Abstract
Several viruses, including members of the gammaherpesvirus family, encode proteins that are secreted into the extracellular environment. We have identified an abundant 44-kDa secreted protein that is present in the supernatant of fibroblasts infected with murine gammaherpesvirus 68 (γHV68; also referred to as MHV-68) but not in that of uninfected fibroblasts. Sequence analysis of the amino terminus and of internal peptides revealed that this protein is encoded by the γHV68 M3 open reading frame (ORF). The amino-terminal sequence of the secreted protein starts at residue 25 of the M3 ORF, consistent with the first 24 residues functioning as a signal peptide. Northern blot analysis revealed a single abundant ∼1.4-kb early-late lytic transcript encoded by the M3 ORF. Analysis of a partial cDNA clone and subsequent analyses of products of rapid amplification of cDNA ends coupled with S1 nuclease protection assays demonstrate that the M3 protein is encoded by an unspliced, polyadenylated mRNA initiating at bp 7294 and terminating at bp 6007 of the γHV68 genome. The 3′ end of the M3 transcript maps 9 bp downstream of a consensus polyadenylation signal. Thus, the predicted M3 ORF is a functional gene that encodes an abundant secreted protein which is a candidate for interacting with host cellular receptors or cytokines.
Gammaherpesviruses are characterized biologically by their ability to establish latency in lymphocytes and by an association with tumors in immunosuppressed hosts. To help elucidate the pathogenesis of acute and chronic gammaherpesvirus infection, a mouse model of gammaherpesvirus infection has recently been established. Murine gammaherpesvirus 68 (γHV68; also referred to as MHV-68) is a natural pathogen of wild murid rodents (2), capable of infecting both outbred and inbred mice (3, 8, 11, 15, 21, 22). Viral genome structure and sequence analysis indicate that γHV68 is related to the primate gammaherpesviruses herpesvirus saimiri (HVS), Kaposi’s sarcoma-associated herpesvirus (KSHV), and Epstein-Barr virus (EBV) (1, 6, 7, 16, 23).
Many viruses encode proteins that are secreted into the extracellular milieu; the majority of these serve to modulate the host immune response. Notably, within the gammaherpesvirus family, EBV encodes both an interleukin-10 (IL-10) homolog (12) and a soluble colony-stimulating factor-1 receptor (20), while KSHV encodes an IL-6 homolog and three chemokine homologs (18) and HVS encodes an IL-17 homolog (26). In this report, we identify an abundant γHV68 secreted protein encoded by the M3 open reading frame (ORF) and define the structure of the transcript encoding this protein.
Detection of an abundantly secreted protein encoded by the M3 ORF of γHV68.
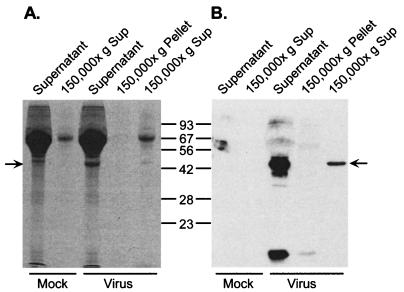
To assess the possibility that γHV68 encodes a secreted protein(s), murine NIH 3T12 fibroblast cells were infected with γHV68 at a multiplicity of infection (MOI) of 5. At 24 h postinfection the culture supernatant was harvested, filtered, and concentrated. It should be noted that there was little evidence of viral cytopathic effect at 24 h postinfection. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of mock-infected and infected culture supernatants revealed the presence of an ∼44-kDa protein present only in the media of γHV68-infected cells which could be detected on Coomassie blue-stained gels (Fig. 1A). To further assess whether this protein was associated with cellular debris or viral particles, the concentrated supernatants were centrifuged at 150,000 × g at 4°C for 3 h to pellet any residual membranes or membrane-associated proteins (Fig. 1). Notably, the 44-kDa virus-specific band could be detected in the 150,000 × g supernatant fraction but was not visible in the 150,000 × g pellet fraction, consistent with this being a secreted protein.
FIG. 1.
Detection of an abundant secreted protein in supernatants of γHV68 infected fibroblasts. A murine 3T12 fibroblast cell line was either mock infected or infected at an MOI of 5 with γHV68 virus (WUMS strain) in Dulbecco’s modified Eagle medium (DMEM) supplemented with 1% fetal calf serum (FCS). After 1 h, the monolayer was washed with phosphate-buffered saline, fresh DMEM containing 1% FCS was added back to the monolayer, and the infection was allowed to proceed for 23 h. The culture supernatant was passed through a 0.2-μm-pore-size filter to remove any cellular debris and was concentrated at 4°C by using an Amicon (Beverly, Mass.) Centriprep-10 concentrator (from 15 ml to 600 μl). In addition, the concentrated supernatants were centrifuged at 150,000 × g for 3 h to remove any remaining aggregated material and residual free virus present in the supernatant samples. The concentrated supernatant samples, as well as the high-speed supernatants and the pellets obtained from the 150,000 × g spin, were analyzed on SDS–12% polyacrylamide gels. (A) SDS-PAGE analysis and staining with Coomassie brilliant blue R250. Note the prominent ca. 45-kDa band in the γHV68-infected precentrifugation and γHV68 ultracentrifugation supernatant lanes and the absence of this band in both the mock-infected supernatant and the virus-infected high-speed pellet. (B) Immunoblot analysis of fractions shown in panel A, probed with a rabbit polyclonal anti-γHV68 antiserum (25) (1:2,000 dilution), followed by a 1:5,000 dilution of horseradish peroxidase-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch, West Grove, Pa.), and visualized with a chemiluminescence detection kit (Amersham, Arlington Heights, Ill.). No reactive species were detected when normal rabbit serum was used as a control primary antibody (data not shown). Molecular sizes in kilodaltons are given in the center. The arrow indicates the migration of the virus-specific band. The prominent band above the virus-specific band is serum albumin.
To address whether the 44-kDa virus-induced protein might be virally encoded, the supernatant fractions were analyzed by immunoblotting using a rabbit polyclonal anti-γHV68 antiserum (Fig. 1B). The immunoblot demonstrated that the 44-kDa protein is recognized by the rabbit antiserum (Fig. 1B) but not by normal rabbit serum (data not shown). In addition, consistent with the Coomassie staining results, the 44-kDa band was not detectable in the 150,000 × g pellet fraction (Fig. 1B). Based on these results, the 44-kDa protein was isolated from concentrated culture media of virus-infected cells by fractionation on an SDS-PAGE gel and blotting to a polyvinylidene difluoride (PVDF) membrane. After the membrane was stained with Coomassie blue, the 44-kDa band was excised and eluted from the membrane. Both amino-terminal sequencing and sequencing of high-pressure liquid chromatography (HPLC)-purified tryptic peptides were carried out. This analysis unambiguously identified the secreted protein as being encoded by the γHV68 M3 ORF (23). The amino-terminal sequence, as well as the sequences of internal tryptic peptides, are outlined in Fig. 2 on the deduced amino acid sequence of the γHV68 M3 ORF. Notably, the amino terminus of the secreted 44-kDa protein starts 25 residues in from the predicted methionine initiation codon. This suggests that the first 24 residues, which are largely hydrophobic, act as a signal peptide. The size of the secreted protein is very consistent with the predicted size (42 kDa) of the M3-encoded protein lacking the first 24 residues and indicates that the M3 protein is unlikely to be glycosylated. This is consistent with a lack of consensus N- or O-linked glycosylation sites in the predicted ORF.
FIG. 2.
Microsequence analysis reveals that the abundant secreted protein is encoded by the γHV68 M3 ORF. Supernatant from γHV68-infected cells was recovered, electrophoresed through an SDS–10% PAGE gel, and transferred to a PVDF membrane. The membrane was stained with Coomassie blue, and the virus-specific band was cut out. The sample was eluted from the PVDF membrane, and 8 to 10 rounds of Edman degradation were performed on either the intact sample or four HPLC-purified tryptic peptides in order to obtain protein sequence information (13). Recovered sequences were compared to the GenBank protein sequence database by use of the program BLASTp (1a). This analysis revealed that the virus-specific secreted protein is encoded by the γHV68 M3 ORF. Sequences recovered by amino-terminal and internal peptide sequencing are boxed. Outlined letters represent amino acids that could not be resolved by microsequencing. The sequence shown in lowercase at the predicted amino terminus of M3 is presumed to encode a signal peptide which is cleaved from the mature secreted protein.
Characterization of the M3 transcript.
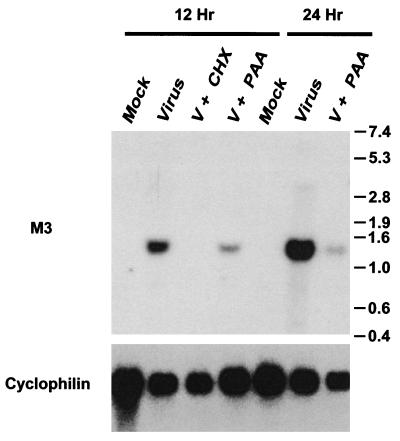
Herpesvirus gene expression follows a pattern of sequential expression of three classes of RNA (9). Immediate-early transcripts require only preformed host factors for expression and are therefore expressed in the presence of drugs that inhibit host cell translation, such as cycloheximide. Early transcripts require the products of immediate-early transcripts for expression, while late transcripts require viral DNA synthesis for maximal expression. To assess the kinetic class of M3 gene transcripts, NIH 3T12 fibroblasts were either mock infected or infected with γHV68 at an MOI of 5 in the presence or absence of inhibitors of protein synthesis (cycloheximide and anisomycin) or viral DNA polymerase activity (phosphonoacetic acid). Total cellular RNA was prepared from mock-infected and γHV68-infected cells either 12 or 24 h postinfection and analyzed by Northern blot hybridization. Hybridization with an M3-specific probe revealed the presence of a single predominant ca. 1.4-kb transcript (Fig. 3). The 1.4-kb transcript was not detected when protein synthesis inhibitors were present but was detected in the cells treated with phosphonoacetic acid (Fig. 3). However, addition of phosphonoacetic acid did significantly diminish the level of M3 transcript detected, indicating that transcription of the M3 gene is not completely independent of viral DNA replication. Thus, we have classified the M3 transcript as an early-late transcript. To control for the loading of RNA on the Northern blot, the blot was stripped and rehybridized with a probe for the cellular cyclophilin transcript (5) (Fig. 3).
FIG. 3.
Northern blot analysis of M3 transcripts reveals that M3 encodes a single abundant early-late mRNA. NIH 3T12 fibroblasts were either mock infected or infected at an MOI of 5 in a volume of 10 ml of DMEM containing 10% FCS for 1 h. After a 1-h incubation with virus, an additional 15 ml of medium was added (with or without the indicated inhibitors), and the flasks were incubated at 37°C under a 5% CO2 atmosphere for 12 or 24 h prior to harvesting of the cells and preparation of RNA. The infected fibroblasts were either left untreated or treated with inhibitors. γHV68 DNA synthesis was inhibited by the addition of phosphonoacetic acid (PAA) to a final concentration of 200 μg/ml, and protein synthesis was inhibited by adding a combination of cycloheximide to a final concentration of 40 μM and anisomycin to a final concentration of 10 μM (CHX). V, virus. Total cellular RNA was harvested by the single-step guanidinium thiocyanate-phenol method (14) and analyzed by Northern blot hybridization. The blot was probed for rat cyclophilin (5) to assess loading and RNA quality. The cyclophilin probe was radiolabeled with the Megaprime DNA Labeling System (Amersham) according to the manufacturer’s protocol. The M3-specific probe was generated by radiolabeling the oligonucleotide 5′-CAAACCTAGAGTAAGGCTCTCAGCCAATCCTCCTGCCAAC-3′, corresponding to the region of the γHV68 genome from bp 7197 to bp 7231 (see Fig. 4), with polynucleotide kinase.
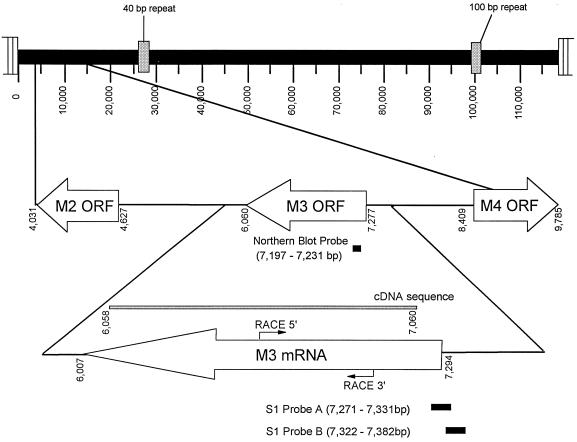
Analysis of ATG-initiated ORFs in the γHV68 genome indicates that the M3 ORF extends from bp 7277 to bp 6060 in the viral genome (Fig. 4). Notably, there is a candidate polyadenylation signal closely linked to the 3′ end of the M3 ORF at bp 6016 in the viral genome (23). Two approaches were used to characterize the structure of the M3 transcript (Fig. 4). Initially a cDNA library, prepared from mRNA isolated from γHV68-infected NIH 3T12 fibroblast cells, was screened with an M3 probe (bp 7176 to 6262). Positive plaques were isolated and sequenced, and the sequences were compiled. This analysis indicated that the entire M3 ORF is present in the mature transcript and is not spliced (Fig. 4). Rapid amplification of cDNA ends (RACE) was performed (17) to map the 5′ and 3′ ends of the M3 transcript. Sequence analysis of the recovered RACE products, in conjunction with the cDNA analysis, demonstrates that M3 encodes an unspliced transcript which is polyadenylated 53 nucleotides downstream of the translation stop codon.
FIG. 4.
Characterization of the structure of the M3-encoded transcript. Analysis of the γHV68 genome indicates that the M3 ORF extends from bp 7277 to bp 6060 of the γHV68 genome (23). Two approaches were used to characterize the mRNA structure of M3. First, a cDNA library (generated from pooled mRNA isolated from γHV68-infected 3T12 cells at 8 h [in the presence of cycloheximide and anisomycin], 12 h [no inhibitors], and 24 h [no inhibitors]) was screened with a labeled M3 probe (the labeled fragment contained the region from bp 6262 to bp 7176 of the γHV68 genome). The M3 fragment was radiolabeled with the Megaprime DNA Labeling System (Amersham) according to the manufacturer’s protocol. Positive plaques were isolated and sequenced by using primers internal to the M3 ORF. The compiled sequence indicated no splicing in the interior of the M3 ORF. In order to further characterize the 5′ and 3′ ends of the transcript, RACE was performed (17) with the Marathon cDNA Amplification Kit (Clontech Laboratories, Palo Alto, Calif.) according to the manufacturer’s protocol. Polyadenylated mRNA species were purified by oligo(dT) column chromatography (mRNA purification kit; Clontech Laboratories) from total γHV68-infected RNA harvested as described above. The PCR primers utilized for the 5′ and 3′ reactions were 5′-GCAGAGACATCTTTTCCATGCCAG-3′ and 5′-GTGGATGATTGACATT CCCAAATC-3′, respectively. Sequence analysis of the recovered RACE products indicated a single predominant unspliced transcript, containing a poly(A) tail added 53 nucleotides downstream of the translational stop signal. In the 3′ RACE reaction, a minor larger product which has not been fully characterized was noted.
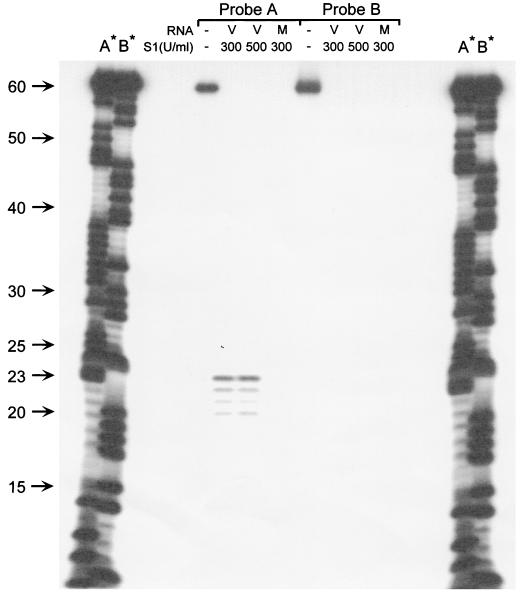
Since RACE analysis cannot precisely map the 5′ end of a transcript, the transcriptional start site was determined by S1 nuclease protection (Fig. 5). End-labeled, overlapping 60-mer oligonucleotides (see Fig. 4) were hybridized to RNA isolated from γHV68-infected NIH 3T12 cells. Notably, neither probe was protected from digestion by S1 nuclease when RNA isolated from mock-infected cells was used in the hybridization (Fig. 5). In contrast, the first 23 nucleotides of probe A were protected from S1 nuclease digestion by RNA from γHV68-infected cells (Fig. 5). This analysis provides strong evidence that the transcriptional start site is between 20 and 25 nucleotides upstream of the site of translation initiation. The fact that probe B, positioned slightly farther upstream of the translation initiation site, was not protected by γHV68 RNA, further supports this conclusion (Fig. 5).
FIG. 5.
S1 nuclease mapping of the 5′ end of the M3 transcript. S1 nuclease analysis was carried out with 40 μg of total RNA isolated from either mock-infected or γHV68-infected NIH 3T12 cells. RNA was hybridized overnight at 37°C with 4 ng of either probe A or probe B, which had been 32P labeled at the 5′ end in a volume of 10 μl of polynucleotide kinase buffer (Boehringer Mannheim, Indianapolis, Ind.). The sequences of the S1 probes used were 5′-AGGCCATGGCTGACGCTCTCCCAGAGTCGCAGGGAGACCCTCCTTAAATATGCTCCATGG-3′ for probe A and 5′-TGCTCCATGGTTTGGCAAAGCCTGCCCAGGCCACCTCAACACAACACTTTCTGTGGTGCC-3′ for probe B. The genomic locations of the S1 probes used are given in Fig. 4 below the schematic illustration of the M3-encoded transcript. After hybridization of the single-stranded oligonucleotide probes to the RNA, S1 nuclease (Promega, Madison, Wis.) in 300 μl of S1 reaction buffer (0.28 M NaCl, 50 mM sodium acetate, 4.5 mM ZnSO4) was added at a final concentration of 300 or 500 U/ml and the reaction mixture was incubated at 37°C for 30 min. The protected products were recovered as previously described (14) and analyzed by electrophoresis on a 10% denaturing acrylamide gel. Chemical cleavages of probe A and B (G+A reaction) were run as size markers in lanes marked A* and B*. V, RNA prepared from virus-infected cells; M, RNA prepared from mock-infected cells.
Conclusions.
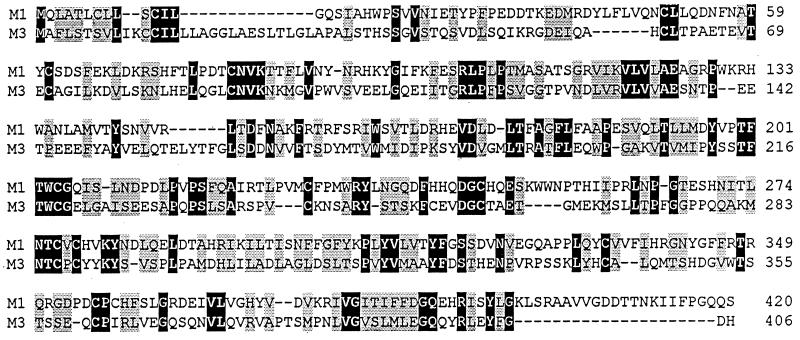
BLASTp analysis of the M3 protein failed to identify any clear homology to known cellular or viral gene products, with one exception. The exception is the γHV68 putative M1 gene product, which displays significant extended homology to the M3 protein (Fig. 6). The region of homology extends over nearly the entire M3 protein (from residue 59 through residue 404). Overall, there is 25% identity and 45% similarity between the M1 and M3 sequences. As such, it seems likely that these genes arose from a gene duplication event. Notably, BLASTp analysis of the putative M1-encoded protein identifies significant homology to the SPI-1 poxvirus serpin conserved among several pox viruses (4, 23), although the functionally important hinge domain of the serpin superfamily does not appear to be conserved in the M1 protein (4). Thus, the significance of the homology between the putative M1 and SPI-1 proteins is unclear. Furthermore, there is no significant homology between the M3 protein and SPI-1.
FIG. 6.
Alignment of the γHV68 M3 protein and putative M1 protein. Residues that match exactly are displayed as white letters on a black background, while residues which are structurally related are displayed on a shaded background. Alignments were analyzed by using the DNASTAR (Madison, Wis.) suite of programs. Alignments were performed with MegAlign, by using the PAM250 residue weight table.
Analysis of the M3 protein sequence predicts an amino-terminal signal peptide, consistent with M3 being a secreted protein, as shown here. In addition, a number of potential casein kinase 2 and protein kinase C phosphorylation sites are present in the M3 protein. However, the functional significance of such sites in a secreted protein is unclear. Finally, there is a potential RGD motif located at residues 52 to 54 in the M3 protein, which could mediate interaction of M3 with a cellular integrin.
Notably, the analysis of γHV68 gene expression in latently infected tissue has provided evidence that M3 may be expressed during viral latency. We readily detected transcripts from the M3 region of the viral genome in latently infected splenocytes isolated from B-cell-deficient mice 42 to 46 days postinfection, and to a lesser extent in RNA isolated from latently infected peritoneal cells obtained from the same animals (24). An independent analysis using in situ hybridization detected M3 transcription as late as 21 days postinfection in spleens of BALB/c mice (19). Thus, it is intriguing to speculate that the secreted M3 protein might play a role in modulating the host immune response, facilitating the establishment of the long-term γHV68 latency reservoir.
In summary, we have identified an abundant secreted protein present in the media of γHV68-infected cells. Sequence analysis of the protein demonstrated that it is encoded by the viral M3 ORF. Analysis of the M3 transcript by Northern blotting, cDNA analysis, RACE, and S1 nuclease protection indicates that M3 encodes a single, unspliced ca. 1.4-kb early-late lytic mRNA. Future studies will address whether the M3 protein is involved in modulating the host immune response against γHV68 infection.
Nucleotide sequence accession number.
The deduced sequence of M3 has been deposited in GenBank under accession no. AF127083.
Acknowledgments
We thank Jim Gould for generating the γHV68 polyclonal antibody, Carl Liu for the cDNA library, Linda Van Dyk and Sharook Kapadia for the generation of RNA, and Lina Yoo for advice on S1 nuclease protection assays. We also thank David Leib and members of his laboratory, as well as members of the Virgin and Speck laboratories, for helpful comments during the course of this research.
This work was supported by NIH grant CA74730 to H.W.V. and S.H.S. In addition, H.W.V. was supported by NIH grant AI39616 and ACS grant RP6-97-134-01-MBC, S.H.S. was supported by NIH grants CA43143, CA52004, and CA58524, and V.v.B. was supported by NIH grant GM07200.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Blaskovic D, Stancekova M, Svobodova J, Mistrikova J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980;24:468. [PubMed] [Google Scholar]
- 3.Blaskovic D, Stanekova D, Rajcani J. Experimental pathogenesis of murine herpesvirus in newborn mice. Acta Virol. 1984;28:225–231. [PubMed] [Google Scholar]
- 4.Bowden R J, Simas J P, Davis A J, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol. 1997;78:1675–1687. doi: 10.1099/0022-1317-78-7-1675. [DOI] [PubMed] [Google Scholar]
- 5.Danielson P E, Forss-Petter S, Brow M A, Calavetta L, Douglass J, Milner R J, Sutcliffe J G. p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA. 1988;7:261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 6.Efstathiou S, Ho Y M, Hall S, Styles C J, Scott S D, Gompels U A. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J Gen Virol. 1990;71:1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- 7.Efstathiou S, Ho Y M, Minson A C. Cloning and molecular characterization of the murine herpesvirus 68 genome. J Gen Virol. 1990;71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- 8.Ehtisham S, Sunil-Chandra N P, Nash A A. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J Virol. 1993;67:5247–5252. doi: 10.1128/jvi.67.9.5247-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honess R W, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaschka-Dierich C, Werner F J, Bauer I, Fleckenstein B. Structure of nonintegrated, circular Herpesvirus saimiri and Herpesvirus ateles genomes in tumor cell lines and in vitro-transformed cells. J Virol. 1982;44:295–310. doi: 10.1128/jvi.44.1.295-310.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mistrikova J, Blaskovic D. Ecology of the murine alphaherpesvirus and its isolation from lungs of rodents in cell culture. Acta Virol. 1985;29:312–317. [PubMed] [Google Scholar]
- 12.Moore K W, Vieira P, Fiorentino D F, Trounstine M L, Khan T A, Mosmann T R. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRF1. Science. 1990;248:1230–1234. doi: 10.1126/science.2161559. [DOI] [PubMed] [Google Scholar]
- 13.Moos M. Isolation of proteins for microsequence analysis. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1996. pp. 10.19.1–10.19.12. [Google Scholar]
- 14.Puglielli M T, Woisetschlaeger M, Speck S H. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J Virol. 1996;70:5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajcani J, Blaskovic D, Svobodova J, Ciampor F, Huckova D, Stanekova D. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 1985;29:51–60. [PubMed] [Google Scholar]
- 16.Russo J J, Bohenzky R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer B C. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem. 1995;227:255–273. doi: 10.1006/abio.1995.1279. [DOI] [PubMed] [Google Scholar]
- 18.Schulz T F. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 19.Simas J P, Swann D, Bowden R A, Efstathiou S. Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J Gen Virol. 1999;80:75–82. doi: 10.1099/0022-1317-80-1-75. [DOI] [PubMed] [Google Scholar]
- 20.Strockbine L D, Cohen J I, Farrah T, Lyman S D, Wagener F, DuBose R F, Armitage R J, Spriggs M K. The Epstein-Barr virus BARF1 gene encodes a novel, soluble colony-stimulating factor-1 receptor. J Virol. 1998;72:4015–4021. doi: 10.1128/jvi.72.5.4015-4021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunil-Chandra N P, Efstathiou S, Arno J, Nash A A. Virological and pathological features of mice infected with murine gammaherpesvirus 68. J Gen Virol. 1992;73:2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 22.Sunil-Chandra N P, Efstathiou S, Nash A A. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol. 1992;73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 23.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virgin H W, Presti R M, Li X-Y, Liu C, Speck S H. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J Virol. 1999;73:2321–2332. doi: 10.1128/jvi.73.3.2321-2332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weck K E, Dal Canto A J, Gould J D, O’Guin A K, Roth K A, Saffitz J E, Speck S H, Virgin H W. Murine gammaherpesvirus 68 causes large vessel arteritis in mice lacking interferon-gamma responsiveness: a new model for virus-induced vascular disease. Nat Med. 1997;3:1346–1353. doi: 10.1038/nm1297-1346. [DOI] [PubMed] [Google Scholar]
- 26.Yao Z, Fanslow W C, Seldin M F, Rousseau A-M, Painter S L, Comeau M R, Cohen J I, Spriggs M K. Herpesvirus saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. Immunity. 1995;3:811–821. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]