ABSTRACT
The human fecal and oral microbiome may play a role in the etiology of breast cancer through modulation of endogenous estrogen metabolism. This study aimed to investigate associations of circulating estrogens and estrogen metabolites with the fecal and oral microbiome in postmenopausal African women. A total of 117 women with fecal (N = 110) and oral (N = 114) microbiome data measured by 16S rRNA gene sequencing, and estrogens and estrogen metabolites data measured by liquid chromatography tandem mass spectrometry were included. The outcomes were measures of the microbiome and the independent variables were the estrogens and estrogen metabolites. Estrogens and estrogen metabolites were associated with the fecal microbial Shannon index (global P < 0.01). In particular, higher levels of estrone (β = 0.36, P = 0.03), 2-hydroxyestradiol (β = 0.30, P = 0.02), 4-methoxyestrone (β = 0.51, P = 0.01), and estriol (β = 0.36, P = 0.04) were associated with higher levels of the Shannon index, while 16alpha-hydroxyestrone (β = −0.57, P < 0.01) was inversely associated with the Shannon index as indicated by linear regression. Conjugated 2-methoxyestrone was associated with oral microbial unweighted UniFrac as indicated by MiRKAT (P < 0.01) and PERMANOVA, where conjugated 2-methoxyestrone explained 2.67% of the oral microbial variability, but no other estrogens or estrogen metabolites were associated with any other beta diversity measures. The presence and abundance of multiple fecal and oral genera, such as fecal genera from families Lachnospiraceae and Ruminococcaceae, were associated with several estrogens and estrogen metabolites as indicated by zero-inflated negative binomial regression. Overall, we found several associations of specific estrogens and estrogen metabolites and the fecal and oral microbiome.
IMPORTANCE Several epidemiologic studies have found associations of urinary estrogens and estrogen metabolites with the fecal microbiome. However, urinary estrogen concentrations are not strongly correlated with serum estrogens, a known risk factor for breast cancer. To better understand whether the human fecal and oral microbiome were associated with breast cancer risk via the regulation of estrogen metabolism, we conducted this study to investigate the associations of circulating estrogens and estrogen metabolites with the fecal and oral microbiome in postmenopausal African women. We found several associations of parent estrogens and several estrogen metabolites with the microbial communities, and multiple individual associations of estrogens and estrogen metabolites with the presence and abundance of multiple fecal and oral genera, such as fecal genera from families Lachnospiraceae and Ruminococcaceae, which have estrogen metabolizing properties. Future large, longitudinal studies to investigate the dynamic changes of the fecal and oral microbiome and estrogen relationship are needed.
KEYWORDS: estrogens, fecal microbiome, oral microbiome, postmenopausal African women
INTRODUCTION
The human microbiome may play a role in the etiology of breast cancer. Several cross-sectional studies have reported associations between the human microbiome and breast cancer risk (1–3). One potential role of the human gut microbiome in breast cancer development is the regulation of estrogen metabolism, especially in postmenopausal women (4). Elevated circulating estrogens and estrogen metabolites are associated with a higher risk of postmenopausal breast cancer (5, 6). Estrogen metabolism via hydroxylation or methylation can lead to variations in bioavailability (7). The parent estrogens and estrogen metabolites can be also conjugated with sulfate to create a reservoir in the body for future use, or conjugated with glucuronic acid to enable excretion in bile and feces or excretion in urine (8). The circulating estrogens/estrogen metabolites consist of both unconjugated and conjugated forms.
It has been reported that the human gut microbiome modulates endogenous estrogen metabolism through bacterial beta-glucuronidase and beta-glucosidase enzyme that deconjugate estrogens, leading to their reabsorption into circulation (4, 9). The fecal Clostridia class, especially genera in the Ruminococcaceae family, contains beta-glucuronidase enzyme activity and has been observed to have a higher relative abundance in breast cancer cases (1, 10). Fecal Clostridia was also reported to be positively associated with levels of urinary estrogens and their metabolites (11, 12). There is also evidence that the oral microbiome is associated with breast cancer risk (13); however, no studies have investigated associations between estrogens and their metabolites and the oral microbiome.
To explore the interrelationships among estrogen metabolism and the human microbiome, we investigated the associations of circulating estrogens and estrogen metabolites with fecal and oral microbiome in postmenopausal women from the Ghana Breast Health Study, a population-based case-control study of breast cancer conducted in Accra and Kumasi, Ghana, West Africa (14).
RESULTS
As shown in Table 1, the mean age at blood draw was 57.9 (standard deviation 8.2) years. Most participants were never smokers (99.1%), never consumed alcohol (61.5%), never used oral contraceptives (86.3%), did not having a history of diabetes (88.0%), and reported not using antibiotics within 30 days of blood draw (67.5%).
TABLE 1.
Descriptive characteristics (N = 117)
| Covariates | N | Percent |
|---|---|---|
| Age at blood draw Mean (± SD) | 57.9 ± 8.2 | |
| Study site | ||
| KBTH | 6 | 5.1 |
| KATH | 67 | 57.3 |
| PLA | 44 | 37.6 |
| Year at blood draw | ||
| 2014 | 27 | 23.1 |
| 2015 | 90 | 76.9 |
| Time since menopause (yrs) | ||
| ≤2 | 17 | 14.5 |
| 3-5 | 15 | 12.8 |
| 6-10 | 28 | 23.9 |
| >10 | 43 | 36.8 |
| Missing | 14 | 12.0 |
| BMI (kg/m2) | ||
| Underweight (17–18.5) | 6 | 5.1 |
| Healthy wt (18.5–24.9) | 43 | 36.8 |
| Overweight (25.0–29.9) | 35 | 29.9 |
| Obese (30–67) | 32 | 27.4 |
| Unknown/Missing | 1 | 0.9 |
| Smoking status | ||
| Never | 116 | 99.1 |
| Unknown | 1 | 0.9 |
| Alcohol consumption | ||
| Yes | 38 | 32.5 |
| No | 72 | 61.5 |
| Unknown/Missing | 7 | 6.0 |
| Diabetes | ||
| Ever | 13 | 11.1 |
| Never | 103 | 88.0 |
| Unknown | 1 | 0.9 |
| Oral contraceptive use | ||
| Ever | 16 | 13.7 |
| Never | 101 | 86.3 |
| Antibiotic use | ||
| More than 30 days of blood draw | 79 | 67.5 |
| Within the last 30 days of blood draw | 28 | 23.9 |
| Unknown/Missing | 10 | 8.5 |
The associations between estrogens and estrogen metabolites with fecal microbial alpha-diversity are presented in Table 2. Overall, estrogens and estrogen metabolites were associated with the fecal microbial Shannon index (global P < 0.01) in the mutually adjusted model. In particular, concentrations of estrone (β = 0.36, P = 0.03), 2-hydroxyestradiol (β = 0.30, P = 0.02), 4-methoxyestrone (β = 0.51, P = 0.01), and estriol (β = 0.36, P = 0.04) were positively associated with the fecal Shannon index, while the association with 16-alpha-hydroxyestrone was inverse (β=-0.57, P < 0.01). In the individual hormone models, estrone (β = 0.22, P = 0.02), 4-methoxyestrone (β = 0.42, P < 0.01), and estriol (β = 0.25, P = 0.04) were positively associated with the fecal microbial Shannon index, and additionally, positive associations were also found for estradiol (β = 0.25, P = 0.01), 17-epiestriol (β = 0.28, P = 0.03), conjugated estrone (β = 0.20, P = 0.01), conjugated estradiol (β = 0.11, P = 0.02), and conjugated estriol (β = 0.21, P = 0.02). However, only 4-methoxyestrone had an FDR adjusted P-value less than 5%. No statistically significant associations were found for fecal microbial observed ASVs and Faith’s PD in both mutually adjusted and individual hormone models. The measured estrogens/estrogen metabolites were not associated with oral microbial alpha-diversity with all global P-values and FDR adjusted P-values greater than 0.05 (Table S1).
TABLE 2.
Association between estrogens and estrogen metabolites with fecal microbial alpha diversity (N = 110)a
| Estrogens and estrogen metabolites | Mutually adjusted modelb |
Individual hormone modelc |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed ASVs |
Faith's PD |
Shannon index |
Observed ASVs |
Faith's PD |
Shannon index |
|||||||
| (global p-value = 0.07) |
(global p-value = 0.22) |
(global p-value < 0.01) |
||||||||||
| β | P value | β | P value | β | P value | β | P value | β | P value | β | P value | |
| Parent Estrogens | ||||||||||||
| Estrone | 14.14 | 0.15 | 0.70 | 0.25 | 0.36 | 0.03 | 10.17 | 0.05 | 0.65 | 0.04 | 0.22 | 0.02 |
| Unconjugated Estrone | — | — | — | — | — | — | 6.33 | 0.40 | 0.39 | 0.39 | 0.19 | 0.15 |
| Conjugated Estrone | — | — | — | — | — | — | 10.31 | 0.02 | 0.67 | 0.01 | 0.20 | 0.01 |
| Estradiol | 1.72 | 0.86 | −0.05 | 0.94 | 0.15 | 0.35 | 10.42 | 0.05 | 0.63 | 0.05 | 0.25 | 0.01 |
| Unconjugated Estradiol | — | — | — | — | — | — | 6.08 | 0.28 | 0.41 | 0.24 | 0.16 | 0.10 |
| Conjugated Estradiol | — | — | — | — | — | — | 5.21 | 0.05 | 0.31 | 0.05 | 0.11 | 0.02 |
| 2-Hydroxylation Pathway Metabolites | ||||||||||||
| 2-Hydroxyestrone | −3.42 | 0.84 | 0.42 | 0.70 | −0.49 | 0.10 | 17.52 | 0.07 | 1.23 | 0.03 | 0.30 | 0.08 |
| 2-Hydroxyestradiol | 11.76 | 0.13 | 0.51 | 0.29 | 0.30 | 0.02 | 6.39 | 0.38 | 0.34 | 0.43 | 0.19 | 0.13 |
| 2-Hydroxyestrone-3-methyl ether | 1.35 | 0.90 | −0.11 | 0.86 | −0.05 | 0.77 | 10.78 | 0.14 | 0.46 | 0.30 | 0.14 | 0.27 |
| 2-Methoxyestrone | −1.28 | 0.90 | −0.12 | 0.84 | 0.15 | 0.35 | 6.67 | 0.33 | 0.43 | 0.30 | 0.19 | 0.11 |
| Unconjugated 2-Methoxyestrone | — | — | — | — | — | — | 0.28 | 0.97 | 0.06 | 0.87 | 0.07 | 0.55 |
| Conjugated 2-Methoxyestrone | — | — | — | — | — | — | 1.55 | 0.68 | 0.13 | 0.57 | 0.06 | 0.34 |
| 2-Methoxyestradiol | -7.52 | 0.47 | -0.27 | 0.68 | -0.11 | 0.52 | −6.49 | 0.42 | −0.26 | 0.58 | −0.07 | 0.64 |
| Unconjugated 2-Methoxyestradiol | — | — | — | — | — | — | −2.83 | 0.75 | −0.27 | 0.62 | 0.01 | 0.95 |
| Conjugated 2-Methoxyestradiol | — | — | to | — | — | — | −5.75 | 0.31 | −0.24 | 0.48 | −0.06 | 0.56 |
| 4-Hydroxylation Pathway Metabolites | ||||||||||||
| 4-Hydroxyestrone | −5.58 | 0.59 | −0.26 | 0.69 | −0.24 | 0.17 | 9.62 | 0.13 | 0.50 | 0.19 | 0.16 | 0.15 |
| 4-Methoxyestrone | 16.07 | 0.15 | 0.67 | 0.33 | 0.51 | 0.01 | 17.99 | 0.02 | 0.89 | 0.06 | 0.42 | <0.01* |
| 4-Methoxyestradiol | 0.67 | 0.95 | −0.30 | 0.66 | 0.14 | 0.45 | −8.07 | 0.35 | −0.74 | 0.16 | −0.09 | 0.55 |
| 16-alpha-Hydroxylation Pathway Metabolites | ||||||||||||
| Estriol | 14.70 | 0.16 | 0.94 | 0.15 | 0.36 | 0.04 | 12.69 | 0.05 | 0.81 | 0.04 | 0.25 | 0.03 |
| Unconjugated Estriol | — | — | — | — | — | — | −4.49 | 0.65 | −0.28 | 0.64 | 0.08 | 0.65 |
| Conjugated Estriol | — | — | — | — | — | — | 10.81 | 0.04 | 0.69 | 0.03 | 0.21 | 0.02 |
| 16-alpha-Hydroxyestrone | −23.19 | 0.02 | −0.99 | 0.10 | −0.57 | <0.01 | −1.59 | 0.76 | −0.01 | 0.97 | −0.01 | 0.91 |
| 16-Ketoestradiol | 8.26 | 0.44 | 0.13 | 0.85 | 0.19 | 0.28 | 5.89 | 0.36 | 0.28 | 0.47 | 0.11 | 0.35 |
| 16-Epiestriol | −4.55 | 0.60 | −0.29 | 0.58 | −0.09 | 0.53 | 7.15 | 0.24 | 0.37 | 0.31 | 0.13 | 0.25 |
| 17-Epiestriol | 15.53 | 0.09 | 0.98 | 0.09 | 0.15 | 0.33 | 18.46 | 0.01 | 0.96 | 0.02 | 0.28 | 0.03 |
Linear regression model that includes all metabolites with additional adjustment for age and BMI; the missing BMI value of one woman was replaced by the median category; ASVs, amplicon sequence variants; Faith's PD, Faith’s phylogenetic diversity.
As the concentration of the 5 conjugated metabolites are calculated as the difference between the combined estrogen measurement and the unconjugated estrogen measurement (e.g., conjugated estrone = estrone − unconjugated estrone), the unconjugated and conjugated analytes were not included in the mutually adjusted model.
The False discovery rate (FDR) adjusted P value was calculated for the individual hormone models to adjust for multiple testing; *FDR adjusted P value ≤0.05; The P values present in the table are all normal P values.
In the beta-diversity analyses, conjugated 2-methoxyestrone was associated with oral microbial unweighted UniFrac (Table S2), but estrogens or estrogen metabolites were not associated with the remaining beta diversity matrices of the fecal and oral microbiome as indicated by MiRKAT models. Consistent results were observed by PERMANOVA, where conjugated 2-methoxyestrone explained 2.67% of the oral microbial variability in unweighted UniFrac (FDR adjusted P-value <0.05) (Fig. S1).
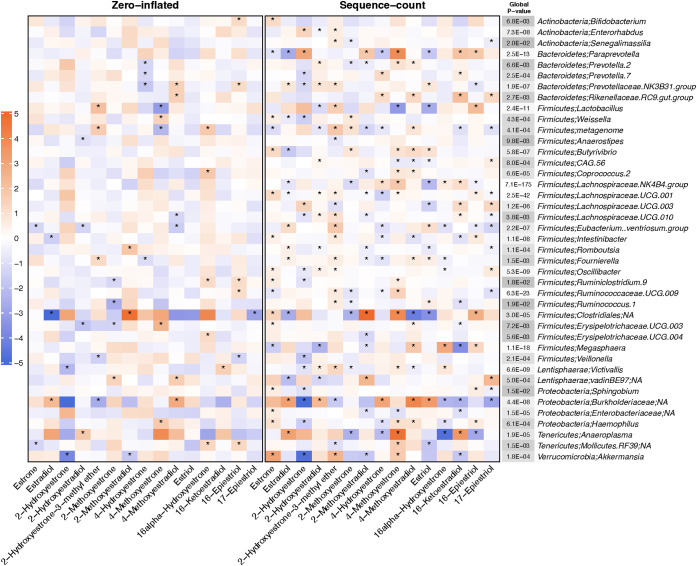
Fig. 1 presents results for the associations of estrogens and estrogen metabolites with fecal genera from the zero-inflated negative binomial regression analysis at an FDR of 5% or less for the global P-value (results for all genera are included in Table S3). Overall, estrogens and estrogen metabolites were significantly associated with 41 fecal genera. Eighteen out of these 41 genera belong to the order Clostridiales, including nine genera from the family Lachnospiraceae and five genera from the family Ruminococcaceae. For the individual associations of the estrogen or estrogen metabolites with these fecal genera, the sequence-count (number of reads for each genus) contributed to more associations with estrogen and estrogen metabolites than the zero-inflated part (presence of each genus).
FIG 1.
Results from the zero-inflated negative binomial regression for the associations of estrogens and estrogen metabolites with fecal genera with an FDR adjusted global P-value less than 0.05. The zero-inflated model indicates the associations for the genus-level presence, and the sequence-count model indicates the associations for the number of reads for each genus. The color of the cells (orange to blue) indicates the strength of the association (model estimates, β). The “*” in the cell indicates that the corresponding P-value is less than 0.05. The color of the “Global P value” column (white to gray) indicates the scale of the global P-value and the exact number in the cell indicates the corresponding global P-value.
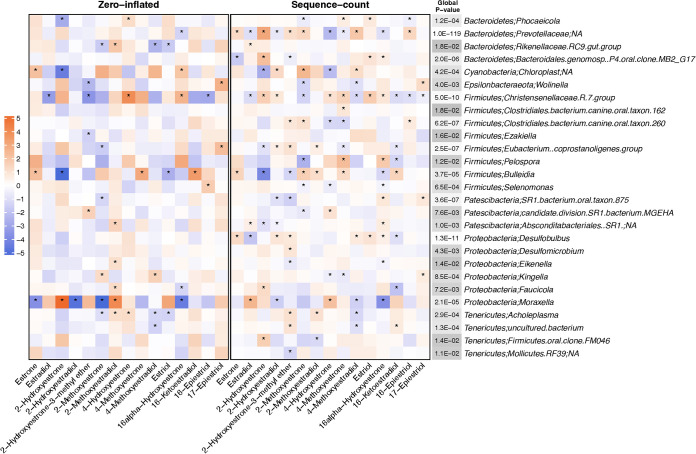
Fig. 2 presents results for the associations of estrogens and estrogen metabolites with oral genera from the zero-inflated negative binomial regression analysis at an FDR of 5% or less for the global P-value (results for all genera are included in Table S4). Estrogens and estrogen metabolites were positively or inversely associated with 27 oral genera. Similar to the fecal microbiome results, the sequence-count also contributed to more associations compared to the zero-inflated part of the model. Of the genus-level associations, the counts for Christensenellaceae.R.7.group were associated with 12 out of the 15 estrogens and estrogen metabolites (sequence-count β ranging from −1.40 to 1.78), while the presence of Moraxella was strongly associated with most estrogens and estrogen metabolites (zero-inflated β ranging from −1.35 to 1.27).
FIG 2.
Results from the zero-inflated negative binomial regression for the associations of estrogens and estrogen metabolites with oral genera with an FDR adjusted global P-value less than 0.05. The zero-inflated model indicates the associations for the genus-level presence, and the sequence-count model indicates the associations for the number of reads for each genus. The color of the cells (orange to blue) indicates the strength of the association (model estimates, β). The “*” in the cell indicates that the corresponding P-value is less than 0.05. The color of the “Global P value” column (white to gray) indicates the scale of the global P-value and the exact number in the cell indicates the corresponding global P-value.
In sensitivity analyses excluding women who had diabetes or used antibiotics within the last 30 days before blood draw, the alpha-diversity associations were similar to those for the overall population. The associations of Shannon index were stronger among women who were menopausal for less than 10 years, whereas the associations of observed ASVs were stronger among women who were menopausal for greater than 10 years (Table S5).
DISCUSSION
In this study of the association of estrogen metabolism and the fecal and oral microbiome in postmenopausal African women in the Ghana Breast Health Study, we found that parent estrogens and several estrogen metabolites, including 2-hydroxyestradiol, 4-methoxyestrone, estriol, and 17-epiestriol, were positively associated with fecal microbial Shannon index (alpha-diversity), while 16alpha-hydroxyestrone was inversely associated with this same measure. In contrast, estrogens and estrogen metabolites were not associated with oral microbial alpha-diversity. Estrogens and estrogen metabolites were not associated with the fecal or oral microbial communities as measured by beta-diversity in general, although conjugated 2-methoxyestrone was associated with oral microbial unweighted UniFrac. Estrogens and estrogen metabolites were associated with the presence and abundance of multiple fecal and oral genera, such as fecal genera from families Lachnospiraceae and Ruminococcaceae, which have estrogen metabolizing properties.
The associations between estrogens and the fecal microbiome have been reported by previous cross-sectional studies. Similar to our results, one Korean study that included 26 women aged 25 to 65 years found that women in a high estradiol group had a greater average fecal microbial Shannon index and Simpson index than the medium and low group, but no significant differences in the richness determined by Chao1 (15), suggesting that estrogen levels may be related to both microbiome richness and evenness (as measured by Shannon index or Simpson index) but not microbiome richness alone (as measured by observed ASVs, Faith’s PD, or Chao1). In contrast, results of another study in Spain that included 15 women with polycystic ovary syndrome, 16 nonhyperandrogenic female and 15 male controls, found an inverse association between serum estradiol and the fecal microbial Shannon Index and Chao1 in a combined analysis of all participants (16). However, these inconsistent results are not surprising since these studies were small and included participants with different health conditions.
A few studies also investigated the associations between estrogen metabolites and the fecal microbiome, but only considered urinary estrogen metabolites. It is difficult to compare our results with these previous studies since estrogen metabolite concentrations have not been observed to be strongly correlated between urine and serum (17). One study conducted within 25 men and 7 postmenopausal women in the United States reported that levels of total urinary estrogens, and most individual estrogen metabolites, including the 16-pathway estrogen metabolites, were positively associated with fecal microbial observed species and the Shannon index. This study also found no association of estrogens and fecal microbial beta-diversity (11). In a cross-sectional study within 60 healthy postmenopausal women enrolled in the Kaiser Permanente of Colorado health plan in the United States, total urinary estrogens and estrogen metabolites tended to nonstatistically significantly increase over quintiles of alpha-diversity measures; the ratio of metabolites to parent estrogen and the ratio of 2- to 16-hydroxylation pathway metabolites showed a statistically significant increasing trend across quintiles of Faith’s PD, but not for the other alpha-diversity measures (12). Another cross-sectional study among 48 postmenopausal breast cancer cases and 48 matched controls also from Kaiser Permanente in Colorado found that Faith’s PD was weakly correlated with total urinary estrogens and the estrogen metabolite to parent estrogen ratio in controls (1, 10).
We found multiple associations of estrogen and estrogen metabolites with the presence and abundance of multiple fecal and oral genera. Among the 100 fecal genera that were included in our analysis, 41 were associated with circulating estrogens and estrogen metabolites based on the FDR of 5% or less for the global P-value, with almost half of these genera (18 out of 41) belonging to the order Clostridiales. In previous studies of urinary estrogen metabolism, the relative abundance of Clostridiales was positively associated with urine estrogen and the ratio of metabolites to parent estrogens among postmenopausal women and men (11, 12). The order Clostridiales has been found to be associated with breast cancer risk (1, 3), where cases had a higher relative abundance of the family Ruminococcaceae and a lower relative abundance of the family Lachnospiraceae. In our study, Ruminococcaceae and Lachnospiraceae were the families that contained the most fecal genera associated with estrogen metabolism, although there was a mix of positive and inverse associations across estrogens and estrogen metabolites and the genera. One of the mechanisms by which the fecal microbiome may be related to breast cancer is through the beta-glucuronidase and beta-glucosidase enzyme activity of some bacteria which enables them to deconjugate conjugated estrogens, leading to their reabsorption into circulation (4, 9). In this study, we found some genera previously noted to contain beta-glucuronidase and/or beta-glucosidase (18), including bacteria belonging to the order Clostridiales such as the genus Coprococcus within the Lachnospiraceae family and the genus Ruminococcus within the Ruminococcaceae family, were associated estrogen metabolism. Also, there were many other bacteria that have not been reported to contain genes encoding beta-glucuronidase and/or beta-glucosidase also associated with estrogen metabolism in our analyses, such as the genus Intestinibacter within the Clostridiales order or the genus Sphingobium within the Sphingomonadales order. Further studies are needed to replicate our findings and better understand the underlying mechanisms for the observed associations.
The oral microbiome has been found to be associated with breast cancer in our previous study, where alpha-diversity was inversely associated with breast cancer, and associations were observed for beta-diversity and multiple taxa (13). However, we did not find associations between estrogens and estrogen metabolites with oral microbial alpha-diversity, and only conjugated 2-methoxyestrone was associated with oral microbial unweighted UniFrac, suggesting that the association between the oral microbial composition and breast cancer may not be related to estrogen metabolism. Some previous studies have suggested that having a history of periodontal disease, a chronic inflammatory condition considered to be caused by periodontal pathogens (19, 20), is associated with breast cancer risk (21, 22). None of the 27 oral genera associated with estrogens and estrogen metabolites belong to the red- or orange-complex of periodontal pathogens (two groups of bacteria strongly associated with periodontal disease) (19), which also suggests that the possible association between periodontal pathogens and breast cancer may be independent of alterations in estrogen metabolism.
Our study has several important strengths. It is the first to investigate the associations between estrogen metabolism with the fecal and oral microbiome in an African population. We obtained data on both the fecal and oral microbiome, and we used a high-performance LC-MS/MS assay to evaluate individual estrogens and estrogen metabolites with high reliability, sensitivity, and specificity. However, limitations of this study should also be noted. First, this study is cross-sectional, therefore we were unable to evaluate the dynamic change of the microbiome and estrogen interrelationships or the causality of their associations (i.e., whether the microbiome plays a role in estrogen metabolism, or microbial changes are a result of circulating estrogen levels). Second, since 16S rRNA gene sequencing is unable to detect the microbial functional genes, this study could not identify whether there are any microbial pathways involved in estrogen metabolism. Finally, the sample size of our study is relatively small and therefore limited our statistical power to identify small differences in associations.
In conclusion, we found positive associations of estrogens and estrogen metabolites, including 2-hydroxyestradiol, 4-methoxyestrone, estriol, and 17-epiestriol, and inverse associations of 16alpha-hydroxyestrone, with the fecal microbiome. Multiple associations with individual genera for both fecal and oral microbiome were also identified. Larger studies are needed to confirm our findings. Longitudinal studies to investigate the dynamic changes of the fecal and oral microbiome and estrogen interrelationships are also needed.
MATERIALS AND METHODS
Study population selection.
The Ghana Breast Health Study has been described in detail previously (14). In brief, breast cancer cases or nonmalignant breast disease cases were identified from three hospitals in Ghana: Korle Bu Teaching Hospital in Accra and Komfo Anoyke Teaching Hospital and Peace and Love Hospital in Kumasi. Population controls were frequency matched by age group to cases and identified using a household census of randomly selected enumeration areas giving rise to the cases. Study subjects had to have resided for at least 1 year in the study areas.
After providing informed consent, participants responded to a standardized interview-based questionnaire that focused on demographic characteristics and breast cancer risk factors, and provided saliva, stool, and blood samples. Previously, 392 breast cancer cases, 100 nonmalignant breast disease cases, and 433 controls with available fecal and oral samples were selected to study the associations of the fecal or oral microbiome with breast cancer and nonmalignant breast disease (3, 13). In addition, blood samples from 635 postmenopausal controls not using exogenous hormones at blood draw were selected to measure the concentrations of the estrogens and their metabolites (23, 24).
In this study, 117 controls with both microbiome (fecal and oral) and circulating estrogen data were included. To be consistent with the rarefaction rate of previous analyses (3, 13), we excluded individuals whose oral samples had <20,000 reads (N = 3) or fecal samples with <6,250 reads (N = 7), which yielded samples sizes of 110 women and 114 women in the fecal and oral microbiome analyses, respectively.
Fecal and oral sample processing and bioinformatics.
The methods for DNA extraction, PCR amplification, and sequencing of the fecal and oral samples were described previously (3, 13). In brief, DNA was extracted from the fecal samples using the MO-BIO PowerMag Soil DNA isolation kit and DNA was extracted from the oral samples using the DSP DNA Virus Pathogen kit (Qiagen) in two separate laboratories. The V4 region of the 16S rRNA gene was PCR amplified for both sample types. On the Illumina MiSeq, 2 × 150 bp paired end sequencing was conducted for the fecal samples and 2 × 250 bp paired end sequencing was conducted for the oral samples.
Sequencing data from both fecal and oral samples were processed using QIIME 2 version 2019.1 (25). Sequence variants were generated using the DADA2 plugin. Taxonomy was assigned to the resulting amplicon sequence variants (ASVs) using SILVA classifier version v132 and relative abundances of taxa were estimated (26). Based on rarefaction curves in the previous analyses (3, 13), we chose a rarefaction rate of 6,250 reads/sample for the fecal microbiome and 20,000 reads/sample for the oral microbiome. Alpha-diversity measures (i.e., observed ASVs, Faith’s Phylogenetic Diversity [PD], and the Shannon index) and beta-diversity measures (i.e., Bray-Curtis, Jaccard, weighted Unifrac, and unweighted Unifrac) were calculated from the rarefied data using QIIME 2 (25).
Serum estrogen assay.
The methods for serum estrogen measurement have been described previously (27–29). Briefly, stable isotope dilution liquid chromatography tandem mass spectrometry (LC-MS/MS) was used to quantify 15 serum estrogens and estrogen metabolites, including estrone, estradiol, 2-hydroxylation pathway metabolites (2-hydroxyestrone, 2-hydroxyestradiol, 2-hydroxyestrone-3-methyl ether, 2-methoxyestrone, and 2-methoxyestradiol); 4-hydroxylation pathway metabolites (4-hydroxyestrone, 4-methoxyestrone, and 4-methoxyestradiol); and 16-alpha-hydroxylation pathway metabolites (estriol, 16alpha-hydroxyestrone, 16-ketoestradiol, 16-epiestriol, and 17-epiestriol,). Five of the estrogens/metabolites (estrone, estradiol, estriol, 2-methoxyestrone, and 2-methoxyestradiol) were also measured in their unconjugated forms, and then the conjugated concentrations of these estrogens/metabolites were calculated by subtracting the unconjugated from the combined concentration. Estrogens and estrogen metabolites were expressed in picomoles per L. Quality control of the serum estrogen assay has been reported elsewhere (23).
Statistical analysis.
Given their right-skewed distributions, the circulating estrogen measures were log-transformed for analysis. Linear regression models were used to calculate the associations between estrogens and estrogen metabolites as the predictor with fecal and oral microbial alpha-diversity as the outcome using two approaches. First, parent estrogens and estrogen metabolites were all analyzed jointly in the same model (i.e., mutually adjusted). Then, individual estrogen/estrogen metabolite levels (including unconjugated and calculated conjugated concentrations) were further evaluated in separate models. A false discovery rate (FDR) was calculated for the individual hormone models to adjust P-values due to multiple testing (N = 25 tests).
For beta-diversity, the association between the estrogens and estrogen metabolites and overall beta-diversity matrices was tested using Microbiome Regression-Based Kernel Association Test (MiRKAT) (30). Permutational Multivariate Analysis of Variance (PERMANOVA) for the beta-diversity matrices was used to calculate the distance-based coefficient of determination, R2, to quantify the percentage of microbial variability explained by estrogens and estrogen metabolites. The FDR adjusted P-value (N = 25 tests) was calculated for both MiRKAT and PERMANOVA.
For the taxonomic analyses, we restricted to genera present in 5% to 80% of the population. A series of zero-inflated negative binomial regression models were used to examine associations of overall estrogens and estrogen metabolites with the presence/absence of (zero-inflated) and number of reads (sequence-count) for specific genera. A global P-value and an FDR adjusted global P-value (N fecal microbiome = 100 tests; N oral microbiome = 67 tests) were calculated for each model. Individual estimates and the corresponding P-values for each estrogen or estrogen metabolite were reported for the genera.
All models were adjusted for age (continuous) and body mass index (BMI, <18.5,18.5 to 24.9, 24.9-29.9, and ≥30 kg/m2). For one woman with missing BMI, the median category was used. Additional adjustment for other potential confounding variables, including study site, year at blood draw, time since menopause, alcohol consumption, diabetes, oral contraceptive use, and antibiotic use did not substantially change the estimates, and therefore only results from the minimally adjusted models are presented. In sensitivity analyses we restricted the study population to participants who never had diabetes, did not use antibiotics within the last 30 days, were menopausal for <10 years or ≥10 years. All statistical analyses were conducted using R version 3.6.2.
Ethics statement.
All participants provided written informed consent. This study was approved by the Special Studies Institutional Review Board of the National Cancer Institute (NCI; Rockville, MD, USA; FWA number 00005897 and IORG number 00010), the Ghana Heath Service Ethical Review Committee and Institutional Review Boards at the University of Ghana Noguchi Memorial Institute for Medical Research (Accra, Ghana; FWA number 00001824 and IORG number 0000908), the Kwame Nkrumah University of Science and Technology (Kumasi, Ghana), and the School of Medical Sciences at Komfo Anokye Teaching Hospital (Kumasi, Ghana).
Data availability.
The sequencing data are available on the Sequence Read Archive (NCBI SRA) under BioProject ID PRJNA658160 for fecal microbiome data and BioProject ID PRJNA767189 for oral microbiome data. Further data are available from Thomas U. Ahearn (thomas.ahearn@nih.gov) upon request.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program in the Division of Cancer Epidemiology and Genetics, the US National Institutes of Health (NIH), National Cancer Institute (NCI). The success of this investigation would not have been possible without exceptional teamwork and the diligence of the field staff who oversaw the recruitment, interviews, and collection of data from study subjects. Special thanks are due to the following individuals: Korle Bu Teaching Hospital, Accra—Adu-Aryee, Obed Ekpedzor, Angela Kenu, Victoria Okyne, Naomi Oyoe Ohene Oti, Evelyn Tay; Komfo Anoyke Teaching Hospital, Kumasi— Marion Alcpaloo, Bernard Arhin, Emmanuel Asiamah, Isaac Boakye, Samuel Ka-chungu and; Peace and Love Hospital, Kumasi—Samuel Amanama, Emma Abaidoo, Prince Agyapong, Thomas Agyei-Ansong, Debora Boateng, Margaret Frempong, Bridget Nortey Mensah, Richard Opoku, and Kofi Owusu Gyimah. The study was further enhanced by surgical expertise provided by Lisa Newman of the University of Michigan and by pathological expertise provided by Stephen Hewitt and Petra Lenz of the National Cancer Institute Maire A. Duggan from the Cumming School of Medicine, University of Calgary, Canada. Study management assistance was received from Ricardo Diaz, Shelley Niwa, Usha Singh, Ann Truelove, and Michelle Brotzman at Westat, Inc. Appreciation is also expressed to the many women who agreed to participate in the study and to provide information and biospecimens in hopes of preventing and improving outcomes of breast cancer in Ghana. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
We have no conflicts of interest to declare.
This work was supported by the Intramural Research Program in the Division of Cancer Epidemiology and Genetics, the US National Institutes of Health (NIH), National Cancer Institute (NCI).
Footnotes
Supplemental material is available online only.
Contributor Information
Emily Vogtmann, Email: emily.vogtmann@nih.gov.
Sangeeta Khare, U. S. Food and Drug Administration.
REFERENCES
- 1.Goedert JJ, Jones G, Hua X, Xu X, Yu G, Flores R, Falk RT, Gail MH, Shi J, Ravel J, Feigelson HS. 2015. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J Natl Cancer Inst 107:djv147. doi: 10.1093/jnci/djv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J, Liao M, Yao Z, Liang W, Li Q, Liu J, Yang H, Ji Y, Wei W, Tan A, Liang S, Chen Y, Lin H, Zhu X, Huang S, Tian J, Tang R, Wang Q, Mo Z. 2018. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome 6:136. doi: 10.1186/s40168-018-0515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrd DA, Vogtmann E, Wu Z, Han Y, Wan Y, Clegg-Lamptey JN, Yarney J, Wiafe-Addai B, Wiafe S, Awuah B, Ansong D, Nyarko K, Hullings AG, Hua X, Ahearn T, Goedert JJ, Shi J, Knight R, Figueroa JD, Brinton LA, Garcia-Closas M, Sinha R. 2021. Associations of fecal microbial profiles with breast cancer and non-malignant breast disease in the Ghana Breast Health Study. Int J Cancer 148:2712–2723. doi: 10.1002/ijc.33473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parida S, Sharma D. 2019. The microbiome-estrogen connection and breast cancer risk. Cells 8:1642. doi: 10.3390/cells8121642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemons M, Goss P. 2001. Estrogen and the risk of breast cancer. N Engl J Med 344:276–285. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- 6.Sampson JN, Falk RT, Schairer C, Moore SC, Fuhrman BJ, Dallal CM, Bauer DC, Dorgan JF, Shu XO, Zheng W, Brinton LA, Gail MH, Ziegler RG, Xu X, Hoover RN, Gierach GL. 2017. Association of estrogen metabolism with breast cancer risk in different cohorts of postmenopausal women. Cancer Res 77:918–925. doi: 10.1158/0008-5472.CAN-16-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samavat H, Kurzer MS. 2015. Estrogen metabolism and breast cancer. Cancer Lett 356:231–243. doi: 10.1016/j.canlet.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raftogianis R, Creveling C, Weinshilboum R, Weisz J. 2000. Estrogen metabolism by conjugation. J Natl Cancer Inst Monogr 2000:113–124. doi: 10.1093/oxfordjournals.jncimonographs.a024234. [DOI] [PubMed] [Google Scholar]
- 9.Kwa M, Plottel CS, Blaser MJ, Adams S. 2016. The intestinal microbiome and estrogen receptor-positive female breast cancer. J Natl Cancer Inst 108:djw029. doi: 10.1093/jnci/djw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goedert JJ, Hua X, Bielecka A, Okayasu I, Milne GL, Jones GS, Fujiwara M, Sinha R, Wan Y, Xu X, Ravel J, Shi J, Palm NW, Feigelson HS. 2018. Postmenopausal breast cancer and oestrogen associations with the IgA-coated and IgA-noncoated faecal microbiota. Br J Cancer 118:471–479. doi: 10.1038/bjc.2017.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flores R, Shi J, Fuhrman B, Xu X, Veenstra TD, Gail MH, Gajer P, Ravel J, Goedert JJ. 2012. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med 10:253. doi: 10.1186/1479-5876-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuhrman BJ, Feigelson HS, Flores R, Gail MH, Xu X, Ravel J, Goedert JJ. 2014. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J Clin Endocrinol Metab 99:4632–4640. doi: 10.1210/jc.2014-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Z, Byrd DA, Wan Y, Ansong D, Clegg-Lamptey JN, Wiafe-Addai B, Edusei L, Adjei E, Titiloye N, Dedey F, Aitpillah F, Oppong J, Vanderpuye V, Osei-Bonsu E, Dagnall CL, Jones K, Hutchinson A, Hicks BD, Ahearn TU, Shi J, Knight R, Biritwum R, Yarney J, Wiafe S, Awuah B, Nyarko K, Figueroa JD, Sinha R, Garcia-Closas M, Brinton LA, Vogtmann E. 2022. The oral microbiome and breast cancer and nonmalignant breast disease, and its relationship with the fecal microbiome in the Ghana Breast Health Study. Int J Cancer 151:1248–1260. doi: 10.1002/ijc.34145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinton LA, Awuah B, Nat Clegg-Lamptey J, Wiafe-Addai B, Ansong D, Nyarko KM, Wiafe S, Yarney J, Biritwum R, Brotzman M, Adjei AA, Adjei E, Aitpillah F, Edusei L, Dedey F, Nyante SJ, Oppong J, Osei-Bonsu E, Titiloye N, Vanderpuye V, Brew Abaidoo E, Arhin B, Boakye I, Frempong M, Ohene Oti N, Okyne V, Figueroa JD. 2017. Design considerations for identifying breast cancer risk factors in a population-based study in Africa. Int J Cancer 140:2667–2677. doi: 10.1002/ijc.30688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin JH, Park YH, Sim M, Kim SA, Joung H, Shin DM. 2019. Serum level of sex steroid hormone is associated with diversity and profiles of human gut microbiome. Res Microbiol 170:192–201. doi: 10.1016/j.resmic.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Insenser M, Murri M, Del Campo R, Martinez-Garcia MA, Fernandez-Duran E, Escobar-Morreale HF. 2018. Gut microbiota and the polycystic ovary syndrome: influence of sex, sex hormones, and obesity. J Clin Endocrinol Metab 103:2552–2562. doi: 10.1210/jc.2017-02799. [DOI] [PubMed] [Google Scholar]
- 17.Coburn SB, Stanczyk FZ, Falk RT, McGlynn KA, Brinton LA, Sampson J, Bradwin G, Xu X, Trabert B. 2019. Comparability of serum, plasma, and urinary estrogen and estrogen metabolite measurements by sex and menopausal status. Cancer Causes Control 30:75–86. doi: 10.1007/s10552-018-1105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dabek M, McCrae SI, Stevens VJ, Duncan SH, Louis P. 2008. Distribution of beta-glucosidase and beta-glucuronidase activity and of beta-glucuronidase gene gus in human colonic bacteria. FEMS Microbiol Ecol 66:487–495. doi: 10.1111/j.1574-6941.2008.00520.x. [DOI] [PubMed] [Google Scholar]
- 19.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Jr, 1998. Microbial complexes in subgingival plaque. J Clin Periodontol 25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 20.Teles R, Teles F, Frias-Lopez J, Paster B, Haffajee A. 2013. Lessons learned and unlearned in periodontal microbiology. Periodontol 2000 62:95–162. doi: 10.1111/prd.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung SD, Tsai MC, Huang CC, Kao LT, Chen CH. 2016. A population-based study on the associations between chronic periodontitis and the risk of cancer. Int J Clin Oncol 21:219–223. doi: 10.1007/s10147-015-0884-6. [DOI] [PubMed] [Google Scholar]
- 22.Freudenheim JL, Genco RJ, LaMonte MJ, Millen AE, Hovey KM, Mai X, Nwizu N, Andrews CA, Wactawski-Wende J. 2016. Periodontal Disease and breast cancer: prospective cohort study of postmenopausal women. Cancer Epidemiol Biomarkers Prev 25:43–50. doi: 10.1158/1055-9965.EPI-15-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geczik AM, Falk RT, Xu X, Ansong D, Yarney J, Wiafe-Addai B, Edusei L, Dedey F, Vanderpuye V, Titiloye N, Adjei E, Aitpillah F, Osei-Bonsu E, Oppong J, Biritwum R, Nyarko K, Wiafe S, Awuah B, Clegg-Lamptey JN, Ahearn TU, Figueroa J, Garcia-Closas M, Brinton LA, Trabert B. 2022. Measured body size and serum estrogen metabolism in postmenopausal women: the Ghana Breast Health Study. Breast Cancer Res 24:9. doi: 10.1186/s13058-022-01500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geczik AM, Falk RT, Xu X, Wiafe-Addai B, Yarney J, Awuah B, Biritwum R, Vanderpuye V, Dedey F, Adjei E, Aitpillah F, Osei-Bonsu E, Oppong J, Titiloye N, Edusei L, Nyarko K, Clegg-Lamptey JN, Wiafe S, Ansong D, Ahearn TU, Figueroa J, Garcia-Closas M, Brinton LA, Trabert B. 2022. Relation of circulating estrogens with hair relaxer and skin lightener use among postmenopausal women in Ghana. J Expo Sci Environ Epidemiol 33:301–310. doi: 10.1038/s41370-021-00407-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Roman JM, Issaq HJ, Keefer LK, Veenstra TD, Ziegler RG. 2007. Quantitative measurement of endogenous estrogens and estrogen metabolites in human serum by liquid chromatography-tandem mass spectrometry. Anal Chem 79:7813–7821. doi: 10.1021/ac070494j. [DOI] [PubMed] [Google Scholar]
- 28.Brinton LA, Trabert B, Anderson GL, Falk RT, Felix AS, Fuhrman BJ, Gass ML, Kuller LH, Pfeiffer RM, Rohan TE, Strickler HD, Xu X, Wentzensen N. 2016. Serum estrogens and estrogen metabolites and endometrial cancer risk among postmenopausal women. Cancer Epidemiol Biomarkers Prev 25:1081–1089. doi: 10.1158/1055-9965.EPI-16-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trabert B, Brinton LA, Anderson GL, Pfeiffer RM, Falk RT, Strickler HD, Sliesoraitis S, Kuller LH, Gass ML, Fuhrman BJ, Xu X, Wentzensen N. 2016. Circulating estrogens and postmenopausal ovarian cancer risk in the Women's Health Initiative Observational Study. Cancer Epidemiol Biomarkers Prev 25:648–656. doi: 10.1158/1055-9965.EPI-15-1272-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao N, Chen J, Carroll IM, Ringel-Kulka T, Epstein MP, Zhou H, Zhou JJ, Ringel Y, Li H, Wu MC. 2015. Testing in microbiome-profiling studies with MiRKAT, the Microbiome Regression-Based Kernel Association Test. Am J Hum Genet 96:797–807. doi: 10.1016/j.ajhg.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download spectrum.01572-23-s0001.pdf, PDF file, 0.08 MB (86.1KB, pdf)
Tables S1 to S5. Download spectrum.01572-23-s0002.xlsx, XLSX file, 0.08 MB (81.9KB, xlsx)
Data Availability Statement
The sequencing data are available on the Sequence Read Archive (NCBI SRA) under BioProject ID PRJNA658160 for fecal microbiome data and BioProject ID PRJNA767189 for oral microbiome data. Further data are available from Thomas U. Ahearn (thomas.ahearn@nih.gov) upon request.