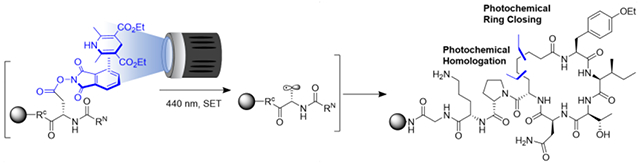
Abstract
Forging new C(sp3)-C(sp3) bonds to central positions within a peptide backbone is critical for the development of new therapeutics and chemical probes. Currently, there are no methods to decarboxylate Asp and Glu sidechains solid-phase photochemically, or use such radicals to form peptide macrocycles. Herein, electron-donor-acceptor (EDA) complexes between Hantzsch ester and on-resin peptide N-hydroxyphthalimide radical precursors are used to access these radicals, demonstrated with two-carbon homologations and homologation cyclizations of Atosiban and RGDf.
Graphical Abstract
Peptides are a critical class of therapeutics, as they occupy a chemical space regime between small molecules and large biologics and can modulate a vast array of biological processes, including protein-protein interactions. With access to a substantial pool of chiral building blocks, large peptide libraries have been developed, allowing synthetically easy access to diverse structures.1,2 However, the flexibility of peptides lowers their binding affinity, selectivity, permeability, and metabolic stability, minimizing their efficacy.2,3 To explore this chemical space more effectively, new strategies must be developed to stabilize polypeptide conformations. The most common strategy to reinforce the natural conformational bias of a given peptide sequence is covalent macrocyclization.
The most prevalent methods of creating peptide macrocycles are disulfide bond formation, lactamization, and lactonization.4 Therapeutics such as Atosiban, Oxytocin, and Vasostrict are all disulfide-cyclized peptides with an N-terminal linkage to a central position on the peptide chain. Replacing disulfide and other polar macrocycle linkages with C(sp3)-C(sp3) bonds serves to enhance permeability as well as proteolytic and chemical stability of therapeutics.5,6 Currently, solid-phase ruthenium-catalyzed ring-closing metathesis (RCM) followed by olefin reduction is the only strategy to synthesize secondary C(sp3)-C(sp3)-cyclized peptides at internal amino-acid positions.5 Furthermore, no methods have been reported to homologate Asp and Glu sidechains.
Previous reports of photochemical peptide cyclizations have required activated radical precursors, preventing the use of Asp and Glu as chiral radical sources within a peptide.7–9 The most common photochemical cyclization strategy is thus to perform a Giese-type reaction using radicals generated by oxidative decarboxylation of a free C-terminal Gly, limiting the overall structure of the macrocycles generated.10,11 We envisioned a direct route to a series of C(sp3)-C(sp3)-linked macrocycles from a common intermediate utilizing natural amino acids via tandem photochemical couplings on unactivated primary radicals. To accomplish this, electron-donor electron-acceptor (EDA) complexes were employed to access alaninyl- and homoalaninyl radicals (Figure 1, A). Such radicals, embedded within peptides, could then be homologated intermolecularly with allyl acrylate or cyclized onto an acrylamide (Figure 1, B, C).
Figure 1.
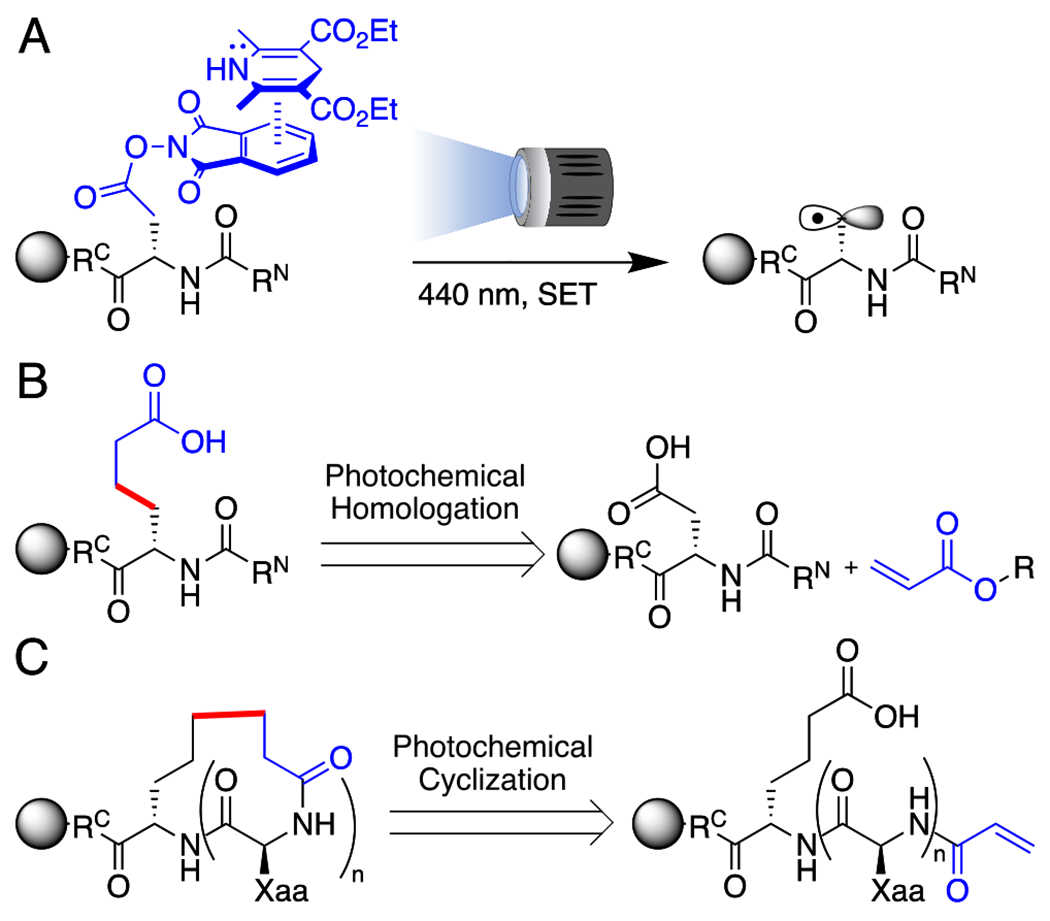
(A) Photochemical generation of an alaninyl radical by EDA complex and single electron transfer (SET) between Hantzsch ester and NHPI-peptide. (B) Retrosynthetic analysis of photochemical homologation. (C) Retrosynthetic analysis of photochemical cyclization.
Several precedents suggested the feasibility of this approach. Baran et al. demonstrated that an N-hydroxyphthalimide (NHPI) radical precursor could be formed on-resin and used to generate a stabilized α-prolinyl radical via nickel-catalyzed decarboxylative hydroalkylation.12 Furthermore, they carried out solution phase studies demonstrating the α-prolinyl radical cyclization onto an i-1 Lys(acrylamide). Additionally, there are examples of Asp or Glu amino acids being used as photochemical radical precursors to make α-selenoamino acids.13 In that work, NHPI-functionalized Asp and Glu amino acids were used to generate alanyl- and homoalanyl radicals using photoexcited [Ru(bpy)3]Cl2, the radicals generated being captured by diselenides. Recently, we reported the first example of solid-phase photochemical modification of peptides, in which we developed conditions for the on-resin intermolecular hydroalkylation of peptide-bound enamides.14 Shortly after, solid-phase deaminative photochemical Giese additions using Katritzky salts were also demonstrated on peptides.15
The photochemical radical pathway has several advantages over RCM and nickel-catalyzed decarboxylative hydroalkylation, including being able to: 1) use Asp and Glu as inexpensive chiral radical precursors. 2) modify a common intermediate for late-stage transformations. 3) use light-mediated transformations that are mild, often compatible with aqueous buffers, and typically operate within minutes under ambient conditions. 4) avoid organometallic catalysts by using photoinduced electron transfer activation from EDA complexes. 5) incorporate multicomponent reactions via dual photocatalytic cycles, ideal for the synthesis of diverse peptide macrocycles.16 6) provide flexibility for use of both RCM and radical chemistry in parallel to synthesize peptides with multiple cyclizations.
Atosiban, an Oxytocin antagonist, is a hormone clinically used as a tocolytic therapeutic to prevent premature birth.17 Atosiban is typically administered intravenously and has a poor metabolic half-life (16-18 min).18 The short half-life has been partly attributed to the disulfide linkage within the macrocycle. Utilizing RCM, Vederas et al. demonstrated that replacement of the disulfide linkage with all C(sp3)-C(sp3) bonds leads to a 2-3-fold increase in the half-life; with nearly identical activity.19 Herein, the synthesis of Atosiban is used to demonstrate the viability of using solid-phase photochemical peptide homologation and cyclization to construct peptide macrocycles.
Initial efforts focused on optimizing formation of the NHPI redox active ester (RAE) using solid-phase techniques. To do so requires activation of Asp or Glu sidechains, making them prone to intramolecular cyclization to form aspartimide or glutarimide. Studies were begun on a model di-peptide Ac-Glu-Phe-Rink amide polystyrene resin. Phenylalanine was incorporated to increase the retention time on the HPLC column and to provide a chromophore for UV/Vis detection. Glutamic acid was chosen as the radical precursor to minimize formation of the less favorable 6-membered cyclization compared to aspartic acid’s 5-membered cyclization. Previous work to install the NHPI RAE solid-phase utilized 1,3-diisopropylcarbodiimide (DIC) as the coupling reagent and required reaction for 2 h at 37 °C. We observed the desired product by LCMS along with a substantial peak consistent with the peptide backbone-cyclized side-product (SI, S5). Using hexafluorophosphate azabenzotriazole tetramethyl uronium (HATU) for 45 min under ambient conditions decreased the cyclized side-product by 91% and led to an overall cleaner reaction (SI, S6). Freshly synthesized RAE di-peptides were treated with allyl acrylate (10 equiv) and Hantzsch ester (HE, 10 equiv) at 0.2 M and irradiated with blue LEDs for 2 h (Scheme 1). All starting material was consumed, and crude HPLC yields were obtained for both Glu (36%) and Asp (41%) homologation reactions. Interestingly, side-products of the reaction were hydrodecarboxylation of Asp and Glu, giving alanine and homoalanine, respectively.. For the Ac-Asp-Phe-resin substrate, increasing the reaction concentration to 0.3 M and 0.5 M increased the yield to 70% and 56%, respectively. Substitution of NMP for DMF decreased the yield from 70% to 48%. Peptide Ac-Glu-Phe was reacted on a 50 μmol scale without allyl acrylate, cleanly affording homoalanine with an HPLC isolated yield of 22% (SI, S16). In our previous work on solid-phase Giese addition, we reported the compatibility of Trp, His, Cys, and Met with the overall transformation involving on-resin enamides and 10 equiv of RAE.14 In the current work, we performed the homologation on peptides Ac-Xaa-Gly-Glu-resin examining the same functional groups. All peptides exhibited comparable isolated yields of 6-16 % (SI, S35-S37). Met was not stable in the presence of allyl acrylate. We also sought to capture the radical with styrene to make phenylalanine homologues. Although styrene is less radicophilic with electron-rich radicals than acrylates, we still obtained appreciable yields of approximately 1 mg on a 25 μmol scale using both Asp and Glu sidechains. Interestingly, dicarbo-functionalized peptide was found to be an additional product isolated with comparable yields. This provides evidence that the newly generated α-radical may be further captured for other dicarbo-functionalization reactions.16,20,21
Scheme 1.
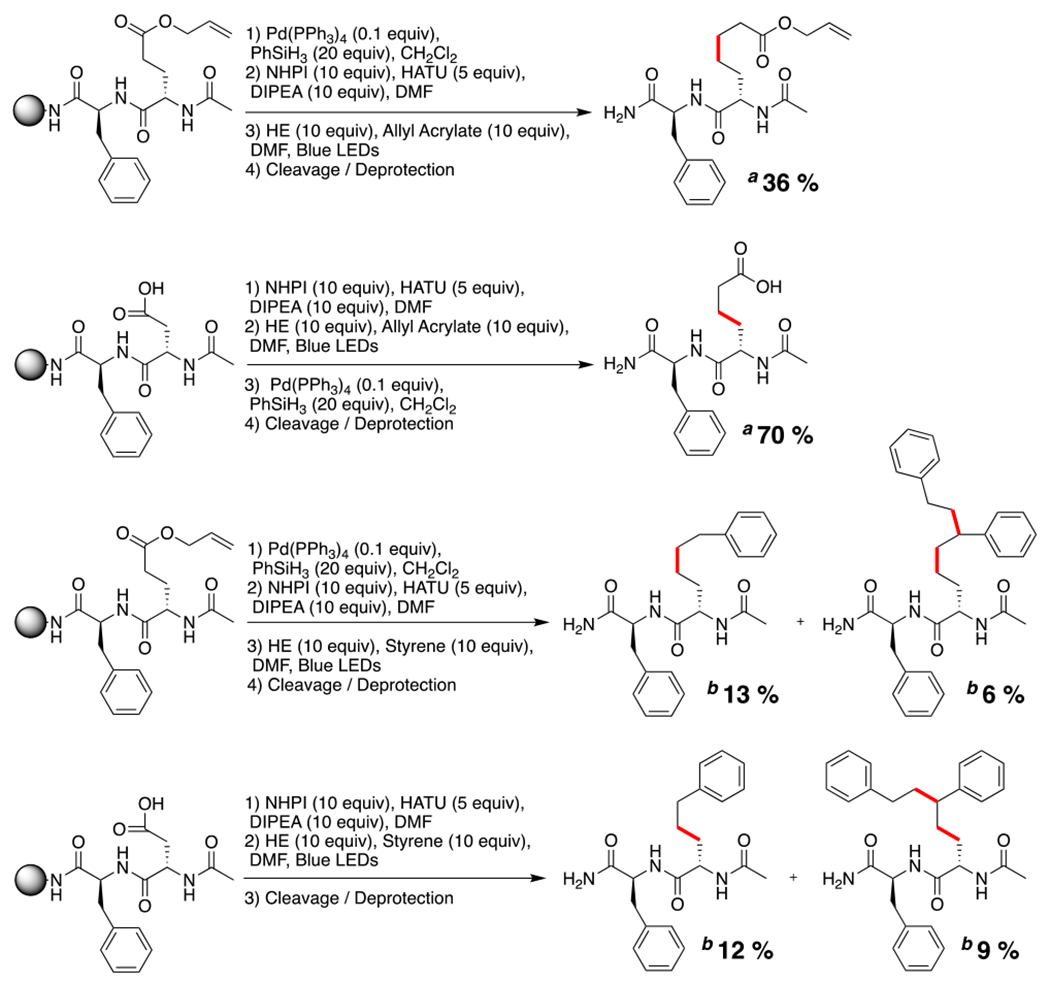
On-Resin Homologations of Asp and Glu-Containing Dipeptides.
aQuantitative HPLC yields reported. bIsolated yields reported.
Having optimized conditions for both on-resin RAE formation and homologation, we began the synthesis of Atosiban on a 100 μmol resin scale, reporting overall yields based on label loading. For accuracy, we relied on crude HPLC yields and standard curves derived from NMR. After peptide elongation, 5 μmol of resin was cleaved, worked up, and found to contain 4.5 μmol of Peptide-1 by quantitative HPLC (SI, S17) (Scheme 2, A). For formation of Atosiban-3, 20 μmol of resin was acylated with acrylic acid and then treated with Pd(0) to remove the allyl group. The aspartic acid sidechain was converted to the NHPI ester and reacted with Hantzsch ester under 440 nm light for 2 h, leading to a 12% overall yield by HPLC and an isolated yield of 5% (2.6 mg). Upon cyclization, a large upfield shift was observed in the 1H NMR spectra for Asp (linker) (0.38 ppm), Thr (0.49 ppm), and Ile (0.31 ppm) amides, all amino acids within the cycle. Half the resin of Peptide-1 (50 μmol) was protected on the N-terminus, using trityl chloride to avoid potential issues with Fmoc incompatibility in photochemistry. A small amount of resin (~3 mg) was subjected to acylation conditions, where the trityl-protection was found to proceed to 100% conversion (SI, S18). The allyl group was deprotected, and Asp was homologated, leading to Peptide-3 in 27% yield (SI, S19). The hydrodecarboxylated peptide (alanine) was again found as the major side product in the reaction as evident by MS and an additional methyl doublet in the 1HNMR spectra (SI, S52). Cyclization of Peptide-3 on a 5 μmol scale using PyAOP led to a clean 18% overall yield of Atosiban-3 (SI, S21). Alternatively, 25 μmol of Peptide-3 was acylated on the N-terminus, then subjected to Pd(0), revealing the free carboxylic acid. The peptide was then activated, converted to the NHPI ester, and cyclized to Atosiban-5 in 12% overall yield by HPLC and an isolated yield of 5% (1.4 mg). Cyclization led to a large upfield shift in the 1H NMR spectra for the linker (0.14 ppm), Thr (0.46 ppm), Ile (0.35 ppm), and Asn (0.44 ppm) amides, all amino acids within the cycle. 1H NMR assignments based on TOCSY spectra are consistent with literature precedents (SI, S47). The high cyclization yield of 47% for Atosiban-5 is similar to both MacMillan’s and Xu’s findings on photochemical C-N cyclizations of peptides.10,11
Scheme 2.
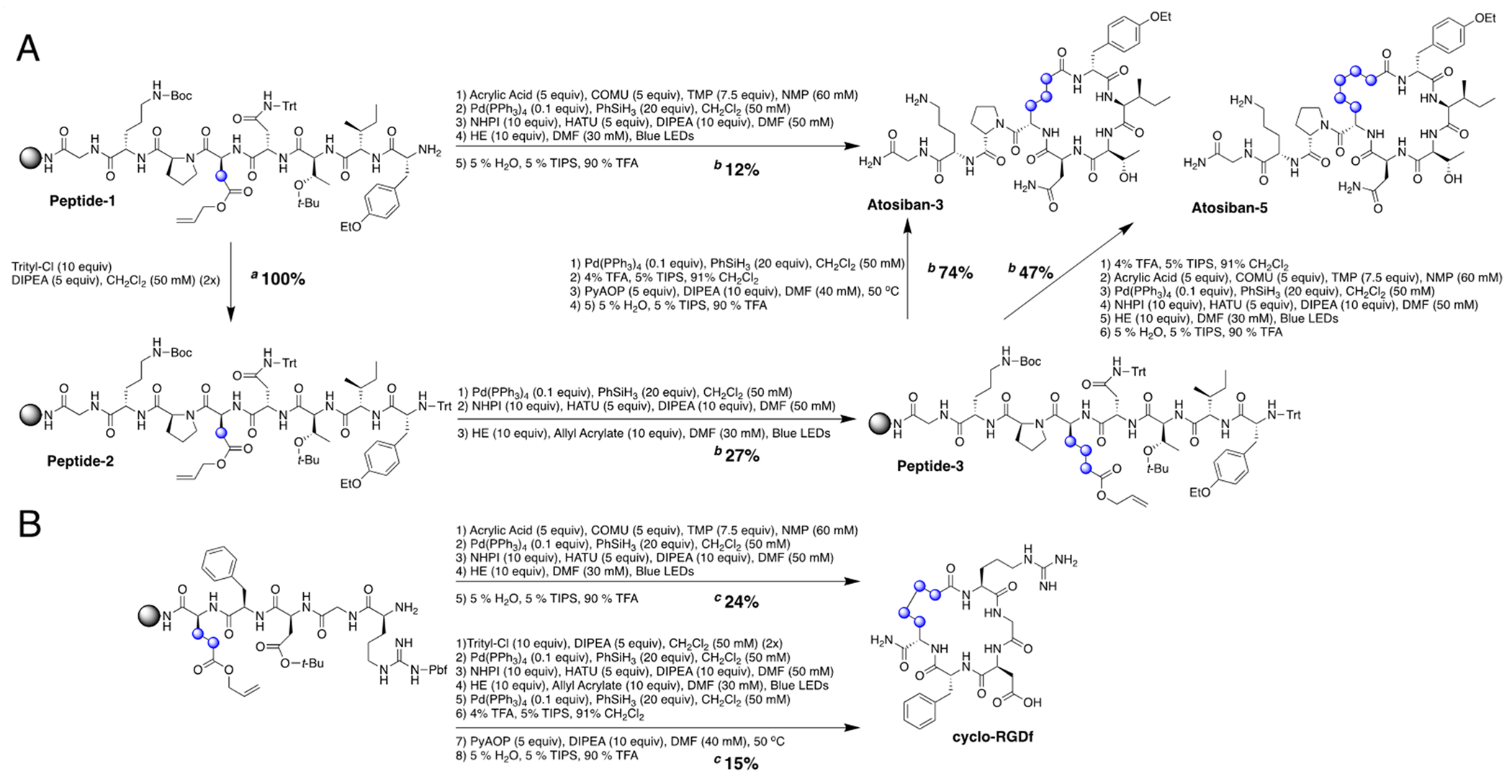
Synthetic Scheme for the Late-Stage modification of Atosiban (A) and Cyclo-RGDf (B) Peptides for Photochemical Homologation and Cyclizations
Concentrations and equivalents are based on resin loading. a Conversion determined by acylation and crude HPLC. b Crude HPLC yields are reported and obtained by standard curves based on quantitative 1H NMR. c Isolated yields reported.
To examine the homologation/cyclization efficiency on a different 5-amino acid peptide substrate, we performed a C-N cyclization on Integrin ligand cyclo-RGDf peptide (Scheme 2, B).22 Performing the photochemical intermolecular homologation followed by PyAOP cyclization led to an isolated yield of 15%, similar to that found for Atosiban-3, suggesting that structure does not significantly impact the transformation. Performing the photochemical cyclization led to a greater isolated yield of 24%, comparable to that of Atosiban-5. Given the similar reactivities of Glu and Asp and its homolog, the position of the radical within the peptide appears to matter less than the ring strain. For macrocycles with less ring-strain, the intramolecular cyclization will likely be higher-yielding than the intermolecular homologation. For macrocycles with greater ring-strain, the intermolecular homologation followed by PyAOP cyclization would be preferred.
In conclusion, we have demonstrated the solid-phase use of Asp and Glu sidechains to generate alaninyl- and homoalaninyl radicals by photochemical EDA decarboxylation. The method was used to homologate peptide precursors to Atosiban and cyclo-RGDf, which was subsequently cyclized to the target macrocycle. Reported are the first conditions to offer a route to a series of C(sp3)-C(sp3)-linked macrocycles, solid-phase, as a late-stage modification from a common intermediate. The chemistry is operationally simple, using natural amino acids under ambient conditions and expedient reaction times. The research described herein foretells the potential for other solid-phase peptide radical transformations beyond decarboxylative hydroalkylation, providing new strategies to access this unique regime of chemical space.23
Supplementary Material
ACKNOWLEDGMENT
The authors are grateful for the financial support provided by NIGMS (R35 GM 131680) and NSF (CHE-1952583) to G.M., NIH supplement awards 3R01GM118510-03S1 and 3R01GM087605-06S1, and Vagelos Institute for Energy Science and Technology for supporting the purchase of the NMRs used in this study. We thank Dr. Charles W. Ross, III (University of Pennsylvania) for obtaining HRMS data.
Footnotes
Supporting Information
The Supporting Information is available free of charge at http://pubs.acs.org./doi/***
Preparation of starting materials, optimization, control studies, and characterization data for products [NMR and LC/mass spectrometry] (PDF).
The authors declare no competing financial interest.
Contributor Information
Michael B. Elbaum, Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania 19104-6323, United States
Mahmoud A. Elkhalifa, Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania 19104-6323, United States
Gary A. Molander, Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania 19104-6323, United States
David M. Chenoweth, Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia, Pennsylvania 19104-6323, United States
REFERENCES
- (1).Sohrabi C; Foster A; Tavassoli A Methods for Generating and Screening Libraries of Genetically Encoded Cyclic Peptides in Drug Discovery. Nat. Rev. Chem 2020, 4 (2), 90–101. [DOI] [PubMed] [Google Scholar]
- (2).Lian W; Upadhyaya P; Rhodes CA; Liu Y; Pei D Screening Bicyclic Peptide Libraries for Protein-Protein Interaction Inhibitors: Discovery of a Tumor Necrosis Factor-α Antagonist. J. Am. Chem. Soc 2013, 135 (32), 11990–11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Vinogradov AA; Yin Y; Suga H Macrocyclic Peptides as Drug Candidates: Recent Progress and Remaining Challenges. J. Am. Chem. Soc 2019, 141 (10), 4167–4181. [DOI] [PubMed] [Google Scholar]
- (4).White CJ; Yudin AK Contemporary Strategies for Peptide Macrocyclization. Nat. Chem 2011, 3 (7), 509–524. [DOI] [PubMed] [Google Scholar]
- (5).Walensky LD; Bird GH Hydrocarbon-Stapled Peptides: Principles, Practice, and Progress. J. Med. Chem 2014, 57 (15), 6275–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Tian Y; Jiang Y; Li J; Wang D; Zhao H; Li Z Effect of Stapling Architecture on Physiochemical Properties and Cell Permeability of Stapled α-Helical Peptides: A Comparative Study. ChemBioChem 2017, 18 (21), 2087–2093. [DOI] [PubMed] [Google Scholar]
- (7).Bottecchia C; Noël T Photocatalytic Modification of Amino Acids, Peptides, and Proteins. Chem. - A Eur. J 2019, 25 (1), 26–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Raynal L; Rose NC; Donald JR; Spicer CD Photochemical Methods for Peptide Macrocyclisation. Chem. - A Eur. J 2021, 27 (1), 69–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Liu JQ; Shatskiy A; Matsuura BS; Kärkäs MD Recent Advances in Photoredox Catalysis Enabled Functionalization of α-Amino Acids and Peptides: Concepts, Strategies and Mechanisms. Synth. 2019, 51 (14), 2759–2791. [Google Scholar]
- (10).McCarver SJ; Qiao JX; Carpenter J; Borzilleri RM; Poss MA; Eastgate MD; Miller MM; MacMillan DWC Decarboxylative Peptide Macrocyclization through Photoredox Catalysis. Angew. Chemie - Int. Ed 2017, 56 (3), 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wang M; Wang C; Huo Y; Dang X; Xue H; Liu L; Chai H; Xie X; Li Z; Lu D; Xu Z Visible-Light-Mediated Catalyst-Free Synthesis of Unnatural α-Amino Acids and Peptide Macrocycles. Nat. Commun 2021, 12 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Qin T; Malins LR; Edwards JT; Merchant RR; Novak AJE; Zhong JZ; Mills RB; Yan M; Yuan C; Eastgate MD; Baran PS Nickel-Catalyzed Barton Decarboxylation and Giese Reactions: A Practical Take on Classic Transforms. Angew. Chemie 2017, 129 (1), 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Jiang M; Yang H; Fu H Visible-Light Photoredox Synthesis of Chiral α-Selenoamino Acids. Org. Lett 2016, 18 (9), 1968–1971. 10.1021/acs.orglett.6b00489. [DOI] [PubMed] [Google Scholar]
- (14).Elkhalifa M; Elbaum MB; Chenoweth DM; Molander GA Solid-Phase Photochemical Decarboxylative Hydroalkylation of Peptides. Org. Lett 2021, 23 (21), 8219–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Openy J; Amrahova G; Chang J-Y; Noisier A; ’t Hart P Solid-phase Peptide Modification via Deaminative Photochemical Csp3-Csp3 Bond Formation Using Katritzky Salts. Chem. – A Eur. J 2022, No. Scheme 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Badir SO; Molander GA Developments in Photoredox/Nickel Dual-Catalyzed 1,2-Difunctionalizations. Chem 2020, 6 (6), 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Melin P; Trojnar J; Johansson B; Vilhardt H; Akerlund M Synthetic Antagonists of the Myometrial Response to Vasopressin and Oxytocin. J. Endocrinol 1986, 111 (1), 125–131. [DOI] [PubMed] [Google Scholar]
- (18).Tsatsaris V; Carbonne B; Cabrol D Atosiban for Preterm Labour. Drugs 2004, 64 (4), 375–382. [DOI] [PubMed] [Google Scholar]
- (19).Stymiest JL; Mitchell BF; Wong S; Vederas JC Synthesis of Oxytocin Analogues with Replacement of Sulfur by Carbon Gives Potent Antagonists with Increased Stability. J. Org. Chem 2005, 70 (20), 7799–7809. [DOI] [PubMed] [Google Scholar]
- (20).Dhungana RK; KC S; Basnet P; Giri R Transition Metal-Catalyzed Dicarbofunctionalization of Unactivated Olefins. Chem. Rec 2018, 18 (9), 1314–1340. [DOI] [PubMed] [Google Scholar]
- (21).Derosa J; Apolinar O; Kang T; Tran VT; Engle KM Recent Developments in Nickel-Catalyzed Intermolecular Dicarbofunctionalization of Alkenes. Chem. Sci 2020, 11 (17), 4287–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kapp TG; Rechenmacher F; Neubauer S; Maltsev OV; Cavalcanti-Adam EA; Zarka R; Reuning U; Notni J; Wester HJ; Mas-Moruno C; Spatz J; Geiger B; Kessler H A Comprehensive Evaluation of the Activity and Selectivity Profile of Ligands for RGD-Binding Integrins. Sci. Rep 2017, 7 (January), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Malins LR Decarboxylative Couplings as Versatile Tools for Late-Stage Peptide Modifications. Pept. Sci 2018, 110 (3), 1–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


