Jones and Drummond et al. identify the C1 domain from the polarity kinase aPKC as a membrane recruitment module. Regulation of C1-mediated membrane recruitment by several other domains, including aPKC’s catalytic module, leads to polarized membrane targeting.
Abstract
Recruitment of the Par complex protein atypical protein kinase C (aPKC) to a specific membrane domain is a key step in the polarization of animal cells. While numerous proteins and phospholipids interact with aPKC, how these interactions cooperate to control its membrane recruitment has been unknown. Here, we identify aPKC’s C1 domain as a phospholipid interaction module that targets aPKC to the membrane of Drosophila neural stem cells (NSCs). The isolated C1 binds the NSC membrane in an unpolarized manner during interphase and mitosis and is uniquely sufficient among aPKC domains for targeting. Other domains, including the catalytic module and those that bind the upstream regulators Par-6 and Bazooka, restrict C1’s membrane targeting activity—spatially and temporally—to the apical NSC membrane during mitosis. Our results suggest that aPKC polarity results from cooperative activation of autoinhibited C1-mediated membrane binding activity.
Introduction
The Par complex polarizes animal cells by excluding specific cortical factors from the Par cortical domain (Lang and Munro, 2017; Venkei and Yamashita, 2018). During polarization, the proteins Par-6 and atypical protein kinase C (aPKC), which make up the complex, are recruited to a specific, continuous region of the cell membrane, such as the apical surface of epithelia (Tepass, 2012), the anterior hemisphere of the C. elegans zygote (Nance and Zallen, 2011), or the apical hemisphere of Drosophila neural stem cells (NSCs; Prehoda, 2009). Factors that are directly polarized by the Par complex, such as Miranda and Numb, are aPKC substrates (Atwood and Prehoda, 2009; Smith et al., 2007). Phosphorylation is coupled to removal from the Par domain, causing these substrates to localize to a complementary membrane domain (Bailey and Prehoda, 2015). Thus, the pattern of Par-polarized factors is ultimately determined by the mechanisms that specify aPKC’s membrane recruitment and activation.
Many aPKC interactions with proteins and phospholipids have been identified (Fig. 1 A), although how they collaborate to polarize aPKC remains poorly understood. Bazooka (Baz aka Par-3) and Par-6 form direct physical contacts with aPKC, and each protein has possible pathways for membrane recruitment: Baz through direct interactions with the membrane and Par-6 through interactions with prenylated Cdc42 (Joberty et al., 2000; Lin et al., 2000; Krahn et al., 2010). Several direct interactions with phospholipids have also been reported, including with ceramide (Wang et al., 2005), sphingosine-1-phosphate (Kajimoto et al., 2019), and phosphoinositides (Dong et al., 2020; Standaert et al., 1997). The aPKC catalytic domain may also play a role in membrane recruitment as perturbations in this domain can cause aPKC to become depolarized (Rodriguez et al., 2017; Hannaford et al., 2019).
Figure 1.
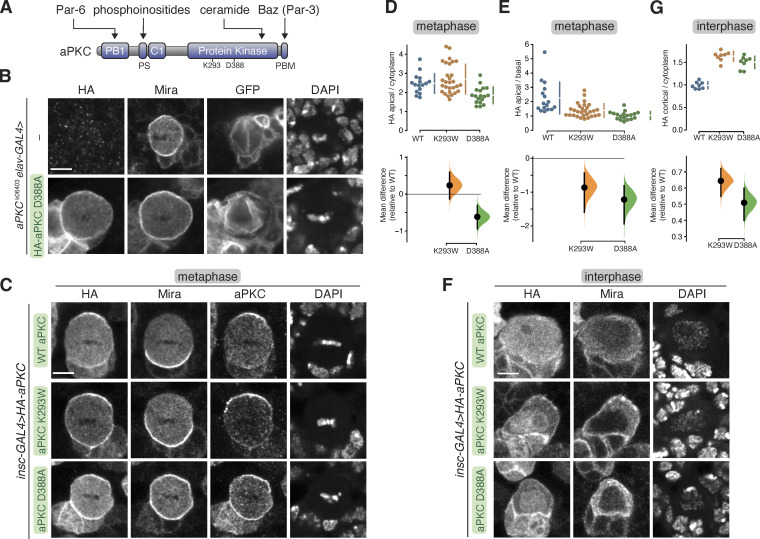
Localization of aPKC with kinase inactivating mutations in larval brain NSCs. (A) Domain structure of aPKC showing the location of PB1, PS (pseudosubstrate), C1, kinase domain, PBM (PDZ binding motif), along with the location of K293 and D388 residues. (B) Localization of HA-tagged aPKC harboring the D388A kinase inactivating mutation in metaphase, positively marked (mCD8-GFP) aPKCk06403 mutant larval brain NSC with an aPKCK06403 mutant larval brain NSC shown for comparison. Nucleic acids are shown with DAPI. The scale bar is 5 µm in all panels. (C) Localization of HA-tagged aPKC harboring either the D388A or K293W kinase inactivation mutations in metaphase larval brain NSCs with endogenous aPKC. The basal cortical marker Miranda, total aPKC (“aPKC,” endogenous and exogenously expressed) and nucleic acid (DAPI) are also shown. (D and E) Gardner-Altman estimation plots of the effect of the D388A and K293W mutations on metaphase aPKC membrane recruitment. Apical cortical to cytoplasmic (D) and apical/basal (E) signal intensities of anti-HA signals are shown for individual metaphase NSCs expressing either HA-WT or HA-D388A or HA-K293W aPKC. The error bar in the upper graph represents one standard deviation (gap is mean); the error bar in the lower graph represents bootstrap 95% confidence interval; n = 16 (from six distinct larval brains), 29 (8), 18 (4) for WT, K293W, D388A, respectively. (F) Localization of HA-tagged aPKC harboring either the D388A or K293W kinase inactivation mutations in interphase larval brain NSCs with endogenous aPKC. The basal cortical marker Miranda and nucleic acid (DAPI) are also shown. (G) Gardner–Altman estimation plot of the effect of the D388A and K293W mutations on interphase aPKC membrane recruitment. Cortical to cytoplasmic cortical signal intensities of anti-HA signals are shown for individual metaphase NSCs expressing either HA-WT or HA-D388A or HA-K293W aPKC. The error bar in upper graph represents one standard deviation (gap is mean); the error bar in the lower graph represents bootstrap 95% confidence interval; n = 8 for WT (from three distinct larval brains), K293W (3), and D388A (2).
While interactions that could potentially recruit aPKC to the membrane have been identified, what is missing is an understanding of how the interactions function together to polarize aPKC at the proper time. One possibility is an avidity model where each interaction is weak, unable to recruit aPKC to the membrane on its own, but the energy provided by multiple interactions allows for recruitment. Alternately, one or more interactions could be sufficient for recruitment, but somehow regulated to ensure that targeting only occurs when the appropriate cues are present. These models could be distinguished by determining if any interactions are sufficient for targeting.
We used Drosophila NSCs to investigate how aPKC is recruited to the membrane during polarization (Homem and Knoblich, 2012). During interphase, aPKC is cytoplasmic in NSCs but becomes targeted to the apical hemisphere early in mitosis, ultimately concentrating near the apical pole before depolarizing and returning to the cytoplasm as the division completes (Oon and Prehoda, 2019). The highly dynamic nature of the NSC polarity cycle makes it possible to assess both spatial (polarized, depolarized, or cytoplasmic) and temporal (interphase or mitotic) aspects of aPKC membrane recruitment regulation.
Results
Mutations that inactivate catalytic activity depolarize aPKC
We began our examination of aPKC membrane targeting mechanisms in NSCs by evaluating the role of the catalytic domain. At metaphase, aPKC is highly enriched at the apical membrane of larval brain NSCs (Rolls et al., 2003). We examined the effect of mutations that inactivate aPKC’s catalytic activity on its localization in cells that lacked endogenous aPKC (apkcK06403 in positively marked clones; Lee and Luo, 1999). Besides aPKC’s localization, we also examined its activity in these metaphase NSCs by determining the localization of Miranda (Mira), an aPKC substrate that is normally restricted to the basal cortex by apical aPKC activity (Fig. 1 B; Atwood and Prehoda, 2009; Ikeshima-Kataoka et al., 1997). Consistent with previous observations, we found that in metaphase apkcK06403 NSCs, Mira was depolarized (Fig. 1 B). Expression of wild-type aPKC restores the apical aPKC and basal Mira localization found in normally functioning NSCs to positively marked aPKCK06403 metaphase null clones (Holly et al., 2020).
To examine the effect of the perturbing catalytic activity, we expressed aPKC harboring a mutation (D388A) that does not have detectable activity in an in vitro protein kinase assay (Holly and Prehoda, 2019). This mutation alters a residue that coordinates the γ-phosphate of ATP and is thought to allow ATP to bind but prevent phosphotransfer (Cameron et al., 2009). Unlike wild-type aPKC, which is restricted to the apical domain at metaphase, we found that aPKC D388A was largely depolarized, localizing along the entire cortex of positively marked aPKCK06403 metaphase null clones (Fig. 1 B). The membrane localization was somewhat unevenly distributed, possibly owing to the presence of a fine membrane structure (LaFoya and Prehoda, 2021, 2023). Mira was also depolarized in these cells, confirming that aPKC D388A is inactive both in vitro (Holly and Prehoda, 2019) and in vivo (Fig. 1 B). We conclude that inactivation of the aPKC catalytic domain depolarizes aPKC in metaphase NSCs.
Kinase inactive aPKC is not polarized by endogenous aPKC
Our results indicate that mutations that perturb aPKC’s catalytic activity also influence its localization. Previous examinations of chemically inhibited aPKC also found that perturbing catalytic activity caused aPKC to localize to the membrane but in a depolarized manner (Rodriguez et al., 2017; Hannaford et al., 2019). The depolarization caused by perturbations to the kinase domain could be explained if aPKC’s catalytic activity directly participated in its own localization (e.g., by a feedback mechanism). Alternately, perturbations in the kinase domain could alter other aPKC functions. Besides catalyzing phosphotransfer, the aPKC kinase domain also binds a pseudosubstrate in its NH2 terminal region, causing autoinhibition (Graybill et al., 2012). Mutations or small molecules that influence the active site could perturb the intramolecular interaction in addition to inhibiting catalytic activity. To determine whether the loss of aPKC’s catalytic activity is responsible for the localization defects of aPKC D388A, we examined whether the presence of wild-type, endogenous aPKC with its normal level of catalytic activity could restore aPKC D388A polarity. We also tested the localization of a well-characterized kinase inactive mutation K293W, which blocks ATP binding (Graybill et al., 2012). In NSCs containing endogenous aPKC, both aPKC D388A and K293W remained enriched at the membrane (Fig. 1, C and D) but depolarized (Fig. 1, C and E), indicating that aPKC catalytic activity is not sufficient to restore polarity to these proteins. Interestingly, in cells expressing aPKC K293W, Mira was basally polarized, but in cells expressing aPKC D388A, it was depolarized, suggesting that aPKC D388A influences the localization or activity of endogenous aPKC (Fig. 1 C). We do not know the origin of the differential effects of aPKC K293W and aPKC D388A on Mira localization, but it may arise from differences in the amounts of the two proteins and how endogenous aPKC is affected.
We also examined the localization of the aPKC variants during interphase, when membrane-bound aPKC is normally not detectable, and found that the kinase-inactivating mutations were predominantly found in the nucleus, presumably due to an embedded nuclear localization signal (Perander et al., 2001; Seidl et al., 2012). However, we also found that the aPKC kinase domains variants were enriched at the membrane relative to the cytoplasm (at sites away from progeny cell contacts) although at a level somewhat lower than apical WT protein at metaphase (Fig. 1, F and G).
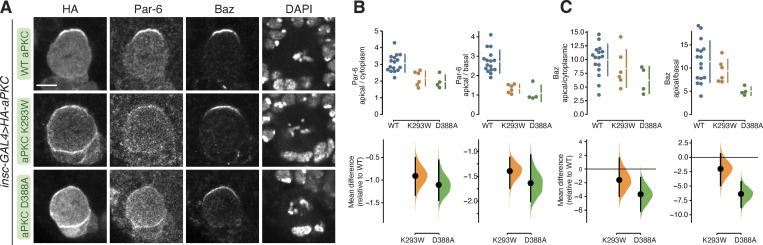
Baz and Par-6 are normally found at the apical membrane with aPKC at metaphase. We examined whether the kinase-inactive aPKC mutants influence Baz or Par-6 localization (Fig. 2). We found that Baz remained apically polarized in cells expressing aPKC, as well as those expressing the aPKC D388A or aPKC K293W variants, although the intensity of this crescent was slightly reduced in aPKC D388A (Fig. 2, A and C). While Baz localization was unperturbed by the expression of the kinase-inactive aPKC variants, Par-6 expanded into the basal domain like the localization of aPKC D388A and aPKC K293W (Fig. 2, A and B). We conclude that expression of kinase-inactive aPKC leads to loss of Par-6 polarity but does not influence the localization of Baz. Depolarization of Par-6 but not Baz has also been observed when aPKC was chemically inhibited (Hannaford et al., 2019).
Figure 2.
Localization of Bazooka and Par-6 in larval brain NSCs expressing kinase-inactive aPKCs. (A) Localization of Par-6 and Bazooka (Baz) in metaphase larval brain NSCs expressing HA-tagged aPKC D388A or aPKC K293W. Nucleic acids are shown with DAPI. Scale bar is 5 µm. (B and C) Gardner–Altman estimation plots of the effect of expressing aPKC D388A or aPKC K293W on Par-6 (B) or Baz (C) cortical localization and polarity. Apical cortical to cytoplasmic or basal cortical signal intensities of cortical and cytoplasmic signals are shown for individual metaphase NSCs expressing either HA-WT or HA-D388A or HA-K293W aPKC. The error bar in the upper graphs represents one standard deviation (gap is mean); the error bar in lower graphs represents bootstrap 95% confidence interval; n = 16 (from three distinct larval brains), 6 (3), 4 (3) for WT, K293W, D388A, respectively (Par-6) and 16, 6, 4 for Baz.
Kinase inactive aPKC membrane recruitment is less sensitive to the loss of Cdc42 and Bazooka
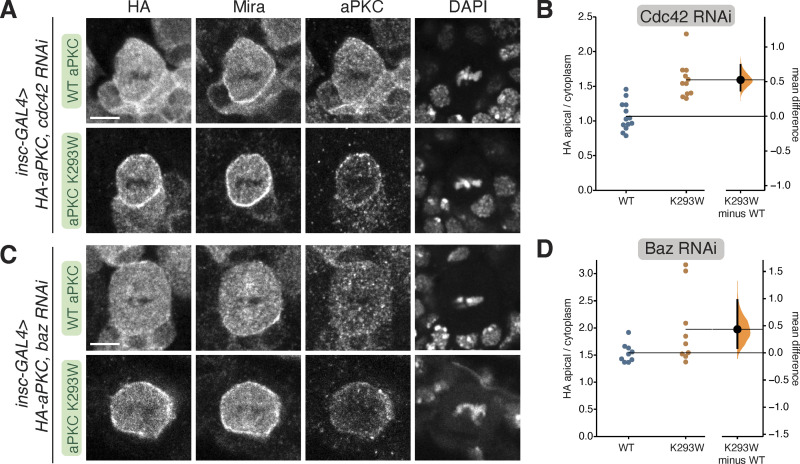
Membrane targeting of aPKC normally requires the activities of Baz and the small GTPase Cdc42 (Wodarz et al., 2000; Rolls et al., 2003; Atwood et al., 2007). We tested whether these upstream regulators are required for membrane localization of kinase-inactive aPKC by examining the localization of aPKC K293W in NSCs expressing Baz or Cdc42 RNAi. We found that wild-type aPKC membrane enrichment was reduced in these contexts, as previously reported (Fig. 3, A–D; Atwood et al., 2007; Rolls et al., 2003). We also observed a reduction of aPKC K293W on the membrane in NSCs expressing Cdc42 or Baz RNAi, but less so than for WT aPKC (Fig. 3, A–D), suggesting that aPKC K293W membrane recruitment is less sensitive to the loss of Cdc42 or Baz.
Figure 3.
Cortical localization of kinase-inactive aPKC in NSCs lacking Bazooka or Cdc42. (A) Localization of HA-tagged aPKC K293W in metaphase larval brain NSCs expressing an RNAi directed against Cdc42. The scale bar is 5 µm in all panels. (B) Gardner–Altman estimation plots of the effect of expressing Cdc42 RNAi on WT and K293W aPKC cortical localization. Apical cortical to cytoplasmic signal intensities of anti-HA signals are shown for individual metaphase NSCs expressing either HA-WT or HA-K293W aPKC. Error bar represents bootstrap 95% confidence interval; n = 13 (from five distinct larval brains), 11 (5) for WT, and K293W, respectively. (C) Localization of HA-tagged aPKC K293W in metaphase larval brain NSCs expressing an RNAi directed against Bazooka. (D) Gardner–Altman estimation plots of the effect of expressing Baz RNAi on WT and K293W aPKC cortical localization. Apical cortical to cytoplasmic signal intensities of anti-HA signals are shown for individual metaphase NSCs expressing either HA-WT or HA-K293W aPKC. Error bar represents bootstrap 95% confidence interval; n = 9 (from two distinct larval brains), 9 (6) for WT and K293W, respectively.
The aPKC C1 domain is a membrane-targeting module
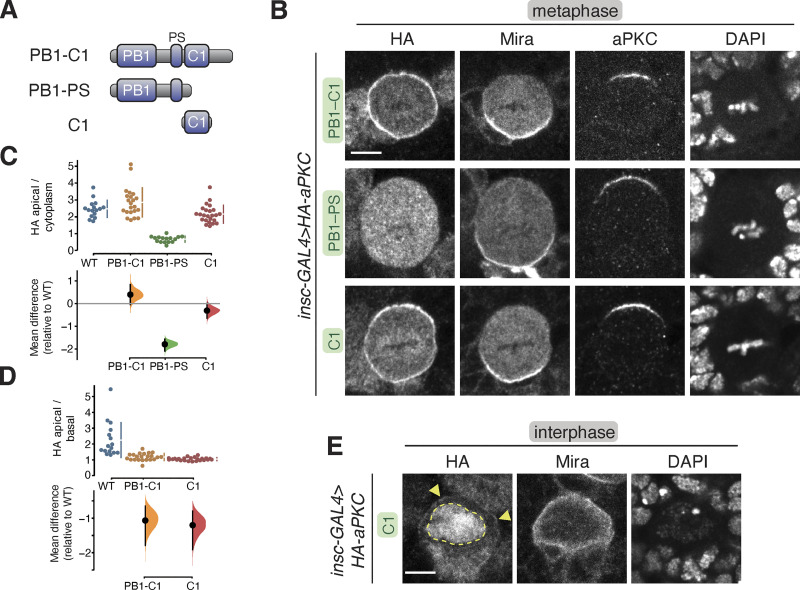
Our results indicate that the depolarized membrane localization of aPKC with inactive kinase domains (e.g., K293W) could be at least partially independent of both Baz and Cdc42. One model consistent with these results is that aPKC contains a membrane targeting module that is regulated by its kinase domain, along with Baz and Cdc42 binding. To identify the putative module, we first examined whether removing the kinase domain leads to the same depolarized membrane localization phenotype. We found that aPKC PB1-C1 (aka ∆KD; Fig. 4 A) was enriched at the membrane and not polarized (Fig. 4, B–D). A similar localization pattern has been reported for aPKC PB1-C1 expressed in cultured cells (Dong et al., 2020). Thus, aPKC’s NH2-terminal regulatory region, consisting of PB1, PS, and C1 domains, is responsible for membrane binding. The PB1 could mediate interaction with the membrane via protein–protein interactions with Par-6 (Atwood et al., 2007; Petronczki and Knoblich, 2001), the PS domain through direct interactions with lipids (Dong et al., 2020), or the C1 domain, which serves as a membrane-targeting module in other PKCs (Colón-González and Kazanietz, 2006) but has not been reported to do so in aPKCs. Removal of the C1 domain from the regulatory domain (aPKC PB1-PS) leaving the PB1 and PS domains (Fig. 4 A) resulted in a protein that was not enriched at the membrane (Fig. 4, B–D). Thus, the PB1 and PS domains are not sufficient for membrane targeting in NSCs. We next tested the C1 domain alone and found that it was enriched at the membrane at metaphase (Fig. 4, B–D). We also observed C1 enrichment at the membrane relative to the cytoplasm during interphase (Fig. 4 E), although the domain was predominantly found in the nucleus, presumably due to an embedded nuclear localization signal (Perander et al., 2001; Seidl et al., 2012). We conclude that the aPKC C1 domain is a membrane-targeting module.
Figure 4.
Localization of the aPKC regulatory domain in larval brain NSCs. (A) aPKC regulatory domain fragments. (B) Localization of HA-tagged aPKC regulatory domain fragments in metaphase larval brain NSCs. The basal marker Miranda, endogenous aPKC (using an antibody that does not react with the regulatory domain), and nucleic acids (DAPI) are shown for comparison. Scale bar is 5 µm. (C and D) Gardner–Altman estimation plot of aPKC regulatory domain cortical localization (C) and polarity (D). Apical cortical to cytoplasmic (C) and apical cortical to basal cortical signal intensity ratios (D) of anti-HA signals are shown for individual metaphase NSCs expressing either aPKC PB1-C1, PB1-PS, or C1 regulatory domain fragments. The data for wild type is the same as in Fig. 1. Apical to basal ratios are only shown for proteins with detectable membrane signals. Error bar in the upper graphs represents one standard deviation (gap is mean); the error bar in the lower graphs represents bootstrap 95% confidence interval; n = 16 (from six distinct larval brains), 22 (6), 16 (5), 24 (9) for WT, PB1-C1, PB1-PS, and C1, respectively. (E) Localization of the HA-tagged aPKC C1 domain in interphase larval brain NSCs. Arrowheads highlight the membrane signal, and the nuclear signal is outlined by a dashed line.
C1 is a lipid-binding module that is required for aPKC membrane recruitment
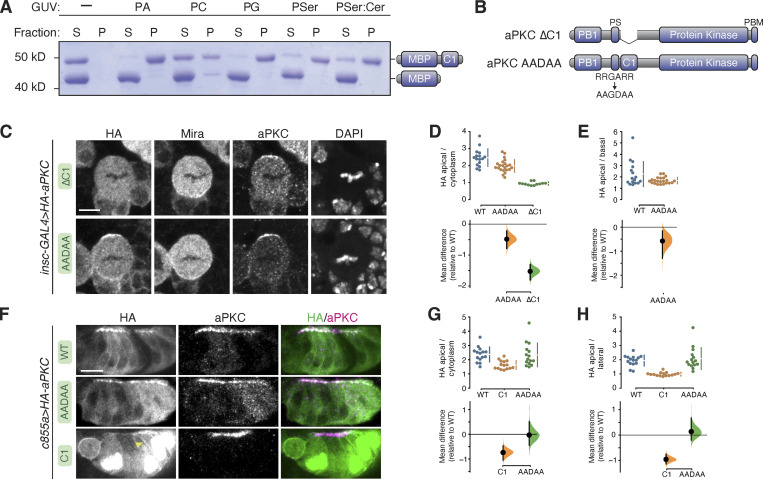
How might the C1 domain mediate interaction with the NSC membrane? C1 domains from canonical PKCs bind diacylglycerol (DAG), although the aPKC C1 domain does not bind DAG (Colón-González and Kazanietz, 2006). However, we sought to determine if aPKC’s C1 domain binds other phospholipids. We used a vesicle pelleting assay in which Giant Unilamellar Vesicles (GUVs) with varying phospholipid compositions were mixed with purified aPKC C1 domain. The vesicles were separated from the soluble phase by ultracentrifugation and any associated C1 was identified by protein gel electrophoresis. We observed varying degrees of C1 binding to a broad array of phospholipids (Fig. 5 A), suggesting that the C1 is a nonspecific phospholipid binding module.
Figure 5.
Phospholipid binding of aPKC C1 domain and role of C1 and PS domains in aPKC localization in larval brain NSCs and epithelia. (A) Binding of a maltose binding protein (MBP) fusion of the aPKC C1 domain to phospholipids. Supernatant (S) and pellet (P) fractions from cosedimentation with Giant Unilamellar Vesicles (GUVs) of the indicated phospholipid composition are shown (PA, phosphatidic acid; PC, phosphatidyl choline; PG, phosphatidyl glycerol; PSer, phosphatidyl serine; PSer:Cer, phosphatidyl serine mixture with ceramide). MBP alone is included as an internal negative control. (B) Schematics of ∆C1 and AADAA aPKC variants. (C) Localization of HA-tagged aPKC ∆C1 and AADAA variants in metaphase larval brain NSCs. The basal marker Miranda, total aPKC (expressed variant and endogenous), and nucleic acids (DAPI) are shown for comparison. The scale bar is 5 µm. (D and E) Gardner–Altman estimation plots of aPKC AADAA and ∆C1 cortical localization in NSCs. Apical cortical to cytoplasmic (D) or apical to basal (E) signal intensity ratios of anti-HA signals are shown for individual metaphase NSCs expressing either aPKC AADAA or ∆C1. The data for wild type is the same as in Fig. 1. Apical to basal ratios are only shown for proteins with detectable membrane signals. Error bar in the upper graphs represents one standard deviation (gap is mean); error bar in the lower graphs represents bootstrap 95% confidence interval; n = 16 (from six distinct larval brains), 22 (8), and 12 (5) for WT, AADAA, and ∆C1, respectively. (F) Localization of HA-tagged aPKC ∆C1 and AADAA variants in larval brain inner proliferation center (IPC) epithelium. Arrowhead highlights aPKC C1 localization at the lateral membrane. As in interphase NSC cells, the C1 is highly enriched in the epithelial nuclei. Scale bar is 5 µm. (G and H) Gardner–Altman estimation plots of aPKC AADAA and C1 cortical localization in IPC epithelial cells. Apical cortical to cytoplasmic (D) or apical to lateral (E) signal intensity ratios of anti-HA signals are shown for individual epithelial cells from the IPC expressing either aPKC AADAA or C1. Error bar in upper graphs represents one standard deviation (gap is mean); error bar in lower graphs represents bootstrap 95% confidence interval; n = 16 (from three distinct larval brains), 15 (3), and 15 (3) for WT, C1, and AADAA, respectively. Source data are available for this figure: SourceData F5.
To better understand the role of the C1 in aPKC polarity, we examined the effect of removing it (aPKC ∆C1; Fig. 5 B) on aPKC’s localization. We found that aPKC ∆C1 remained in the cytoplasm and was not enriched at the membrane (Fig. 5, C–E), leading us to conclude that the C1 is required for aPKC membrane targeting in NSCs. Interestingly, Mira localization was also disrupted in NSCs expressing aPKC ∆C1, suggesting that the C1 also plays a role in regulating aPKC’s protein kinase activity in NSCs. The displacement of cortical Mira in NSCs expressing aPKC ∆C1 is consistent with the increase in catalytic activity observed in in vitro measurements of this protein (Graybill et al., 2012; Zhang et al., 2014).
The PS domain has been reported to be a membrane binding module required for the membrane recruitment of aPKC in cultured cells and epithelia (Dong et al., 2020). Our results suggest that the PS is not sufficient for localization to the NSC membrane as aPKC PB1-PS remains in the cytoplasm. We tested whether the PS is required for NSC aPKC membrane recruitment by examining the localization of aPKC in which the positively charged, basic residues were removed and a negatively charged side chain added (aPKC AADAA; Fig. 5 B). This mutation has been reported to abrogate membrane binding in contexts where the PS is required (Dong et al., 2020). We found that the apical localization of aPKC AADAA in NSCs was slightly reduced compared with WT aPKC, suggesting that the PS contributes to the recruitment process but is not absolutely required for recruitment or polarization in this cellular context (Fig. 5, C–E). Mira localization was cytoplasmic in aPKC AADAA-expressing NSCs, however, consistent with the increase in kinase activity that is expected when the autoinhibitory PS is inactivated (Graybill et al., 2012). We also assessed the localization of aPKC AADAA in the epithelium of the larval brain inner proliferation center by expressing it with the c855a-GAL4 driver. We found this protein to be enriched at the apical membrane of interphase epithelial cells in a similar pattern to wild-type aPKC (Fig. 5, F–H). We also examined the localization of the isolated C1 in interphase epithelial cells and observed a similar pattern to NSCs—predominantly nuclear but also enriched at the membrane relative to the cytoplasm (Fig. 5, F–H).
Regulation of membrane recruitment by the PB1 domain and its interaction with Par-6
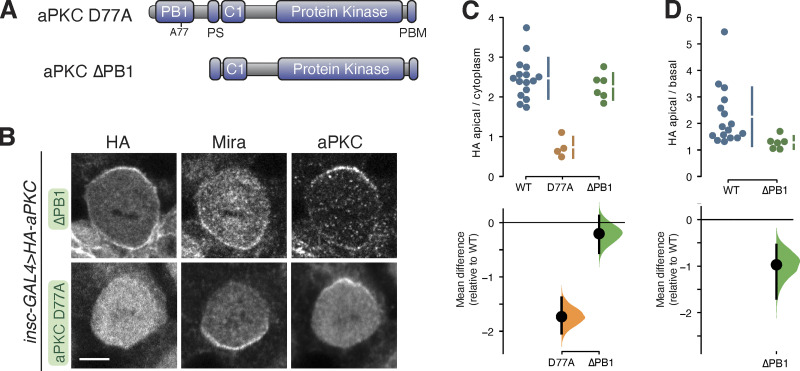
Our results indicate that the recruitment of aPKC to the membrane is mediated primarily by the C1 domain. The membrane binding of the isolated C1 suggests that other aPKC domains regulate the C1 through autoinhibition to yield the dynamic membrane localization of full-length aPKC. In this model, perturbation of a regulatory domain could lead to either cytoplasmic or depolarized, membrane-bound aPKC if the perturbation were to disrupt C1 activation or repression, respectively. Cytoplasmic aPKC has been observed upon inactivation of aPKC’s PDZ binding motif (PBM; Holly et al., 2020) and kinase domain active site perturbations lead to depolarized membrane binding (Fig. 1; Hannaford et al., 2019; Rodriguez et al., 2017). We determined if aPKC’s PB1 domain (Fig. 1 A) participates in C1 regulation by examining the localization of aPKC D77A, a PB1 point mutation that disrupts interaction with Par-6’s PB1 domain (Hirano et al., 2004), and aPKC ∆PB1, which lacks the PB1 entirely (Fig. 6 A). Disrupting the interaction with Par-6 leads to cytoplasmic aPKC in cultured cells (Dong et al., 2020), and we also observed cytoplasmic localization of aPKC D77A in neuroblasts (Fig. 6, B–D). Intriguingly, complete deletion of the PB1 caused a different effect, with aPKC ∆PB1 localizing to the membrane in an unpolarized manner (Fig. 6, B–D). The distinct localization resulting from these two perturbations indicates that the PB1 is required to repress aPKC membrane recruitment, and PB1’s interaction with Par-6 is required to overcome this regulation.
Figure 6.
Localization of aPKC with PB1 domain perturbations in larval brain NSCs. (A) Schematics of D77A and ∆PB1 aPKC variants. (B) Localization of HA-tagged aPKC D77A and ∆PB1 variants in metaphase larval brain NSCs. The basal marker Miranda, and total aPKC (expressed variant and endogenous), are shown for comparison. Scale bar is 5 µm. (C and D) Gardner-Altman estimation plots of aPKC D77A and ∆PB1 cortical localization. Apical cortical to cytoplasmic (C) or apical to basal (D) signal intensity ratios of anti-HA signals are shown for individual metaphase NSCs expressing either aPKC D77A or ∆PB1. The data for wild type is the same as in Fig. 1. Apical to basal ratios are only shown for proteins with detectable membrane signal. The error bar in the upper graphs represents one standard deviation (gap is mean); the error bar in lower graphs represents bootstrap 95% confidence interval; n = 16 (from six distinct larval brains), 4 (1), and 6 (4) for WT, D77A, and ∆PB1, respectively.
Discussion
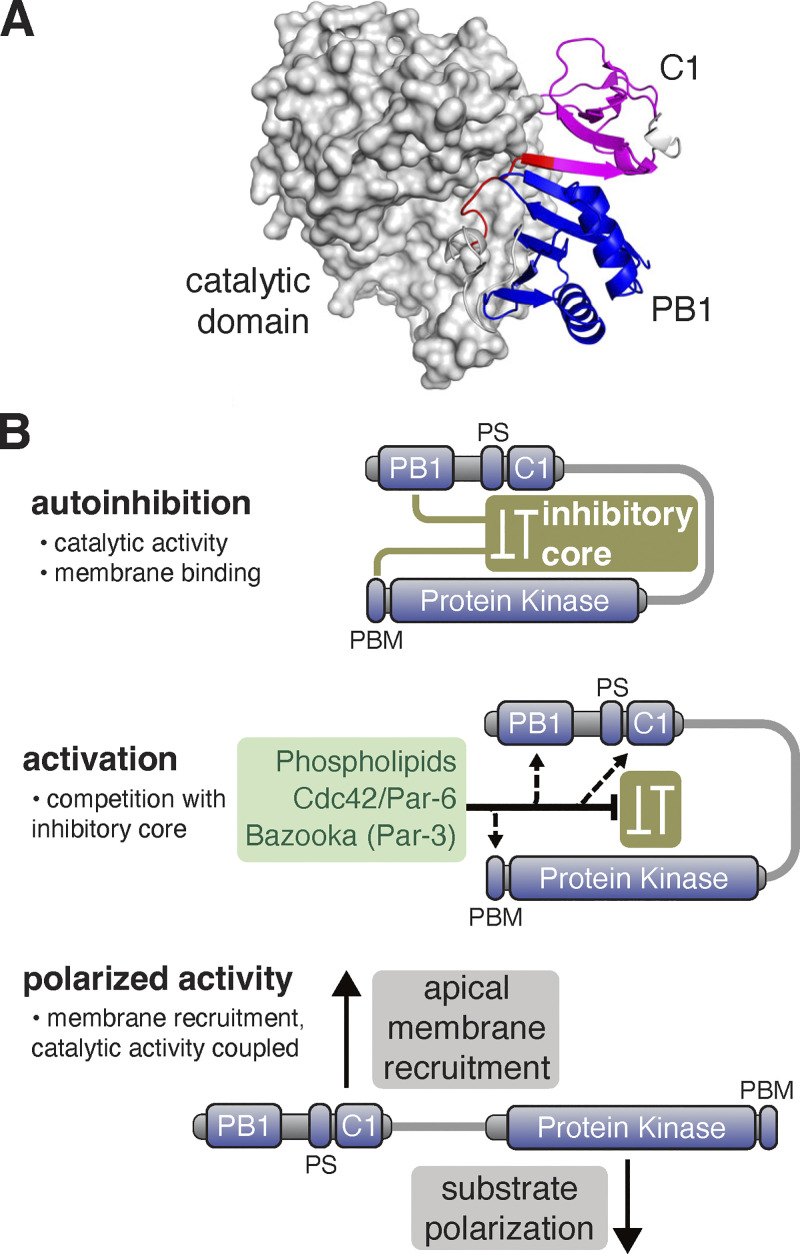
The Par complex component aPKC undergoes a dynamic localization cycle in NSCs, targeting the apical membrane briefly in mitosis and returning to the cytoplasm as division completes (Oon and Prehoda, 2019). The function of interphase, cytoplasmic localization is unknown, but the apical localization of aPKC during mitosis is necessary for the polarization of fate determinants (Atwood et al., 2007; Prehoda, 2009; Rolls et al., 2003), a prerequisite for asymmetric cell division. Regulated membrane recruitment of aPKC is a central aspect of Par-mediated polarity and many physical interactions between aPKC and proteins and phospholipids have been identified. Conceptually, targeting could occur through the concerted action of multiple weak interactions (i.e., avidity). However, we discovered that the aPKC C1 domain is a phospholipid binding module sufficient for membrane recruitment, whereas domains that mediate protein–protein interactions or other interactions with phospholipids are not sufficient for aPKC targeting. As the C1 is the only domain within aPKC with this capability in NSCs, our results suggest that other aPKC domains, including the catalytic domain, function, at least in part, to regulate the membrane recruitment activity of the C1. While the PS and C1 domains are both known to form intramolecular interactions with the catalytic domain (Graybill et al., 2012; Zhang et al., 2014), a predicted structure of aPKC suggests that there are also significant intramolecular interactions within the regulatory PB1-PS-C1 module (Fig. 7 A; Jumper et al., 2021; Varadi et al., 2022). Taken collectively, we propose that cooperative activation of the C1 leads to the spatially and temporally controlled localization of aPKC observed in many animal cells.
Figure 7.
Model for regulation of aPKC activity and membrane recruitment. (A) Alphafold database structure (UNIPROT: A1Z9X0) of aPKC showing the putative supramolecular architecture of PB1 (blue), PS (red), and C1 (magenta) domains and interaction of both domains with the catalytic domain. (B) Model for cooperative polarization and activation of aPKC. An inhibitory core couples repression of catalytic activity (protein kinase domain) to membrane localization (C1 with some contribution from PS). The PB1 and PBM are also coupled to the inhibitory core to allow for cooperative activation by Cdc42/Par-6 binding to the PB1 and Baz binding to the PBM. Disruption of the inhibitory core leads to spatially (apical) and temporally (mitotic) regulated localization and activation of catalytic activity.
The C1 is unique in its ability to promote aPKC membrane recruitment in NSCs. While numerous interactions between phospholipids and domains outside the C1 have been reported (Wang et al., 2005; Ivey et al., 2014; Kajimoto et al., 2019; Dong et al., 2020), our results suggest that they are not sufficient for membrane recruitment in NSCs. Similarly, the domains that mediate protein–protein interactions, such as the PB1 that binds Par-6 and the PBM that binds Baz, are also not sufficient for aPKC recruitment. While the isolated Par-6 and aPKC PB1 domains bind one another in vitro, our data indicate that the aPKC PB1 domain is not sufficient for membrane targeting, perhaps because Par-6 itself requires aPKC for membrane targeting (Rolls et al., 2003).
The unpolarized nature of C1 membrane binding raises the question of how its activity is regulated to yield aPKC’s precise spatially and temporally controlled localization. The aPKC catalytic domain forms intramolecular interactions with the PS and C1 that repress kinase activity (Graybill et al., 2012; Zhang et al., 2014), forming an “inhibitory core” (Fig. 7 B). Our results suggest the inhibitory core is coupled to C1 membrane binding. We observed C1 localization at the membrane throughout the cell cycle and depolarized membrane binding in mitosis when aPKC is normally restricted to the apical hemisphere. We also observed unpolarized membrane recruitment in aPKC variants where the catalytic domain was perturbed, suggesting that it is required for the repression of C1 activity. We suggest that perturbations to the catalytic domain that influence protein kinase activity can also disrupt the inhibitory core, consistent with the complex allosteric pathways in eukaryotic protein kinase domains (Ahuja et al., 2019). It has been proposed that the PS plays a central role in membrane recruitment and coupling localization to the inhibitory core through its interactions with the catalytic domain (Dong et al., 2020). Our results suggest that the PS is not sufficient for membrane recruitment (Fig. 4), but it remains possible that the PS is autoinhibited by the aPKC PB1 domain. Consistent with this possibility, an interaction between the PS and a PB1 has been reported (Tsai et al., 2015). The PS could also cooperate with the C1 to mediate membrane binding, and aPKC regulatory module localization in cultured cells supports this model (Cobbaut et al., 2023). The PS interaction with the aPKC catalytic domain is mostly dispensable for aPKC’s polarization in NSCs or epithelia, however, because aPKC AADAA is polarized in these cells. We suggest that the PS plays a more significant role in regulating catalytic activity than localization in these tissues.
Given that the C1 appears to be autoinhibited by the catalytic domain, how might it become activated? We previously found that inactivation of the aPKC PBM (aPKC V606A), which binds Baz, leads to cytoplasmic aPKC localization (Holly et al., 2020). Taken with our current results, we suggest that the interaction with Baz is required for membrane recruitment not because Baz directly recruits aPKC (e.g., aPKC ∆C1 is cytoplasmic) but because the Baz interaction is required for disruption of the inhibitory core and activation of the C1. Similarly, in NSCs lacking the PB1-binding Par-6 protein, aPKC also remains in the cytoplasm, even though Baz remains properly polarized and could potentially bind aPKC’s PBM (Rolls et al., 2003). These observations underlie our emphasis on the cooperative nature of aPKC membrane recruitment—activation of the C1 by at least Par-6 and Baz leads to the complex localization dynamics of aPKC observed in NSCs. Future work will be directed at understanding how the aPKC PB1 and PBM might be coupled to the inhibitory core and activation of aPKC membrane binding.
Materials and methods
Drosophila
Flies were grown at indicated temperatures on standard cornmeal/yeast media. Both male and female larvae were used in this study. Transgenic constructs were cloned into the pUAST attB vector (GenBank: EF362409.1) that was modified to include an N-terminal 3xHA or 1xHA tag. Integration of the vectors was done using standard Phi-C31 integration into an attP landing site on the third chromosome (attP2) by Rainbow Genetics or BestGene Inc. Positive insertion was determined by the presence of colored eyes after backcrossing to y,w stock. For Gal4-UAS line generation, insc-Gal4 virgins were crossed to males containing an aPKC transgene on the third chromosome or an RNAi on the second chromosome and an aPKC transgene on the third chromosome. Crosses were laid in vials for 24 h at ∼20°C. The resulting embryos were incubated at 30°C until larvae reached the third instar wandering larva stage. For optic lobe experiments, c855a-Gal4 virgins were crossed to males containing an aPKC transgene on the third chromosome. Crosses were laid in vials for 7 d (∼20°C), and the resulting progeny were incubated at 30°C for 24 h. Wandering third instar larvae were selected for dissection.
To create MARCM larval NSC clones, FRT-G13, aPKCK06403/CyO virgins were crossed with 3xHA aPKC D388A males. The subsequent progeny were allowed to grow to adulthood and were screened for the absence of the CyO marker. Males lacking CyO were crossed to elav-Gal4, UAS-mCD8:GFP, hs:flp; FRT-G13, and tubPGal80 virgins. After 24 h, the resulting embryos were incubated at room temperature (∼20°C) for an additional 24 h. These vials were then heat shocked at 37°C for 90 min. Another heat shock was possible within 18 h. Larvae were allowed to grow at room temperature or 18°C until the third instar wandering larva stage before analysis. Key resources are shown in Table 1.
Table 1.
Key resources table
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-aPKC | SCBT | Mouse Anti-PKC zeta (H1); SC-17781 | 1:1,000 |
| Antibody | Anti-Par-6 | Alpha diagnostic | N/A | Rat Anti-Par-6 (polyclonal custom antibody); 1:500 |
| Antibody | Anti-Mira | Abcam | Rat Anti-Mira; Ab197788 | 1:500 |
| Antibody | Anti-HA | Cell Signaling Tech. | Rabbit Anti-HA (C29F4); 3724 | 1:1,000 |
| Antibody | Anti-HA | Covance | Mouse Anti-HA; MMS-101P | 1:500 |
| Antibody | Anti-Baz | C.Q.Doe Lab | N/A | Guinea Pig Anti-Baz (polyclonal custom antibody); 1:2,000 |
| Antibody | Anti-GFP | Abcam | Chicken Anti-GFP; Ab13970 | 1:500 |
| Antibody | Anti-rat Cy3 secondary | Jackson Immuoresearch Lab. | Donkey Anti-Rat Cy3; 712-165-153 | 1:500 |
| Antibody | Anti-rabbit 647 secondary | Jackson Immunoresearch Lab. | Donkey Anti-Rabbit 647; 711-605-152 | 1:500 |
| Antibody | Anti-mouse 647 secondary | Jackson Immunoresearch Lab. | Donkey Anti-Mouse 647; 715-605-151 | 1:500 |
| Antibody | Anti-mouse 488 secondary | Jackson Immunoresearch Lab. | Donkey Anti-Mouse 488; 715-545-151 | 1:500 |
| Antibody | Anti-chicken 488 secondary | Jackson Immunoresearch Lab. | Donkey Anti-Chicken 488; 703-545-155 | 1:500 |
| Antibody | Anti-guinea pig 405 secondary | Jackson Immunoresearch Lab. | Donkey Anti-Guinea Pig 405; 706-475-148 | 1:500 |
| Genetic reagent (Drosophila melanogaster) | insc-GAL4 | Bloomington Drosophila Stock Center | ;insc-GAL4 | RRID:BDSC_8751 |
| Genetic reagent (Drosophila melanogaster) | elav-GAL4 | Bloomington Drosophila Stock Center | elav-Gal4, UAS-mCD8:GFP, hs:flp; FRT-G13, tubPGal80 | RRID:BDSC_5145 |
| Genetic reagent (Drosophila melanogaster) | c855a-Gal4 | Bloomington Drosophila Stock Center | ;;c855a-GAL4 | RRID:BDSC_6990 |
| Genetic reagent (Drosophila melanogaster) | aPKCK06403 | C.Q. Doe Lab | ; FRT-G13, aPKCK06403/CyO | |
| Genetic reagent (Drosophila melanogaster) | aPKC D388A | This study | ;;3xHA-aPKC D388A (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC D388A | This study | ;;1xHA-aPKC D388A (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC WT | This study | ;;1xHA-aPKC 1-606 (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC K293W | This study | ;;1xHA-aPKC K293W (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC PB1-C1 | This study | ;;1xHA-aPKC 1-195 (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC PB1-PS | This study | ;;1xHA-aPKC 1-141 (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC C1 | This study | ;;1xHA-aPKC 139-195 (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC ∆C1 | This study | ;;1xHA-aPKC 1-606 ∆141-196 (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC AADAA | This study | ;;1xHA-aPKC R131A, R132A, A134D, R135A, R136A (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC ∆PB1 | This study | ;;1xHA-aPKC 107-606 (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | aPKC D77A | This study | ;;1xHA-aPKC D77A (aPKC-PA) | |
| Genetic reagent (Drosophila melanogaster) | Baz RNAi | Bloomington Drosophila Stock Center | ;UAS-Baz RNAi | RRID:BDSC_39072 |
| Genetic reagent (Drosophila melanogaster) | Cdc42 RNAi | Bloomington Drosophila Stock Center | ;UAS-Cdc42 RNAi | RRID:VDRC_100794 |
| Chemical compound, drug | Amylose resin | NEB | E8021L | |
| Chemical compound, drug | Schneider’s Insect Medium (SIM) | Sigma-Aldrich | S0146 | |
| Chemical compound, drug | SlowFade Diamond Antifade Mountant with DAPI | Invitrogen | S36964 | |
| Chemical compound, drug | Phosphatidylserine (PS) | Avanti Polar Lipids | L-α-phosphatidylserine; 840032C | |
| Chemical compound, drug | Phosphatidylcholine (PC) | Avanti Polar Lipids | L-α-phosphatidylcholine; 840051C | |
| Chemical compound, drug | Phosphatidic acid (PA) | Avanti Polar Lipids | L-α-phosphatidic acid; 840101C | |
| Chemical compound, drug | Phosphatidylglycerol (PG) | Avanti Polar Lipids | L-α-phosphatidylglycerol; 841138P | |
| Chemical compound, drug | Ceramide | Avanti Polar Lipids | C12 ceramide; 860512P |
Immunofluorescence
For analysis of neural stem cells, larval brains were dissected and the tissue was incubated in 4% PFA fixative for 20 min within 20 min of dissection. This and all subsequent wash steps involved agitation by placing on a nutator. After fixation, brains were rinsed and washed three times for 15 min each in PBST (1xPBS with 0.3% Triton X-100). Brains were stored for up to 3 d at 4°C before staining. Before staining, brains were blocked for 30′ in PBSBT (PBST with 1% BSA) and then incubated with primary antibody solution overnight at 4°C. Brains were subsequently rinsed and washed three times for 15 min each in PBSBT and incubated with secondary antibody (conjugated to Cy3, AlexaFluor 647, AlexaFluor 488, or AlexaFluor 405 as appropriate) for 2 h in a vessel that protected the sample from light. Brains were subsequently rinsed and washed three times for 15 min each in PBST followed by storage in SlowFade w/DAPI at least overnight before imaging. Brains were imaged at room temperature on an upright Leica TCS SPE confocal using an ACS APO 40 × 1.15 NA Oil CS objective or an Olympus FluoView FV1000 upright laser scanning confocal with PlanApo N 60 × 1.42 NA oil objective. The acquisition was controlled with Leica LAS X or FluoView software, respectively.
Membrane enrichment and polarization quantification
A 10-pixel-wide line from apical to basal membrane in the medial optical section was used to measure membrane signals in metaphase NSCs. The signal at the edge of the cell was used as the membrane signal and the cytoplasmic signal was taken as the average of 20 data points located 10 points from the apical peak. For interphase NSCs and epithelial cells, the cortical intensity was measured by tracing the cortex with a 3px line, while the cytoplasmic signal was taken as an average of the entire cytoplasmic signal. All images were analyzed using Fiji and statistical analysis was done using DABEST python package (https://github.com/ACCLAB/DABEST-python). Figures were assembled using Adobe Illustrator.
Vesicle cosedimentation assay
MBP-C1 was purified using amylose agarose affinity purification as previously described (Graybill et al., 2012). Briefly, lysates of BL21 E. coli containing MBP-C1 expressed from the pMAL-C2(aPKC C1) plasmid in MBP lysis buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM DTT) were incubated with amylose coupled agarose resin. The MBP-C1 fusion protein was eluted from the resin with MBP elution buffer (20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, 5 mM maltose) and dialyzed at 4°C overnight in 20 mM Tris-HCl, pH 7.5, 50 mM NaCl.
For Giant Unilamellar Vesical (GUV) production, 50 μl of the specified lipids at 10 mg/ml in chloroform was dried in a test tube under an N2 stream and then in a vacuum chamber to ensure all chloroform was removed. Lipids were resuspended in a 0.2 M sucrose solution to a final concentration of 0.5 mg/ml and heated in a water bath at 50°C for 5 h with occasional agitation. All lipids were stored at 4°C and used within 3 d. All spins were carried out using an Optima MAX-TL Ultracentrifuge with a TLA-100 rotor at 65,000 at 4°C.
MBP-C1 protein was diluted to 50 μM in 20 mM HEPES, pH 7.5, 50 mM NaCl, 1 mM DTT, and pre-cleared for 30′. Reaction conditions were as follows: 20 mM HEPES, pH 7.5, 50 mM NaCl, 1 mM DTT, 0.25 mg/ml GUVs, and 5 μM MBP-C1. The reaction was carried out at room temperature for 15′ and then spun down for 30′. The supernatant fraction was removed and the pellet was resuspended in an equivalent volume of 1X Dilution buffer. Both the supernatant and pellet samples were mixed with 6× loading dye and separated using 12.5% SDS-PAGE. Gels were stained with Coomassie and imaged using a scanner.
Supplementary Material
is the source file for Fig. 5.
Acknowledgments
This work was supported by the National Institutes of Health grant GM127092 (K.E. Prehoda).
Author contributions: K.A. Jones, M.L. Drummond: Conceptualization, methodology, validation, formal analysis, investigation, writing, and visualization. R.R. Penkert: Investigation, editing, and visualization. K.E. Prehoda: Conceptualization, methodology, formal analysis, writing, visualization, supervision, project administration, and funding acquisition.
Data availability
The confocal microscopy data are openly available in the Dryad data repository at https://doi.org/10.5061/dryad.34tmpg4qz.
References
- Ahuja, L.G., Taylor S.S., and Kornev A.P.. 2019. Tuning the “violin” of protein kinases: The role of dynamics-based allostery. IUBMB Life. 71:685–696. 10.1002/iub.2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood, S.X., Chabu C., Penkert R.R., Doe C.Q., and Prehoda K.E.. 2007. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J. Cell Sci. 120:3200–3206. 10.1242/jcs.014902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood, S.X., and Prehoda K.E.. 2009. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol. 19:723–729. 10.1016/j.cub.2009.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, M.J., and Prehoda K.E.. 2015. Establishment of Par-polarized cortical domains via phosphoregulated membrane motifs. Dev. Cell. 35:199–210. 10.1016/j.devcel.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron, A.J.M., Escribano C., Saurin A.T., Kostelecky B., and Parker P.J.. 2009. PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat. Struct. Mol. Biol. 16:624–630. 10.1038/nsmb.1606 [DOI] [PubMed] [Google Scholar]
- Cobbaut, M., McDonald N.Q., and Parker P.J.. 2023. Control of atypical PKCι membrane dissociation by tyrosine phosphorylation within a PB1-C1 interdomain interface. J. Biol. Chem. 299:104847. 10.1016/j.jbc.2023.104847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-González, F., and Kazanietz M.G.. 2006. C1 domains exposed: From diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta. 1761:827–837. 10.1016/j.bbalip.2006.05.001 [DOI] [PubMed] [Google Scholar]
- Dong, W., Lu J., Zhang X., Wu Y., Lettieri K., Hammond G.R., and Hong Y.. 2020. A polybasic domain in aPKC mediates Par6-dependent control of membrane targeting and kinase activity. J. Cell Biol. 219:e201903031. 10.1083/jcb.201903031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybill, C., Wee B., Atwood S.X., and Prehoda K.E.. 2012. Partitioning-defective protein 6 (Par-6) activates atypical protein kinase C (aPKC) by pseudosubstrate displacement. J. Biol. Chem. 287:21003–21011. 10.1074/jbc.M112.360495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannaford, M., Loyer N., Tonelli F., Zoltner M., and Januschke J.. 2019. A chemical-genetics approach to study the role of atypical protein kinase C in Drosophila. Development. 146:dev170589. 10.1242/dev.170589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, Y., Yoshinaga S., Ogura K., Yokochi M., Noda Y., Sumimoto H., and Inagaki F.. 2004. Solution structure of atypical protein kinase C PB1 domain and its mode of interaction with ZIP/p62 and MEK5. J. Biol. Chem. 279:31883–31890. 10.1074/jbc.M403092200 [DOI] [PubMed] [Google Scholar]
- Holly, R.W., and Prehoda K.E.. 2019. Phosphorylation of Par-3 by Atypical Protein Kinase C and Competition between Its Substrates. Dev. Cell. 49:678–679. 10.1016/j.devcel.2019.05.002 [DOI] [PubMed] [Google Scholar]
- Holly, R.W., Jones K., and Prehoda K.E.. 2020. A conserved PDZ-binding motif in aPKC interacts with Par-3 and mediates cortical polarity. Curr. Biol. 30:893–898.e5. 10.1016/j.cub.2019.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem, C.C.F., and Knoblich J.A.. 2012. Drosophila neuroblasts: A model for stem cell biology. Development. 139:4297–4310. 10.1242/dev.080515 [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka, H., Skeath J.B., Nabeshima Y., Doe C.Q., and Matsuzaki F.. 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 390:625–629. 10.1038/37641 [DOI] [PubMed] [Google Scholar]
- Ivey, R.A., Sajan M.P., and Farese R.V.. 2014. Requirements for pseudosubstrate arginine residues during autoinhibition and phosphatidylinositol 3,4,5-(PO₄)₃-dependent activation of atypical PKC. J. Biol. Chem. 289:25021–25030. 10.1074/jbc.M114.565671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty, G., Petersen C., Gao L., and Macara I.G.. 2000. The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2:531–539. 10.1038/35019573 [DOI] [PubMed] [Google Scholar]
- Jumper, J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature. 596:583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimoto, T., Caliman A.D., Tobias I.S., Okada T., Pilo C.A., Van A.A.N., Andrew McCammon J., Nakamura S.-I., and Newton A.C.. 2019. Activation of atypical protein kinase C by sphingosine 1-phosphate revealed by an aPKC-specific activity reporter. Sci. Signal. 12:eaat6662. 10.1126/scisignal.aat6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn, M.P., Klopfenstein D.R., Fischer N., and Wodarz A.. 2010. Membrane targeting of Bazooka/PAR-3 is mediated by direct binding to phosphoinositide lipids. Curr. Biol. 20:636–642. 10.1016/j.cub.2010.01.065 [DOI] [PubMed] [Google Scholar]
- LaFoya, B., and Prehoda K.E.. 2023. Consumption of a polarized membrane reservoir drives asymmetric membrane expansion during the unequal divisions of neural stem cells. Dev. Cell. 58:993–1003.e3. 10.1016/j.devcel.2023.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFoya, B., and Prehoda K.E.. 2021. Actin-dependent membrane polarization reveals the mechanical nature of the neuroblast polarity cycle. Cell Rep. 35:109146. 10.1016/j.celrep.2021.109146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, C.F., and Munro E.. 2017. The PAR proteins: From molecular circuits to dynamic self-stabilizing cell polarity. Development. 144:3405–3416. 10.1242/dev.139063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T., and Luo L.. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22:451–461. 10.1016/s0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- Lin, D., Edwards A.S., Fawcett J.P., Mbamalu G., Scott J.D., and Pawson T.. 2000. A mammalian PAR-3-PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat. Cell Biol. 2:540–547. 10.1038/35019582 [DOI] [PubMed] [Google Scholar]
- Nance, J., and Zallen J.A.. 2011. Elaborating polarity: PAR proteins and the cytoskeleton. Development. 138:799–809. 10.1242/dev.053538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oon, C.H., and Prehoda K.E.. 2019. Asymmetric recruitment and actin-dependent cortical flows drive the neuroblast polarity cycle. Elife. 8:e45815. 10.7554/eLife.45815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perander, M., Bjorkoy G., and Johansen T.. 2001. Nuclear import and export signals enable rapid nucleocytoplasmic shuttling of the atypical protein kinase C lambda. J. Biol. Chem. 276:13015–13024. 10.1074/jbc.M010356200 [DOI] [PubMed] [Google Scholar]
- Petronczki, M., and Knoblich J.A.. 2001. DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat. Cell Biol. 3:43–49. 10.1038/35050550 [DOI] [PubMed] [Google Scholar]
- Prehoda, K.E. 2009. Polarization of Drosophila neuroblasts during asymmetric division. Cold Spring Harb. Perspect. Biol. 1:a001388. 10.1101/cshperspect.a001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, J., Peglion F., Martin J., Hubatsch L., Reich J., Hirani N., Gubieda A.G., Roffey J., Fernandes A.R., St Johnston D., et al. 2017. aPKC cycles between functionally distinct PAR protein assemblies to drive cell polarity. Dev. Cell. 42:400–415.e9. 10.1016/j.devcel.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls, M.M., Albertson R., Shih H.-P., Lee C.-Y., and Doe C.Q.. 2003. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 163:1089–1098. 10.1083/jcb.200306079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl, S., Braun U.B., and Leitges M.. 2012. Functional comparison of protein domains within aPKCs involved in nucleocytoplasmic shuttling. Biol. Open. 1:436–445. 10.1242/bio.2012505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.A., Lau K.M., Rahmani Z., Dho S.E., Brothers G., She Y.M., Berry D.M., Bonneil E., Thibault P., Schweisguth F., et al. 2007. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 26:468–480. 10.1038/sj.emboj.7601495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standaert, M.L., Galloway L., Karnam P., Bandyopadhyay G., Moscat J., and Farese R.V.. 1997. Protein kinase C-zeta as a downstream effector of phosphatidylinositol 3-kinase during insulin stimulation in rat adipocytes. Potential role in glucose transport. J. Biol. Chem. 272:30075–30082. 10.1074/jbc.272.48.30075 [DOI] [PubMed] [Google Scholar]
- Tepass, U. 2012. The apical polarity protein network in Drosophila epithelial cells: Regulation of polarity, junctions, morphogenesis, cell growth, and survival. Annu. Rev. Cell Dev. Biol. 28:655–685. 10.1146/annurev-cellbio-092910-154033 [DOI] [PubMed] [Google Scholar]
- Tsai, L.-C.L., Xie L., Dore K., Xie L., Del Rio J.C., King C.C., Martinez-Ariza G., Hulme C., Malinow R., Bourne P.E., and Newton A.C.. 2015. Zeta inhibitory peptide disrupts electrostatic interactions that maintain atypical protein kinase C in its active conformation on the Scaffold p62. J. Biol. Chem. 290:21845–21856. 10.1074/jbc.M115.676221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi, M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. 2022. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50:D439–D444. 10.1093/nar/gkab1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkei, Z.G., and Yamashita Y.M.. 2018. Emerging mechanisms of asymmetric stem cell division. J. Cell Biol. 217:3785–3795. 10.1083/jcb.201807037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Silva J., Krishnamurthy K., Tran E., Condie B.G., and Bieberich E.. 2005. Direct binding to ceramide activates protein kinase Czeta before the formation of a pro-apoptotic complex with PAR-4 in differentiating stem cells. J. Biol. Chem. 280:26415–26424. 10.1074/jbc.M501492200 [DOI] [PubMed] [Google Scholar]
- Wodarz, A., Ramrath A., Grimm A., and Knust E.. 2000. Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J. Cell Biol. 150:1361–1374. 10.1083/jcb.150.6.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Neimanis S., Lopez-Garcia L.A., Arencibia J.M., Amon S., Stroba A., Zeuzem S., Proschak E., Stark H., Bauer A.F., et al. 2014. Molecular mechanism of regulation of the atypical protein kinase C by N-terminal domains and an allosteric small compound. Chem. Biol. 21:754–765. 10.1016/j.chembiol.2014.04.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 5.
Data Availability Statement
The confocal microscopy data are openly available in the Dryad data repository at https://doi.org/10.5061/dryad.34tmpg4qz.