Abstract
Summary
Oxford Nanopore Technologies’ (ONT) sequencing platform offers an excellent opportunity to perform real-time analysis during sequencing. This feature allows for early insights into experimental data and accelerates a potential decision-making process for further analysis, which can be particularly relevant in the clinical context. Although some tools for the real-time analysis of DNA-sequencing data already exist, there is currently no application available for differential transcriptome data analysis designed for scientists or physicians with limited bioinformatics knowledge. Here, we introduce NanopoReaTA, a user-friendly real-time analysis toolbox for RNA-sequencing data from ONT. Sequencing results from a running or finished experiment are processed through an R Shiny-based graphical user interface with an integrated Nextflow pipeline for whole transcriptome or gene-specific analyses. NanopoReaTA provides visual snapshots of a sequencing run in progress, thus enabling interactive sequencing and rapid decision making that could also be applied to clinical cases.
Availability and implementation
Github https://github.com/AnWiercze/NanopoReaTA; Zenodo https://doi.org/10.5281/zenodo.8099825.
1 Introduction
In standard sequencing experiments, practical steps and data analysis are usually performed independently, with the latter initiated by bioinformatics experts once after sequencing is complete. Nowadays, new technologies such as Oxford Nanopore Technologies (ONT) offer a unique opportunity to start downstream analysis while sequencing is still ongoing (Amarasinghe et al. 2020, Wang et al. 2012). Some platforms, such as EPI2ME from ONT (https://labs.epi2me.io/) or minoTour (https://github.com/minoTour/minoTour, Munro et al. 2022), already provide real-time pipelines for rapid ONT data acquisition integrated into a user interface (UI), and thus accessible to users with limited bioinformatics skills. However, as these platforms’ focus mainly lies on the analysis of DNA-sequencing data, there is a lack of real-time applications in the field of transcriptomics. Here, we introduce NanopoReaTA, an on-demand toolbox for real-time transcriptomic analysis that provides rapid insight on RNA-sequencing data from ONT. Users receive transcriptome-wide and gene-specific information directly while sequencing is still running, such as differences between conditions or expression levels of individual genes. In addition, implemented quality control features allow the user to monitor data variability during the ongoing sequencing process. Ultimately, the tool can provide frequent biologically relevant snapshots of the current sequencing run, which in turn can enable interactive fine-tuning of the sequencing run itself, facilitate decisions to abort the ongoing run to save time and material, e.g. when sufficient accuracy is achieved, or even accelerate the resolution of clinical cases with high urgency.
2 Material and methods
2.1 Test data
NanopoReaTA has been tested on self-generated direct cDNA-sequencing data from Hek293 and HeLa cells (Supplementary Table S1 and Supplementary Figs S1–S9).
2.2 Usage
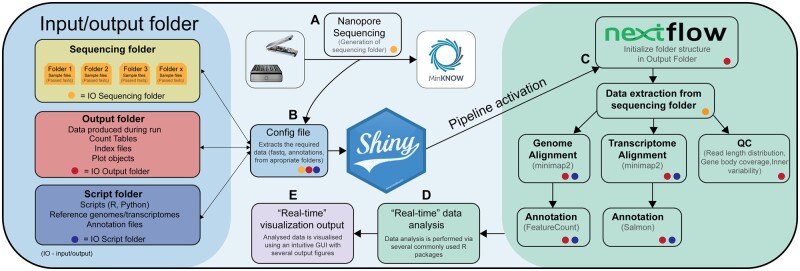
NanopoReaTA can be launched directly after starting a sequencing run of cDNA or direct RNA via ONT’s sequencing software MinKNOW (Fig. 1A and Supplementary Fig. S1). Within NanopoReaTA’s UI, the user will be guided through several configuration settings to extract all information required for data processing such as reference sequences, annotation files, output directory defined in MinKNOW (into which sequencing output is written), and more (Fig. 1B and Supplementary Fig. S2A–C). Preprocessing of basecalled reads from a running or completed experiment is integrated into a Nextflow pipeline and can be started via a one-button-click within the UI (Fig. 1C and Supplementary Fig. S2D; Di Tommaso et al. 2017). As soon as sequencing data are generated, the Nextflow pipeline automatically updates generated files, including gene counts or mapping files. Based on the output files from preprocessing, downstream analyses can be performed within the following tabs integrated into NanopoReaTA: “Overview,” “Gene-wise Analysis,” and “Differential Expression Analysis” (Supplementary Figs S4–S6). The resulting figures can be constantly updated during sequencing (Supplementary Figs S7 and S8). See more details in the Supplementary Information.
Figure 1.
NanopoReaTA workflow visualized as a graphical sketch. (A) Sequencing start; (B) Configuration of settings in NanopoReaTA’s UI; (C) Preprocessing pipeline by Nextflow; (D) Transcriptional analysis and (E) visualization of results in NanopoReaTA’s UI. Detailed information on the individual modules is given in the Supplementary Information, as well as in the user manual on the GitHub repository: https://github.com/AnWiercze/NanopoReaTA.
2.3 Preprocessing via Nextflow
The Nextflow pipeline takes all fastq files that pass the quality threshold defined in MinKNOW and performs genome and transcriptome alignment using minimap2 (Li 2018) as well as feature quantification using FeatureCounts (Liao et al. 2014) and Salmon (Patro et al. 2017). In addition, we incorporated a quality control utility extracting sample- and group-wise read length distribution, variability measurements, genome/transcriptome coverage based on RSeQC (Wang et al. 2012), and gene count per iteration, enabling the assessment of specific quality metrics over time (Supplementary Fig. S7). See more details in the Supplementary Information.
2.4 Downstream analyses based on R
The subsequent downstream analyses are based on commonly used R packages such as DESeq2 (Love et al. 2014) for principal component analysis and differential expression analysis of gene and transcript expression, and DEXSeq (Anders et al. 2012) and DRIMSeq (Nowicka and Robinson 2016) for differential transcript usage (Fig. 1D and E). In addition, gene body coverage and counts per sample and condition can be visualized for a subset of genes of interest (Fig. 1E). All tables and figures can be downloaded via button clicks (Supplementary Figs S3–S6). See more details in the Supplementary Information.
2.5 Installation and requirements
NanopoReaTA can be installed on Linux and Windows via docker by pulling a prebuild docker image containing all package requirements. For installation, requirements, and user manual, visit https://github.com/AnWiercze/NanopoReaTA.
Hardware: 64GB RAM, 16 threads. Software: Docker.
3 Discussion
NanopoReaTA represents a real-time analysis toolbox that allows users to perform interactive transcriptional analyses of cDNA and direct RNA-sequencing data in real-time via a user-friendly and intuitive UI based on R Shiny. We aim to provide a tool that supports users from biological research and clinical diagnostics of transcriptomics by accelerating decision-making processes of future experiments or patient treatment, especially when time and money are limiting factors. For future perspectives, we envision that additional functions such as novel transcript detection, RNA modification detection, and integration of multi-omics levels in real-time can be integrated. NanopoReaTA is open source to also enable the scientific community to contribute such enhancements.
Supplementary Material
Contributor Information
Anna Wierczeiko, Institute of Human Genetics, University Medical Center of the Johannes Gutenberg University Mainz, Mainz 55131, Germany.
Stefan Pastore, Institute of Pharmaceutical and Biomedical Sciences, Johannes Gutenberg-University Mainz, Mainz 55128, Germany.
Stefan Mündnich, Institute of Pharmaceutical and Biomedical Sciences, Johannes Gutenberg-University Mainz, Mainz 55128, Germany.
Anne M Busch, Institute of Human Genetics, University Medical Center of the Johannes Gutenberg University Mainz, Mainz 55131, Germany.
Vincent Dietrich, Institute of Human Genetics, University Medical Center of the Johannes Gutenberg University Mainz, Mainz 55131, Germany.
Mark Helm, Institute of Pharmaceutical and Biomedical Sciences, Johannes Gutenberg-University Mainz, Mainz 55128, Germany.
Tamer Butto, Institute of Pharmaceutical and Biomedical Sciences, Johannes Gutenberg-University Mainz, Mainz 55128, Germany.
Susanne Gerber, Institute of Human Genetics, University Medical Center of the Johannes Gutenberg University Mainz, Mainz 55131, Germany.
Author contributions
T.B. and S.G. conceived and supervised the project. A.W. and S.P. designed, implemented, and tested the GUI. A.W., S.P., V.D., and A.M.B. implemented GUI updates. S.M. performed the RNA isolation from Hek293 and HeLa and T.B. performed the direct cDNA library preparation. T.B., A.W., and S.P. wrote the manuscript. S.G., M.H., and S.M. edited the manuscript and provided valuable input and feedback in various discussions. All authors read and approved the final manuscript.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Funding
T.B. and S.G. acknowledge funding by the Landes Initiative Rheinland‐Pfalz and the Resilience, Adaptation, and Longevity (ReALity) initiative of the Johannes Gutenberg University of Mainz. V.D. and S.G. acknowledge funding by SFB 1551 Project No. 464588647 of the Deutsche Forschungsgemeinschaft (DFG). The work of M.H. and S.M. has been funded by the DFG (German Research Foundation)—Project-ID 439669440—TRR 319 (C01).
References
- Amarasinghe SL, , SuS, , Dong X et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol 2020;21:30. 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res 2012;22:2008–17. 10.1101/GR.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P, Chatzou M, Floden EW et al. Nextflow enables re-producible computational workflows. Nat Biotechnol 2017;35:316–9. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018;34:3094–100. 10.1093/BIOINFORMATICS/BTY191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. 10.1093/BIOINFORMATICS/BTT656. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550–21. 10.1186/S13059-014-0550-8/FIGURES/9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro R, Santos R, Payne A et al. minoTour, real-time monitoring and analysis for nanopore sequencers. Bioinformatics 2022;38:1133–5. 10.1093/BIOINFORMATICS/BTAB780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka M, Robinson MD. DRIMSeq: a Dirichlet-multinomial framework for multivariate count outcomes in genomics. F1000Research 2016;5:1356. 10.12688/F1000RESEARCH.8900.2/DOI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI et al. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–9. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 2012;28:2184–5. 10.1093/BIOINFORMATICS/BTS356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.