Abstract
The mammalian SWI-SNF complex is a chromatin-remodelling machinery involved in the modulation of gene expression. Its activity relies on two closely related ATPases known as brm/SNF2α and BRG-1/SNF2β. These two proteins can cooperate with nuclear receptors for transcriptional activation. In addition, they are involved in the control of cell proliferation, most probably by facilitating p105Rb repression of E2F transcriptional activity. In the present study, we have examined the ability of various brm/SNF2α deletion mutants to reverse the transformed phenotype of ras-transformed fibroblasts. Deletions within the p105Rb LXCXE binding motif or the conserved bromodomain had only a moderate effect. On the other hand, a 49-amino-acid segment, rich in lysines and arginines and located immediately downstream of the p105Rb interaction domain, appeared to be essential in this assay. This region was also required for cooperation of brm/SNF2α with the glucocorticoid receptor in transfection experiments, but only in the context of a reporter construct integrated in the cellular genome. The region has homology to the AT hooks present in high-mobility-group protein I/Y DNA binding domains and is required for the tethering of brm/SNF2α to chromatin.
The compaction of genomic DNA into chromatin fibers forms a potent obstacle to transcription and replication in eucaryotic cells. In recent years, the characterization of several large multisubunit protein complexes which are able to modify locally the structure of nucleosomes has shed light on how the cell may regulate the accessibility of chromatin-embedded promoters. These chromatin-remodelling complexes include the SWI-SNF complex (18, 35), initially identified in yeast but also present in Drosophila and in mammals; the RSC complex (6), identified in yeast; the NURF (51, 52), CHRAC (54), and ACF (21) complexes, characterized in Drosophila; and finally the NRD-NuRD complexes (49, 55, 61), detected in Xenopus and mammals. While these complexes differ in subunit composition, they all harbor one subunit containing a helicase-like domain with DNA-dependent ATPase activity. In each complex, this protein (SWI2-SNF2 in the SWI-SNF complex; STH1 in RSC; ISWI in NURF, CHRAC, and ACF; and CHD family members in NRD-NuRD) is likely to be the subunit responsible for the actual nucleosome perturbation, powered by ATP hydrolysis (for reviews, see references 5, 22, 24, 34, 53, and 58).
In the mammalian SWI-SNF complex, the ATPase activity is provided by either brm or BRG-1. These two highly homologous proteins (more than 80% identical) are also known as SNF2α and SNF2β, respectively (8, 25, 33). Unlike other related proteins, the homology of brm and BRG-1 to the yeast SWI2-SNF2 ATPase is not restricted to the helicase-like domain, suggesting that they may be the functional counterparts of the yeast protein in higher eucaryotes. The two proteins have been extensively characterized in the last few years, both individually and in the context of the mammalian SWI-SNF complex. The brm and BRG-1 proteins appear to be associated with the SWI-SNF complex in a mutually exclusive manner. Purification of the complex from tumor-derived cell lines failing to express the two proteins has further shown that a partial SWI-SNF complex can still assemble in their absence (57). During interphase, the brm and BRG-1 proteins are tightly associated with chromatin and a subfraction is also bound to the nuclear matrix (39). At the G2/M transition, the proteins are phosphorylated, leading first to decreased chromatin affinity and then to exclusion from the condensed mitotic chromosomes (30, 42). Several functional assays to monitor brm and BRG-1 activity have been developed. In transient-transfection assays, the two proteins can function as coactivators for nuclear receptors (8, 33, 57), and a ligand-dependent interaction between the estrogen receptor and the mammalian SNF2 proteins has also been reported (20). The brm or BRG-1 protein may also cooperate with members of the retinoblastoma (Rb) family of tumor suppressors to control cell growth. The p105Rb, p107, and p130 pocket proteins all are able to interact directly with brm or BRG-1 through an LXCXE sequence similar to the Rb binding motif present in several viral oncogenes, including papillomavirus E7, adenovirus E1a, and simian virus 40 large T antigen. In addition, the brm and BRG-1 proteins, when transiently transfected in SW13 cells, can cooperate with p105Rb to induce the formation of flat, growth-arrested cells (12, 43, 47). Cotransfection studies also show a cooperation between brm and p105Rb for the repression of E2F-activated transcription (50). Consistent with these observations, the brm protein was found to accumulate in quiescent cells (27, 29). In contrast, the level of this protein is down-regulated upon serum stimulation or transformation by an activated ras oncogene. Reexpression of brm in ras-transformed cells leads to a partial reversion of the transformed phenotype (29).
A mutation in the ATP binding site of the helicase-like domain of brm (ATPmut) strongly impairs the ability of the protein to revert the phenotype of ras-transformed fibroblasts, and it is clear that the helicase-like domain plays a central role in the activity of brm. However, other protein motifs have been identified in brm, including a bromodomain (16, 23) located in the C-terminal region. To determine if sequences outside the helicase-like domain were important for brm to affect cell growth, we examined the growth properties of ras-transformed NIH 3T3 cells expressing different mutant brm proteins. The mutations affected either the bromodomain, the LXCXE Rb binding sequence (referred to here as the E7 homology region), or a short C-terminal region characterized by a high lysine and arginine content (hereafter called the KR region). Surprisingly, this latter region appeared to be the most important for the activity of brm in our assay. Transfection experiments with a cell line carrying an integrated mouse mammary tumor virus (MMTV) chloramphenicol acetyltransferase (CAT) reporter construct further showed that in the context of chromatin, deletion of this region prevented cooperative transcriptional activation by brm and the glucocorticoid receptor (GR). Biochemical studies revealed that the KR region had DNA binding activity and that deletion of this region decreased the affinity of brm for chromatin.
MATERIALS AND METHODS
Cell culture and preparation of stable cell lines.
C33A, NIH 3T3, DT, and DT-derived cell lines were maintained at 37°C under 7% CO2 in Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 7% fetal calf serum unless otherwise indicated. Stable cell lines were established in DT or C33A cells as previously described (29, 36).
Plasmid constructs.
The T7-CMV-GR and the MMTV-CAT reporter constructs have been described previously (7, 14). Wild-type (WT) and mutant human brm (hbrm) constructs were inserted as EcoRI fragments downstream of the Moloney murine leukemia virus long terminal repeat (LTR) of pVLMPN1 (28) for stable expression as hemagglutinin (HA)-tagged proteins. The WT, ATPmut, and ΔCter(1337) hbrm inserts have been described previously (29, 31, 33). The ΔE7, ΔKR, and ΔBromo expression constructs were derived from WT hbrm and contain deletions from amino acids (aa) 1264 to 1337, 1342 to 1400, and 1401 to 1463, respectively. The ΔCter(1393) construct carries an out-of-frame mutation in codon 1394. The WT glutathione S-transferase (GST)-hbrm fusion was constructed by inserting a PCR fragment encoding the C-terminal end of the protein starting from aa 1188 into pGEX2T (Pharmacia). The GST-hbrm deletion mutants were generated in a similar way with the above-described deletion mutants as templates for the PCRs. The DGD point mutation was introduced into the GST-hbrm expression construct by using the QuikChange site-directed mutagenesis kit from Stratagene. All constructs containing PCR products were verified by DNA sequencing.
Transient-transfection assays.
Transient transfections were performed as described previously (33), with 50 ng of the GR expression vector and 3 μg of expression vector containing the hbrm-derived constructs. When the GR expression vector was used, the cells were treated with 10−6 M dexamethasone.
Cellular fractionation and immunoblotting.
Chromatin fractionation and high-salt isolation of the nuclear matrix were performed as described previously (39). The volume of each fraction was adjusted to 300 μl, and an equal volume of each fraction was used for analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (6% polyacrylamide). Immunoblotting was carried out by standard procedures (39) with the purified polyclonal rabbit anti-hbrm or anti-BRG-1 antibodies (30). Enhanced chemiluminescence reagents were used for detection.
Electrophoretic mobility shift assays (EMSAs).
GST fusion proteins were expressed in Escherichia coli BL21 and purified essentially as described previously (32), except that washes and elution were performed in A250 buffer (25 mM Tris [pH 7.5], 15 mM MgCl2, 15 mM EGTA, 10% glycerol, 0.3% Triton X-100, 250 mM NaCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). Mobility shift assays were performed in binding buffer (25 mM HEPES [pH 7.9], 2 mM MgCl2, 0.1 mM ZnCl, 40 mM KCl, 1 mM dithiothreitol, 0.25% milk) in the presence of ∼1 μg of fusion protein and 100,000 cpm of 32P-labelled probe. The 337-nucleotide fragment of genomic Drosophila was amplified from the SNR-1 gene with oligonucleotides GCGGATCCTCGCTCGTCGACCAGGTC and CAGAATTCAGTTGTGGTATTGGCCAGTC. The 0AT, 10AT, and 24AT oligonucleotides had the sequences GATCCGAGTCGCGCTGCAGCTCGCTCGTCGCA, GATCCGAGTCGCATATATATATGCTCGTCGCA, and GATCCATATATATATATATATATATATATGCA, respectively. When indicated, distamycin A (2 μM) or double-stranded poly(dA-dT) or poly(dG-dC) (300 ng) was added to the reaction mixtures. Samples were loaded on a 5% polyacrylamide gel in 0.25× Tris-borate-EDTA (TBE).
RESULTS
The C-terminal region of brm is necessary for reversion of ras transformation in mouse fibroblasts.
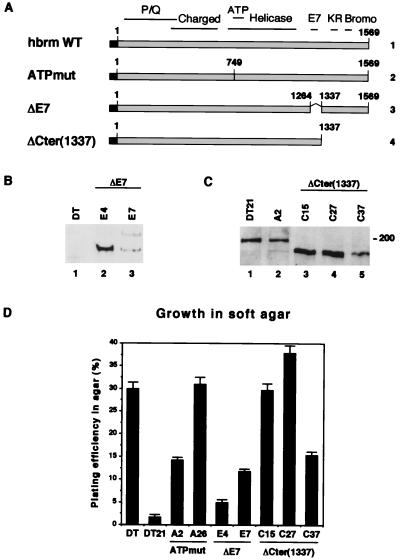
In a recent study, we showed that NIH 3T3 cells express easily detectable levels of both brm and BRG-1. On the other hand, NIH 3T3 cells transformed with an activated Ki-ras gene (DT cells) contain normal levels of BRG-1 but no detectable brm. Reintroduction of a cDNA encoding hbrm into DT cell leads to partial reversion of the transformed phenotype and prevents the cells from forming colonies in soft agar. A point mutation in the ATP binding site of hbrm (ATPmut) (Fig. 1A, line 2) abolishes the effect of hbrm on DT cell growth (29). To allow comparison, these results are included in Fig. 1D (compare the plating efficiencies of DT, DT21, A2, and A26). To investigate the role of regions other than the helicase-like domain, we transfected other mutant hbrm cDNAs into DT cells and isolated clones stably expressing the encoded proteins. Like WT hbrm, a protein deleted in the pRb binding LXCXE motif (ΔE7) (Fig. 1A, line 3, and Fig. 1B, lanes 2 and 3) led to decreased ability of DT cells to grow in soft agar. However, the effect was not as pronounced as in cells expressing the wild-type protein (Fig. 1D, compare DT21, E4, and E7), indicating that the ΔE7 mutant was moderately impaired in its ability to revert the transformed phenotype of DT cells. Deletion of the last 232 aa of hbrm [ΔCter(1337)] (Fig. 1A, line 4) had a more drastic effect. Of the three tested cell lines expressing this construct (Fig. 1C, lanes 3 to 5), two had plating efficiencies in soft agar similar to that of the original DT cells (Fig. 1D, C15 and C27). The third cell line was still eightfold more efficient in this assay than was the cell line expressing WT hbrm (Fig. 1D, C37). The expression level of the hbrm construct was higher in clones C15 and C27 than in clone C37. It was also moderately higher than in clones DT21 and A2, which express WT hbrm and ATPmut, respectively (Fig. 1C). Interestingly, the plating efficiencies for clones C15 and C27 were higher than for clone C37, suggesting that expression of ΔCter may favor rather than inhibit colony formation (compare the expression levels and plating efficiencies of C27 and C37 in Fig. 1C and D). An hbrm-derived construct with a deletion in the N-terminal part (aa 69 to 686 deleted) was also transfected into DT cells. However, we were unable to identify clones stably expressing this construct, suggesting that it may be toxic for normal cell proliferation.
FIG. 1.
The C-terminal region of brm is required for reversion of the ras-induced transformed phenotype of DT cells. (A) Schematic representation of the various hbrm-derived constructs expressed in DT cells. The solid box represents the HA tag. (B and C) Extracts from DT-derived cell lines expressing either ΔE7 (clones E4 and E7) (B) or ΔCter(1337) (clones C15, C27, and C37) (C) were resolved by SDS-PAGE and analyzed by Western blotting with anti-hbrm antibodies. To allow comparison, panels B and C show levels of brm in the parental DT cells as well as in DT21 and A2 cells, which express WT hbrm and ATPmut, respectively. (D) Growth in soft agar. Parental DT cells or DT cells expressing either WT hbrm, ATPmut, ΔE7, or ΔCter(1337) were plated in 60-mm dishes (102 or 103 cells per dish). The total number of visible colonies was scored after 15 days in culture and compared to the total number of cells initially seeded (plating efficiency, expressed as a percentage). The results shown here are averages and standard deviations from three independent experiments.
A region rich in arginine and lysine residues is required for hbrm activity.
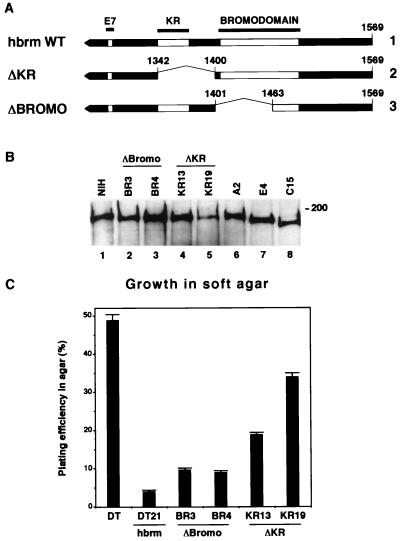
The observations described above indicate that the C-terminal part of hbrm is critical for the activity of this protein. The region deleted in the ΔCter(1337) construct encompasses at least two sequences of potential interest: the bromodomain and a short sequence rich in arginines and lysines (the KR region), located between the LXCXE motif and the bromodomain. To determine if either of these two regions was responsible for the loss of activity observed upon deletion of the C-terminal region of hbrm, we established two sets of DT-derived clones, one expressing an hbrm protein with aa 1342 to 1400 deleted and missing the KR region and one expressing an hbrm protein with aa 1401 to 1463 deleted and lacking helices A and B of the canonical bromodomain (ΔKR and ΔBromo respectively, Fig. 2A and B). These clones were assayed for growth in soft agar. The ΔBromo clones showed a plating efficiency twofold higher than the reference clone expressing WT hbrm (Fig. 2C, BR3 and BR4). The plating efficiencies of the ΔKR clones were five- to eightfold higher than that of the reference clone but still did not reach the plating efficiency of the original DT cells (Fig. 2C, KR13 and KR19). To allow comparison of the levels of expression of the different mutant proteins, we included in the Western blot in Fig. 2B three lanes previously shown in Fig. 1B or C. Taken together, the data presented in Fig. 2C suggest that the function of the C-terminal region of hbrm should be attributed to more than one protein domain. They also define the KR region as a novel hbrm sequence necessary for reversion of the transformed phenotype induced by ras.
FIG. 2.
Reversion of the ras-transformed phenotype by exogenous hbrm is dependent on the KR region. (A) Schematic representations of the C-terminal region of either WT hbrm (line 1), ΔKR (line 2), or ΔBromo (line 3). In all constructs, the N-terminal region that is not shown is WT. (B) Extracts from DT-derived cell lines expressing either ΔBromo or ΔKR were resolved by SDS-PAGE and analyzed by Western blotting with anti-hbrm antibodies. To allow comparison, expression levels in NIH 3T3 cells as well as in A2, E4, and C15 cells, expressing ATPmut, ΔE7, and ΔCter(1337), respectively, are also shown. (C) Growth in soft agar. Parental DT cells or DT cells expressing either WT hbrm, ΔBromo, or ΔKR were plated in 60-mm dishes (102 or 103 cells per dish). The total number of visible colonies was scored after 15 days in culture and compared to the total number of cells initially seeded (plating efficiency, expressed as a percentage). The results shown here are averages and standard deviations from three independent experiments.
The KR region is required for transcriptional synergy between hbrm and GR in the context of chromatin.
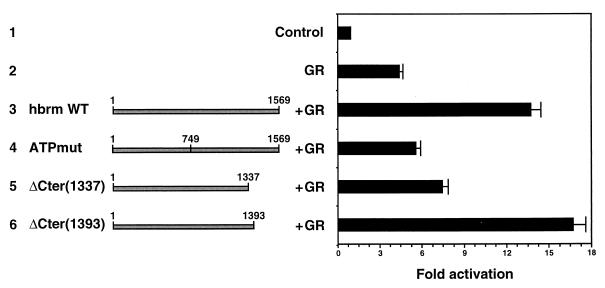
In transient-transfection assays, the hbrm protein can cooperate with the GR for transcriptional activation of the MMTV LTR or synthetic promoters containing GR-responsive elements. We therefore wished to investigate whether the KR region defined above was also necessary for this activity of hbrm. In an earlier study, we showed that a reporter construct cotransfected with hbrm and GR expression vectors is activated at least as efficiently by the ΔCter(1337) mutant as by WT hbrm in the presence of GR (33). The KR region that is absent in the ΔCter(1337) mutant therefore appeared to be unnecessary for cooperation between hbrm and GR. Several studies have, however, shown a clear difference in the chromatin structure of the MMTV LTR reporter constructs when transiently transfected into cells and when stably integrated in the cellular genome (1, 26). We hypothesized that the absent or poorly organized chromatin present on transfected templates rendered the KR region dispensable in the cooperation between hbrm and GR and that an effect of this region would be visible only on stably integrated nucleosomal templates. To test this hypothesis, we prepared a C33A-derived cell line containing an integrated MMTV LTR upstream of a CAT reporter gene. This cell line, which expresses no endogenous hbrm and low levels of BRG-1, was used for cotransfection assays with a GR expression vector and several constructs expressing hbrm mutants. The stimulation of GR activation by hbrm was significantly lower under these conditions than in transient transfections, essentially because all the cells contained the reporter construct and expressed CAT mRNA at a basal level whereas only 1 to 5% of the transfected cells expressed the reporter gene at activated levels. This situation resulted in a large increase in the background CAT activity in the assays and led us to repeat all the transfections at least four times to obtain reliable results. In the absence of hbrm, GR activated the integrated MMTV promoter about fivefold (Fig. 3, line 2). This activation was further stimulated threefold in the presence of WT hbrm (line 3). As in the transient transfections, this stimulation of GR activity was almost completely abolished by a mutation in the ATP binding site of hbrm (ATPmut) (line 4). Interestingly, under these conditions, the ΔCter(1337) mutant also had low activity (line 5). On the other hand, an hbrm construct longer by 56 aa and containing the KR region regained its activity in this assay [ΔCter(1393) mutant] (line 6). The ΔBromo mutant was also fully active under these conditions (data not shown). These experiments demonstrate that the KR region is required for transcriptional stimulation by the hbrm protein, but only in the context of chromatin.
FIG. 3.
The KR region is required for transcriptional synergy between hbrm and GR in the context of chromatin. A C33A-derived cell line containing an integrated MMTV CAT reporter construct was transfected with the vector without the insert (line 1) or with the GR expression vector either in the absence (line 2) or in the presence of an expression vector for WT hbrm (line 3), ATPmut (line 4), ΔCter(1337) (line 5), or ΔCter(1393) (line 6). The cells were harvested 36 h posttransfection, and CAT activity was measured. The results are shown as fold activation above CAT activity obtained with the vector without the insert and are compiled from seven independent experiments.
The KR region can function as an NLS.
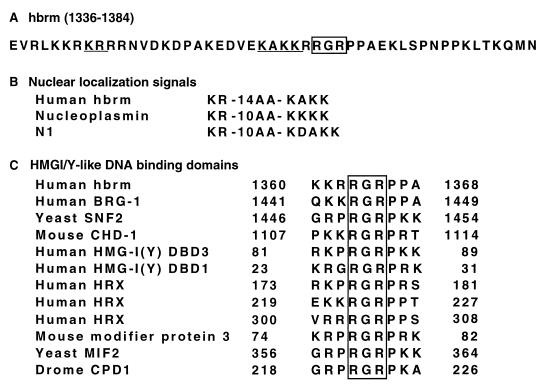
Examination of the primary sequence of the KR region shows the presence of two clusters of basic residues spaced by 14 aa (Fig. 4). This motif is strongly reminiscent of a bipartite nuclear localization signal (NLS) as initially defined in nucleoplasmin (Fig. 4B) (see references 10 and 60 for reviews). In transient-transfection assays, the KR region (aa 1336 to 1569) was sufficient to target a β-galactosidase (β-gal) fusion protein to the nucleus, further suggesting that the hbrm protein may rely on this region for its nuclear import. However, immunofluorescent staining of the DT-derived cell lines used in this study showed that all the hbrm mutants, including the ΔCter(1337) and the ΔKR mutants, had a strictly nuclear localization. In addition, we found that another region rich in arginines and lysines, located between aa 541 and 564, was able to target a β-gal fusion protein to the nucleus. Finally, a β-gal protein fused to the conserved helicase domain (aa 740 to 1334) localized both in the cytoplasm and in the nucleus (data not shown). These observations strongly suggested that loss of activity of the ΔKR mutant is not a consequence of improper cellular localization and that the hbrm protein contains several signals allowing its nuclear import.
FIG. 4.
Potential NLS and HMGI-like DNA binding domain within the KR region. (A) Amino acid sequence of hbrm between positions 1336 and 1384 of the published sequence. The putative bipartite NLS is underlined. The conserved motif found in the HMGI DNA binding domain is boxed. (B) Alignment of the putative NLS present in the KR region with the well-characterized NLS sequences from nucleoplasmin and N1 proteins. (C) Alignment of the KR region of hbrm with several proteins known or predicted to contain an HMGI-like DNA binding domain, also known as an AT hook.
The KR region harbors DNA binding activity.
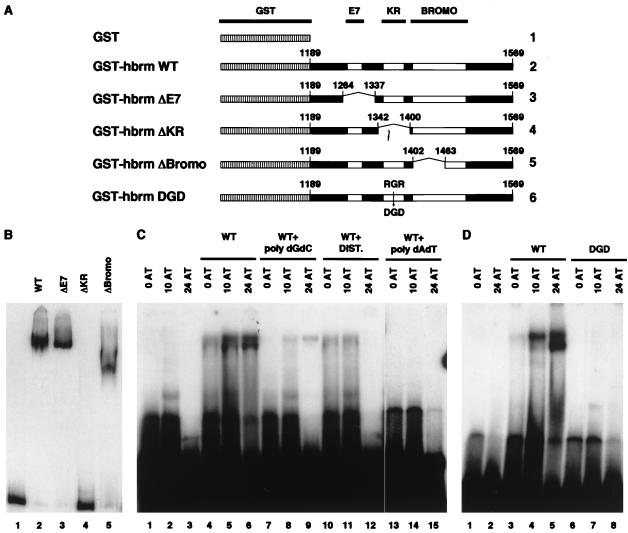
Further examination of the KR region as depicted in Fig. 4A (aa 1336 to 1384) revealed homology to the DNA binding domains found in proteins such as human High-Mobility-Group Protein I/Y (HMGI/Y) and HRX/ALL-1 or Drosophila D1 (3, 15, 44). These domains, known as AT hooks, bind the minor grove of DNA with a preference for AT-rich sequences. The presence of an AT hook has been suggested previously for the yeast SWI2-SNF2 protein in a region corresponding to the hbrm KR sequence (19). This prompted us to test whether the KR region could function as a DNA binding domain. To address this issue, we constructed a series of plasmids bearing DNA encoding aa 1189 to 1569 of hbrm fused to GST. The constructs contained a WT hbrm sequence or a sequence with deletion of either the E7 homology, KR, or Bromodomain region (GST-hbrm WT, GST-ΔE7, GST-ΔKR, and GST-ΔBromo, respectively) (Fig. 5A). All the constructs were expressed in E. coli and used for electrophoretic mobility shift assays (EMSA). WT GST-hbrm bound efficiently to a 300-bp fragment of randomly chosen Drosophila genomic DNA. This binding was not affected by deletion of the E7 homology region but was completely abolished by deletion of the KR region. Deletion of the bromodomain did not affect the ability of the protein to bind DNA, but it modified its gel mobility. This change in mobility may reflect a modified tertiary structure of this mutant protein (Fig. 5B). The GST-hbrm fusion protein also bound cruciform structures in EMSA, but with lower affinity than to the double-stranded DNA fragment (data not shown). The 300-nucleotide DNA fragment used for binding assays contained several stretches of four to five consecutive A · T base pairs, all located at one end of the fragment. When the 300-nucleotide DNA fragment was cleaved approximately in half by restriction digestion, the portion containing the stretches of A · T base pairs was bound as efficiently as the initial fragment. On the other hand, the other portion was bound with lower affinity (data not shown). These observations suggested that the binding of GST-hbrm was dependent on the A+T content rather than on the length of the DNA fragment. To further investigate this issue, we assayed the binding of WT GST-hbrm to 32-mer oligonucleotides containing either 0, 10, or 24 A · T base pairs. We observed a 10-fold increased binding to the 24AT oligonucleotide compared to the 0AT nucleotide (Fig. 5C, compare lanes 4 and 6). In addition, binding to the 24AT oligonucleotide was competed by a 100-fold excess of poly(dA-dT) but was partially resistant to the same amount of poly(dG-dC) double-stranded DNA (lanes 9 and 15). Furthermore, the binding to the 24AT oligonucleotide was inhibited in the presence of distamycin, suggesting that, like HMGI/Y, hbrm binds to the minor groove of DNA (lane 12). To confirm that the DNA binding of hbrm was mediated by the putative AT hook, we expressed a GST-hbrm fusion with a double point mutation changing the conserved arginine-glycine-arginine (RGR) motif (defined in Fig. 4C) into aspartic acid-glycine-aspartic acid (DGD). This mutant fusion protein was unable to bind any of the 32-mer oligonucleotides (Fig. 5D, lanes 6 to 8).
FIG. 5.
The KR region mediates hbrm DNA binding. (A) Schematic of the GST-hbrm fusion proteins used to assay the DNA binding properties of the hbrm KR region. (B) EMSA was performed with a 337-bp randomly chosen fragment of genomic Drosophila DNA and either GST alone (lane 1), wild-type GST-hbrm (lane 2), GST-ΔE7 (lane 3), GST-ΔKR (lane 4), or GST-ΔBromo (lane 5). (C) As in panel B, EMSA was performed with either GST alone (lanes 1 to 3) or WT GST-hbrm (lanes 4 to 15) with a 32-mer double-stranded oligonucleotide containing either 0, 10, or 24 A · T base pairs. When indicated, 300 ng of double-stranded poly(dG-dC) or poly(dA-dT) or 2 μM distamycin A was added to the reaction mixtures. (D) EMSA with either GST alone (lanes 1 and 2), WT GST-hbrm, or GST-hbrm DGD point mutant, using 32-mer double-stranded oligonucleotides containing either 0, 10, or 24 A · T base pairs as indicated.
Deletion of the KR region modifies the affinity of hbrm for chromatin fractions.
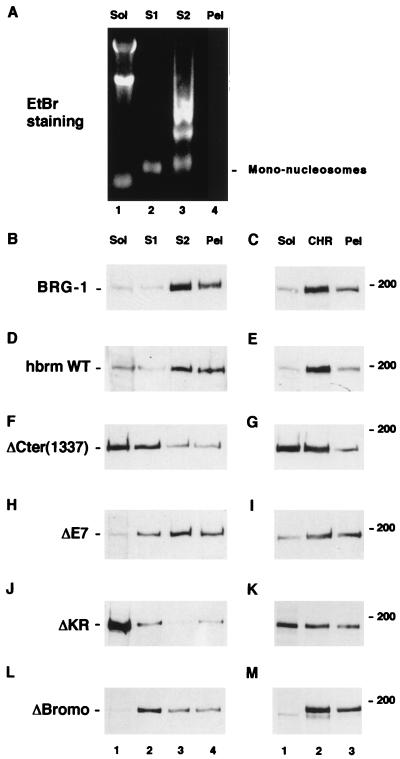
As described above, the KR region of hbrm is able to bind DNA in vitro. To determine if this region is also able to mediate the association with chromatin in vivo, we used a previously described technique that divides the cellular components into four fractions (40). In the first fractionation step, a detergent treatment (0.3% Nonidet P-40) lyses the cytoplasmic membrane but spares the nuclear envelope. The resulting fraction contains cytoplasmic proteins and RNA (Fig. 6A, lane 1) as well as nuclear proteins not attached to nuclear structures. The second fraction (fraction S1) is obtained after mild digestion of the nuclei by micrococcal nuclease. This releases DNA fragments corresponding to mononucleosomes (lane 2). The fraction contains easily accessible chromatin and factors attached thereto. The third fraction (S2) is obtained after further subjecting the nuclei to osmotic shock. This fraction contains larger DNA fragments corresponding to dinucleosomes, trinucleosomes, etc. (lane 3), as well as proteins attached to less accessible chromatin. The pellet remaining after this treatment contains DNA fragments of heterogeneous sizes that remain bound to the nuclear scaffold (insoluble chromatin, representing only a minor fraction of the total DNA). In earlier studies, we showed that hbrm is strongly attached to nuclear structures and cannot be extracted from interphasic cells by treatment with nonionic detergents (30, 39). As expected, fractionation of DT cells stably expressing exogenous WT hbrm showed that this protein was essentially present in the S2 and insoluble fractions (Fig. 6D). In these cells, the endogenous mBRG-1 was found in the same fractions as the reintroduced hbrm protein (Fig. 6B). The distribution of the hbrm was somewhat modified by a mutation in the E7 homology and bromodomain regions, resulting mainly in a redistribution of some of the protein from the insoluble fraction to the S1 fraction (Fig. 6H and L). On the other hand, deletion of the KR region or the entire C-terminal region resulted in an obvious decrease in the affinity for nuclear structures and a large fraction of these mutant proteins were detected in the soluble fraction (Fig. 6F and J, lanes 1). As mentioned above, deletion of the KR region or the C-terminal region did not affect the nuclear localization of the hbrm protein. To confirm the increased solubility of the ΔCter(1337) and ΔKR proteins, we fractionated the cells by the standard method to obtain nuclear matrix (17). Using this technique, we observed that WT hbrm was not extracted by the initial 0.5% Triton X-100 treatment in isotonic buffer (soluble fraction) but was essentially released after digestion with DNase I and extraction with 1 M ammonium sulfate (chromatin fraction) (Fig. 6E). Likewise, the ΔBromo protein resisted the detergent extraction (Fig. 6M). In contrast, the ΔCter(1337) and ΔKR proteins were partially released in the Triton X-100-soluble fraction, confirming a decreased affinity for nuclear structures of these hbrm-derived proteins (Fig. 6G and K, lanes 1). By this method, deletion of the E7 homology region results in some detergent extractability of the mutant protein (Fig. 6I, lane 1). This observation suggests that in vivo, sequences contained in the E7 region may complement the KR region for correct chromatin association of hbrm.
FIG. 6.
Deletion of the KR region of hbrm results in decreased affinity for nuclear structures. DT-derived cell lines expressing either WT hbrm, ΔCter(1337), ΔE7, ΔKR, or ΔBromo were fractionated by two different methods. In method 1 (A, B, D, F, H, J, and L), cells were lysed in a buffer containing 0.3% Nonidet P-40 and separated into supernatant (Sol fraction) and pellet. The pellet was treated with micrococcal nuclease and again centrifuged to separate supernatant (S1 fraction) and pellet. This pellet was further extracted with EDTA and centrifuged to yield supernatant (S2 fraction) and a pellet. This pellet was finally solubilized in 8 M urea (Pel fraction). One-tenth of each fraction was then extracted with phenol-chloroform and analyzed on a 1% agarose gel (A) or resolved directly by SDS-PAGE. For the latter, proteins were visualized by Western blotting with either anti-BRG-1 (B) or anti-hbrm (D, F, H, J, and L) antibodies. In method 2 (C, E, G, I, K, and M), nuclear matrix was prepared by the high-salt method as described in Materials and Methods. Cells were sequentially extracted with 0.5% Triton X-100 (Sol fraction), DNase I and 0.25 M (NH4)2SO4 (CHR fraction), and 2 M NaCl, and the remaining pellet was solubilized in 8 M urea (Pel fraction). As in method 1, 1/10 of each fraction was subjected to SDS-PAGE and immunoblotted with anti-BRG-1 (C) or anti-hbrm (E, G, I, K, and M) antibodies. EtBr, ethidium bromide.
DISCUSSION
Identification of a novel protein region required for the growth-suppressive activity of brm.
In an earlier study, we showed that expression of the brm protein is down-regulated in mouse fibroblasts upon transformation by activated ras. Reintroduction of a brm protein into these cells leads to partial reversion of the ras-transformed phenotype. This reversion could easily be estimated by assaying the ability of the cells expressing exogenous brm to form colonies in soft agar. By this assay, we showed that the ATPase domain of brm was essential for the reversion (33). In the present study, we have used the same assay to identify other regions of brm required for reversion of ras transformation. Surprisingly, deletion of the LXCXE motif, previously described as being required for interaction between brm and pocket proteins (Rb family members), had only a mild effect on reversion. It is possible, as previously suggested, that Rb simultaneously contacts other regions of the brm protein (50). Alternatively, the mammalian SWI-SNF complex may control cell growth through several parallel pathways. For example, it has recently been shown that hbrm and BRG-1 can associate with cyclin E and that this cyclin can rescue BRG-1-induced growth arrest by a mechanism that does not rely on the Rb protein (41).
Deletion of the entire C-terminal region of hbrm completely eliminates the growth-inhibiting effect on DT cells. The deleted region contains two potential sequences of interest: the bromodomain and a region rich in lysines and arginines (the KR region) located just upstream of the bromodomain. Sequences downstream of the bromodomain appear essentially unstructured when analyzed with a protein-structure prediction software (PredictProtein/EMBL). Deletion of the bromodomain had little effect on the ability of the hbrm protein to restrict DT cell growth. By contrast, DT clones expressing the ΔKR protein grew significantly better in soft agar than did clones expressing either the WT or ΔE7 hbrm constructs. These observations define the KR region as a new protein domain necessary for the antitransforming activity of hbrm. However, clones expressing this mutant do not reach the plating efficiency of clones expressing the ΔCter(1337) construct, and, unlike the ΔCter(1337) protein, expression of high levels of ΔKR does not favor colony formation in soft agar. This strongly suggests that C-terminal sequences other than the KR region are also involved in the effect of hbrm on DT cell growth. Alternatively, the large C-terminal deletion could be deleterious for the overall tertiary structure of hbrm, and this truncation may affect the activity of other important protein domains by a mechanism in cis. Detailed mutagenesis studies will be required to address this issue.
The KR region contains an AT-hook-like DNA binding domain.
Gel retardation assays with a C-terminal fragment of hbrm fused to GST showed that the KR region could function as a DNA binding domain. DNA binding activity has been ascribed previously to both the yeast and human SWI-SNF complexes, although the subunits responsible for this binding were not identified (37, 56). The yeast complex was found to bind only some promoter fragments, suggesting at least a moderate sequence specificity. It also showed high affinity for synthetic four-way-junction DNA. These properties are very similar to the DNA binding properties of HMG proteins, which interact with the minor groove of the DNA with low sequence specificity. A member of the HMG family of proteins (BAF57) has been found associated with the mammalian SWI-SNF complex (56). However, complexes containing a BAF57 protein mutated in the HMG domain are still able to bind DNA, suggesting a redundancy of HMG-like binding activities within the complex. Binding assays with GST-hbrm fusion proteins showed that the KR region has affinity for double-stranded DNA, with a preference for AT-rich sequences. The KR region is strongly basic and shows some homology to the DNA binding domains of the chromosomal protein HMGI/Y. This protein belongs to a family of HMG proteins that do not contain an HMG domain but contact DNA through sequences known as AT hooks. Interestingly, HMGI/Y is required for the assembly of higher-order transcription enhancer complexes, most probably by modifying the structure of the promoter DNA (4, 11, 13, 48).
The KR region is a potential chromatin interaction domain.
Our in vitro DNA binding data is highly suggestive of direct contact between the KR region of hbrm and chromatin. A role for this region in chromatin interaction is also suggested by our transfection experiments. We found that the KR region is dispensable for hbrm cooperation with the GR when the promoter construct is cotransfected with the hbrm and GR expression vectors (33). On the other hand, the KR region is required when the promoter is integrated into the cellular genome. A transfected promoter construct is unlikely to present positioned nucleosomes, whereas an integrated promoter will be fully organized into chromatin. Our observation therefore implies that a chromatin-embedded MMTV promoter becomes less accessible for the activating protagonists in the absence of the KR DNA binding domain.
Using two similar but distinct methods, we showed that WT hbrm is retained in the chromatin fractions whereas the ΔKR mutant is partially detergent extractable. This observation further suggests that the KR region is involved in the tethering of the hbrm protein to its target chromatin. In addition, the binding of the KR region to DNA or chromatin may be required for proper activation of the DNA-dependent ATPase activity of hbrm. Interestingly, the coupling of an ATPase domain to a DNA binding domain is reminiscent of the structure of another chromatin-bound mammalian SWI2-SNF2 homologue, known as CHD-1 (9, 45, 46, 59). Like the KR region, the DNA binding domain of this protein contains a potential AT hook surrounded by basic amino acids. We speculate that the DNA binding activities may be involved in similar mechanisms in the two proteins.
A target specificity for brm and BRG-1?
Our recent study on inactivation of the mouse brm (mbrm) gene by homologous recombination has shown that down-regulation of mbrm protein levels leads to accumulation of increased levels of mBRG-1. This new pool of mBRG-1 protein is able to associate with the complexes left vacant by the absent mbrm. The functional compensation of mbrm by mBRG-1 is reflected by the fairly mild phenotype of the mbrm−/− mice. Nonetheless, these mice are 10 to 15% heavier than their WT litermates and show obvious deregulations of the cell cycle checkpoints (38). These observations demonstrate that mBRG-1 and mbrm exhibit distinct functional properties. The DNA binding domain identified in hbrm is most probably also present in BRG-1 (the DNA binding motif is fully conserved). The presence of these DNA binding activities raises the possibility that the two proteins are targeted to specific chromatin regions and that these regions are not identical for brm and BRG. Immunofluorescent staining with brm and BRG-1 antibodies show that the two proteins have a nuclear-diffuse distribution but are also concentrated in spots, giving a microspeckled staining pattern. Interestingly, the brm spots never overlap with the BRG-1 spots (33a). Furthermore, an important region of sequence divergence between the hbrm and the BRG-1 proteins is located just upstream of the KR region, and transcripts with alternative splicing in the E7 and KR regions have been identified for both hbrm and BRG-1 (8, 33a, 47). If target specificity exists for hbrm and BRG-1, it might be determined by this region. The recent identification of a bona fide target gene for the mammalian SWI-SNF complex may allow us to address this issue (2).
ACKNOWLEDGMENTS
We are grateful to M. Noda and B. Wasylyk for the gift of DT cells and to J. Rouvière-Yaniv for four-way junction probes. We also thank J.-C. Reyes, A. Yeivin, and S. Schaper for valuable discussion. Special thanks also go to J. Weitzman and J. Seeler for critical reading of the manuscript.
This work was supported by l’Association pour la Recherche sur le Cancer, La Ligue Nationale Française Contre le Cancer, and the ACC program of the French Ministry of Science.
REFERENCES
- 1.Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong J A, Bieker J J, Emerson B M. A Swi/Snf-related chromatin remodeling complex, E-Rc1, is required for tissue-specific transcriptional regulation by Eklf in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 3.Ashley C T, Pendleton C G, Jennings W W, Saxena A, Glover C V. Isolation and sequencing of cDNA clones encoding Drosophila chromosomal protein D1. A repeating motif in proteins which recognize at DNA. J Biol Chem. 1989;264:8394–8401. [PubMed] [Google Scholar]
- 4.Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 5.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 6.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 7.Cato A C, Skroch P, Weinmann J, Butkeraitis P, Ponta H. DNA sequences outside the receptor-binding sites differently modulate the responsiveness of the mouse mammary tumour virus promoter to various steroid hormones. EMBO J. 1988;7:1403–1410. doi: 10.1002/j.1460-2075.1988.tb02957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delmas V, Stokes D G, Perry R P. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SWI2-like helicase domain. Proc Natl Acad Sci USA. 1993;90:2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 11.Du W, Thanos D, Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 12.Dunaief J L, Strober B E, Guha S, Khavari P A, Ålin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 13.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFN beta gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 14.Godowski P J, Rusconi S, Miesfeld R, Yamamoto K R. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature. 1987;325:365–368. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- 15.Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 16.Haynes S R, Dollard C, Winston F, Beck S, Trowsdale J, Dawid I B. The bromodomain: a conserved sequence found in human, Drosophila and yeast proteins. Nucleic Acids Res. 1992;20:2603. doi: 10.1093/nar/20.10.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He D C, Nickerson J A, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 19.Huth J R, Bewley C A, Nissen M S, Evans J N, Reeves R, Gronenborn A M, Clore G M. The solution structure of an HMG-I(Y)-DNA complex defines a new architectural minor groove binding motif. Nat Struct Biol. 1997;4:657–665. doi: 10.1038/nsb0897-657. [DOI] [PubMed] [Google Scholar]
- 20.Ichinose H, Garnier J M, Chambon P, Losson R. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene. 1997;188:95–100. doi: 10.1016/s0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- 21.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 22.Ito T, Tyler J K, Kadonaga J T. Chromatin assembly factors: a dual function in nucleosome formation and mobilization? Genes Cells. 1997;2:593–600. doi: 10.1046/j.1365-2443.1997.1500348.x. [DOI] [PubMed] [Google Scholar]
- 23.Jeanmougin F, Wurtz J M, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 24.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 25.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 26.Lee H L, Archer T K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeGouy E, Thompson E M, Muchardt C, Renard J P. Differential preimplantation regulation of two mouse homologues of the yeast SWI2 protein. Dev Dyn. 1998;212:38–48. doi: 10.1002/(SICI)1097-0177(199805)212:1<38::AID-AJA4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 28.Marty L, Roux P, Royer M, Piechaczyk M. MoMuLV-derived self-inactivating retroviral vectors possessing multiple cloning sites and expressing the resistance to either G418 or hygromycin B. Biochimie. 1990;72:885–887. doi: 10.1016/0300-9084(90)90007-4. [DOI] [PubMed] [Google Scholar]
- 29.Muchardt C, Bourachot B, Reyes J-C, Yaniv M. ras transformation is associated with decreased expression of the brm/SNF2alpha ATPase from the mammalian SWI-SNF complex. EMBO J. 1998;17:223–231. doi: 10.1093/emboj/17.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muchardt C, Reyes J C, Bourachot B, Legouy E, Yaniv M. The hbrm and BRG-1 proteins, components of the human SNF/SWI complex, are phosphorylated and excluded from the condensed chromosomes during mitosis. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 31.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to S. cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muchardt C, Seeler J S, Gaynor R B. Regulation of HTLV-I gene expression by tax and AP-2. New Biol. 1992;4:541–550. [PubMed] [Google Scholar]
- 33.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Muchardt, C. Unpublished data.
- 34.Pazin M J, Kadonaga J T. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- 35.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 36.Pfarr C M, Mechta F, Spyrou G, Lallemand D, Carillo S, Yaniv M. Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell. 1994;76:747–760. doi: 10.1016/0092-8674(94)90513-4. [DOI] [PubMed] [Google Scholar]
- 37.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 38.Reyes J C, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reyes J C, Muchardt C, Yaniv M. Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J Cell Biol. 1997;137:263–274. doi: 10.1083/jcb.137.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose S M, Garrard W T. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984;259:8534–8544. [PubMed] [Google Scholar]
- 41.Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. 1999;19:1460–1469. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sif S, Stukenberg P T, Kirschner M W, Kingston R E. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 44.Slany R K, Lavau C, Cleary M L. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stokes D G, Perry R P. DNA-binding and chromatin localization properties of CHD1. Mol Cell Biol. 1995;15:2745–2753. doi: 10.1128/mcb.15.5.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stokes D G, Tartof K D, Perry R P. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc Natl Acad Sci USA. 1996;93:7137–7142. doi: 10.1073/pnas.93.14.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strober B E, Dunaief J L, Guha S, Goff S P. Functional interaction between the hBRM/hBRG-1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 49.Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- 50.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. Rb and hbrm co-operate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 52.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 53.Varga-Weisz P D, Becker P B. Chromatin-remodeling factors: machines that regulate? Curr Opin Cell Biol. 1998;10:346–353. doi: 10.1016/s0955-0674(98)80010-0. [DOI] [PubMed] [Google Scholar]
- 54.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 55.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 56.Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree G R. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 58.Wolffe A P, Wong J, Pruss D. Activators and repressors: making use of chromatin to regulate transcription. Genes Cells. 1997;2:291–302. doi: 10.1046/j.1365-2443.1997.1260323.x. [DOI] [PubMed] [Google Scholar]
- 59.Woodage T, Basrai M A, Baxevanis A D, Hieter P, Collins F S. Characterization of the CHD family of proteins. Proc Natl Acad Sci USA. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoneda Y. How proteins are transported from cytoplasm to the nucleus. J Biochem. 1997;121:811–817. doi: 10.1093/oxfordjournals.jbchem.a021657. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]