Abstract
During human immunodeficiency virus type 1 (HIV-1) assembly, the primer tRNA for the reverse transcriptase-catalyzed synthesis of minus-strand strong-stop cDNA, tRNA3Lys, is selectively packaged into the virus and annealed onto the primer binding site on the RNA genome. Annealing of tRNA3Lys in HIV-1 is independent of polyprotein processing and is facilitated in vitro by p7 nucleocapsid (NCp7). We have previously shown that mutations in clusters of basic amino acids flanking the first Cys-His box in NC sequence inhibit annealing of tRNA3Lys in vivo by 70 to 80%. In this report, we have investigated whether these NC mutations act through Pr55gag or Pr160gag-pol. In vivo placement of tRNA3Lys is measured with total viral RNA as the source of primer tRNA-template in an in vitro reverse transcription assay. Cotransfection of COS cells with a plasmid coding for either mutant Pr55gag or mutant Pr160gag-pol, and with a plasmid containing HIV-1 proviral DNA, shows that only the NC mutations in Pr55gag inhibit tRNA3Lys placement. The NC mutations in Pr55gag reduce viral infectivity by 95% and are trans-dominant-negative, i.e., they inhibit genomic placement of tRNA3Lys even in the presence of wild-type Pr55gag. This dominant phenotype may indicate that the mutant Pr55gag is disrupting an ordered Pr55gag structure responsible for the annealing of tRNA3Lys to genomic RNA.
In human immunodeficiency virus type 1 (HIV-1), tRNA3Lys is the primer for reverse transcription. This tRNA is incorporated into the virion during its assembly and annealed to the primer binding site (PBS) on the viral RNA (8). Annealing is facilitated in vitro by mature nucleocapsid protein (NCp7) (3), but since in vivo genomic placement of primer tRNA occurs independently of proteolytic maturation of either Pr55gag or Pr160gag-pol (11), the NC sequences must act within the context of either the Gag or the Gag-Pol precursor.
We have previously shown that mutations flanking the first Cys-His box in HIV-1 NC sequence, while not inhibiting the selective packaging of tRNALys into HIV-1 (6), inhibit genomic placement of tRNA3Lys in vivo by 70 to 80% (5). These mutations include a triple mutation in basic residues just upstream of the first Cys-His box (R7R10K11 S), as well as mutations within the 7-amino-acid linker sequence between the two Cys-His boxes (R32G and a triple mutation, R32K33K34 S). The mutations are dominant-negative since genomic placement of tRNA3Lys cannot be rescued by cotransfection of COS cells with plasmids coding for either wild-type (wt) Pr55gag or Pr160gag-pol. The mutant NC sequences must be acting within the context of either Pr55gag or Pr160gag-pol, and in this report, we have determined that they act through Pr55gag, and not through Pr160gag-pol.
To measure the amount of tRNA3Lys placed onto the PBS in vivo, total viral RNA is used as the source of primer-template in an in vitro reverse transcription reaction, with exogenous HIV-1, reverse transcriptase (RT), dCTP, dTTP, [α-32P]dGTP, and ddATP (5). Since the sequence of the first six deoxynucleoside triphosphates incorporated is CTGCTA, placed primer tRNA3Lys will be extended by six bases, and the extended tRNA3Lys can be resolved and detected by one-dimensional polyacrylamide gel electrophoresis. The interpretation of the data from such experiments makes the assumption that the primer tRNA in the total viral RNA has been placed upon the PBS in vivo and not in vitro during the reverse transcription reaction. Several pieces of data support this. First, the different degrees of inhibition of genomic placement produced by wt and mutant Pr55gag and Pr160gag-pol (6) must reflect what had occurred in the virus since the total viral RNA used in the reaction has been deproteinized, and none of the mutants tested alter the amount of tRNA3Lys packaged. Second, we have also found that in the RT reaction buffer, the primer tRNA3Lys bound to the template is very heat stable, dissociating at temperatures only above 70°C (data not shown). Third, we have reported that a mutant tRNA3Lys with an altered anticodon sequence (SUU to CUA) is an efficient primer for reverse transcription in vitro when it is heat annealed to genomic RNA. However, while this mutant tRNA is packaged into HIV-1 in vivo, it does not act as a primer tRNA in our RT assay with total viral RNA unless we first heat denature the total viral RNA and allow the tRNA to anneal back to the genomic RNA (7).
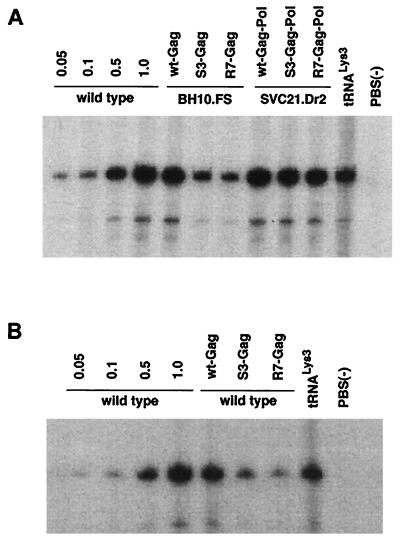
We have measured the ability of either mutant precursor protein to inhibit genomic placement of tRNA3Lys, and the data from cotransfection experiments is shown in Fig. 1 (see also Table 2). Many of the mutant HIV-1 plasmids used here have been described previously, and their properties are listed in Table 1. In Fig. 1, lanes 1 to 4 in panel A or B represent cells transfected with wt HIV-1 proviral DNA (BH10). Increasing amounts of genomic RNA were used to demonstrate the linear increase in the hybridization signal with increasing amounts of placed primer tRNA3Lys. (The amount of genomic RNA in the total viral RNA sample was determined by dot blot hybridization with a genomic RNA probe [12]). In the cotransfection experiments represented in lanes 5 to 12 (Fig. 1A) and 5 to 9 (B), the RT reactions producing the six-base tRNA3Lys extension product use total viral RNA containing 5 × 108 copies of genomic RNA, as in lane 4. In the experiments represented in lanes 5 to 7 (Fig. 1A), BH10.FS is the source of wt Pr160gag-pol, and because frameshifting always occurs during translation, no Pr55gag is coded for by this proviral construct. In these experiments, cotransfection with plasmids coding for mutant Pr55gag shows that the S3(S32-34) or R7R10K11 S NC mutations in Pr55gag molecules can inhibit genomic placement of the tRNA3Lys by approximately 80% (Table 2) and are correlated with a severe reduction in viral infectivity. In lanes 8 to 10 (Fig. 1A), SVC21.Dr2 codes for wt Pr55gag and mutant Pr160gag-pol (mutated in RT sequences), but the mutant Pr160gag-pol is not packaged into the virus. Cotransfection with plasmids coding for Pr160gag-pol molecules mutated in NC sequences shows that the S3(S32-34) or R7R10K11 S NC mutations in Pr160gag-pol do not affect tRNA3Lys placement. Genomic RNA-CAp24 was not determined here since these viral particles are protease negative and do not produce mature CAp24. The lack of infectivity in these viruses is probably due to the lack of viral protease activity.
FIG. 1.
Inhibition of genomic placement of tRNA3Lys by NC mutants in Pr55gag or Pr160gag-pol. Placement was measured by the ability of tRNA3Lys to be extended six bases in an in vitro reverse transcription reaction with HIV-1 RT and total viral RNA as the source of primer tRNA-template. The transfection of COS7 cells, the isolation of HIV-1 and total viral RNA, and the conditions of reverse transcription were as previously described (5). The sequence of the first six deoxynucleoside triphosphates incorporated is CTGCTA. In the presence of dCTP, dTTP, [α-32P]dGTP, and ddATP instead of dATP, extension will therefore terminate after six bases. After incubation, the reaction products were precipitated with 2-propanol and resuspended in loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) and the samples were electrophoresed for 16 h at 4°C, with 11% polyacrylamide–7 M urea. COS cells were either transfected with wt HIV proviral DNA or cotransfected with the following plasmids. Wild type is the HIV-1 BH10 proviral clone; BH10.FS is frameshift positive, i.e., it produces Pr160gag-pol, but not Pr55gag (2); SVC21.Dr2 is the HIV-1 proviral DNA containing a 2-amino-acid insertion in the connection domain and results in the lack of Pr160gag-pol incorporation into the virus (12); wt-Gag and wt-Gag-Pol contain wt sequences of Pr55gag and Pr160gag-pol, respectively, inserted into pBABY vectors (14) and were donated by D. Rekosh, University of Virginia. They contain the RNA recognition element but lack most of the HIV-1 sequences upstream and downstream of the sequences coding for Gag and Gag-Pol, and the mutant genomic RNA is not packaged. S3-Gag, S3-Gag-Pol, R7-Gag, and R7-Gag-Pol represent the wt-Gag and wt-Gag-Pol sequences which have been modified with the S3(S32-34) and R7R10K11 S NC mutations, respectively. Genomic placement of tRNA3Lys was measured in the wt and mutant virus as described in the text. Lanes 5 to 12 in panel A and lanes 5 to 9 in panel B contain the same amount of genomic RNA found in lane 4 of both panels, i.e., 5 × 108 copies. Lanes 1 to 3 in both panels represent fractional amounts of this genomic RNA copy number. Lanes 11 and 8 in panels A and B, respectively, represent the use in the RT reaction of purified human placental tRNA3Lys heat annealed to synthetic genomic RNA, as previously described (7), and used as a size marker. Lanes 12 and 9 in panels A and B, respectively, represent virus whose genome lacks a PBS.
TABLE 2.
Effect of mutant NC upon genomic RNA packaging and annealing of tRNA3Lys to genomic RNA
| Virusa | Genomic RNA packaging (% of wt)b | tRNA3Lys placement (% of wt)c | Infectivityd |
|---|---|---|---|
| wt (BH10) | 100 | 100 | 100 |
| R7R10K11 S | 22 | 24 | 0.44 |
| C15S/C18S | 6 | 54 | 0.11 |
| S3(32-34) | 20 | 25 | 0.49 |
| C36S/C39S | 22 | 105 | 1.0 |
| Cotransfections | |||
| BH10.FS plus | |||
| wt-Gag | 93 | 97 | 98 |
| S3-Gag | 14 | 24 | 0.65 |
| R7-Gag | 17 | 17 | 1.6 |
| SVC21.Dr2 plus | |||
| wt-Gag-Pol | 101 | 0 | |
| S3-Gag-Pol | 87 | 0 | |
| R7-Gag-Pol | 86 | 0 | |
| wt (BH10) plus | |||
| wt-Gag | 70 | 89 | 82 |
| S3-Gag | 35 | 26 | 5 |
| R7-Gag | 49 | 12 | 5 |
The wt and mutant HIV-1 proviral sequences are described in Table 1.
To measure viral RNA packaged per CAp24, virus was first pelleted from culture medium by centrifugation in a Beckman Ti45 rotor at 35,000 rpm for 1 h. The viral pellet was then purified by centrifugation in a Beckman SW41 rotor at 26,500 rpm for 1 h through 15% sucrose onto a 65% sucrose cushion. After resuspension of the viral pellet, CAp24 was measured with a commercial kit available for p24 antigen capture (Abbott Laboratories). Viral genomic RNA was extracted and quantitated as previously described (12), i.e., genomic RNA in total viral RNA samples was quantitated by dot blot hybridization with a radioactive DNA probe specific for genomic RNA, with known amounts of in vitro-transcribed, synthetic genomic RNA as a standard.
tRNA3Lys genomic placement was measured as described in the legend to Fig. 1.
Viral infectivity was measured by the MAGI assay. A total of 0.4 × 105 cells in 1 ml of culture medium (Dulbecco modified Eagle medium) plus 10% fetal calf serum) were plated per well. After 16 h, the medium was removed and replaced with 150 μl of culture medium containing various dilutions of virus. DEAE-Dextran was added to a final concentration of 20 μg/ml, and viral absorption was allowed for 2 h. Then the medium and virus were removed and replaced with 1 ml of fresh culture medium. At 48 h later, the medium was removed, and fixative was added for 5 min (1% formaldehyde and 2.5% gluteraldehyde, in phosphate-buffered saline). Fixative was then removed and replaced with staining solution for 50 min (950 μl of phosphate-buffered saline, 20 μl of 0.2 M potassium ferrocyanide, 20 μl of 0.2 M potassium ferricyanide, 1.0 μl of 2.0 M Mg2Cl, and 10 μl of X-Gal stock [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] [40 mg of dimethyl sulfoxide per ml]). The cells were then washed twice with phosphate-buffered saline, and blue cells per well per equal amounts of p24 were counted.
TABLE 1.
List of HIV-1 proviral viral constructs
| Viral construct | NC mutation | Viral sequencea | Major viral protein(s) | Reference(s) |
|---|---|---|---|---|
| BH10 | None | A | All | 11, 18 |
| R7R10K11 S | R7R10K11 S | A | All | 14 |
| C15S/C18S | C15S/C18S | A | All | 4 |
| S3(S32-34) | R32K33K34 S | A | All | 4 |
| C36S/C39S | C36S/C39S | A | All | 4 |
| BH10.FS | None | B | All except Pr55gag | 1 |
| SVC21.Dr2 | None | C | All except Pr160gag-pol | 13 |
| wt-Gag | None | D | wt-Pr55gag | 16, 17 |
| S3-Gag | R32K33K34 S | D | S3-Pr55gag | 5, 6 |
| R7-Gag | R7R10K11 S | D | R7-Pr55gag | 5, 6 |
| wt-Gag-Pol | None | E | wt-Pr160gag-pol | 16, 17 |
| S3-Gag-Pol | R32K33K34 S | E | S3-Pr160gag-pol | 5, 6 |
| R7-Gag-Pol | R7R10K11 S | E | R7-Pr160gag-pol | 5, 6 |
Viral sequence type: A, full-length HIV-1 proviral DNA. B, full-length HIV-1 proviral DNA but frameshift positive. C, full-length proviral DNA but contains a 6-bp in-frame nucleotide insertion within the connection domain of RT (HXB2 nucleotide position 3715). Mutant Pr160gag-pol is synthesized but not packaged into the virion. D, proviral DNA starting at HXB2 nucleotide 679 (SacI site) and terminating immediately after the Gag open reading frame (HXB2 nucleotide 2428 [BclI site]). Proviral DNA is thus missing all the 5′ long terminal repeat and leader sequences, as well as all coding regions downstream of Gag, but includes the RNA recognition element sequence. Donated by D. Rekosh, University of Virginia. This construct was previously referred to as pSVGAG-RRE-R. E, proviral DNA starting at HXB2 nucleotide 679 (SacI site) and terminating immediately after the Vif open reading frame (HXB2 nucleotide 5785 [SalI site]). Proviral DNA is thus missing all the 5′ long terminal repeat and leader sequences, as well as all coding regions downstream of Vif, but includes the RNA recognition element sequence. It contains an inactive protease (D25G) and a deletion of 5 T’s (nucleotides 2082 to 2086) which results in only Pr160gag-pol, and not Pr55gag, being synthesized. Donated by D. Rekosh, University of Virginia. This construct was previously referred to as pSVFS5TprotD25G.
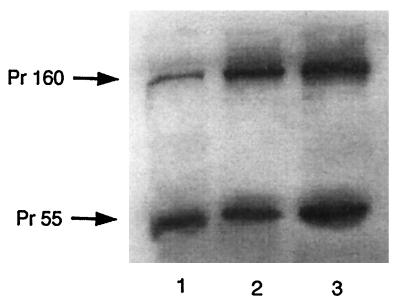
The differences in the abilities of mutant Pr55gag and Pr160gag-pol to inhibit tRNA3Lys annealing to genomic RNA are not due to a greater expression of Gag than of Gag-Pol. Figure 2 shows Western blots of cell lysates probed with antibody to p24. Lane 1 represents cell lysate protein from cells transfected with a vector expressing wt virus, while lanes 2 and 3 represent cell lysate protein from cells cotransfected with vectors expressing both Pr55gag and Pr160gag-pol in the same cell. Lanes 2 and 3 represent, respectively, the cytoplasmic expression of both precursors containing either the R7R10K11 S mutation (lane 2) or the S3(S32-34) mutation (lane 3). It can be seen that both proteins are expressed approximately equally.
FIG. 2.
Cytoplasmic expression of Gag and Gag-Pol precursor proteins. Shown is Western blot analysis of wt and mutant viral proteins. COS7 cells were transfected, or cotransfected, with the following plasmids: BH10 (wt HIV-1) (lane 1), R7-Gag and R7-Gag-Pol (lane 2), and S3-Gag and S3-Gag-Pol (lane 3). At 63 h posttransfection, cells were washed twice with ice-cold phosphate-buffered saline, scraped off the tissue culture dishes, and lysed with radioimmunoprecipitation assay buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 1% deoxycholate, 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 2 mg of aprotinin per ml, 2 mg of leupeptin per ml, 1 mg of pepstatin A per ml, 100 mg of phenylmethylsulfonyl fluoride per ml). Aliquots of total cell lysates were resolved by 10% (wt/vol) sodium dodecyl sulfate–one-dimensional polyacrylamide gel electrophoresis and blotted onto nitrocellulose membranes. Protein detection utilized monoclonal antibodies reactive to HIV-1 p24 and was performed by enhanced chemiluminescence with a goat anti-mouse cross conjugated to horseradish peroxidase as a secondary antibody.
The data in Fig. 1 and 2 indicates that the tRNA3Lys annealing function of the basic amino acid clusters in NC acts through the Gag protein. Mutations at these positions have other reported effects on the virus as well. There is a reduction in the assembly and production of viral particles, and the extracellular particles produced package less genomic RNA (1). The basic amino acid clusters flanking the first Cys-His box have also been associated with the interaction (I) domain in both HIV-1 and Rous sarcoma virus, and it is likely therefore that the particles produced have a less-than-normal density in sucrose gradients (1). Nevertheless, in spite of the pleiotypic effect of these mutations, the inability of these mutations in Gag-Pol to inhibit tRNA3Lys placement points to the tRNA3Lys annealing function of NC acting through Gag only.
The reduced packaging of genomic RNA in the R7-Gag and S3-Gag virus (Table 2) is not likely to be responsible for reducing tRNA3Lys genomic placement. First, the ability of mutations in the sequences flanking the first Cys-His box in HIV-1 NC to directly inhibit the annealing of tRNA3Lys to the PBS is supported by in vitro work in which wt or mutant NCp7 is used to anneal tRNA3Lys to equal amounts of genomic RNA (3). Second, the different degrees of in vivo tRNA3Lys placement detected are all being measured on genomic RNA which does get packaged into virions. Reduction in genomic RNA packaging may be an all-or-nothing phenomenon in which genomic RNA is either packaged as a dimer at normal concentrations in the virus or not at all, and virions lacking genomic RNA will not contribute to our measurements.
In support of this, the data listed in Table 2 indicates a lack of correlation between genomic RNA packaging and genomic placement of tRNA3Lys. The tRNA3Lys placement and genomic RNA packaging of the four NC mutants listed at the top of Table 2 have been previously reported (5, 6). The mutations occur in full-length HIV-1 proviral DNA and are thus found in both Pr55gag and Pr160gag-pol. C15S/C18S and C36S/C39S are mutations found within the first and second Cys-His boxes, respectively. While the 75% reduction in tRNA3Lys placement in the R7R10K11 S (R7) and S3(S32-34) (S3) mutant virus is correlated with an 80% inhibition of genomic RNA packaging, this correlation does not exist for the Cys-His mutations. For example, while genomic RNA packaging is reduced by 94% in the C15S/C18S virion, tRNA3Lys placement is reduced only moderately (54%). On the other hand, tRNA3Lys placement is not inhibited at all in the C36S/C39S virus, which shows an 80% reduction in genomic RNA packaging. Infectivity of all these mutant viruses is severely reduced.
Further evidence for this lack of correlation between genomic RNA packaging and tRNA3Lys placement is shown in the experiments in lanes 5 to 7 of Fig. 1B, the quantitation of which is listed at the bottom of Table 2. In these experiments, cells are transfected with wt HIV-1 proviral DNA (BH10) and with a vector coding for either wt or mutant Gag protein. In all the experiments, BH10 is a source of wt Gag. It is noted that when the cell is cotransfected with a second wt-Gag vector coding for wt Gag, genomic RNA packaging decreases 30%, probably because of the excess number of Gag particles made and the fact that wt-Gag RNA lacks a packaging signal. Even so, tRNA3Lys placement is inhibited by only 11%. Infectivity of these viruses is only moderately reduced (18%), probably because of the Gag particles lacking genomic RNA. When S3-Gag or R7-Gag is cotransfected with BH10, genomic RNA packaging is moderately inhibited, but tRNA3Lys placement is much more severely disrupted. This is especially evident for R7-Gag, which reduces genomic RNA packaging by approximately 50% but reduces tRNA3Lys placement by 88%. Infectivity of these mutant Gag viruses is reduced by 95%.
In these cotransfection experiments, transfection of some cells with only one of the two plasmids utilized will have little effect upon our results. Cells transfected with only wt-Gag-Pol are unable to produce particles (14, 15), while cells transfected with either wt-Gag or wt-Gag-Pol will produce particles without genomic RNA, since the viral RNA produced does not contain genomic RNA packaging signals (14, 15). Although cells transfected with only wt HIV proviral DNA could in principle increase the amount of genomic placement seen, the fact that tRNA3Lys annealing is greatly inhibited in cotransfection experiments with BH10 and mutant Gag argues against the transfection of cells with only wt proviral DNA occurring frequently enough to affect our results.
Our results indicate that NC mutations affecting tRNA3Lys placement exert their effect through the Pr55gag precursor, and not through Pr160gag-pol. Furthermore, this effect is trans-dominant-negative, i.e., annealing of tRNA3Lys to the PBS will be inhibited by mutant Gag even in the presence of wt Gag. This may be indicative of a structured annealing complex composed of Gag which can be disrupted by mutant Gag molecules, and this is supported by the findings that mutations in the basic amino acid clusters in NC disrupt Gag interaction in virions, as manifested by a lighter density of the mutant virus (1). The lack of evidence thus far for Pr160gag-pol being involved in tRNA3Lys placement might explain, in part, why the absence of RT in murine leukemia virus (MuLV) does not affect the placement of tRNAPro onto the PBS (4, 9, 10). But why do RT-negative mutations affect primer tRNA placement in avian retrovirus (4, 13) and HIV-1 (5)? The answer might be a different requirement for selective packaging of primer tRNA, which is inhibited in all three virus families by RT-negative mutants. However, this selective packaging is less striking in murine retrovirus than in avian retrovirus or HIV-1. The changing concentration of primer tRNA relative to the total viral low-molecular-weight RNA in moving from cytoplasm to virus is as follows: tRNAPro (AKR MuLV), 5 to 6 to 12 to 24% (16); tRNATrp (avian myeloblastosis virus), 1.4 to 32% (16); tRNA3Lys (HIV-1), 2 to 20% (11). The tRNAPro concentration required for genomic placement in MuLV may be lower than that required in avian myeloblastosis virus and HIV-1 for tRNATrp or tRNA3Lys, respectively, perhaps because of different affinities between the primer tRNA and the Gag protein within an annealing complex.
Acknowledgments
S.C. and Y.H. contributed equally to the work.
This work was supported by grants from the Medical Research Council and Health Canada. We thank David Rekosh, Mary-Lou Hammarskjöld, Alan Rein, and Robert Gorelick for the gift of plasmids used in this work.
We thank Sandy Fraiberg for assistance in preparation of the manuscript.
REFERENCES
- 1.Bowzard J B, Bennett R P, Krishna N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cherry, E., C. Laing, L. Rong, Y. Quan, P. Inouye, X. Li, N. Morin, M. Kotler, and M. A. Wainberg. Characterization of human immunodeficiency virus type-1 particles that express protease-reverse transcriptase fusion proteins. J. Mol. Biol., in press. [DOI] [PubMed]
- 3.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M-C, Roques B, Darlix J-L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu W, Ortiz-Conde B A, Gorelick R J, Hughes S H, Rein A. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J Virol. 1997;71:6940–6946. doi: 10.1128/jvi.71.9.6940-6946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y, Khorchid A, Gabor J, Wang J, Li X, Darlix J-L, Wainberg M A, Kleiman L. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA3Lys genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J Virol. 1998;72:3907–3915. doi: 10.1128/jvi.72.5.3907-3915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, Khorchid A, Wang J, Parniak M A, Darlix J-L, Wainberg M A, Kleiman L. Effect of mutations in the nucleocapsid protein (NCp7) upon Pr160gag-pol and tRNALys incorporation into human immunodeficiency virus type 1. J Virol. 1997;71:4378–4384. doi: 10.1128/jvi.71.6.4378-4384.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, Shalom A, Li Z, Wang J, Mak J, Wainberg M A, Kleiman L. Effects of modifying the tRNA3Lys anticodon on the initiation of human immunodeficiency virus type 1 reverse transcription. J Virol. 1996;70:4700–4706. doi: 10.1128/jvi.70.7.4700-4706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang M, Mak J, Ladha A, Cohen E, Klein M, Rovinski B, Kleiman L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol. 1993;67:3246–3253. doi: 10.1128/jvi.67.6.3246-3253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin J G, Hu S C, Rein A, Messer L I, Gerwin B I. Murine leukemia virus mutant with a frameshift in the reverse transcriptase coding region: implications for pol gene structure. J Virol. 1984;51:470–478. doi: 10.1128/jvi.51.2.470-478.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levin J G, Seidman J G. Effect of polymerase mutations on packaging of primer tRNAPro during murine leukemia virus assembly. J Virol. 1981;38:403–408. doi: 10.1128/jvi.38.1.403-408.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mak J, Jiang M, Wainberg M A, Hammarskjöld M-L, Rekosh D, Kleiman L. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J Virol. 1994;68:2065–2072. doi: 10.1128/jvi.68.4.2065-2072.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mak J, Khorchid A, Cao Q, Huang Y, Lowy I, Parniak M A, Prasad V R, Wainberg M A, Kleiman L. Effects of mutations in Pr160gag-pol upon tRNALys3 and Pr160gag-pol incorporation into HIV-1. J Mol Biol. 1997;265:419–431. doi: 10.1006/jmbi.1996.0742. [DOI] [PubMed] [Google Scholar]
- 13.Peters G G, Hu J. Reverse transcriptase as the major determinant for selective packaging of tRNA’s into avian sarcoma virus particles. J Virol. 1980;36:692–700. doi: 10.1128/jvi.36.3.692-700.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith A J, Cho M-I, Hammarskjöld M L, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into viruslike particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith A J, Srinivasakumar N, Hammarskjöld M-L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters L C, Mullin B C. Transfer RNA in RNA tumor viruses. Prog Nucleic Acid Res Mol Biol. 1977;20:131–160. doi: 10.1016/s0079-6603(08)60471-7. [DOI] [PubMed] [Google Scholar]