ABSTRACT
While primarily considered a respiratory pathogen, influenza A virus (IAV) is nonetheless capable of spreading to, and replicating in, numerous extrapulmonary tissues in humans. However, within-host assessments of genetic diversity during multicycle replication have been largely limited to respiratory tract tissues and specimens. As selective pressures can vary greatly between anatomical sites, there is a need to examine how measures of viral diversity may vary between influenza viruses exhibiting different tropisms in humans, as well as following influenza virus infection of cells derived from different organ systems. Here, we employed human primary tissue constructs emulative of the human airway or corneal surface, and we infected both with a panel of human- and avian-origin IAV, inclusive of H1 and H3 subtype human viruses and highly pathogenic H5 and H7 subtype viruses, which are associated with both respiratory disease and conjunctivitis following human infection. While both cell types supported productive replication of all viruses, airway-derived tissue constructs elicited greater induction of genes associated with antiviral responses than did corneal-derived constructs. We used next-generation sequencing to examine viral mutations and population diversity, utilizing several metrics. With few exceptions, generally comparable measures of viral diversity and mutational frequency were detected following homologous virus infection of both respiratory-origin and ocular-origin tissue constructs. Expansion of within-host assessments of genetic diversity to include IAV with atypical clinical presentations in humans or in extrapulmonary cell types can provide greater insight into understanding those features most prone to modulation in the context of viral tropism.
IMPORTANCE Influenza A virus (IAV) can infect tissues both within and beyond the respiratory tract, leading to extrapulmonary complications, such as conjunctivitis or gastrointestinal disease. Selective pressures governing virus replication and induction of host responses can vary based on the anatomical site of infection, yet studies examining within-host assessments of genetic diversity are typically only conducted in cells derived from the respiratory tract. We examined the contribution of influenza virus tropism on these properties two different ways: by using IAV associated with different tropisms in humans, and by infecting human cell types from two different organ systems susceptible to IAV infection. Despite the diversity of cell types and viruses employed, we observed generally similar measures of viral diversity postinfection across all conditions tested; these findings nonetheless contribute to a greater understanding of the role tissue type contributes to the dynamics of virus evolution within a human host.
KEYWORDS: influenza, ocular, tropism
INTRODUCTION
While primarily considered a respiratory pathogen in humans, influenza A viruses (IAV) can spread to extrapulmonary organs and be associated with a range of clinical syndromes following infection (1). For example, the human ocular surface represents a secondary mucosal surface that bears permissive receptors for influenza virus, with numerous documented cases of influenza virus infection in humans following ocular exposure (2). Epithelial cells derived from both human respiratory tract and ocular tissues can support productive replication of a range of both human and zoonotic IAV, though viruses may differ in their elicitation of innate host responses depending on the cell type infected (3–6). Of note, ocular-tropic H7 subtype viruses have been shown to downregulate genes associated with innate immune responses compared with respiratory-tropic viruses, with differential induction of cytokines and signaling pathways following IAV infection in human ocular cells compared with those derived from the respiratory tract (3). As host immune responses can serve as selective constraints governing the emergence of viral mutants capable of evading host antiviral responses (7), there is a need to understand how cell type-specific differences may modulate the generation or emergence of viral populations during multicycle replication (8).
Investigations of viral diversity emergence in discrete cell populations have primarily been conducted in respiratory tract cell types and only rarely have investigated cultures derived from multiple organ systems concurrently. A prior study did not observe a selective bottleneck effect during multicycle replication of a 2009 H1N1 pandemic virus in either MDCK or A549 cells (9), but that did not examine additional viruses or segments beyond the hemagglutinin. However, differential bottlenecks were detected in this study during viral amplification in ovo compared with in vitro, highlighting that selective pressures in diverse culture environments can influence resulting viral populations postpropagation. Considering the capacity of IAV to acquire mutations associated with changes in tissue tropism during within-host replication (10), investigating how diverse IAV overcome within-host bottlenecks in different cell types is needed to fully understand the role cellular tropism contributes to selection pressures.
Here, we employed a panel of human and avian IAV associated with either human respiratory disease or conjunctivitis, and we examined changes in genetic diversity following multicycle replication in either human primary airway or corneal tissue constructs. Respiratory-tropic IAV replicated to a higher titer and elicited more robust induction of antiviral host responses in airway-derived cultures compared with ocular-tropic viruses, while generally comparable viral titers and innate immune responses were detected among all strains postinfection in corneal-derived cultures. Despite detection of strain-specific differences in viral diversity metrics, all human and avian IAV were associated with generally similar patterns of mutational frequency during multicycle replication in both culture types, independent of viral tropism.
RESULTS
IAV replication in human airway and corneal epithelial tissue constructs.
IAV isolated from cases or outbreaks associated with either human respiratory disease or conjunctivitis were chosen (Table 1) and are here referred to as “respiratory-tropic” or “ocular-tropic” viruses in this study. To first examine if viruses associated with differential tropism in humans exhibited a tissue-specific replication advantage, we assessed replication kinetics of all viruses in both tissue types to determine the capacity of these cell types to support production of infectious virus. EpiAirway constructs, derived from human tracheal and bronchial epithelial cells and cultured to form a ciliated apical surface and mucociliary epithelium when cultured at an air-liquid interface, were employed to emulate cells derived from the respiratory tract. EpiCorneal constructs, derived from corneal epithelial cells and cultured to form a stratified, squamous epithelium, were employed to emulate cells derived from the ocular system.
TABLE 1.
Influenza A viruses isolated from human cases used in the study
| Virus | Abbreviation | Subtypea | Tropism | Reference |
|---|---|---|---|---|
| A/Netherlands/219/2003 | NL/219 | HPAI H7N7 | Ocularb | 21 |
| A/Netherlands/230/2003 | NL/230 | HPAI H7N7 | Ocular | 21 |
| A/Mexico/InDRE7218/2012 | Mex/7218 | HPAI H7N3 | Ocular | 22 |
| A/Anhui/1/2013 | Anhui/1 | LPAI H7N9 | Respiratoryc | 49 |
| A/Thailand/16/2004 | Thai/16 | HPAI H5N1 | Respiratoryc | 50 |
| A/Panama/2007/1999 | Panama/99 | Seasonal H3N2 | Respiratory | 51 |
| A/Mexico/4482/2009 | Mexico/4482 | pdm09 H1N1 | Respiratory | 52 |
HPAI, highly pathogenic avian influenza; LPAI, low pathogenic avian influenza; pdm09, virus derived from the 2009 pandemic.
NL/219 virus was isolated from a fatal respiratory case but was considered ocular-tropic for the study, as the majority of human cases from this outbreak were associated with conjunctivitis.
Anhui/1 and Thai/16 viruses were isolated from fatal respiratory cases.
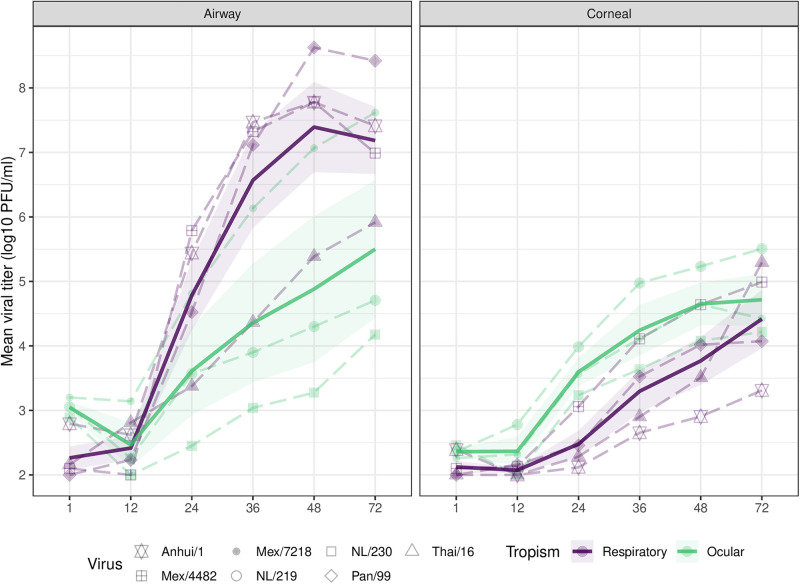
In agreement with previous studies, both EpiAirway and EpiCorneal cultures supported productive replication of avian and human IAV for the duration of the 72-h experiment (4, 11, 12). In EpiAirway cultures, the seasonal H3N2 virus Panama/99 and the 2009 H1N1 pandemic virus Mex/4482 both replicated to significantly higher titers through 72 h postinoculation (p.i.) compared with highly pathogenic avian influenza (HPAI) A viruses from the HPAI H7N7 outbreak in the Netherlands in 2003 and an HPAI H5N1 virus isolated from a fatal respiratory case in 2004 (P ≤ 0.0001) (Fig. 1), highlighting the capacity of well-adapted human viruses to reach high titers in cells derived from the respiratory tract compared with several avian strains. Significant differences in viral titers were not observed among viruses exhibiting different human tropisms in EpiCorneal constructs.
FIG 1.
Replication of IAV in respiratory and ocular cultures. Influenza A viruses classified as respiratory-tropic (Anhui/1 [LPAI H7N9], Mexico/4482 [pdm09 H1N1], Panama/99 [seasonal H3N2], and Thai/16 [HPAI H5N1]; show in purple) or ocular-tropic (Mexico/7218 [HPAI H7N3], NL/219 [HPAI H7N7], and NL/230 [HPAI H7N7]; shown in green) in this study were used to infect EpiAirway or EpiCorneal tissue constructs at an MOI of 0.01 in triplicate. The apical surface of each construct was incubated with 200 μL medium at the indicated times postinfection, and the viral titer was determined by standard plaque assay (limit of detection, 100 PFU). Virus detected at the 1-h time point represents residual viral inoculum following gentle washing of cells. Solid bold lines represent the viral tropism group mean with standard error (shading).
Differences in total cell density between EpiAirway and EpiCorneal tissue constructs precluded direct statistical comparison of replication efficiency for individual viruses based on infected cell type. However, respiratory-tropic viruses possessed a mean total area under the curve values 1.3- to 5.9-fold higher in airway epithelial cultures compared with corneal epithelial cultures (P < 0.0001 for Anhui/1, Panama/99, and Mex/4482 viruses), in contrast with ocular-tropic viruses (differences 0.7- to 1.7-fold between airway and corneal cultures), suggesting that viruses associated with human respiratory disease replicated to higher sustained titers in airway cultures than in corneal cultures. While peak titers varied in magnitude, detection of productive replication of all viruses in both cell types permitted subsequent closer examination of selective pressures based on culture origin.
Differential induction of host responses based on virus tropism and cell type.
Prior studies have supported that elicitation of innate host responses following IAV infection can be differentially modulated between cell types (3, 13), but the extent to which these parameters varied in tissue constructs was not known. To examine differential induction of host antiviral responses elicited following virus infection, we infected EpiAirway and EpiCorneal constructs with either Anhui/1 virus (a LPAI H7N9 virus associated with a fatal respiratory cases) or Mex/7218 virus (an HPAI H7N3 virus associated with human conjunctivitis) at a multiplicity of infection (MOI) of 2 and isolated total RNA from cells 24 h p.i. At this higher MOI, both viruses were detected at comparable titers in EpiCorneal constructs (5.2 ± 0.34 and 5.21 ± 0.05 log10 PFU/mL for Anhui/1 and Mex/7218 viruses, respectively [means ± standard errors]), suggesting that the reduced viral titers observed for Anhui/1 virus relative to Mex/7218 virus in Fig. 1 indicate an impaired ability of Anhui/1 virus to replicate at lower, but not higher, input doses in EpiCorneal constructs. In EpiAirway constructs, Anhui/1 virus replicated to significantly higher titer (8.13 ± 0.3 log10 PFU/mL) than Mex/7218 virus (7.22 ± 0.05 log10 PFU/mL) (P < 0.007), in agreement with enhanced replication of respiratory-tropic viruses in respiratory tract cultures compared with ocular-tropic strains (Fig. 1).
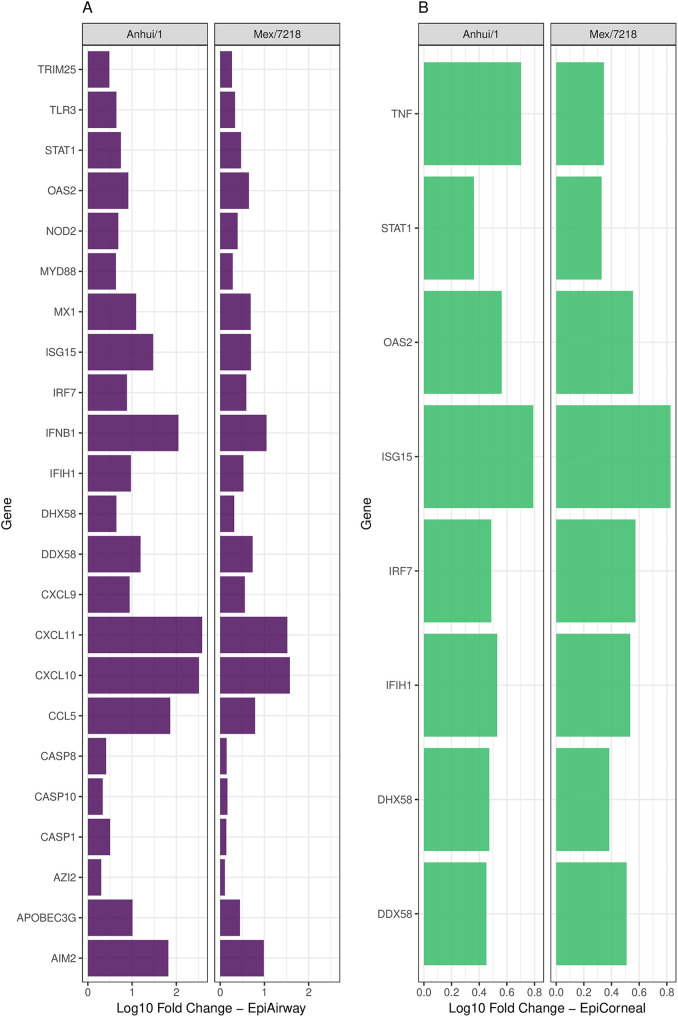
RNA isolated from these cultures was subsequently tested by PCR array to determine the extent to which a panel of genes (n = 84) associated with antiviral responses were upregulated following viral infection relative to that of mock-infected cells. Genes related to antiviral responses that were significantly (P < 0.05) upregulated in each culture relative to mock-infected wells were assessed. In airway tissue constructs, infection with Anhui/1 virus resulted in the upregulation (≥2-fold) of more genes, and to an elevated level, compared with Mex/7218 virus infection (fold upregulation over mock, 2.0 to 382.93 [mean, 47.69] Anhui/1; 1.28 to 37.73 [mean, 6.52] Mex/7218) (Fig. 2A), suggestive of heightened host innate immune responses following infection with a respiratory-tropic compared with an ocular-tropic virus in this tissue (Fig. 2A). In contrast, viral infection of corneal tissue constructs resulted in diminished induction of host innate immune responses, with no appreciable difference based on the tropism of the virus (fold upregulation over mock, 2.3 to 6.18 Anhui/1; 2.13 to 6.71 Mex/7218) (Fig. 2B). Collectively, these results support both virus-specific and tissue-specific differences in elicitation of host responses early after infection.
FIG 2.
Fold regulation of selected genes related to human antiviral responses in respiratory and ocular cultures following infection with H7 subtype IAV. EpiAirway (A) and EpiCorneal (B) tissue constructs were infected with Anhui/1 and Mex/7218 viruses at an MOI of 2 in triplicate. Total RNA was isolated at 24 h p.i. and examined by real-time RT-PCR array analysis. Genes which exhibited a significant (P < 0.05) fold change in expression relative to mock-infected wells and for which at least one of the viruses elicited a fold change of ≥2 are shown. The x-axis scale is the log10 fold change.
Influenza A virus genetic diversity following replication in human airway and corneal epithelial cultures.
To examine if differential induction of host responses represented a selective pressure capable of modulating the scope of genetic diversity among IAV following infection of different human cell types, deep sequencing was performed on cell supernatants collected 72 h p.i. from virus-infected EpiAirway or EpiCorneal tissue constructs. We detected both synonymous and nonsynonymous mutations with all viruses tested compared with input inoculum, with Anhui/1 virus, followed by NL/219 virus, having the most single-nucleotide variants overall (see Table S1 in the supplemental material). Despite differences in replication kinetics discussed above, no discernible patterns were apparent between respiratory-tropic and ocular-tropic viruses at the level of either gene or cell type. The ratio of nonsynonymous to synonymous substitutions (dN/dS) and ratio of the mean number of pairwise differences per site of nonsynonymous to synonymous substitutions (piN/piS) for HA, NA, and PB2 were largely consistent between EpiAirway and EpiCorneal constructs (Table S2). When examining nucleic acid substitutions on the HA gene, transitions from A to G were the most common form of base change mutation across all viruses, followed by C-to-T mutations, with the exception of Pan/99 virus, where G-to-T transversions were more common (Table S3 and Fig. S1). However, substitutions between cell type were not significant from A to G for any virus (t test, P ≥ 0.108); C to T was significant (P = 0.005) only for NL/230 virus (Mex/4482 and Pan/99 viruses were not tested due to low sample size). When examined proportionally (Fig. S2), C-to-T substitutions were higher in cells opposite of virus tropism for Anhui/1, Mex/4482, NL/219, NL/230, and Thai/16 viruses. These findings indicated that base change proportions detectable following multicycle replication with both respiratory-tropic and ocular-tropic viruses were not modulated based on the cell type examined.
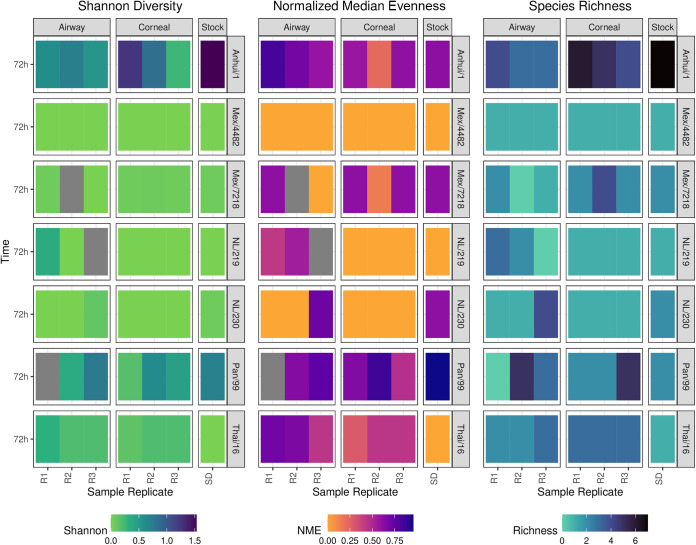
We next examined measures of viral alpha diversity. In agreement with Anhui/1 virus bearing the greatest number of single-nucleotide variants among all viruses evaluated, this virus showed the most significant differences to other viruses for several metrics (Fig. 3, Fig. S3, Table S4, and Table S5). Overall, these metrics varied most by virus with no discernible trends at the level of viral tropism or viral titer postinfection. With few exceptions, divergence in diversity metrics between cell types infected with the same virus was not observed (Table S6). Only NL/219 virus-infected EpiAirway constructs were significantly different from EpiCorneal and stock for normalized mean evenness (NME) (P < 0.001) and species richness (P = 0.029). When NME and richness were broken out by virus and cell type (Fig. S4), we observed higher species richness and NME in EpiAirway cells for NL/219 and NL/230 viruses and higher richness and NME in EpiCorneal constructs for Mex/7218 virus. In contrast, we observed a trend of higher richness in EpiCorneal cultures but higher NME in EpiAirway cultures for Anhui/1 and Thai/16 viruses. Collectively, these results supported that differences in diversity metrics may vary at the level of both viral strain and cell type but are not driven primarily at the level of virus tropism.
FIG 3.
IAV diversity metrics following multicycle replication in respiratory or corneal cultures. Heatmap of Shannon diversity, normalized median evenness, and species richness for each of the seven influenza A viruses for each tissue construct (airway, corneal) at 72 h p.i. and stock viral quasispecies populations. R1 to R3, individual sample replicates; SD, diluted inoculum stock. Dark gray shading represents missing data.
Detection of nonsynonymous mutations 72 h p.i. following IAV replication in human airway and corneal epithelial cultures.
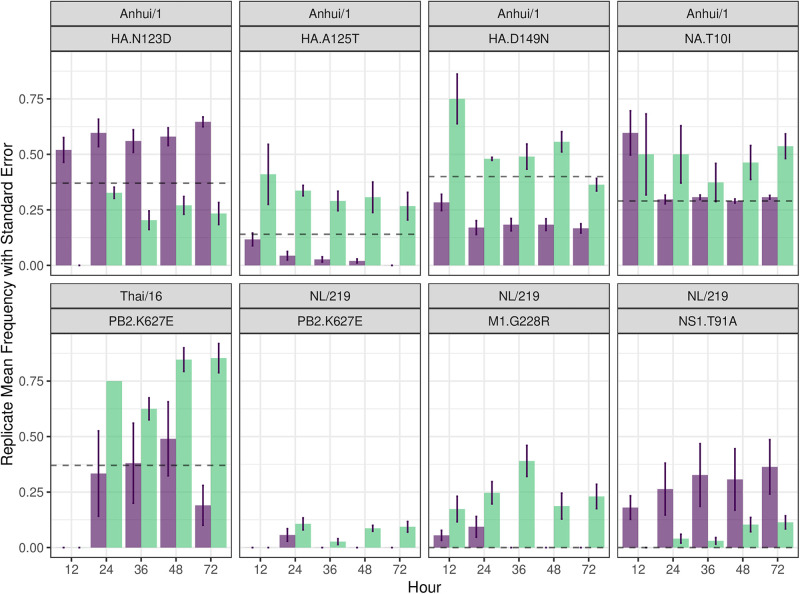
Beyond assessments of viral diversity in general, we examined selection pressures at specific residues to identify variability by cell type or virus tropism by focusing on specific nonsynonymous mutations leading to amino acid substitutions. Analysis of samples collected at 72 h p.i. revealed the presence of many nonsynonymous variants present at ≥5% abundance (Fig. 4). The Anhui/1 virus stock contained a heterogeneous virus population with high nonsynonymous variability concentrated in HA (N123D and D149N), NA (T10I), and PA (N409S) (Table S7). Following multicycle replication in EpiCorneal cultures, Anhui/1 virus showed an increased number of variants with substitutions in HA, NA, PA, and M1 that contrasted with the ones detected following multicycle replication in EpiAirway cultures, though only one variant (HA N123D) was significantly different (P = 0.027) between cell types at 72 h p.i. (Table S8).
FIG 4.
Viral mutation frequency of IAV over time in respiratory or corneal cultures are shown. Selected mutations of interest show mean frequency across all replicates with standard error bars in EpiAiway (purple) and EpiCorneal (green) tissue constructs between 12 and 72 h p.i. The dashed black line represents the stock inoculum mean frequency.
In contrast to Anhui/1 virus, NL/219 virus infection resulted in reduced nonsynonymous genetic variability between EpiAirway and EpiCorneal constructs. While no overall statistical significance was found between cell types at 72 h p.i. (Table S8), an increase in genetic variability was observed in NL/219-virus infected samples collected from EpiCorneal constructs, mostly in M1 (G228R) and NS1 (P216S) (Table S9). Similarly, the percentages of variants obtained from both tissue constructs following Thai/16 virus infection were often similar (Table S10). Both NL/219 and Thai/16 viruses bear a lysine at position 627 in PB2, a mutation associated with host range of IAV (14). At 72 h p.i., higher mean frequencies of a 627E viral subpopulation were detected for both viruses in EpiCorneal compared with EpiAirway cultures; this difference was statistically significant (P = 0.047) for Thai/16 virus only. The remaining viruses examined had no statistically significant differences in variant composition between cell type at 72 h p.i. Collectively, these results supported that IAV infection in either respiratory or ocular cultures does not lead to strikingly different selective pressures following multicycle replication.
Modulation of diversity metrics and mutational frequency during multicycle replication of IAV in human airway and corneal epithelial cultures.
Focusing on three viruses that exhibit different tropisms and disease severities in humans (Anhui/1, NL/219, and Thai/16), we examined if there was selection of overall diversity or virus variants over time between different cell types and, if so, how rapidly these emerged or were maintained over the course of the in vitro infection. Overall, Shannon diversity was largely stable between 12 h and 72 h p.i. for all viruses, with few exceptions. Increased Shannon diversity (alongside other diversity metrics related to species richness and evenness) was noted in EpiCorneal cultures infected with Anhui/1 virus (Fig. S5, Fig. S6, and Table S4). Additionally, increased diversity metrics were detected over time in EpiAirway cultures infected with NL/219 virus. However, consistent trends in these diversity metrics for a specific virus or cell type were not observed.
Following infection with Anhui/1 virus, selected variants showed a pattern of divergence between cell types (Fig. 4, Fig. S7, and Table S7). HA N123D showed a trending increase over stock amount in EpiAirway cultures, while a decreased yet stable trend was observed in EpiCorneal cultures, with statistical significance being observed at 12 h (P = 0.014) and 72 h p.i. time points (P = 0.027) (Table S8). A similar trend was observed for position PA N409S (not present in stock inoculum), with statistical significance detected at the 24-h (P = 0.048) and 36-h time points (P = 0.0358). However, selected variants (e.g., HA positions A125T and D149N) displayed trending increases over time in EpiCorneal but not EpiAirway cells, supporting that divergence from stock inoculum varied at specific positions and not only by tissue origin.
NL/219 virus-infected cultures displayed few distinct differences between culture type over time. Interestingly, similar to Anhui/1 PA N409S, numerous variants emerged following NL/219 virus infection that were not present in stock inoculum. (Fig. 4, Fig. S8, and Table S9). Variant populations were generally higher in EpiCorneal cells with few exceptions (notably, NS1 T91A). NS1 P216S was only observed in EpiCorneal cells. However, only positions M1 G228R at 36 h (P = 0.049) and PB2 K627E at 48 h (P = 0.031) were found to be statistically significant (Table S8). Similarly, variant proportions were largely comparable between cell types following Thai/16 virus infection (Fig. 4, Fig. S9, and Table S10). While limited divergence between cell types was observed at PB2 positions K443R and K627E, with levels generally above those observed in the stock sample in EpiCorneal but not EpiAirway cultures; only PB2 K627E at 72 h (P = 0.047) was statistically significant, as discussed above (Table S8). Collectively, these data indicate the capacity for population divergence at the level of tissue type during multicycle replication, but they further support that viral tropism is not strongly impacting within-host bottlenecks in different cellular environments.
DISCUSSION
IAV are capable of replication in a multitude of discrete cell types within and beyond the human respiratory tract (15). However, assessments of viral evolution during multicycle replication often are conducted in one representative cell type only (16), or a specimen (such as a nasal wash) that is inclusive of virus replication at multiple independent sites (9, 17, 18). Studies that do examine changes in viral quasispecies in multiple independent sites of virus replication over time are typically conducted in vivo, which frequently limits assessments to one virus or virus subtype only (19). To investigate the capacity of cellular origin to modulate within-host viral genetic diversity during multicycle replication, we infected human primary tissue constructs emulative of the human airway or corneal surface with a panel of viruses associated with either respiratory or ocular tropisms in humans. We found that both respiratory-tropic and ocular-tropic viruses exhibited generally similar patterns of synonymous and nonsynonymous mutations following multicycle replication in both cell types, despite differences in selective pressures between respiratory and ocular tissues.
Deep sequencing analyses of IAV following controlled multicycle replication within ocular tissues has not been reported previously. A recent study from our group found that multiple IAV acquire similar mutations in nasal wash specimens during replication in a ferret host following either respiratory or ocular exposure to virus (20), but that study did not sequence ocular specimens specifically, due to relatively low viral titers in this specimen. Clinical specimens collected from the ocular surface (e.g., following swabbing the conjunctiva) have been critical for the isolation of H7 subtype IAV from humans (such as during poultry outbreaks associated with human infection in the Netherlands in 2003, Canada in 2004, and Mexico in 2012) (21–23). Notably, in the Netherlands and Canada outbreaks, sequence differences were noted between specimens collected from respiratory samples compared with conjunctival clinical specimens (23, 24). Understanding if respiratory and ocular cell types support differing levels of within-host diversity during multicycle virus infection thus provides necessary contextual information when examining virus isolated and cultured from different anatomical sites.
The human eye has evolved over time to limit inflammatory processes that could result in damage to vision, a feature known as “immune privilege” (25). A central question of this study was to assess if ocular immune privilege modulated viral diversity over time relative to a different tissue, such as the respiratory tract, that is governed by induction of different host immune responses following challenge by a pathogen. Viral infection of EpiCorneal cells resulted in the induction of fewer antiviral response-associated genes, and to a lesser extent when upregulated, compared to homologous virus infection in EpiAirway cultures, in agreement with prior studies supporting delayed and weakened induction of host responses in other human ocular cell types (3). In contrast, viral infection of EpiAirway cultures resulted in a more robust upregulation of genes associated with inflammatory processes (notably those associated with Toll-like receptor, RIG-I, and interferon signaling pathways [26]), with all genes upregulated to a higher extent following infection a respiratory-tropic H7 virus compared with an ocular-tropic H7 virus, also in agreement with prior studies (3). However, despite disparate elicitations of host responses in respiratory versus ocular cultures following virus infection, this study revealed generally similar viral diversity metrics independent of the infected tissue origin (Fig. 3), suggesting that these selective pressures did not have a profound impact on these parameters.
That said, closer examination of differences in mutational frequencies between cell types infected with the same virus did reveal instances suggestive of tropism-related selection. Most notably, sequencing from EpiCorneal constructs infected with viruses bearing a lysine at PB2 position 627 (NL/219 and Thai/16) both had a higher replicative mean frequency of 627E compared with homologous virus-infected EpiAirway constructs, suggesting that corneal tissue exerts a less selective pressure to “fix” virus with the more pathogenic 627K substitution compared with airway cultures (Fig. 4); this is in agreement with in vivo studies which have shown E-to-K host adaptation in murine respiratory tract tissues following acute HPAI virus infection (27). Further mechanistic studies to investigate ways in which ocular tissues may support differential tolerance of markers associated with mammalian adaptation (and/or other key molecular determinants of virulence which are typically studied only following replication in respiratory tissues) are warranted.
Beyond the well-studied PB2 627, our study identified a capacity for cultures of different tissue origin to support divergent viral populations postinoculation with a homologous challenge input. The high diversity of Anhui/1 stock virus is likely attributable in part to its propagation in eggs, as several of the amino acid substitutions in the HA and NA detected in this study have been reported previously (28). Positions 123, 125, and 149 in the HA in Anhui/1 and other H7 viruses are recognized as sites of high variability with implications in avian host range (29–31). It is of interest that the A125T mutation, known for binding to α2-3-linked sialic acids, was enriched in EpiCorneal cells (the surface of which bear both α2-3 and α2-6 receptors) and not EpiAirway cultures (32). Many of other variants which emerged by 72 h p.i. were at positions in the HA associated with receptor binding or in the polymerase genes associated with host adaptation, though frequencies of these mutations were generally low (Fig. S7 to S9).
It should be noted that while this study represents a necessary first step in elucidating the role of viral tropism in within-host measures of viral diversity, additional refinements and analyses are needed beyond the scope of this work. Tissue constructs cultured at the air-liquid interface are capable of more closely emulating the complex cellular environment found at both locations; in the case of EpiCorneal cultures, these constructs have been shown to possess features (such as membrane-associated mucins) at higher levels than primary corneal epithelial cells, making them well-suited for tropism investigations conducted in vitro (4). However, viral replication and elicitation of innate immune responses can vary between different cell types at the ocular surface (3); a greater understanding of how these selective pressures may modulate diversity metrics following multicycle replication is needed. While a fixed 0.01 MOI and 37°C culture temperature were employed for all infections, both respiratory and ocular cell types can be productively infected with lower, more physiologically relevant challenge doses, and at the cooler temperatures (e.g., 33°C) associated with the upper respiratory tract and human ocular surface (4). As viral input can affect subsequent parameters associated with within-host diversity and cooperative dynamics between quasispecies (8, 33), it is possible that more striking differences would have been present under different experimental conditions.
While this study employed ocular cells as a comparative extrapulmonary site of virus replication, IAV can replicate and elicit proinflammatory responses in numerous systemic tissues (34). Great strides have been made in understanding within-host viral evolution of influenza A and B viruses (8, 35), but it should be noted the vast majority of this work has taken place exclusively within cell or specimen types derived from the respiratory tract. Expanding the scope of these studies to include extrapulmonary sites of viral replication (including but not limited to the ocular milieu) will provide a greater understanding of the role tissue type contributes to the dynamics of virus evolution within a human host.
MATERIALS AND METHODS
Viruses.
Viruses used in this study are listed in Table 1. Virus stocks were generated in either MDCK cells (Mex/4482) or the allantoic cavity of 10-day-old embryonated hens’ eggs (all other viruses), as described previously (36). Avian-origin viruses were handled under biosafety level 3 containment, including enhancements, as required by the U.S. Department of Agriculture and the Federal Select Agent Program (37). All viruses used had a passage history of ≤3 in the matrix specified, with the exception of Panama/99 (passage 6).
Cells and infections.
EpiAirway and EpiCorneal tissue constructs (MatTek Corporation; 0.62-cm2 growth area) were unpacked immediately upon receipt and preequilibrated overnight at 37°C before use the next morning. Constructs arrived and were maintained at the air-liquid interface according to the manufacturer’s instructions, with serum-free basolateral medium. MOIs for tissue constructs were calculated based on the midpoint of the manufacturer’s estimate of cells on the apical surface (200,000 cells for EpiAirway, 75,000 cells for EpiCorneal).
For replication kinetics, tissue constructs (unwashed) were inoculated apically in triplicate at an MOI of 0.01 (150 μL) for 1 h, after which inoculum was removed and cells were gently washed with phosphate-buffered saline before restoring to the air-liquid interface at 37°C. At the indicated time points, 200 μL of supernatant (incubated atop tissue constructs for 20 min before collection) was collected. Samples were frozen at −80°C until titration by standard plaque assay in MDCK cells (limit of detection, 100 PFU) (36). Statistical significance of replication kinetics assessments was determined by calculating the area under the curve for each replicate (12 to 72 h p.i.), followed by a two-way ANOVA of these values using GraphPad Prism, version 7.
For PCR array analyses, tissue constructs were inoculated apically in triplicate at an MOI of 2 as described above with Anhui/1 or Mex/7218 virus or with an equal dilution of normal allantoic fluid to serve as a mock-infected control. At 24 h p.i., after a supernatant time point was collected, total RNA was extracted from all mock-infected or virus-infected cell monolayers, followed by cDNA synthesis. RT2 Profiler PCR array analysis for human antiviral response (Qiagen catalog number PAHS-122Z) was performed on all specimens and analyzed per the manufacturer’s instructions.
RNA extraction and sequencing.
Culture supernatants were inactivated in AVL buffer (Qiagen). Influenza viral RNA was extracted using a QIAamp RNA extraction kit (Qiagen) with a high-throughput automated liquid handler, QIAcube HT (Qiagen). Amplicons for sequencing libraries were generated through multisegment reverse transcription-PCR, a technique used to amplify the influenza genome in a single PCR (38). The resulting amplicons were then quantified using a Quant-iT double-stranded DNA high-sensitivity assay (Invitrogen), and quality of the amplicons was assessed by visualizing the amplicon fragments on the QIAxcel Advanced system (Qiagen) for size confirmation and presence of amplicon segments. Paired-end DNA libraries were produced using the Nextera XT sample prep kit (Illumina) and Nextera XT index kit v2 (Illumina) using half-volume reaction mixtures. The amplicon libraries were purified using 0.8× AMPure XP beads (Beckman Coulter Inc.) on a Zephyr Compact liquid handling workstation (Perkin Elmer). Purified libraries were then normalized and pooled using high-sensitivity Quant-iT dsDNA to assess concentration, and mean library size was assessed by QIAxcel. Six picomolar of the pooled libraries, including 5% PhiX, was loaded into a MiSeq v2 300 cycle kit and MiSeq (Illumina) sequencer.
Bioinformatic and data analysis.
Raw sequencing reads were processed and aligned using IRMA v0.6.8+ (39) to obtain base change counts and variant frequencies for each gene segment filtered with a minimum frequency of 5%. Nucleotide diversity as measures of the dN/dS and piN/piS was calculated in SNPGenie (40) for HA, NA, and PB2 genes, for each sample, for all seven viruses. Alignment files in SAM format from IRMA were then used in CliqueSNV v2.0.3 (41) to reconstruct haplotypes for the HA gene for all virus stocks and 72-h time point and Anhui/1, NL/219, and Thai/16 at 12-h collection. Using the haplotypes and their frequencies, we calculated several metrics for diversity richness and evenness (species richness, Shannon diversity, exponential Shannon, normalized median evenness [NME], Pielou’s evenness, inverse Simpson) with the assistance of R packages vegan v2.6-2 (42) and QSutils v1.8.0 (43). Exponential Shannon and inverse Simpson imdices put more emphasis on the dominate haplotypes, while Shannon, NME, and Pielou’s provide more balanced metrics. Statistical comparisons between groups used a t test with Holm correction for multiple comparisons with significance set at <0.05. Figures were created in R using the packages tidyverse v1.3.2 (44), ggplot2 v3.3.6 (45), scales v1.1.1 (46), and patchwork v1.1.2 (47). All R analyses were conducted using version 4.0.3 (48).
ACKNOWLEDGMENTS
The findings and conclusions are those of the authors and do not necessarily reflect the official position of the Agency for Toxic Substances and Disease Registry, Centers for Disease Control and Prevention (CDC). We thank Jason Wilson and Hannah Creager for helpful discussions and technical assistance. This project was supported in part by an appointment to the Research Participation Program at the CDC administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the CDC (T.J.K.).
Footnotes
Supplemental material is available online only.
Contributor Information
Jessica A. Belser, Email: jbelser@cdc.gov.
Mathilde Richard, Erasmus MC.
REFERENCES
- 1.Uyeki TM, Katz JM, Jernigan DB. 2017. Novel influenza A viruses and pandemic threats. Lancet 389:2172–2174. doi: 10.1016/S0140-6736(17)31274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belser JA, Lash RR, Garg S, Tumpey TM, Maines TR. 2018. The eyes have it: influenza virus infection beyond the respiratory tract. Lancet Infect Dis 18:e220–e227. doi: 10.1016/S1473-3099(18)30102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belser JA, Zeng H, Katz JM, Tumpey TM. 2011. Ocular tropism of influenza A viruses: identification of H7 subtype-specific host responses in human respiratory and ocular cells. J Virol 85:10117–10125. doi: 10.1128/JVI.05101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creager HM, Kumar A, Zeng H, Maines TR, Tumpey TM, Belser JA. 2018. Infection and replication of influenza virus at the ocular surface. J Virol 92. doi: 10.1128/JVI.02192-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan MC, Chan RW, Yu WC, Ho CC, Yuen KM, Fong JH, Tang LL, Lai WW, Lo AC, Chui WH, Sihoe AD, Kwong DL, Wong DS, Tsao GS, Poon LL, Guan Y, Nicholls JM, Peiris JS. 2010. Tropism and innate host responses of the 2009 pandemic H1N1 influenza virus in ex vivo and in vitro cultures of human conjunctiva and respiratory tract. Am J Pathol 176:1828–1840. doi: 10.2353/ajpath.2010.091087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan RW, Kang SS, Yen HL, Li AC, Tang LL, Yu WC, Yuen KM, Chan IW, Wong DD, Lai WW, Kwong DL, Sihoe AD, Poon LL, Guan Y, Nicholls JM, Peiris JS, Chan MC. 2011. Tissue tropism of swine influenza viruses and reassortants in ex vivo cultures of the human respiratory tract and conjunctiva. J Virol 85:11581–11587. doi: 10.1128/JVI.05662-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol Mol Biol Rev 76:159–216. doi: 10.1128/MMBR.05023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xue KS, Moncla LH, Bedford T, Bloom JD. 2018. Within-host evolution of human influenza virus. Trends Microbiol 26:781–793. doi: 10.1016/j.tim.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varble A, Albrecht RA, Backes S, Crumiller M, Bouvier NM, Sachs D, Garcia-Sastre A, tenOever BR. 2014. Influenza A virus transmission bottlenecks are defined by infection route and recipient host. Cell Host Microbe 16:691–700. doi: 10.1016/j.chom.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessiere P, Volmer R. 2021. From one to many: the within-host rise of viral variants. PLoS Pathog 17:e1009811. doi: 10.1371/journal.ppat.1009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell H, Levin D, Forrest S, Beauchemin CA, Tipper J, Knight J, Donart N, Layton RC, Pyles J, Gao P, Harrod KS, Perelson AS, Koster F. 2011. Higher level of replication efficiency of 2009 (H1N1) pandemic influenza virus than those of seasonal and avian strains: kinetics from epithelial cell culture and computational modeling. J Virol 85:1125–1135. doi: 10.1128/JVI.01722-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerlach RL, Camp JV, Chu YK, Jonsson CB. 2013. Early host responses of seasonal and pandemic influenza A viruses in primary well-differentiated human lung epithelial cells. PLoS One 8:e78912. doi: 10.1371/journal.pone.0078912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belser JA, Zeng H, Katz JM, Tumpey TM. 2011. Infection with highly pathogenic H7 influenza viruses results in an attenuated proinflammatory cytokine and chemokine response early after infection. J Infect Dis 203:40–48. doi: 10.1093/infdis/jiq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol 67:1761–1764. doi: 10.1128/JVI.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumlin U, Olofsson S, Dimock K, Arnberg N. 2008. Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respir Viruses 2:147–154. doi: 10.1111/j.1750-2659.2008.00051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Si L, Bai H, Oh CY, Jin L, Prantil-Baun R, Ingber DE. 2021. Clinically relevant influenza virus evolution reconstituted in a human lung airway-on-a-chip. Microbiol Spectr 9:e0025721. doi: 10.1128/Spectrum.00257-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moncla LH, Zhong G, Nelson CW, Dinis JM, Mutschler J, Hughes AL, Watanabe T, Kawaoka Y, Friedrich TC. 2016. Selective bottlenecks shape evolutionary pathways taken during mammalian adaptation of a 1918-like avian influenza virus. Cell Host Microbe 19:169–180. doi: 10.1016/j.chom.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilker PR, Dinis JM, Starrett G, Imai M, Hatta M, Nelson CW, O'Connor DH, Hughes AL, Neumann G, Kawaoka Y, Friedrich TC. 2013. Selection on haemagglutinin imposes a bottleneck during mammalian transmission of reassortant H5N1 influenza viruses. Nat Commun 4:2636. doi: 10.1038/ncomms3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakdawala SS, Jayaraman A, Halpin RA, Lamirande EW, Shih AR, Stockwell TB, Lin X, Simenauer A, Hanson CT, Vogel L, Paskel M, Minai M, Moore I, Orandle M, Das SR, Wentworth DE, Sasisekharan R, Subbarao K. 2015. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 526:122–125. doi: 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belser JA, Sun X, Kieran TJ, Brock N, Pulit-Penaloza JA, Pappas C, Basu Thakur P, Jones J, Wentworth DE, Zhou B, Tumpey TM, Maines TR. 2022. Detection of airborne influenza A and SARS-CoV-2 virus shedding following ocular inoculation of ferrets. J Virol 96:e0140322. doi: 10.1128/jvi.01403-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, Bosman A. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Martinez I, Balish A, Barrera-Badillo G, Jones J, Nunez-Garcia TE, Jang Y, Aparicio-Antonio R, Azziz-Baumgartner E, Belser JA, Ramirez-Gonzalez JE, Pedersen JC, Ortiz-Alcantara J, Gonzalez-Duran E, Shu B, Emery SL, Poh MK, Reyes-Teran G, Vazquez-Perez JA, Avila-Rios S, Uyeki T, Lindstrom S, Villanueva J, Tokars J, Ruiz-Matus C, Gonzalez-Roldan JF, Schmitt B, Klimov A, Cox N, Kuri-Morales P, Davis CT, Diaz-Quinonez JA. 2013. Highly pathogenic avian influenza A(H7N3) virus in poultry workers, Mexico, 2012. Emerg Infect Dis 19:1531–1534. doi: 10.3201/eid1909.130087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, Halpert C, Hirst M, Astell C, Lawrence D, Mak A. 2004. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis 10:2196–2199. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, Koch G, Bosman A, Koopmans M, Osterhaus AD. 2004. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci USA 101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Paiva CS, St Leger AJ, Caspi RR. 2022. Mucosal immunology of the ocular surface. Mucosal Immunol 15:1143–1157. doi: 10.1038/s41385-022-00551-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmolke M, Garcia-Sastre A. 2010. Evasion of innate and adaptive immune responses by influenza A virus. Cell Microbiol 12:873–880. doi: 10.1111/j.1462-5822.2010.01475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Min JY, Santos C, Fitch A, Twaddle A, Toyoda Y, DePasse JV, Ghedin E, Subbarao K. 2013. Mammalian adaptation in the PB2 gene of avian H5N1 influenza virus. J Virol 87:10884–10888. doi: 10.1128/JVI.01016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shcherbik S, Pearce N, Balish A, Jones J, Thor S, Davis CT, Pearce M, Tumpey T, Cureton D, Chen LM, Villanueva J, Bousse TL. 2015. Generation and characterization of live attenuated influenza A(H7N9) candidate vaccine virus based on Russian donor of attenuation. PLoS One 10:e0138951. doi: 10.1371/journal.pone.0138951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pantin-Jackwood MJ, Miller PJ, Spackman E, Swayne DE, Susta L, Costa-Hurtado M, Suarez DL. 2014. Role of poultry in the spread of novel H7N9 influenza virus in China. J Virol 88:5381–5390. doi: 10.1128/JVI.03689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. 2013. Structural analysis of the hemagglutinin from the recent 2013 H7N9 influenza virus. J Virol 87:12433–12446. doi: 10.1128/JVI.01854-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Carney PJ, Donis RO, Stevens J. 2012. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. J Virol 86:8645–8652. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura K, Shirakura M, Suzuki Y, Naito T, Fujisaki S, Tashiro M, Nobusawa E. 2016. Development of a high-yield reassortant influenza vaccine virus derived from the A/Anhui/1/2013 (H7N9) strain. Vaccine 34:328–333. doi: 10.1016/j.vaccine.2015.11.050. [DOI] [PubMed] [Google Scholar]
- 33.Xue KS, Hooper KA, Ollodart AR, Dingens AS, Bloom JD. 2016. Cooperation between distinct viral variants promotes growth of H3N2 influenza in cell culture. Elife 5:e13974. doi: 10.7554/eLife.13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Short KR, Veeris R, Leijten LM, van den Brand JM, Jong VL, Stittelaar K, Osterhaus A, Andeweg A, van Riel D. 2017. Proinflammatory cytokine responses in extra-respiratory tissues during severe influenza. J Infect Dis 216:829–833. doi: 10.1093/infdis/jix281. [DOI] [PubMed] [Google Scholar]
- 35.Valesano AL, Fitzsimmons WJ, McCrone JT, Petrie JG, Monto AS, Martin ET, Lauring AS. 2020. Influenza B viruses exhibit lower within-host diversity than influenza A viruses in human hosts. J Virol 94. doi: 10.1128/JVI.01710-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szretter KJ, Balish AL, Katz JM. 2006. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol Chapter15:Unit5G.1. doi: 10.1002/0471729256.mc15g01s3. [DOI] [PubMed] [Google Scholar]
- 37.Meecham PJ. 2020. Biosafety in microbiological and biomedical laboratories, 6th ed. CDC, Atlanta, GA. [Google Scholar]
- 38.Zhou B, Wentworth DE. 2012. Influenza A virus molecular virology techniques. Methods Mol Biol 865:175–192. doi: 10.1007/978-1-61779-621-0_11. [DOI] [PubMed] [Google Scholar]
- 39.Shepard SS, Meno S, Bahl J, Wilson MM, Barnes J, Neuhaus E. 2016. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics 17:708. doi: 10.1186/s12864-016-3030-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nelson CW, Moncla LH, Hughes AL. 2015. SNPGenie: estimating evolutionary parameters to detect natural selection using pooled next-generation sequencing data. Bioinformatics 31:3709–3711. doi: 10.1093/bioinformatics/btv449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knyazev S, Tsyvina V, Shankar A, Melnyk A, Artyomenko A, Malygina T, Porozov YB, Campbell EM, Switzer WM, Skums P, Mangul S, Zelikovsky A. 2021. Accurate assembly of minority viral haplotypes from next-generation sequencing through efficient noise reduction. Nucleic Acids Res 49:e102. doi: 10.1093/nar/gkab576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oksanen J, Simpson G, Blanchet F, Kindt R, Legendre P, Minchin P, O'Hara R, Solymos P, Stevens M, Szoecs E, Wagner H, Barbour M, Bedward M, Bolker B, Borcard D, Carvalho G, Chirico M, De Caceres M, Durand S, Evangelista H, FitzJohn R, Friendly M, Furneaux B, Hannigan G, Hill M, Lahti L, McGlinn D, Ouellette M, Ribeiro Cunha E, Smith T, Stier A, Ter Braak C, Weedon J. 2022. vegan: Community Ecology Package, v2.6–4. https://CRAN.R-project.org/package=vegan.
- 43.Guerrero-Murillo M, Font J. 2023. QSutils: quasispecies diversity, R package v1.16.0. https://bioconductor.org/packages/release/bioc/html/QSutilis.html.
- 44.Wickham H, Averick M, Bryan J, Chang W, McGowan L, Francois R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen T, Miller E, Bache S, Muller K, Ooms J, Robinson D, Seidel D, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H. 2019. Welcome to the tidyverse. J Open Source Software 4:1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 45.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 46.Wickham H, Seidel D. 2022. scales: scale functions for visualization, v1.2.1. https://CRAN.R-project.org/package=scales.
- 47.Pedersen T. 2022. patchwork: the composer of plots, v1.1.2. https://CRAN.R-project.org/package=patchwork.
- 48.Team RC. 2020. R: a language and environment for statistical computing. R Project for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 49.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 50.Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HH, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol 79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng H, Goldsmith C, Thawatsupha P, Chittaganpitch M, Waicharoen S, Zaki S, Tumpey TM, Katz JM. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type I interferon response in polarized human bronchial epithelial cells. J Virol 81:12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.01166-23-s0001.pdf, PDF file, 0.5 MB (504.6KB, pdf)
Supplemental material. Download spectrum.01166-23-s0002.xlsx, XLSX file, 0.09 MB (93.7KB, xlsx)