ABSTRACT
Petabases of environmental metagenomic data are publicly available, presenting an opportunity to characterize complex environments and discover novel lineages of life. Metagenome coassembly, in which many metagenomic samples from an environment are simultaneously analyzed to infer the underlying genomes’ sequences, is an essential tool for achieving this goal. We applied MetaHipMer2, a distributed metagenome assembler that runs on supercomputing clusters, to coassemble 3.4 terabases (Tbp) of metagenome data from a tropical soil in the Luquillo Experimental Forest (LEF), Puerto Rico. The resulting coassembly yielded 39 high-quality (>90% complete, <5% contaminated, with predicted 23S, 16S, and 5S rRNA genes and ≥18 tRNAs) metagenome-assembled genomes (MAGs), including two from the candidate phylum Eremiobacterota. Another 268 medium-quality (≥50% complete, <10% contaminated) MAGs were extracted, including the candidate phyla Dependentiae, Dormibacterota, and Methylomirabilota. In total, 307 medium- or higher-quality MAGs were assigned to 23 phyla, compared to 294 MAGs assigned to nine phyla in the same samples individually assembled. The low-quality (<50% complete, <10% contaminated) MAGs from the coassembly revealed a 49% complete rare biosphere microbe from the candidate phylum FCPU426 among other low-abundance microbes, an 81% complete fungal genome from the phylum Ascomycota, and 30 partial eukaryotic MAGs with ≥10% completeness, possibly representing protist lineages. A total of 22,254 viruses, many of them low abundance, were identified. Estimation of metagenome coverage and diversity indicates that we may have characterized ≥87.5% of the sequence diversity in this humid tropical soil and indicates the value of future terabase-scale sequencing and coassembly of complex environments.
IMPORTANCE Petabases of reads are being produced by environmental metagenome sequencing. An essential step in analyzing these data is metagenome assembly, the computational reconstruction of genome sequences from microbial communities. “Coassembly” of metagenomic sequence data, in which multiple samples are assembled together, enables more complete detection of microbial genomes in an environment than “multiassembly,” in which samples are assembled individually. To demonstrate the potential for coassembling terabases of metagenome data to drive biological discovery, we applied MetaHipMer2, a distributed metagenome assembler that runs on supercomputing clusters, to coassemble 3.4 Tbp of reads from a humid tropical soil environment. The resulting coassembly, its functional annotation, and analysis are presented here. The coassembly yielded more, and phylogenetically more diverse, microbial, eukaryotic, and viral genomes than the multiassembly of the same data. Our resource may facilitate the discovery of novel microbial biology in tropical soils and demonstrates the value of terabase-scale metagenome sequencing.
KEYWORDS: metagenomics, terabase, tropical soil, redox, rare biosphere
INTRODUCTION
Metagenomics projects are producing terabases (Tbp) of sequence data (1–3), with some 2.5 petabases (Pbp) of metagenome data currently in the NCBI SRA database. These data sets are an opportunity to better understand environments and biogeochemical processes, to probe the rare biosphere (low-abundance microbes [4–6]), and to discover new lineages of life (7). However, using the existing wealth of metagenomic data sets to achieve these goals is difficult because the computational task of reconstructing an environment’s underlying microbes from sequencing reads, known as genome assembly (8), can be prohibitively computationally expensive for large data sets (9). Because of this limitation, the data analysis workflows available to most researchers rely on assembling metagenome samples one at a time, a process called multiassembly, followed by the complicated process of removing redundancy resulting from multiple samples of the same environment. Multiassembly is commonly used because the de Bruijn graph-based assembly software’s (10) memory requirements increase with sequence complexity and are exceeded at the terabase scale on single compute nodes. Resource limitations on sequencing capacity may present researchers with the choice of sequencing more samples at less depth, or fewer samples at more depth. These tradeoffs may result in a shallower accounting of microbial communities’ full complexity, with incomplete, fragmented metagenome-assembled genomes (MAGs); a bias toward the most abundant community members; and the absence of rare biosphere MAGs.
Coassembling (simultaneous assembly of multiple samples) metagenome samples from the same environment, possibly at the terabase scale, is a promising approach (11), provided that the petabytes of memory potentially required by de Bruijn graph assemblers can be accommodated. To this end, we previously developed MetaHipMer (11), a metagenome assembler that runs distributed across hundreds or thousands of compute nodes to coassemble terabases of metagenomic data, with assembly quality comparable to the state-of-the-art assemblers metaSPAdes (12) and MEGAHIT (13). That study (11) provided a proof of principle, illustrating that metagenome coassembly yields more contiguous assemblies representing more of an environment’s underlying genome sequences, with less redundancy, and no requirement for complicated deduplication procedures, than multiassembly (while multiassembly remains useful due to, in addition to its computational feasibility, better characterization of abundant genomes with high strain variation).
In this paper, we present the coassembly, along with the resulting MAGs and functional annotations, of 3.4 Tbp of metagenome sequence data derived from soil in the Luquillo Experimental Forest (LEF) in Puerto Rico, a Long Term Ecological Research Network site (14). Our goal is to demonstrate the potential wealth of biological discoveries that can be driven by large-scale metagenomic sequencing and coassembly and to provide a resource of MAGs and associated functional annotation for an environment in which terabases of sequence are available. The coassembled data came from the Great Redox Experiment (GRE) (15), a study focused on how redox oscillation frames the activities of microbial communities in humid tropical soils. The experiment used replicate soil incubations under oscillating oxic and anoxic conditions, along with 13C plant biomass amendments to perform stable isotope probing (SIP) experiments, resulting in 95 metagenome samples, which were sequenced at the DOE Joint Genome Institute (proposal: Microbial Carbon Transformations in Wet Tropical Soils: Effects of Redox Fluctuation https://doi.org/10.46936/10.25585/60000880). A coassembly of this scale of data was only possible using MetaHipMer on a supercomputing cluster, in this case the Oak Ridge National Laboratory’s Summit system (16). Metagenome binning of the coassembly contigs resulted in MAGs from a broad array of phyla, including some candidate groups, a substantial increase in the number of both MAGs and phyla over the individual metagenome samples’ assemblies (referred to collectively as the multiassembly). Low-abundance microbes and viruses absent in the multiassembly, and a mostly complete fungal genome and several unicellular eukaryotic MAGs, were found in the coassembly. The coassembly, MAGs, and annotations, along with search and analysis tools, are available in the IMG/MER Database (17).
RESULTS AND DISCUSSION
Metagenome coassembly.
A total of 3.4 terabase pairs (Tbp) of metagenome sequence data were coassembled to yield 75 Gbp of sequence in 55 million scaffolds (Table 1). Consistent with our previous finding (11) that coassembly results in improved contiguity, the coassembly scaffold L50 value is >3 times greater, and the number of scaffolds larger than 50 kbp is ~27-fold more, than in the multiassembly (combined individual metaSPAdes [12] assemblies). The coassembly thus has substantially more of the lengths of scaffolds interesting to biologists: those which may harbor genes, operons, and viruses and facilitate more accurate MAG extraction (18). Alignment of 22,666,691,390 reads to coassemble contigs of ≥500 bp resulted in 19,830,702,257 (87.5%) aligned reads (the median for individual assemblies of the GRE data was 51.2%). The length-weighted mean coverage of coassembly contigs was 40×. The alignment statistics indicate that substantially more of the LEF’s underlying microbial communities are being assembled in coasssembly than in multiassembly; nevertheless, 12.5% of the reads are not represented in the coassembly.
TABLE 1.
GRE coassembly summary statisticsa
| Statistic | Coassembly | Multiassembly | Individual assembly (median) |
|---|---|---|---|
| No. of scaffolds | 55,342,847 | 279,810,992 | 2,893,912 |
| Scaffold (bp) | 74,970,251,022 | 146,314,652,061 | 1,479,843,562 |
| Scaffold N50 | 9,058,474 | 70,618,834 | 734,629 |
| Scaffold L50 (bp) | 1,656 | 526 | 517 |
| Longest scaffold (bp) | 1,464,928 | 1,406,223 | 80,051 |
| No. of scaffolds >50 Kbp | 26,818 | 1,000 | 3 |
| Assembled sequence in scaffolds >50 Kbp (%) | 3.25 | 0.05 | 0.015 |
| Avg GC (%) | 64.1 | 65.0 | 65.3 |
Statistics from 95 individual assemblies (collectively referred to as the multiassembly) of the same data are shown for comparison.
We used metaSPAdes rather than MetaHipMer for the individual assemblies because, first, it is the standard Joint Genome Institute (JGI) metagenome assembler. Second, we could not use metaSPAdes for the coassembly, because a regression model based on the memory usage of previous metaSPAdes assemblies indicated that the 265 billion unique 31-mers in the GRE data would require 4.5 TB of memory to coassemble, 3-fold more than the capacity of the largest compute node we have access to. Coasssembling the GRE data with metaSPAdes would also require potentially several weeks of run time, whereas MetaHipMer completed the coassembly in about 1.5 h. We have previously extensively compared metaSPAdes and MetaHipMer and found them to be comparable in resulting assembly quality (11). Moreover, the independent Critical Assessment of Metagenome Interpretation 2 (CAMI2) contest also found MetaHipMer to be at least as good as metaSPAdes in quality (18).
Annotation and MAG recovery.
The ~55 million assembled contigs were annotated and binned using the JGI metagenome workflow (19). In all, 112,151,078 genes were predicted, 99.4% of which are protein-coding genes (the remainder being RNA genes), with nearly all having some predicted function (Table 2). Metagenome binning resulted in 1,321 MAGs, 307 of which were high or medium quality (39 high and 268 medium) (20), and 1,014 of which were of low quality (which, because they are not made available in the IMG/MER Database, are provided in Table S1). The high- and medium-quality MAGs are made available on IMG and are the default set of MAGs discussed in this paper unless otherwise noted. These 307 MAGs represented 23 distinct phyla, including candidate groups Dormibacterota (candidate division AD3), Eremiobacterota (candidate division WPS-2), Dependentiae (candidate division TM6), and Methylomirabilota (candidate division NC10). With the exception of one medium-quality Methylomirabilota MAG in the multiassembly, these candidate phyla were absent from the 294 high- and medium-quality multiassembly MAGs, which in contrast, represented nine phyla (Table 3). Of note, the two Eremiobacterota (21) MAGs were of high quality (both >96% complete and <1% contaminated), indicating the potential of terabase-scale coassembly to produce reference-quality genomes for unculturable microbes.
TABLE 2.
Summary statistics on the functional annotation of coassembly contigs
| Type of gene | No. | Percent of total |
|---|---|---|
| All genes | 112,151,078 | |
| RNA genes | 655,297 | 0.58 |
| rRNA genes | 26,420 | 0.02 |
| 5S rRNA | 5,637 | 0.01 |
| 16S rRNA | 8,150 | 0.01 |
| 18S rRNA | 468 | 0.00 |
| 23S rRNA | 11,438 | 0.01 |
| 28S rRNA | 727 | 0.00 |
| tRNA genes | 628,877 | 0.56 |
| Protein-coding genes | 111,495,781 | 99.42 |
| With product name | 57,522,550 | 51.29 |
| With COG | 53,798,710 | 47.97 |
| With Pfam | 57,522,915 | 51.29 |
| With TIGRfam | 11,086,716 | 9.89 |
| With SMART | 11,999,050 | 10.70 |
| With SUPERFam | 55,768,333 | 49.73 |
| With CATH FunFam | 41,525,148 | 37.03 |
| With KO | 30,524,203 | 27.22 |
| With enzyme | 17,696,960 | 15.78 |
| With MetaCyc | 11,000,722 | 9.81 |
| With KEGG | 17,363,888 | 15.48 |
| COG clusters | 4,617 | 99.70 |
| Pfam clusters | 11,088 | 57.81 |
| TIGRfam clusters | 3,895 | 86.79 |
| CRISPR count | 31,334 |
TABLE 3.
Summary of coassembly MAGs, multiassembly MAGs, and species-level clusters (Mash distance of ≤0.05) per phyluma
| Phylum | Coassembly MAGs (n) | Multiassembly MAGs (n) | Species-level clusters (n) |
|---|---|---|---|
| Acidobacteriota | 59 | 71 | 64 |
| Actinobacteriota | 37 | 22 | 39 |
| Bacteroidota | 13 | 7 | 13 |
| Bdellovibrionota | 2 | 2 | |
| Chloroflexota | 7 | 7 | |
| Cyanobacteria | 1 | 1 | |
| Dependentiae | 1 | 1 | |
| Desulfobacterota | 1 | 1 | |
| Desulfobacterota_B | 6 | 6 | |
| Dormibacterota | 3 | 3 | |
| Eisenbacteria | 2 | 25 | 4 |
| Eremiobacterota | 2 | 2 | |
| Fibrobacterota | 1 | 1 | |
| Gemmatimonadota | 1 | 1 | |
| Methylomirabilota | 2 | 1 | 3 |
| Myxococcota | 29 | 15 | 32 |
| Myxococcota_A | 2 | 32 | 3 |
| Patescibacteria | 5 | 5 | |
| Planctomycetota | 42 | 42 | |
| Proteobacteria | 79 | 111 | 93 |
| Spirochaetota | 1 | 1 | |
| Verrucomicrobiota | 7 | 10 | 8 |
| Verrucomicrobiota_A | 4 | 4 | |
| Total MAGs | 307 | 294 | - |
| Distinct phyla | 23 | 9 | - |
Notice that while the coassembly and multiassembly have similar total numbers of MAGs, the coassembly covers more than twice as many phyla and that species-level clustering of the coassembly and multiassembly MAGs combined reveals the considerable redundancy in the multiassembly (e.g., in the phyla Eisenbacteria and Myxococcota_A). Notice also that for a few phyla (e.g., Acidobacteriota and Proteobacteria) the multiassembly captures some species-level diversity that the coassembly does not. -, not applicable
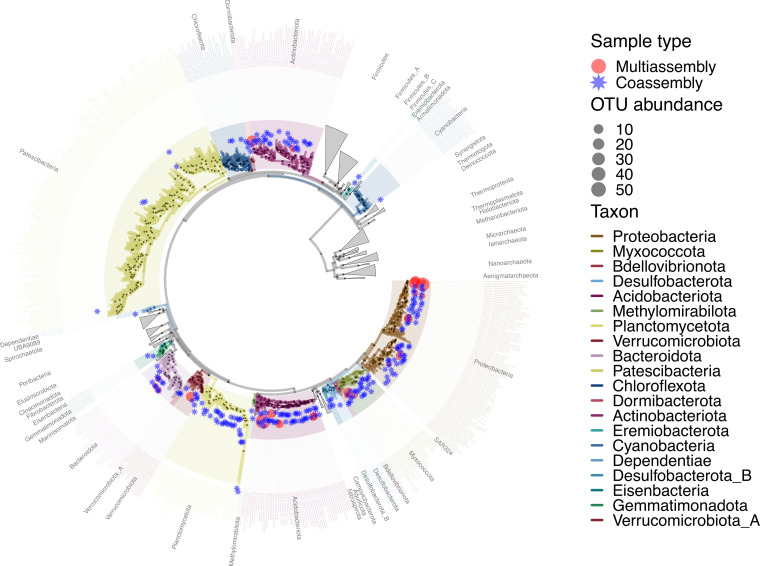
The RNA Pol-based phylogenetic tree in Fig. 1 provides an overview of the distribution of MAGs derived from our MetaHipMer coassembly versus the metaSPAdes multiassembly. Consistent with our finding of a broader array of phyla captured in the coassembly MAGs (Table 3), the phylogenetic diversity (PD) of coassembly MAGs was also far greater than that of the multiassembly MAGs (PD, 17.5 for the coassembly MAGs; PD, 3.4 for the multiassembly MAGs). Mash 0.05 clustering (22) of the 307 coassembly MAGs did not reveal any species-level redundancy, whereas clustering of the multiassembly MAGs reduced the species-level operational taxonomic unit (OTU) count to 38, indicating that the same community members were likely being assembled across samples (indicated by OTU abundance in Fig. 1). Of these 38 species-level OTUs from the multiassembly, 9 were found in the coassembly MAGs (using Mash 0.05 clustering), 298 clustered OTUs remained unique to the coassembly, and 29 OTUs were uniquely found in the multiassembly MAGs. The finding of some MAGs unique to the multiassembly is an expected result, as we previously found (11) that multiassembly better captures abundant, high-strain-variation genomes, while coassembly better captures low-abundance, rare genomes. At the phylum level, however, the coassembly MAGs contain more than twice as many taxa, and no taxa were uniquely found in the multiassembly MAGs.
FIG 1.
Phylogenetic overview of the MAG set (coassembly and multiassembly MAGs combined and dereplicated at 0.05 Mash pairwise distance, or approximately species level) from the Great Redox Experiment (GRE), a study of a humid tropical soil incubated under multiple redox conditions and amended with plant biomass. MAGs derived from either the multiassembly or coassembly are denoted by the red circles and blue stars, respectively. The phylogeny, consisting of MAGs and a reference set of genomes containing roughly genus- to family-level representatives spanning both bacterial and archaeal domains, was constructed by extracting, aligning, and concatenating the alignments of a set of three RNA polymerase subunit genes. Bubble sizes refer to the number of multiassembly MAGs within each Mash 0.05 cluster, noted by “OTU abundance” in the legend, and indicate community members assembled across multiple samples. All 307 coassembly MAGs were distinct species-level OTUs, whereas the 294 multiassembly MAGs contained 38 species-level OTUs. Phylum-level annotations are shown using GTDB-Tk taxon strings.
Rare biosphere.
We hypothesized that low-abundance rare biosphere microbes (4) might be present in the low-quality MAGs (completion, <50%; contamination, <10%) and might have low completeness due to low coverage in the data. Among the low-quality MAGs, we noticed a bin trivially lower than the medium-quality (MQ) completeness cutoff (49.1% completeness, 0.0% contamination) assigned to the candidate phylum FCPU426. FCPU426 has been detected in a 16S rRNA gene amplicon survey (23) and a function-driven single-cell genomics analysis (24) of different hot springs. Both studies estimated FCPU426’s abundance in its environment to be <1% of total microbial community composition. Here, the FCPU426 bin was covered at 23× in the coassembly. None of the 95 samples contributed more than ~1× coverage to the FCPU426 bin, indicating that this microbe would have been impossible to assemble using a multiassembly approach, as optimal microbial genome assembly typically requires >20× coverage (25). The NCBI taxonomy database currently has eight nucleotide sequences belonging to FCPU426, suggesting that, in addition to being low abundance in its environment, this candidate phylum is also poorly represented in genomic repositories.
Two low-quality bins with contamination slightly higher than the MQ cutoff were assigned to the genus Labilithrix (90.5% completeness, 10.32% contamination) and order UTPRO1 of the uncultured phylum Binatota (candidate division UBP10, 65.96% completeness, 10.53% contamination), covered at 19× and 55×, respectively. Labilithrix is a myxobacterium genus of the phylum Myxococcota, isolated from forest soil (26). Labilithrix is typically considered rare and it is of particular interest for secondary metabolite production capabilities along with other Myxococcota (27, 28). Myxobacteria grow as saprophytes on decaying organic matter, but under nutrient-limiting conditions, these bacteria form a cooperative multicellular swarm that becomes predatory (29). Given the supply of plant litter as a carbon source in these samples, it is reasonable to assume this Labilithrix organism was growing in its single cellular saprophyte low-density state; hence its low coverage. Binatota, on the other hand, is largely undescribed. Binatota genomes recovered from metagenome assemblies appear capable of aerobic methylotrophy, alkane degradation, and pigment production, but these have not yet been confirmed (30, 31). None of the 95 samples contributed more than 1× coverage to either of these bins, which again indicates the usefulness of coassembly for rare biosphere discovery.
In this data set, numerous LQ bins nearing the MQ completeness and contamination cutoffs have only phylum-level taxonomic assignment. For some of these unresolved bins, sequencing reads mapped from the 95 samples point to increased abundance specific to one of the redox treatment growth conditions relative to the others, including the carbon isotope supplied. Increased abundance under specific growth conditions could yield clues about the ecological niches these potentially novel taxa occupy that would have been missed with a multiassembly approach.
High- and medium-quality MAGs are often prioritized by biologists, but lower-quality MAGs, or even unbinned contigs, can enable insight into rare biosphere microbes. In the absence of targeted enrichment techniques (24), exhaustive sequencing may be required for sufficient coverage to resolve MAGs for the rarest microbes in an environment, which are often at <1% of total abundance. Our findings demonstrate the utility of massive metagenome sequencing and coassembly of the resulting data sets and suggest the possibility of coassembling agglomerated publicly available data sets (1, 3) for the discovery of rare biosphere microbes.
Eukaryotic MAGs.
We hypothesized that other low-quality MAGs, or MAGs that do not even meet the MIMAG (20) standard for low quality due to high contamination, might be of eukaryotic origin. To test this hypothesis, we ran EukCC (32) to assign a eukaryotic origin to the low-quality MAGs. Applying the MIMAG (20) standards for medium- or higher-quality (completion, ≥50%; contamination, <10%), we identified one MAG at 81.2% completeness and 0.75% contamination, apparently belonging to the fungal class Sordariomycetes. The Sordariomycetes include the model fungus Neurospora crassa (33), important plant pathogens (34), and numerous endophytes (35). BUSCO (36) analysis, which estimates genome completeness based on the presence or absence of conserved single-copy orthologs, estimated the apparent fungal MAG to be 77.5% and 74.7% complete with respect to the fungal and Sordariomycetes lineages, respectively, roughly consistent with the EukCC completeness estimate. BLAST searches against reference databases indicated that this fungus is likely in the Coniochaeta clade (37), which includes fungal species found in decaying wood, leaf litter, and soil and likely represents a fungus occupying a saprotrophic role in the LEF soils. The fungal MAG’s coverage was 13× across all 95 samples, with no individual sample contributing more than 4× coverage; thus, it is unlikely this fungus could have been assembled using a multiassembly approach.
At lower completeness levels (10 to 50%), 30 additional partial eukaryotic MAGs were found, including multiple MAGs from the protist orders Tintinnida (5 MAGs), Diplonemea (5 MAGs), Diplomonadida (4 MAGs), and Hymenostomatida (2 MAGs). These may represent starting points for more detailed analyses of unicellular eukaryotes in LEF soils.
Viruses.
To detect viral genomes in the GRE coassembly, assembled contigs were processed with a novel bioinformatic pipeline, geNomad (38). A total of 22,254 viral sequences were identified, of which 239 (1.1%) were found to represent high-quality genomes (estimated completeness, ≥90%) and 470 (2.1%) corresponded to medium-quality genomes (estimated completeness, 50 to 90%). Cross-referencing the identified viruses with medium- and high-quality MAGs revealed 44 proviruses integrated into 35 different genomes, mostly assigned to Proteobacteria (17 MAGs), Planctomycetota (8 MAGs), and Acidobacteriota (4 MAGs). Clustering of coassembly viruses together with similarly detected multiassembly viruses resulted in 25,355 species-level virus taxonomic units (vOTUs); 85.6% of these were unique to the coassembly, 12.3% were unique to the multiassembly, and the remaining 2.1% were shared. When clustering at the genus or family levels, the proportion of vOTUs unique to the coassembly remained roughly the same, while the proportion of vOTUs unique to the multiassembly shrank (4.2% genus level and 3.2% family level). These results indicate that, while multiassembly still captures some potentially valuable species-level metagenomic diversity, coassembly captures substantially more viral contigs from a broader phylogenetic distribution. Moreover, the read coverage of viruses unique to the coassembly was substantially lower than in those viruses found in both the coassembly and multiassembly, indicating, as with microbial MAGs, that the GRE coassembly may contain potentially rare, low-abundance viruses.
Sequencing coverage and diversity.
To assess how thoroughly 3.4 Tbp of sequencing covers the LEF microbial communities, we applied Nonpareil (39) to the 95 GRE metagenomes. The fraction of a metagenomic community covered by a sequencing effort may bias the results due to insufficient coverage of less abundant microbes, resulting in fragmented, incomplete assemblies; thus, an assessment of the coverage of a sequencing effort given diversity in the community is useful. Nonpareil samples reads and analyzes the redundancy of k-mers to estimate coverage and predict the amount of sequencing effort required to achieve >95% of the diversity in an environment. The median diversity index, Nd, computed by Nonpareil for the GRE data sets was 22.8, consistent with soil samples analyzed in reference 39. As expected, the bulk samples, which are not biased by stable isotope probing (SIP) fractionating, have a higher Nd (median, 23.1) than the SIP fractions (median, 22.7). Nonpareil’s estimate of the projected sequencing to cover 95% of the sequence diversity (median of 1.25 Tbp based on the bulk samples) is exceeded by the 3.4 Tbp we have from the GRE. Allowing for bias that may have been introduced by the redox and SIP experiments, the GRE metagenomes may cover the majority of the microbial diversity in the LEF.
The implication that 3.4 Tbp of sequencing is possibly enough to cover 95% of the diversity in LEF soils is tempered by our finding that 87.5% of the reads mapped to contigs, leaving 12.5% of the reads unaccounted for. The JGI assembly release process excludes all scaffolds of <500 bp, and it is possible that a substantial fraction of the unmapped reads would map to these excluded contigs. It could also be that some rare biosphere microbes in the LEF are at such low abundance that 3.4 Tbp of sequencing still does not provide sufficient coverage to assemble them.
Conclusions.
In summary, we present the coassembly, MAGs, and functional annotations of 3.4 Tbp of tropical soil metagenome data. We suggest that terabase-scale metagenome coassembly captures substantially more high-quality MAGs from a broader array of phyla than multiassembly of the same samples and is a useful tool for characterizing an environment’s full taxonomic diversity, including eukaryotic organisms and rare, low-abundance microbes and viruses. Multiassembly remains a valuable approach as, in addition to its computational feasibility, it may recover more species- and strain-level variation than coassembly. Thus, coassembly and multiassembly are complementary strategies for metagenome analysis, and we suggest that the optimal results can be achieved when both can be performed. As sequencing of environments goes deeper and bioinformatics methods improve, we may enter the era of the reference metagenome, in which high-quality MAGs may be assembled for the majority of a microbial community, and provide broad utility for the interpretation of experiments such as the GRE.
MATERIALS AND METHODS
Sample collection.
Several kilos of soil was collected from the Luquillo Experimental Forest soil, homogenized, and incubated as described in reference 15. Briefly, we collected surface soil from a mid-ridge position in the LEF near the El Verde Field Station and then conducted a well-replicated resolution redox oscillation study in the laboratory at Lawrence Livermore National Laboratory (LLNL), the Great Redox Experiment (GRE). These soils are pH 5 clay-rich volcanoclastic oxisols overlain by a tabunuco forest and receive 2,000 to 6,000 mm in annual rainfall. In 178 microcosms, soils were amended with 13C-labeled or unlabeled (12C) plant litter and incubated for 44 days under four treatments: (i) static anoxic, (ii) static oxic, (iii) flux 4-day (4 days anoxic, 4 days oxic), and (iv) flux 8-day (4 days anoxic, 8 days oxic). DNA was extracted from replicate microcosms harvested at 44 days using a modified Griffith’s protocol (40). DNA was then density fractionated on the LLNL HT-SIP pipeline (41). A total of 10 unfractionated bulk samples (2 replicates from each redox treatment and 2 time-zero original samples) and 86 stable isotope probing density fractions (2 samples per treatment/isotope, and the 5 to 6 heaviest fractions per sample) were submitted to the JGI for metagenomic sequencing. One sample failed during library prep and was abandoned.
Genome sequencing.
Libraries were generated using either using the Kapa Biosystems library preparation kit (Roche) or the Nextera XT kit (Illumina), depending on available mass. For the Kapa Biosystems libraries, 200 ng of DNA was sheared to approximately 500 bp using an LE220 focused-ultrasonicator (Covaris). The sheared DNA fragments were size selected by double-solid phase reversible immobilization (SPRI), and then the selected fragments were end-repaired, A-tailed, and ligated with Illumina-compatible sequencing adaptors from IDT containing a unique molecular index barcode for each sample library. For the Nextera XT libraries, 2 ng of DNA was fragmented and adapter ligated. The ligated DNA fragments were enriched with 9 to 12 cycles of PCR and purified using SPRI beads (Beckman Coulter or Omega Bio-tek). Quantitative PCR (qPCR) was used to determine the concentration of the libraries using a LightCycler 480 real-time PCR instrument (Roche). Sequencing of the flow cell was performed on the NovaSeq (Illumina) sequencer using NovaSeq XP V1 reagent kits, S4 flow cell, following a 2 × 151-indexed run recipe.
Metagenome assembly.
For the individual metaSPAdes assemblies, paired-end Illumina reads were trimmed and screened according to the documentation for BBTools (42) filtered reads and were read corrected using BFC (43) version r181 (bfc -1 -s 10g -k 21 -t 10). Reads with no mate pair were removed. The resulting reads were then assembled using metaSPAdes version 3.12.0 (12) using a range of kmers (spades.py -m 2000 –only-assembler -k 33,55,77,99,127 –meta -t 32 -1 -2). The entire filtered read set was mapped to the final assembly, and coverage information was generated using BBMap version 38.22 using default parameters except for “ambiguous=random.”
To generate the coassembly, 95 FASTQ files totaling 7.74 TB of filtered reads, representing 3.40 Tbp of sequence and 265 billion unique 31-mers, were coassembled with MetaHipMer2 (11) version 2.0.1.v2.0.0-110-gb87d7c1-Issue69 (mhm2.py -v –post-asm-align –post-asm-abd –checkpoint=yes –checkpoint-merged=no –pin=none –ranks-per-gpu = 7) on 512 nodes on the Oak Ridge National Laboratory (ORNL) Summit supercomputer, taking approximately 1 h 24 m. Following assembly, contigs smaller than 500 bp were removed. Alignment coverage information was computed internally in MetaHipMer2 using a method similar to merAligner (44) during postassembly processing. Alignments for binning and annotation purposes were computed with BBTools (42) version 38.90 (bbmap.sh nodisk=true interleaved=true ambiguous=random mappedonly=t trimreaddescriptions=t usemodulo=t fast=t). Coverage information was determined using BBTools version 38.79 (pileup.sh) and default parameters on the combined read set, and again for each of the 95 individual read sets. Bin- or contig-wise coverage information reported in this paper was taken from the Avg_fold field output by pileup.sh.
Annotation.
Feature prediction and functional annotation of the assembled contigs were performed with version 5.0.24 of the IMG Annotation Pipeline (available for data set submission at https://img.jgi.doe.gov/submit). CRISPR elements were predicted via an in-house modified version of CRT-CLI version 1.2 (45) using the following search criteria: an element needs to have at least three repeats, search window size is set to 7 bp, minimum and maximum spacer length is set to 20 and 60 bp, respectively, and minimum and maximum repeat lengths are set to 20 and 50 bp, respectively. rRNA genes (5S, 16S, 23S), RNA regulatory features, and noncoding RNA genes were identified by comparing the contigs against the Rfam version 13.0 database (46) via cmsearch from the Infernal version 1.1.3 package (47) using the trusted cutoffs parameter (–cut_tc). Prediction of tRNAs was performed using the “bacterial” and “archaeal” search modes (-B/-A) of tRNAscan-SE version 2.0.8 (48). A combination of GeneMarkS-2 version 1.05 (49) (–Meta mgm_11.mod –incomplete_at_gaps 30) and Prodigal version 2.6.3 (50) (-p meta -m) was used to predict protein-coding genes.
Thereafter, the protein-coding genes were associated with functional annotations. KEGG Orthology (KO) terms and Enzyme Commission (EC) numbers were derived by running lastal 1066 from the LAST package (51) with default parameters against a reference database of isolate proteomes (IMG-NR 20190607). Comparison of the protein sequences against the remaining databases was performed with the HMMER version 3.1b2 (52) package, and specifically, via a thread-optimized version of hmmsearch (53). Assignments to version 15.0 of the TIGRFAM database (54), the frozen set of version 4.2.0 of the CATH-FunFam database (55), version 1.75 of the SUPERFAMILY database (56), version 01_06_2016 of the SMART database (57), and the updated 2014 COG models (58) were derived using a per-domain E value cutoff of 0.01 (–domE 0.01), whereas for Pfam-A assignments, the proteins were compared to version 30 of the Pfam database (59) using model-specific trusted cutoffs (–cut_tc).
Based on the name of their associated protein family, protein product names were then assigned in the order of priority: KO term > TIGRFAM > COG > Pfam. If none of the mentioned annotations were assigned, proteins were annotated as “hypothetical protein.”
Overlap resolution, filtering, and postprocessing of the various steps were executed as detailed in reference 19.
Binning and MAG extraction.
MetaBAT version 2:2.15 (60) was used to generate the depths from the per-sample alignment files (jgi_summarize_bam_contig_depths –outputDepth) and generate the bins (metabat2 -m 3000 –minS 80 –maxEdges 500 –seed 1000), completeness and contamination were assessed with CheckM version 1.1.3 (checkm lineage_wf -x fa) (61), and taxonomy was assigned using GTDB-Tk version 1.3.0 and GTDB database release 95 (gtdbtk classify_wf –extension fa) (62). Bins were assigned as high, medium, or low quality based on the MiMAG standards (20). The multiassembly MAGs were generated by applying the same procedure to each of the individual added metaSPAdes assemblies, using each assembly’s read alignment file to generate the depths for MetaBAT. The 294 medium- and high-quality multiassembly MAGs are provided in Table S2.
Construction of a concatenated marker gene phylogeny.
A three-subunit RNA polymerase (COG00085, COG00086, and COG0202) concatenated marker gene tree was constructed by combining a set of reference genomes spanning the bacterial and archaeal domains together with the GRE query genomes, i.e., the GRE coassembly and multiassembly MAGs. The set of reference genomes was collected by clustering the full set of public Integrated Microbial Genomes (IMG) isolate genomes using Cd-hit (63) version 4.8.1 to cluster the RNA polymerase gene (rpoB) at 80%, producing a set of references that was roughly unique at the genus to family taxonomic level. The GRE MAGs were dereplicated by grouping MAGs into species-level groups using a Mash (22) version 2.0 cutoff distance of 0.05, followed by clustering with mcl (64) version 14-137 with an inflation parameter of 1.5. The dereplicated set of MAGs and the reduced set of archaeal and bacterial reference genomes were passed through the SGTree version 0.0.10 pipeline (65). Briefly, this pipeline extracts the set of three marker genes from the set of query and reference genomes using hmmsearch version 3.1b2 (52), performs alignments of each marker with MAFFT (66) version v7.490 (2021/Oct/30) using the mafft-linsi option, trims alignments with trimAl version 1.4 (67), and removes sites when more than 90% of taxa contain a gap. The presence of all 3 subunits was required for a reference and/or query genome to be included into the tree; thus, due to various levels of MAG completeness, not all MAGs are included in Fig. 1. Finally, individual protein alignments were concatenated, followed by maximum likelihood tree construction with IQ-TREE (68) multicore version 1.6.1 using the WAG substitution model with 1,000 bootstraps. Trees were visualized with ggtree (69) version ggtree_3.2.1, and Faith’s phylogenetic diversity (PD, the sum of all branch lengths separating taxa in a community) was computed using the R Picante package (70) version picante_1.8.2. Phylum-level taxonomic designations were assigned using GTDB-Tk (62).
Rare biosphere microbes.
Taxonomic assignment of low-quality MAGs was performed with GTDB-Tk version 1.5.1 (gtdbtk classify_wf –extension fa) (62). Completeness and contamination metrics were calculated with CheckM during the binning procedure detailed above. Bin contig coverage information was calculated with BBTools as described above in the metagenome assembly details. Coassembly and multiassembly coverage of rare biosphere bins as a whole were calculated as contig length-weighted means of Avg_fold values for each contig.
Eukaryotic MAGs.
To detect possible eukaryotic MAGs, EukCC version 2.1.2 (32) (eukcc folder –threads 32 –suffix fa) was run on the nucleotide sequences of the low-quality bins (completion, <50%; contamination, <10%) using the database downloaded from http://ftp.ebi.ac.uk/pub/databases/metagenomics/eukcc/.
The fungal MAG’s completeness was estimated using BUSCO version 5.4.3 (36) (busco -m genome -c 32 -q -f) and the fungi_odb10 database and was inferred to be in the Coniochaeta clade based on BLAST searches against the MycoCosm (71) and NCBI nonredundant (nr) (72) databases.
Virus identification and clustering.
To identify and taxonomically classify viral genomes in the multiassembly and coassembly, contigs with at least 3,000 bp were processed with geNomad version 1.1.0 (available at https://github.com/apcamargo/genomad) (38). Composition-based score calibration was employed to set the false-discovery rate to 5% (–enable-score-calibration –max-fdr 0.05), and the presence of at least one virus hallmark was required (–min-virus-hallmarks 1). Genome completeness was estimated using CheckV version 1.0.1 (database version 1.4) (73).
To cluster the viral genomes identified across all the assemblies into viral taxonomic units (vOTUs), we first performed an all-versus-all BLAST (74) search (version 2.13.0+, parameters: -task megablast -evalue 1e-5 -max_target_seqs 20000) to align pairs of similar sequences. Next, the average nucleotide identity (ANI) and the aligned fraction (AF) of each pair were computed from the alignments, and a graph was constructed by connecting viral contigs with an ANI of ≥95% and AF of ≥85% (75). Last, sequences were clustered into vOTUs using the Leiden algorithm (76) (from the igraph Python library, version 0.9.10, resolution parameter = 1.0). The code used for vOTU-level clustering can be found at https://github.com/apcamargo/bioinformatics-snakemake-pipelines/tree/main/genome-ani-leiden-clustering-pipeline. Genomes were also clustered at the genus and family levels using pairwise average amino acid identities (AAI) computed from the output of an all-versus-all DIAMOND (version 2.0.15.153) search, as described previously (77). Scripts used for AAI-based clustering can be found at https://github.com/snayfach/MGV/tree/master/aai_cluster.
Metagenome coverage and diversity.
To estimate metagenome coverage and diversity, we used Nonpareil3 version 3.3.03 (39) (nonpareil -T kmer -f fastq -t 32 -R 110000). FASTQ files were pooled into 23 groups according to the SIP/bulk experiment (Table S3) and split by read pair using BBTools version 38.75 (reformat.sh int=t). Because Nonpareil analyzes only one member of each paired read, and because ours is a paired-read data set, we take the computed LRstar of 1.25 Tbp to obtain our estimate of how much sequencing would be required to cover 95% of the diversity in the LEF.
Data availability.
The GRE coassembly, high- and medium-quality MAGs, and annotations are available in the IMG/MER Database under the taxon object ID 3300047160. The corresponding data for the 95 individual metagenomes that were individually assembled for this work can be found by following the related sample links from the microbiome details page. The metadata for these samples can be found in the GOLD database (https://gold.jgi.doe.gov/) under the analysis project identifier Ga0500728. Raw reads can be found in the NCBI SRA database using the accession numbers provided in Table S4.
ACKNOWLEDGMENTS
The work (proposal 10.46936/10.25585/60000880), conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under contract no. DE-AC02-05CH11231. Samples were produced via a U.S. Department of Energy Early Career Award to J.P.-R. (SCW1478) administered by the Office of Biological and Environmental Research (OBER), Genomic Sciences Program. Sequencing was conducted via the Joint Genome Institute Community Sequencing Program, proposal ID 502924. Work at LLNL was supported by DOE award SCW1632 and performed under the auspices of the U.S. Department of Energy under contract DE-AC52-07NA27344. This research used resources of the National Energy Research Scientific Computing Center (NERSC) (https://ror.org/05v3mvq14), a U.S. Department of Energy Office of Science User Facility located at Lawrence Berkeley National Laboratory. Both are supported by the Office of Science of the U.S. Department of Energy operated under contract no. DE-AC02-05CH11231. This research was also supported by the Exascale Computing Project (17-SC-20-SC), a collaborative effort of two U.S. Department of Energy organizations (the Office of Science and the National Nuclear Security Administration) responsible for the planning and preparation of a capable exascale ecosystem, including software, applications, hardware, advanced system engineering, and early testbed platforms, in support of the nation’s exascale computing imperative. This research used resources of the Oak Ridge Leadership Computing Facility at the Oak Ridge National Laboratory (https://ror.org/01qz5mb56), which is supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC05-00OR22725 and the resources of the National Energy Research Scientific Computing Center (NERSC), a U.S. Department of Energy Office of Science User Facility located at Lawrence Berkeley National Laboratory, operated under contract no. DE-AC02-05CH11231.
R.R., R.M.B., A.P.Camargo., M.K., and A.Clum. wrote the paper; A.N.Campbell. and J.P.-R. generated the samples; R.R., R.M.B., A.P.Camargo., R.E., B.F., S.H., M.H., M.K., J.A.K., and N.J.V. analyzed data; E.A.E.-F., J.P.-R., and A.Clum. conceived and supervised the project. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Robert Riley, Email: rwriley@lbl.gov.
Erik F. Y. Hom, University of Mississippi
REFERENCES
- 1.Sunagawa S, Acinas SG, Bork P, Bowler C, Eveillard D, Gorsky G, Guidi L, Iudicone D, Karsenti E, Lombard F, Ogata H, Pesant S, Sullivan MB, Wincker P, de Vargas C, Tara Oceans Coordinators . 2020. Tara Oceans: towards global ocean ecosystems biology. Nat Rev Microbiol 18:428–445. doi: 10.1038/s41579-020-0364-5. [DOI] [PubMed] [Google Scholar]
- 2.He S, Malfatti SA, McFarland JW, Anderson FE, Pati A, Huntemann M, Tremblay J, Glavina del Rio T, Waldrop MP, Windham-Myers L, Tringe SG. 2015. Patterns in wetland microbial community composition and functional gene repertoire associated with methane emissions. mBio 6:e00066-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Integrative HMP (iHMP) Research Network Consortium. 2019. The Integrative Human Microbiome Project. Nature 569:641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch MDJ, Neufeld JD. 2015. Ecology and exploration of the rare biosphere. Nat Rev Microbiol 13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 5.Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pascoal F, Costa R, Magalhães C. 2021. The microbial rare biosphere: current concepts, methods and ecological principles. FEMS Microbiol Ecol 97:fiaa227. doi: 10.1093/femsec/fiaa227. [DOI] [PubMed] [Google Scholar]
- 7.Danczak RE, Johnston MD, Kenah C, Slattery M, Wrighton KC, Wilkins MJ. 2017. Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 5:112. doi: 10.1186/s40168-017-0331-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapidus AL, Korobeynikov AI. 2021. Metagenomic data assembly: the way of decoding unknown microorganisms. Front Microbiol 12:613791. doi: 10.3389/fmicb.2021.613791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 10.Compeau PEC, Pevzner PA, Tesler G. 2011. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol 29:987–991. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmeyr S, Egan R, Georganas E, Copeland AC, Riley R, Clum A, Eloe-Fadrosh E, Roux S, Goltsman E, Buluç A, Rokhsar D, Oliker L, Yelick K. 2020. Terabase-scale metagenome coassembly with MetaHipMer. Sci Rep 10:10689. doi: 10.1038/s41598-020-67416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 14.National Science Foundation. 2022. Long Term Ecological Research Network. https://lternet.edu.
- 15.Bhattacharyya A, Campbell AN, Tfaily MM, Lin Y, Kukkadapu RK, Silver WL, Nico PS, Pett-Ridge J. 2018. Redox fluctuations control the coupled cycling of iron and carbon in tropical forest soils. Environ Sci Technol 52:14129–14139. doi: 10.1021/acs.est.8b03408. [DOI] [PubMed] [Google Scholar]
- 16.Oak Ridge National Laboratory. 2018. Summit: Oak Ridge Leadership Computing Facility. https://www.olcf.ornl.gov/summit.
- 17.Chen IMA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M, Hajek P, Ritter S, Varghese N, Seshadri R, Roux S, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides N. 2021. The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res 49:D751–D763. doi: 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer F, Fritz A, Deng ZL, Koslicki D, Lesker TR, Gurevich A, Robertson G, Alser M, Antipov D, Beghini F, Bertrand D, Brito JJ, Brown CT, Buchmann J, Buluç A, Chen B, Chikhi R, Clausen PTLC, Cristian A, Dabrowski PW, Darling AE, Egan R, Eskin E, Georganas E, Goltsman E, Gray MA, Hansen LH, Hofmeyr S, Huang P, Irber L, Jia H, Jørgensen TS, Kieser SD, Klemetsen T, Kola A, Kolmogorov M, Korobeynikov A, Kwan J, LaPierre N, Lemaitre C, Li C, Limasset A, Malcher-Miranda F, Mangul S, Marcelino VR, Marchet C, Marijon P, Meleshko D, Mende DR, Milanese A, et al. 2022. Critical Assessment of Metagenome Interpretation: the second round of challenges. Nat Methods 19:429–440. doi: 10.1038/s41592-022-01431-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clum A, Huntemann M, Bushnell B, Foster B, Foster B, Roux S, Hajek PP, Varghese N, Mukherjee S, Reddy TBK, Daum C, Yoshinaga Y, O’Malley R, Seshadri R, Kyrpides NC, Eloe-Fadrosh EA, Chen IMA, Copeland A, Ivanova NN, Segata N. 2021. DOE JGI Metagenome Workflow. mSystems 6:e00804-20. doi: 10.1128/mSystems.00804-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowers RM, Kyrpides NC, Stepanauskas R, Harmon-Smith M, Doud D, Reddy TBK, Schulz F, Jarett J, Rivers AR, Eloe-Fadrosh EA, Tringe SG, Ivanova NN, Copeland A, Clum A, Becraft ED, Malmstrom RR, Birren B, Podar M, Bork P, Weinstock GM, Garrity GM, Dodsworth JA, Yooseph S, Sutton G, Glöckner FO, Gilbert JA, Nelson WC, Hallam SJ, Jungbluth SP, Ettema TJG, Tighe S, Konstantinidis KT, Liu WT, Baker BJ, Rattei T, Eisen JA, Hedlund B, McMahon KD, Fierer N, Knight R, Finn R, Cochrane G, Karsch-Mizrachi I, Tyson GW, Rinke C, Lapidus A, Meyer F, Yilmaz P, Parks DH, Eren AM, Genome Standards Consortium , et al. 2017. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol 35:725–731. doi: 10.1038/nbt.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji M, Williams TJ, Montgomery K, Wong HL, Zaugg J, Berengut JF, Bissett A, Chuvochina M, Hugenholtz P, Ferrari BC. 2021. Candidatus Eremiobacterota, a metabolically and phylogenetically diverse terrestrial phylum with acid-tolerant adaptations. ISME J 15:2692–2707. doi: 10.1038/s41396-021-00944-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Youssef N, Steidley BL, Elshahed MS. 2012. Novel high-rank phylogenetic lineages within a sulfur spring (Zodletone Spring, Oklahoma), revealed using a combined pyrosequencing-Sanger approach. Appl Environ Microbiol 78:2677–2688. doi: 10.1128/AEM.00002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doud DFR, Bowers RM, Schulz F, De Raad M, Deng K, Tarver A, Glasgow E, Vander Meulen K, Fox B, Deutsch S, Yoshikuni Y, Northen T, Hedlund BP, Singer SW, Ivanova N, Woyke T. 2020. Function-driven single-cell genomics uncovers cellulose-degrading bacteria from the rare biosphere. ISME J 14:659–675. doi: 10.1038/s41396-019-0557-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desai A, Marwah VS, Yadav A, Jha V, Dhaygude K, Bangar U, Kulkarni V, Jere A. 2013. Identification of optimum sequencing depth especially for de novo genome assembly of small genomes using next generation sequencing data. PLoS One 8:e60204. doi: 10.1371/journal.pone.0060204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto E, Muramatsu H, Nagai K. 2014. Vulgatibacter incomptus gen. nov., sp. nov. and Labilithrix luteola gen. nov., sp. nov., two myxobacteria isolated from soil in Yakushima Island, and the description of Vulgatibacteraceae fam. nov., Labilitrichaceae fam. nov. and Anaeromyxobacteraceae fam. nov. Int J Syst Evol Microbiol 64:3360–3368. doi: 10.1099/ijs.0.063198-0. [DOI] [PubMed] [Google Scholar]
- 27.Mulwa L, Jansen R, Praditya D, Mohr K, Wink J, Steinmann E, Stadler M. 2018. Six Heterocyclic Metabolites from the Myxobacterium Labilithrix luteola. Molecules 23:542. doi: 10.3390/molecules23030542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li N, Li X, Zhang HJ, Fan XY, Liu YK. 2021. Microbial community and antibiotic resistance genes of biofilm on pipes and their interactions in domestic hot water system. Sci Total Environ 767:144364. doi: 10.1016/j.scitotenv.2020.144364. [DOI] [PubMed] [Google Scholar]
- 29.Muñoz-Dorado J, Marcos-Torres FJ, García-Bravo E, Moraleda-Muñoz A, Pérez J. 2016. Myxobacteria: moving, killing, feeding, and surviving together. Front Microbiol 7:781. doi: 10.3389/fmicb.2016.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy CL, Sheremet A, Dunfield PF, Spear JR, Stepanauskas R, Woyke T, Elshahed MS, Youssef NH. 2021. Genomic analysis of the yet-uncultured binatota reveals broad methylotrophic, alkane-degradation, and pigment production capacities. mBio 12:e00985-21. doi: 10.1128/mBio.00985-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camargo AP, de Souza RSC, Jose J, Gerhardt IR, Dante RA, Mukherjee S, Huntemann M, Kyrpides NC, Carazzolle MF, Arruda P. 2023. Plant microbiomes harbor potential to promote nutrient turnover in impoverished substrates of a Brazilian biodiversity hotspot. ISME J 17:354–370. doi: 10.1038/s41396-022-01345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saary P, Mitchell AL, Finn RD. 2020. Estimating the quality of eukaryotic genomes recovered from metagenomic analysis with EukCC. Genome Biol 21:244. doi: 10.1186/s13059-020-02155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker SE, Schackwitz W, Lipzen A, Martin J, Haridas S, LaButti K, Grigoriev IV, Simmons BA, McCluskey K. 2015. Draft genome sequence of Neurospora crassa strain FGSC 73. Genome Announc 3:e00074-15. doi: 10.1128/genomeA.00074-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuomo CA, Güldener U, Xu J-R, Trail F, Turgeon BG, Di Pietro A, Walton JD, Ma L-J, Baker SE, Rep M, Adam G, Antoniw J, Baldwin T, Calvo S, Chang Y-L, Decaprio D, Gale LR, Gnerre S, Goswami RS, Hammond-Kosack K, Harris LJ, Hilburn K, Kennell JC, Kroken S, Magnuson JK, Mannhaupt G, Mauceli E, Mewes H-W, Mitterbauer R, Muehlbauer G, Münsterkötter M, Nelson D, O'donnell K, Ouellet T, Qi W, Quesneville H, Roncero MIG, Seong K-Y, Tetko IV, Urban M, Waalwijk C, Ward TJ, Yao J, Birren BW, Kistler HC. 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- 35.Franco MEE, Wisecaver JH, Arnold AE, Ju Y-M, Slot JC, Ahrendt S, Moore LP, Eastman KE, Scott K, Konkel Z, Mondo SJ, Kuo A, Hayes RD, Haridas S, Andreopoulos B, Riley R, LaButti K, Pangilinan J, Lipzen A, Amirebrahimi M, Yan J, Adam C, Keymanesh K, Ng V, Louie K, Northen T, Drula E, Henrissat B, Hsieh H-M, Youens-Clark K, Lutzoni F, Miadlikowska J, Eastwood DC, Hamelin RC, Grigoriev IV, U’Ren JM. 2022. Ecological generalism drives hyperdiversity of secondary metabolite gene clusters in xylarialean endophytes. New Phytol 233:1317–1330. doi: 10.1111/nph.17873. [DOI] [PubMed] [Google Scholar]
- 36.Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. 2021. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 38:4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiménez DJ, Hector RE, Riley R, Lipzen A, Kuo RC, Amirebrahimi M, Barry KW, Grigoriev IV, van Elsas JD, Nichols NN. 2017. Draft genome sequence of Coniochaeta ligniaria NRRL 30616, a Lignocellulolytic fungus for bioabatement of inhibitors in plant biomass hydrolysates. Genome Announc 5:e01476-16. doi: 10.1128/genomeA.01476-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camargo AP, Roux S, Schulz F, Babinski M, Xu Y, Hu B, Chain PSG, Nayfach S, Kyrpides NC. 2023. You can move, but you can’t hide: identification of mobile genetic elements with geNomad. bioRxiv. doi: 10.1101/2023.03.05.531206. [DOI] [PMC free article] [PubMed]
- 39.Rodriguez-R LM, Gunturu S, Tiedje JM, Cole JR, Konstantinidis KT. 2018. Nonpareil 3: fast estimation of metagenomic coverage and sequence diversity. mSystems 3:e00039-18. doi: 10.1128/mSystems.00039-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffiths RI, Whiteley AS, O’Donnell AG, Bailey MJ. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl Environ Microbiol 66:5488–5491. doi: 10.1128/AEM.66.12.5488-5491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nuccio EE, Blazewicz SJ, Lafler M, Campbell AN, Kakouridis A, Kimbrel JA, Wollard J, Vyshenska D, Riley R, Tomatsu A, Hestrin R, Malmstrom RR, Firestone M, Pett-Ridge J. 2022. HT-SIP: a semi-automated stable isotope probing pipeline identifies cross-kingdom interactions in the hyphosphere of arbuscular mycorrhizal fungi. Microbiome 10. doi: 10.1186/s40168-022-01391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bushnell B. 2022. BBMap. DOE Joint Genome Institute. https://sourceforge.net/projects/bbmap/.
- 43.Li H. 2015. BFC: correcting Illumina sequencing errors. Bioinformatics 31:2885–2887. doi: 10.1093/bioinformatics/btv290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georganas E, Buluç A, Chapman J, Oliker L, Rokhsar D, Yelick K. 2015. merAligner: a fully parallel sequence aligner, p 561–570. In 2015 IEEE International Parallel and Distributed Processing Symposium. Institute of Electrical and Electronics Engineers (IEEE), New York, NY, USA. doi: 10.1109/IPDPS.2015.96. [DOI] [Google Scholar]
- 45.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P. 2007. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD, Petrov AI. 2018. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res 46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nawrocki EP, Eddy SR. 2013. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol 21962:1–14. doi: 10.1007/978-1-4939-9173-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lomsadze A, Gemayel K, Tang S, Borodovsky M. 2018. Modeling leaderless transcription and atypical genes results in more accurate gene prediction in prokaryotes. Genome Res 28:1079–1089. doi: 10.1101/gr.230615.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res 21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res 41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arndt W. 2018. Modifying HMMER3 to run efficiently on the cori supercomputer using OpenMP tasking, p 239–246. In 2018 IEEE International Parallel and Distributed Processing Symposium Workshops (IPDPSW). IEEE. Institute of Electrical and Electronics Engineers (IEEE), New York, NY, USA. doi: 10.1109/IPDPSW.2018.00048. [DOI] [Google Scholar]
- 54.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. 2012. TIGRFAMs and Genome Properties in 2013. Nucleic Acids Res 41:D387–D395. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sillitoe I, Dawson N, Lewis TE, Das S, Lees JG, Ashford P, Tolulope A, Scholes HM, Senatorov I, Bujan A, Ceballos Rodriguez-Conde F, Dowling B, Thornton J, Orengo CA. 2019. CATH: expanding the horizons of structure-based functional annotations for genome sequences. Nucleic Acids Res 47:D280–D284. doi: 10.1093/nar/gky1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gough J, Karplus K, Hughey R, Chothia C. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 57.Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z. 2019. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 64.Van Dongen S. 2008. Graph clustering via a discrete uncoupling process. SIAM J Matrix Anal Appl 30:121–141. doi: 10.1137/040608635. [DOI] [Google Scholar]
- 65.Schulz F, Walker EW, Jungbluth S. 2019. SGTree. DOE Joint Genome Institute. https://pypi.org/project/sgtree/.
- 66.Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu G. 2020. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinform 69:e96. doi: 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
- 70.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 71.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T, Nordberg H, Dubchak I, Shabalov I. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res 42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, Connor R, Funk K, Kelly C, Kim S, Madej T, Marchler-Bauer A, Lanczycki C, Lathrop S, Lu Z, Thibaud-Nissen F, Murphy T, Phan L, Skripchenko Y, Tse T, Wang J, Williams R, Trawick BW, Pruitt KD, Sherry ST. 2022. Database resources of the national center for biotechnology information. Nucleic Acids Res 50:D20–D26. doi: 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC. 2021. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol 39:578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, Ye W, Zhang Y, Xu Y. 2015. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res 43:7762–7768. doi: 10.1093/nar/gkv784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, Kuhn JH, Lavigne R, Brister JR, Varsani A, Amid C, Aziz RK, Bordenstein SR, Bork P, Breitbart M, Cochrane GR, Daly RA, Desnues C, Duhaime MB, Emerson JB, Enault F, Fuhrman JA, Hingamp P, Hugenholtz P, Hurwitz BL, Ivanova NN, Labonté JM, Lee KB, Malmstrom RR, Martinez-Garcia M, Mizrachi IK, Ogata H, Páez-Espino D, Petit MA, Putonti C, Rattei T, Reyes A, Rodriguez-Valera F, Rosario K, Schriml L, Schulz F, Steward GF, Sullivan MB, Sunagawa S, Suttle CA, Temperton B, Tringe SG, Thurber RV, Webster NS, Whiteson KL, et al. 2019. Minimum Information about an Uncultivated Virus Genome (MIUViG). Nat Biotechnol 37:29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Traag VA, Waltman L, van Eck NJ. 2019. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 9:5233. doi: 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nayfach S, Páez-Espino D, Call L, Low SJ, Sberro H, Ivanova NN, Proal AD, Fischbach MA, Bhatt AS, Hugenholtz P, Kyrpides NC. 2021. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat Microbiol 6:960–970. doi: 10.1038/s41564-021-00928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download spectrum.00200-23-s0001.txt, TXT file, 10.7 MB (10.7MB, txt)
Table S2. Download spectrum.00200-23-s0002.txt, TXT file, 2.1 MB (2.1MB, txt)
Table S3. Download spectrum.00200-23-s0003.txt, TXT file, 0.00 MB (3.6KB, txt)
Table S4. Download spectrum.00200-23-s0004.txt, TXT file, 0.00 MB (5.1KB, txt)
Fig. S1. Download spectrum.00200-23-s0005.eps, EPS file, 1.5 MB (1.5MB, eps)
Data Availability Statement
The GRE coassembly, high- and medium-quality MAGs, and annotations are available in the IMG/MER Database under the taxon object ID 3300047160. The corresponding data for the 95 individual metagenomes that were individually assembled for this work can be found by following the related sample links from the microbiome details page. The metadata for these samples can be found in the GOLD database (https://gold.jgi.doe.gov/) under the analysis project identifier Ga0500728. Raw reads can be found in the NCBI SRA database using the accession numbers provided in Table S4.