Summary
Meiotic crossovers are required for accurate chromosome segregation and producing new allelic combinations. Meiotic crossover numbers are tightly regulated within a narrow range, despite an excess of initiating DNA double-strand breaks. Here, we reveal the tumor suppressor FANCM as a meiotic anti-crossover factor in mammals. We use unique large-scale crossover analyses with both single-gamete sequencing and pedigree-based bulk-sequencing datasets to identify a genome-wide increase in crossover frequencies in Fancm-deficient mice. Gametogenesis is heavily perturbed in Fancm loss-of-function mice, which is consistent with the reproductive defects reported in humans with biallelic FANCM mutations. A portion of the gametogenesis defects can be attributed to the cGAS-STING pathway after birth. Despite the gametogenesis phenotypes in Fancm mutants, both sexes are capable of producing offspring. We propose that the anti-crossover function and role in gametogenesis of Fancm are separable and will inform diagnostic pathways for human genomic instability disorders.
Graphical abstract
Highlights
-
•
FancmΔ2/Δ2 limits meiotic crossovers in mammals and crossover interference is unaffected
-
•
Despite dramatic defects in gametogenesis, FancmΔ2/Δ2 mice can produce live offspring
-
•
FancmΔ2/Δ2 mice are sub-fertile in a strain-dependent manner
-
•
cGAS-STING drives spermatogonial defects in FancmΔ2/Δ2 mice
Tsui and Lyu et al. demonstrate dual roles of Fancm in mice using a combination of wet- and dry-lab techniques. They show that Fancm-deficient mice have significantly increased meiotic crossovers but are still able to generate offspring despite perturbed gametogenesis.
Introduction
Fanconi anemia (FA) is a rare genetic condition that affects multiple systems in the body. Individuals with FA are predisposed to health issues such as cancers, progressive bone marrow failure, birth defects, and reduced fertility.1,2 The genes that are mutated in individuals with FA act in a common genetic and biochemical pathway. The FA pathway is a DNA repair mechanism best known for its role in the repair of DNA interstrand cross-links in somatic cells. There are 22 FANC and FANC-like genes: FANCA, FANCB, FANCC, FANCD1, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCJ, FANCL, FANCM, FANCN, FANCO, FANCP, FANCQ, FANCR, FANCS, FANCT, FANCU, FANCV, and FANCW.3 FANCM is a DNA translocase, which can remodel branched DNA structures and provides a physical connection between multiple genomic stability-related protein complexes. FANCM orthologs are widely conserved across eukaryotes and prokaryotes. However, vertebrates tend to have an additional ERCC4 domain at the carboxyl terminus compared with other taxa.4 The significance of the ERCC4 in vertebrates is not clear, as it appears to lack catalytic activity.
In humans and mice, FANCM is required for normal fertility and gametogenesis.5,6,7,8,9,10,11,12 Reports of male patients with biallelic mutations in FANCM revealed sterility characterized by non-obstructive azoospermia and Sertoli cell-only seminiferous epithelium.8,9,13 Females with a homozygous pathogenic FANCM mutation experience premature ovarian insufficiency and therefore early menopause.7 It was previously reported that a reduction in the number of primordial germ cells occurs prior to birth in Fancm-mutant mice of both sexes.6 Similar losses of primordial germ cells have been observed in other Fanc mouse models.14,15,16 In Fancm-deficient mice, at the molecular level, a significant portion of this reduction was attributed to the ATM-p53-p21 axis, most notably in the testis. Further, female Fancm mutants were born at sub-Mendelian ratios,5,6,10 and this was attributed to embryonic death induced by genomic instability activating inflammatory pathways.10 Male pups, on the other hand, were born at normal frequencies.10
A key process within germ cell development is meiosis. At meiosis, crossovers—large reciprocal exchanges between homologous chromosomes—are necessary for balanced chromosome segregation at the first division. Strict mechanisms exist to ensure that each pair of homologous chromosomes forms at least one “obligate” crossover.17 An absence of crossovers between homologs can lead to aneuploid karyotypes and conditions such as trisomy 21.18,19 The obligate crossover is linked to a phenomenon known as crossover interference, which results in relatively even spacing of multiple crossovers on the same chromosome.20,21,22,23 Additional layers of control of crossover distribution are determined by PRDM9, through the direction of the initiating DNA double-strand breaks (DSBs),24,25,26 as well as separate mechanisms that influence megabase-scale crossover patterning.27,28,29,30
Meiotic crossovers are formed via the highly regulated and well-conserved meiotic DSB repair pathway.17 Programmed meiotic DNA DSBs are generated by SPO11.31,32,33 The DSBs are resected to generate 3′ single-strand overhangs.34,35,36,37 The recombinases RAD51 and DMC1 are recruited to these single-stranded tails and form nucleoprotein filaments, which promote repair that uses the homolog as a template to fill in missing genetic information. The single-stranded nucleoprotein filament displaces one strand of the homolog, forming a displacement loop (D loop).37,38,39 Second-end capture can progress the D loop toward “joint molecules” such as a double Holliday junction, which is dependent on a group of proteins collectively referred to as the ZMMs.40,41,42 The ZMMs are required for most crossovers in animals, budding yeast, and plants, and these crossovers are sensitive to interference and often referred to as “class I” crossovers.41,43,44,45,46 The number of class I crossovers is positively correlated with synaptonemal complex length.47,48,49,50 An alternative “class II” crossover pathway produces a smaller proportion of crossovers that are not sensitive to interference.45,51,52,53,54,55,56,57
SPO11-dependent meiotic DSBs occur in excess to the number of crossovers in diverse species, and therefore conserved mechanisms likely exist that cap the number of crossovers per meiosis.22,23 When programmed meiotic DSBs are repaired with a homologous template, joint molecules are formed as intermediates prior to a crossover or non-crossover outcome,42,43,58,59,60,61,62,63 and therefore joint molecules present a key step at which crossover rates can be altered. FANCM family members have previously been shown to dissociate joint molecules with high efficiency via helicase activity in yeast64,65 and translocase activity in mammals.66 In addition, FANCM, along with co-factors MHF1 and MHF2, has been found to limit the number of meiotic crossovers in Arabidopsis, fission yeast, Drosophila, canola, rice, and pea.67,68,69,70,71,72,73,74 This crossover-limiting function of FANCM may occur through the ability to dissociate joint molecules during meiotic DSB repair, but is difficult to demonstrate directly. However, any potential role of Fancm in meiotic crossover regulation in mammals remains unexplored.
Previous studies have used cytological approaches to test for roles of FANC genes at meiosis and crossover formation in mice, in particular, immunofluorescence localization.6,15,16,75,76,77,78,79,80,81 However, it can be unclear what the meiotic consequences of any cytological phenotypes are because immunofluorescence data—while extremely useful and used in this study—represent static time points in a dynamic process. To the best of our knowledge, this is the first study, using genetic approaches through analysis of haploid meiotic products or live offspring, to demonstrate a role for a FANC gene in meiotic crossover formation in a species that has a bona fide FA pathway and therefore tangible implications for medical research and reproductive health.
Genomic instability has been connected to the activation of innate immunity.82,83,84 One way that innate immunity is activated is through the action of a protein called cyclic GMP-AMP synthase (cGAS), which binds to double-stranded DNA in the cytosol and triggers a type I interferon response through another protein called stimulator of interferon genes (STING).85 However, the exact relationship between reduced fertility resulting from genomic instability and the activation of innate immunity is not yet fully understood.
Here we reveal an anti-crossover role during meiosis for Fancm in mammals using traditional pedigree-based techniques and single-sperm sequencing. We also present the application of new bioinformatics and wet-lab tools, sgcocaller, comapr, and SSNIP-seq,86,87 in this area, in particular in the use of a droplet-based single-sperm sequencing technique. Further, genomic analysis of products of meiosis is achieved using both pedigree-based and sperm-based sequencing approaches. Side-by-side comparisons of these traditional and novel approaches are largely unexplored in mammals.88 We also build on previous findings on the role of Fancm in fertility and gametogenesis. We reveal a role for the cGAS-STING innate immune response in driving the loss of germ cells in males, which indicates that the cGAS-STING pathway may be a target for ameliorating the impaired gametogenesis that occurs in patients with genomic instability.
Results
Generation of Fancm-mutant mice
To investigate a potential function for Fancm in crossover control in mammals, we generated two mouse models. Fancm-knockout mice were generated independently—with two separate targeting events—in different genetic backgrounds, FVB/N and C57BL/6J, herein referred to as FVB.FancmΔ2/Δ2 and B6.FancmΔ2/Δ2, respectively (Figure S1). Two single-guide RNAs (sgRNAs) were generated to target the second exon of FANCM, similar to a previous approach.5 Targeted resequencing of the exon 2 borders confirmed the deletion (Figure S1). The Fancm heterozygous founders were backcrossed three times to their respective strains of FVB/N or C57BL/6J to increase the probability of eliminating any possible off-target mutations that may have been introduced by Cas9. FancmΔ2/Δ2 mice were viable in both strains. Null mice of both strains were outwardly normal except for a slight reduction in the body weight of female B6.FancmΔ2/Δ2 mice (p < 0.0001; Figure S2) and sub-fertility. Complete blood counts were similar to those of the wild-type controls (Figure S2). Cancers were not detected in any mice in the study; however, mice used in the study were typically young adults. The loss of Fancm resulted in phenotypes consistent with what has been observed previously.5,6,8 We observed a dose-dependent increase in chromosome fragility in lymphocytes following exposure to the cross-linking drug mitomycin C (Figures S3A and S3B). We also observed an increase in micronucleated red blood cells under basal conditions, similar to previous findings6 (Figure S3C). Finally, we observed a loss of FANCD2 monoubiquitination in splenic B cells under basal conditions, under replication stress (hydroxyurea), and when treated with a DNA cross-linker (mitomycin C), consistent with loss of function of Fancm and loss of the catalytic function of the E3 ligase complex that it anchors to somatic DNA damage5,89 (Figure S3D).
Fancm-mutant mice are sub-fertile
Human studies have reported that loss of FANCM is associated with infertility in males and premature menopause in females.7,8 Therefore, to extend our understanding of the origins of this pathology using mouse models, we set up breeding pairs with either a female or a male FancmΔ2/Δ2 mouse with a Fancm+/+ or Fancm+/Δ2 partner. Both male and female FancmΔ2/Δ2 produced pups that appeared outwardly normal. Average litter sizes were significantly reduced in male and female B6.FancmΔ2/Δ2 mice (p = 0.004 and p = 0.022, respectively) compared with B6.Fancm+/Δ2 controls (Figure S4A). The average litter sizes were not different between Fancm loss-of-function genotypes for FVB/N and F1 (FVB/N × C57BL/6J) strains. There was no difference observed in the average time between litters for all strains (Figure S4B). When investigating the average number of pups born per litter, we found that female FVB and B6.FancmΔ2/Δ2 mice produced fewer litters, which were also smaller (Figures S4C and S4D), suggesting that these mice could be useful models for the investigation of premature ovarian insufficiency seen in humans with biallelic mutations in FANCM.7 This aside, the fertility of the mice provided an opportunity to use genetic approaches to study meiotic outcomes in a FancmΔ2/Δ2 mammal.
Fancm limits meiotic crossovers in mammals
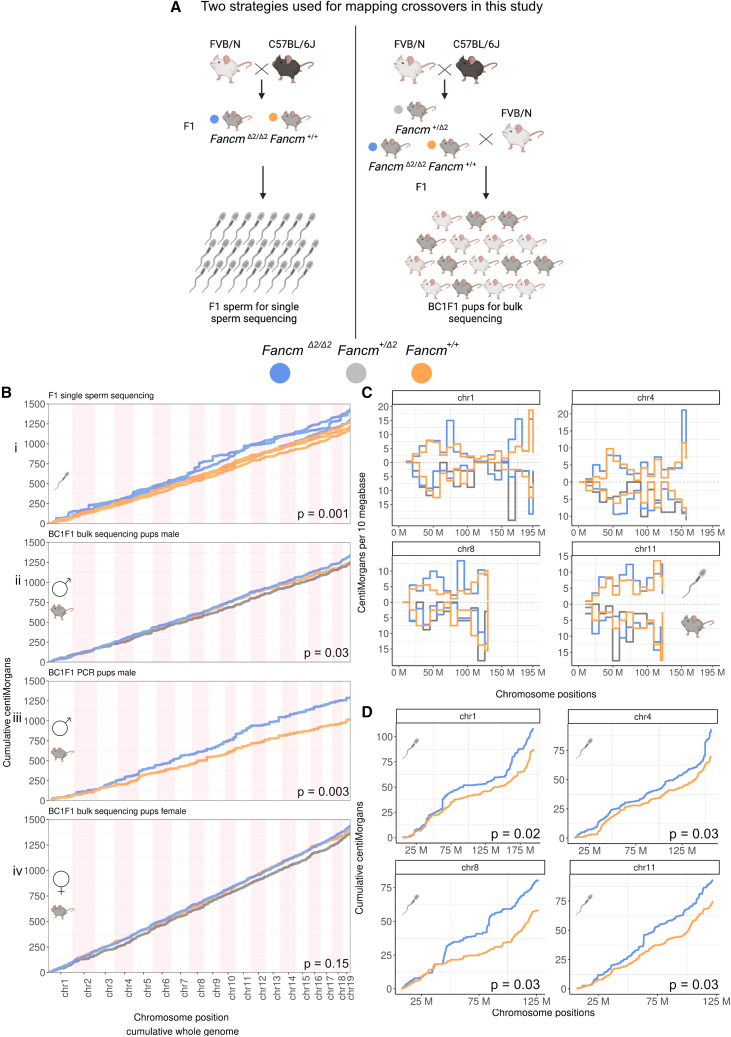
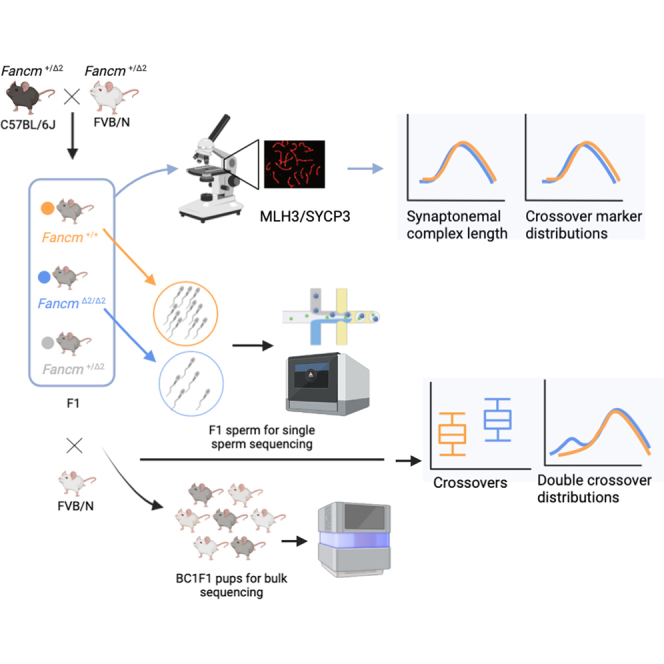
We characterized the effects of Fancm loss on meiotic crossing-over in F1 hybrids using three different methods. We began with male F1 mice, which were backcrossed to the FVB/N strain to generate BC1F1 offspring (Figure 1A). Using a PCR-based genotyping method with a set of 271 informative SNP markers across 56 mice (n = 31 FancmΔ2/Δ2, n = 25 Fancm+/+), we determined that F1.FancmΔ2/Δ2 have a significant increase in crossover numbers and therefore total genetic map length. The genetic maps for FancmΔ2/Δ2 and Fancm+/+ were 1,305 and 1,030 cM, respectively (p = 0.003, permutation test [B = 3,000]) (Figure 1Biii).
Figure 1.
Fancm limits meiotic crossovers as measured using different generations and technologies
(A) Crossovers were assayed using high-throughput sequencing of individual sperm from F1 mice, bulk whole-genome DNA sequencing of BC1F1 animals, and PCR of selected SNPs.
(B) Cumulative centimorgans calculated using data from three different assays: (i) F1 single-sperm sequencing (n = 3 animals per genotype; n = 218 sperm from FancmΔ2/Δ2, 190 sperm from Fancm+/+), (ii) high-throughput sequencing with BC1F1 pups (n = 98 mice from FancmΔ2/Δ2, 34 mice from Fancm+/Δ2, 40 mice from Fancm+/+), and (iii) PCR-based markers with BC1F1 pups (n = 31 FancmΔ2/Δ2, 25 Fancm+/+ independent animals per genotype). (iv) 200 BC1F1 pups sequenced to derive female genetic distances (n = 100 FancmΔ2/Δ2, 50 Fancm+/Δ2, 50 Fancm+/+ independent animals per genotype). Observed crossover fractions were used as input and converted into genetic distances in centimorgans using the Kosambi mapping function and presented as cumulative centimorgans across the genome. All three approaches demonstrate a significant increase in total genetic map length in male FancmΔ2/Δ2 mutants compared with that measured in Fancm+/+ littermates. Note: for PCR genotyping, chromosome 10 is not presented as full length due to lower marker coverage.
(C and D) Four selected chromosomes, 1, 4, 8, and 11, which drove a significant portion of the increase in map length in the mutants are plotted. (C) Genetic distances were plotted in 10 Mb chromosome bins for four selected chromosomes for two assays (F1 single-sperm sequencing [top] and bulk DNA sequencing of BC1F1 pups [bottom]). No significant differences were observed in genotype groups in each bin.
(D) Cumulative genetic distances per chromosome from F1 single-sperm sequencing data with permutation p values printed for each chromosome after multiple testing correction.
Using the same breeding strategy (Figure 1A), we extended the crossover analysis with high-throughput “bulk” DNA sequencing with BC1F1 individual offspring and generated DNA sequencing datasets with 1×–5× whole-genome coverage for each BC1F1 pup (total n = 372, with 200 from female F1 and 172 from male F1). Approximately 4 million informative SNP markers between C57BL/6J and FVB/N were assayed to detect the strain origins in the BC1F1 genomes to call crossovers. With increased marker density, more crossovers were detected in the wild-type mice, and we observed an increase in total genetic distances in FancmΔ2/Δ2 compared with Fancm+/+ and Fancm+/Δ2 pooled together (p = 0.03, permutation test [B = 1,000], with 1,339 and 1,249 cM, respectively) (Figures 1Bii and S5), and the hypercrossover phenotype due to the loss of Fancm was recessive (p = 0.45, permutation test [B = 1,000], 1,242 cM Fancm+/Δ2 and 1,256 cM Fancm+/+). Female genetic distances were higher in FancmΔ2/Δ2 compared with Fancm+/+ and Fancm+/Δ2, but not significantly different (p = 0.15, permutation test [B = 1,000], 1,438 and 1,385 cM) (Figure 1Biv), and crossover distributions were unaffected (Figure S6).
Next, we assayed crossover frequency and distributions with single-gamete (haploid) sequencing in F1 males (total n = 408 gametes from three animals per genotype). We used a droplet-based method of partitioning individual gametes coupled with low-level whole-genome amplification (STAR Methods). Total map lengths per sample within genotype groups were reproducible (Figures 1Bi, S7A, and S7B) and no bias in marker segregation was observed (Figures S7C and S7D). Crossover frequencies across the genome were reproducible between single-gamete and bulk sequencing assays (Figures 1C and S8). Further, the total map lengths of all chromosomes for all conditions had a minimum approximate length of 50 cM, similar to the BC1F1 sequencing data (Figure S7B). These observations suggest that single-gamete sequencing can be reliably used for crossover detection. Differences in genetic distances along chromosomes in bins of 10 Mb were tested in genotype groups, but no significant differences were observed for both assays for any one interval (Figure 1C). To call crossovers using single-sperm sequencing data and downstream analyses, we developed an open-source bioinformatic tool kit containing sgcocaller and comapr86 (see STAR Methods). Consistent with the bulk-sequencing and PCR-based methods, an increase in total crossover frequency and map length was observed in FancmΔ2/Δ2 compared with Fancm+/+ (p = 0.001, permutation test [B = 1,000], 1,347 and 1,178 cM, respectively) (Figure 1Bi). In conclusion, the triangulation of complementary and independent methods of crossover detection supports the finding that FANCM limits crossover formation in mammals, with the complementary strategies revealing an effect of a similar order of magnitude when considering sampling variance, and that marker densities vary substantially between the techniques used.
While genome-wide crossover numbers were elevated in Fancm-deficient mice, the general crossover distribution in FancmΔ2/Δ2 appeared to be comparable to that of the wild type. The regions that had an increase in crossover frequency did not have a detectable unifying genomic feature (Figures S9–S11). Nevertheless, the data from individual sperm show a significant increase in total crossovers per chromosome that was most notable for chromosomes 1, 4, 8, and 11 (Figure 1) (one-sided t test with multiple testing correction “fdr” with a total number of 19 tests). Crossover frequencies were also converted to genetic distances in centimorgans per chromosome. The differences in total centimorgans between genotypes per chromosome were tested using permutation-based methods, which showed a significant increase in FancmΔ2/Δ2 for chromosomes 1, 2, 4, 8, 9, 11, 13, and 18 before multiple testing correction (permutation p < 0.05, B = 1,000), while chromosomes 1, 2, 4, 8, 11, and 13 remained significant after multiple testing correction (using fdr for a total number of 19 tests) (Figure 1D). An increase in crossover frequency on chromosome 11 in sperm from FancmΔ2/Δ2 mice was detected with all three methods, and an attempt to understand this phenomenon was pursued. Previous studies have highlighted how chromosome 11 has a particularly high crossover density per megabase in the wild type and have developed predictive models of crossover distributions.30,90 We compared the predicted crossover densities from the fitted linear regression model from Pratto et al.,30 which used the physical chromosome size, meiotic DSB frequency, and replication speed on C57BL/6 × CAST,90 with our observed crossover densities. The model predicts lower crossover densities generally than are observed for the C57BL/6 × FVB/N data (this study), and predictions were better for FancmΔ2/Δ2 than for Fancm+/+. Chromosome 11 fit relatively well with the model in FancmΔ2/Δ2 in both the pedigree- and the sperm-based approaches, with a much higher crossover density (Figure S12), revealing that chromosome size, DSB number, and replication speed are good chromosome-scale crossover predictors for Fancm-deficient mice. We interpreted this fit to indicate that the extra crossovers in Fancm loss-of-function mice had a relative distribution between chromosomes comparable to that of Fancm+/+.
During the development of our single-sperm sequencing and crossover mapping methods we attempted to identify crossovers with single-sperm RNA-sequencing (RNA-seq) data. Despite careful data generation and analysis, however, this was unsuccessful. Cytoplasmic sharing of sperm is documented,91 but we sought to test whether a sufficient proportion of non-shared transcripts could be used for crossover mapping. We sorted haploid sperm cells from F1.FancmΔ2/Δ2 and F1.Fancm+/+ to use as input for library preparation with the 10× Genomics single-cell gene expression protocol (Single Cell 3′ v.2). CellRanger (cellranger-mm10-3.0.0, GRCm38, annotation version 93) called over 15,000 barcodes per input sample as high-confidence single cells. However, even after rigorous quality control of called cells using best-practice workflows,92 we observed frequent and extensive regions of heterozygosity (variant allele frequencies 0.5) for putative haploid cells. The extent of heterozygosity was too high to allow mapping of crossovers from droplets containing one spermatid. The frequent detection of heterozygosity suggests that the degree of cytoplasmic transcript-sharing levels between spermatids prevents the accurate haplotyping of gametes that are needed for crossover mapping. Nevertheless, we could reproduce previous findings93 that there exists a subset of transcripts that are more informative than average as to the nuclear genotype of a spermatid (Figure S13). Taken together, these results suggest that “single-cell” sequencing of RNA from mouse sperm should be considered as a mixture of spermatogonial transcripts that are partitioned into the products of meiosis and that do not necessarily represent the nuclear genome of post-meiotic sperm cells.
Crossover interference and the obligate crossover are unaffected by a hyperrecombination phenotype
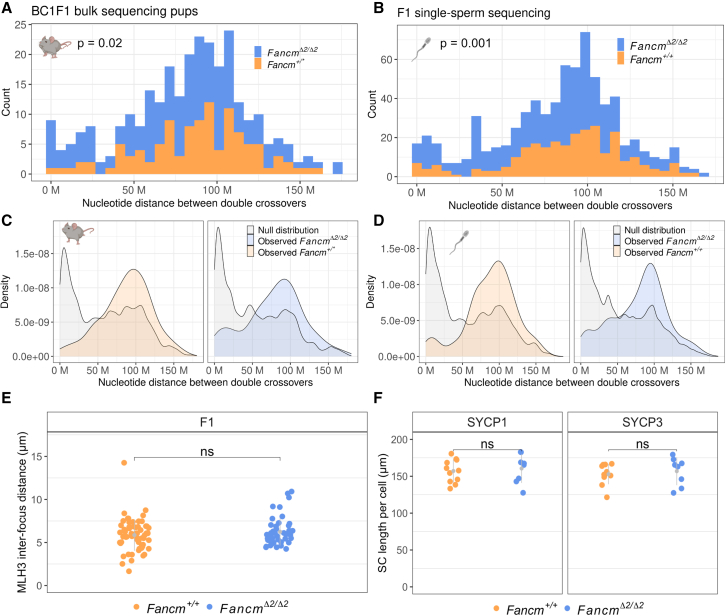
As we observed an increase in crossover frequency in Fancm-deficient mice, we sought to test how these extra crossovers are generated and if they show characteristics of the class I and/or class II pathways. Crossovers from the class I pathway are typically sensitive to crossover interference, meaning that they are more distantly spaced than would occur by chance. Therefore, we analyzed spacing between two crossover events on the same chromosome. These intercrossover distances from our genetic data revealed that crossovers were more distantly spaced than random in all datasets analyzed where sample size permitted (Figures 2A and 2B) for both the wild type and the mutant. The random distributions of intercrossover distances were generated through permutation via label swapping (Figures 2C and 2D and supplemental information).
Figure 2.
Extra crossovers in Fancm are consistent with the characteristics of class II crossovers
Interference is detected in Fancm-deficient male mice, but extra crossovers fit with a more random distribution and are not labeled by MLH1/3.
(A and B) Intercrossover distances were quantified in the wild type and the mutant from the BC1F1-derived bulk sequencing and sperm sequencing data by including only chromosomes with exactly two crossovers. Printed p values are from Wilcoxon rank-sum test. (A) n = (175 FancmΔ2/Δ2 and 109 Fancm+/∗ double crossovers per group. Fancm+/∗ indicates pooling of Fancm+/+ and Fancm+/Δ2 data, as their crossover rates were shown not to be significantly different). (B) n = (453 FancmΔ2/Δ2 and 276 Fancm+/+).
(C and D) Crossover interference is observed in the wild type and the mutant, as the observed intercrossover distributions are more evenly spaced than an expected random distribution generated via permuting genotype group labels (B = 1,000).
(E) Cytological measurement of interference used MLH3 interfocus distances in both the wild type and the mutant. n = 2 F1 (FVB/N × C57BL/6J) mice per genotype. Total cells analyzed: 62 F1.Fancm+/+ and 47 F1.FancmΔ2/Δ2. Gray bars indicate mean ± SD.
(F) Synaptonemal complex length was assessed by measuring SYCP1 and SYCP3 length in micrometers per nucleus in pachytene spermatocytes. Total cells analyzed: 16 F1.Fancm+/+ and 22 F1.FancmΔ2/Δ2. Gray bars indicate mean ± SD.
Given that there is an increase in crossover number in the Fancm-deficient mice, we tested whether the proportion of class I and class II crossovers was affected. In the BC1F1 bulk sequencing experiment, the distribution of intercrossover distances was significantly different between the mutant and the wild type (p = 0.02, Wilcoxon rank-sum test with continuity correction, n = 175 FancmΔ2/Δ2 and 109 Fancm+/∗ double crossovers per group), with the difference being attributable to a portion of closer double crossovers (Figure 2A). The increased proportion of physically closer double crossovers in Fancm-deficient mice is consistent with the idea that there was an increase in class II crossovers. The same pattern was observed in the single-gamete sequencing data (p = 0.001, Wilcoxon rank-sum test with continuity correction, n = 453 FancmΔ2/Δ2 and 276 Fancm+/+ double crossovers per group) (Figure 2B). We used a complementary approach to test for characteristics of class I and class II crossovers in the Fancm-deficient mice by analyzing MLH1 and MLH3 focus distributions, which are required for, and localize to, the sites of class I crossovers.57,94,95,96,97,98 Using labeling of MLH3 and MLH1 as a proxy for class I crossovers revealed no difference in event spacing and that crossover interference is present in the wild type and the mutant (Figure 2E). The length of the synaptonemal complex is positively correlated with the number of class I crossovers.47,48,49,50 Therefore, we measured the length of the synaptonemal complex in the absence of Fancm by measuring the central and axial elements, SYCP1 and SYCP3, respectively. Synaptonemal complex length appeared unchanged in pachytene in the mutant compared with the wild type (Figure 2F). No differences in prophase I progression were observed in F1.FancmΔ2/Δ2 compared with the wild type, with all meiotic prophase sub-stages from leptotene to diakinesis being observed in similar proportions (Table S3). These genetic and cytological data are consistent with an increase in class II non-interfering crossovers but an unchanged class I crossover number and distribution.
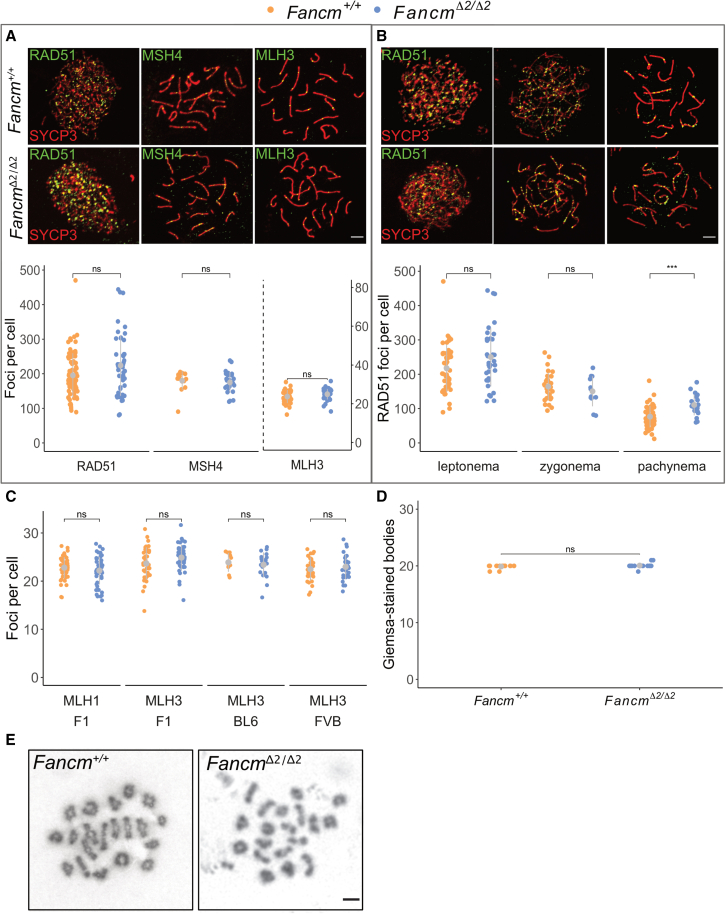
As F1.FancmΔ2/Δ2 males have increased crossover rates, we sought to cytologically investigate markers of DSB formation and repair in mouse spermatocytes using chromosome spreads. To analyze DSB formation we quantified RAD51 foci throughout early meiotic prophase I (Figures 3A and 3B). The progressive decrease in variation and number of RAD51 focus counts in the wild type was consistent with previous work.23 In FancmΔ2/Δ2 spermatocytes, we found that RAD51 foci were slightly increased at pachytene in the mutant (p < 0.001) in both the F1 hybrid and the FVB/N inbreds, suggesting a delay in processing of joint molecules that are processed in the absence of Fancm (Figure 3B). Elevated RAD51 at pachytene was also observed in Fancm homozygotes previously.6 To assess whether the elevated RAD51 focus numbers may contribute to DSB repair failure, we analyzed γH2AX localization. Using γH2AX, sex body formation appeared normal in F1.FancmΔ2/Δ2, and no persistent γH2AX was observed on autosomes at pachytene using meiotic chromosome spreads. We next sought to analyze DSB repair and pathway choice. We analyzed the loading of the ZMM MSH4, which initially localized to a majority of recombination sites and, along with its binding partner MSH5, is required for normal synapsis and crossover formation.99,100,101 MSH4 focus numbers were unchanged between Fancm genotypes in recombination intermediates labeled by MSH4 (Figure 3A). Similarly, MLH1 and MLH3 focus numbers, markers of class I crossovers, were unchanged in Fancm-deficient mice (Figures 3A and 3C). We assessed whether the extra crossovers in FancmΔ2/Δ2 affect meiotic completion. The obligate crossover was unaffected in FancmΔ2/Δ2, as bivalent formation was unchanged when analyzed with Giemsa-stained male meiotic metaphase I chromosomes (Figures 3D and 3E). Similar quantification of meiotic metaphase I on histological sections revealed no detectable difference in unbalanced univalents between genotypes (Table S4). Taken together, these data from different steps of DSB repair and crossover formation suggest that the extra crossovers in a mammalian Fancm mutant are not class I crossovers and do not dramatically affect meiotic progression.
Figure 3.
Meiotic double-strand break repair was relatively normal in FancmΔ2/Δ2 spermatocytes
(A) Representative images of F1.Fancm+/+ and F1.FancmΔ2/Δ2 spermatocyte chromosome spreads from meiotic prophase, stained for the indicated proteins (top), and quantification of foci per nucleus in wild-type and FancmΔ2/Δ2 spermatocytes. Each data point is a count from one cell. Scale bar, 5 μm. Gray bars indicate mean SD. F1.Fancm+/+: RAD51 foci at leptonema (217.65 68.8), MSH4 foci at zygonema (164.2 40.4), and MLH3 foci at pachynema (23.7 3.3). F1.FancmΔ2/Δ2: RAD51 foci at leptonema (250.0 85.5), MSH4 foci at zygonema (150.3 43.6), and MLH3 foci at pachynema (24.9 2.7). Total RAD51-labeled cells analyzed: 79 F1.Fancm+/+ and 46 F1.FancmΔ2/Δ2. Total MSH4-labeled cells analyzed: 16 F1.Fancm+/+ and 26 F1.FancmΔ2/Δ2. Total MLH3-labeled cells analyzed: 40 F1.Fancm+/+ and 47 F1.FancmΔ2/Δ2.
(B) RAD51 focus numbers are higher at pachynema in FancmΔ2/Δ2. Gray bars indicate mean SD. Each data point indicates a count from one nucleus. F1.Fancm+/+: RAD51 foci at leptonema (217.7 68.8), zygonema (164.2 40.4), and pachynema (76.5). FancmΔ2/Δ2: RAD51 foci at leptonema (250.0 85.5), zygonema (150.3 43.6), and pachynema (110.6 30.3). Scale bar, 5 μm (∗∗∗p 0.001 [unpaired t test], two F1 mice analyzed per genotype at each meiotic phase).
(C) Variation in MLH3 foci in B6, FVB, and F1 backgrounds. Eighty-nine F1.Fancm+/+, 101 F1.FancmΔ2/Δ2, 10 B6.Fancm+/+, 20 B6.FancmΔ2/Δ2, 32 FVB.Fancm+/+, and 32 FVB.FancmΔ2/Δ2 cells were analyzed. Gray bars indicate mean ± SD.
(D) The number of Giemsa-stained bodies per spermatocyte in Fancm+/+ and FancmΔ2/Δ2. Thirty-one Fancm+/+ and 19 FancmΔ2/Δ2 cells were analyzed. Gray bars indicate mean ± SD.
(E) Representative meiotic metaphase I spreads for B6.Fancm+/+ and B6.FancmΔ2/Δ2. Scale bar, 5 μm.
Spermatogenesis and oogenesis are perturbed in FancmΔ2/Δ2 mice
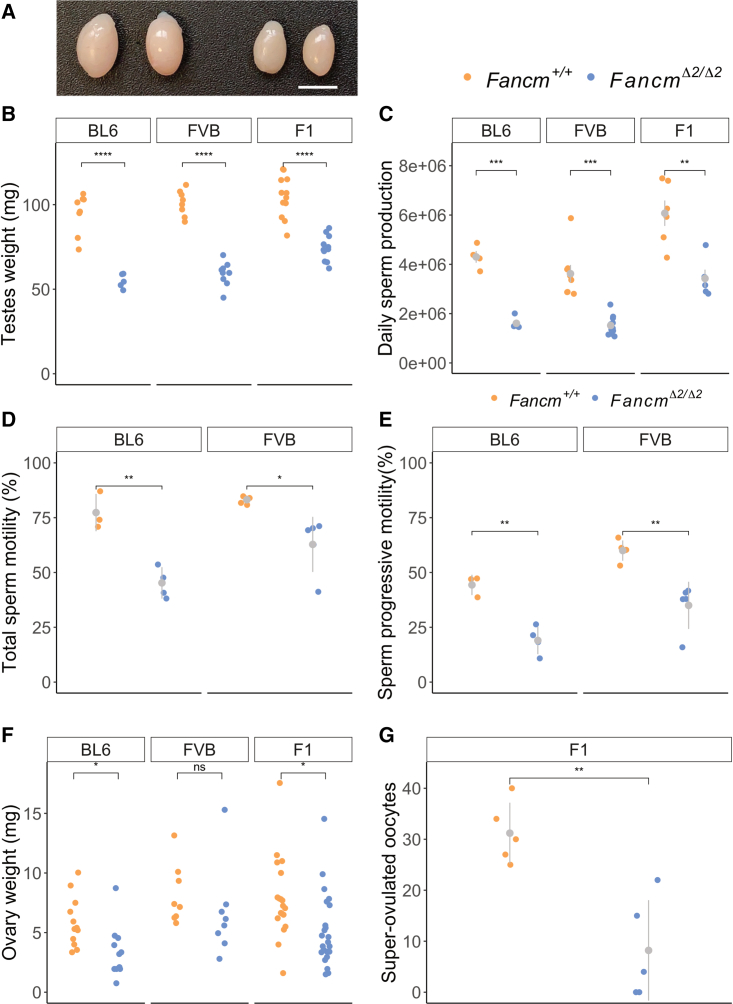
One of the few unifying phenotypes of mouse models from the FA pathway, including Fancm-deficient mice,5,6,8 is abnormal gonad development and sub-fertility.102 We explored this further and found that FancmΔ2/Δ2 testicular weight was reduced (Figures 4A and 4B). The reduction in testes weight was significant in all strains: FVB.Fancm, B6.Fancm, and F1.Fancm (Figure 4B). The reduction in the F1.Fancm, however, was not as strong as the reduction in testicular weight in the two inbred strains. In all three strains, daily sperm production was reduced in the mutant, indicating that Fancm is required for normal sperm production levels (Figure 4C). Further, computer-assisted sperm analysis revealed that, for those sperm that were produced, both B6.Fancm and FVB.Fancm had a decrease in the percentage of motile and progressively motile sperm (Figures 4D and 4E); however, this resulted in reduced litter sizes only in B6.Fancm. In females, ovary weights were reduced in B6.Fancm; however, the difference was less dramatic than what was seen for male gonads. FVB.Fancm and F1.Fancm ovary weights were lower than those of their wild-type littermates on average but did not reach statistical significance (Figure 4F). However, a dramatic reduction in the number of follicles per ovary was observed in Fancm-deficient FVB/N, C57BL/6J, and F1 mice, consistent with a premature ovarian insufficiency phenotype (Figures S14 and S4). Further, superovulated F1 mutants had a reduction in oocyte numbers (Figure 4G).
Figure 4.
Hypogonadism and gametogenesis defects in both sexes of Fancm-deficient mice
(A) Testes from B6.Fancm+/+ (left) and B6.FancmΔ2/Δ2 (right) mice. Scale bar, 5 mm.
(B) Six- to 14-week-old testes weights were lower in the mutants in all strains. Gray bars indicate mean ± SD.
(C) Quantification of daily sperm production. Gray bars indicate mean ± SD.
(D and E) Total sperm motility (D) and sperm progressive motility (E) were assessed with computer-assisted sperm analysis from 10-week-old mice. Gray bars indicate mean ± SD.
(F) Ovary weights were quantified across the three strains used from 5-week or older mice. Gray bars indicate mean ± SD.
(G) F1 females were superovulated and Fancm-deficient mice produced fewer oocytes. Gray bars indicate mean ± SD. Five mice per genotype; ∗p 0.05, ∗∗p 0.01, ∗∗∗p 0.001, and ∗∗∗∗p 0.0001.
cGAS-STING drives spermatogonial defects in Fancm mice
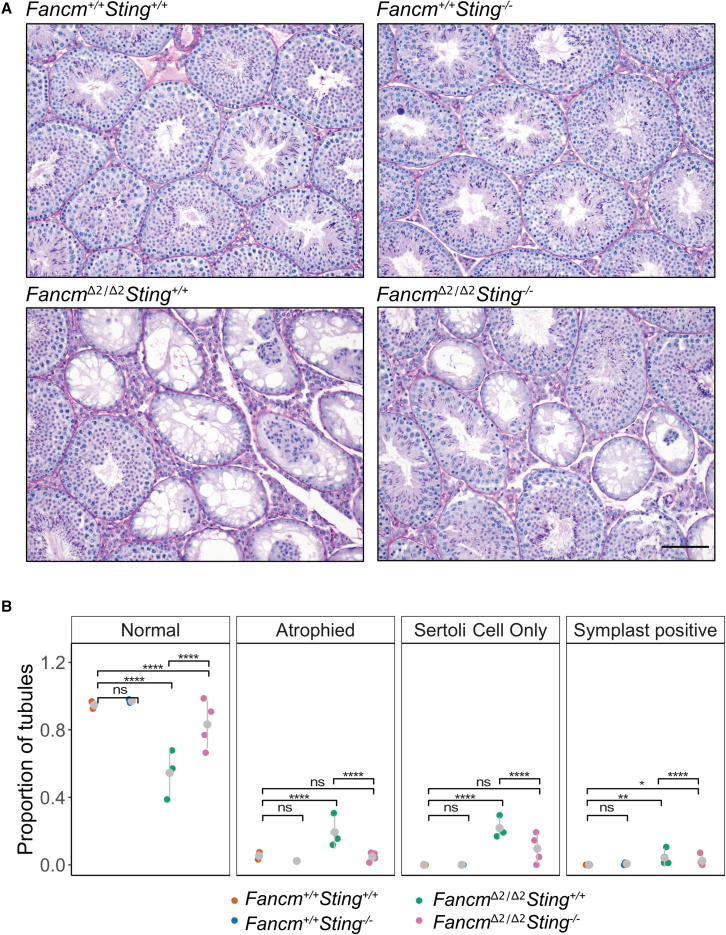
A mouse model with a point mutation in Fancm has previously been shown to have smaller testes due to impaired primordial germ cell proliferation, in addition to a progressive loss of male germ cells in adulthood.6 To investigate the origin of the sub-fertility of our B6.FancmΔ2/Δ2 males and the reduced daily sperm production across all Fancm-deficient strains, we analyzed the seminiferous tubules of wild-type and mutant mice. Histological analysis of the FVB.FancmΔ2/Δ2 strain showed no overt differences in the testicular histology between mutant and wild-type littermates (Figure S19). FVB.FancmΔ2/Δ2 seminiferous tubules appeared replete with all germ cell types present, suggesting the strong reductions in testes weight and daily sperm production (Figures 4B–4E) are likely driven by deficient primordial germ cell proliferation as described previously.6 Histological analysis of B6.FancmΔ2/Δ2, however, showed a number of defects. Most notably, and consistent with the progressive loss of germ cells seen previously due to Fancm deficiency,6 a small but significant subset of seminiferous tubules was characterized by either partial or complete absence of germ cells from the epithelium (Sertoli cell only; Figures 5A, 5B, S15, and S19). In the least severely affected of these, vacuoles between adjacent Sertoli cells were observed in addition to areas of Sertoli cell cytoplasm devoid of germ cells (Figure 5B). In more moderately affected tubules, whole populations of specific germ cell types were depleted (Figures 5B and S15). The most severely affected tubules were characterized by a Sertoli cell-only phenotype, wherein the loss of integrity of the seminiferous epithelium ultimately led to the depolarization of Sertoli cells from the basement membrane and their collapse into multinucleated symplasts (Figures 5A, 5B, and S15).
Figure 5.
Sting depletion partially rescues histological defects in the seminiferous tubules in Fancm-deficient mice
(A) H&E stain of seminiferous tubules in B6.Fancm+/+ and B6.FancmΔ2/Δ2 mice. Scale bar, 100 μm. Wild-type tubules are relatively replete in their appearance. Mutant mice have a heterogeneous reduction in spermatocytes (hypospermatogenesis) at the level of individual tubules. A mixture of normal, atrophied, symplast-positive, and Sertoli cell-only tubules is observed.
(B) FancmΔ2/Δ2Sting−/− testes have more full tubules than the Fancm single mutant (p-adjusted < 0.0001). Quantification of the PAS-stained seminiferous tubule cross sections. Each data point represents data from one mouse shown as a proportion for the given classification (∗p 0.05, ∗∗p 0.01, ∗∗∗∗p 0.0001 (pairwise proportion test with multiple testing correction fdr). A total of 290 Fancm+/+Sting+/+, 305 Fancm+/+Sting−/−, 479 FancmΔ2/Δ2Sting+/+, and 605 FancmΔ2/Δ2Sting−/− tubules were analyzed. Gray bars indicate mean ± SD.
An initial analysis of apoptosis with histological sections from C57BL/6J and FVB/N testes sections was performed by immunolabeling cleaved caspase-3 and -7 (Figure S16). We did not detect a change in the number of caspase-3/7-positive germ cells per seminiferous tubule in the mutants compared with the wild type. More refined analysis of the B6.FancmΔ2/Δ2 seminiferous epithelium suggested that there were at least two possible points of reduced daily sperm production, in addition to the well-documented reduced proliferation of primordial germ cells via the p21-p53-ATM axis.6 As shown in Figure S15B, in the B6.FancmΔ2/Δ2 seminiferous tubules characterized by progressive germ cell loss, pachytene spermatocytes often appeared to be the first germ cell population to be absent. Stage V–VIII tubules that harbored their characteristic populations of spermatids, spermatogonia, and other spermatocyte types103 were often either partially or completely devoid of pachytene spermatocytes (chi-squared = 19.314, df = 1, p < 0.001; Figure S17 and Table S5). This phenotype appeared to be driven by a significant increase in the death of spermatocytes at mid-pachytene. Indeed, analysis of stage V seminiferous tubules revealed a significant increase in the rate of pachytene spermatocytes with periodic acid-Schiff (PAS)-positive cytoplasm—PAS is suggested to stain apoptotic cells104—in B6.FancmΔ2/Δ2 testes compared with wild-type controls (p < 0.05; Figure S17F).
A second point of loss may be at the first meiotic division in metaphase I-anaphase I. At stage XII of spermatogenesis when the meiotic divisions are normally detectable on histological sections, the number of cells in metaphase I-anaphase I was comparable between B6.FancmΔ2/Δ2 and littermate controls (Figure S17C). The number of metaphase I-anaphase I spermatocytes that remained in the succeeding stage I, wherein meiosis is normally complete, was also comparable between genotypes (Figure S17C). Alignment of bivalents on the metaphase plate and their segregation also appeared unchanged (Figures 3 and S17C). However, a strong and significant increase in PAS-positive metaphase I-anaphase I spermatocytes, indicative of meiosis arrest, was observed in stage XII tubules (Figure S17). Likewise, a more than 2-fold increase in PAS-positive metaphase I-anaphase I or pyknotic cells was observed in stage I tubules (Welch two-sample t test, p < 0.0001, n = 4 mice per genotype; Figure S17E). However, no abnormalities were detected with either SYCP3 staining meiotic prophase sub-stages nor two-dimensional metaphase I chromosome spreads (Figure 3 and see Discussion). Histological analysis of spermatogenesis detected occasional binucleated round spermatids indicative of failed cytokinesis in B6.FancmΔ2/Δ2, but not in wild-type controls. In conclusion, within B6.FancmΔ2/Δ2 mice, the loss of FANCM resulted in at least three detectable defects using histology. This aside, sperm were produced and the mice retained some level of fertility.
We next analyzed the innate immune response, as previous studies have revealed that genomic instability in mouse models can also activate the cGAS-STING pathway,82,84 which is a part of the innate immune system that detects cytosolic DNA.105,106,107 We sought to investigate whether the cGAS-STING pathway could be driving the reduction in germ cells in the B6.FancmΔ2/Δ2 mice. Fancm Sting double mutants, compared with Fancm single mutants, had an increase of approximately 20% in testicular weight, while epididymis weight and daily sperm production were unchanged and far inferior to wild-type levels (Figure S18). Next we performed histological analyses with the seminiferous tubules. This revealed that the Fancm Sting double mutants, compared with Fancm, had a statistically significant increase in seminiferous tubules containing complete spermatogenesis (p < 0.0001, pairwise proportion test after multiple testing correction) (Figures 5 and S19). In contrast, folliculogenesis in Fancm-deficient mice was not ameliorated by the removal of the Sting pathway (Figure S21).
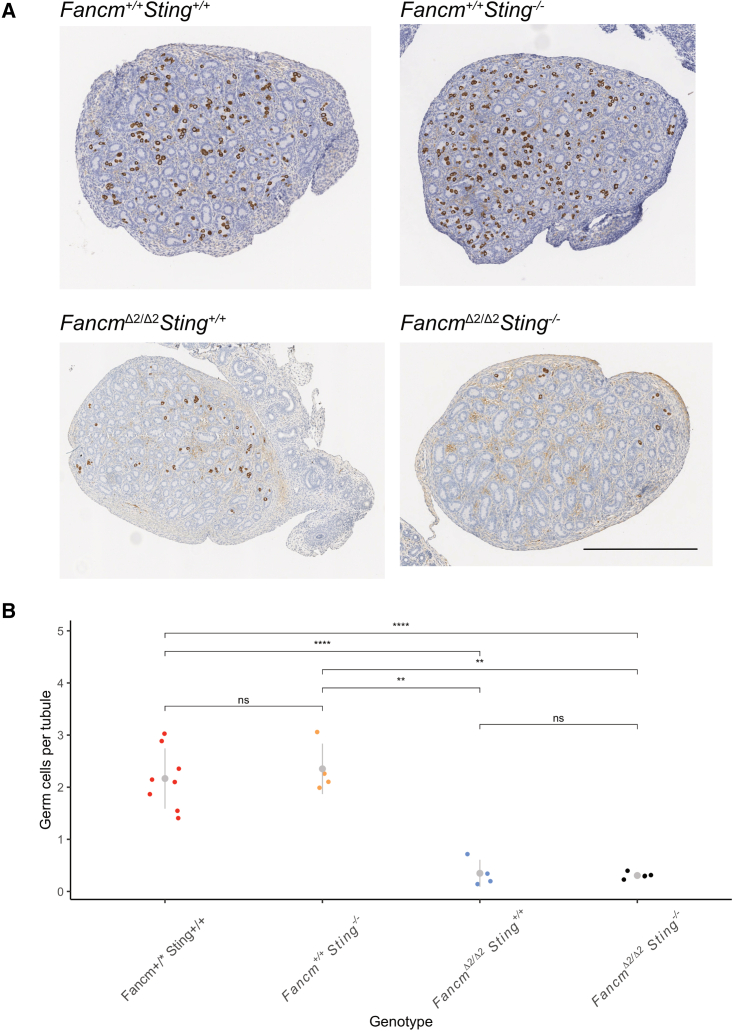
We next aimed to determine whether the STING pathway affects depletion of germ cells in Fancm-deficient mice during embryogenesis or after. Our results showed that the number of germ cells in Sting-deficient newborn male mice was similar to that of wild-type mice (Figures 6 and S20). However, Fancm-deficient mice had significantly fewer germ cells, as reported previously.6 Germ cell numbers in Fancm Sting double mutants were similar to those in Fancm single mutants. Therefore, it appears that the STING-dependent depletion of a portion of germ cells in Fancm-deficient mice occurs after birth.
Figure 6.
Sting depletion does not rescue germ cell numbers in newborn male Fancm-deficient mice
(A) Representative images of testis cross sections stained with anti-MVH. Scale bar, 500 μm.
(B) Germ cells per tubule were quantified in the indicated genotypes using anti-MVH. Each data point represents average germ cells per tubule for a unique mouse. A minimum of 500 tubules were counted per mouse (∗∗p 0.01, ∗∗∗∗p 0.0001). Gray bars indicate mean ± SD.
Given the interaction between Fancm and the innate immune response via the cGAS-STING pathway, we sought to test alternative inflammatory pathways. Previous work has revealed that there is a female-biased embryonic lethality in the absence of Fancm.10,108 During the course of our experiments we observed an underrepresentation of B6.FancmΔ2/Δ2 mice, which was more severe for females (Table 1). Previous studies have observed sub-Mendelian birth rates of female Fancm-deficient mice5,6,10; however, with a larger sample size in our study, we showed that this occurs for B6.FancmΔ2/Δ2 males, too. FVB.FancmΔ2/Δ2 were not underrepresented in either sex, however, and neither were F1 mutants descended from a cross between B6.Fancm+/Δ2 and FVB.Fancm+/Δ2 (Tables S1 and S2). These data suggest that the sub-Mendelian birth rates of Fancm mutants in our experiments are specific to the C57BL/6J background and are a recessive trait. Previous observations of bias in lethality of female embryos were shown to be due to inflammation caused by genomic instability and could be alleviated by supplementing the drinking water of pregnant mothers with the anti-inflammatory drug ibuprofen.10 We tested whether ibuprofen reversed the seminiferous tubule defects of Fancm-deficient mice; however, there was no effect compared with controls (Figure S22).
Table 1.
Offspring of B6.FancmΔ2/+ × B6.FancmΔ2/+ intercrosses
| Male |
Female |
|||
|---|---|---|---|---|
| Observed | Expected | Observed | Expected | |
| Fancm+/+ | 64 | 57 | 63 | 57 |
| Fancm+/Δ2 | 137 | 114 | 118 | 114 |
| FancmΔ2/Δ2 | 45∗ | 57 | 29∗∗∗ | 57 |
| Total | 246 | – | 210 | – |
∗p < 0.05 (chi-squared test).
∗∗∗p < 0.0005 (chi-squared test).
Discussion
Fancm performs distinct functions during reproduction
Fancm is essential for efficient homologous recombination by directing the outcomes of DNA repair intermediates. This study identifies Fancm as a rate-limiting step during mammalian meiotic crossover formation. This study also extends the understanding of the role of Fancm in fertility. We propose that these two functions are distinct, as meiotic outcomes appear relatively unaffected at a chromosomal and molecular scale, whereas gamete numbers are affected in males and females.
We suggest that the meiotic hypercrossover and other spermatogenesis phenotypes that occur due to the loss of Fancm are separable for several reasons: (1) there are no notable defects in bivalent formation compared with the wild type that could account for such a dramatic reduction in spermatogenesis and the requisite stem cells. The meiotic program progresses normally when considering direct immunofluorescence readouts of the synaptonemal complex formation and meiotic DSB repair. There are more meiotic DSBs that persist at pachytene than in the wild type (Luo et al.6 and this study), but given the rate of daily sperm production in mutants and number of healthy pups that were born, it does not seem that these lead to major meiotic errors and perhaps simply reflect altered and later processing of a subset of meiotic DSBs. (2) The heterogeneous Sertoli cell-only phenotype of some seminiferous tubules may be due to stochastic DNA repair-related aberrations that occurred during the establishment of the germ cell lineages early in embryo development and perhaps also during failed maintenance of spermatogonia that achieve only a limited number of rounds of spermatogenesis, which would be consistent with other mice lacking the FA pathway.6,102 Similarly, some tubules have a “lacy” phenotype that may be due to the loss of individual spermatogonial stem cells. Phenotypes that are driven by strong meiotic DSB repair defects have a homogeneous arrest at the pachytene and typically produce no round spermatids or spermatozoa as assessed with histological sections of the seminiferous tubules.109,110 No such pachytene arrest is observed in F1.FancmΔ2/Δ2, neither with surface spreads that visualize the synaptonemal complex nor with histological analysis. However, we do not exclude the possibility that the hypercrossover Fancm could make a minor contribution to reproductive defects. (3) Recent work with Fanci has shown that the gene function was required for primordial germ cell development, but not postnatally.111 Fancm mutants were also shown to have a reduction in primordial germ cell numbers6 and an increase in specific mutational signatures,112 which may account for the reduction in gamete numbers and litter sizes. One possibility of how these two functions co-exist is through the vertebrate-specific addition of the ERCC4 domain. The translocase domain would be the domain that remodels DSB repair intermediates to limit crossover rates. However, the ERCC4 domain may be involved in FA pathway activation and in turn protect stem cells from damage. In summary, we propose that these observations support the hypothesis that the meiotic anti-crossover function of FANCM can be uncoupled from its role in promoting genomic stability and efficient spermatogenesis.
FANCM is an evolutionarily conserved crossover-limiting factor
Here we demonstrate that Fancm limits meiotic crossovers in mammals. This is the first report of a species with a bona fide FA pathway that has been shown to use FANCM to limit meiotic crossover rates. This result was demonstrated with novel large-scale pedigree and single-sperm sequencing approaches: SSNIP-seq, sgcocaller, and comapr.86,87 These novel approaches complement methods for sperm sequencing analysis.113,114,115,116 As crossover rates and the occurrence of trisomies are correlated,19 it is tempting to speculate that the Fancm mechanism of limiting crossovers may in some cases contribute to rare instances of chromosome mis-segregation. Typically, it is a reduction in crossover rates that is associated with meiotic non-disjunction and trisomies, but it is possible that deregulated and aberrant crossover positioning could also lead to chromosome segregation defects.
The limited shuffling of genetics enforced by Fancm is conserved across diverse eukaryotes, including mammals (this study), plants, fission yeast, and fly.69,70,74,117,118 This raises the question of whether FANCM achieves its crossover-limiting function with other components of the FA pathway, such as the FANCC-FANCE-FANCF complex,119 which will be interesting for future studies to discern. Crossover formation is a tightly regulated and well-conserved process. DNA DSBs outnumber crossovers consistently across diverse species, which suggests that there are conserved DNA repair factors that keep crossover numbers below their possible upper bounds. This study and previous studies highlight Fancm as a major driver of this phenomenon.69,70,74,117,118 Why crossover numbers per cell are limited is difficult to discern. One hypothesis is that a baseline level of crossovers at meiosis—and therefore only once per generation—has a net positive effect on a population in a changing environment. However, there will also likely be negative consequences for a number of individuals associated with the decoupling of favorable combinations of linked alleles that have been selected for on evolutionary timescales120; therefore, on average, parental combinations of alleles could be more beneficial than excessive shuffling of allelic combinations. Another hypothesis is that excessive crossovers impede correct segregation of chromosomes; however, this appears unlikely as a universal rule, given that significant increases in crossover frequency did not affect chromosome segregation in this study, in mutants of Arabidopsis, or in Drosophila mutants.70,73,117 Also significant differences in crossover rates are normal within species, such as differences in crossover rates between males and females,121,122 and some species naturally have very high levels of crossovers per chromosome.46
Several crossover-limiting factors have been identified, many of which play important roles in maintaining genomic stability.45,67,71,73,123 The molecular function of FANCM is consistent with an anti-crossover factor, as it can act on a diverse range of branched DNA substrates, many of which are produced during meiotic DSB repair, such as D loops and Holliday junctions that could progress to crossover formation. Therefore, the loss of FANCM function would force the repair of joint molecules to occur using alternate repair pathways and non-crossover outcomes. All studies of the anti-crossover role of FANCM have characterized loss-of-function alleles, and Fancm has not been identified in studies that screened for natural variants that influence crossover rates, to the best of our knowledge. Other meiotic crossover-limiting factors that tend to be characterized only as knockouts are Sgs1/BLM, RTEL-1 homologs, and their binding partners,61,70,71,73,124,125,126 which are also required for genomic stability.127,128,129,130 Perhaps the loss-of-function phenotypes are identified only in laboratory settings because their somatic functions tolerate little to no variation, which can be seen in their respective chronic pediatric diseases FA, Bloom syndrome, and dyskeratosis congenita. This contrasts with natural alleles of genes such as PRDM9, RNF212, HEI10, and REC8 that are associated with the location of recombination hotspots, meiotic chromosome axis structure, and global crossover rates.24,26,131,132
The extra crossovers in F1.FancmΔ2/Δ2 were not linked to overt changes with respect to relative distributions of crossovers along chromosomes. Further, these extra crossovers in Fancm-deficient mice were not labeled by MLH1/3 and were distributed with less even spacing than occurred in the wild type. This is consistent with an increase in non-interfering (class II) crossovers. These findings are consonant with what was observed in Arabidopsis.67 A difference between Arabidopsis and mouse Fancm knockouts, however, is that in the mouse, FANCM limits crossovers in interspecific F1 crosses; the strong anti-crossover activity of AtFancm is limited to inbreds or long runs of homozygosity.67,72,123 We speculate that these differences in crossover formation in interspecific hybrids may reflect alternate regulation of the early stages of strand invasion. The crossover density prediction model that we implemented from Pratto et al.30 suggests that physical chromosome size, replication speed, and DSBs are good predictors of the extra crossovers that occur in the absence of Fancm (class II crossovers), just as they were good predictors of class I crossovers in the wild-type cross between C57BL/6 and CAST mice in Pratto et al.30 Therefore, we propose that: (1) in mammals, Fancm limits class II crossovers and (2) while class I and class II crossovers have differing genetic requirements, class I and II crossover formation pathways have similar characteristics with respect to the genomic features that influence crossover formation.
Reproductive defects occur at distinct periods of development in Fancm mice
Humans and mice that lack proper Fancm function are infertile or have severely impacted reproductive capacity.5,6,7,8,9 This is different from other species, in which loss of Fancm has no effect on gamete numbers despite leading to massive increases in total crossover numbers. These differences and other data that we present about meiotic outcomes further support the idea that the reproductive defects in the absence of Fancm are not tied to the anti-crossover role at meiosis.
The gonads of male and female Fancm knockouts were reduced in weight in two inbred strains and the F1 and are consistent with hypogonadism phenotypes in human FA patients and other Fanc gene mouse models.5,6,8,15,102,133 Similarly, a consistent reduction in daily sperm production in Fancm-deficient mice was observed in all strains. Histological analyses of the testis revealed a dramatic phenotype in the C57BL/6J strain on seminiferous tubule composition, notably, a progression toward a Sertoli cell-only phenotype.
Direct analysis of markers of DSB formation and repair did not suggest a strong, or any, defect in meiotic DSB repair in the absence of Fancm. Similarly, chromosome dynamics were unchanged when considering synapsis and chromosome segregation. Nevertheless, histological analyses showed unambiguous phenotypes when whole tubules were considered, as opposed to individual meiocytes, such as a complete absence of large groups of pachytene cells that would be found in comparable tubules in the wild type. Considering these data together, it seems most plausible that any recombination defect that may exist in Fancm-deficient mice is mild, particularly given that daily sperm production is measured in the millions, and litter sizes from male mutants are relatively unchanged. However, an alternative interpretation could be that a particularly mild recombination defect triggers a checkpoint arrest, which is reflected by the PAS-positive metaphase I-anaphase I cells. In B6.Fancm we found that a portion of the defects in spermatogenesis is driven by the cGAS/STING-dependent pathway. However, we did not observe the same effect in newborn males, suggesting that STING promotes the death of spermatogonial stem cells in Fancm-deficient mice after birth, but not before. This is supported by our data on female gonads, where no difference in ovarian follicle numbers was observed between B6.Fancm and B6.Fancm Sting, as females cannot make more oocytes after birth, whereas sperm production can be altered postnatally. Our findings suggest that an inhibitor of the cGAS/STING pathway could improve fertility rates in males with genetic conditions rooted in genomic instability, potentially beyond the FA pathway. Further, our data support the idea that STING promotes reproductive cell death after birth, rather than during embryogenesis. This may inform opportunities for testing STING inhibitors after birth for specific manifestations of germline genomic instability conditions like FA. However, we note that daily sperm production was not altered in the Fancm Sting double mutants compared with Fancm-deficient mice, and more levels of regulation of spermatogenesis will need to be deciphered.
In contrast to B6.Fancm, for FVB.Fancm or F1.Fancm, a lesser effect on gonad weight, daily sperm, and seminiferous tubule composition was detected in experiments with histopathology. These data suggest that the B6.Fancm seminiferous tubule phenotype is recessive in the F1.Fancm and the causal variant(s) can likely be positionally cloned in future studies to identify modifiers of genomic instability phenotypes. The differing phenotypes of C57BL/6J and FVB/N seminiferous tubule phenotypes reveal a difference between the two strains that appears unlinked to Fancm; however, the relevance of this is unclear and could be investigated in future studies.
In summary, despite the defects in gametogenesis (this study and Bakker et al.,5 Luo et al.,6 Fouquet et al,7 Yin et al.,8 and Kasak et al.9), we show that Fancm knockouts are capable of producing offspring, which we could leverage to reveal the anti-crossover function of mammalian FANCM. The capacity of mouse knockouts to produce a tractable number of offspring has been shown for multiple Fanc gene knockout mice, highlighting the utility of these DNA damage response-defective models to inform clinical fertility research for patients with chronic genetic conditions. Further, as human infertility can present before other serious symptoms caused by genetic conditions, there are opportunities to improve diagnostic pathways for life-limiting diseases by considering reproductive status.102,134
Limitations of the study
Our sequencing-based approaches provided detailed analyses on crossover profiles in Fancm-deficient mice. However, sample throughput was constrained by costs and analytic complexities, and as such, we were unable to obtain sufficient read coverage to call short/non-crossovers. Further, although there are advancements in strain-specific reference genomes, some regions of the genome remain unresolved, and this may have impeded our ability to call crossovers in these regions. Our study made use of two mouse strains to use the SNPs to identify crossovers. As some of our results have shown, different genetic strains with the same genetic mutation in Fancm had variable fertility phenotypes and therefore may not be generalized to other mouse strains. Other limitations include the lack of antibodies that worked in our hands for mouse Fancm and for cytological markers of class II crossovers. Future studies could also consider assays that increase the sensitivity to detect class II chiasmata by crossing Fancm-deficient mice with ZMM mutants.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal FANCD2 | Novus Biologicals | NB100-182; RRID:AB_10002867 |
| Mouse ɑ-tubulin | Cell Signaling | #3873; RRID:AB_1904178 |
| Cleaved caspase-3 | Cell Signaling | #9664; RRID:AB_2070042 |
| Cleaved caspase-7 | Cell Signaling | #9491; RRID:AB_2068144 |
| Deposited data | ||
| Single-cell DNA sequencing of sperm cells and bulk DNA sequencing of BC1F1 pups | This paper | European Nucleotide Archive https://www.ebi.ac.uk/ena/browser/home ENA: PRJEB57095 |
| Analysis pipelines and processed data files | This paper | https://doi.org/10.5281/zenodo.7939158 |
| FVB/N SNP variants | Mouse Genome Project https://www.sanger.ac.uk/data/mouse-genomes-project/ |
https://ftp.ebi.ac.uk/pub/databases/mousegenomes/REL-1505-SNPs_Indels/strain_specific_vcfs/ |
| Mouse reference genome Genome Reference Consortium Mouse Build 38 (mm10) | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/ |
| GC Percent track GRCm38 | University of California, Santa Cruz | https://hgdownload.soe.ucsc.edu/goldenPath/mm10/bigZips/ |
| RepeatMasker | Smit et al.135 | http://www.repeatmasker.org |
| Experimental models: Organisms/strains | ||
| Fancm C57BL/6J | This paper | N/A |
| Fancm FVB/N | This paper | N/A |
| C57BL/6J | This paper | N/A |
| FVB/N | This paper | N/A |
| Oligonucleotides | ||
|
Fancm forward primer 5′ CGGGGCGG AATGCTAAACTT 3′ |
This paper | N/A |
|
Fancm reverse primer #1 5′ ACACACA GGGACAGAGAACACTC 3′ |
This paper | N/A |
|
Fancm reverse primer #2 5′ GGAAAAG AGAAAAGAAAAGGGGGA 3′ |
This paper | N/A |
|
Sting forward primer #1 5′ GCTGGGAA TTGAACGTAGGA 3′ |
This paper | N/A |
| Sting forward primer #2 5′ 3′ | This paper | N/A |
|
Sting reverse primer 5′ GAGGAGACAA AGGCAAGCAC 3′ |
This paper | N/A |
| Software and algorithms | ||
| R (3.5.1) | R Core Team, 2021 | https://www.r-project.org/ |
| lme4 | Bates et al.136 | https://cran.r-project.org/web/packages/lme4/index.html |
| cellranger-dna cnv (v1.1.0) | 10X Genomics | https://support.10xgenomics.com/single-cell-dna/software/pipelines/latest/using/cnv |
| glmmTMB | Brooks et al.137 | https://cran.r-project.org/web/packages/glmmTMB/index.html |
| sgcocaller and comapr | Lyu et al.86 |
https://gitlab.svi.edu.au/biocellgen-public/sgcocaller https://bioconductor.org/packages/release/bioc/html/comapr.html |
| Gviz | Hahne et al.138 | https://bioconductor.org/packages/release/bioc/html/Gviz.html |
| biomRt | Durinck et al.139 | http://www.bioconductor.org/packages/release/bioc/html/biomaRt.html |
| Fastp | Chen et al.140 | https://github.com/OpenGene/fastp |
| minimap2-v2.7 | Li, Heng141 | https://github.com/lh3/minimap2 |
| gatk(v4.0) | McKenna et al.142 | https://gatk.broadinstitute.org/hc/en-us |
| code repository | This paper | https://gitlab.svi.edu.au/biocellgen-public/fancm-crossovers-2022 |
| Fiji (ImageJ-win64) | Schindelin et al.143 | https://fiji.sc/ |
| ImageJ Scripts for axis detection and foci analysis | Boekhout et al.144 | https://github.com/Boekhout/ImageJScripts |
| synapsis | McNeill, L. | http://bioconductor.org/packages/release/bioc/html/synapsis.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Wayne Crismani (wcrismani@svi.edu).
Materials availability
Mouse lines in this study have been cryopreserved and deposited to the Australian Phenome Facility.
Method details
Ethics statement
All animal procedures were approved by the Animal Ethics Committees at St Vincent’s Hospital Melbourne and Monash University and conducted in accordance with Australian NHMRC Guidelines on Ethics in Animal Experimentation.
Mouse generation and genotyping
The FancmΔ2/Δ2 mice were generated independently in FVB/N and C57BL/6 backgrounds. The same guide RNAs and Cas9 were used to target exon 2 of Fancm. For genotyping, wild-type and mutant alleles were amplified with oligos forward primer (5ʹ CGGGGCGGAATGCTAAACTT 3ʹ), reverse primer 1 (5ʹ ACACACAGGGACAGAGAACACTC 3ʹ, spans deleted region) and reverse primer 2 (5ʹ GGAAAAGAGAAAAGAAAAGGGGGA 3ʹ) and produced bands of 311 bp and 279 bp respectively that were visualised on a 2% agarose gel. Sting-/- mice have been described previously145 and the Sting-/- mice were backcrossed for at least 10 generations on a C57BL/6 background by.146 The oligos used for genotyping were forward primer 1 (5ʹ GCTGGGAATTGAACGTAGGA 3ʹ), forward primer 2 (5ʹ GTGCCCAGTCATAGCCGAAT 3ʹ, spans deleted region) and reverse primer (5ʹ GAGGAGGCAAGCAC 3ʹ).
Genomic DNA preparation and sequencing
Genomic DNA was extracted using the phenol-chloroform method. Briefly, a 0.5 mm of tail was placed in a 1.5 mL microfuge tube with 500 μL of tissue digestion buffer (100 mM Tris-Cl pH 8.0, 5 mM EDTA pH 8.0, 200 mM NaCl, proteinase K 0.4 mg/mL) overnight on a shaking platform at 55 C. The next day, 1 μL of 10 mg/mL RNAse A was added to each sample and incubated at 55 C for 30 min. 0.7 mL of neutralized phenol/chloroform/iso-amyl alcohol (25:24:1) was added to each sample and mixed vigorously at 4 C on a clinical rotor for 1 hour. Samples were spun in a bench top centrifuge at max speed (21,000 x g) for 5 min and the upper aqueous phase containing DNA was transferred to a new 1.5 mL microfuge tube containing 1 mL of 100% ethanol to precipitate the DNA. The samples were spun at max speed for 5 min and supernatant was removed. DNA pellet was washed with cold 70% ethanol and spun again at max speed for 5 min. 1 X TE buffer was added to the samples to resuspend the DNA and placed on a 55 C block to evaporate residual ethanol. All samples were checked for protein contamination by using Nanodrop A260/A280 and DNA quality by running samples on a 0.8% agarose gel to ensure there was no DNA shearing.
For bulk sequencing, genomic DNA was sent to BGI (China) or AGRF (Australia) and sequenced with DNBSeq or Illumina platforms respectively with between 1X and 5X coverage.
Haploid nucleic isolation for droplet-based single-cell library preparation
To obtain single haploid cells, testes were harvested from male mice and processed as described in the SSNIP-seq protocol.87 Briefly, on ice, the tunica albuginea was removed to release the seminiferous tubules into a 1.5 mL microfuge tube and incubated on ice for 30 min. The spleen was also harvested and homogenised through a 40 μm strainer to act as a diploid control for flow cytometry.
600 μL of testes or 300 μL of splenic homogenate was transferred to a new 1.5 mL LoBind microfuge tube. 1 mL of chilled Nuclei EZ Lysis buffer (Sigma) was added to each sample, inverted twice, and incubated on ice for 2 min. Samples were centrifuged at 500 x g for 5 min at 4 C and supernatant was removed, ensuring that the pellet was still immersed. Samples were resuspended with 1 mL of Nuclei EZ Lysis buffer, inverted several times and incubated at 500 x g for 5 min at 4 C. Samples were centrifuged at 500 x g for 5 min at 4 C and supernatant was removed. 1 mL of Nucleic Wash and Resuspension Buffer (NWRB; PBS with 1% BSA) was added slowly to the samples to avoid disruption of the pellet and allow buffer interchange. Samples were centrifuged at 500 x g for 5 min at 4 C and resuspended with 1 mL of NWRB, and this was repeated. Samples were centrifuged at 500 x g for 5 min at 4 C and supernatant removed. Sample was resuspended with 1 mL NWRB with DAPI (10 μg/mL). The samples were then filtered through using a 40 μm Flowmi cell strainer into a 5 mL round bottom polystyrene tube.
Using the spleen sample as a control for the diploid peak, 100 000 haploid nuclei was sorted into a round-bottom 96-well plate with 50 μL of NRWB + 0.4% BSA. Sorting was performed using an BD FACSARIA II cell sorter. After sorting, samples were visualised under an epifluorescent microscope to confirm the integrity and morphology of cells. The nucleic samples were then processed using a 10X Genomics scCNV library preparation kit following the manufacturer’s instructions.
Genomic DNA data processing
The paired-end DNA sequencing dataset of bulk samples were first preprocessed for filtering out low-quality reads, and trimming adapter bases using fastp140 with default parameters. The filtered and trimmed sequences were then mapped to the mouse reference genome mm10 using minimap2-v2.7141 with options -ax sr. Duplicated reads were marked using MarkDuplicates from gatk(v4.0).142
Single-cell DNA sequencing reads were processed using cellranger-dna cnv (v1.1.0) pipeline that demultiplexed single-cell reads and, and aligned reads to the mouse reference genome (mm10).
Finding informative SNP markers for crossover detection
Variants in FVB/N mouse genome were downloaded from Mouse Genome Project (FVB_NJ.mgp.v5.snps.dbSNP142.vcf),147 which contains the variants found in the FVB mouse genome when comparing to the reference mouse genome mm10 (C57BL/6 strain). The informative SNP markers for calling crossovers are SNPs with heterozygous genotypes in F1. To get the list of informative SNP markers, the downloaded variants were further filtered for variants with homozygous alternative genotype (GT==1/1).
Calling crossovers
Crossovers were detected by finding genotype or haplotype shifts in the BC1F1 or sperm cell genomes through modelling DNA read counts observed per sample or per cell. With DNA reads from each sample mapped and sequenced, the haplotype can be inferred by looking at the alleles carried by the DNA reads across the list of informative SNPs. To account for technical artifact (from sequencing and mapping), Hidden Markov Model based methods were implemented for crossover detection in BC1F1 samples and sperm cells with different settings (Supplemental information).
Hidden Markov Model
A Hidden Markov model (HMM) with binomial emission probabilities was constructed for finding crossover positions in mouse genomes while accounting for the technical artefacts including mapping errors. The observable variable in the model is the allele specific counts across the list of informative SNP sites, whereas the genotype of SNP sites is a hidden factor to be inferred. Allele read counts per sample across all informative SNP positions were obtained using bcftools (v.1.9)148 with the above filtered VCF file as the region file specified via the -R option for each BC1F1 sample using their mapped DNA reads. Customized R scripts were implemented for applying the HMM on BC1F1 samples and inferred the sequence of hidden states against the list of SNP markers for each chromosome. Crossover detection from the single-sperm sequencing data was also by applying HMM but with different HMM configurations. Allele read counts for single cells were obtained using customized software tool sgcocaller86 that summaries the allele counts across all cells and SNP positions into count matrices per sample and crossovers were called with options –cmPmb 0.0001 –maxDP 6 –maxTotalDP 30.
Analyzing crossover frequencies and distributions
Output files from sgcocaller are further parsed and analysed for crossover interval identification and crossover profile comparison. With the sequence of hidden states inferred, the crossover interval locations were called by applying functions implemented in R package comapr.86 The construction of genetic distance maps and crossover distributions were also performed using comapr. Crossover frequencies were converted to genetic distances in centiMorgans using the Kosambi mapping function149 via calling the calGeneticDist function. Permutation testing function (permuteDist) was performed to test whether there is statistical difference in total or binned genetic distances between genotype groups (FancmΔ2/Δ2 versus Fancm+/∗) (Supplemental information).
The effect of genomic features including SNP density, GC content, and TSS (transcription start site) density were visualized by plotting the genomic feature tracks with crossover density tracks using R package Gviz.138 The GC percent track summary for mm10 was downloaded from UCSC Genome Browser, and TSS annotation was obtained from the R package bioMart139 (STAR Methods and deposited code repository of this paper.)
The correlation of crossovers sites with genomics features was also investigated by plotting the distribution of distances of identified crossovers sites to genomic features including meiotic DSBs (data from113), H3K4me3 markers (data from113), transcription start sites, DNA transposons, long interspersed nuclear elements, long terminal repeat retrotransposons, low complexity repeats are AT-rich or GC-rich regions, retrotransposons, satellite repeats, short interspersed nuclear elements, and simple repeats (genomic annotations from RepeatMasker135)
Regression modeling of crossover densities
A linear regression model was fitted to predict crossover distribution using the physical chromosome size, meiotic DSB frequency and replication speed using public data90 as described.30 The predicted crossover densities from the previous model were compared with observed crossover densities in this study using linear regression.
Meiotic chromosome spreads with immunofluorescence at prophase I
Meiotic chromosome spreads from adults testes (>8 weeks) were prepared as previously described.150 Antibodies and dilutions used were: SYCP3 (Santa Cruz, sc-74569 at 1:100); RAD51 (EMD PC130 at 1:250); MSH4 (Abcam ab58666 at 1:200); MLH1 (BD Biosciences 51-1327GR at 1:50), MLH3 (gifted, 1:500); γH2AX (Millipore at 1:500); FANCD2 (Novus Biologicals at 1:50). After immunostaining, slides were countered stained with DAPI (10 μg/mL in 1xPBS) and mounted in Dako Fluorescence mounting medium. Secondary fluorescent antibodies were: goat anti-mouse Alexa Fluor 488, donkey anti-mouse Alexa Fluor 488, goat anti-mouse Alexa Fluor 594, donkey anti-rabbit Alexa Fluor 568.
Image analysis was performed using Fiji (ImageJ-win64)143 with a macro developed by Boekhout et al.144, synapsis151 and manual counts. For cell scoring, the following criteria were used to classify the prophase stage: (i) leptotene: Short, discontinuous SYCP3 signals. (ii) Early zygotene: Long, discontinuous SYCP3 signals, more than 1 but fewer than 10 autosome pairs synapsing. (iii) Mid zygotene: Between 10 and 16 autosome pairs synapsed. (iv) Late zygotene: At least 17 but fewer than 19 autosome pairs synapsing, no clear XY body. (v) Early pachytene: 19 synapsed autosome pairs, clear XY body, thicker lateral element complex (as visualized by SYCP3 staining). (vi) Mid pachytene: 19 synapsed autosome pairs, clear XY body, elongated SYCP3 signal (as visualized by SYCP3 staining). (vii) Late pachytene: 19 synapsed autosome pairs, clear XY body, elongated SYCP3 signal, ending in knob-like structures (as visualized by SYCP3 staining). (viii) diplotene: At least one autosome pair starting to de-synapse.
Meiotic metaphase I chromosome spreads
Meiotic chromosome spreads from adult testes (>8 weeks) were prepared following the methods described in.152 For staining, slides were incubated in 10% Giemsa for 10 minutes, washed briefly with de-ionised water, air dried and mounted with DPX mounting medium.
Blood collection and micronucleus assay
Blood was collected into heparin solution. Red and white blood cells were counted using a Beckman-Coulter haemtology machine. The micronuclei assays were performed using mouse erythrocytes as described in.153
Fertility characterization
FancmΔ2/Δ2 male or female mice were assessed for fertility defects using the strategy as previously described.154 Briefly, 8 to 12-week old mice were mated with wild-type mice for 6 months or until each pair had dropped 5 litters. Genetic combinations assessed included Fancm+/+ male x FancmΔ2/Δ2 female, FancmΔ2/Δ2 male x Fancm+/+ female, and FancmΔ2/+ male x FancmΔ2/+ female breeding pairs. Litter sizes were recorded and sex ratios were monitored.
Testes were harvested from adult mice and fixed in Bouin’s solution (Amber Scientific) for 5 hours at room temperature and alcohol processed for histopathology into paraffin wax and sectioned using standard methods. Sectioned dewaxed testis sections were stained using periodic acid-Schiff’s reagent and haemotoxylin reagents. All slides were dehydrated and mounted under a coverslip with DPX (Sigma-Aldrich, USA).
Testis daily sperm production (DSP), was determined as previously described.155 Briefly for each animal, one testis was harvested, weighed, snap-frozen and stored at -80°C. After thawing, the testis were homogenized by sonication using a Model 150VT ultrasonic homogenizer in DSP buffer (0.15 M NaCl, 0.01% NaN3, 0.05% Triton X-100) to lyse all cells except the condensed spermatid nuclei. Three 10 second pulses at 30% amplitude were applied with the sonicator.
The number of elongated spermatids per testis were determined using a Neubauer haemocytometer. Samples were first briefly vortexed before 10 μL was loaded into the haemocytometer. The number of sperm per 10 μL was determined by counting the number of sperm heads contained within five 0.20 mm x 0.20 mm x 0.1 mm squares. For each animal two replicates were counted and averaged. The average number of sperm per 20 μl was multiplied by 50,000 to determine the average number of sperm per 1 ml and then multiplied by the volume of homogenate (1 mL + sample weight) to determine the number of sperm per homogenate. The number of sperm per testis was then calculated by first determining the number of sperm per gram of testis ((n/homogenate) / sample weight (g) = n/g [sperm/ g of testis]) and then multiplying this by the whole testis weight. The DSP of the testis was then calculated by dividing the number of sperm (n) per testis by the time divisor 4.84, which corresponds to the estimated number of days that an elongated spermatid remains with the testis.156
These calculations can be expressed as follows: 1. (n/homogenate) / sample weight (g) = n/g [sperm/ g of testis] 2. (n/g) x testis weight (g) = total/testis [sperm/testis] 3. (total/testis) / 4.84 = DSP for 1 testis
Seminiferous tubule analysis was performed on Bouin’s fixed and periodic acid-Schiff’s reagent and haemaotoxylin stained testis sections using FIJI software. Image acquisition was performed with a Leica Thunder microscope and LASX software.
Computer-assisted sperm analysis (CASA)
Analysis of sperm motility was conducted as previously described.157 Briefly, cauda epididymal spermatozoa were collected via backflushing of 10-12 week old mice. Sperm were transferred into modified Tyrode’s 6 medium and loaded into 80 μm deep CASA slides. Movement parameters were measured using the MouseTraxx CASA system (Hamilton-Thorne, USA). Rapid movement was classified as velocity over 35 μm/s, medium motility was classified as 10–35 μm/s, slow motility was classified as 10 μm/s and static was 0 μm/s. At least 1000 sperm were measured per animal with a minimum of four mice per genotype and age.
Direct follicle counts
Ovarian follicles were quantified as previously described.158 Briefly, ovaries were harvested and fixed in either 10% (v/v) neutral buffered formalin solution (#ANBFC, Australian Biostain) or Bouin’s solution (#HT10132, Sigma-Aldrich) for 24 hours. Tissue was embedded in paraffin and was exhaustively serially sectioned at 5 μm with a MicroTec Cut 4060 paraffin microtome and every ninth section was collected and stained with periodic acid-Schiff’s reagent and haematoxylin. Slides were scanned at 20x using an Aperio Digital Pathology Slide Scanner (Leica Biosystems). The Aperio Imagescope program was used to quantify every primordial, primary, secondary, antral and atretic follicle based on morphology159 and for which an oocyte nucleus was clearly visible, to obtain raw counts of oocytes sampled. The total follicle number was determined by multiplying the raw counts by 9 to correct for the sections not counted. Direct follicle count data was analysed using Graphpad Prism 8 (Version 8.0.2). These data are presented as mean ± SEM and p-values for each graph are specified following unpaired t-tests.
Oocyte collection
For superovulation, adult female mice were injected subcutaneously with 5 IU equine chorionic gonadotropin, followed 44 to 48 h later with 5 IU human chorionic gonadotropin. Oocytes were harvested from oviducts 12 to 14 h later.
Histology and immunohistochemistry
Germ cell apoptosis
Germ cell apoptosis was evaluated by immunostaining of cleaved caspase-3 (Cell signalling #9664, 1:100) and -7 (Cell signalling #9491, 1:500). 5 μm testis sections were dewaxed and antigen retrieval was performed using citrate buffer pH 6.0. Sections were blocked for endogenous peroxidase with 3% H2O2 for 5 min and then washed with 1xPBS twice. Sections were then blocked with Cas-Block™ (ThermoFisher #008120) for a minimum of 30 minutes at room temperature. Sections were then incubated in vprimary antibodies in Dako antibody diluent) overnight at 4 C. The next day, sections were washed three times for 5 min. Secondary anti-rabbit Dako Envision Dual Link-HRP (#K4063) was incubated on the sections for 30 min at room temperature, then washed three times for 5 min with 1xPBS. DAB Dako liquid chromagen was applied for 1 min, then removed and washed with water for 5 min. Sections were counterstained with Mayer’s Haemotoxlyin for 4 min and washed briefly with water. Scott’s Tap Water was applied to the sections for 1 min then washed briefly with water. Sections were then dehydrated, cleared and mounted with DPX mounting medium.
Sections were then scanned and the number of cleaved caspase-positive cells in a minimum of 90 randomly selected seminiferous tubules per mouse were counted (n = 3 per genotype).
Statistical analysis of germ cell apoptosis data was then conducted in R version 3.5.1 (R Core Team, 2021). Generalised linear mixed (GLM) models are used to compare the number of caspase-positive cells per tubule across genotypes, with animal ID included as a random effect to account for repeated measures per individual. For each model, Akaike information criteria (AIC) estimates are used to select the most appropriate error distribution and link function (i.e. Poisson, negative binomial, zero-inflated Poisson, zero-inflated negative binomial) using the glmer function (lme4 package;136 ) and glmmTMB function (glmmTMB, package;137). For all models, a zero-inflated negative binomial distribution (fitted with glmmTMB, using the ziformula argument) was selected as the most appropriate error distribution and link function (i.e. had the lowest AIC score).
Germ cells in newborn male mice
To determine if Sting ameliorated loss of FancmΔ2/Δ2 fertility by increasing the number of germ cell numbers, we time mated Fancm+/Δ2 Sting-/- with Fancm+/Δ2 Sting-/- or Fancm+/Δ2 Sting+/+ with Fancm+/Δ2 Sting+/+ and collected testes from male pups at post-natal days 1–3. All Histology processing was performed by the Monash Histology Platform. Briefly, one formalin fixed paraffin embedded testis from each animal was exhaustively sectioned at 5 μm, and every 10th section was collected onto super-frost slides. Deparaffinisation, rehydration and targeted Citrate retrieval at 98 °C was performed in a Dako PT Link. All immunohistochemistry staining steps and washes were then performed in the Dako Autostainer Plus using a routine Immunohistochemistry protocol, with the addition of a MOM IgG block (Jackson ImmunoResearch Cat# 115-007-003, 1:50) to stop non-specific binding, followed by a germ cell-specific primary antibody against DDX4 (mAbcam27591), diluted in DAKO diluent (1:3000) followed by Dako Anti-Mouse HRP (K4001) and 3,3 -diaminobenzidine (DAB) (Dako K3468). Sections were then counterstained with haematoxylin (Dako S3301) and cover slipped. Slides were scanned at 40x objective using the Aperio Digital Pathology Slide Scanner (Leica Biosystems). The images were then visualised by a blinded assessor using Aperio ImageScope software (Leica Biosystems) and all stained germ cells counted in every section.
Germ cells in newborn male mice were counted from every section Slides are single testes were serially sectioned at 5 μm and every 10th section was collected and stained with anti-MVH (ab13840).
Tubules and MVH-positive cells were quantified using FIJI. All sections of all testes were counted except for 14 of 234 experimental slides (6%) which had weak anti-MVH signal. Quantification was performed with the genotypes blinded to the counter.
MMC-induced chromosomal breakage analysis
MMC-induced chromosomal breakage tests were assessed using primary splenic B lymphocytes from Fancm mice using methods from.160,161 After culturing B cells in the presence of LPS for 24 hours, mitomycin C was added at varying concentrations (10, 50 and 100 ng/mL). After 72 hours, cells were given fresh media containing colcemid (Karyomax, 0.2 μg/mL) for two hours. Following colcemid treatment, cells were harvested, washed and resuspended in 1x PBS. Pre-warmed swelling buffer (75 mM KCl) was added by dropwise addition while gently vortexing. Cells were incubated at 37 C for 10 mins and a couple of drops of chilled fixation buffer (1 methanol : 1 glacial acetic acid) was added. Cells were spun down and supernatant removed. Cells were resuspended in chilled fixative buffer with constant agitation three times before dropping spreading onto slides.
Delivery of ibuprofen in drinking water
Ibuprofen treatment was performed following methods described in McNairn et al.10. Fancm+/Δ2 males were mated to Fancm+/Δ2 females. The parents and pups were provided with water bottles containing ibuprofen (children’s Advil), 5 mL (100 mg) in 250 mL. They were allowed to drink ad libitum (50–80 mg per kg per day) at all stages of development. Newborn mice were genotyped for the Fancm allele. Gonads and blood were collected from 8-week old pups for analysis.
Acknowledgments
We are grateful to Chloé Girard, Andrew Lloyd, and Raphaël Mercier for helpful comments on the manuscript. Florencia Pratto, Kevin Brick, and Phil Jordan provided assistance and helpful feedback with the crossover density predictive modeling. The surface spreading technique was generously taught by Rocío Gómez. W.C. and D.M. receive fellowships and funding related to this work from the Australian National Health and Medical Research Council and Victorian Cancer Agency (GNT1129757, GNT1112681, GNT1185387, MCRF21006). R. Lyu and V.T. are recipients of a Research Training Program scholarship from the Australian Government and the University of Melbourne and SVI Foundation Top-Up scholarship from St Vincent’s Institute. R. Lyu receives a Xing Lei PhD Top-Up scholarship in Mathematics and Statistics. V.T. receives a St Vincent’s Institute (SVI) Top-Up scholarship. SVI receives Operational Infrastructure Support from the Victorian State Government. L.M. received a fellowship from the Lorenzo and Pamela Galli Research Trust. The anti-MLH3 antibody was a gift from Corinne Grey, Bernard de Massy, and Valérie Borde. The Sting mouse line was a gift from Benjamin Kile. Casey Anttila, Peter Hickey, and Stephen Wilcox from the WEHI Cellular Genomics Projects and Sequencing Teams assisted with scRNA-seq experiments. The authors would like to acknowledge the technical support of the Monash Animal Research Platform and Monash Histology Platform, Monash University. BioRender was used for illustrations.
Author contributions
Studies were conceived and designed by V.T., R. Lyu, L.G.M., J.M.S., E.R.H., M.K.O., K.H., A.J.D., J.H., D.J. McCarthy, and W.C. Experiments were performed by V.T., S.N., J.M.S., J.E.M.D., D.J. Merriner, E.G., T.S., A.L., L.G.M., and N.Z. Data analyses and plot generation were performed by R. Lyu, V.T., R. Liu, J.M.S., J.E.M.D., T.S., D.J.M., and W.C. R. Lyu implemented the computational workflow and statistical methods for crossover detection, performed bioinformatics analysis, generated plots, and wrote and edited the manuscript. W.C. and D.J.M. supervised the work.
Declaration of interests
The authors declare no competing interests.
Published: June 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2023.100349.
Contributor Information
Davis J. McCarthy, Email: dmccarthy@svi.edu.au.
Wayne Crismani, Email: wcrismani@svi.edu.au.
Supplemental information
Data and code availability
The single-cell DNA sequencing samples and bulk DNA-sequencing datasets are publicly available and can be accessed from ENA (European Nucleotide Archive) with accession number ENA: PRJEB57095. The computational workflows, original codes, and reports of computational analyses of crossovers are publicly available and can be found in the Zenodo frozen repository, Zenodo: https://zenodo.org/record/7939158#.ZGavGexBz0o (key resources table in STAR Methods). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Shimamura A., Alter B.P. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24:101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter B.P., Giri N., Savage S.A., Rosenberg P.S. Cancer in the national cancer institute inherited bone marrow failure syndrome cohort after fifteen years of follow-up. Haematologica. 2018;103:30–39. doi: 10.3324/haematol.2017.178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niraj J., Färkkilä A., D’Andrea A.D. The fanconi anemia pathway in cancer. Annu. Rev. Cell Biol. 2019;3:457–478. doi: 10.1146/annurev-cancerbio-030617-050422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basbous J., Constantinou A. A tumor suppressive DNA translocase named FANCM. Crit. Rev. Biochem. Mol. Biol. 2019;54:27–40. doi: 10.1080/10409238.2019.1568963. [DOI] [PubMed] [Google Scholar]
- 5.Bakker S.T., van de Vrugt H.J., Rooimans M.A., Oostra A.B., Steltenpool J., Delzenne-Goette E., van der Wal A., van der Valk M., Joenje H., te Riele H., de Winter J.P. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum. Mol. Genet. 2009;18:3484–3495. doi: 10.1093/hmg/ddp297. [DOI] [PubMed] [Google Scholar]
- 6.Luo Y., Hartford S.A., Zeng R., Southard T.L., Shima N., Schimenti J.C. Hypersensitivity of primordial germ cells to compromised replication-associated DNA repair involves ATM-p53-p21 signaling. PLoS Genet. 2014;10:e1004471. doi: 10.1371/journal.pgen.1004471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouquet B., Pawlikowska P., Caburet S., Guigon C., Mäkinen M., Tanner L., Hietala M., Urbanska K., Bellutti L., Legois B., et al. A homozygous FANCM mutation underlies a familial case of non-syndromic primary ovarian insufficiency. Elife. 2017;6:e30490. doi: 10.7554/eLife.30490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin H., Ma H., Hussain S., Zhang H., Xie X., Jiang L., Jiang X., Iqbal F., Bukhari I., Jiang H., et al. A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet. Med. 2019;21:62–70. doi: 10.1038/s41436-018-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasak L., Punab M., Nagirnaja L., Grigorova M., Minajeva A., Lopes A.M., Punab A.M., Aston K.I., Carvalho F., Laasik E., Smith L.B., GEMINI Consortium. Conrad D.F., Laan M. Biallelic recessive loss-of-function variants in FANCM cause non-obstructive azoospermia. Am. J. Hum. Genet. 2018;103:200–212. doi: 10.1016/j.ajhg.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNairn A.J., Chuang C.H., Bloom J.C., Wallace M.D., Schimenti J.C. Female-biased embryonic death from inflammation induced by genomic instability. Nature. 2019;567:105–108. doi: 10.1038/s41586-019-0936-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruth K.S., Day F.R., Hussain J., Martínez-Marchal A., Aiken C.E., Azad A., Thompson D.J., Knoblochova L., Abe H., Tarry-Adkins J.L., et al. Genetic insights into biological mechanisms governing human ovarian ageing. Nature. 2021;596:393–397. doi: 10.1038/s41586-021-03779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Li P., Liu N., Jing T., Ji Z., Yang C., Zhao L., Tian R., Chen H., Huang Y., et al. Novel bi-allelic variants of fancm cause sertoli cell-only syndrome and non-obstructive azoospermia. Front. Genet. 2021;12:799886. doi: 10.3389/fgene.2021.799886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Encarnación J., Cerezuela P., Español I., García M., Manso C., De la Fuente I., Garrigós N., Viney A., Minguillon J., Surrallés J. Fanconi-like anemia related to a FANCM mutation. Eur. J. Med. Genet. 2022;65:104399. doi: 10.1016/j.ejmg.2021.104399. [DOI] [PubMed] [Google Scholar]
- 14.Nadler J.J., Braun R.E. Fanconi anemia complementation group C is required for proliferation of murine primordial germ cells. Genesis. 2000;27:117–123. doi: 10.1002/1526-968X(200007)27:3<117::AID-GENE40>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 15.Wong J.C.Y., Alon N., Mckerlie C., Huang J.R., Meyn M.S., Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum. Mol. Genet. 2003;12:2063–2076. doi: 10.1093/hmg/ddg219. [DOI] [PubMed] [Google Scholar]
- 16.Kato Y., Alavattam K.G., Sin H.S., Meetei A.R., Pang Q., Andreassen P.R., Namekawa S.H. FANCB is essential in the male germline and regulates H3K9 methylation on the sex chromosomes during meiosis. Hum. Mol. Genet. 2015;24:5234–5249. doi: 10.1093/hmg/ddv244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter N. Meiotic recombination : the essence of heredity. Cold Spring Harbor Perspect. Biol. 2015;7 doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warren A.C., Chakravarti A., Wong C., Slaugenhaupt S.A., Halloran S.L., Watkins P.C., Metaxotou C., Antonarakis S.E. Evidence for reduced recombination on the nondisjoined chromosomes 21 in down syndrome. Science. 1987;237:652–654. doi: 10.1126/science.2955519. [DOI] [PubMed] [Google Scholar]
- 19.Sherman S.L., Takaesu N., Freeman S.B., Grantham M., Phillips C., Blackston R.D., Jacobs P.A., Cockwell A.E., Freeman V., Uchida I., et al. Trisomy 21: association between reduced recombination and nondisjunction. Am. J. Hum. Genet. 1991;49:608–620. [PMC free article] [PubMed] [Google Scholar]
- 20.Sturtevant A.H. The linear arrangement of six sex linked factors in Drosophila, as shown by their mode of association. J. Exp. Zool. 1913;14:43–59. doi: 10.1002/jez.1400140104. [DOI] [Google Scholar]
- 21.Broman K.W., Weber J.L. Characterization of human crossover interference. Am. J. Hum. Genet. 2000;66:1911–1926. doi: 10.1086/302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martini E., Diaz R.L., Hunter N., Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole F., Kauppi L., Lange J., Roig I., Wang R., Keeney S., Jasin M. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 2012;14:424–430. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers S., Bowden R., Tumian A., Bontrop R.E., Freeman C., MacFie T.S., McVean G., Donnelly P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parvanov E.D., Petkov P.M., Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science (New York, N.Y.) 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., Przeworski M., Coop G., De Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McVean G.A.T., Myers S.R., Hunt S., Deloukas P., Bentley D.R., Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 28.Myers S., Bottolo L., Freeman C., McVean G., Donnelly P. Genetics: a fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 29.Pratto F., Brick K., Khil P., Smagulova F., Petukhova G.V., Camerini-Otero R.D. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pratto F., Brick K., Cheng G., Lam K.-W.G., Cloutier J.M., Dahiya D., Wellard S.R., Jordan P.W., Camerini-Otero R.D. Meiotic recombination mirrors patterns of germline replication in mice and humans. Cell. 2021;184:4251–4267.e20. doi: 10.1016/j.cell.2021.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keeney S., Giroux C.N., Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/S0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 32.Romanienko P.J., Camerini-Otero R.D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/S1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 33.Grelon M., Vezon D., Gendrot G., Pelletier G. Atspo11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-M. [DOI] [PubMed] [Google Scholar]
- 35.Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-I. [DOI] [PubMed] [Google Scholar]
- 36.Sun H., Treco D., Szostak J.W. Extensive 3’-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 37.Bishop D.K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-J. [DOI] [PubMed] [Google Scholar]
- 38.Shinohara A., Ogawa H., Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-K. [DOI] [PubMed] [Google Scholar]
- 39.Cloud V., Chan Y.-L., Grubb J., Budke B., Bishop D.K. Rad51 is an accessory factor for dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–1225. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lao J.P., Oh S.D., Shinohara M., Shinohara A., Hunter N. Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pyatnitskaya A., Borde V., De Muyt A. Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma. 2019;128:181–198. doi: 10.1007/s00412-019-00714-8. [DOI] [PubMed] [Google Scholar]
- 42.Peterson S.E., Keeney S., Jasin M. Mechanistic insight into crossing over during mouse meiosis. Mol. Cell. 2020;78:1252–1263.e3. doi: 10.1016/j.molcel.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Börner G.V., Kleckner N., Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 44.Lynn A., Soucek R., Börner G.V. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 45.Holloway J.K., Booth J., Edelmann W., McGowan C.H., Cohen P.E. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008;4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mercier R., Mézard C., Jenczewski E., Macaisne N., Grelon M. The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 2015;66:297–327. doi: 10.1146/annurev-arplant-050213-035923. [DOI] [PubMed] [Google Scholar]
- 47.Lynn A., Koehler K.E., Judis L., Chan E.R., Cherry J.P., Schwartz S., Seftel A., Hunt P.A., Hassold T.J. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science. 2002;296:2222–2225. doi: 10.1126/science.1071220. [DOI] [PubMed] [Google Scholar]
- 48.Tease C., Hultén M.A. Inter-sex variation in synaptonemal complex lengths largely determine the different recombination rates in male and female germ cells. Cytogenet. Genome Res. 2004;107:208–215. doi: 10.1159/000080599. [DOI] [PubMed] [Google Scholar]
- 49.Giraut L., Falque M., Drouaud J., Pereira L., Martin O.C., Mézard C. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gruhn J.R., Rubio C., Broman K.W., Hunt P.A., Hassold T. Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS One. 2013;8:e85075. doi: 10.1371/journal.pone.0085075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith G.R., Boddy M.N., Shanahan P., Russell P. Fission yeast mus81· eme1 holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osman F., Dixon J., Doe C.L., Whitby M.C. Generating crossovers by resolution of nicked holliday junctions: a role for mus81-eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/S1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 53.de los Santos T., Hunter N., Lee C., Larkin B., Loidl J., Hollingsworth N.M. The mus81/mms4 endonuclease acts independently of double-holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guillon H., Baudat F., Grey C., Liskay R.M., De Massy B. Crossover and noncrossover pathways in mouse meiosis. Mol. Cell. 2005;20:563–573. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 55.De Boer E., Stam P., Dietrich A.J.J., Pastink A., Heyting C. Two levels of interference in mouse meiotic recombination. Proc. Natl. Acad. Sci. USA. 2006;103:9607–9612. doi: 10.1073/pnas.0600418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berchowitz L.E., Francis K.E., Bey A.L., Copenhaver G.P. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3:e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Svetlanov A., Baudat F., Cohen P.E., De Massy B. Distinct functions of MLH3 at recombination hot spots in the mouse. Genetics. 2008;178:1937–1945. doi: 10.1534/genetics.107.084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwacha A., Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 59.Collins I., Newlon C.S. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell. 1994;76:65–75. doi: 10.1016/0092-8674(94)90173-2. [DOI] [PubMed] [Google Scholar]
- 60.Hunter N., Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 61.Oh S.D., Lao J.P., Hwang P.Y.-H., Taylor A.F., Smith G.R., Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zakharyevich K., Tang S., Ma Y., Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandhu R., Monge Neria F., Monge Neria J., Chen X., Hollingsworth N.M., Börner G.V. DNA helicase Mph1FANCM ensures meiotic recombination between parental chromosomes by dissociating precocious displacement loops. Dev. Cell. 2020;53:458–472.e5. doi: 10.1016/j.devcel.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun F., Handel M.A. Regulation of the meiotic prophase I to metaphase I transition in mouse spermatocytes. Chromosoma. 2008;117:471–485. doi: 10.1007/s00412-008-0167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prakash R., Satory D., Dray E., Papusha A., Scheller J., Kramer W., Krejci L., Klein H., Haber J.E., Sung P., Ira G. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gari K., Décaillet C., Delannoy M., Wu L., Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. USA. 2008;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crismani W., Girard C., Froger N., Pradillo M., Santos J.L., Chelysheva L., Copenhaver G.P., Horlow C., Mercier R. FANCM limits meiotic crossovers. Science. 2012;336:1588–1590. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- 68.Knoll A., Higgins J.D., Seeliger K., Reha S.J., Dangel N.J., Bauknecht M., Schröpfer S., Franklin F.C.H., Puchta H. The fanconi anemia ortholog FANCM ensures ordered homologous recombination in both somatic and meiotic cells in <i>Arabidopsis</i>. Plant Cell. 2012;24:1448–1464. doi: 10.1105/tpc.112.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lorenz A., Osman F., Sun W., Nandi S., Steinacher R., Whitby M.C. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science. 2012;336:1585–1588. doi: 10.1126/science.1220111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuo H.K., McMahan S., Rota C.M., Kohl K.P., Sekelsky J. Drosophila FANCM helicase prevents spontaneous mitotic crossovers generated by the MUS81 and SLX1 nucleases. Genetics. 2014;198:935–945. doi: 10.1534/genetics.114.168096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Girard C., Crismani W., Froger N., Mazel J., Lemhemdi A., Horlow C., Mercier R. FANCM-associated proteins MHF1 and MHF2, but not the other Fanconi anemia factors, limit meiotic crossovers. Nucleic Acids Res. 2014;42:9087–9095. doi: 10.1093/nar/gku614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziolkowski P.A., Berchowitz L.E., Lambing C., Yelina N.E., Zhao X., Kelly K.A., Choi K., Ziolkowska L., June V., Sanchez-Moran E., et al. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. Elife. 2015;4:1–29. doi: 10.7554/eLife.03708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Séguéla-Arnaud M., Crismani W., Larchevêque C., Mazel J., Froger N., Choinard S., Lemhemdi A., Macaisne N., Van Leene J., Gevaert K., et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc. Natl. Acad. Sci. USA. 2015;112:4713–4718. doi: 10.1073/pnas.1423107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mieulet D., Aubert G., Bres C., Klein A., Droc G., Vieille E., Rond-Coissieux C., Sanchez M., Dalmais M., Mauxion J.-P., et al. Unleashing meiotic crossovers in crops. Native Plants. 2018;4:1010–1016. doi: 10.1038/s41477-018-0311-x. [DOI] [PubMed] [Google Scholar]
- 75.Xu X., Aprelikova O., Moens P., Deng C.X., Furth P.A. Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development. 2003;130:2001–2012. doi: 10.1242/dev.00410. [DOI] [PubMed] [Google Scholar]
- 76.Sharan S.K., Pyle A., Coppola V., Babus J., Swaminathan S., Benedict J., Swing D., Martin B.K., Tessarollo L., Evans J.P., et al. BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development. 2004;131:131–142. doi: 10.1242/dev.00888. [DOI] [PubMed] [Google Scholar]
- 77.Kuznetsov S., Pellegrini M., Shuda K., Fernandez-Capetillo O., Liu Y., Martin B.K., Burkett S., Southon E., Pati D., Tessarollo L., et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J. Cell Biol. 2007;176:581–592. doi: 10.1083/jcb.200608130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crossan G.P., Van Der Weyden L., Rosado I.V., Langevin F., Gaillard P.H.L., McIntyre R.E., Sanger Mouse Genetics Project. Gallagher F., Kettunen M.I., Lewis D.Y., Brindle K., et al. Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 2011;43:147–152. doi: 10.1038/ng.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simhadri S., Peterson S., Patel D.S., Huo Y., Cai H., Bowman-Colin C., Miller S., Ludwig T., Ganesan S., Bhaumik M., et al. Male fertility defect associated with disrupted BRCA1-PALB2 interaction in mice. J. Biol. Chem. 2014;289:24617–24629. doi: 10.1074/jbc.M114.566141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alavattam K.G., Kato Y., Sin H.S., Maezawa S., Kowalski I.J., Zhang F., Pang Q., Andreassen P.R., Namekawa S.H. Elucidation of the fanconi anemia protein network in meoisis and its function in the regulation of histone modifications. Cell Rep. 2016;17:1141–1157. doi: 10.1016/j.celrep.2016.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun X., Brieño-Enríquez M.A., Cornelius A., Modzelewski A.J., Maley T.T., Campbell-Peterson K.M., Holloway J.K., Cohen P.E. FancJ (Brip1) loss-of-function allele results in spermatogonial cell depletion during embryogenesis and altered processing of crossover sites during meiotic prophase I in mice. Chromosoma. 2016;125:237–252. doi: 10.1007/s00412-015-0549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brégnard C., Guerra J., Déjardin S., Passalacqua F., Benkirane M., Laguette N. Upregulated LINE-1 activity in the fanconi anemia cancer susceptibility syndrome leads to spontaneous pro-inflammatory cytokine production. EBioMedicine. 2016;8:184–194. doi: 10.1016/j.ebiom.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mackenzie K.J., Carroll P., Martin C.-A., Murina O., Fluteau A., Simpson D.J., Olova N., Sutcliffe H., Rainger J.K., Leitch A., et al. Cgas surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Heijink A.M., Talens F., Jae L.T., van Gijn S.E., Fehrmann R.S.N., Brummelkamp T.R., van Vugt M.A.T.M. Brca2 deficiency instigates cgas-mediated inflammatory signaling and confers sensitivity to tumor necrosis factor-alpha-mediated cytotoxicity. Nat. Commun. 2019;10:100. doi: 10.1038/s41467-018-07927-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Decout A., Katz J.D., Venkatraman S., Ablasser A. The cgas–sting pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 2021;21:548–569. doi: 10.1038/s41577-021-00524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lyu R., Tsui V., Crismani W., Liu R., Shim H., McCarthy D.J. Sgcocaller and comapr: personalised haplotype assembly and comparative crossover map analysis using single-gamete sequencing data. Nucleic Acids Res. 2022;50:e118. doi: 10.1093/nar/gkac764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Novakovic S., Tsui V., Semple T., Martelotto L., McCarthy D.J., Crismani W. Ssnip-seq: a simple and rapid method for isolation of single-sperm nucleic acid for high-throughput sequencing. PLoS One. 2022;17 doi: 10.1371/journal.pone.0275168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou Y., Shen B., Jiang J., Padhi A., Park K.-E., Oswalt A., Sattler C.G., Telugu B.P., Chen H., Cole J.B., et al. Construction of prdm9 allele-specific recombination maps in cattle using large-scale pedigree analysis and genome-wide single sperm genomics. DNA Res. 2018;25:183–194. doi: 10.1093/dnares/dsx048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deans A.J., West S.C. FANCM connects the genome instability disorders bloom’s syndrome and fanconi anemia. Mol. Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 90.Yin Y., Jiang Y., Lam K.W.G., Berletch J.B., Disteche C.M., Noble W.S., Steemers F.J., Camerini-Otero R.D., Adey A.C., Shendure J. High-throughput single-cell sequencing with linear amplification. Mol. Cell. 2019;76:676–690.e10. doi: 10.1016/j.molcel.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ventelä S., Toppari J., Parvinen M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Mol. Biol. Cell. 2003;14:2768–2780. doi: 10.1091/mbc.e02-10-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amezquita R.A., Lun A.T.L., Becht E., Carey V.J., Carpp L.N., Geistlinger L., Marini F., Rue-Albrecht K., Risso D., Soneson C., et al. Orchestrating single-cell analysis with bioconductor. Nat. Methods. 2020;17:137–145. doi: 10.1038/s41592-019-0654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bhutani K., Stansifer K., Ticau S., Bojic L., Villani A.-C., Slisz J., Cremers C.M., Roy C., Donovan J., Fiske B., Friedman R.C. Widespread haploid-biased gene expression enables sperm-level natural selection. Science. 2021;371 doi: 10.1126/science.abb1723. [DOI] [PubMed] [Google Scholar]
- 94.Baker S.M., Plug A.W., Prolla T.A., Bronner C.E., Harris A.C., Yao X., Christie D.-M., Monell C., Arnheim N., Bradley A., et al. Involvement of mouse mlh1 in dna mismatch repair and meiotic crossing over. Nat. Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 95.Anderson L.K., Reeves A., Webb L.M., Ashley T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics. 1999;151:1569–1579. doi: 10.1093/genetics/151.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lipkin S.M., Moens P.B., Wang V., Lenzi M., Shanmugarajah D., Gilgeous A., Thomas J., Cheng J., Touchman J.W., Green E.D., et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 2002;31:385–390. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 97.Marcon E., Moens P. Mlh1p and mlh3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 2003;165:2283–2287. doi: 10.1093/genetics/165.4.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kan R., Sun X., Kolas N.K., Avdievich E., Kneitz B., Edelmann W., Cohen P.E. Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biol. Reprod. 2008;78:462–471. doi: 10.1095/biolreprod.107.065771. [DOI] [PubMed] [Google Scholar]
- 99.Kneitz B., Cohen P.E., Avdievich E., Zhu L., Kane M.F., Hou H., Kolodner R.D., Kucherlapati R., Pollard J.W., Edelmann W. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14:1085–1097. doi: 10.1101/gad.14.9.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reynolds A., Qiao H., Yang Y., Chen J.K., Jackson N., Biswas K., Holloway J.K., Baudat F., De Massy B., Wang J., et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 2013;45:269–278. doi: 10.1038/ng.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Milano C.R., Holloway J.K., Zhang Y., Jin B., Smith C., Bergman A., Edelmann W., Cohen P.E. Mutation of the ATPase domain of mutS homolog-5 (MSH5) reveals a requirement for a functional MutSG complex for all crossovers in mammalian meiosis. G3. 2019;9:1839–1850. doi: 10.1534/g3.119.400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tsui V., Crismani W. The fanconi anemia pathway and fertility. Trends Genet. 2019;35:199–214. doi: 10.1016/j.tig.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 103.Hess R.A., de Franca L.R. In: Cheng C.Y., editor. Springer New York; 2008. Spermatogenesis and cycle of the seminiferous epithelium; pp. 1–15. (Molecular Mechanisms in Spermatogenesis). [Google Scholar]
- 104.Brinkworth M.H., Weinbauer G.F., Schlatt S., Nieschlag E. Identification of male germ cells undergoing apoptosis in adult rats. Reproduction. 1995;105:25–33. doi: 10.1530/jrf.0.1050025. [DOI] [PubMed] [Google Scholar]
- 105.Wu J., Sun L., Chen X., Du F., Shi H., Chen C., Chen Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Diner E.J., Burdette D.L., Wilson S.C., Monroe K.M., Kellenberger C.A., Hyodo M., Hayakawa Y., Hammond M.C., Vance R.E. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bakker S.T., Van De Vrugt H.J., Visser J.A., Delzenne-Goette E., Van Der Wal A., Berns M.A.D., Van De Ven M., Oostra A.B., De Vries S., Kramer P., et al. Fancf-deficient mice are prone to develop ovarian tumours. J. Pathol. 2012;226:28–39. doi: 10.1002/path.2992. [DOI] [PubMed] [Google Scholar]
- 109.Pittman D.L., Cobb J., Schimenti K.J., Wilson L.A., Cooper D.M., Brignull E., Handel M.A., Schimenti J.C. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for dmc1, a germline-specific reca homolog. Mol. Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 110.Edelmann W., Cohen P.E., Kneitz B., Winand N., Lia M., Heyer J., Kolodner R., Pollard J.W., Kucherlapati R. Mammalian muts homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 1999;21:123–127. doi: 10.1038/5075. [DOI] [PubMed] [Google Scholar]
- 111.Yang Y., Xu W., Gao F., Wen C., Zhao S., Yu Y., Jiao W., Mi X., Qin Y., Chen Z.-J., Zhao S. Transcription–replication conflicts in primordial germ cells necessitate the fanconi anemia pathway to safeguard genome stability. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.220320811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bloom J., Schimenti J. Genes and Development; 2020. Sexually Dimorphic DNA Damage Responses and Mutation Avoidance in the Mouse Germline; pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hinch A.G., Zhang G., Becker P.W., Moralli D., Hinch R., Davies B., Bowden R., Donnelly P. Factors influencing meiotic recombination revealed by whole-genome sequencing of single sperm. Science. 2019;363 doi: 10.1126/science.aau8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yin Y., Jiang Y., Lam K.-W.G., Berletch J.B., Disteche C.M., Noble W.S., Steemers F.J., Camerini-Otero R.D., Adey A.C., Shendure J. High-throughput single-cell sequencing with linear amplification. Mol. Cell. 2019;76:676–690.e10. doi: 10.1016/j.molcel.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bell A.D., Mello C.J., Nemesh J., Brumbaugh S.A., Wysoker A., McCarroll S.A. Insights into variation in meiosis from 31,228 human sperm genomes. Nature. 2020;583 doi: 10.1038/s41586-020-2347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carioscia S.A., Weaver K.J., Bortvin A.N., Ariad D., Bell A.D., McCoy R.C. Strict adherence to mendel’s first law across a large sample of human sperm genomes. bioRxiv. 2021 doi: 10.1101/2021.11.19.469261. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Crismani W., Mercier R. What limits meiotic crossovers? Cell Cycle. 2012;11:3527–3528. doi: 10.4161/cc.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Blary A., Gonzalo A., Eber F., Bérard A., Bergès H., Bessoltane N., Charif D., Charpentier C., Cromer L., Fourment J., et al. FANCM limits meiotic crossovers in Brassica crops. Front. Plant Sci. 2018;9:368. doi: 10.3389/fpls.2018.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Singh D.K., Gamboa R.S., Singh A.K., Walkemeier B., Van Leene J., De Jaeger G., Siddiqi I., Guerois R., Crismani W., Mercier R. The FANCC–FANCE–FANCF complex is evolutionarily conserved and regulates meiotic recombination. Nucleic Acids Research. 2023;51:2516–2528. doi: 10.1093/nar/gkac1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barton N.H., Charlesworth B. Why sex and recombination? Science. 1998;281:1986–1990. doi: 10.1126/science.281.5385.1986. [DOI] [PubMed] [Google Scholar]
- 121.Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G., et al. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 122.Paigen K., Szatkiewicz J.P., Sawyer K., Leahy N., Parvanov E.D., Ng S.H.S., Graber J.H., Broman K.W., Petkov P.M. The recombinational anatomy of a mouse chromosome. PLoS Genet. 2008;4:e1000119. doi: 10.1371/journal.pgen.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Girard C., Chelysheva L., Choinard S., Froger N., Macaisne N., Lemhemdi A., Mazel J., Crismani W., Mercier R. AAA-ATPase FIDGETIN-LIKE 1 and helicase FANCM antagonize meiotic crossovers by distinct mechanisms. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Holloway J.K., Morelli M.A., Borst P.L., Cohen P.E. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J. Cell Biol. 2010;188:779–789. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Youds J.L., Mets D.G., McIlwraith M.J., Martin J.S., Ward J.D., ONeil N.J., Rose A.M., West S.C., Meyer B.J., Boulton S.J. RTEL-1 enforces meiotic crossover interference and homeostasis. Science (New York, N.Y.) 2010;327:1254–1258. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kohl K.P., Jones C.D., Sekelsky J. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science. 2012;338:1363–1365. doi: 10.1126/science.1228190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gangloff S., McDonald J.P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391-8398.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Watt P.M., Louis E.J., Borts R.H., Hickson I.D. Sgs1: a eukaryotic homolog of E. coil RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 129.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 130.Ballew B.J., Yeager M., Jacobs K., Giri N., Boland J., Burdett L., Alter B.P., Savage S.A. Germline mutations of regulator of telomere elongation helicase 1, RTEL1, in Dyskeratosis congenita. Hum. Genet. 2013;132:473–480. doi: 10.1007/s00439-013-1265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sandor C., Li W., Coppieters W., Druet T., Charlier C., Georges M. Genetic variants in REC8, RNF212, and PRDM9 influence male recombination in cattle. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ziolkowski P.A., Underwood C.J., Lambing C., Martinez-Garcia M., Lawrence E.J., Ziolkowska L., Griffin C., Choi K., Franklin F.C.H., Martienssen R.A., Henderson I.R. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 2017;31:306–317. doi: 10.1101/gad.295501.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cheng N.C., van de Vrugt H.J., van der Valk M.A., Oostra A.B., Krimpenfort P., de Vries Y., Joenje H., Berns A., Arwert F. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum. Mol. Genet. 2000;9:1805–1811. doi: 10.1093/hmg/9.12.1805. [DOI] [PubMed] [Google Scholar]
- 134.Punjani N., Lamb D.J. Canary in the coal mine? Male infertility as a marker of overall health. Annu. Rev. Genet. 2020;54:465–486. doi: 10.1146/annurev-genet-022020-023434. [DOI] [PubMed] [Google Scholar]
- 135.Smit A., Hubley R., Green P. 2013-2015. Repeatmasker open-4.0. [Google Scholar]
- 136.Bates D.W., Mächler M., Bolker B., Walker S. Fitting linear mixed-effects models using lme4. BMJ Qual. Saf. 2015;24:1–3. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 137.Brooks M., Kristensen K., Benthem K., Magnusson A., Berg C., Nielsen A., Skaug H., Mächler M., Bolker B. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. The R Journal. 2017;9:378–400. doi: 10.32614/RJ-2017-066. [DOI] [Google Scholar]
- 138.Hahne F., Ivanek R. Springer; 2016. Visualizing Genomic Data Using Gviz and Bioconductor. [DOI] [PubMed] [Google Scholar]
- 139.Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 140.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Boekhout M., Karasu M.E., Wang J., Acquaviva L., Pratto F., Brick K., Eng D.Y., Xu J., Camerini-Otero R.D., Patel D.J., Keeney S. REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol. Cell. 2019;74:1053–1068.e8. doi: 10.1016/j.molcel.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jin L., Hill K.K., Filak H., Mogan J., Knowles H., Zhang B., Perraud A.-L., Cambier J.C., Lenz L.L. Mpys is required for ifn response factor 3 activation and type i ifn production in the response of cultured phagocytes to bacterial second messengers cyclic-di-amp and cyclic-di-gmp. J. Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.White M.J., McArthur K., Metcalf D., Lane R.M., Cambier J.C., Herold M.J., Van Delft M.F., Bedoui S., Lessene G., Ritchie M.E., et al. Apoptotic caspases suppress mtdna-induced sting-mediated type i ifn production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kosambi D.,D. The estimation of map distances from recombination values. The Annals of Eugenics. 1944;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- 150.Peters A.H., Plug A.W., van Vugt M.J., de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1016/0039-9140(75)80147-0. [DOI] [PubMed] [Google Scholar]
- 151.McNeill L. 2021. Synapsis: An R Package to Automate the Analysis of Double-Strand Break Repair during Meiosis. R Package Version 1.0.0. [DOI] [Google Scholar]
- 152.Yun Y., Ito M., Sandhu S., Hunter N. Homologous Recombination. Springer; 2021. Cytological monitoring of meiotic crossovers in spermatocytes and oocytes; pp. 267–286. [DOI] [PubMed] [Google Scholar]
- 153.Balmus G., Karp N.A., Ng B.L., Jackson S.P., Adams D.J., McIntyre R.E. A high-throughput in vivo micronucleus assay for genome instability screening in mice. Nat. Protoc. 2015;10:205–215. doi: 10.1038/nprot.2015-010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Houston B.J., Conrad D.F., O’Bryan M.K. A framework for high-resolution phenotyping of candidate male infertility mutants: from human to mouse. Hum. Genet. 2021;140:155–182. doi: 10.1007/s00439-020-02159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dunleavy J.E.M., O’Connor A.E., Okuda H., Merriner D.J., O’Bryan M.K. Katnb1 is a master regulator of multiple katanin enzymes in male meiosis and haploid germ cell development. Development. 2021;148:dev199922. doi: 10.1242/dev.199922. [DOI] [PubMed] [Google Scholar]
- 156.Russell L.D., Ettlin R.A., Hikim A.P.S., Clegg E.D. Histological and histopathological evaluation of the testis. Int. J. Androl. 1993;16:83. doi: 10.1111/j.1365-2605.1993.tb01156.x. [DOI] [Google Scholar]
- 157.Houston B.J., Nagirnaja L., Merriner D.J., O’Connor A.E., Okuda H., Omurtag K., Smith C., Aston K.I., Conrad D.F., O’Bryan M.K. The sertoli cell expressed gene secernin-1 (scrn1) is dispensable for male fertility in the mouse. Dev. Dyn. 2021;250:922–931. doi: 10.1002/dvdy.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sarma U.C., Winship A.L., Hutt K.J. Comparison of methods for quantifying primordial follicles in the mouse ovary. J. Ovarian Res. 2020;13:121. doi: 10.1186/s13048-020-00724-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Myers M., Britt K.L., Wreford N.G.M., Ebling F.J.P., Kerr J.B. Methods for quantifying follicular numbers within the mouse ovary. Reproduction. 2004;127:569–580. doi: 10.1530/rep.1.00095. [DOI] [PubMed] [Google Scholar]
- 160.Auerbach A.D. Diagnosis of fanconi anemia by diepoxybutane analysis. Curr. Protoc. Hum. Genet. 2015;85:8.7.1–8.7.17. doi: 10.1002/0471142905.hg0807s85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wong D.M., Li L., Jurado S., King A., Bamford R., Wall M., Walia M.K., Kelly G.L., Walkley C.R., Tarlinton D.M., et al. The transcription factor asciz and its target dynll1 are essential for the development and expansion of myc-driven b cell lymphoma. Cell Rep. 2016;14:1488–1499. doi: 10.1016/j.celrep.2016.01.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single-cell DNA sequencing samples and bulk DNA-sequencing datasets are publicly available and can be accessed from ENA (European Nucleotide Archive) with accession number ENA: PRJEB57095. The computational workflows, original codes, and reports of computational analyses of crossovers are publicly available and can be found in the Zenodo frozen repository, Zenodo: https://zenodo.org/record/7939158#.ZGavGexBz0o (key resources table in STAR Methods). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.