Abstract
Integrase of human immunodeficiency virus type 1 (HIVIN) consists of 288 amino acids, and its minimum DNA-binding domain (MDBD) (amino acids [aa] 220 to 270) is required for the integration reaction. We produced and characterized four murine monoclonal antibodies (MAbs) to the MDBD of HIVIN (strain LAI). Immunoblot and enzyme-linked immunosorbent assays with truncated HIVINs showed that those MAbs recognized sequential epitopes within the MDBD (aa 228 to 236, 237 to 252, 253 to 261, and 262 to 270). Their binding to HIVIN inhibited terminal cleavage and strand transfer activities but not disintegration activity in vitro. This collection of MAbs is useful for studying the structure and function of the MDBD by complementing mutational analyses and other biochemical studies.
Integration of a DNA copy of the viral RNA genome into a chromosome of the host cell is an essential step in the retroviral life cycle (4, 21, 41). The viral enzyme integrase (IN) catalyzes the process in three steps (5, 19). First, two nucleotides are removed from the 3′ ends of the viral DNA (in a process known as terminal cleavage [TC]). Second, the recessed 3′ ends of the viral DNA are then joined to 5′ staggered sites in the target DNA in a concerted cleavage and ligation reaction (in a process known as strand transfer [ST]). Finally, integration is completed by repair of the short gaps flanking the viral DNA intermediate. The TC and ST reactions can be reproduced in vitro with purified IN and double-stranded oligonucleotide substrates that mimic the ends of viral DNA (6, 8, 23, 38). Furthermore, IN catalyzes a reversal of the ST reaction in vitro (disintegration) with a branched-DNA substrate (Y-mer) that mimics the product of the ST reaction (11).
Biochemical analysis of IN from human immunodeficiency virus type 1 (HIV-1) has revealed that the C-terminal region (amino acids [aa] 160 to 288) contains nonspecific DNA-binding activity (18, 31, 40, 42), which is mapped to aa 220 to 270 (the minimum DNA-binding domain [MDBD]) (30, 31). Analyses by nuclear magnetic resonance also revealed that the MDBD consists of a five-stranded β-barrel similar to that of Src homology region 3 domains forming a homodimer (14, 29). Mutational analysis showed that the MDBD is essential for TC and ST activities of HIV-1 IN (HIVIN) (7, 13, 15, 16, 26), whereas it is dispensable for disintegration activity (13, 30, 37, 39, 40). Mutations in this region abolish viral DNA synthesis (reverse transcription), implying that HIVIN interacts with reverse transcriptase (RT) (17, 27, 32). Moreover, substitution of the W235 residue within the MDBD does not affect in vitro TC and ST activities, whereas the virus mutants carrying those substitution mutations cannot replicate (9, 10, 27, 28). But the function of the MDBD is not well characterized by monoclonal antibodies (MAbs), partly because few MAbs to the MDBD have been cloned (3, 33). This study presents a collection of MAbs reactive against the MDBD and demonstrates the effects of MAb binding on various in vitro activities, such as TC and ST activities, and on the capability of HIV-1 to interact with RT.
Production of MAbs against IN.
Female BALB/c mice were immunized primarily with HIVIN fused to Escherichia coli maltose-binding protein (MBP) and thereafter with HIVIN, with the N-terminal 20 aa residues containing a hexahistidine tag. MBP-HIVIN and hexahistidine-HIVIN were expressed and purified as described in references 7, 34, and 36, with equipment from New England Biolabs, (NEB), Beverly, Mass., and Novagen, Madison, Wis. Spleen cells of the mice were fused with P3-X63-Ag8.653 mouse myeloma cells (22, 24). Screening culture supernatants by enzyme-linked immunosorbent assay (ELISA) with hexahistidine-HIVIN identified about 50 hybridomas. Twelve hybridomas were successfully subcloned by limiting dilution and then injected into the peritoneal cavity of pristane-primed female BALB/c mice to obtain ascites fluid containing MAbs. Nine hybridomas produced enough ascites fluid for further analyses. Each MAb was purified with a HiTrap protein A column (Amersham Pharmacia Biotech Ltd., Little Chalfont, United Kingdom) followed by subsequent dialysis against 20 mM HEPES–Na (pH 7.5). Immunoblot analysis with MBP-HIVIN and six-His-tagged HIVIN (data not shown) showed that six clones (7-19, 8-6, 2-19, 8-22, 4-20, and 6-19) were specific to HIVIN (Fig. 1), whereas the others were specific to the hexahistidine tag.
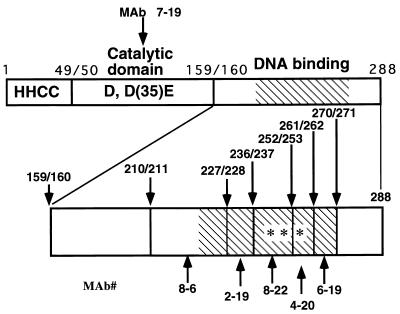
FIG. 1.
Schematic representation of epitopes recognized by the MAbs. The upper part of the figure shows a linear map of HIVIN. The N terminus (aa 1 to 49) with the HHCC motif, the central catalytic core (aa 50 to 159) with the DD(35)E motif, and the C terminus (aa 160 to 288) with DNA-binding activity domains are shown. A hatched box shows the MDBD (aa 220 to 270). The epitope regions recognized by the individual MAbs are shown below the IN map with the clone numbers. The lower part of the figure shows a linear map of HIVIN spanning from aa 160 to 288. Five MAbs recognized epitopes within this region. The numbers above the map indicate amino acid positions. Individual MAbs are shown by clone numbers below the IN map, with arrows indicating epitope regions. The three asterisks in the map indicate predicted β-strands, which may form an interface of homodimerization in a structure of triple-stranded antiparallel β-sheets (14, 29).
These MAbs displayed two isotypes (Table 1): immunoglobulin G2b (IgG2b) (clones 7-19, 2-19, and 6-19) and IgG1 (clones 8-6, 8-22, and 4-20). Semiquantitative ELISA (3) (Table 1) demonstrated that the titers of MBP-HIVIN varied about 150-fold: the minimal antibody concentration required for detection of IN with MAb 7-19 was the highest, whereas that with MAb 6-19 was the lowest.
TABLE 1.
MAbs against HIVIN produced by six independent hybridoma clonesa
| MAb | Isotype | Titer of MAb on MBP-HIV-1 IN (ng/ml) |
|---|---|---|
| 7-16 | IgG2b | 3 |
| 8-6 | IgG1 | 2 |
| 2-19 | IgG2b | 20 |
| 8-22 | IgG1 | 100 |
| 4-20 | IgG1 | 100 |
| 6-19 | IgG2b | 300 |
Titers are expressed as the minimum concentration of each MAb in nanograms per milliliter that gives an optical density at 492 nm fivefold greater than the background value (without primary antibody) (3). Each value was rounded to the nearest whole number.
Epitope mapping.
The epitopes for the purified MAbs were determined by reactivity to HIV/Rous sarcoma virus (RSV) chimera INs and HIVIN deletion mutants by immunoblot analysis and ELISA (summarized in Fig. 1; also see Table 2 and Fig. 2 and 3). RSV (strain CS8) IN fused to MBP was described previously (25). Similarly, mutants of HIVIN with deletions from aa 271 to 288 (HIVIN ending at aa 270 [HIVIN270]), HIVIN261, HIVIN252, HIVIN236, HIVIN227, HIVIN210, and HIVIN185 were expressed and purified as MBP fusion proteins. HIV/RSV chimera INs {HIVIN aa 1 to 236 with RSV aa 235 to 286 [H(1-236)R(235-286)], H(1-159)R(160-286), and R(1-36)H(40-288)} were also obtained as MBP fusion proteins.
TABLE 2.
ELISA with various deletion and chimeric mutants of HIVIN
| MBP-IN | Activity with anti-HIVIN MAb
|
|||||
|---|---|---|---|---|---|---|
| 7-19 | 8-6 | 2-19 | 8-22 | 4-20 | 6-19 | |
| HIVIN288a | + | + | + | + | + | + |
| HIVIN270 | NA | NA | + | + | + | + |
| HIVIN261 | NA | NA | + | + | + | − |
| HIVIN252 | NA | NA | + | + | − | NA |
| HIVIN236 | NA | + | + | − | − | NA |
| HIVIN227 | NA | + | − | NA | NA | NA |
| HIVIN210 | NA | − | − | NA | NA | NA |
| HIVIN185 | + | − | − | − | − | − |
| H(1-236)R(235-286) | + | + | + | − | − | − |
| H(1-159)R(160-286) | + | − | − | − | − | − |
| R(1-36)H(40-288) | + | + | + | + | + | + |
| RSV IN | − | − | − | − | − | − |
| HIVINNL4-3 | NA | NA | + | + | + | + |
| HIVINBH10 | NA | NA | + | + | + | + |
| W235A | NA | NA | − | + | NA | NA |
| W235E | NA | NA | − | + | NA | NA |
| W235AKGA | NA | NA | − | + | NA | NA |
| W235EKGE | NA | NA | − | + | NA | NA |
HIVIN288 is the wild-type IN whose binding activity was normalized to 100%; activities are reported as follows: +, positive (10 to 100%); −, negative (below 10%); NA, not assayed.
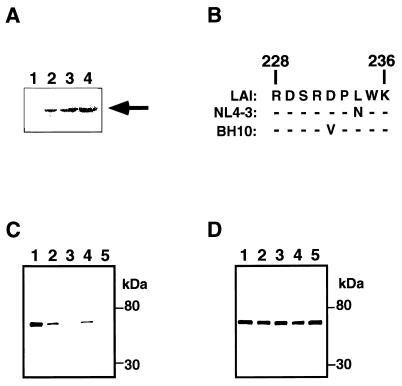
FIG. 2.
Effect of amino acid change on affinity of MAb 2-19. (A) Immunoblot analysis of various INs with MAb 2-19. Recombinant INs expressed in E. coli were separated in an SDS–10% polyacrylamide gel and blotted onto a nitrocellulose membrane. The proteins were probed with MAb 2-19. Lane 1, MBP-LacZα; lane 2, MBP-IN (LAI); lane 3, MBP-IN (NL4-3); lane 4, MBP-IN (BH10). The arrow shows the position of MBP-HIVIN. (B) Amino acid sequences from aa 228 to 236 of HIVIN strains LAI, NL4-3, and BH10 are shown. Dashes indicate that the amino acid residues are the same as the corresponding residues of the IN of the LAI strain. There are no amino acid changes in the region from aa 237 to 270 among those strains. A panel of MBP-IN proteins were separated by SDS-PAGE and blotted onto a nitrocellulose membrane. The blotted proteins were analyzed with MAb 2-19 (C) or MAb 8-22 (D). Lanes 1, MBP-IN (wild-type LAI); lanes 2, MBP-IN (W235A); lanes 3, MBP-IN (W235AKGA); lanes 4, MBP-IN (W235E); lanes 5, MBP-IN (W235EKGE). The positions of size markers are shown to the right of the panels.
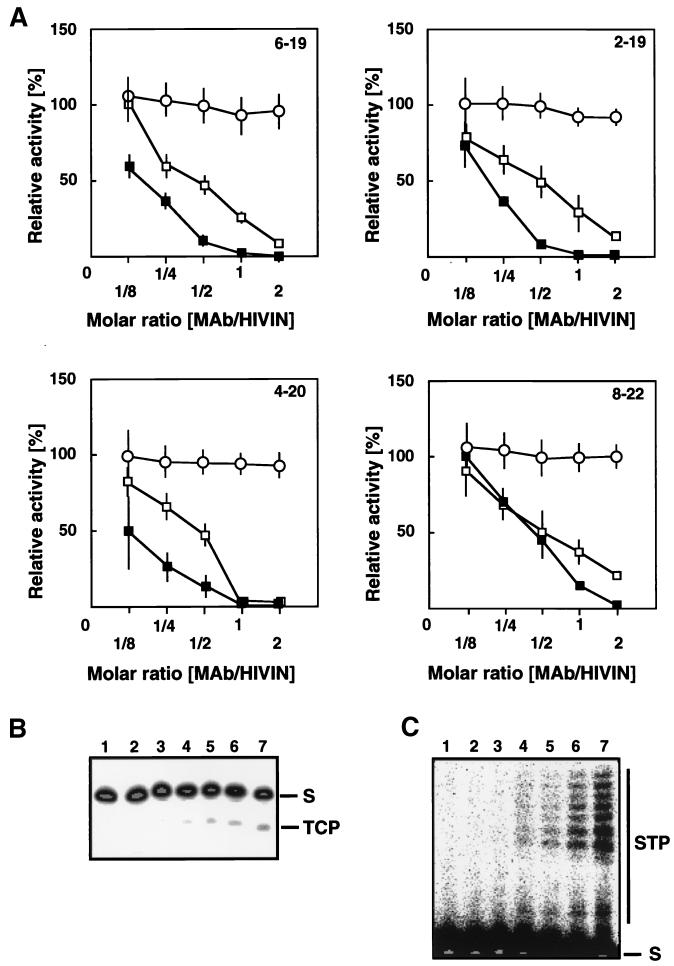
FIG. 3.
Effect of MAb binding on in vitro activities of HIVIN. (A) TC (open square), ST (solid square), and disintegration (open circle) activities were assayed with the blunt-ended, precleaved, and Y-mer oligonucleotide substrates, respectively. Each MAb tested is indicated in each panel. The effect of MAb binding on HIVIN activities was measured relative to the result obtained in a reaction with MAb reactive to the hexahistidine tag (100% activity). Results are based on the average of at least three independent assays (error bar, standard error). (B) Typical gel image obtained in TC assay. IN was incubated on ice prior to addition of radiolabeled oligonucleotide substrates with MAb 2-19 at MAb/IN molar ratios of 2 (lane 2), 1 (lane 3), 0.5 (lane 4), 0.25 (lane 5), and 0.125 (lane 6). Lane 1, without IN; lane 7, without MAb 2-19 but with anti-hexahistidine tag antibody as an unrelated antibody. S, substrates; TCP, TC products. (C) Typical gel image obtained in ST assay. The lane arrangements are the same as in panel B. S, substrates; STP, ST products.
The results of ELISA (Table 2) and immunoblot analysis (Fig. 2) showed that MAbs 7-16 and 8-6 recognized an epitope within the central catalytic domain of HIVIN and the region from aa 211 to 227, respectively. Furthermore, we inferred that the epitopes for MAbs 2-19, 8-22, 4-20, and 6-19 are likely to be contained in a simple linear sequence in tandem within the MDBD (Fig. 1 and Table 2). None of these MAbs recognized RSV IN (Table 2) or HIV-2 IN (data not shown). The epitopes for MAbs 2-19 (from aa 228 to 236) and 6-19 (from aa 261 to 270) are likely to protrude outwards, whereas those for MAbs 8-22 (from aa 237 to 252) and 4-20 (from aa 253 to 261) form a homodimer interface (14, 29).
Effects of amino acid changes on capability of MAbs to bind mutant HIVINs.
To test whether the MAbs recognize changes in the amino acid sequence of HIVIN we prepared several IN mutants. The unique BsmFI-BseRI region of the IN open reading frame of pLAI was replaced with synthetic double-stranded oligonucleotides; the wild-type sequence (5′-CACTTTGGAAAGGAC-3′) was changed to 5′-CACTTGCCAAAGGAC-3′, 5′-CACTTGAAAAAGGAC-3′, 5′-CACTTGCCAAAGGAGCCAAAGGAC-3′, or 5′-CACTTGAAAAAGGAGAAAAAGGAC-3′ to generate W235A, W235E, W235AKGA, or W235EKGE, respectively. All the mutant INs as well as the wild-type INs of NL4-3 (1) and BH10 (35) were produced as MBP fusion proteins as was HIV INLAI and subjected to ELISA and immunoblot analysis. First, we found that MAb 2-19 as well as the other MAbs to MDBD (8-22, 4-20, and 6-19) interacted to a similar extent not only with HIVINLAI but also with HIVINNL4-3 and HIVINBH10, both in ELISA (Table 2) and in an immunoblot analysis (Fig. 2A). Apparently, the amino acid changes within the region from aa 228 to 236 did not affect the binding capability of MAb 2-19 much (Fig. 2B). Secondly, we observed that MAb 2-19 was capable of interacting with W235A and W235E in an immunoblot analysis (Fig. 2C) but not in an ELISA (Table 2). In contrast, MAb 2-19 did not interact with W235AKGA or W235KGE in either assay (Fig. 2C and Table 2). The data imply that W235A and W235E show a subtle, yet distinct structural change and agree with the fact that they retain in vitro integration activity but lack in vivo infectivity (9, 27). The data suggest that the structural change in W235 IN is probably in the epitope for the MAb.
Effects of MAb binding on in vitro activities of IN.
To test whether the anti-MDBD MAbs interfere with the in vitro activities of HIVIN, we assayed TC, ST, and disintegration activities (12, 15) in the presence of each MAb. Unlike ELISA or immunization, these assays utilized MBP-free HIVIN prepared by cleavage of MBP-HIVIN with factor Xa (NEB) to minimize the unexpected effects, if any, of the N-terminal tag. Briefly, each purified MAb was preincubated with 7.5 pmol of purified HIVIN at various MAb/IN molar ratios on ice for 60 min in 14.5 μl of solution containing 20 mM sodium 3-N-morpholino propanesulfonate (MOPS-Na, pH 7.0), 10 mM MnCl2, 45 mM NaCl, 0.1% IGEPAL-CA630 (Sigma), and 1 mM dithiothreitol (DTT). Reactions were initiated by adding 32P-labeled substrates (0.2 pmol; specific activity, ∼106 cpm/pmol) in 0.5 μl of solution containing 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 100 mM NaCl followed by incubation at 37°C for 20 min. The integration products were separated by denaturing polyacrylamide gel electrophoresis (PAGE) and analyzed by autoradiography with an image analyzer (Fuji Photo Film Co., Ltd., Tokyo, Japan).
The effects of MAb binding on the TC, ST, and disintegration activities of IN are shown in Fig. 3. Both TC and ST activities were inhibited by all the four MAbs reactive to the MDBD, whereas the disintegration activity was much less affected. This is compatible with the results of genetic analyses of HIVIN (31, 40) and HIV-2 IN (31). Moreover, this agrees with the reports that MAb (33) and Fab (2) reactive to the region from aa 262 to 273 of HIVIN significantly inhibited both TC and ST activities and showed little inhibitory effect on disintegration activity. We concluded that the MDBD is essential to TC and ST activities but not to disintegration activity.
Effects of MAb binding on interaction between IN and RT.
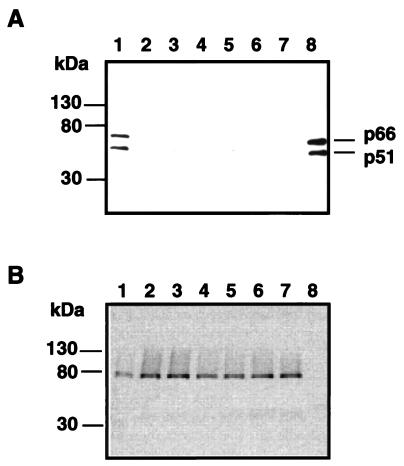
Hoping to demonstrate the usefulness of the MAbs in studying IN function, we performed a pull-down experiment with MAb 2-19 and found that it inhibited RT-IN interaction. Briefly, MBP-HIVIN (5 μg; wild type or mutant) immobilized on amylose resin (5 μl; NEB) in buffer B (20 mM HEPES-Na [pH 7.2], 0.1 M NaCl, 5 mM DTT, 5 mM MgCl2, 1 mM EDTA, 1% bovine serum albumin) was incubated with endonuclease (Benzonase; 50 U/ml; Sigma) at 37°C for 10 min followed by an extensive wash with buffer B. The immobilized MBP-HIVIN was incubated with MAb 2-19 or MAb 8-16 on ice for an hour and thereafter with 50 ng of RT (F. Hoffman-La Roche Ltd., Basel, Switzerland) on ice for an additional hour. The resin was washed extensively with a solution containing 50 mM HEPES-Na (pH 7.2), 50 mM NaCl, 5 mM DTT, 1 mM EDTA, 1% bovine serum albumin and 0.25% IGEPAL-CA630. The bound proteins were separated by sodium dodecyl sulfate (SDS)-PAGE and analyzed by Western blot analysis with an anti-RT (p66/p51) antibody (mouse MAb; Advanced Biotechnologies). MAb 2-19 as well as 8-22 inhibited RT-IN interaction (Fig. 4A, lanes 1, 6, and 7). Furthermore, HIVINs with an amino acid substitution or insertion at W235 lost the ability to find RT (Fig. 4A, lanes 2 to 5). The results suggest that the region containing the epitope of MAb 2-19 is responsible for RT-IN interaction and that W235 mutants lack some structure required for that interaction.
FIG. 4.
Interaction of IN with RT. MBP-HIVINs of W235 mutants as well as wild-type LAI were immobilized on amylose resin in the presence or absence of MAb 2-19 or 8-22 and were incubated with RT. The bound proteins were separated by SDS-PAGE and blotted onto a nitrocellulose membrane. The blotted proteins were probed with an anti-RT (A) or an anti-MBP (B) antibody. Incubated with 0.5 μg of RT were HIVINs of the wild-type LAI (lanes 1, 6, and 7), W235A (lanes 2), W235E (lanes 3), W235 AKGA (lanes 4), and W235EKGE (lanes 5). For assaying the effect of MAb binding, MBP-HIVIN was incubated for an hour on ice with MAb 2-19 (lanes 6) or MAb 8-22 (lanes 7) prior to incubation with RT. Lanes 8, RT alone as a positive control. (A) Bars show the positions of p66 and p51 subunits of RT.
Our current working hypothesis is that MAb 2-19 recognizes a conformational motif in the MDBD which is conserved across different primary amino acid sequences, because MAb 2-19 interacted with INs of HIV-1LAI, HIV-1NL4-3, and HIV-1BH10 to a comparable extent (Fig. 2A) although their amino acid sequences (aa 228 to 236) are different (Fig. 2B). This seems to be consistent with a report that mouse MAbs reactive to epitopes within the carboxyl region of HIVIN cross-reacted with HIV-2 IN (33) and with an observation on an anti-RT intracellular antibody (20). We speculate that W235A and W235E mutants contain such a subtle and local structural change in the above-mentioned conformational motif that MAb 2-19 detected those W235 mutants in the immunoblot analysis (Fig. 2C). This is compatible with the fact that W235A and W235E retain in vitro integration activity (9, 10, 27, 28). But we also infer that the local conformational change around W235 is so definite that IN with a W235E or W235A substitution could not be detected by MAb 2-19 in ELISA (Table 2) nor could these mutants interact with RT (Fig. 4). This inference agrees with the report that pooled sera from HIV-1-positive patients cannot recognize mutants carrying a substitution at residue W235 of the HIVIN (10). This probable definite, yet subtle structural change may account for the incompetence of W235A and W235E mutants in replication (9, 10, 27, 28).
In summary, we have described a collection of MAbs with sequential epitopes on the MDBD of HIVIN demonstrating that the MDBD is essential for TC and ST activities but dispensable for disintegration and have shown that the MDBD seems to contain a structural motif common to various strains of HIV-1. These MAbs should prove useful for further studies of the structure and function of HIVIN and the molecular design of inhibitors to HIVIN.
Acknowledgments
We thank Keith Peden for pLAI. We thank Tadahito Kanda, Kunito Yoshiike, and Hiroshi Yoshikura for critical reading of the manuscript.
This work was supported by a grant for AIDS Research from the Ministry of Health and Welfare of Japan and the Program to Develop Countermeasures for Health Management and Decreasing Immunity in Relation to Public Hygiene from the Japan Health Sciences Foundation.
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barsov E V, Huber W E, Marcotrigiano J, Clark P K, Clark A D, Arnold E, Hughes S H. Inhibition of human immunodeficiency virus type 1 integrase by the Fab fragment of a specific monoclonal antibody suggests that different multimerization states are required for different enzymatic functions. J Virol. 1996;70:4484–4494. doi: 10.1128/jvi.70.7.4484-4494.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bizub-Bender D, Kulkosky J, Skalka A M. Monoclonal antibodies against HIV type 1 integrase: clues to molecular structure. AIDS Res Hum Retroviruses. 1994;10:1105–1115. doi: 10.1089/aid.1994.10.1105. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O. Integration of retroviral DNA. In: Swanstrom R, Vogt P K, editors. Retroviruses. Strategies of replication. New York, N.Y: Springer-Verlag; 1990. pp. 19–48. [Google Scholar]
- 5.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bushman F D, Craigie R. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc Natl Acad Sci USA. 1991;88:1339–1343. doi: 10.1073/pnas.88.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bushman F D, Fujiwara T, Craigie R. Retroviral DNA integration directed by HIV integration protein in vitro. Science. 1990;249:1555–1558. doi: 10.1126/science.2171144. [DOI] [PubMed] [Google Scholar]
- 9.Cannon P M, Byles E D, Kingsman S M, Kingsman A J. Conserved sequences in the carboxyl terminus of integrase that are essential for human immunodeficiency virus type 1 replication. J Virol. 1996;70:651–657. doi: 10.1128/jvi.70.1.651-657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon P M, Wilson W, Byles E, Kingsman S M, Kingsman A J. Human immunodeficiency virus type 1 integrase: effect on viral replication of mutations at highly conserved residues. J Virol. 1994;68:4768–4775. doi: 10.1128/jvi.68.8.4768-4775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chow S A, Vincent K A, Ellison V, Brown P O. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science. 1992;255:723–726. doi: 10.1126/science.1738845. [DOI] [PubMed] [Google Scholar]
- 12.Craigie R, Hickman A B, Engelman A. Integrase. In: Karn J, editor. HIV: a practical approach. Vol. 2. Oxford, United Kingdom: IRL Press; 1995. pp. 53–71. [Google Scholar]
- 13.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 14.Eijkelenboom A P, Lutzke R A, Boelens R, Plasterk R H, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 15.Engelman A, Bushman F D, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelman A, Englund G, Orenstein J M, Martin M A, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelman A, Hickman A B, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 20.Gargano N, Biocca S, Bradbury A, Cattaneo A. Human recombinant antibody fragments neutralizing human immunodeficiency virus type 1 reverse transcriptase provide an experimental basis for the structural classification of the DNA polymerase family. J Virol. 1996;70:7706–7712. doi: 10.1128/jvi.70.11.7706-7712.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goff S P. Genetics of retroviral integration. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 22.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 23.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 24.Kearney J F, Radbruch A, Liesegang B, Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- 25.Kitamura Y, Lee Y M, Coffin J M. Nonrandom integration of retroviral DNA in vitro: effect of CpG methylation. Proc Natl Acad Sci USA. 1992;89:5532–5536. doi: 10.1073/pnas.89.12.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leavitt A D, Robles G, Alesandro N, Varmus H E. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J Virol. 1996;70:721–728. doi: 10.1128/jvi.70.2.721-728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leavitt A D, Shiue L, Varmus H E. Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J Biol Chem. 1993;268:2113–2119. [PubMed] [Google Scholar]
- 29.Lodi P J, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 30.Lutzke R A, Plasterk R H. Structure-based mutational analysis of the C-terminal DNA-binding domain of human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J Virol. 1998;72:4841–4848. doi: 10.1128/jvi.72.6.4841-4848.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutzke R A, Vink C, Plasterk R H. Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res. 1994;22:4125–4131. doi: 10.1093/nar/22.20.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masuda T, Planelles V, Krogstad P, Chen I S Y. Genetic analysis of human immunodeficiency virus type 1 integrase and the U3 att site: unusual phenotype of mutants in the zinc finger-like domain. J Virol. 1995;69:6687–6696. doi: 10.1128/jvi.69.11.6687-6696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nilsen B M, Haugan I R, Berg K, Olsen L, Brown P O, Helland D E. Monoclonal antibodies against human immunodeficiency virus type 1 integrase: epitope mapping and differential effects on integrase activities in vitro. J Virol. 1996;70:1580–1587. doi: 10.1128/jvi.70.3.1580-1587.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okui N, Kobayashi N, Kitamura Y. Production of uninfectious human immunodeficiency virus type 1 containing viral protein R fused to a single-chain antibody against viral integrase. J Virol. 1998;72:6960–6964. doi: 10.1128/jvi.72.8.6960-6964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 36.Riggs P. Expression and purification of maltose-binding protein fusions. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 16.6.1–16.6.14. [DOI] [PubMed] [Google Scholar]
- 37.Schauer M, Billich A. The N-terminal region of HIV-1 integrase is required for integration activity, but not for DNA-binding. Biochem Biophys Res Commun. 1992;185:874–880. doi: 10.1016/0006-291x(92)91708-x. [DOI] [PubMed] [Google Scholar]
- 38.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vincent K A, Ellison V, Chow S A, Brown P O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993;67:425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vink C, Oude Groeneger A M, Plasterk R H. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitcomb J M, Hughes S H. Retroviral reverse transcription and integration: progress and problems. Annu Rev Cell Biol. 1992;8:275–306. doi: 10.1146/annurev.cb.08.110192.001423. [DOI] [PubMed] [Google Scholar]
- 42.Woerner A M, Marcus-Sekura C J. Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res. 1993;21:3507–3511. doi: 10.1093/nar/21.15.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]