ABSTRACT
This study investigated the resistance mechanisms and the distribution and proportions of virulence genes, including exoU, in 182 imipenem-nonsusceptible Pseudomonas aeruginosa (INS-PA) strains collected from China in 2019. There was no obvious prevalent sequence type or concentrated evolutionary multilocus sequence typing (MLST) type on the INS-PA phylogenetic tree in China. All of the INS-PA isolates harbored β-lactamases with/without other antimicrobial mechanisms, such as gross disruption of oprD and overexpression of efflux genes. Compared with exoU-negative isolates, exoU-positive isolates (25.3%, 46/182) presented higher virulence in A549 cell cytotoxicity assays. The southeast region of China had the highest proportion (52.2%, 24/46) of exoU-positive strains. The most frequent exoU-positive strains belonged to sequence type 463 (ST463) (23.9%, 11/46) and presented multiple resistance mechanisms and higher virulence in the Galleria mellonella infection model. The complex resistance mechanisms in INS-PA and the emergence of ST463 exoU-positive, multidrug-resistant P. aeruginosa strains in southeast China indicated a challenge that might lead to clinical treatment failure and higher mortality.
IMPORTANCE This study investigates the resistance mechanisms and distribution and proportions of virulence genes of imipenem-nonsusceptible Pseudomonas aeruginosa (INS-PA) isolates in China in 2019. Harboring PDC and OXA-50-like genes is discovered as the most prevalent resistance mechanism in INS-PA, and the virulence of exoU-positive INS-PA isolates was significantly higher than that of exoU-negative INS-PA isolates. There was an emergence of ST463 exoU-positive INS-PA isolates in Zhejiang, China, most of which presented multidrug resistance and hypervirulence.
KEYWORDS: Pseudomonas aeruginosa, imipenem nonsusceptible, resistance mechanisms, exoU, ST463
INTRODUCTION
Pseudomonas aeruginosa is one of the most common nosocomial pathogens worldwide. Carbapenem-resistant P. aeruginosa (CRPA) has been listed as a critical priority pathogen by the World Health Organization (WHO), and new antibiotics are urgently required to treat infected patients due to high global mortality (1). In 2020, China Antimicrobial Surveillance Network (CHINET) surveillance data revealed that the percentages of imipenem (IPM)-resistant and meropenem (MEM)-resistant P. aeruginosa isolates were 23.2% and 19.3%, respectively (2). The acquisition of carbapenem-hydrolyzing β-lactamases (such as Klebsiella pneumoniae carbapenemases [KPC], AIM, DIM, GIM, IMP, NDM, SPM, VIM, and OXA), overexpression of chromosome-encoded AmpC β-lactamase, acquisition of extended-spectrum AmpC cephalosporinases (ESACs), reduction of permeability of the outer membrane protein OprD, and overexpression of the major resistance-nodulation-division (RND) efflux pump systems (MexAB-OprM, MexCD-OprJ, MexEF-OprN, and MexXY-OprM) are all involved in carbapenem resistance in P. aeruginosa (3). The most common mechanism of imipenem resistance in P. aeruginosa is a combination of chromosomal AmpC production and a porin change. Indeed, a low level of AmpC enzyme production does not result in high-level carbapenem resistance, due to their limited potential to hydrolyze carbapenem drugs. However, their overproduction, together with reduced outer membrane porin permeability and/or efflux pump overexpression, contributes to high-level carbapenem resistance in this pathogen. P. aeruginosa can also obtain other β-lactamases by horizontal genetic transfer, including extended-spectrum β-lactamases (ESBLs), KPC, VIM, and metallo-β-lactamases (MBLs). The combination of these enzymes leads to high rates of carbapenem resistance in P. aeruginosa isolates (4).
Moreover, many Gram-negative bacteria, including P. aeruginosa, possess type III secretion systems (T3SS), which they utilize to introduce virulence factors directly into host cells. In P. aeruginosa, T3SS transport four secreted factors, exoU, exoS, exoY, and exoT. However, all of these factors may not be common to all P. aeruginosa strains. The exoU gene encodes a cytotoxic protein that rapidly destroys the cell membranes of mammalian cells by using its phospholipase activity (5). These virulence factors play important roles that may be involved in the genesis of acute lung injury, bacteremia, sepsis, and invasion of tissues. P. aeruginosa possesses a T3SS virulence mechanism (6).
Both carbapenem resistance and hypervirulence would probably result in poor clinical outcomes for infected patients. Therefore, this research is aimed at investigating the resistance mechanisms and distribution and proportions of virulence genes using whole-genome-sequencing techniques.
RESULTS
General information on P. aeruginosa isolates in 2019.
Of the isolates collected in 2019, 35 strains were proved to be contaminated in the process of whole-genome sequencing (WGS) and were not included in this analysis. The final 522 P. aeruginosa isolates that were taken into consideration were collected from intraabdominal tract infections (IAIs) (57/522), urinary tract infections (UTIs) (45/522), respiratory tract infections (RTIs) (374/522), and bloodstream infections (BSIs) (46/522) in 6 regions of China: central (45/522), east (104/522), northeast (63/522), south (77/522), southeast (153/522), and southwest (80/522). There were 340 isolates (patient age, 60.4 ± 18.7 years [mean ± standard deviation]) that were susceptible to imipenem and 182 isolates (patient age, 64.4 ± 16.8 years) that were nonsusceptible to imipenem. For imipenem-susceptible P. aeruginosa (IS-PA) isolates, 237 isolates were collected from males and 103 from females. For imipenem-nonsusceptible P. aeruginosa (INS-PA), 139 isolates were collected from males and 43 from females.
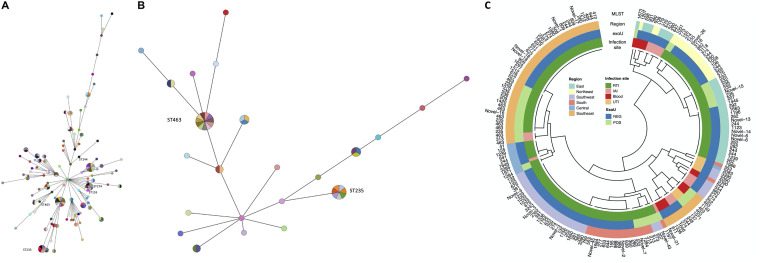
According to the WGS results, there were 107 multilocus sequence typing (MLST) types detected among the 182 INS-PA isolates, with sequence type 463 (ST463) being the most frequently isolated (11/182), followed by ST244 (9/182), ST235 (7/182), and ST274 (7/182). The isolates did not present a concentrated evolutionary MLST type on the phylogenetic tree (Fig. 1A). There was no obvious prevalent sequence type in China in INS-PA isolates currently. Detailed information (STs, virulence genes, different regions, and infections) on all INS-PA isolates is shown in Fig. 1C.
FIG 1.
Phylogenetic trees of STs of all of the strains (A) and of exoU-positive strains (B) and of STs, virulence genes, different regions, and infection sites (C). (A and B) Minimum spanning trees of STs as determined by MLST, colored by strains. The size of each node reflects the number of isolates contained within the same clade. Each color represents one isolate. (C) MLST types of all of the 182 INS-PA isolates are listed outside the circle. MLST types, regions, exoU-positive or -negative status, and infection sites are marked with different colors from outside to inside. MLST, multilocus sequence typing; IAI, intra-abdominal tract infection; RTI, respiratory tract infection; UTI, urinary tract infection; NEG, negative; POS, positive.
Antimicrobial susceptibility of P. aeruginosa isolates.
The antimicrobial susceptibilities of tested strains to common antibacterial agents are shown in Table 1. For IS-PA isolates, all of the antibacterial agents tested showed >70% susceptibility, except for piperacillin-tazobactam (TZP) and aztreonam (ATM). The rate of susceptibility of IS-PA isolates to aztreonam was 69.1%. Lower rates of susceptibility (<70%) were found for INS-PA isolates for all of the antibacterial agents tested, except for ceftolozane-tazobactam (C/T), amikacin (AMK), and tobramycin (TOB). However, INS-PA isolates from RTIs had higher rates of resistance to various antibacterial agents than INS-PA isolates from other infection sites on a numerical basis.
TABLE 1.
Susceptibility rates and MIC distributions of common antibacterial agents against INS-PA and IS-PA isolates by infection site
| Group of isolates, druga | Value forb: |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All indicated isolates |
Indicated isolates from: |
||||||||||||||
| BSI |
IAI |
RTI |
UTI |
||||||||||||
| S (%) | MIC50 | MIC90 | S (%) | MIC50 | MIC90 | S (%) | MIC50 | MIC90 | S (%) | MIC50 | MIC90 | S (%) | MIC50 | MIC90 | |
| INS-PA | n = 182 | n = 14 | n = 18 | n = 142 | n = 8 | ||||||||||
| Ceftazidime | 46.7 | 16 | >32 | 50 | 8 | >32 | 56.3 | 8 | >32 | 44.4 | 16 | >32 | 50 | 8 | >32 |
| Cefepime | 51.6 | 8 | >32 | 57.1 | 8 | >32 | 56.3 | 8 | >32 | 50 | 8 | >32 | 50 | 8 | >32 |
| Piperacillin-Tazobactam | 24.7 | >64 | >64 | 21.4 | >64 | >64 | 18.8 | >64 | >64 | 23.9 | >64 | >64 | 37.5 | >64 | >64 |
| Ceftolozane-Tazobactam | 75.3 | 1 | >32 | 78.6 | 1 | >32 | 87.5 | 1 | >32 | 74.6 | 1 | >32 | 50 | 1 | >32 |
| Meropenem | 20.3 | 8 | >32 | 28.57 | 8 | >32 | 25 | 8 | 16 | 18.31 | 8 | >32 | 12.5 | 4 | >32 |
| Aztreonam | 30.2 | >16 | >16 | 21.4 | >16 | >16 | 31.3 | 16 | >16 | 31 | >16 | >16 | 12.5 | >16 | >16 |
| Levofloxacin | 31.3 | 2 | >4 | 21.4 | 2 | >4 | 43.8 | 2 | >4 | 31 | 4 | >4 | 12.5 | >4 | >4 |
| Amikacin | 86.3 | <4 | >32 | 100 | <4 | 8 | 93.8 | <4 | 16 | 85.2 | <4 | >32 | 62.5 | 8 | >32 |
| Tobramycin | 84.1 | <0.5 | >8 | 100 | <0.5 | 1 | 87.5 | <0.5 | >8 | 83.1 | 1 | >8 | 62.5 | 1 | >8 |
| Colistin | 81.3 | 2 | 4 | 64.3 | 2 | 4 | 93.8 | 2 | 2 | 81.7 | 2 | 4 | 87.5 | 2 | 4 |
| IS-PA | n = 340 | n = 32 | n = 39 | n = 232 | n = 37 | ||||||||||
| Ceftazidime | 75.6 | 4 | >32 | 90.6 | 4 | 8 | 76.9 | 4 | 32 | 72.4 | 4 | >32 | 81.1 | 4 | 32 |
| Cefepime | 84.1 | 4 | 16 | 96.9 | 2 | 8 | 87.2 | 4 | 16 | 80.6 | 4 | 16 | 91.9 | 4 | 8 |
| Piperacillin-Tazobactam | 58.8 | 8 | >64 | 81.3 | 8 | >64 | 64.1 | 8 | >64 | 53.4 | 8 | >64 | 67.6 | 8 | >64 |
| Ceftolozane-Tazobactam | 94.7 | 1 | 4 | 100 | 0.5 | 1 | 94.9 | 1 | 2 | 93.5 | 1 | 4 | 97.3 | 1 | 1 |
| Meropenem | 93.8 | 0.5 | 2 | 0.5 | 1 | <0.25 | 0.5 | 4 | <0.25 | 0.5 | 2 | <0.25 | 0.5 | 2 | <0.25 |
| Aztreonam | 69.1 | 8 | >16 | 90.6 | 8 | 8 | 69.2 | 8 | >16 | 63.8 | 8 | >16 | 83.8 | 8 | 16 |
| Levofloxacin | 77.9 | <0.5 | 4 | 90.6 | <0.5 | 1 | 89.7 | <0.5 | 2 | 73.7 | <0.5 | 4 | 81.1 | <0.5 | >4 |
| Amikacin | 99.4 | <4 | 8 | 100 | <4 | <4 | 100 | <4 | <4 | 99.1 | <4 | 8 | 100 | <4 | 8 |
| Tobramycin | 98.2 | <0.5 | 1 | 100 | <0.5 | 1 | 100 | <0.5 | 1 | 97.8 | <0.5 | 1 | 97.3 | <0.5 | 1 |
| Colistin | 82.4 | 2 | 4 | 81.3 | 2 | 4 | 89.7 | 2 | 4 | 81.5 | 2 | 4 | 81.1 | 2 | 4 |
INS-PA, imipenem-nonsusceptible Pseudomonas aeruginosa; IS-PA, imipenem-susceptible Pseudomonas aeruginosa.
S, susceptible; BSI, bloodstream infection; IAI, intraabdominal tract infection; RTI, respiratory tract infection; UTI, urinary tract infection.
Resistance mechanism proportions and antimicrobial resistance gene distributions of INS-PA isolates by region.
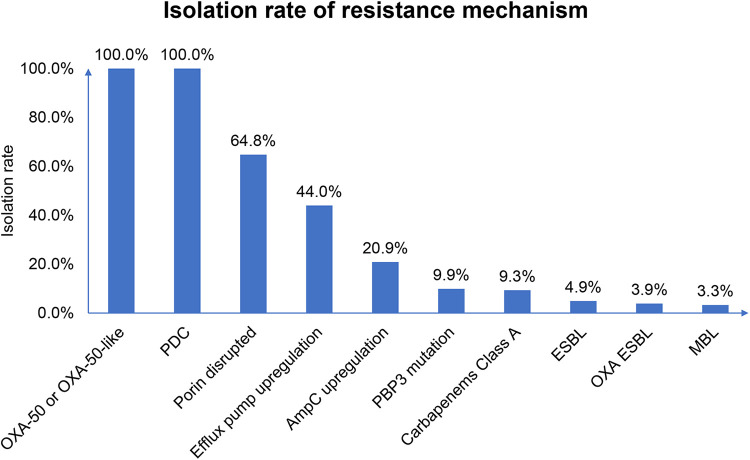
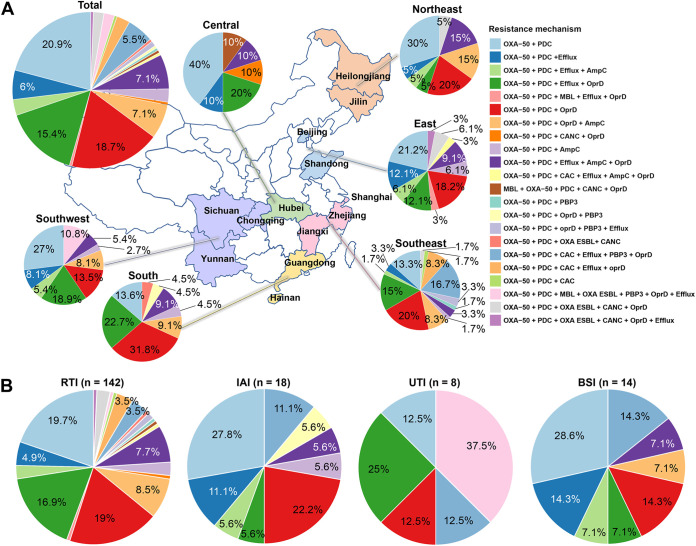
Antimicrobial resistance (AMR) gene analysis showed that all of the INS-PA isolates harbored blaPDC and blaOXA-50 or blaOXA-50-like (Fig. 2). In addition, 79.1% (144/182) of INS-PA strains harbored other AMR genes, such as blaKPC-2, oprD with a gross disruption (oprD gross disruption), or mexR or nalD loss/truncation, which indicated efflux upregulation combined with AMR genes blaPDC and blaOXA-50 or blaOXA-50-like. Except for the generally encoded combination of blaPDC and blaOXA-50 or blaOXA-50-like (PDC+OXA-50/OXA-50-like) (38/182, 20.9%), the most common resistance mechanism combination was blaPDC, blaOXA-50-like, and oprD gross disruption (PDC+OXA-50-like+oprD gross disruption) (34/182, 18.7%), followed by PDC+OXA-50-like+oprD gross disruption+efflux upregulation (28/182, 15.4%). The distributions and proportions, however, were different in each region (Fig. 3). For the southeast region, 16.7% (10/60) of isolates had AMR genes comprising class A carbapenemase or ESBL genes (blaKPC-2, blaGES-1, or blaGES-5), blaPDC, blaOXA-50, or blaOXA-50-like, penicillin-binding protein 3 gene with mutation (PBP3 gene mutation), efflux upregulation genes, and oprD gross disruption simultaneously.
FIG 2.
Proportion of each type of resistance mechanism. WGS was conducted to detect AMR and resistance mechanisms, while rates of upregulation were determined from publications in the literature that have demonstrated the loss or mutation of certain regulatory genes that cause upregulation. ESBL, extended-spectrum β-lactamase; OXA ESBL, OXA-10 or OXA-10-like; MBL, metallo β-lactamase.
FIG 3.
Distribution of resistance mechanisms by region (A) and infection site (B). Each color represents a combination of different kinds of resistance mechanisms. MBL, metallo β-lactamase; CAC, class A carbapenemase; OXA ESBL, OXA-10 or OXA-10-like; CANC, class A noncarbapenemase; RTI, respiratory tract infection; IAI, intraabdominal tract infection; UTI, urinary tract infection; BSI, bloodstream infection.
Virulence gene analysis.
Among 182 INS-PA isolates, there were 46 strains that were exoU positive (25.3%). The distributions of exoU-positive isolates exhibited significant differences between regions (Table 2). The southeast region had the highest proportion and isolation rate of exoU-positive strains, which was significantly different from the other regions (P = 0.001). The proportion of exoU-positive isolates with OXA-50-like+PDC+class A carbapenemase+efflux regulation+oprD gross disruption with or without the PBP3 gene mutation was significantly higher than that of exoU-negative isolates (P ≤ 0.001) (Table 3).
TABLE 2.
Comparison of characteristics between exoU-positive and exoU-negative isolates
| Characteristica | No. (%) of isolates that were: |
P value | |
|---|---|---|---|
| exoU negative | exoU positive | ||
| n = 136 | n = 46 | ||
| Patient age (yrs) | |||
| <18 | 2 (1.47) | 1 (2.17) | |
| ≥18 | 134 (98.53) | 45 (97.83) | 1.000 |
| Patient gender | |||
| Female | 33 (24.26) | 10 (21.7) | |
| Male | 103 (75.74) | 36 (78.3) | 0.730 |
| Source | |||
| BSI | 10 (7.35) | 4 (8.70) | 1.000 |
| IAI | 12 (8.82) | 4 (8.70) | 1.000 |
| RTI | 107 (78.68) | 35 (76.09) | 0.714 |
| UTI | 5 (3.68) | 3 (6.52) | 0.691 |
| Region | |||
| Central | 8 (5.88) | 2 (4.35) | 0.984 |
| East | 24 (17.65) | 9 (19.57) | 0.770 |
| Northeast | 18 (13.24) | 2 (4.35) | 0.096 |
| South | 16 (11.76) | 6 (13.04) | 0.818 |
| Southeast | 36 (26.47) | 24 (52.17) | 0.001 |
| Southwest | 34 (25) | 3 (6.52) | 0.007 |
| Department | |||
| Emergency room | 7 (5.15) | 2 (4.35) | 1.000 |
| ICU, general, unspecified | 12 (8.82) | 4 (8.70) | 1.000 |
| Medicine, general | 56 (41.18) | 14 (30.43) | 0.195 |
| Medicine, ICU | 13 (9.56) | 9 (19.57) | 0.072 |
| None given | 2 (1.47) | 0 (0) | 0.993 |
| Pediatric, general | 1 (0.74) | 0 (0) | 1.000 |
| Surgery, general | 34 (25.00) | 13 (28.26) | 0.662 |
| Surgery, ICU | 11 (8.09) | 4 (8.70) | 1.000 |
BSI, bloodstream infection; IAI, intraabdominal tract infection; RTI, respiratory tract infection; UTI, urinary tract infection; ICU, intensive care unit.
TABLE 3.
Comparison of resistance mechanisms between exoU-positive and exoU-negative INS-PA isolatesa
| Resistance mechanisms | Value for isolates that were (n = 182): |
P value | |||
|---|---|---|---|---|---|
|
exoU positive (n = 46) |
exoU negative (n = 136) |
||||
| No. | % | No. | % | ||
| OXA-50 + PDC | 4 | 8.7 | 34 | 25.0 | 0.019 |
| OXA-50 + PDC + efflux | 3 | 6.5 | 8 | 5.9 | 0.875 |
| OXA-50 + PDC + efflux + ampC | 0 | 0.0 | 6 | 4.4 | 0.332 |
| OXA-50 + PDC + efflux + oprD | 6 | 13.0 | 22 | 16.2 | 0.661 |
| OXA-50 + PDC + MBL + efflux + oprD | 1 | 2.2 | 0 | 0.0 | 0.568 |
| OXA-50 + PDC + oprD | 7 | 15.2 | 27 | 19.9 | 0.486 |
| OXA-50 + PDC + oprD + ampC | 0 | 0.0 | 13 | 9.6 | 0.065 |
| OXA-50 + PDC + ESBL + OprD | 0 | 0.0 | 1 | 0.7 | 1.000 |
| OXA-50 + PDC + ampC | 0 | 0.0 | 5 | 3.7 | 0.425 |
| OXA-50 + PDC + efflux + ampC + oprD | 3 | 6.5 | 10 | 7.4 | 0.850 |
| OXA-50 + PDC + class A carbapenemase + efflux + ampC + oprD | 1 | 2.2 | 0 | 0.0 | 0.568 |
| OXA-50 + PDC + MBL + ESBL + oprD | 0 | 0.0 | 1 | 0.7 | 1.000 |
| OXA-50 + PDC + PBP3 | 0 | 0.0 | 1 | 0.7 | 1.000 |
| OXA-50 + PDC + oprD + PBP3 | 1 | 2.2 | 0 | 0.0 | 0.568 |
| OXA-50 + PDC + oprD + PBP3 + efflux | 2 | 4.3 | 0 | 0.0 | 0.104 |
| OXA-50 + PDC + OXA ESBL + ESBL | 1 | 2.2 | 0 | 0.0 | 0.568 |
| OXA-50 + PDC + class A carbapenemase + efflux + PBP3 + oprD | 10 | 21.7 | 0 | 0.0 | <0.001 |
| OXA-50 + PDC + class A carbapenemase + efflux + oprD | 5 | 10.9 | 0 | 0.0 | 0.001 |
| OXA-50 + PDC + class A carbapenemase | 0 | 0.0 | 1 | 0.7 | 1.000 |
| OXA-50 + PDC + MBL + OXA ESBL + PBP3 + OprD + efflux | 0 | 0.0 | 4 | 2.9 | 0.552 |
| OXA-50 + PDC + OXA ESBL + ESBL + oprD | 2 | 4.3 | 2 | 1.5 | 0.569 |
| OXA-50 + PDC + OXA ESBL + ESBL + oprD + efflux | 0 | 0.0 | 1 | 0.7 | 1.000 |
INS-PA, imipenem-nonsusceptible Pseudomonas aeruginosa.
Virulence of exoU-positive strains.
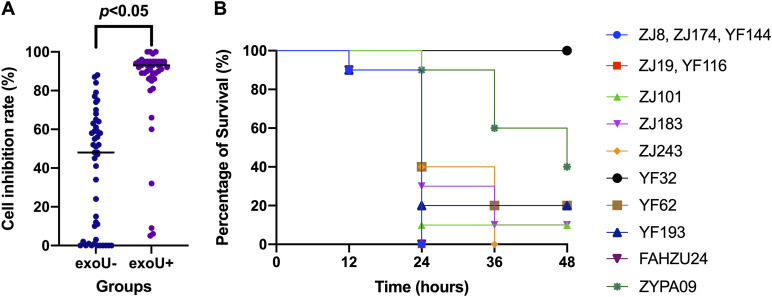
The cytotoxicity assay (Fig. 4) showed that the cell inhibition rates of exoU-negative strains were significantly lower than those of exoU-positive strains (P < 0.05). Most cell inhibition rates of exoU-positive strains were >92% (the inhibition rate of the hypervirulent control strain, FAHZU24), and some even reached 100%. The cell inhibition rates of exoU-negative strains were mostly <78% (the inhibition rate of the hypovirulent control strain, ZYPA09), with some even reduced to 0%.
FIG 4.
Virulence tests of ST463 strains with or without exoU. (A) Cell inhibition rates of 46 exoU-positive and 46 exoU-negative strains in A549 human pulmonary adenocarcinoma cell cytotoxicity assays. (B) Probability of survival of larvae infected with 11 ST463 exoU-positive strains from Zhejiang in the Galleria mellonella larva infection model. Strain FAHZU24 was the hypervirulent control, and strain ZYPA09 the hypovirulent control.
Virulence and resistance analysis of ST463 strains.
All of the ST463 strains (n = 11) collected from Zhejiang province were exoU positive. Six of the 11 ST463 strains were collected from intensive care units (ICUs), and the patient ages ranged from 42 to 72 years (Table 4). Two of the 11 ST463 strains were collected from blood samples, which was significantly higher than for non-ST463 strains (12/171, P < 0.05). All of these ST463 isolates harbored blaOXA-486, blaPDC-8 combined with PBP3 gene mutation, oprD gross disruption, and efflux pump upregulation, and 10 of them harbored blaKPC-2 (Table 4). The imipenem MICs of the 11 strains were all >32 μg/mL, presenting high-level resistance to imipenem.
TABLE 4.
Characteristics of the 11 ST463 strains
| Strain | Patient |
Sourcea | Department | Beta-lactamases | PBP3 mutation | AmpC upregulation | oprD | Efflux pump upregulation | |
|---|---|---|---|---|---|---|---|---|---|
| Age | Gender | ||||||||
| 1 | 66 | Male | RTI | Surgery, ICU | OXA-486, PDC-8 | F533Lb | No | Gross disruption | Yes (mexZ, mexXY) |
| 2 | 48 | Female | IAI | Surgery, general | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes (mexZ, mexXY) |
| 3 | 74 | Male | RTI | Medicine, ICU | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes (mexZ, mexXY) |
| 4 | 44 | Male | BSI | Medicine, general | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes (mexZ, mexXY) |
| 5 | 72 | Female | UTI | Medicine, ICU | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes (mexZ, mexXY) |
| 6 | 59 | Male | RTI | Medicine, ICU | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes (mexZ, mexXY) |
| 7 | 54 | Male | RTI | Surgery, ICU | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes, (mexZ, mexXY) |
| 8 | 72 | Female | RTI | Emergency room | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes, (mexZ, mexXY) |
| 9 | 56 | Male | RTI | Medicine, ICU | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes, (mexZ, mexXY) |
| 10 | 42 | Female | IAI | Surgery, general | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes, (mexZ, mexXY) |
| 11 | 69 | Female | BSI | Medicine, general | KPC-2, OXA-486, PDC-8 | F533L | No | Gross disruption | Yes, (mexZ, mexXY) |
RTI, respiratory tract infection; IAI, intraabdominal tract infection; BSI, bloodstream infection; UTI, urinary tract infection.
F533L, a change of F to L at position 533.
In the Galleria mellonella larva infection model, the virulence of 63.6% (7/11) ST463 exoU-positive strains showed no significant difference (P < 0.05) from that of the hypervirulent control strain, FAHZU24 (Fig. 4), and that of 36.4% (4/11) showed significant differences (P < 0.05) from the virulence of both FAHZU24 and the hypovirulent control, ZYPA09. In all, 63.6% (7/11) of ST463 exoU-positive strains were as virulent as the hypervirulent control, FAHZU24, and 27.3% (3/11) showed medium virulence (between that of the hypervirulent control, FAHZU24, and the hypovirulent control, ZYPA09). Only 9.1% (1/11) of the ST463, exoU-positive strains showed hypovirulence similar to that of the hypovirulent control, ZYPA09.
Most of the ST463 exoU-positive strains collected from Zhejiang presented carbapenem resistance and hypervirulence. However, the evolutionary relationship was not obviously concentrated, based on the phylogenetic tree (Fig. 1B).
DISCUSSION
The resistance mechanisms of INS-PA are complex and associated with several AMR genes or resistance mechanisms. Because all of the isolates in this research were imipenem-nonsusceptible strains rather than meropenem- or ertapenem-nonsusceptible isolates, the proportion of porin loss was higher than that of efflux pump upregulation numerically. The rate of metallo-β-lactamase (MBL)-harboring isolates was low, and most of the MBL-harboring isolates were collected from the southwest (4/6).
The sequence type (ST) distribution varied from region to region. ST235 has been reported as the most prevalent sequence type in single-center research conducted in the southwest over a 10-year period (7). It is noteworthy that ST235 was not detected in the southwest in the present study but was found in 4 isolates from the southeast and 3 from the east. ST235 with the hypervirulence gene is the most prevalent sequence type, with clones categorized as high-risk and widespread associated with poor clinical outcomes. In part, this is due to multilevel and high-level antibiotic resistance (8). In the present research, the ST463 strains that were isolated from Zhejiang Province had the highest proportion. Other research from Zhejiang also reported that ST463 with coexistence of exoS and exoU was the most prevalent ST type (9) and presented both multidrug resistance (MDR) and hypervirulence (10). Ten of the 11 ST463 isolates in this research harbored blaKPC-2, resulting in high MICs for most of the β-lactam antibiotics tested, such as ceftazidime (CAZ) (>32 mg/L), cefepime (FEP) (>32 mg/L), piperacillin-tazobactam (>64 mg/L), ceftolozane-tazobactam (>32 mg/L), and meropenem (>32 mg/L, except for one isolate). As there was no obvious evolutionary relationship observed, the ST463 strains collected from the same place tend to be the prevalent type rather than outbreak strains. The clone of ST463 should be noted in clinical practice as high risk, because it contains both the virulence gene and AMR genes.
The present study found that there was no prevalent ST clone of INS-PA nationwide. Therefore, INS-PA is not spreading as a resistant clone in China. However, it should be pointed out that the higher proportions of exoU-positive isolates and ST463 strains in the southeast region could indicate that a resistant clone is spreading within certain regions. This finding was different from reported results from other countries, which had prevalent clones of resistant or high-risk P. aeruginosa, such as ST111, ST175, and ST235 (11). For example, ST175 is the most frequent extremely drug-resistant (XDR) high-risk clone detected in Spanish hospitals (12). However, the emergence of ST463 exoU-positive, multidrug-resistant P. aeruginosa strains in east China also indicates a challenge that may lead to failures of clinical treatment and a higher mortality rate. One retrospective cohort study in eastern China found that ST463 was predominant (48.0%) among 50 CRPA BSI cases and that the 28-day mortality was significantly higher for ST463 cases than for non-ST463 cases (66.7% versus 33.3%, P = 0.03) (13, 14). The reason given for the presence of ST463 with poorer outcomes was explained in the publication (14). Infections related to exoU-producing strains have been found to be associated with more severe clinical symptoms and poorer outcomes than infections caused by exoS-positive (exoU-negative) isolates (6). Moreover, drug-resistant P. aeruginosa strains, especially CRPA, contributed to poorer clinical outcomes simultaneously. One meta-analysis demonstrated a >2-fold-increased risk of mortality with multidrug-resistant P. aeruginosa (MDR-PA) (relative risk [RR], 2.34; 95% confidence interval [CI], 1.53 to 3.57) and a prolonged length of hospitalization compared to the risk of mortality and length of hospitalization from infections with susceptible P. aeruginosa strains (15). Thus, exoU and exoS virulence genes coexisting with the blaKPC resistance gene in ST463 CRPA may be an important intrinsic cause of the poor prognosis of clinical P. aeruginosa BSIs (14). Therefore, infections caused by hypervirulent and carbapenem-resistant organisms have clinical and economic consequences. Hospital-acquired resistant and MDR P. aeruginosa infections will probably result in poorer clinical outcomes, and it will be necessary to rigorously monitor patients in clinical practice.
The INS-PA strains were isolated from 13 provinces, which may not represent the whole of China, because both sequencing types and resistance mechanisms are different across regions. The IS-PAs were not tested by WGS, which may make the relationship between genotype and phenotype unreliable.
MATERIALS AND METHODS
Isolates from SMART in 2019.
All of the P. aeruginosa isolates were collected during the Study for Monitoring Antimicrobial Resistance Trends (SMART) (16), which isolated pathogens from abdominal, urinary tract, blood, and respiratory tract specimens of patients from 16 hospitals in six regions (central, east, northeast, south, southeast, and southwest) across China in 2019. The central clinical microbiology laboratory of the Peking Union Medical College Hospital reidentified all of the isolates using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Vitek MS; bioMérieux, France). The Human Research Ethics Committee of Peking Union Medical College Hospital approved the study protocols (no. S-K238).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed at the Peking Union Medical College Hospital central laboratory using panels purchased from Thermo Fisher Scientific (Cleveland, OH, USA). MICs were interpreted using the CLSI breakpoints (17) (all antimicrobial agents except colistin [COL]) or the EUCAST breakpoint (colistin) (18).
The antimicrobial agents tested included imipenem (IPM), meropenem (MEM), ceftazidime (CAZ), cefepime (FEP), piperacillin-tazobactam (TZP), ceftolozane-tazobactam (C/T), aztreonam (ATM), levofloxacin (LVX), amikacin (AMK), tobramycin (TOB), and colistin (COL). P. aeruginosa ATCC 27853 was used as the quality control (QC) strain for each batch of MIC tests.
Cytotoxicity assay.
Cytotoxicity assays using A549 human pulmonary adenocarcinoma cells were conducted on all 46 exoU-positive strains and 46 exoU-negative strains selected according to region to evaluate the virulence. The high-cytotoxicity control was the exoU-positive/exoS-negative, ST235 Pseudomonas aeruginosa strain FAHZU24, and the low-cytotoxicity control strain was the exoU-negative/exoS-positive, ST236 strain ZYPA09. The cells were cultured in F-12K medium with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. Amounts of 100 μL of fresh medium with 6 × 103 cells/well were plated in 96-well plates and cultured for 24 h. Then, overnight cultures of single colonies from agar plates were diluted 103 times (3 × 106 CFU/mL) with F-12K medium (including 10% FBS). One hundred microliters diluted bacterial culture was added into each well (multiplicity of infection of 50) and cultured at 37°C with 5% CO2 for 3 h. Then, the supernatant was discarded and the cells washed with 100 μL fresh medium. Finally, 100 μL fresh F-12K medium (including 10% FBS) and 10 μL cell counting kit-8 (CCK-8) solution was added. The optical density at 450 nm (OD450) was detected after 2.5 h of culture. Each group had 6 compound wells. The inhibition rate was calculated as follows: (Acontrol − Aexperiment)/(Acontrol − Ablank) × 100, where A is absorbance. A higher inhibition rate indicated stronger cytotoxicity. GraphPad Prism 9 software was employed for statistical analysis, and a P value of <0.05 was considered significant.
Galleria mellonella larva infection model.
The virulence of exoU-positive ST463 strains was also tested by the Galleria mellonella larva infection model. Normal saline was used to adjust the bacterial suspension to 1 × 106 CFU/mL, and 10 μL bacterial suspension was injected into each larva (n = 10 larvae/strain). Then, the larvae were incubated at 35°C and the number of surviving larvae was recorded once every 12 h for 48 h. Strain FAHZU24 is referred to as the hypervirulent control, and ZYPA09 as the hypovirulent control (10).
WGS.
All imipenem-nonsusceptible isolates were sent for whole-genome sequencing (WGS). Bacteria cultured to stationary phase from single colonies were pelleted in 1× Tris-EDTA buffer. DNA isolation used magnetic bead chemistry, and library preparation used the in-house method of Beijing Genomics Institute (BGI, Wuhan, China). Libraries were sequenced on a high-throughput Illumina sequencer in a 2 × 150-bp paired-end configuration to a calculated coverage depth of ×100.
WGS analysis.
The CLC Genomics Workbench (Qiagen) was used for WGS analysis. On acquisition of fastq files, reads were trimmed for quality and adapter sequences, sampled to approximately ×100 coverage depth, and assembled de novo for downstream analysis. Genes encoding β-lactamases were identified by screening assemblies using the ResFinder database (https://cge.food.dtu.dk/services/ResFinder/), downloaded on 9 July 2020. Coverage and identity cutoffs were set to ≥35% and ≥72%, respectively, though any positively identified antimicrobial resistance gene that was <100% for either parameter was confirmed by read mapping and/or examination at the amino acid level to identify a β-lactamase variant. Various genes of interest were analyzed for gross disruptions or previously characterized mutations by pairwise alignment to a reference sequence. oprD was tested for permeability, and the PBP3 gene (ftsI) was tested for target mutation. For detecting ampC regulation, ampD, ampDh2, ampDh3, dacB (pbp4), mpl, nuoN, and ampR were analyzed. To detect efflux regulation, nalD, mexR, nalC, and mexZ were analyzed. The exoU gene was tested for hypervirulence (19–29). Multilocus sequence typing determination was performed using CLC Genomics with schema downloaded from Pubmlst.org on 4 November 2020. The phylogenetic tree was constructed based on MLST of each isolate using GrapeTree (version 1.4.0, https://github.com/achtman-lab/GrapeTree) and clustered based on MLST, region, exoU-positive/-negative status, and infection site using the circlize package in R Studio (version 4.0.5).
Statistical analysis.
SPSS (version 17.0; IBM) was used for all statistical analysis. Descriptive analysis was performed to calculate the susceptibility and proportion of each resistance mechanism. Student’s t test was performed for the cytotoxicity assay for comparison between the cell inhibition rates of the exoU-positive group and the exoU-negative group. The Gehan-Breslow-Wilcoxon test was performed on the survival curves of clinical strains and control strains. Comparison of the resistance mechanisms and clinical characteristics between the exoU-positive and exoU-negative groups was evaluated using the chi-square test. P values of <0.05 were considered to be statistically significant findings.
ACKNOWLEDGMENTS
We were solely responsible for the conception and implementation of this study and for writing the manuscript. We are truly grateful to Yunsong Yu from Sir Run Run Shaw Hospital, Zhejiang University, for the virulence control strains, ZYPA09 and FAHZU24.
Yue Kang is an employee of MSD China. The other authors declare that they have no competing interests.
This work was supported by the National Key Research and Development Program of China under grants number 2021YFC2301002 and 2018YFE0101800, the National Natural Science Foundation of China under grants number 82272380 and 82072318, and National High Level Hospital Clinical Research Funding under grant number 2022-PUMCH-B-028. The study was also supported by funding from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA.
Contributor Information
Yingchun Xu, Email: xycpumch@139.com.
Qiwen Yang, Email: yangqiwen81@vip.163.com.
Krisztina M. Papp-Wallace, JMI Laboratories
REFERENCES
- 1.Mohamed A, Daef E, Nafie A, Shaban L, Ibrahim M. 2021. Characteristics of carbapenem-resistant gram-negative bacilli in patients with ventilator-associated pneumonia. Antibiotics (Basel) 10:1325. doi: 10.3390/antibiotics10111325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CHINET. 2020. China surveillance of bacteria resistance: results of 2020. China Antimicrobial Surveillance Network. http://www.chinets.com/Document. Accessed 30 December 2021.
- 3.Kao C-Y, Chen S-S, Hung K-H, Wu H-M, Hsueh P-R, Yan J-J, Wu J-J. 2016. Overproduction of active efflux pump and variations of OprD dominate in imipenem-resistant Pseudomonas aeruginosa isolated from patients with bloodstream infections in Taiwan. BMC Microbiol 16:107. doi: 10.1186/s12866-016-0719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santajit S, Indrawattana N. 2016. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016:2475067. doi: 10.1155/2016/2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subedi D, Vijay AK, Kohli GS, Rice SA, Willcox M. 2018. Association between possession of ExoU and antibiotic resistance in Pseudomonas aeruginosa. PLoS One 13:e0204936. doi: 10.1371/journal.pone.0204936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elabbadi A, Pont S, Verdet C, Plésiat P, Cretin F, Voiriot G, Fartoukh M, Djibré M. 2020. An unusual community-acquired invasive and multi systemic infection due to ExoU-harboring Pseudomonas aeruginosa strain: clinical disease and microbiological characteristics. J Microbiol Immunol Infect 53:647–651. doi: 10.1016/j.jmii.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Feng W, Huang Q, Wang Y, Yuan Q, Li X, Xia P, Sun F. 2021. Changes in the resistance and epidemiological characteristics of Pseudomonas aeruginosa during a ten-year period. J Microbiol Immunol Infect 54:261–266. doi: 10.1016/j.jmii.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 8.Treepong P, Kos VN, Guyeux C, Blanc DS, Bertrand X, Valot B, Hocquet D. 2018. Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin Microbiol Infect 24:258–266. doi: 10.1016/j.cmi.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Hu Y, Peng W, Wu Y, Li H, Wang Q, Yi H, Zhang R, Shao B, Zhu K. 2021. A potential high-risk clone of Pseudomonas aeruginosa ST463. Front Microbiol 12:670202. doi: 10.3389/fmicb.2021.670202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang P, Wang J, Li Y, Shi W, Cai H, Yang Q, Li X, Yu Y, Qu T, Jiang Y. 2022. Emergence of bla(KPC-33)-harboring hypervirulent ST463 Pseudomonas aeruginosa causing fatal infections in China. J Infect 85:e86–e88. doi: 10.1016/j.jinf.2022.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Pérez A, Gato E, Pérez-Llarena J, Fernández-Cuenca F, Gude MJ, Oviaño M, Pachón ME, Garnacho J, González V, Pascual Á, Cisneros JM, Bou G. 2019. High incidence of MDR and XDR Pseudomonas aeruginosa isolates obtained from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother 74:1244–1252. doi: 10.1093/jac/dkz030. [DOI] [PubMed] [Google Scholar]
- 12.Del Barrio-Tofiño E, Zamorano L, Cortes-Lara S, López-Causapé C, Sánchez-Diener I, Cabot G, Bou G, Martínez-Martínez L, Oliver A, GEMARA-SEIMC/REIPI Pseudomonas Study Group . 2019. Spanish nationwide survey on Pseudomonas aeruginosa antimicrobial resistance mechanisms and epidemiology. J Antimicrob Chemother 74:1825–1835. doi: 10.1093/jac/dkz147. [DOI] [PubMed] [Google Scholar]
- 13.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H, ESGEM Study Group . 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu H, Zhang Y, Zhang P, Wang J, Yuan Q, Shi W, Zhang S, Feng H, Chen Y, Yu M, Chen H, Jiang Y, Yang Q, Qu T. 2021. Bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing P. aeruginosa sequence type 463, associated with high mortality rates in China: a retrospective cohort study. Front Cell Infect Microbiol 11:756782. doi: 10.3389/fcimb.2021.756782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. 2014. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control 3:32. doi: 10.1186/2047-2994-3-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lob SH, Karlowsky JA, Young K, Motyl MR, Hawser S, Kothari ND, Sahm DF. 2020. In vitro activity of imipenem-relebactam against resistant phenotypes of Enterobacteriaceae and Pseudomonas aeruginosa isolated from intraabdominal and urinary tract infection samples—SMART Surveillance Europe 2015–2017. J Med Microbiol 69:207–217. doi: 10.1099/jmm.0.001142. [DOI] [PubMed] [Google Scholar]
- 17.CLSI. 2023. M100. Performance standards for antimicrobial susceptibility testing, 33rd ed. CLSI, Wayne, PA. [Google Scholar]
- 18.EUCAST. 2023. Breakpoint tables for interpretation of MICs and zone diameters, version 13.0. https://www.eucast.org/clinical_breakpoints.
- 19.Quale J, Bratu S, Gupta J, Landman D. 2006. Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 50:1633–1641. doi: 10.1128/AAC.50.5.1633-1641.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torrens G, Hernández SB, Ayala JA, Moya B, Juan C, Cava F, Oliver A. 2019. Regulation of AmpC-driven beta-lactam resistance in Pseudomonas aeruginosa: different pathways, different signaling. mSystems 4:e00524-19. doi: 10.1128/mSystems.00524-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kos VN, McLaughlin RE, Gardner HA. 2016. Elucidation of mechanisms of ceftazidime resistance among clinical isolates of Pseudomonas aeruginosa by using genomic data. Antimicrob Agents Chemother 60:3856–3861. doi: 10.1128/AAC.03113-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsutsumi Y, Tomita H, Tanimoto K. 2013. Identification of novel genes responsible for overexpression of ampC in Pseudomonas aeruginosa PAO1. Antimicrob Agents Chemother 57:5987–5993. doi: 10.1128/AAC.01291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabot G, Ocampo-Sosa AA, Domínguez MA, Gago JF, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L, Oliver A, Spanish Network for Research in Infectious Diseases (REIPI) . 2012. Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56:6349–6357. doi: 10.1128/AAC.01388-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caille O, Zincke D, Merighi M, Balasubramanian D, Kumari H, Kong K-F, Silva-Herzog E, Narasimhan G, Schneper L, Lory S, Mathee K. 2014. Structural and functional characterization of Pseudomonas aeruginosa global regulator AmpR. J Bacteriol 196:3890–3902. doi: 10.1128/JB.01997-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balasubramanian D, Kumari H, Mathee K. 2015. Pseudomonas aeruginosa AmpR: an acute-chronic switch regulator. Pathog Dis 73:1–14. doi: 10.1111/2049-632X.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morita Y, Cao L, Gould VC, Avison MB, Poole K. 2006. nalD encodes a second repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa. J Bacteriol 188:8649–8654. doi: 10.1128/JB.01342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hay T, Fraud S, Lau CHF, Gilmour C, Poole K. 2013. Antibiotic inducibility of the mexXY multidrug efflux operon of Pseudomonas aeruginosa: involvement of the MexZ anti-repressor ArmZ. PLoS One 8:e56858. doi: 10.1371/journal.pone.0056858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han S, Zaniewski RP, Marr ES, Lacey BM, Tomaras AP, Evdokimov A, Miller JR, Shanmugasundaram V. 2010. Structural basis for effectiveness of siderophore-conjugated monocarbams against clinically relevant strains of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 107:22002–22007. doi: 10.1073/pnas.1013092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.del Barrio-Tofiño E, López-Causapé C, Cabot G, Rivera A, Benito N, Segura C, Montero MM, Sorlí L, Tubau F, Gómez-Zorrilla S, Tormo N, Durá-Navarro R, Viedma E, Resino-Foz E, Fernández-Martínez M, González-Rico C, Alejo-Cancho I, Martínez JA, Labayru-Echverria C, Dueñas C, Ayestarán I, Zamorano L, Martinez-Martinez L, Horcajada JP, Oliver A. 2017. Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob Agents Chemother 61:e01589-17. doi: 10.1128/AAC.02352-17. [DOI] [PMC free article] [PubMed] [Google Scholar]