Abstract
In plant cells, a large pool of iron (Fe) is contained in the nucleolus, as well as in chloroplasts and mitochondria. A central determinant for intracellular distribution of Fe is nicotianamine (NA) generated by NICOTIANAMINE SYNTHASE (NAS). Here, we used Arabidopsis thaliana plants with disrupted NAS genes to study the accumulation of nucleolar iron and understand its role in nucleolar functions and more specifically in rRNA gene expression. We found that nas124 triple mutant plants, which contained lower quantities of the iron ligand NA, also contained less iron in the nucleolus. This was concurrent with the expression of normally silenced rRNA genes from nucleolar organizer regions 2 (NOR2). Notably, in nas234 triple mutant plants, which also contained lower quantities of NA, nucleolar iron and rDNA expression were not affected. In contrast, in both nas124 and nas234, specific RNA modifications were differentially regulated in a genotype dependent manner. Taken together, our results highlight the impact of specific NAS activities in RNA gene expression. We discuss the interplay between NA and nucleolar iron with rDNA functional organization and RNA methylation.
Keywords: Iron, methylation, nicotianamine, nucleolus, rDNA, redox
Low iron in the nucleolus and RNA modification changes suggest a central role of nicotianamine synthesis in rRNA gene expression.
Introduction
The nucleolus is a multifunctional structure linked to ribosome biogenesis, assembly of ribonucleoprotein complexes, nuclear chromatin organization, sequestering of proteins, and stress response (Boulon et al., 2010; Hernandez-Verdun et al., 2010; Stepinski, 2014; Tsai and Pederson, 2014; Pederson and Powell, 2015; Tsekrekou et al., 2017).
The plant nucleolus has an additional, and less investigated, role as a pool of iron and for the intracellular distribution of that iron (Roschzttardtz et al., 2011). Iron is one of the most important micronutrients in plants, implicated in almost every cellular process. Iron excess or deprivation lead to severe growth phenotypes in plants, decrease in biomass production, and chlorosis of leaves (Briat et al., 2015). Accumulation of iron in the nucleolus is intriguing also from the perspective that several neurodegenerative diseases are related to nucleolar iron in human cells (Quintana et al., 2006; Heeney and Finberg, 2014; Kumar et al., 2016). In Alzheimer’s disease, iron damages DNA (Henle et al., 1996, 1999) and binds directly to ribosomal RNA (rRNA), as well as ribosomes, leading to their oxidation and impacting translation efficiency (Honda et al., 2005). Despite these deleterious effects of nucleolar iron, recent studies point out a regulatory role of iron on ribosome assembly and function under specific cellular conditions. The ribosome is particularly dependent on Mg2+ to fold and maintain stability due to the negative charge of the rRNA backbone. However, ribosomal Mg2+ can be substituted with other divalent cations, including Fe2+ and the ribosome can competently mediate translation in this state (Bray et al., 2018; Smethurst et al., 2020; Smethurst and Shcherbik, 2021). Notably, while Fe2+ interactions with the ribosome were relevant in ancient ribosomes (containing Mg+2 before the evolution of photosynthesis and the increase in molecular oxygen), little is known how iron impact nucleolus organization and ribosome synthesis in present-day oxidative environmental conditions.
Nicotianamine (NA) is a key ligand for essential metals, including iron (Curie and Briat, 2003). NA is an amino acid derivative, synthesized from three S-adenosyl-methionine (SAM) moieties by NICOTIANAMINE SYNTHASE (NAS) [Fig. 1A and references in (Klatte et al., 2009)]. The genome of A. thaliana contains four NAS genes (Bauer et al., 2004). In the quadruple nas mutant (nas4x-2), NA is not detectable in rosette leaves; nas4x-2 also shows a chloronerva-like phenotype and is sterile (Klatte et al., 2009).
Fig. 1.
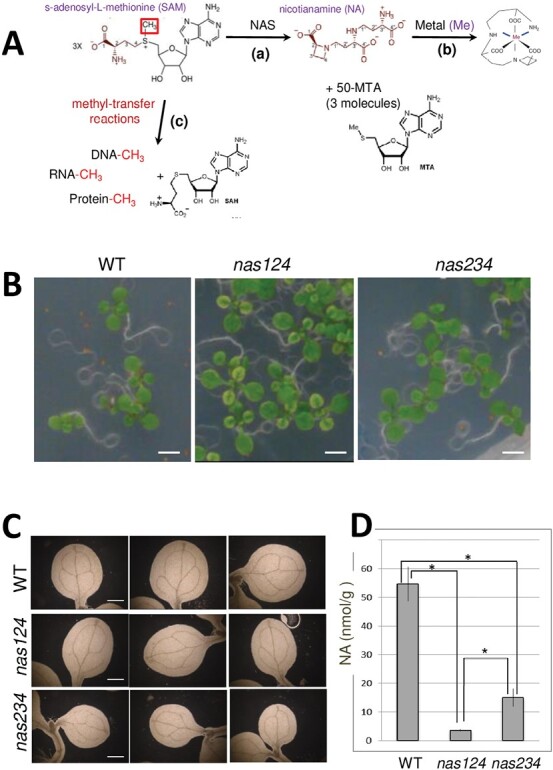
Nicotianamine synthesis and nas124 and nas234 plants. (A) The synthesis of NA by NAS from three molecules of SAM includes two carboxypropyl group transfers and one azetidine ring formation, with three molecules of 50-methylthioadenisine (50-MTA) released. NA chelates metal ions including iron. SAM is also a methyl donor in methylation reactions including methylation of DNA, RNA, and proteins. (B) WT, nas124, and nas234 plants grown for 12 d in vitro. Bar=2.5 mm. (C) Cleared cotyledon leaves of WT and nas124 and nas234 plants, viewed under dark-field illumination. Scale bar=0.4 mm. (D) Nicotianamine (NA) content in 15-day-old in vitro grown WT, nas124, and nas234 plants. Data are means ±SE of n=3 independent pools of three plants each, * indicates statistical difference with a Wilcoxon test, P<0.1.
Here, we demonstrate that in specific Arabidopsis nas mutant plants the nucleolar localization of iron is inhibited. Concomitantly, in these nas mutants, rDNA genes from normally silenced NOR2 are expressed and RNA and methylation profiles changed. We discuss how inhibition of NAS activity might affect rDNA gene expression and/or methylation of RNAs.
Materials and methods
Plant material and growth conditions
All lines were derived from A. thaliana Columbia-0 ecotype. Seeds corresponding to nas1-2 (SALK_082174), nas1-1 (GK223A09), nas2-2 (SALK_066962), nas3-2 (SALK_106467), and nas4-1 (SALK_135507) mutant plants were obtained from the Nottingham Arabidopsis Stock Centre (https://arabidopsis.info/). Crosses between single homozygous T-DNA insertion lines were performed to generate triple homozygous mutants, and were identified by PCR and RT–PCR (Supplementary Fig. S1). The mutant nuc1-2 was previously described (Pontvianne et al., 2010; Durut et al., 2014).
Plants were grown either in soil for 21 d, or in vitro on 1× Murashige and Skoog medium (Duchefa Biochemie M0231), including Gamborg B5 vitamins, supplemented with 0.05% (w/v) 2-(N-morpholino)ethanesulfonic acid (MES) and 1% (w/v) plant agar (pH was adjusted to 5.7 with KOH) for 15 d. For the soil-grown plants, only the aerial parts were collected for analysis, whereas under the in vitro conditions the whole seedlings were analysed. Plants were grown under an 8 h dark/16 h light photoperiod at 21°C.
Nicotianamine (NA) analysis
Approximately 0.1 g of fresh 15-day-old in vitro plants was ground to a fine powder in liquid nitrogen and analysed for NA amount by reversed-phase high-performance liquid chromatography (RP-HPLC) with pre-column derivatization with o-phthalaldialdehyde, as previously described (Klatte et al., 2009).
Molecular cloning and plasmid constructs
For construction of the Fib2-roGFP plasmid, the GAR domain (amino acids 1–73) of the Arabidopsis Fibrillarin 2 (Fib2) gene was PCR amplified with Phusion polymerase (Finnzymes) using the primers Fib2_KpnI and Fib2_BamH1 to add KpnI and BamHI restriction sites (Supplementary Table S1). The PCR product was cloned blunt-end into the vector pGEM-T (Promega). The GAR domain was then subcloned into the expression vector pBinAR-GRX1-roGFP2 (Marty et al., 2009) with KpnI and BamHI. The construct was transformed into Agrobacterium strain C58C1 and used for Arabidopsis transformation.
Confocal laser-scanning microscopy and redox ratio analysis
Confocal microscopic observations were carried out using an Axio observer Z1 microscope with a LSM 700 scanning module and ZEN 2010 software (Zeiss). Excitation of roGFP2 was performed at 488 and 405 nm and a bandpass (BP) 490–555 nm emission filter was used to collect the roGFP2 signal. For background subtraction, the signal was recovered using a BP 420–480 nm emission filter during excitation at 405 nm. Image analyses and quantifications were performed as previously described (Gutscher et al., 2008), using the public domain image analysis program ImageJ 1.52i (https://imagej.nih.gov/ij/).
Cleared cotyledons and Perls staining coupled to diaminobenzidine (DAB) intensification
To observe vein patterning, cotyledons from 10-day-old WT, nas124, and nas234 in vitro seedlings were cleared in a 3:1 (v/v) ethanol:acetic acid solution, rinsed and conserved in 70% ethanol, and observed under dark-field illumination. For Perls/DAB staining, leaf fragments from 15-day-old in vitro seedlings were vacuum-infiltrated with a fixation solution containing 2% (w/v) paraformaldehyde, 1% (v/v) glutaraldehyde, 1% (w/v) caffeine in 100 mM phosphate buffer (pH 7). The fixed samples were treated and analysed for Fe histochemical staining according to Roschzttardtz et al. (2009). Vein pattern and Pearls/DAB observations were performed using LEICA MZ12 and DFC425 digital microscope camera systems.
Transmission electron microscopy (TEM)
Roots from 5-, 10-, and 15-day-old plants, in vitro grown, were first fixed with 3% (v/v) glutaraldehyde in 0.025 M cacodylate buffer (pH 7.3) at room temperature. After washing, the samples were post-fixed with 1% OsO4 in the same buffer. The samples were then dehydrated in a methanol series (30, 50, 70, and 100%) at room temperature. The samples were acetylated and methylated with a freshly prepared 5:1 (v/v) methanol/acetic anhydride mixture at 25°C. Samples were then washed in pure methanol and embedded in Epon 812 resin (Sigma). Ultrathin sectioning was performed on an ultramicrotome (Leica Ultracut), and counterstained with uranyl acetate and lead citrate before being observed using a 7500 Hitachi TEM.
Fluorescence in situ hybridization (FISH)
The FISH probe containing intergenic spacer (IGS) and 5ʹ external transcribed spacer (5ʹETS) rDNA sequences (-220/+250) was amplified with primers o112/o113 (Supplementary Table S1) and cloned into a pGEM-T® vector (Promega) and amplified by PCR using universal primers T7/M13R, and supplying biotin-16-dUTP (Roche) to the reaction. The analysis was performed using nuclei from leaves of 21-day-old plants grown in soil. The biotin-labelled probe was detected using digoxigenin (1:200; Roche) followed by a sheep anti-digoxigenin antibody conjugated with the fluorochrome Alexa 480 (1:200; Invitrogen). Slides were prepared using Vectashield (Vector Laboratories) mounting medium supplemented with 1 μg/mL 4ʹ,6-diaminido-2-phenylindole (DAPI) and then observed by confocal microscopy (Zeiss LSM700 scanning microscope).
DNA and RNA isolation
DNA was extracted from liquid nitrogen ground powder by using a DNeasy® Plant Mini Kit (Qiagen). Alternatively, DNA for bisulfite analysis was extracted with illustra™ Nucleon™ PhytoPure™ (GE Healthcare). For RNA, around 800 µl of frozen powder were supplemented with 5 ml of Tri Reagent® (Molecular Research Center, Inc.), then 1 ml of cold chloroform was added. After a 3 min incubation, the mix was centrifuged at 8000 g for 15 min at 4°C. Then 3 ml of cold isopropanol was added to the conserved aqueous phase and the resulting product was then centrifuged at 8000 g for 30 min at 4°C. Isopropanol was replaced by 75% ethanol and the mix was incubated overnight at –20°C. The pellet was dried and resuspended in 65°C diethylpyrocarbonate (DEPC)-treated water. DNA was removed from samples by using the TURBO DNA-freeTM kit (Invitrogen), according to the manufacturer’s instructions.
PCR, RT–PCR, and qPCR
RT–PCR reactions were performed using SuperSript IV Reverse Transcriptase (ThermoFisher Scientific). For cDNA production we simultaneously used random and oligo dT primers. The protocol was performed following the manufacturer’s instructions. The cDNAs obtained were used for later PCR experiments. PCR amplifications were carried out with GoTaq® DNA Polymerase and 5× GoTaq® Green Buffer (Promega). 3ʹETS products from RT–PCR and PCR reactions were loaded on an agarose gel (2% agarose) in 0.5× Tris-acetate-EDTA (TAE) supplemented with Gel Red® (Interchim, Roche). Quantitative PCR (qPCR) was performed using a LightCycler 480 system and TaykonTM qPCR for SYBR® Assays (Eurogentec) following the manufacturer’s instructions. All primer pairs used in RT–PCR, PCR, and qPCR are provided in Supplementary Table S1.
Bisulfite DNA treatment and analysis
DNA was converted (me5C to T) using the EpiTect Plus Bisulfite Conversion Kit (Qiagen). The target region was then amplified by PCR using the Ex Taq DNA Polymerase, Hot Start Version Kit (TaKaRa Bio Inc.) and oligo primers o112/o113 and o124/o125 (Supplementary Table S1) to amplify promoter/5ʹETS and 3ʹETS, respectively. 3ʹETS PCR products were run on an agarose gel, sliced from the gel, and purified using a GeneClean® Turbo Kit (MP Biomedicals). PCR products were cloned into a pGEM®-T easy vector (Promega), transformed into Escherichia coli DH5α, and sequenced using T7 and SP6 primers. The sequences obtained were mapped to a reference sequence and the alignment was sent to CyMATE to analyse methylation sites in the sequence. The results were analysed using the prop.test function in R software to verify significant differences with R software (Pontvianne et al., 2012).
RiboMethSeq analysis and detection of nucleosides using the multiple reaction monitoring (MRM) method in mass spectrometry
RiboMethSeq analysis was performed using the method described in Marchand et al. (2016) and recently used in Arabidopsis plants (Azevedo-Favory et al., 2021).
Enzymatic processing of RNA.
Total RNA (~400 ng) from 15-day-old in vitro grown WT, nas124, and nas234 plants was digested with 0.001 U of Nuclease P1 (Sigma, N8630) and 3 µl of 0.1 M ammonium acetate (pH 5.3) for 2 h at 42 °C. Then a dephosphorylation of nucleosides was performed with 0.001 U of alkaline phosphatase (Sigma, P4252) and 3 µl of 1 M ammonium acetate for 2 h at 37°C. Next, the mixture was diluted twice and was filtered with 0.22 µm filters (Millex®-GV, Millipore, SLGVR04NL). Five microlitres of each sample was injected in triplicate into a liquid chromatography coupled to tandem mass spectrometry (LC-MSMS) and then methylated/unmethylated ratios were estimated. Since the adenosine signal was saturated in the samples, we used uridine (U) to estimate ratios.
RNA-seq and bioinformatics analyses
Total RNA from either 15-day-old in vitro grown WT or nas124 mutant plants was prepared to generate three biological replicates per sample. Sequencing was performed by the Plateforme transcriptOmique de l'iPS2 (POPS) Facility (Institut de Génomique-CNS, Evry, France) from stranded ribozero RNA-seq libraries and using NextSeq500 (Illumina) and Hiseq2000 to generate 2 × 75-bp-long reads. Raw reads were trimmed using Trimmomatic v0.39 (Bolger et al., 2014). Trimmed reads corresponding to mitochondrial, chloroplastic, and rRNA sequences were filtered out using bowtie2 v2.3.5 (Langmead and Salzberg, 2012) in ‘sensitive-local’ mode. Read mapping against the TAIR10 genome with the Araport11 gtf file was performed using Hisat2 v2.1.0 (Kim et al., 2015). Read counting was performed using htseq-count v0.12.4 (Anders et al., 2015) in ‘union’ mode and normalized by total of mapped reads (reads per millions, rpm). Differential analysis was performed using the Bioconductor R v4.1.2 package DESeq2 (Love et al., 2014) with a false discovery rate of 0.05. P values were corrected for multiple tests by the Benjamini–Hochberg rule (adjusted P value). The up-regulated genes were defined with a fold change minimum to 2 and the down-regulated genes with a fold change less than 1. The heat map was obtained using the heatmap.2 function of the gplots R package (https://CRAN.R-project.org/package=gplots) using Pearson distance and an average method for the hierarchical clustering. To identify differentially expressed long non-coding RNAs (DE lncRNAs), Col-0, and nas124, Illumina reads were aligned to the Arabidopsis Transcriptome ReconstructoR pipeline (Ivanov et al., 2021) All WT and nas124 sequences were submitted to the Sequence Read Archive (SRA): PRJNA782822 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA782822?reviewer=ee044uijk32bst5p1v52bogou9). Gene Ontology (GO) analysis was performed using PANTHER (http://www.pantherdb.org/).
Enzyme activity measurements and detection of reactive oxygen species
To measure enzyme activity, plantlets (~200 mg) were harvested, immediately frozen in liquid nitrogen, and conserved at -80°C until assay. Plant material was ground in liquid nitrogen (CryoMill, Retsch), resuspended in 400 µl of cold buffer (phosphate buffer 50 mM pH 7,2, 1 mM EDTA, 2% of polyvinyl pyrrolidone, 1 tablet of complete protease inhibitor cocktail (Roche) for 10 ml) before high-speed centrifugation (21000 g, 4°C, 10 min). The supernatant was conserved on ice until enzyme analysis (Veljovic-Jovanovic et al., 2001). CAT (catalase) activity was measured according to Cakmak and Marschner (1992). The assay depends on the decrease in absorbance at 240 nm as H2O2 is degraded. The reaction mixture 1 ml containing 25 mM sodium phosphate buffer (pH 7.0), 10 mM H2O2 and 0.1 ml enzyme extract. The reaction was started by adding H2O2. APX (ascorbate peroxidase) activity was measured according to Nakano and Asada (1981). The assay depends on the decrease in absorbance at 290 nm as ascorbate is oxidized. The 1 ml reaction mixture contained 25 mM sodium phosphate buffer (pH 7.0), 0.5 mM ascorbate, 0.1 mM H2O2, 0.1 mM EDTA and 0.1 ml enzyme extract. The reaction was started by adding H2O2.
Detection of ROS was performed as previously described (Mhamdi et al., 2010). Staining was performed at room temperature on Col-0 and nas124 7-day-old plants. For the detection of H2O2, plants were vacuum infiltrated in the nitro blue tetrazolium (NBT) staining medium [10 mM phosphate buffer pH 7.5, 10 mM Na azide, 1 tablet of NBT (10 mg Sigma)]. For the detection of superoxide, plants were vacuum infiltrated in 5 mM DAB at pH 3.8. For both staining, plants were then incubated in the same medium until the coloration was observed and then chlorophyll was removed in 95% ethanol before pictures taking.
Primer extension and northern blot analysis
Total RNA extractions and 5' end-labelling of oligo probes (p23, p43, p5 and p6, tis, p, u3, p18S and p25S; see Supplementary Table S1) were performed as described previously (Pontvianne et al., 2010). Northern blots and primer extension gels were performed using respectively 3 μg and 15 μg of total RNA from 21-day-old plants grown in soil. Two northern blot membranes were pre-incubated in PerfectHyb Plus hybridization buffer (Sigma) for at least 3 h at 42 °C. Labelled probes p5, p23, p43, p18S and p25S were then added (1µl at 10µM) and incubated overnight at 42 °C. Membranes were washed at 50 °C during 15 min with 2X SSC 0.1% SDS, then with 0.5X SSC, 0.1% SDS, and finally with 0.1X SSC 0.1% SDS. Primer extension and dideoxy sequencing reactions were performed according to Pontvianne et al. (2010). Northern blots and primer extension reactions were analysed on a Personal Molecular Imager (PMI, BioRad) and quantified using Quantity One software.
Results
Plant phenotype and NA accumulation in nas124 and nas234 plants
The genome of A. thaliana contained four NAS genes that were differentially expressed. NAS1, NAS2, and NAS4 were essentially expressed in roots, whereas NAS3 was expressed at low levels in roots and leaves. Single T-DNA insertion lines did not show any visible phenotype (Bauer et al., 2004) and had WT-nicotianamine levels (Klatte et al., 2009). We first characterized the nas124 and nas234 triple mutant plants obtained from single nas1-2 (SALK_082174), nas2-2 (SALK_066962), nas3-2 (SALK_106467), and nas4-1 (SALK_135507) mutant plants (Supplementary Fig. S1A, B).
The nas124 and nas234 triple mutants grown in soil did not show growth and/or plant developmental defects compared to the WT plants, though the first leaves from in vitro grown nas124 displayed a chloronerva-like phenotype (Fig. 1B). Cleared cotyledons from nas124 and nas234 showed disturbed vein patterns (Fig. 1C). Furthermore, nas124 and nas234 contained 6.5% and 27%, respectively, of the NA concentration compared to the WT (Fig. 1D). It was expected that residual NA detected in nas124 came from low levels of NAS3 expressed in roots and leaves, whereas NA detected in nas234 came from highly expressed NAS1 in roots compared to leaves (Supplementary Fig. S1C).
Loss of nucleolar iron in nas124
NA is a central ligand for intracellular iron (Bauer et al., 2004; Klatte et al., 2009). Therefore, we determined the impact of reduced NA content on the accumulation of nucleolar iron in nas124 and nas234 plants. We performed Perls associated with diaminobenzidine (Perls/DAB) staining (Fig. 2; Supplementary Fig. S2A). The Perls/DAB staining detected iron in the nucleolus of WT and nas234 mesophyll cells (black staining, upper and lower panels). However, no iron was detected in nas124 nucleoli (middle panel; Fig. 2). Similarly, Perls/DAB staining of roots cells detected iron in WT and nas234 nucleoli but not in nas124 nucleoli (Supplementary Fig. S3). We also observed that iron formed aggregates in the xylem vessels of nas124 that might correspond to iron-ferritin complexes or Fe precipitates of unknown origin (Supplementary Fig. S2). In addition, increased ferritin protein levels were detected in nas124 (Supplementary Fig. S2B). Altogether these results show that simultaneous disruption of NAS1, NAS2, and NAS4 genes, and decreased NA amount provoked loss of nucleolar iron.
Fig. 2.
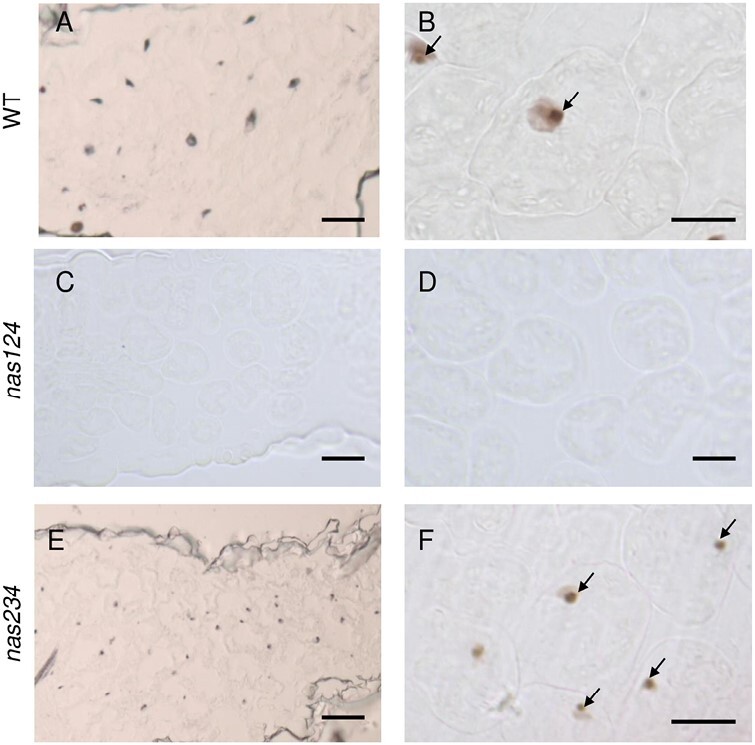
Perls/DAB staining to detect nucleolar iron. Nucleolar iron in nas124 and nas234 leaf sections showing mesophyll cells from 15-day-old WT (A and B), nas124 (C and D), and nas234 (E and F) plants grown in vitro were stained with Perls/DAB. Arrows indicate nucleolar iron in WT and nas234. Scale bars=40 µm for A, C, and E, and 20 µm for B, D, and F.
Absence of iron in nas124 nucleoli does not change nucleolar/nuclear redox state
As the ferrous ion (Fe2+) is highly reactive with H2O2 to produce hydroxide (OH-) and hydroxyl (OH.) radicals through Fenton reactions, we reasoned that the contrasted iron accumulation in the nucleolus between WT and nas124 plants might change the redox state of the nucleolus or the surrounding nucleoplasm. To monitor the nuclear redox state in WT and nas124 plants, we used the redox sensor roGFP2 (Fig. 3A) in which the roGFP2 is fused to the glutaredoxin GRX1 (Gutscher et al., 2008) and indicates the redox state by excitation at different wavelengths. This construct is expressed in the cytosol and the nuclear compartments but is excluded from the nucleolus (Fig. 3B). To also monitor the redox state in the nucleolus, we fused GRX1-roGFP2 with the GAR domain of the Fibrillarin2 (Fib2) protein which targets the protein to the nucleolus (Barneche et al., 2000) and documents the redox state in the nucleolus and in the nucleoplasm (Fig. 3B). The system was calibrated with H2O2 and DTT, respectively, to fully oxidize or reduce roGFP2 and Fib2-roGFP2, and confirmed that both sensors were similarly responsive to the redox environment (Fig. 3C-E). When comparing the fluorescence ratio (405/488 nm) in WT and nas124 plants, we observed that the ratio was close to the ratio measured under DTT treatment, suggesting that in both genetic backgrounds the redox sensor is fully reduced in the nucleolus and the nucleus (Fig. 3D-E). Therefore, we concluded that different iron contents in WT and nas124 nucleoli do not impact the nucleolar or the nuclear redox state.
Fig. 3.
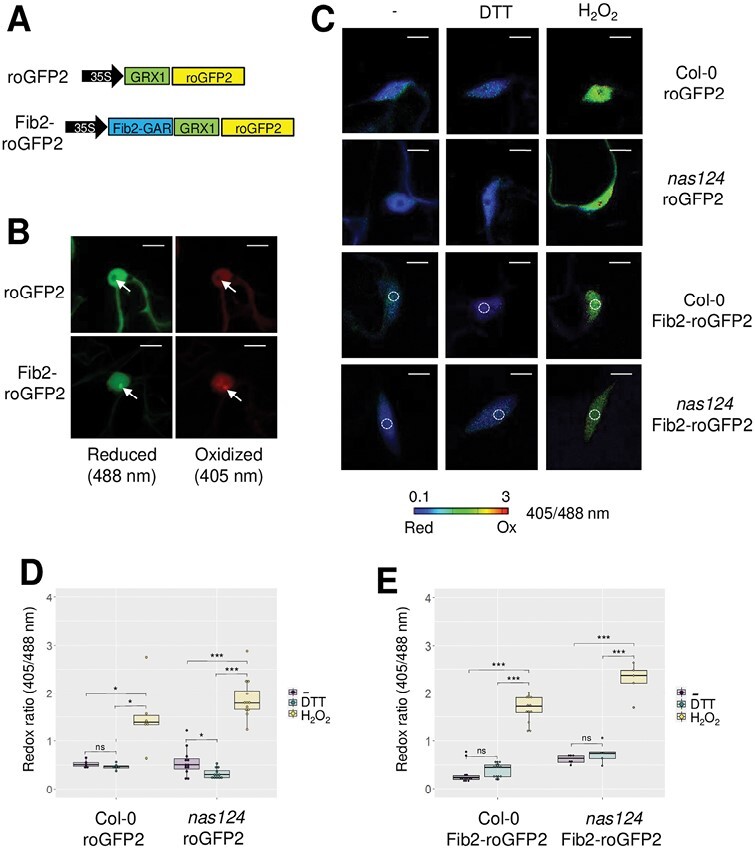
In vivo monitoring of the nucleolar/nuclear redox state in nas124. (A) Gene structure of the roGFP2 constructs used to transform WT and nas124 plants. Both constructs were under the control of the 35S-CaMV promoter. In Fib2-roGFP2, GRX1 was fused to roGFP2 as in roGFP2, but the GAR domain (1–73 N-terminal amino acids) of Fib2 was additionally fused to GRX1-roGFP2. (B) Confocal images of nuclei of WT cotyledons expressing roGFP2 or Fib2-roGFP2. RoGFP2 was excited at 488 nm or 405 nm to monitor the reduced or oxidized forms, respectively. Arrows indicate the nucleolus. Scale bars=10 µm. (C) Steady state ratio images of WT or nas124 plants expressing roGFP2 or Fib2-roGFP2 and calculated as the 405/488 nm fluorescence. To fully reduce or oxidize the sensor, seedlings were immersed in 10 mM DTT or 100 mM H2O2, respectively. Control samples were immersed in MS/2 liquid medium only. False colours indicate the fluorescence ratio on a scale from blue (reduced) to red (oxidized). Scale bars=10 µm. White circles indicate the location of the nucleolus. (D-E) Fluorescence ratio of roGFP2 (D) and Fib2-roGFP2 (E) calculated from ratio images. Ratios were calculated from six to 16 nuclei per sample. Asterisks indicate a significant difference calculated by Student’s t-test (* P≤0.05, ** P≤0.01, and *** P≤0.001, ns P>0.05).
Furthermore, we measured reactive oxygen species (ROS) accumulation in planta by staining nas124 plants with 3,3ʹ-diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) that react with H2O2 and O2.-, respectively. Similar staining was found in nas124 and WT plants (Supplementary Fig. S4A). Major ROS detoxification enzymes, catalases, and ascorbate peroxidases also had similar activities in WT and nas124 plants (Supplementary Fig. S4B). In summary, nas124 mutants did not experience oxidative stress due to perturbed iron distribution.
Transcriptome analysis of nas124 plants
Disruption of nucleolar organization affects nuclear gene expression in A. thaliana (Pontvianne et al., 2016). To investigate the impact of reduced nucleolar iron and/or decreased NA on global gene expression, we performed RNAseq analysis of WT and nas124 plants, with good correlation between two replicates (Fig. 4; Supplementary Fig. S5A; Supplementary Table S2A). We identified 60 differentially expressed (DE) genes with a false discovery rate (FDR) lower than 5%. Among them were 28 up-regulated genes with fold-change (log2) values (nas124/WT) ≥2, and 32 down-regulated genes with fold-change (log2) values ≤0.5 (Fig. 4A; Supplementary Table S2B). The most down-regulated gene was NAS1 (–8.1-fold) and the most up-regulated gene encoded for the hypothetical protein At1g64795 (8.1-fold). The NAS2 and NAS4 genes did not appear among the 60 DE genes, nevertheless a decreased number of NAS2 and NAS4 reads (–2.2- and –1.6-fold, respectively) and a slightly increased number of NAS3 reads (1.3-fold) were observed in nas124 plants (Fig. 4B; Supplementary Table S2). Down-regulated genes other than NAS were the iron metabolism-related gene FERRETIN 1 (At5g01600) and defence or pathogen response genes, including the S-adenosyl-l-methionine-dependent methyltransferase superfamily protein gene (At2g32160). Among the detected up-regulated genes were OLIGOPEPTIDE TRANSPORTER, OPT3 (At4g16370), IRON REGULATED TRANSPORTER 3, IRT3 (At1g60960), and IRONMAN 2 (At1g47395) (Supplementary Table S2B). Furthermore, a GO (GO-PANTHER) analysis of protein classes highlighted two categories: ‘metabolism interconversion enzymes’ (six DE genes) and ‘transporters’ (six DE genes), whereas the GO-Slim molecular function and biological processes highlighted the categories ‘catalytic activity’ (12 DE genes), ‘binding’ (eight DE genes), and ‘cellular processes’ (13 DE genes) (Fig. 4C; Supplementary Table S3). Lastly, further analysis to determine de-regulation of non-coding RNA did not reveal significant transcriptomic variations of small RNAs (snoRNA, snRNA, tRNA) or long non-coding RNAs in nas124 plants (Supplementary Fig. S5B). Taken together, RNAseq analysis shows that simultaneous disruption of NAS1, NAS2, and NAS4 affects expression of genes involved in metal metabolism and/or transport, and accumulation of transcripts associated with plant defence or pathogen response.
Fig. 4.
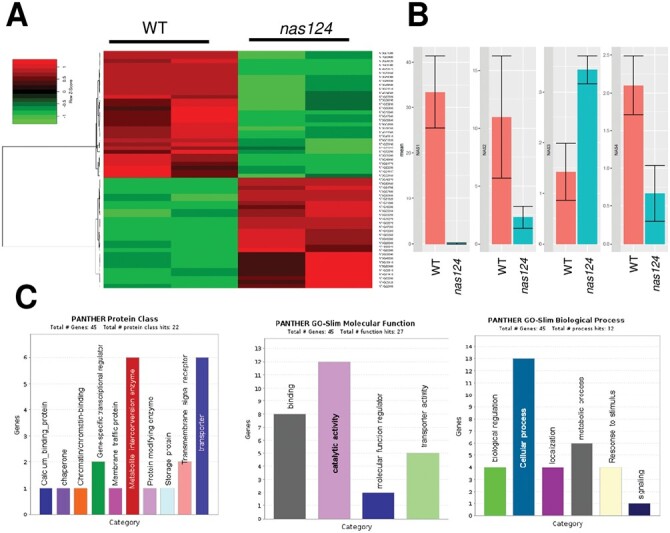
Transcriptomic analysis of nas124. (A) Heat map of 60 DE genes in 15-day-old in vitro grown nas124 compared to WT plants. The colour key, histogram, and values are shown. Down- and up-regulated genes are in green and red, respectively. (B) Graph of normalized reads (rpkm) in WT and nas124 plants for NAS1 (At5g04950), NAS2 (At5g56080), NAS3 (At1g09240), and NAS4 (At1g09240) transcripts, respectively, from left to right on the graph. Graphs and plots were generated using RStudio, Ri 386 version 4.1.1. (C) GO analysis for protein classes, -Slim molecular function, and -Slim biological process.
Functional organization of nas124 nucleoli
We carried out observations by TEM to investigate if reduced amounts of nucleolar iron in nas124 plants could impact nucleolar structural and/or functional organization (Fig. 5). In the nucleolus, three major functional structures are observed: fibrillar centres (FC), where transcriptionally active 45S rDNA localizes, dense fibrillar components (DFC), the place of primary rRNA processing events, and the granular component (GC), mainly composed of pre-ribosome particles. Transcription of rDNA by RNA Pol I occurs at the interface of the FC and the DFC.
Fig. 5.
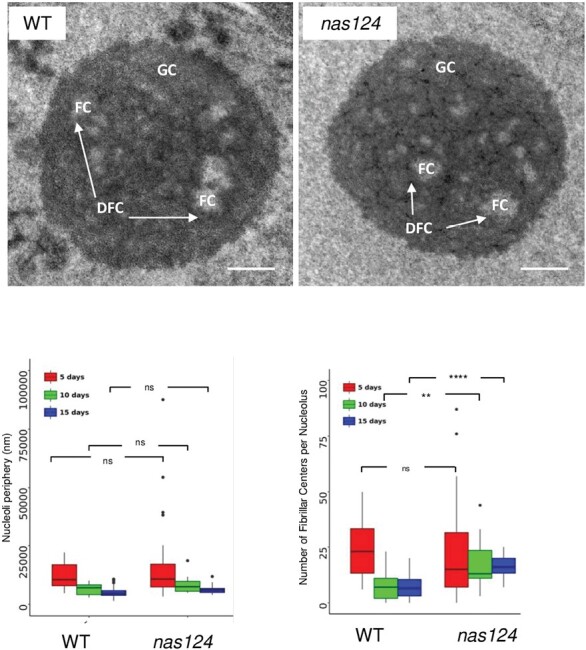
Nucleolus organization in nas124. Top, TEM images of root cells from 10-day-old WT and nas124 plants grown in vitro. The Fibrillar Centers (FC) are surrounded by the Dense Fibrillar Component (DFC, indicated by white arrows) and embedded in the Granular Component (GC). Scale bar=500 nm. Bottom, boxplot graphs of nucleoli periphery (left) and number of FC structures per nucleolus (tight) in 5-, 10-, and 15-day-old root cells. The dots indicate the outlier samples. 28 (5-day-old), 13 (10-day-old), and 46 (15-day-old) WT nucleoli and 35 (5-day-old), 19 (10-day-old), and 16 (15-day-old) nas124 nucleoli were analysed. Asterisks indicate a significant difference calculated by Student’s t-test: ns P>0.05, ** P≤0.01, and **** P≤0.001.
In plant cells, there are two types of FCs: homogeneous FCs, small, numerous, and associated with active nucleoli, and heterogeneous FCs, fewer and larger, associated with low rates of nucleolar activity (Saez-Vasquez and Medina, 2008). To address this issue in nas124, we performed TEM of nucleoli from 5-, 10-, and 15-day-old WT and nas124 plants. Then we measured nucleoli periphery and determined the number of FC per nucleolus (left and right graphs respectively in Fig. 5). In 5-day-old plants, nucleoli from both genotypes were larger than nucleoli from 10- and 15-day-old plants (Fig. 5). Also 5-day-old plant nucleoli from WT and nas124 were similar in size and shape, containing between 5 and 30 FC. In 10-day-old plants, WT and nas124 nucleoli also had similar size and functional (FC, DFC, and GC) organization. However, nas124 nucleoli contained around twice the number of FC structures than WT nucleoli. Likewise, nucleoli from 15-day-old WT and nas124 plants displayed similar sizes, but a higher number of FC was observed in nas124 (between 12 and 30 FC) than in WT (≤15 FC). Taken together, the TEM results suggested that nas124 nucleoli were more active compared to WT nucleoli.
NOR organization and expression in nas124
The genome of A. thaliana Col-0 contains around 1500 copies of 45S rDNA units per diploid genome (Saez-Vasquez and Gadal, 2010; Layat et al., 2012; Mohannath et al., 2016). They localize in the nucleolar organizer regions (NORs) from chromosomes 2 and 4 (NOR2 and NOR4) (Copenhaver and Pikaard, 1996a, b); however, only rDNA (VAR2 and 3) from NOR4 is transcribed in most plant organs. The rDNA (VAR1) from NOR2 is repressed by epigenetic mechanisms and expressed mainly early during seed germination and in specific mutant plants with nucleolar disorganization (Pontvianne et al., 2010), lower copy numbers (Pontvianne et al., 2013), or altered proportion of rDNA variants (Durut et al., 2014).
Active NORs are associated to the nucleolus while inactive NORs remain at the nuclear periphery (Sáez-Vásquez and Delseny, 2019). To examine nucleolus association/dissociation of NORs in nas124 plants, we performed FISH with a probe homologous to the 45S rDNA (Fig. 6A). In WT nuclei, the FISH probe detected a single and strong signal (active NOR4) associated with the nucleolus, whereas two weak signals close to the nuclear border reflected the non-associated, inactive NOR2. In contrast, in nas124 nuclei, the FISH probe detected three to four strong signals associated with the nucleolus. The analysis based on 32 WT and 18 nas124 nuclei revealed around 2-fold more nucleolus-associated and around 3-fold less non-associated signals in nas124 nuclei compared to WT. This increased association between the nucleolus and NOR2 indicates transcriptional activation of this usually silent rDNA locus.
Fig. 6.
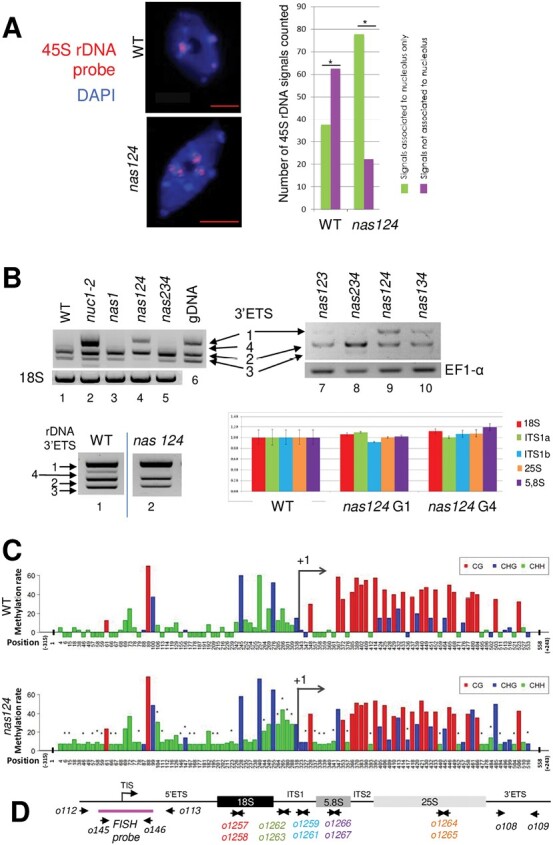
NOR expression in nas124. (A) FISH analysis of 45S rDNA performed on leaf cells from 21-day-old WT and nas124 plants grown in soil. 45S-labelled probes hybridized to NOR2 and NOR4 (red signals). DNA in the nucleoplasm is stained in blue with DAPI and the black area in the nucleus corresponds to the nucleolus that is not stained because the rDNA chromatin is decondensed and too thin to be stained by DAPI (Pontvianne et al., 2016a). Scale bars=5 µm. The histogram shows the number of 45S FISH signals in 32 WT and 18 nas124 cells. (B) Top panels, RT–PCR to detect 45S pre-rRNA variants in leaves from 21-day-old WT (lane 1), nuc1-2 (lane 2), nas1 (lane 3), nas124 (lanes 4 and 9), nas234 (lanes 5 and 8), nas123 (lane 7), and nas134 (lane 10) plants grown in soil. Amplification of 18S (lanes 1–5) and EF1-alpha (lanes 7–10) RNA transcripts was performed to verify the similar amount of RNA in each sample. Amplification of genomic DNA (lane 6) shows up to four rDNA variants in A. thaliana Col-0 plants. Bottom panels, PCR (left) analysis of 3ʹETS rDNA in WT (lane 1) and nas124 (lane 2) plants and RT–qPCR (right) of 18S, Internal Transcribed Spacer 1 (ITS1a and ITS1b), 25S and 5.8S rDNA sequences in WT and the nas124 siblings (G1 and G4). (C) Bisulfite analysis of promoter/5ʹETS (from –315 to +243) rDNA sequences from WT and nas124 plants. The graph bars show CG, CHG, and CHH methylation levels at different positions in the rDNA. The tiny dark stars over the graph bars indicate sites with significant (P ≤0.05) difference of methylation rate between WT and nas124 for each position. (D) The schematic of a 45S rDNA unit shows the position of primers used in bisulfite sequencing analysis (o112/o113) to amplify rDNA for the FISH probe (o145/o146) to detect 3ʹETS (o108/o109) by PCR, and 18S (o1257/o1258), ITS1 (o1262/o1263 and o1259/o1261), 5.8S (o1266/o1267), and 25S (o1264/o1265) by qPCR.
To verify expression of NOR2 and NOR4 in nas124 plants, we performed RT–PCR analysis using specific primers that amplify the 3ʹ external transcribed spacer (3ʹETS) rDNA from NOR2 (VAR1) and/or from NOR4 (VAR2 and 3) (Pontvianne et al., 2010). RT–PCR amplification of the 3ʹETS generated specific product sizes for each rRNA gene variant (Fig. 6B) and confirmed expression of VAR1 in nuc1-2 plants with disrupted nucleolin 1 (NUC1), as reported (Pontvianne et al., 2010; Durut et al., 2014) and used as a positive control here. The analysis detected expression of rRNA VAR1 in nas124, but not in WT or nas234 plants. In nas124 plants, rRNA VAR2 expression was unaltered, whereas VAR3 was at same point repressed. We also detected, but at lower level, expression of rRNA VAR1 in nas123 and nas134. Finally, rRNA VAR1 was not expressed in nas1 plants indicating that disruption of NAS1 alone was not sufficient to induce expression of NOR2.
Furthermore, to determine rDNA copy number variations and/or organization that could have affected expression of rDNA variants, we performed PCR and qPCR using genomic DNA from WT and nas124, including a fourth generation of amplified homozygous mutants (Fig. 6B). Except for a lower PCR signal corresponding to rDNA VAR4 observed in nas124 compared to WT, this analysis shows no obvious differences in the ratio of rDNA variants VAR1-3 in nas124 compared to WT plants. Similarly, RT–qPCR of ITS1 (Internal Transcribed Spacer 1), 18S, 5.8S, and 25S rDNA sequences excluded significant differences in the total copy number of rDNA in nas124, from the 1st to the 4th generation.
Transcriptionally active rDNA is hypomethylated and associated with acetylated histones (H3Ac and H4Ac) or dimethylated histone H3 at lysine 4 (H3K4me2), whereas inactive rDNA is hypermethylated and associated with dimethylated histone H3 at lysine 9 (H3K9me2) (Probst et al., 2004; Espada et al., 2007; Earley et al., 2010). Therefore, we analysed the epigenetic state of 45S rDNA in nas124 plants. We performed bisulfite analysis of promoter and 5ʹETS rDNA (–315 to +243) sequences (Fig. 6C). Notably, CG sites were more present downstream of Transcription Initiation Site at +1 (TIS/+1), whereas CHG and CHH sites were enriched upstream of TIS/+1. Bisulfite analysis revealed a slight hypermethylation in nas124 at specific CHG and CHH sites in the 5ʹETS, whereas the CG methylation in nas124 remained similar to WT. Notably, in the promoter sequence, there was higher CHG and CHH methylation in nas124 plants compared to WT. Silencing of 45S rRNA genes involves long non-coding RNA (Schmitz et al., 2010) or small interfering RNA (siRNA) that direct specific DNA methylation (Costa-Nunes et al., 2010). Northern blot experiments did not reveal significant differences in the accumulation of promoter siRNA 45S and siR759, or IGS rRNA sequences (Supplementary Fig. S6).
To determine if the CHG and CHH hypermethylation was specific for either VAR1, and/or VAR2, and/or VAR3, we performed bisulfite analysis of the 3ʹETS rDNA region (Supplementary Fig. S7). In contrast to the promoter and 5ʹETS regions, CG, CHG, and CHH sites were more equally distributed in the 3ʹETS of all rDNA variants. The bisulfite analysis did not reveal significant differences in the methylation state between rDNA variants from nas124 and WT plants. Only one CG site at position 44 in VAR1 was hypomethylated in nas124.
We also examined histone marks by Chromatin Immuno-Precipitation (ChIP) followed by qPCR analysis (Supplementary Fig. S8). Primers matching mature 25S and non-coding (IGS, 5ʹETS, and 3ʹETS) rDNA sequences did not detect changes of either active (H3Ac and H3K4me2) or inactive (H3K9me2) histone marks in nas124 compared to WT plants.
Together these analyses revealed that among the epigenetic parameters investigated, changes in DNA methylation at rDNA promoter sequences are most evident with expression of rDNA from NOR2 in the nas124 mutant. In contrast, in the nas234 mutant we do not observe NOR2 activity and/or nucleolus association or epigenetic changes (Supplementary Fig. S9).
RNA methylation in nas124 and nas234 plants
rRNA is modified concomitantly or immediately after RNA pol I transcription. Reported rRNA modifications include sugar and base methylation and/or acetylation, and serve to stabilize structures of the rRNA scaffold and ensure efficiency and accuracy of translation (Sharma and Lafontaine, 2015; Sloan et al., 2017). Since S-adenosyl-methionine (SAM) moieties used by NAS enzymes to synthesise NA are also donors in methylation reactions, including methylation of DNA, RNA, and proteins (Fig. 1), we questioned whether reduced NAS activity in the nas124 and nas234 plants might impact availability of SAM and subsequently methylation of coding and non-coding RNAs. Here, we studied the impact of NAS reduction (in nas124 and nas234) and/or the expression of rRNA from NOR2 (in nas124) on RNA base modifications and on 2ʹ-O-methylation (2ʹ-O-Me), the most abundant rRNA modification (Azevedo-Favory et al., 2021; Wu et al., 2021).
We performed the MRM methodology in mass spectrometry to analyse 18 different RNA base modifications (Fig. 7; Supplementary Fig. S10), including m1A, m5C, m7G, m3A, and ac4C identified in rRNA from yeast and animal cells (Sharma and Lafontaine, 2015; Sloan et al., 2017). The foremost observation was a significant (≤0.05) increase of methylation of m1A, m1G, m2,7,7G, m3C, m5C, m7G, and also ac4C in nas124. No decreased methylation of specific RNA modifications was observed in nas124. In contrast, methylation of m1A, m3C, m5C, m7G, m2,2G, m3U, mcm5s2U/U, and ncm5U/U was decreased in nas234, whereas methylation of m2,7,7G and m6A increased. No significant changes were detected for I, m2G, m6Am, A, oxo8G, and/or pseudouridylation (Psi) in nas124 or nas234 (Fig. 7; Supplementary Table S4).
Fig. 7.

RNA modifications in nas124 and nas234. Graphs show ratio of ac4C/C, I/U, m1A/U, m1G/G, m2,2,7G/G, m2,2G/G, m2G/G, m3C/C, m3U/U, m5C/C, A/U, m6A/U, m6Am/U, m7G/G, mcm5s2U/U, ncm5U/U, oxo8G/G, and Psi/U detected by the LC-MRM method in WT and nas124. P values are indicated for each methylated/non-methylated nucleotide ratio. Asterisks indicate a significant difference calculated by Student’s t-test: ns >0.05, * P≤0.05, ** P≤0.01, and ***P≤0.001. Statistical differences with Wilcoxon test, P≤0.1 are provided for m6,6A/U, mcm5s2U/U, ncm5U/U.
We performed RiboMethSeq analysis to determine 2ʹ-O-Me profiles of 18S, 5.8S, and 25S rRNA in nas124 and nas234 compared to WT plants. No significant changes were detected in nas124 or nas234 plants (Supplementary Fig. S10A; Supplementary Table S5).
Together, these analyses revealed specific RNA base methylation changes in nas124 and nas234. Notably RNA base methylation changes detected in nas124 were opposite to, or are not affected in, nas234.
Discussion
The nas124 and nas234 plants described here each had a reduced concentration of NA; however, specific cellular and molecular features were detected in these mutants. Under standard growth conditions, a reduced concentration of NA did not have an evident impact on nas124 or nas234 plant growth or development phenotypes, except for mildly disturbed vein patterns in cotyledons in both genotypes and chloronerva-like phenotypes in primary leaves in nas124 (Fig. 1). Leaf chlorosis was also observed in the fertile and quadruple mutant nas4x-1. However, this chlorosis was minor at the vegetative stage and intensified at the reproductive stage (Klatte et al., 2009).
Nevertheless, the most outstanding observation was that only nas124 contains low or undetectable iron in the nucleolus (Fig. 2; Supplementary Figs S2, S3). We cannot yet explain the chloronerva-like and low nucleolar iron phenotypes in nas124, which do not occur in nas234 plants. The lower amount of total NA in nas124 and the lack of NAS activity in roots might have an impact in iron transport from tissue to tissue, cell to cell, and at last to the nucleolus. However, inhibition of iron uptake or increased iron availability did not affect nas124 growth (root phenotype) compared to WT plants; neither media composition change (MS and/or sugar) affected chlorosis in nas124 (Supplementary Fig. S11).
Transcriptome analysis did not explain the phenotypes of the nas124 plants (Fig. 4; Supplementary Fig. S5). However, mRNA transcripts encoding proteins involved in iron metabolism or transport were de-regulated in nas124 plants, indicating a disturbed sensing of iron availability status in these plants. In addition, variations of transcripts associated with plant defence or pathogen response indicate that nas124 might be also sensitive to biotic stresses. This is in agreement with the reported role of iron in the plant pathogenesis response (reviewed in Liu et al., 2021).
How reduced NA inhibits nucleolar accumulation of iron in nas124, but not in nas234, remains an open question. However, the finding of low or undetectable iron in the nucleolus in nas124 allowed us to address the role of nucleolar iron in plants.
The functional and structural organization of the nucleolus is linked to ribosome biogenesis (Saez-Vasquez and Medina, 2008; Boulon et al., 2010). In nas124, the lack of nucleolar iron induces increased number of FC likely due to rRNA VAR1 expression from normally silenced NOR2. It may be expected that rDNA VAR1 expression in nas124 is a consequence of failed repression rather than activation of NOR2. This is in agreement with steady rDNA VAR1 expression in nas124 early during seedling establishment (Supplementary Fig. S12). Indeed, in early states of seedling establishment, NOR2 and NOR4 were both expressed in WT plants. Later, NOR2 becomes progressively silenced (Earley et al., 2010; Benoit et al., 2013; Durut et al., 2014). Expression of NOR2 and low nucleolar iron and the presence of aggregates in the xylem vessels was also observed nuc1-2 plants (Supplementary Fig. S2). However, in contrast to nas124, disruption of NUC1 also induced nucleolus disorganization, chromatin de-condensation, and rDNA hypomethylation (Pontvianne et al., 2007, 2010), indicating that loss of nucleolar iron and NOR2 expression are triggered by different mechanisms.
Expression of rRNA VAR1 is associated with rDNA CpG hypomethylation, rDNA copy number or ratio, rDNA chromatin decondensation, and/or modifications of histone methylation and/or acetylation (Earley et al., 2010; Mozgova et al., 2010; Pontvianne et al., 2010, 2012, 2013; Durut et al., 2014). None of these features were affected in nas124 (Fig. 6). In contrast, CHG and CHH methylation on the rDNA promoter were increased in nas124. This is unexpected as increased DNA methylation is generally correlated with gene repression (Probst et al., 2004; Earley et al., 2006; Espada et al., 2007). In this context, the conserved iron-sulfur cluster assembly protein MET18 could play a key role. Indeed, in Arabidopsis, MET18 gene disruption causes DNA hypermethylation in particular in the CHH context (Wang et al., 2016). Interestingly, similarly to nas124 the rRNA VAR1 accumulated in met18 plants (Supplementary Fig. S13). Therefore, in nas124, lack of nucleolar iron might inhibit MET18 causing CHH hypermethylation of rDNA.
In animal cells there is a subset of rRNA genes that are transcriptionally inactive but are poised for transcription activation. Such poised rDNA promoters are unmethylated and are marked by both euchromatic and heterochromatic histone modifications (Xie et al., 2012; Grummt and Langst, 2013). Therefore, we can speculate that hypermethylation in nas124 takes place on ‘poised rDNA promoters’ rather than on transcriptionally active rDNA. Furthermore, higher numbers of FC in nas124 is a likely consequence of nucleolar association of NOR2 and an increased number of transcribed rRNA genes. However, we predict a low RNA pol I transcription activity in these rRNA genes since accumulation of rRNA precursors and/or mature 18S, 5.8S, and 25S rRNA in nas124 is similar to WT and nas234 plants (Supplementary Fig. S14). Controlling rRNA synthesis by modulating RNA pol I loading on rDNA and activity has been reported in yeast and animal cells (French et al., 2003; Goodfellow and Zomerdijk, 2013; Darriere et al., 2019).
NAS enzymes use SAM to produce NA (Fig. 1) and it is possible that reduced rates of NA synthesis in nas124 and nas234 might fine-tune SAM availability and subsequently affect methylation of RNA, DNA, and/or proteins. Remarkably, in contrast to the lack of nucleolar iron and NOR2 gene expression detected in nas124, RNA methylation modifications are impacted in both nas124 and nas234 (Fig. 7). However, the specific and contrasting changes of RNA modifications levels in nas124 and nas234 are intriguing. For instance, whereas increased level of RNA modifications (including m1A, m5C, m7G) could be correlated with a differential rRNA gene expression in nas124, this is not the case for a decreased level of these same RNA modifications in nas234. The identification of differentially methylated RNAs in nas124 and nas124 requires further investigation. However, detected modifications m1A, m5C, m7G and ac4C are likely associated with rRNA because they are the most abundant cellular RNAs and the first to be detected by LC-MSMS. These modifications occur co-transcriptionally in the nucleolus (Sharma and Lafontaine, 2015; Sloan et al., 2017), and it is tempting to propose that lack of nucleolar iron might affect the activity of iron-sulfur (Fe-S) protein factors involved in depositing RNA modifications. Notably, several rRNA modifications are introduced by RNA modifying enzymes containing (Fe-S) clusters (Kimura and Suzuki, 2015). However, to our knowledge, rRNA modifying enzymes containing (Fe-S) clusters have not yet been reported in plants.
Redox-active iron [Fe2+, iron (II)] has the potential to produce a highly reactive.OH radicals by the Fenton reaction when it meets hydrogen peroxide (Smethurst and Shcherbik, 2021). However, nuclear/nucleolar redox state seems unaffected in nas124, suggesting that nucleolar iron accumulating in WT plants is likely unavailable for Fenton reactions. We did not observe more pronounced ROS accumulation, indicating that nas124 does not experience oxidative stress due to perturbed cellular/or subcellular iron distribution (Fig. 3; Supplementary Fig. S4). Nevertheless, it is of particular interest to highlight that the ribosome, which generally depends on Mg2+, might also contain several Fe2+ at specific sites (Smethurst et al., 2020). Therefore, further characterization of nas124 plants should help us to understand the role of iron in ribosome assembly and/or activity specifically under oxidative conditions.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Identification of triple nas mutant plants.
Fig. S2. Nucleolar iron and ferritin in nas124 and nuc1 mutant plants.
Fig. S3. Nucleolar iron in roots from nas124 and nas234 plants.
Fig. S4. Redox activity in nas124.
Fig. S5. Transcriptomic analysis of nas124.
Fig. S6. Northern blots to detect long and small non-coding RNA in nas124 and nas234.
Fig. S7. DNA methylation in nas124.
Fig. S8. H3 histone marks in nas124.
Fig. S9. NOR/ rDNA functional organization in nas234.
Fig. S10. RNA methylation changes in nas124 and nas234 plants.
Fig. S11. Iron and MS/sucrose impact on nas124 and nas234 plants.
Fig. S12. rRNA expression in nas124 during seedling establishment.
Fig. S13. Expression of rRNA variants in met18.
Fig. S14. Transcription and processing of pre-rRNAs in nas124 and nas234.
Table S1. List of primers used in this work.
Table S2. RNAseq analysis of nas124.
Table S3. GO analysis of protein genes de-regulated in nas124.
Table S4. LC-MRM method data for WT, nas124, and nas234 plants.
Table S5. RiboMethSeq data for WT, nas124, and nas234 plants.
Acknowledgements
The authors thank Michel Delseny and Ortrun Mittelsten Scheid for critical reading of the manuscript and Hervé Moreau for facilitating access to the BioPIC platform. The authors also thank Thomas Roscoe for comments and correction of the English language; Karl Ravet for antibodies against Arabidopsis ferritin and Sophie Brando and Michèle Laudie for technical assistance.
Glossary
Abbreviations
- NA
nicotianamine
- NAS
NICOTIANAMINE SYNTHASE
- NORs
nucleolar organizer regions
- SAM
S-adenosyl-methionine
- DAB
3,3'-diaminobenzidine
- ROS
reactive oxygen species
- GRX1
glutaredoxin 1
- Fib2
fibrillarin 2
- NBT
nitro blue tetrazolium chloride
- OPT3
OLIGOPEPTIDE TRANSPORTER
- IRT3
IRON REGULATED TRANSPORTER 3
- TEM
transmission electron microscopy
- FC
fibrillar centres
- DFC
dense fibrillar component
- GC
granular component
- ribosomal RNA and DNA
rRNA and rDNA
- FISH
fluorescence in situ hybridization
- ChIP
chromatin immunoprecipitation
- LC-MRM
liquid chromatography-multiple reaction monitoring
- RiboMethSeq
RNA 2ʹ-O-methylation sequencing
Contributor Information
Charlotte Montacié, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Christophe Riondet, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Lili Wei, Institut Agro, BPMP, CNRS, INRAE, Université Montpellier, 34060 Montpellier, France.
Tommy Darrière, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Alizée Weiss, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Frédéric Pontvianne, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Marie-Line Escande, Observatoire Océanologique de Banyuls s/ mer, CNRS, 66650 Banyuls-sur-mer, France; BioPIC Platform of the OOB, 66650 Banyuls-sur-mer, France.
Anne de Bures, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Edouard Jobet, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Adrien Barbarossa, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Marie-Christine Carpentier, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Mark G M Aarts, Laboratory of Genetics, Wageningen University & Research, 6700AA Wageningen, Netherlands.
Aurore Attina, INSERM, CHU Montpellier, CNRS, IRMB, Université Montpellier, 34090 Montpellier, France.
Christophe Hirtz, INSERM, CHU Montpellier, CNRS, IRMB, Université Montpellier, 34090 Montpellier, France.
Alexandre David, IGF, CNRS, INSERM, Université Montpellier, 34090 Montpellier, France.
Virginie Marchand, Epitranscriptomics and RNA Sequencing (EpiRNA-Seq) Core Facility, CNRS, INSERM, IBSLor (UMS2008/US40), Université de Lorraine, F-54000 Nancy, France.
Yuri Motorin, Epitranscriptomics and RNA Sequencing (EpiRNA-Seq) Core Facility, CNRS, INSERM, IBSLor (UMS2008/US40), Université de Lorraine, F-54000 Nancy, France; CNRS, IMoPA (UMR 7365), Université de Lorraine, F-54000 Nancy, France.
Catherine Curie, Institut Agro, BPMP, CNRS, INRAE, Université Montpellier, 34060 Montpellier, France.
Stéphane Mari, Institut Agro, BPMP, CNRS, INRAE, Université Montpellier, 34060 Montpellier, France.
Jean-Philippe Reichheld, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Julio Sáez-Vásquez, Laboratoire Génome et Développement des Plantes (LGDP), UMR 5096, CNRS, 66860 Perpignan, France; LGDP, UMR 5096, Université Perpignan Via Domitia, 66860 Perpignan, France.
Pablo Manavella, Instituto de Agrobiotecnología del Litoral, Argentina.
Author contributions
CM, CR, LW, FP, TD, AB, EJ, MLE, AW, and J-PR performed the experiments; MCC performed the bioinformatics analysis; MGMA obtained the nas triple mutant lines; AA, CH, and AD performed the LC-MRM; VM and YM performed the RiboMetSeq; AdB provided technical assistance; SM and CC supervised the experiments; JS-V, CM, and J-PR analysed the data and wrote the manuscript; and all authors approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest in relation to this work.
Funding
This work was supported by the CNRS, the UPVD (fellowships to CM), and by grants ANR SUBCELIF 087217, ANR MetRibo 210877, and ANR RoxRNase 210880, and Labex AGRO (under the I-Site Muse framework) coordinated by the Agropolis Fondation (grant no. Flagship Project 1802-002—CalClim). This study is set within the framework of the ‘Laboratoires d’Excellences (LABEX)’ TULIP (ANR-10-LABX-41) and of the ‘Ecole Universitaire de Recherche (EUR)’ TULP-GS (ANR-18-EURE-00019).
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.
References
- Anders S, Pyl PT, Huber W.. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo-Favory J, Gaspin C, Ayadi L, Montacie C, Marchand V, Jobet E, Rompais M, Carapito C, Motorin Y, Saez-Vasquez J.. 2021. Mapping rRNA 2ʹ-O-methylations and identification of C/D snoRNAs in Arabidopsis thaliana plants. RNA Biology 18, 1760–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barneche F, Steinmetz F, Echeverria M.. 2000. Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. Journal of Biological Chemistry 275, 27212–27220. [DOI] [PubMed] [Google Scholar]
- Bauer P, Thiel T, Klatte M, Bereczky Z, Brumbarova T, Hell R, Grosse I.. 2004. Analysis of sequence, map position, and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiology 136, 4169–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit M, Layat E, Tourmente S, Probst AV.. 2013. Heterochromatin dynamics during developmental transitions in Arabidopsis - a focus on ribosomal DNA loci. Gene 526, 39–45. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI.. 2010. The nucleolus under stress. Molecular Cell 40, 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray MS, Lenz TK, Haynes JW, Bowman JC, Petrov AS, Reddi AR, Hud NV, Williams LD, Glass JB.. 2018. Multiple prebiotic metals mediate translation. Proceedings of the National Academy of Sciences, USA 115, 12164–12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Dubos C, Gaymard F.. 2015. Iron nutrition, biomass production, and plant product quality. Trends in Plant Science 20, 33–40. [DOI] [PubMed] [Google Scholar]
- Cakmak I, Marschner H.. 1992. Magnesium deficiency and high light intensity enhance activities of superoxide dismutase, ascorbate peroxidase, and glutathione reductase in bean leaves. Plant Physiology 98, 1222-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS.. 1996a. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. The Plant Journal 9, 259–272. [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS.. 1996b. Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. The Plant Journal 9, 273–282. [DOI] [PubMed] [Google Scholar]
- Costa-Nunes P, Pontes O, Preuss SB, Pikaard CS.. 2010. Extra views on RNA-dependent DNA methylation and MBD6-dependent heterochromatin formation in nucleolar dominance. Nucleus 1, 254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Briat JF.. 2003. Iron transport and signaling in plants. Annual Review of Plant Biology 54, 183–206. [DOI] [PubMed] [Google Scholar]
- Darriere T, Pilsl M, Sarthou MK, et al. 2019. Genetic analyses led to the discovery of a super-active mutant of the RNA polymerase I. PLoS Genetics 15, e1008157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durut N, Abou-Ellail M, Pontvianne F, et al. 2014. A duplicated NUCLEOLIN gene with antagonistic activity is required for chromatin organization of silent 45S rDNA in Arabidopsis. The Plant Cell 26, 1330–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS.. 2006. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes & Development 20, 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS.. 2010. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes & Development 24, 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J, Ballestar E, Santoro R, et al. 2007. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Research 35, 2191–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL.. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Molecular and Cellular Biology 23, 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow SJ, Zomerdijk JC.. 2013. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcellular Biochemistry 61, 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Langst G.. 2013. Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochimica et Biophysica Acta 1829, 393–404. [DOI] [PubMed] [Google Scholar]
- Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP.. 2008. Real-time imaging of the intracellular glutathione redox potential. Nature Methods 5, 553–559. [DOI] [PubMed] [Google Scholar]
- Heeney MM, Finberg KE.. 2014. Iron-refractory iron deficiency anemia (IRIDA). Hematology/Oncology Clinics of North America 28, 637–652. [DOI] [PubMed] [Google Scholar]
- Henle ES, Han Z, Tang N, Rai P, Luo Y, Linn S.. 1999. Sequence-specific DNA cleavage by Fe2+-mediated Fenton reactions has possible biological implications. Journal of Biological Chemistry 274, 962–971. [DOI] [PubMed] [Google Scholar]
- Henle ES, Luo Y, Gassmann W, Linn S.. 1996. Oxidative damage to DNA constituents by iron-mediated Fenton reactions. The deoxyguanosine family. Journal of Biological Chemistry 271, 21177–21186. [PubMed] [Google Scholar]
- Hernandez-Verdun D, Roussel P, Thiry M, Sirri V, Lafontaine DL.. 2010. The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdisciplinary Reviews: RNA 1, 415–431. [DOI] [PubMed] [Google Scholar]
- Honda K, Smith MA, Zhu X, et al. 2005. Ribosomal RNA in Alzheimer disease is oxidized by bound redox-active iron. Journal of Biological Chemistry 280, 20978–20986. [DOI] [PubMed] [Google Scholar]
- Ivanov M, Sandelin A, Marquardt S.. 2021. TrancriptomeReconstructoR: data-driven annotation of complex transcriptomes. BMC Bioinformatics 22, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nature Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Suzuki T.. 2015. Iron-sulfur proteins responsible for RNA modifications. Biochimica et Biophysica Acta 1853, 1272–1283. [DOI] [PubMed] [Google Scholar]
- Klatte M, Schuler M, Wirtz M, Fink-Straube C, Hell R, Bauer P.. 2009. The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiology 150, 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Rizek P, Jog M.. 2016. Neuroferritinopathy: pathophysiology, presentation, differential diagnoses and management. Tremor and Other Hyperkinetic Movements 6, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layat E, Saez-Vasquez J, Tourmente S.. 2012. Regulation of Pol I-transcribed 45S rDNA and Pol III-transcribed 5S rDNA in Arabidopsis. Plant and Cell Physiology 53, 267–276. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kong D, Wu HL, Ling HQ.. 2021. Iron in plant-pathogen interactions. Journal of Experimental Botany 72, 2114–2124. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Blanloeil-Oillo F, Helm M, Motorin Y.. 2016. Illumina-based RiboMethSeq approach for mapping of 2'-O-Me residues in RNA. Nucleic Acids Research 44, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzlander M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld JP, Hell R.. 2009. The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 9109–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A, Hager J, Chaouch S, et al. 2010. Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiology 153, 1144–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohannath G, Pontvianne F, Pikaard CS.. 2016. Selective nucleolus organizer inactivation in Arabidopsis is a chromosome position-effect phenomenon. Proceedings of the National Academy of Sciences, USA 113, 13426–13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I, Mokros P, Fajkus J.. 2010. Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. The Plant Cell 22, 2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Asada K.. 1981. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant and Cell Physiology 22, 867–880. [Google Scholar]
- Pederson T, Powell K.. 2015. Thoru Pederson: Spotting novel roles for the nucleolus. Journal of Cell Biology 208, 384–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Abou-Ellail M, Douet J, et al. 2010. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genetics 6, e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Feng W, Stroud H, Jacobsen SE, Michaels SD, Pikaard CS.. 2012. Histone methyltransferases regulating rRNA gene dose and dosage control in Arabidopsis. Genes & Development 26, 945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, et al. 2013. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes & Development 27, 1545–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Carpentier MC, Durut N, et al. 2016. Identification of nucleolus-associated chromatin domains reveals a role for the nucleolus in 3D organization of the A. thaliana genome. Cell Reports 16, 1574–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Saez-Vasquez J.. 2007. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Molecular Biology of the Cell 18, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Fagard M, Proux F, et al. 2004. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. The Plant Cell 16, 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana C, Bellefqih S, Laval JY, Guerquin-Kern JL, Wu TD, Avila J, Ferrer I, Arranz R, Patino C.. 2006. Study of the localization of iron, ferritin, and hemosiderin in Alzheimer’s disease hippocampus by analytical microscopy at the subcellular level. Journal of Structural Biology 153, 42–54. [DOI] [PubMed] [Google Scholar]
- Roschzttardtz H, Fuentes I, Vásquez M, Corvalán C, León G, Gómez I, Araya A, Holuigue L, Vicente-Carbajosa J, Jordana X.. 2009. A nuclear gene encoding the iron-sulfur subunit of mitochondrial complex II is regulated by B3 domain transcription factors during seed development in Arabidopsis. Plant Physiology 150, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Grillet L, Isaure MP, Conejero G, Ortega R, Curie C, Mari S.. 2011. Plant cell nucleolus as a hot spot for iron. Journal of Biological Chemistry 286, 27863–27866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Vásquez J, Delseny M.. 2019. Ribosome biogenesis in plants: from functional 45S ribosomal DNA organization to ribosome assembly factors. The Plant Cell 31, 1945–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Vasquez J, Gadal O.. 2010. Genome organization and function: a view from yeast and Arabidopsis. Molecular Plant 3, 678–690. [DOI] [PubMed] [Google Scholar]
- Saez-Vasquez J, Medina FJ.. 2008. The plant nucleolus. In: Kader J-C, Delseny M, eds. Botanical research: Incorporating advances in plant pathology, Vol.47. San Diego: Elsevier Academic Press Inc., 1–46. [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I.. 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes & Development 24, 2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Lafontaine DL.. 2015. ‘View from a bridge’: A new perspective on eukaryotic rRNA base modification. Trends in Biochemical Sciences 40, 560–575. [DOI] [PubMed] [Google Scholar]
- Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, Bohnsack MT.. 2017. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biology 14, 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smethurst DGJ, Kovalev N, McKenzie ER, Pestov DG, Shcherbik N.. 2020. Iron-mediated degradation of ribosomes under oxidative stress is attenuated by manganese. Journal of Biological Chemistry 295, 17200–17214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smethurst DGJ, Shcherbik N.. 2021. Interchangeable utilization of metals: New perspectives on the impacts of metal ions employed in ancient and extant biomolecules. Journal of Biological Chemistry 297, 101374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepinski D. 2014. Functional ultrastructure of the plant nucleolus. Protoplasma 251, 1285–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, Pederson T.. 2014. Connecting the nucleolus to the cell cycle and human disease. The FASEB Journal 28, 3290–3296. [DOI] [PubMed] [Google Scholar]
- Tsekrekou M, Stratigi K, Chatzinikolaou G.. 2017. The nucleolus: In genome maintenance and repair. International Journal of Molecular Sciences 18, 1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veljovic-Jovanovic SD, Pignocchi C, Noctor G, Foyer CH.. 2001. Low ascorbic acid in the vtc-1 mutant of Arabidopsis is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiology 127, 426–435. [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li Q, Yuan W, Cao Z, Qi B, Kumar S, Li Y, Qian W.. 2016. The cytosolic Fe-S cluster assembly component MET18 is required for the full enzymatic activity of ROS1 in active DNA demethylation. Scientific Reports 6, 26443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wang Y, Wang J, Li X, Li J, Ye K.. 2021. Profiling of RNA ribose methylation in Arabidopsis thaliana. Nucleic Acids Research 49, 4104–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W.. 2012. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proceedings of the National Academy of Sciences, USA 109, 8161–8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary materials published online.


