Abstract
Serum response factor (SRF) plays a central role in the transcriptional response of mammalian cells to a variety of extracellular signals. It is a key regulator of many cellular early response genes which are believed to be involved in cell growth and differentiation. The mechanism by which SRF activates transcription in response to mitogenic agents has been extensively studied; however, significantly less is known about regulation of the SRF gene itself. Previously, we identified distinct regulatory elements in the SRF promoter that play a role in activation, including a consensus ETS domain binding site, a consensus overlapping Sp/Egr-1 binding site, and two SRF binding sites. We further showed that serum induces SRF by a mechanism that requires an intact SRF binding site, also termed a CArG box. In the present study we demonstrate that in response to stimulation of cells by a purified growth factor, basic fibroblast growth factor (bFGF), the SRF promoter is upregulated by a complex pathway that involves at least two independent mechanisms: a CArG box-independent mechanism that is mediated by an ETS binding site, and a novel CArG box-dependent mechanism that requires both an Sp factor binding site and the CArG motifs for maximal stimulation. Our analysis indicates that the CArG/Sp element activation mechanism is mediated by distinct signaling pathways. The CArG box-dependent component is targeted by a Rho-mediated pathway, and the Sp binding site-dependent component is targeted by a Ras-mediated pathway. Both SRF and bFGF have been implicated in playing an important role in mediating cardiogenesis during development. The implications of our findings for SRF expression during development are discussed.
Serum response factor (SRF) is a member of the MADS (MCM1, Agamous and Deficiens, and SRF) box family of transcription factors that is an important regulator of many genes associated with cell growth and differentiation. SRF was first identified based on its ability to mediate serum and growth factor activation of the c-fos proto-oncogene (reviewed in reference 58). Subsequently, it was found that SRF and/or SRF binding sites (CC[A/T]6GG), termed CArG boxes, regulate expression of a wide variety of serum-responsive genes (8, 9, 15, 34, 35, 55, 59). In addition to mediating activation of transcription by serum growth factors, SRF also regulates transcription mediated by treatment of cells with neurotrophins (51, 61), neurotransmitters and agents that raise intracellular calcium levels (3, 39, 41), stress agents, and viral activators (2, 23, 24). SRF has also been implicated in playing a regulatory role in cell cycle progression and myogenic differentiation (25, 60) and in development (1). The diversity of stimuli that activate SRF-dependent expression indicates that SRF is likely a common nuclear target of multiple, distinct signaling pathways.
The mechanism by which SRF mediates transactivation has been extensively studied in murine fibroblasts treated with serum growth factors that activate members of the mitogen-activated protein kinase (MAPK) family of inducible Ser/Thr kinases (for a review, see reference 56). In this case, a complex of SRF and a member of the Elk-1 subfamily of ETS oncoproteins, also referred to as ternary complex factors (TCFs), is targeted by activated MAPK family members. Phosphorylation of TCF on conserved serine residues is responsible for transactivation.
SRF can also activate gene expression in a non-TCF-dependent manner (32), although this mechanism is much less well understood. In serum-stimulated murine fibroblasts, non-TCF-dependent activation can occur in a Ras-independent manner and can be mediated by members of the Rho-dependent family of low-molecular-weight G proteins (22, 29, 30). In neuron-like PC12 cells and primary hippocampal neurons, stimuli that elevate intracellular calcium concentrations can also activate SRF-dependent gene expression in a non-TCF-dependent manner (3, 41). This activation can also occur in a Ras-independent manner (40).
In addition to the TCF family of SRF-associated factors, SRF has been shown to functionally interact with a variety of other factors, including YY1 (44), ATF6 (63), and homeodomain-containing proteins such as Phox-1 (28) and tinman (Csx/Nkx-2.5) (13). It has been hypothesized that tissue-specific CArG box-dependent gene expression may be mediated by SRF interaction with cell-type-specific transcription factors. The findings that the cardiac tissue-specific homeobox factor tinman (11–13), which plays an essential role in establishing myogenic lineages (6), and that myogenic basic-loop-helix transcription factors (27) directly interact with SRF to regulate cardiac and myogenic gene expression support this hypothesis and point to a critical role for SRF in myogenesis.
While the mechanism by which SRF mediates gene expression has been the object of significant attention, regulation of expression of the SRF gene itself is far less well characterized. Previously, we (42) and others (45) have demonstrated that in response to serum stimulation, the SRF gene is rapidly induced at the transcriptional level in a protein synthesis-independent manner. In murine fibroblasts, SRF gene expression is transient. SRF message levels peak at approximately 90 to 120 min after serum stimulation and return to nearly basal levels by 4 to 6 h after stimulation (42). In recent studies, we have shown that maximal serum responsiveness of the SRF promoter requires two SRF binding sites located within the first 63 nucleotides (nt) upstream of the start site of transcriptional initiation and a GC box, containing overlapping Sp/Egr-1 binding sites, located at 83 nt upstream of the start site (53).
In cell culture, SRF protein has been detected in all cell types examined, making a role for newly expressed SRF protein unclear. However, in developing avian embryos, SRF is expressed primarily in striated and smooth muscle tissues (11). Belaguli et al. (4) have also shown that in the adult mouse, SRF mRNA shows significant enrichment in cell types derived from embryonic mesoderm, such as cardiac, smooth, and skeletal muscle, as well as to a lesser extent in cell types of neuroectodermal origin. Consistent with this observation, during avian development SRF protein becomes detectable exclusively in the myocardium (18) coincident with the appearance of basic fibroblast growth factor (bFGF) upon fusion of the myocardial tubes (46). SRF recruits the tinman homologue Nkx-2.5, which is a critical mediator of cardiac development, to cardiac muscle-specific promoters. This finding together with the observations that FGF signaling is important for the development of numerous organ systems (for a review, see reference 5) and that FGF is essential for cardiac development (54) suggests that one possible mechanism by which FGF signaling contributes to cardiac specification is by inducing SRF gene expression.
The mechanism by which the SRF gene is regulated during development is not known. Recent studies indicate that SRF binding sites and a proximal GC box motif located in the SRF gene promoter are important for developmentally regulated expression (4). This possibility is consistent with our previous studies which show that a GC box is involved in serum growth factor-mediated stimulation of the SRF gene in mouse fibroblasts and suggests that in addition to SRF, members of the Sp family of transcription factors may play a significant role in mediating regulation of the SRF gene (53). However, the SRF GC boxes contain overlapping Sp1 and Egr-1 binding sites, and studies performed to date examining the role of the GC box in SRF gene regulation have not distinguished between Sp factor and Egr-1 binding. Therefore, whether Sp1 or other Sp factors mediate regulation of the SRF gene has not been addressed by previous studies, nor has the role of other upstream regulatory elements.
In this study, we address the role of three distinct upstream SRF promoter regulatory elements, an ETS binding site, the proximal GC box, and the CArG boxes, in mediating activation by bFGF. Our analysis indicates that in murine fibroblasts, bFGF regulates the SRF promoter by a complex mechanism that targets distinct regulatory elements through multiple signaling pathways. Maximal bFGF-mediated activation of the SRF gene occurs by two independent mechanisms: a CArG box-independent pathway that involves an ETS binding site, and a novel CArG box-dependent pathway that requires both the Sp binding portion of the GC box and the SRF binding sites for maximal stimulation. Furthermore, our results indicate that CArG/GC box-mediated activation is targeted by two distinct signaling pathways. The CArG box-dependent mechanism is targeted by Rho pathway-mediated signals, and the Sp/GC box-dependent mechanism is targeted by a Ras-mediated signaling.
MATERIALS AND METHODS
Construction of SRF chimeric reporter plasmids and mutagenesis.
The SRF–c-fos chimeric reporter −322SRF/c-fos was created by replacing the c-fos promoter in plasmid pF4 (human c-fos genomic clone [57]) with 322 nt of the SRF gene promoter. Recently, Belaguli et al. (4) reported that in mouse embryos the major start site of SRF transcription is 4 nt downstream from the start site previously published for the human cDNA (45). We mapped the SRF start site of transcription in NIH 3T3 cells and found that the major start site corresponded to the human start site and a minor start site corresponded to the start site reported by Belaguli et al. To construct the SRF–c-fos chimeric reporter, the human c-fos gene promoter plus 45 nt of the 5′ untranslated region was excised from the pF4 c-fos genomic plasmid by EcoRI (5′ multiple cloning site) and NotI (+45) restriction digestion. This was replaced with a 367-bp fragment of the murine SRF gene generated by PCR. This fragment included 322 nt of sequence upstream of the initiation site. Primer sequences used in the PCR were 5′-CCGGAATTCCTGCAGTCCTCTCC-3′ and 5′-ATAAGAATGCGGCCGCGAG GGGCCGGGAC-3′. PCR fragments were verified by double-stranded sequencing. The serum response element (SRE) minimal promoter −63SRF/c-fos chimeric reporter was also generated by PCR. Primer sequences used were 5′-CCGGAATTCCTCGCCATATAAGGAGCGG-3′ and 5′-ATAAGAATGCGGCC GCGAGGGGCCGGGAC-3′. Mutagenesis to disrupt transcription factor binding sites was performed by the method of Deng and Nickoloff (19) as previously described (53). Primer sequences used to generate point mutations of the proximal GC box that distinguished between Sp1 and Egr-1 binding were 5′-GCGCCCCCGCTTTCATTGGTCCG-3′, which disrupts the Sp1 site, and 5′-CGAGC CCCCAGTTTCCCCGCCCC-3′, which disrupts the Egr-1 binding site (underlining indicates mutated bases). A mutation which disrupts both Sp1 and Egr-1 binding to the proximal GC box has been previously described (53). The SRF–c-fos chimeric reporter plasmids containing point mutations were constructed as described for −322SRF/c-fos.
Cell culture and transfections, RNase protections, and luciferase assays.
NIH 3T3 fibroblasts were cultured and transfected as previously described (53), with the following modifications. For RNase protection experiments, cells were seeded at a density of 1 × 106 to 2 × 106 cells/100-mm-diameter dish 18 to 24 h before transfection with 10 μg of wild-type or mutant SRF–c-fos chimeric reporter plasmid. As a control for transfection efficiency, 0.5 μg of plasmid SV-α-globin was included in the transfection mixtures. Total DNA was brought to 15 μg with pBluescript unless otherwise noted. For transfections involving dominant inhibitory members of the Ras family of low-molecular-weight GTP binding proteins, 5-μg aliquots of dominant inhibitory expression plasmids were included in the transfection mixtures. The dominant inhibitory constructs used were pRSV-RasN17 (30), pCEV29 RhoAN19 (17), and pCEV29 Rac1N19 (17). The Rac and Rho constructs have been previously described and their expression has been characterized (17). Twelve to 16 h after transfection, cells were washed with warm phosphate-buffered saline and then placed for 24 to 36 h in starvation medium containing 0.5% calf serum-supplemented Dulbecco modified Eagle medium (DMEM). Cells were stimulated with either 20% fetal bovine serum-supplemented DMEM (HyClone) or 50 ng of bFGF (Promega) per ml for 30 min unless otherwise noted. Total RNA was isolated by the RNeasy mini kit protocol (Qiagen) or Trizol reagent (Gibco) as directed by the manufacturer and subsequently DNase treated for 30 min at 37°C. Synthesis of 32P-riboprobes, hybridization, RNase treatment, and electrophoresis were performed as described elsewhere (53). The human c-fos probe protects a fragment of 251 nt from the transfected SRF–c-fos reporters and a 65-nt fragment from the endogenous murine c-fos RNA. The α-globin probe protects a fragment of 133 nt. Data were quantitated with a Molecular Dynamics PhosphorImager. Transfections for luciferase assays were performed as previously described (53), with the following modifications. Cells were grown in 10% calf serum-supplemented DMEM for 24 to 48 h after transfection before harvesting. For transfections involving constitutively active forms of Ras, Rac1, and RhoA, 0.5 μg of the activator was used in the transfection. Activator plasmids used in this study were pZIP KRasV12 (49), pcDNAIIIB Rac1QL (17) and pcDNAIIIB RhoAQL (17), and Gal4–Elk-1 (38). Expression levels of the constitutively active Ras family members have been previously characterized (17). The Gal4-Sp1 expression construct, containing the full-length Sp1 fused to a Gal4 DNA binding site, was a gift of Robert Tjian. Gal4-MEF2C contains full-length MEF2C fused to a Gal4 DNA binding domain and was a gift of J. Molkentin and E. Olson (43). Luciferase assays were performed as described by Promega. Luciferase activity was measured as previously described (53).
Nuclear extract preparation and electrophoretic mobility shift assays.
Nuclear extracts from NIH 3T3 fibroblasts were prepared by the method of Dignam et al. (20) except that the final dialysis buffer D contained 10 μM ZnCl2. Radioactively labeled DNA probes were prepared as previously described (53). Briefly, binding conditions in a 20-μl volume were 1× Dignam buffer D (20) supplemented with 10 μM ZnCl2, 2 μg of poly(dI-dC), 1 μg of sheared herring sperm DNA (Sigma), and 0.1 to 1 ng of DNA probe. Approximately 6 μg of protein was used in each binding reaction. Protein was incubated for 30 min on ice before the addition of labeled probe. For antibody shift reactions, antibody (1:50 dilution) was incubated for 10 min before electrophoresis. Antibodies used were Sp1 (PEP 2)-G sc-59 X (Santa Cruz) and Egr-1 (C-17) sc-354 X (Santa Cruz). For experiments using recombinant protein, 10 to 40 ng of human Sp1 (Promega) and/or 0.5 μl of partially purified bacterially expressed Egr-1 was used. Binding conditions were as described by Khachigian et al. (33).
Northern blot analysis.
Total cellular RNA was isolated as described above, and electrophoresis was performed as described by Sambrook et al. (48). SRF RNA was detected by using a radiolabeled DNA probe consisting of a 305-bp HindIII-DdeI restriction fragment isolated from the human SRF cDNA pT7ΔATG (45). Labeling was performed by the method of Feinberg and Vogelstein (21). Hybridization was performed in a Hybaid rotary oven under conditions described by Church and Gilbert (16). Data were quantitated as described above.
RESULTS
Expression of the SRF gene is stimulated by bFGF.
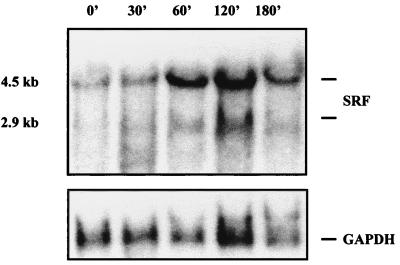
Previously, we and others have shown by Northern blot analysis that expression of the SRF gene is inducible when serum-starved mammalian cells are stimulated with serum (42, 45) or purified growth factors, including nerve growth factor and forskolin (41a). Peak accumulation of SRF RNA occurs between 90 and 120 min poststimulation and is approximately 15-fold above levels in unstimulated cells. To determine whether the SRF gene was induced similarly with bFGF, serum-starved NIH 3T3 fibroblasts were treated for various times with 50 ng of bFGF per ml. Northern analysis of total cellular RNA was then performed with an SRF-specific probe. In Fig. 1, increases in SRF message are seen within 30 min, peak expression occurs at 60 to 120 min after stimulation, and expression begins to decline by 180 min after stimulation. This time course of expression mimics that seen for serum as well as lysophosphatidic acid (53a). Normalization against the constitutively expressed GAPDH gene reveals that SRF message levels are induced up to fivefold above basal levels. Consistent with the induction of SRF RNA levels, the level of SRF protein in the cell is also increased, with an approximately 5-fold increase in SRF-specific SRE DNA binding activity seen by 45 to 60 min after bFGF stimulation (data not shown).
FIG. 1.
The SRF gene is transiently expressed in bFGF-treated fibroblasts. Serum-starved NIH 3T3 fibroblasts were stimulated by the addition of bFGF, and total RNA was isolated at various time points (minutes). Northern blot analysis was then performed with a DNA probe specific to the C-terminal portion of the SRF protein. The blot was stripped and reprobed with a DNA probe corresponding to the constitutively expressed GAPDH gene. Positions of the 4.5- and 2.9-kb SRF mRNAs are indicated.
The SRF promoter can direct transient expression of a c-fos reporter gene.
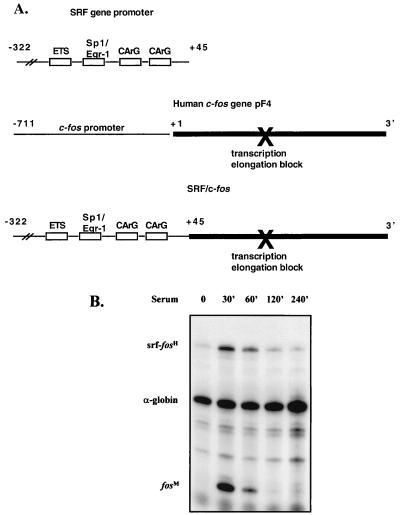
To analyze carefully the contribution of SRF promoter elements responsible for mediating bFGF responsiveness, we wanted to develop a sensitive RNase protection assay that would enhance detection of SRF promoter activity. We initially were interested in developing a protection assay in which the SRF promoter controlled expression of a reporter gene containing genomic SRF coding sequences. This approach proved problematic, and we were unable to develop a reliable assay, presumably due to the high G+C content of the SRF gene (45). Expression of the human c-fos gene has been extensively studied by RNase protection analysis (57); we therefore decided to study the SRF promoter by using the human c-fos gene as a reporter. To do this, we constructed a series of chimeric SRF–c-fos reporters, based on the parent chimeric depicted in Fig. 2A, in which various SRF promoter mutants were fused to nearly the entire transcribed portion of the human c-fos gene. To determine whether these chimeric reporters would respond similarly to other SRF-driven luciferase reporter systems studied previously (53), the ability of a chimeric reporter containing the wild-type SRF promoter to respond to serum was analyzed.
FIG. 2.
The SRF promoter directs transient expression of a human c-fos gene reporter. (A) Schematic representation of derivation of the SRF–c-fos chimeric reporter. (B) NIH 3T3 fibroblasts were transfected with −322SRF/c-fos and serum starved. Cells were serum stimulated for the times (minutes) indicated, and total RNA was isolated. RNase protection assays were performed. Data were quantitated by PhosphorImager analysis. Levels of expression of the −322SRF/c-fos reporter were normalized for transfection efficiency against the level of expression of the transfected α-globin gene. Positions of the protected RNA species are indicated (transfected SRF-human c-fos [srf-fosH] = 251 nt; globin = 133 nt; endogenous mouse c-fos [fosM] = 65 nt).
In Fig. 2B, a time course analysis of serum-stimulated expression of one of these constructs, −322SRF/c-fos, is shown. We have previously shown that the SRF promoter fragment used in this construct contains all upstream sequences necessary for proper serum-mediated regulation of the SRF gene (53). Using the SRF–c-fos reporter, we found that in unstimulated cells the reporter message is virtually undetectable. Message levels peak by 30 min after serum stimulation and return to basal levels by 120 to 240 min after stimulation. Maximal stimulated expression is 16-fold above unstimulated levels. In other experiments, fold stimulation ranged from 10- to 50-fold (data not shown). The transient and robust nature of stimulation from the SRF–c-fos reporters suggested that this system would allow us to very sensitively measure activity of the SRF promoter.
Maximal accumulation of message from the chimeric construct occurs significantly earlier than maximal accumulation of message from the endogenous SRF gene (reference 42 and Fig. 1), and more closely parallels accumulation of message from the endogenous c-fos gene. The basis for the delayed accumulation of the endogenous SRF message relative to the c-fos message has not been carefully investigated; however, the results presented here suggest that this difference is dependent on the nature of the transcribed sequences (see Discussion).
Maximal bFGF-mediated activation of the SRF promoter occurs through multiple distinct mechanisms that involve the ETS, Sp1, and SRF binding sites.
Previously we investigated the role of distinct SRF promoter regulatory elements in serum responsiveness of the SRF gene (53). The 322-nt region upstream of the transcription initiation site that contained regulatory elements necessary for maximal serum-stimulated expression of the gene included a GC box, containing overlapping Sp1/Egr-1 binding sites, located 83 nt upstream from the start site of transcriptional initiation, two adjacent SRF binding sites (CArG boxes) starting at 43 and 63 nt upstream from the initiation site, and a consensus ETS protein binding site 103 nt upstream from start site of initiation. Our previous studies (53) demonstrated that (i) the SRF binding sites are sufficient to mediate serum-inducible expression of the SRF promoter, although weakly; (ii) maximal serum responsiveness of the promoter is dependent on the −83 site and the two SRF binding sites being intact, suggesting that the −83 site and the SRF binding sites cooperate to mediate maximal induction; (iii) the ETS binding site at −103 appeared to be dispensable for serum-mediated activation.
To investigate the mechanism by which bFGF activates the SRF promoter, we first used a series of luciferase reporters driven by SRF promoter deletions (53). The results from these experiments showed that sequences between −322 and −2500 did not significantly affect bFGF-mediated expression (data not shown). We therefore concentrated on studying the role of elements within the −322 region that may be important for SRF gene regulation, the −103 ETS binding site, the −83 site, and CArG boxes. Point mutations that abolished binding to cognate transcription factors were introduced into each of these elements in the context of an SRF–c-fos chimeric reporter plasmid that contained 322 nt of the SRF promoter. We then introduced mutant reporter constructs into NIH 3T3 cells and measured the ability of each to respond to bFGF stimulation of serum-starved cells in a sensitive protection assay as described for Fig. 2.
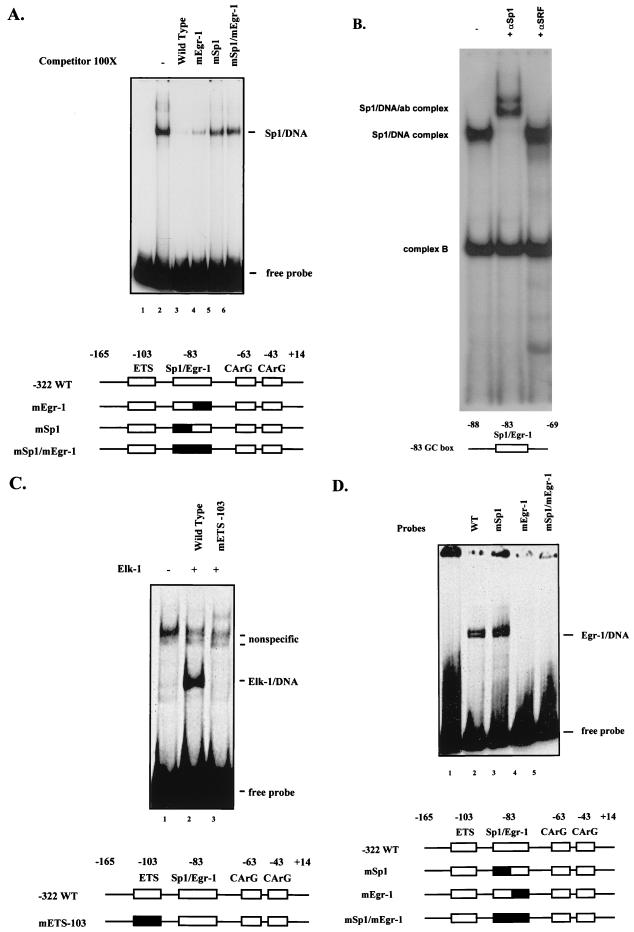
Previously we have shown that the two SRF CArG boxes can bind both purified SRF and SRF from NIH 3T3 cell nuclear extracts with high efficiency (53). As shown in Fig. 3A and B, both purified Sp1 and Sp1 from NIH 3T3 cell nuclear extracts can bind the SRF −83 site. Furthermore, Sp1 binding to the −83 GC box is specific to the Sp1 binding motif of the overlapping Sp1/Egr-1 site. To demonstrate this, the −83 site was altered such that either the Sp1 or Egr-1 binding portion of the element was disrupted. As shown in Fig. 3A and D, the Sp1 binding-site mutation used in these studies specifically abolished binding of the Sp1 binding portion of the overlapping site and left Egr-1 binding intact. In contrast, an Egr-1 motif mutation abolished Egr-1 binding but left Sp1 binding intact.
FIG. 3.
SRF promoter elements bind Elk-1, Sp1, and Egr-1 in vitro. (A) Gel mobility shift assays were performed with recombinant human Sp1 and a 32P-labeled DNA probe corresponding to positions −165 to +14 of the SRF promoter. The indicated molar excess of unlabeled probe containing a mutation of the Sp1 binding portion of the −83 site (mSp1), Egr-1 binding portion of the −83 site (mEgr-1), or both was included in the binding reaction. The positions of the free probe and Sp1-DNA complex are indicated. WT, wild type. (B) Gel mobility shift assays were performed with nuclear extracts from NIH 3T3 cells, using a 32P-labeled DNA probe corresponding to positions −88 to −69 of the SRF promoter containing the wild-type −83 site. Positions of the Sp1-DNA complex and a nonspecific complex, labeled complex B, are indicated. Complex B can be specifically competed with an Egr-1 oligonucleotide (not shown). An Sp1-specific antibody (ab) (αSp1) was added to the second lane, and the position of the shifted complex is indicated; a nonspecific control polyclonal SRF-specific antibody (αSRF) (42) was added to the third lane. (C) Gel mobility shift assays were performed with in vitro-translated Elk-1 and a 32P-labeled DNA probe corresponding to positions −165 to +14 of the wild-type SRF promoter (lanes 1 and 2) or a mutant of the ETS −103 site (lane 3). The positions of free probe, Elk-1–DNA complex, and nonspecific complexes are indicated. (D) Gel mobility shift assays were performed with bacterial extracts containing the Egr-1 protein and 32P-labeled DNA probes corresponding to positions −165 to +14 of the SRF promoter containing the wild-type −83 site or mutation of the Sp1 portion, Egr-1 portion, or both. The positions of free probe and Egr-1–DNA complexes are indicated. The double-banding pattern observed is consistent with that observed previously (14). Structures of the relevant DNA binding probes or competitors are shown schematically at the bottom of each panel.
The SRF promoter also has a high-affinity ETS binding site that is identical to the binding site for the product of the Drosophila E74 gene. This site has previously been shown to have high affinity for ETS proteins including E74 and Elk-1 subfamily members of the ETS family (64). To disrupt the ETS binding site, this motif was converted to a PstI restriction site. There is no detectable binding of endogenous factors found in NIH 3T3 cell extracts when this mutant site is used as a probe in DNA mobility shift assays (data not shown). As shown in Fig. 3C, relative to the wild-type site, promoter fragments containing the mutant ETS site fail to bind with high affinity to the ETS subfamily member Elk-1. The SRF binding site was also mutated, and characterization of double-point mutations in each of the SRF binding sites that abolished in vitro SRF binding have been previously described (53). The ability of these mutants to respond to bFGF stimulation was addressed next.
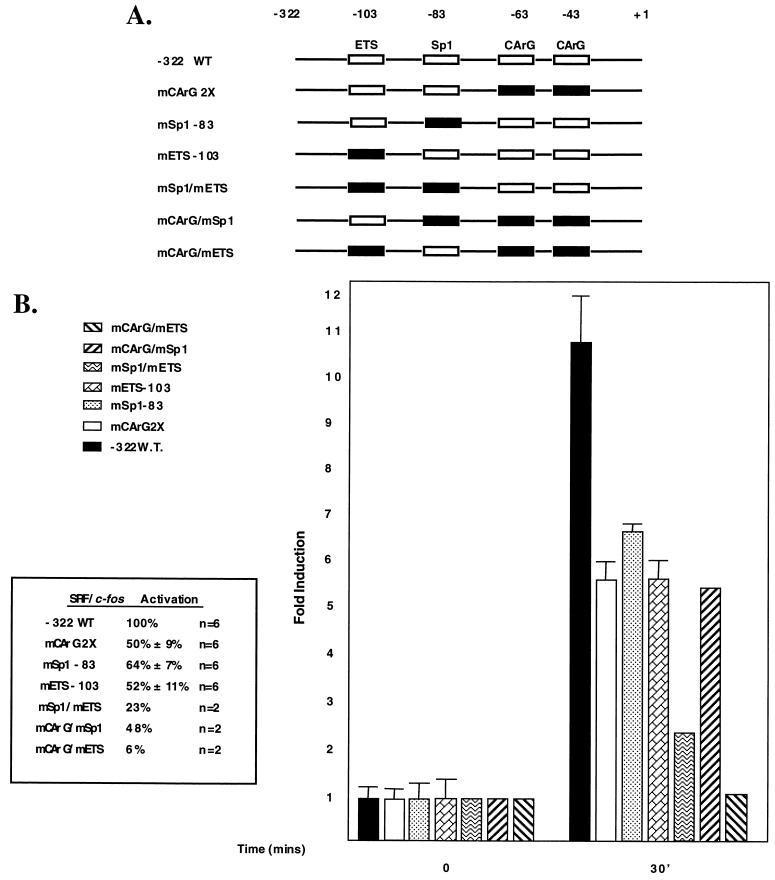
Figure 4 shows that a reporter construct containing a double mutation that abrogates SRF binding to both CArG boxes results in an approximately 50% decrease in bFGF responsiveness, indicating that SRF binding likely plays an important role but is not necessary for bFGF-mediated activation of the SRF promoter. Single mutations in the −83 Sp1 binding site or the ETS binding site result in a 64 to 52% decrease in bFGF responsiveness. It should be pointed out that the Sp1 binding-site mutant left the Egr-1 binding ability of this reporter intact. Experiments in which the Egr-1 site was mutated so as to leave the Sp1 site intact showed no effect on expression (data not shown). A double mutant containing both a mutant −83 site and a mutant ETS site further reduces responsiveness to approximately 23% of that of the intact −322 promoter, which is similar to that seen from a minimal promoter construct containing only two SRF binding sites (not shown). A minimal TATA-only construct is totally unresponsive (not shown). A double-mutant promoter, containing mutant SRF binding sites and a mutant −83 site, reduces expression to a level similar to that for the single mutant construct containing mutant SRF binding sites. In contrast, bFGF-mediated expression from a double-mutant reporter containing both mutant SRF binding sites and a mutant ETS site is nearly completely abolished.
FIG. 4.
bFGF mediates SRF gene activation by distinct regulatory elements. NIH 3T3 fibroblasts were transfected with a −322SRF/c-fos reporter containing the wild-type (WT) sequence or in-context mutations of the Sp1, CArG, and ETS sequences alone or in combination. Cells were serum starved for 36 h and stimulated for 30 min by the addition of bFGF to 50 ng/ml (final concentration). Total RNA was isolated, and RNase protection assays were performed. Data were quantitated by PhosphorImager analysis and normalized for transfection efficiency. Activation was determined by comparing levels of human c-fos RNA from unstimulated and stimulated cells. The activation of the −322 wild-type reporter was set to 100%. Levels of activation of the mutants are expressed as a percentage of the level of activation of the wild-type reporter. The basal level of expression between constructs in uninduced cells differed by less than 32% from experiment to experiment. For the experiments where n = 2, the percent activation is an average and was less than 6% different between experiments.
Together, the results in Fig. 4 indicate that bFGF stimulates the SRF promoter through multiple distinct mechanisms. First, bFGF-mediated signaling pathways can activate the SRF promoter by a CArG box-dependent mechanism that requires an intact SRF binding site. The −83 site can potentiate activation by this mechanism but is not on its own sufficient to mediate activation. Second, bFGF-mediated signaling pathways can activate the SRF promoter by an SRF binding-site-independent mechanism. This mechanism appears to require an intact ETS binding site located at −103. Furthermore, since the SRF binding-site mutant and the SRF binding site/−83 double mutant respond to a similar extent, this finding suggests that the ETS binding site operates by a mechanism that is independent of the −83 site as well. Maximal SRF promoter responsiveness to bFGF requires that both mechanisms be fully functional and that the SRF binding sites, the −83 site, and the −103 ETS binding sites be intact.
Constitutively expressed Sp1 is able to rescue the bFGF response of −83 GC box binding-site mutant reporters.
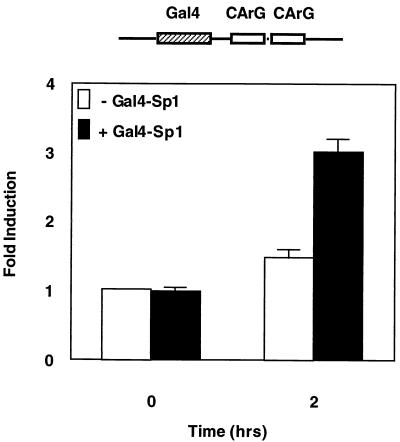
Since the results from the above experiments suggested that factors that bind to the Sp factor binding portion of the −83 GC box are involved in mediating a significant portion of the bFGF response, we next directly investigated whether Sp1 could rescue the bFGF response from a mutant reporter. To address this issue, we targeted a Gal4-Sp1 fusion protein to an SRF-luciferase reporter which contained a single Gal4 site 20 bp upstream of the natural SRF binding sites. The ability of this reporter to respond to bFGF stimulation in the presence or absence of the Gal4-Sp1 fusion construct was then measured.
As seen in Fig. 5, in serum-starved cells a constitutively expressed Gal4-Sp1 fusion construct, which contains full-length Sp1 fused to a Gal4 DNA binding domain, fails to stimulate the Gal4/CArG box-containing promoter above basal levels. However, when cells are stimulated for 2 h with bFGF, there is a modest stimulation of expression from the reporter, which is enhanced nearly twofold in the presence of the Gal4-Sp1 construct. The magnitude of this enhancement is similar to the magnitude of the loss of expression seen in the −83 GC box Sp1 binding-site mutant reporter in Fig. 4, indicating that Sp1 can rescue the −83 GC box mutation. This result together with the binding data presented in Fig. 3 supports the idea that Sp1 or related factors may be acting through this site in vivo. Since an SRF promoter containing an intact −83 GC box and mutant SRF and ETS binding sites fails to support bFGF activation (Fig. 4), these results further suggest that Sp1 functionally cooperates with SRF or other CArG box binding factors to mediate a portion of the bFGF response.
FIG. 5.
Sp1 can contribute to the bFGF response. NIH 3T3 fibroblasts were transfected with a luciferase reporter containing a Gal4 binding site centered 20 nt upstream of the −63 CArG box of the SRF gene promoter. Where indicated, a plasmid expressing a Gal4-Sp1 fusion protein was included. Cells were starved for 36 h and stimulated with bFGF for 2 h, and luciferase assays were performed. Fold induction was determined by comparing the serum-starved level of expression with the stimulated level of expression. The level of expression in unstimulated cells of the reporter in the absence of Gal4-Sp1 was set to a value of 1.0. For each point, assays were performed in triplicate and values were corrected for transfection efficiency. Results from at least three independent experiments are shown. Expression of the Gal4 DNA binding domain alone did not enhance bFGF-stimulated expression (not shown).
Members of the Ras superfamily regulate SRF gene expression.
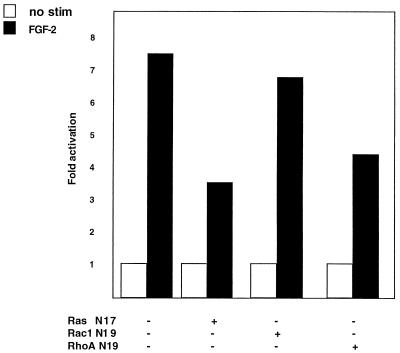
We next wanted to investigate signaling pathways that target the SRF promoter. Since bFGF has been shown to activate signaling molecules that are controlled by the Ras family of monomeric GTPases, we investigated whether the SRF promoter was regulated by various Ras family members, using both dominant inhibitory and constitutively active constructs. In the first set of experiments, we cotransfected constructs that constitutively express dominant inhibitory versions of RasN17, Rac1N19, and RhoAN19 with a SRF–c-fos reporter and measured the ability of the reporter to be activated by bFGF by an RNase protection assay. As can be seen in Fig. 6, when a RasN17 dominant inhibitory construct is cotransfected with a wild-type SRF promoter-driven reporter, bFGF-mediated activation is inhibited greater than 50%. Similarly, the RhoAN19 construct inhibits the response roughly 40%. In contrast, another Rho family member, Rac1N19, fails to show any significant inhibition.
FIG. 6.
Ras-related signaling pathways mediate bFGF activation of the SRF promoter. NIH 3T3 fibroblasts were transfected with the −322SRF/c-fos reporter. Where indicated, a dominant inhibitory form of Ras, Rac1, or RhoA was included in the transfection. Cells were serum starved for 36 h and stimulated with bFGF for 30 min. RNA was harvested, and RNase protections were performed. Data were quantitated by PhosphorImager analysis and normalized for transfection efficiency. Data were normalized between each set of transfections for a given inhibitory construct, and fold activation was determined by comparing the levels of RNA expressed in unstimulated (no stim) and stimulated cells. Fold activation of the −322SRF/c-fos reporter in the absence of cotransfected inhibitor was set to 100%. The results presented are averages of two independent determinations and varied by less than 6% between experiments. Note that the Rac/Rho inhibitory constructs do not inhibit the α-globin reference gene used as a transfection control. The inhibitory Ras constructs reproducibly inhibited the transfection control. As a consequence, fold activation in the Ras experiments should not be directly compared to that in the Rac/Rho experiments.
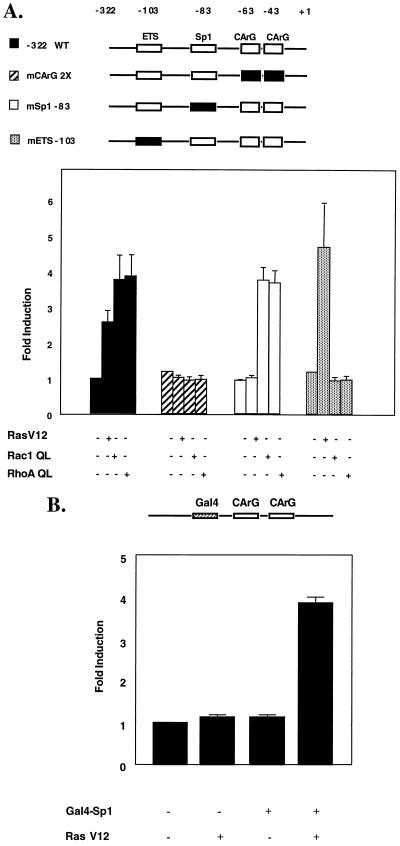
To determine directly whether activation of these pathways is sufficient to activate the SRF promoter, we next cotransfected a constitutively active version of Ras, Rac1, or RhoA with either a wild-type or mutant SRF promoter-luciferase reporter construct and measured luciferase activity 36 to 48 h after transfection. As seen in Fig. 7A, constitutively active Ras, Rac1, and RhoA are each able to upregulate the SRF promoter. Activation by each is dependent on intact SRF binding sites, since a mutant reporter that cannot bind SRF fails to respond to any of the three constructs. However, a mutant reporter in which the −83 GC box is altered so that it can no longer bind Sp1 can still support Rac1- and RhoA-mediated activation. This result is consistent with previous reports which indicate that at the c-fos promoter, SRF is a target for Rho-dependent signaling (29, 30). In contrast, the −83 GC box Sp factor binding-site mutant fails to support activation mediated by constitutively active Ras. Interestingly, Rac- and RhoA-mediated activation of the SRF promoter appears to require both an intact CArG box and an intact −103 ETS site, since mutation of the ETS site or the CArG boxes leads to an unresponsive reporter. In contrast, Ras-mediated activation of the promoter appears to be independent of the ETS site. These results demonstrate that the ability of chronically upregulated individual GTPases to stimulate expression of each reporter differs depending on which promoter element is mutated, suggesting the pathways preferentially target different combinations of elements. Together, the results in Fig. 6 and 7A indicate that the CArG box-dependent mechanism for activation of the SRF promoter involves multiple signaling pathways targeting distinct cis-acting regulatory elements.
FIG. 7.
Ras-, Rac1-, and RhoA-dependent signaling events require specific cis-acting elements in the SRF promoter for regulation. (A) NIH 3T3 fibroblasts were transfected with −322SRF-luciferase (wild type [WT]) or the mutant indicated. Where indicated, a constitutively active form of Ras, Rac1, or RhoA was included. Cells were grown for 36 to 48 h and harvested. Luciferase assays were performed and normalized to total protein content. Activation was determined by comparing the level of luciferase activity in the absence of activator with the level of luciferase activity in the presence of activator. The level of expression of the −322SRF-luciferase reporter in the absence of activator was assigned a value of 1.0. In all cases, values were determined in triplicate and the data are representative of three independent experiments. (B) NIH 3T3 fibroblasts were transfected with a luciferase reporter containing a Gal4 binding site centered 20 nt upstream of the two CArG boxes of the SRF gene promoter. Where indicated, a plasmid expressing a Gal4-Sp1 fusion protein and a plasmid expressing constitutively active RasV12 were included in the transfection. Cells were grown for 36 h, and luciferase assays were performed. Activation was determined by comparing the level of luciferase activity in the absence of RasV12 and Gal4-Sp1 with the level of activity in the presence of RasV12 and Gal4-Sp1. The level of expression of the reporter in the absence of Gal4-Sp1 and RasV12 was set to a value of 1.0. For each point, assays were done in triplicate and normalized to total protein content. Results from at least three independent experiments are shown. Expression of the Gal4 DNA binding domain alone did not enhance Ras-activated expression (not shown).
The results from the −83 GC box mutant in Fig. 7A suggest that Sp1 or related factors may be involved in mediating the Ras-dependent upregulation of SRE-dependent activation. To test this hypothesis directly, we cotransfected constitutively active Ras with a Gal4-Sp1 expression plasmid and a luciferase reporter containing the SRF CArG boxes and a single Gal4 binding site. As seen in Fig. 7B, in the absence of Gal4-Sp1, constitutively active Ras failed to upregulate the minimal Gal4-CArG-driven reporter. However, when Gal4-Sp1 is added, the reporter is upregulated nearly fourfold, to a level similar to that seen with the wild-type promoter.
DISCUSSION
In the present study, we have examined the mechanism by which bFGF and Ras-related signaling GTPases regulate expression of the SRF gene. Our results indicate that maximal activation is mediated by three promoter regulatory elements: a consensus ETS binding site located at −103, an Sp factor binding site located at −83, and two CArG boxes located at −43 and −63 upstream of the start site of transcriptional initiation. Using the CArG boxes as a reference point, we defined two distinct mechanisms of bFGF-mediated activation, one CArG box dependent and the other CArG box independent. The CArG box-independent mechanism is mediated by the −103 ETS binding motif. For the CArG box-dependent mechanism, intact SRF binding sites are both necessary and sufficient to mediate activation. CArG box-mediated activation is potentiated by a −83 GC box Sp factor binding motif; however, the −83 GC box is not sufficient to mediate activation.
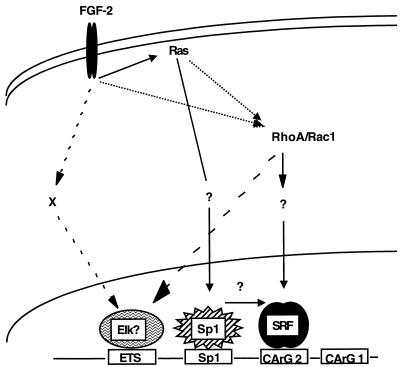
As summarized in Fig. 8, our results also indicate that distinct signaling pathways target these elements to mediate activation. Using constitutively active versions of Ras, RhoA, and Rac1, we have demonstrated that the SRF CArG box-dependent mechanism is targeted by Rho-dependent pathways and the −83 GC box-dependent mechanism is targeted by a Ras-dependent pathway. Since constitutively active mutants of Ras, Rac1, or RhoA are insufficient to activate a CArG mutant SRF promoter that is otherwise intact, this finding suggests that SRF is necessary for activation by all three pathways. In contrast to signaling by constitutively active GTPases, signaling by bFGF appears to be more complex. bFGF can activate the SRF promoter in a CArG-independent manner, mediated by the ETS site. This finding suggests that activation through the ETS site is targeted by yet another pathway and that bFGF activates additional pathways. Further experiments need to be performed to address the nature of the mechanism by which the ETS site mediates bFGF activation and to determine which signaling pathways target which promoter elements upon bFGF treatment of cells. In the case of serum-mediated activation of the SRF promoter, our experiments indicate that dominant inhibitory Ras constructs specifically block the −83 GC box-dependent mechanism. In contrast, inhibition of serum activation by dominant negative Rho constructs is independent of the −83 site (53a).
FIG. 8.
Schematic representation of the CArG box-dependent and independent mechanisms of bFGF mediated activation of the SRF promoter. For discussion, see the text.
In the experiments presented here, to measure SRF promoter activity, we have used a sensitive reporter system that relies on the SRF promoter driving expression of the human c-fos gene. The fold activation of the SRF–c-fos chimeric constructs is significantly higher than that of the endogenous SRF gene, being stimulated 16-fold above the basal level (although fold stimulation ranged from 10 to 50 in different experiments), compared to approximately 5-fold for the endogenous gene. The time course of accumulation of the message from this chimeric construct mimic that of the endogenous c-fos gene. While the precise basis for this discrepancy has not been carefully investigated, based on other systems that use the c-fos gene as a reporter (52), the difference can be explained by the relative instability of the c-fos reporter RNA. The c-fos message has a half-life of approximately 15 min in NIH 3T3 cells, and the message stability determinants of the gene are located in the transcribed region (52). We have not directly measured SRF message half-life. However, in a previous study (42) we showed by nuclear runoff assays that upon serum stimulation the SRF gene is transiently induced at the transcriptional level with a time course similar to that for the c-fos gene, although the peak of SRF message accumulation is significantly delayed relative to the c-fos message (42). Together, these observations strongly suggest that the SRF message is more stable than the c-fos message. Therefore, relative to the endogenous SRF gene, an SRF–c-fos chimera would be expected to result in dramatically lower steady-state levels of reporter message than the endogenous SRF gene in unstimulated cells. Consistent with this interpretation, we find that unstimulated cells always exhibit the presence of some endogenous SRF message (Fig. 1) yet there are very low or undetectable levels of message from the SRF–c-fos chimera. The artificially low basal level of accumulation of the chimeric gene makes the resulting RNase protection analysis a very sensitive measure of SRF promoter activity. Consistent with the idea that the stability of the reporter message governs fold stimulation, we find while stimulation of the chimeric SRF–c-fos constructs with serum results in 10- to 50-fold induction, when the same SRF promoter fragment is fused to a more stable luciferase reporter, only a 4- to 5-fold induction is seen.
It has been previously suggested (4) that inducible SRF expression may be important for regulating SRF gene expression since the SRF gene can be autoregulated. Misra et al. also suggested that induction of SRF may be important for downregulating expression of inducible SRE-dependent gene expression (42). Belaguli et al. demonstrated that overexpression of SRF could upregulate the SRF promoter during myogenesis in culture or in primary myocytes and that expression of a dominant inhibitory version of SRF could block SRF promoter expression (4). Their results demonstrated a positive role for SRF in SRF gene regulation. We have also observed that in bFGF-stimulated NIH 3T3 cells, SRF protein accumulation is delayed relative to promoter activation (data not shown). We have also previously shown that the SRF gene can be induced in fibroblasts in the absence of new protein synthesis by serum growth factors. Together, these observations suggest that increases in SRF protein levels are not necessary for significant SRF gene activation in this system. However, it is possible that in cases where SRF protein is limiting, such as in other tissue types or during early development, an increase in SRF protein synthesis is important for positive autoregulation of the gene. The idea that this may be a significant mechanism of regulation is consistent with the observation that SRF expression can vary dramatically between tissue types (4).
What transcription factors are required for mediating bFGF-stimulated activation of the SRF promoter? Our analysis indicates that factors that bind to the −103 ETS box, the −83 GC box, and the SRF binding sites are critical for mediating activation. We have previously shown that SRF is a major factor that binds to the CArG boxes in the SRF promoter and that at least one intact CArG box is sufficient to mediate serum induction (53). In addition to SRF, the SRF CArG boxes also contain consensus YY1 binding sequences (62), raising the possibility that YY1 contributed to the response seen here. The point mutants that we have introduced in the SRF CArG boxes are located in the 3′ portion of the element well outside the YY1 consensus site. If YY1 were sufficient to mediate the responses we observe, we would expect that disruption of the SRF binding site alone would not have an effect. Consequently, our results strongly implicate SRF as the factor that regulates CArG-dependent SRF gene expression. However, since it has been observed that YY1 can facilitate SRF binding (44), we cannot rule out the possibility that YY1 is involved in the SRF-dependent response. In the present work, we used reporter constructs that contained subtle point mutations in both CArG boxes that we previously showed abolished SRF binding in vitro; as a consequence, we have not directly addressed the issue of whether a single CArG box is sufficient to mediate bFGF activation. However, since we previously showed that SRF binding to each of the CArG boxes is mutually exclusive (53), it is likely that a single intact CArG box is also sufficient to support bFGF-mediated activation.
Recently, Sealy and coworkers found that p35C/EBPβ plays a role in the serum response mediated by the c-fos SRE and that this activation occurs in a TCF-independent, SRF-dependent manner (50). They showed that C/EBPβ binds to a region overlapping and immediately 3′ to the c-fos CArG box. We have not directly investigated whether C/EBPβ can bind to the SRF promoter. However, computer searches of the Transfac databases using SIGSCAN software (52a) did not reveal any consensus C/EBPβ binding sites in the SRF CArG boxes or nearby. Since Sealy and coworkers found that C/EBPβ stimulates the c-fos SRE in an SRF-dependent manner, even if C/EBPβ was found to interact with the SRF promoter, their observations would still be consistent with our conclusion that the SRF CArG box-dependent mechanism for activating the SRF gene requires SRF binding sites.
The SRF promoter −83 GC box contains consensus overlapping binding sites for Sp1 and Egr-1. This overlapping motif is seen in a number of promoters, suggesting that it has a conserved function (33). The observation that the −83 GC box is important for mediating part of the CArG box-dependent response suggests that Sp1, other Sp family members, or Egr-1 is involved in regulation. Since in our experiments we have specifically mutated the Egr-1 binding portion of the −83 GC box, leaving the Sp binding site intact, it is unlikely that Egr-1 or related factors are required for activation. Consistent with a role for Sp factors, we have found that in NIH 3T3 nuclear extracts, Sp1 can bind the −83 site and that a Gal4-Sp1 fusion can rescue a −83 GC box mutant in which the overlapping Sp/Egr-1 binding site is replaced with a Gal4 binding site. It has been suggested that in some promoters, Egr-1 can also serve as a repressor (37). It has also been shown that expression of the Egr-1 gene is inducible by serum growth factors (7). Since we have observed in in vitro assays that binding of Egr-1 and binding of Sp1 to the −83 GC box are mutually exclusive, one possibility is that upon stimulation of cells, newly synthesized Egr-1 displaces Sp1 to help downregulate activation. However, we have seen no difference in message accumulation when stimulated cells containing an SRF–c-fos chimeric reporter construct that contains a mutant Egr-1 binding site in the −83 GC box or wild-type reporters are analyzed in a time course experiment or when a constitutively expressed Egr-1 construct is cotransfected with a wild-type SRF promoter reporter (data not shown).
Our results clearly point to a role for Sp1 or related factors in mediating activation of the SRF gene. Furthermore, this Sp-dependent mechanism requires an intact CArG box and is under control of Ras-mediated signaling pathways. How this Sp factor-dependent mechanism operates, however, is unclear. One possibility is that Ras-activated kinases target Sp1 or other Sp factors. However, while viral Ras oncogenes can activate promoters in an Sp1-dependent manner (10), to our knowledge there are no reports of Ras-dependent inducible phosphorylation or activation of Sp factors. However, it was recently found that in rat ventricular myocytes, Sp1 is involved in calcium-mediated atrial natriuretic factor gene expression in response to pacing (39), and it was suggested that this may occur through a JNK-dependent pathway. Our observation that the −83 GC box is unable to mediate activation of the SRF promoter in the absence of an intact CArG box suggests that direct targeting of Sp1 is not sufficient and may not be necessary to activate transcription. Consistent with this interpretation, we found that in the absence of SRF binding sites, Gal4-Sp1 did not enhance Ras-dependent upregulation of a luciferase reporter, although on its own it was capable of constitutively activating the reporter (data not shown).
Another possibility is that Sp factors are permissive for a complex which consists of at least SRF and Sp1. In this scenario, Ras-dependent signaling may directly target either SRF or another, yet to be identified member of this complex. Consistent with this idea, it has previously been shown that SRF is a target for Ras/MAPK-regulated RSK family members and that it can be inducibly phosphorylated on Ser-103 (47). The role of Ser-103 has been examined in the context of mitogen-mediated activation of the c-fos proto-oncogene (31). While there is no evidence that Ser-103 phosphorylation is required in that context, it may be possible that in a different promoter architecture, such as that found in the SRF gene promoter, Ser-103 phosphorylation is important. Support for the idea that Sp1 and SRF can exist in a complex has come from our observation that SRF and Sp1 can associate in vivo, as determined in a yeast two-hybrid assay system (33a). The role of phosphorylation in this interaction has yet to be investigated. Alternatively, a variety of other transcription factors, including YY1, ATF6, and Phox-1 and other homeodomain proteins, and basic helix-loop-helix proteins, have been shown to interact with SRF to potentiate transcription, and it is possible that Sp1 interacts with one of these in a Ras-dependent manner.
Our results also clearly show that a high-affinity ETS binding site at −103 is sufficient to mediate bFGF-dependent activation. The −103 ETS site is identical to the Drosophila E74 binding site that has previously been shown to bind ETS family members with high affinity. Since it has previously been shown that 3T3 cells contain a binding activity that is related to the Elk-1 subfamily (26, 36), it is possible that Elk-1 subfamily members mediate the −103 responses. Alternatively, other ETS family members may also be involved. While we have not been able to determine which factor binds to this site in vivo, we have shown that a Gal4–Elk-1 chimera can modestly potentiate bFGF-dependent activation of the −83Gal4SRF reporter, supporting the idea that Elk or a related factor binds here in vivo (53a). Further experiments remain to be performed to determine which ETS family member binds the ETS site in vivo. In the c-fos promoter, Elk-related factors mediate activation in response to MAPK-dependent signals by a mechanism that involves ternary complex formation with SRF bound to the SRE. In contrast, we have previously found that Elk-1 does not form a detectable ternary complex in the SRF promoter (53). Paradoxically, we find that while upon bFGF stimulation the SRF promoter can be induced in the absence of an intact −103 ETS site in a Rho/Rac-dependent manner, when the promoter is stimulated by an upregulated Rho or Rac, the −103 ETS site is indispensable. The basis for this observation is unclear, but it supports a model for Rho/Rac-dependent activation that relies on SRF cooperating with additional factors, in this case an ETS binding-site factor.
The observation that the ETS site is sufficient to mediate bFGF-dependent activation of the SRF gene may shed light on how SRF gene expression is upregulated during development. We have previously shown that SRF expression is autoregulated in response to serum, requiring at least one functional SRF binding site. However, since SRF is not detected during the early stages of development, how is the SRF gene upregulated? During avian development, SRF becomes detectable exclusively in the myocardium at stage 10 (18), coincident with the appearance of FGF upon fusion of the myocardial tubes (46). Our results raise the possibility that early in development, bFGF stimulates the SRF-independent pathway, thereby activating SRF gene expression, possibly being mediated by the ETS site. Upon expression of new SRF protein, the SRF gene would then be responsive to additional signaling pathways, including Ras and Rho signals. Such a scenario would imply that expression of the SRF gene is controlled by specific promoter regulatory complexes that are targeted in a developmentally regulated manner. This may afford a mechanism by which levels of SRF gene expression can be regulated in a tissue-restricted manner, without the need to invoke tissue-specific transcription factors. We are currently investigating this hypothesis.
The study reported here was done exclusively with mouse NIH 3T3 cells; therefore, the question arises as to whether the SRF gene is regulated in a similar manner in other cell types. In one other careful study that investigated the regulation of SRF gene expression, Belaguli et al. (4) found that the SRF promoter is upregulated as myoblasts differentiate to myotubes in culture. While their study suggests that SRF binding sites are critical for this upregulation, they did not directly investigate the cis-acting regulatory sites necessary for expression during myogenesis in vivo. However, previous antibody microinjection studies have shown that SRF is necessary for myogenesis in culture (60). Belaguli et al. did report that SRF binding sites are critical for expression of the SRF promoter in primary skeletal muscle cells, which is consistent with our observation that SRF binding sites play a central role in regulating SRF gene expression. They also found that a distal Sp1 binding site, located at −254, is critical for high levels of SRF promoter activity in muscle cells. However, unlike our studies (reference 53 and this work), their studies examined only constitutive expression of the SRF gene in cell culture. Also, they did not investigate the signaling pathways that may be involved in targeting the SRF promoter. As a consequence, the pathways and promoter elements involved in muscle-specific and developmental regulation of the SRF gene remain to be investigated.
ACKNOWLEDGMENTS
This work was supported by the following to R.P.M.: an American Cancer Society Institutional Seed Grant Award from the Medical College of Wisconsin Cancer Center; a grant-in-aid from the American Heart Association, Wisconsin Division; and a Shannon Award (R55 GM/OD51856) and a FIRST award (R29 NS36256) from the National Institutes of Health.
We thank J. Silvio Gutkind for his kind gift of Rho family inhibitory and activator plasmids, and we thank Robert Tjian for the Gal4-Sp1 expression plasmid.
REFERENCES
- 1.Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avantaggiati M L, Natoli G, Balsano C, Chirillo P, Artini M, DeMarzio E, Collepardo D, Levrero M. The hepatitis B virus (HBV) pX transactivates the c-fos promoter through multiple cis-acting elements. Oncogene. 1993;8:1567–1574. [PubMed] [Google Scholar]
- 3.Bading H, Ginty D D, Greenberg M E. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- 4.Belaguli N S, Schildmeyer L A, Schwartz R J. Organization and myogenic restricted expression of the murine serum response factor gene. J Biol Chem. 1997;272:18222–18231. doi: 10.1074/jbc.272.29.18222. [DOI] [PubMed] [Google Scholar]
- 5.Bikfalvi A, Klein S, Pintucci G, Rifkin D B. Biological roles of fibroblast growth factor-2. Endocrine Rev. 1997;18:26–45. doi: 10.1210/edrv.18.1.0292. [DOI] [PubMed] [Google Scholar]
- 6.Bodmer R. The gene tinman is required for speciation of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 7.Cao X, Mahendran R, Guy G R, Tan Y H. Detection and characterization of cellular EGR-1 binding to its recognition site. J Biol Chem. 1993;268:16949–16957. [PubMed] [Google Scholar]
- 8.Changelian P S, Feng P, King T C, Milbrandt J. Structure of the NGFI-A gene and detection of upstream sequences responsible for its transcriptional induction by nerve growth factor. Proc Natl Acad Sci USA. 1989;86:377–381. doi: 10.1073/pnas.86.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chavrier P, Janssen-Timmen U, Mattei M-G, Zerial M, Bravo R, Charnay P. Structure, chromosome location, and expression of the mouse zinc finger gene Krox-20: multiple gene products and coregulation with the proto-oncogene c-fos. Mol Cell Biol. 1989;9:787–797. doi: 10.1128/mcb.9.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen B K, Liu Y W, Yamamoto S, Chang W C. Overexpression of Ha-ras enhances the transcription of human arachidonate 12-lipoxygenase promoter in A431 cells. Biochim Biophys Acta. 1997;1344:270–277. doi: 10.1016/s0005-2760(96)00151-8. [DOI] [PubMed] [Google Scholar]
- 11.Chen C Y, Croissant J, Majesky M, Topouzis S, McQuinn T, Frankovsky M J, Schwartz R J. Activation of the cardiac α-actin promoter depends upon serum response factor, Tinman homologue, Nkx-2.5, and intact serum response elements. Dev Genet. 1996;19:119–130. doi: 10.1002/(SICI)1520-6408(1996)19:2<119::AID-DVG3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 12.Chen C Y, Schwartz R J. Competition between negative acting YY1 versus positive acting serum response factor and tinman homologue Nkx-2.5 regulates cardiac α-actin promoter activity. Mol Endocrinol. 1997;11:812–822. doi: 10.1210/mend.11.6.0015. [DOI] [PubMed] [Google Scholar]
- 13.Chen C Y, Schwartz R J. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol Cell Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christy B, Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Mol Cell Biol. 1989;86:8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christy B, Nathans D. Functional serum response elements upstream of the growth factor-inducible gene zif268. Mol Cell Biol. 1989;9:4889–4895. doi: 10.1128/mcb.9.11.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coso O A, Chiariello M, Yu J-C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-Binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 18.Croissant J D, Kim J H, Eichele G, Goering L, Lough J, Prywes R, Schwartz R J. Avian serum response factor expression restricted primarily to muscle cell lineages is required for α-actin gene transcription. Dev Biol. 1996;177:250–264. doi: 10.1006/dbio.1996.0160. [DOI] [PubMed] [Google Scholar]
- 19.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique restriction site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 20.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;5:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 22.Fromm C, Coso O A, Montaner S, Xu N, Gutkind J S. The small GTP-binding protein Rho links G protein-coupled receptors and G-α12 to the serum response element and to cellular transformation. Proc Natl Acad Sci USA. 1997;94:10098–10103. doi: 10.1073/pnas.94.19.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujii M, Tsuchiya H, Chuhjo T, Akizawa T, Seiki M. Interaction of HTLV-1 Tax-1 with p67SRF causes the aberrant induction of cellular immediate early genes through CArG boxes. Genes Dev. 1992;6:2066–2067. doi: 10.1101/gad.6.11.2066. [DOI] [PubMed] [Google Scholar]
- 24.Fujii M, Tsuchiya H, Chujo T, Minamino T, Miyamoto K, Seiki M. Serum response factor has functional roles both in indirect binding to the CArG box and in the transcriptional activation function of human T-cell leukemia virus type I Tax. J Virol. 1994;68:7275–7283. doi: 10.1128/jvi.68.11.7275-7283.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gauthier-Rouviere C, Cavadore J-C, Blanchard J-M, Lamb N J C, Fernandez A. p67SRF is a constitutive nuclear protein implicated in the modulation of genes required throughout the G1 period. Cell Regul. 1991;2:575–588. doi: 10.1091/mbc.2.7.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham R, Gilman M Z. Distinct protein targets for signals acting at the c-fos serum response element. Science. 1991;251:189–192. doi: 10.1126/science.1898992. [DOI] [PubMed] [Google Scholar]
- 27.Groisman R, Masutani H, Leibovitch M-P, Robin P, Soudant I, Trouche D, Harel-Bellan A. Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J Biol Chem. 1996;271:5258–5264. doi: 10.1074/jbc.271.9.5258. [DOI] [PubMed] [Google Scholar]
- 28.Grueneberg D A, Natesan S, Alexandre C, Gilman M Z. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of the serum response factor. Science. 1992;257:1089–1095. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- 29.Hill C S, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 31.Hill C S, Wynne J, Treisman R. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 1994;13:5421–5432. doi: 10.1002/j.1460-2075.1994.tb06877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansen F E, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khachigian L M, Williams A J, Collins T. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. J Biol Chem. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 33a.Krainc, D., and R. Misra. Unpublished observations.
- 34.Latinkic B, O’Brien T, Lau L. Promoter function and structure of the growth factor-inducible immediate early gene cyr61. Nucleic Acids Res. 1991;19:3261–3267. doi: 10.1093/nar/19.12.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latinkic B V, Lau L F. Transcriptional activation of the immediate early gene pip92 by serum growth factors requires both Ets and CArG-like elements. J Biol Chem. 1994;269:23163–23170. [PubMed] [Google Scholar]
- 36.Liao J, Hodge C, Meyer D, Ho P S, Rosenspire K, Schwartz J. Growth hormone regulates ternary complex factors and serum response factor associated with the c-fos serum response element. J Biol Chem. 1997;272:25951–25958. doi: 10.1074/jbc.272.41.25951. [DOI] [PubMed] [Google Scholar]
- 37.Liu C, Calogero A, Ragona G, Adamson E, Mercola D. EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-β1 is postulated to account for this suppressor activity. Crit Rev Oncog. 1996;7:101–125. [PubMed] [Google Scholar]
- 38.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 39.McDonough P M, Hanford D S, Sprenkle A B, Mellon N R, Glembotski C C. Collaborative roles for c-Jun N-terminal kinase, c-Jun, serum response factor, and Sp1 in calcium-regulated myocardial gene expression. J Biol Chem. 1997;272:24046–24053. doi: 10.1074/jbc.272.38.24046. [DOI] [PubMed] [Google Scholar]
- 40.Miranti C K, Ginty D D, Huang G, Chatila T, Greenberg M E. Calcium activates serum response factor-dependent transcription by a Ras- and Elk-1-independent mechanism that involves a CaM kinase. Mol Cell Biol. 1995;15:3672–3684. doi: 10.1128/mcb.15.7.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misra R, Bonni A, Miranti C K, Rivera V M, Sheng M, Greenberg M E. L-type voltage sensitive calcium channel activation stimulates gene expression by a serum response factor-dependent pathway. J Biol Chem. 1994;269:25483–25493. [PubMed] [Google Scholar]
- 41a.Misra, R. P. Unpublished observations.
- 42.Misra R P, Rivera V M, Wang J M, Fan P-D, Greenberg M E. The serum response factor is extensively modified by phosphorylation following its synthesis in serum-stimulated fibroblasts. Mol Cell Biol. 1991;11:4545–4554. doi: 10.1128/mcb.11.9.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molkentin J D, Black B L, Martin J F, Olson E N. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol Cell Biol. 1996;16:2627–2636. doi: 10.1128/mcb.16.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natesan S, Gilman M. YY1 facilitates the association of serum response factor with the c-fos serum response element. Mol Cell Biol. 1995;15:5975–5982. doi: 10.1128/mcb.15.11.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 46.Parlow M H, Bolender D L, Kokan-Moore N P, Lough J. Localization of bFGF-like proteins as punctate inclusions in the preseptation myocardium of the chicken embryo. Dev Biol. 1991;146:139–147. doi: 10.1016/0012-1606(91)90454-b. [DOI] [PubMed] [Google Scholar]
- 47.Rivera V M, Miranti C K, Misra R P, Ginty D D, Chen R-H, Blenis J, Greenberg M E. A growth factor induced kinase phosphorylates the serum response factor at a site that regulates its DNA binding activity. Mol Cell Biol. 1993;13:6260–6273. doi: 10.1128/mcb.13.10.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 49.Sassone-Corsi P, Der C J, Verma I M. Ras-induced neuronal differentiation of PC12 cells: possible involvement of Fos and Jun. Mol Cell Biol. 1989;9:3174–3183. doi: 10.1128/mcb.9.8.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sealy L, Malone D, Pawlak M. Regulation of c-fos serum response element by C/EBPβ. Mol Cell Biol. 1997;17:1744–1755. doi: 10.1128/mcb.17.3.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheng M, Dougan S T, McFadden G, Greenberg M E. Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol Cell Biol. 1988;8:2787–2796. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shyu A-B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 52a.SIGSCAN Website. [Online.] Transfac database, version 2.5. http://bimas.dcrt.nih.gov/molbio/signal/. [27 March 1999, last date accessed.] [D. S. Prestridge. 1991. A computer program that scans DNA sequences for eukaryotic transcriptional elements. CABIOS 7:203–206.] 1991. [DOI] [PubMed] [Google Scholar]
- 53.Spencer J A, Misra R P. Expression of the serum response factor gene is regulated by serum response factor binding sites. J Biol Chem. 1996;271:16535–16543. doi: 10.1074/jbc.271.28.16535. [DOI] [PubMed] [Google Scholar]
- 53a.Spencer, J. A., and R. P. Misra. Submitted for publication.
- 54.Sugi Y, Sasse J, Lough J. Inhibition of precardiac mesoderm cell proliferation by antisense oligodeoxynucleotide complementary to fibroblast growth factor-2 (FGF-2) Dev Biol. 1993;157:28–37. doi: 10.1006/dbio.1993.1109. [DOI] [PubMed] [Google Scholar]
- 55.Treisman R. The SRE: a growth factor responsive transcriptional regulator. Cancer Biol. 1990;1:47–58. [PubMed] [Google Scholar]
- 56.Treisman R. Ternary complex factors: growth factor regulated transcriptional activators. Curr Opin Genet Dev. 1994;4:96–101. doi: 10.1016/0959-437x(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 57.Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 58.Treisman R, Ammerer G. The SRF and MCM1 transcription factors. Curr Opin Genet Dev. 1992;2:221–226. doi: 10.1016/s0959-437x(05)80277-1. [DOI] [PubMed] [Google Scholar]
- 59.Tsai-Morris C-H, Cao X, Sukhatme V P. 5′ flanking sequence and genomic structure of Egr-1, a murine mitogen inducible zinc finger encoding gene. Nucleic Acids Res. 1988;16:8835–8846. doi: 10.1093/nar/16.18.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vandromme M, Gauthier-Rouviere C, Gilles C, Lamb N, Fernandez A. Serum response factor p67SRF is expressed and required during myogenic differentiation of both mouse C2 and rat L6 muscle cell lines. J Cell Biol. 1992;118:1489–1500. doi: 10.1083/jcb.118.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Visvader J, Sassone-Corsi P, Verma I. Two adjacent promoter elements mediate nerve growth factor activation of the c-fos gene and bind distinct nuclear complexes. Proc Natl Acad Sci USA. 1988;85:9474–9478. doi: 10.1073/pnas.85.24.9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yant S R, Zhu W, Millinoff D, Slightom J L, Goodman M, Gumucio D L. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human β-globin cluster. Nucleic Acids Res. 1995;23:4353–4362. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu C, Johansen F E, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zinck R, Hipskind R A, Pingoud V, Nordheim A. c-fos transcriptional activation and repression correlate temporally with the phosphorylation status of TCF. EMBO J. 1993;12:2377–2387. doi: 10.1002/j.1460-2075.1993.tb05892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]