 promoted browning in WAT, indicating an inhibitory effect of Gp130 on thermogenic programs (36). The authors showed that oncostatin M (OSM), an IL-6 family cytokine, suppressed browning of white adipocytes via Gp130-STAT3 signaling (36). Thus, the CNTF tripartite receptor complex negatively regulates thermogenesis in adipocytes. Consistent with their inhibitory roles, expression of CNTF tripartite receptor components was significantly decreased during thermogenic activation. Moreover, we found that CNTFR had higher expression in BAT than in iWAT, partially explaining why CLCF1 induces dysfunction in BAT but has a minimal effect on WAT. Through binding the CNTF receptor, CLCF1 turns on JAK/STAT3 signaling. We confirmed that STAT3 was required for the effect of CLCF1. Consistently, an in vitro study showed that tofacitinib, an inhibitor of JAK, stably induced a brown-like phenotype in adipocytes (37). More importantly, we revealed a mechanism by which CLCF1 leads to thermogenic dysfunction via STAT3. STAT3 binds to the promoters of PGC-1α and PGC-1β and transcriptionally inhibits their expression. STAT3 is the key mediator of IL-6-type cytokine signal transduction, including IL-6, ciliary neurotrophic factor (CNTF), CLCF1, and OSM. However, both IL-6 and CNTF exhibit antiobesity actions (38–41), which are opposite to the metabolically unfavorable effect of CLCF1. CNTF and IL-6 exert antiobesity effect mainly through their central effect on reducing food intake, rather than directly through adipose tissues (38–42). Some beneficial function of CNTF, for example in muscle, are STAT3-independent (43). Thus, both the target tissues and the mediated signaling determine the different effects of IL-6-type cytokines.
CLCF1 was originally identified in lymphoma cells (20). Despite its higher expression in CD45+ immune cells in BAT, CLCF1 levels in immune cells remained unchanged but decreased in mature adipocytes after cold stimulation. BAT is composed of 65% fat cells and 35% nonfat cells (44), so there are even fewer immune cells. Thus, cold-induced reduction of CLCF1 in BAT most likely was attributed to mature adipocytes. The functions of CLCF1 in immunoregulation and tumor development have been verified, but its role in metabolic regulation remains poorly understood. Here, we identified CLCF1 as an adipocyte-derived cytokine and revealed that CLCF1 autocrine signaling was a pivotal suppressor of thermogenic capacity, thereby leading to metabolic disorders. In addition, a recent study revealed that CLCF1 was an intrahepatic cholangiocyte-derived secreted factor that was elevated in the liver in nonalcoholic steatohepatitis (NASH) patients, and it performed a protective role during NASH pathogenesis (45). According to our findings and those of others, CLCF1 expression is increased in both adipose tissues and the liver under metabolic stress conditions, but CLCF1 has an adverse effect on adipocyte thermogenesis and a favorable effect in hepatic metabolism, illustrating the complex role of CLCF1 in maintaining tissue homeostasis. Despite the defined autocrine and paracrine signals, the endocrine action of CLCF1 is still unknown. We found that CLCF1 had higher expression in both BAT and spleen, implying BAT seems not the unique source of circulating CLCF1. The endocrine function of CLCF1 from BAT or spleen waits for clarification.
In summary, we proposed a model by which CLCF1 signaling repressed thermogenesis in brown fat (SI Appendix, Fig. S9). CLCF1 was down-regulated during activation of thermogenic adipocytes and up-regulated in adipocytes in obesity. Secreted CLCF1 activated the CNTFR-STAT3 signaling pathway in adipocytes and subsequently inhibited PGC-1α and PGC-1β, which in turn gave rise to decreased mitochondrial biogenesis. Consequently, CLCF1-TG mice displayed severely impaired brown fat activity.
Materials and Methods
Animal model, cell culture, AAV administration, oxygen consumption rates measurements, indirect calorimetric assessment, cold tolerance test, GTT, ITT, mtDNA copy number, RNA sequencing, qPCR, western blot, chromatin immunoprecipitation, luciferase reporter assays, transmission electron microscopy analysis, H&E staining and immunofluorescence assay, and statistical analysis were described in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by National Key R&D Program of China Grant 2018YFA0800400 (to Q.-q.T.), Shanghai Rising-Star Program Grant 22QA1402100 (to Y.L.), National Natural Science Foundation of China Grants 82170884 and 81970744 (to Y.L.), scientific research projects of Shanghai Health Commission (20204Y0116), the Scientific Research Foundation provided by Pudong Hospital affiliated to Fudan University (YJYJRC202109), and fund of peak disciplines (type IV) of institutions of higher learning in Shanghai.
Author contributions
M.D., D.-n.P., Y.L., and Q.-q.T. designed research; M.D., H.-y.X., W.-y.Z., Y.-f.X., B.-y.L., Y.-j.S., X.D., Q.-q.Y., S.-w.Q., Y.T., and Y.L. performed research; M.D. and Y.L. analyzed data; Y.L. and Q.-q.T. conceived and directed the project; and Y.L. and Q.-q.T. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. S.K. is a guest editor invited by the Editorial Board.
Contributor Information
Yang Liu, Email: yangliuly@fudan.edu.cn.
Qi-qun Tang, Email: qqtang@shmu.edu.cn.
Data, Materials, and Software Availability
RNA sequencing data generated by this study have been deposited in the Gene Expression Omnibus under accession number GSE235476 (46). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Auger C., Kajimura S., Adipose tissue remodeling in pathophysiology. Annu. Rev. Pathol. 18, 71–93 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakers A., De Siqueira M. K., Seale P., Villanueva C. J., Adipose-tissue plasticity in health and disease. Cell 185, 419–446 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chouchani E. T., Kazak L., Spiegelman B. M., New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metab. 29, 27–37 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Shamsi F., Wang C. H., Tseng Y. H., The evolving view of thermogenic adipocytes - ontogeny, niche and function. Nat. Rev. Endocrinol. 17, 726–744 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen P., Kajimura S., The cellular and functional complexity of thermogenic fat. Nat. Rev. Mol. Cell Biol. 22, 393–409 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cypess A. M., et al. , Activation of human brown adipose tissue by a beta3-adrenergic receptor agonist. Cell Metab. 21, 33–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becher T., et al. , Brown adipose tissue is associated with cardiometabolic health. Nat. Med. 27, 58–65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funcke J. B., Scherer P. E., Beyond adiponectin and leptin: adipose tissue-derived mediators of inter-organ communication. J. Lipid Res. 60, 1648–1684 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheja L., Heeren J., The endocrine function of adipose tissues in health and cardiometabolic disease. Nat. Rev. Endocrinol. 15, 507–524 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Kazak L., et al. , A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163, 643–655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding M., et al. , CHCHD10 Modulates thermogenesis of adipocytes by regulating lipolysis. Diabetes 71, 1862–1879 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Yau W. W., et al. , Thyroid hormone (T(3)) stimulates brown adipose tissue activation via mitochondrial biogenesis and MTOR-mediated mitophagy. Autophagy 15, 131–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chella Krishnan K., et al. , Sex-specific genetic regulation of adipose mitochondria and metabolic syndrome by Ndufv2. Nat. Metab. 3, 1552–1568 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai L., et al. , Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 22, 1948–1961 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uldry M., et al. , Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 3, 333–341 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Lin J., et al. , Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell 119, 121–135 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Popov L. D., Mitochondrial biogenesis: An update. J. Cell Mol. Med. 24, 4892–4899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puigserver P., et al. , A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92, 829–839 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Guo Y. Y., et al. , Cdo1 promotes PPARgamma-mediated adipose tissue lipolysis in male mice. Nat. Metab. 4, 1352–1368 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Senaldi G., et al. , Novel neurotrophin-1/B cell-stimulating factor-3: a cytokine of the IL-6 family. Proc. Natl. Acad. Sci. U.S.A. 96, 11458–11463 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vlotides G., Zitzmann K., Stalla G. K., Auernhammer C. J., Novel neurotrophin-1/B cell-stimulating factor-3 (NNT-1/BSF-3)/cardiotrophin-like cytokine (CLC)–a novel gp130 cytokine with pleiotropic functions. Cytokine Growth Factor Rev. 15, 325–336 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Song M., et al. , Cancer-associated fibroblast-mediated cellular crosstalk supports hepatocellular carcinoma progression. Hepatology 73, 1717–1735 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Zou X., et al. , Neonatal death in mice lacking cardiotrophin-like cytokine is associated with multifocal neuronal hypoplasia. Vet. Pathol. 46, 514–519 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Kim J. W., et al. , Antitumor activity of an engineered decoy receptor targeting CLCF1-CNTFR signaling in lung adenocarcinoma. Nat. Med. 25, 1783–1795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn A. F., et al. , Cold-induced sweating syndrome: CISS1 and CISS2: Manifestations from infancy to adulthood. Four new cases. J. Neurol. Sci. 293, 68–75 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Rousseau F., et al. , Inactivation of cardiotrophin-like cytokine, a second ligand for ciliary neurotrophic factor receptor, leads to cold-induced sweating syndrome in a patient. Proc. Natl. Acad. Sci. U.S.A. 103, 10068–10073 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emont M. P., et al. , A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crisponi L., Buers I., Rutsch F., CRLF1 and CLCF1 in development, health and disease. Int. J. Mol. Sci. 23, 992 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailly C., Toward a repositioning of the antibacterial drug nifuroxazide for cancer treatment. Drug. Discov. Today 24, 1930–1936 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Lee P., et al. , Irisin and FGF21 are cold-induced endocrine activators of brown fat function in humans. Cell Metab. 19, 302–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosen E. D., Spiegelman B. M., What we talk about when we talk about fat. Cell 156, 20–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittle A. J., et al. , BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell 149, 871–885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y., et al. , Hepatic small ubiquitin-related modifier (SUMO)-specific protease 2 controls systemic metabolism through SUMOylation-dependent regulation of liver-adipose tissue crosstalk. Hepatology 74, 1864–1883 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Sun W., et al. , snRNA-seq reveals a subpopulation of adipocytes that regulates thermogenesis. Nature 587, 98–102 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Song A., et al. , Low- and high-thermogenic brown adipocyte subpopulations coexist in murine adipose tissue. J. Clin. Invest. 130, 247–257 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Krieken P. P., et al. , Oncostatin M suppresses browning of white adipocytes via gp130-STAT3 signaling. Mol. Metab. 54, 101341 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moisan A., et al. , White-to-brown metabolic conversion of human adipocytes by JAK inhibition. Nat. Cell Biol. 17, 57–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lambert P. D., et al. , Ciliary neurotrophic factor activates leptin-like pathways and reduces body fat, without cachexia or rebound weight gain, even in leptin-resistant obesity. Proc. Natl. Acad. Sci. U.S.A. 98, 4652–4657 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sadagurski M., et al. , Human IL6 enhances leptin action in mice. Diabetologia 53, 525–535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinberg G. R., et al. , Ciliary neurotrophic factor suppresses hypothalamic AMP-kinase signaling in leptin-resistant obese mice. Endocrinology 147, 3906–3914 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Wallenius V., et al. , Interleukin-6-deficient mice develop mature-onset obesity. Nat. Med. 8, 75–79 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Bluher S., et al. , Ciliary neurotrophic factorAx15 alters energy homeostasis, decreases body weight, and improves metabolic control in diet-induced obese and UCP1-DTA mice. Diabetes 53, 2787–2796 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Watt M. J., et al. , CNTF reverses obesity-induced insulin resistance by activating skeletal muscle AMPK. Nat. Med. 12, 541–548 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Roh H. C., et al. , Simultaneous transcriptional and epigenomic profiling from specific cell types within heterogeneous tissues in vivo. Cell Rep. 18, 1048–1061 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu T., et al. , Intrahepatic paracrine signaling by cardiotrophin-like cytokine factor 1 ameliorates diet-induced NASH in mice. Hepatology, 10.1002/hep.32719 (2022). [DOI] [PMC free article] [PubMed]
- 46.Ding M., et al. , CLCF1 signaling restrains thermogenesis and disrupts metabolic homeostasis by inhibiting mitochondrial biogenesis in brown adipocytes. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE235476. Deposited 22 June 2023. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
RNA sequencing data generated by this study have been deposited in the Gene Expression Omnibus under accession number GSE235476 (46). All study data are included in the article and/or SI Appendix.