Abstract
The Influenza A (H1N1) pdm09 virus caused a global pandemic in 2009 and has circulated seasonally ever since. As the continual genetic evolution of hemagglutinin in this virus leads to antigenic drift, rapid identification of antigenic variants and characterization of the antigenic evolution are needed. In this study, we developed PREDAC-H1pdm, a model to predict antigenic relationships between H1N1pdm viruses and identify antigenic clusters for post-2009 pandemic H1N1 strains. Our model performed well in predicting antigenic variants, which was helpful in influenza surveillance. By mapping the antigenic clusters for H1N1pdm, we found that substitutions on the Sa epitope were common for H1N1pdm, whereas for the former seasonal H1N1, substitutions on the Sb epitope were more common in antigenic evolution. Additionally, the localized epidemic pattern of H1N1pdm was more obvious than that of the former seasonal H1N1, which could make vaccine recommendation more sophisticated. Overall, the antigenic relationship prediction model we developed provides a rapid determination method for identifying antigenic variants, and the further analysis of evolutionary and epidemic characteristics can facilitate vaccine recommendations and influenza surveillance for H1N1pdm.
Keywords: H1N1pdm virus, Antigenic relationship prediction, Antigenic evolution, Vaccine recommendation
Highlights
-
•
A well-performed model was developed to predict variant strains for H1N1pdm viruses.
-
•
Differences in substitution preference between H1N1pdm and the former seasonal influenza H1N1 were found.
-
•
Localized epidemic pattern of the H1N1pdm was more obvious than that of the former seasonal H1N1.
1. Introduction
Influenza viruses are long-term threats to human society since they cause excessive morbidity and mortality, leading to substantial massive economic costs. Influenza virus A (H1N1) is one of the main subtypes responsible for seasonal epidemics of disease; it was first detected in the human population in 1918 (Frost, 1920) and has circulated ever since. It disappeared in 1957 after the influenza A (H2N2) outbreak (Kilbourne, 2006), and re-emerged in 1977 (World Health Organization, 1978). In 2009, a novel swine-origin H1N1 influenza virus caused a global pandemic, and soon after the preceding seasonal H1N1 viruses gradually diminished (World Health Organization, 2010). To date, the novel H1N1 (H1N1pdm) virus has circulated as a seasonal virus, and experienced gradual genetic and antigenic evolution.
Due to the high mutation rate of hemagglutinin, which allows the virus to escape from host immunity, the effectiveness of the influenza virus vaccine decreases over time, thus there is a need to update vaccine strain every season. Vaccine recommendations are mainly based on the antigenic characteristics of circulating influenza viruses, as tested within the WHO Global Influenza Surveillance and Response System (GISRS) (Monto, 2018). Antigenic drift in circulating influenza viruses is primarily analyzed using hemagglutinin inhibition (HI) assays on infected ferret serum (Sandbulte et al., 2011), which is the gold standard for influenza vaccine selection. Liu et al. pointed out that HI titers of antisera from infected ferrets were significantly affected by antigenic site Sa, while in adult humans, antigenic sites Sb and Sa were immunodominant (Liu et al., 2018). In last several years, the WHO has also assessed antigenic characteristics with pre- and post-vaccination human sera. However, those results sometimes showed discrepancies with results from ferret antisera, and the results from people of different ages could also experience some inconformity.
Except for the labor-intensive and time-consuming HI assay used to determine the antigenicity of influenza viruses, many computational methods have been developed to determine the antigenic relationships or antigenic distances between influenza viruses and predict the evolution of influenza. These methods combine techniques and concepts from multivariate statistics, population genetics, epidemiological modelling and phylogenetic theory (Klingen et al., 2018). With the speed and ability in determining antigenicity-altering sites, they generate competitive predictive accuracies in vaccine strain prediction. Most computational works have focused on H3N2 virus strains (Adabor, 2021; Du et al., 2012; Lee et al., 2020; Liao et al., 2008; Xia et al., 2021), since the H3N2 subtype has circulated extensively in the past years, and the immunological assay data were sufficient to generate more reliable predictive models. Models to predict the antigenicity of H1N1 viruses were also developed (Anderson et al., 2018; Li et al., 2020; Yin et al., 2018), and the performance varied due to the different methods and datasets employed. In recent years, some computational works have tended to develop universal models (Peng et al., 2017; Qiu et al., 2022) to predict the antigenicity of different subtypes of influenza virus with universal features, and the performances of these models for H1 were typically not as strong as H3.
Despite all these methods to predict antigenic variants for influenza, the performance of these methods on H1N1pdm dataset has not been reported, and the antigenic evolution of H1N1pdm viruses haven't been systematically investigated. In this study, we modified our previously developed model that mapped the antigenic patterns and evolution of H1N1 viruses (Liu et al., 2015), and tested the performance of this method on H1N1pdm viruses. The results showed good performance in predicting antigenic relationships for H1N1pdm viruses, especially in identifying antigenic variants. With the predicted antigenic relationships between all available H1N1pdm viruses, we further inferred antigenic clusters of H1N1pdm viruses until now. These results allowed us to systematically investigate the antigenic evolution of H1N1pdm, and prediction of antigenic variants may further support influenza surveillance and vaccine recommendations for H1N1pdm.
2. Materials and methods
2.1. Influenza HA sequence acquisition and sequence analysis
Influenza HA sequence data (nucleotide and amino acid) were downloaded from Global Initiative on Sharing All Influenza Data (GISAID) (Shu and Mccauley, 2017) on August 17th, 2022. The sequences with missing or aberrant amino acids (i.e., “X”, “-”) in the HA1 domain were removed in the following antigenic modeling analysis. HA1 sequences were aligned with ClustalW (Larkin et al., 2007). Representative HA1 nucleotide sequences were chosen to generate a phylogenetic tree using CD-HIT (Li and Godzik, 2006) with a 99% identity threshold. The phylogenetic tree was built using FastTree (Price et al., 2009) based on the maximum likelihood model with 1000 bootstrap replicates. Time-scaled phylogenetic analysis was performed through a Markov chain Monte Carlo (MCMC) framework in BEAST v2.7.3 (Bouckaert et al., 2019). We chose the GTR substitution model, a coalescent Bayesian skyline tree prior, and a strict molecular clock model. MCMC chains were run for 10 million steps, and TreeAnnotator was used to analyze the maximum clade credibility (MCC) tree with mean heights, following the burn-in of the first 10% of trees. The tree was visualized using FigTree v1.4.4 and Dendroscope (Huson et al., 2007).
2.2. Hemagglutination inhibition (HI) data collection
Antigenic relationships between H1N1 viruses pre-2009 (former seasonal influenza H1N1) were inferred from HI assay data of former seasonal influenza A (H1N1) viruses collected from Harvey et al.'s work (Harvey et al., 2016). This dataset comprises 19,905 individual measurements of cross-reactivity between viruses. To obtain a more credible training dataset, we collected only those virus pairs with more than one measurement; if there were two measurements between one specific virus pair, we collected the antigenic relationship only if the two results were consistent (antigenically similar or distinct); and if there were more than two measures between one virus pair, we chose the majority consistent results to determine the antigenic relationship between the two viruses. In total, we get a training dataset of 239 antigenically similar pairs and 336 antigenically variant pairs of viruses (see Supplementary Table S1 for detail).
HI data of H1N1 viruses post-2009 were collected from the Weekly Epidemiological Record of the WHO. Virus pairs were considered antigenically variant if their HI titers differed by four dilutions or more. If the HI titer of strain i relative to antisera raised against strain j and the HI titer of strain j relative to antisera raised against strain i both exist, then the antigenic relationship between viruses i and j was calculated by the Archetti-Horsfall distance (dAH) (Archetti and Horsfall, 1950) as follows:
| (1) |
A pair of viruses was considered antigenically similar if dAH <4, otherwise they were considered antigenically different. In total, we obtained 267 pairs of antigenic relationships, including 145 antigenically similar pairs and 122 antigenically variant pairs (see Supplementary Table S2 for detailed information).
2.3. Antigenic relationship prediction model for H1N1pdm viruses
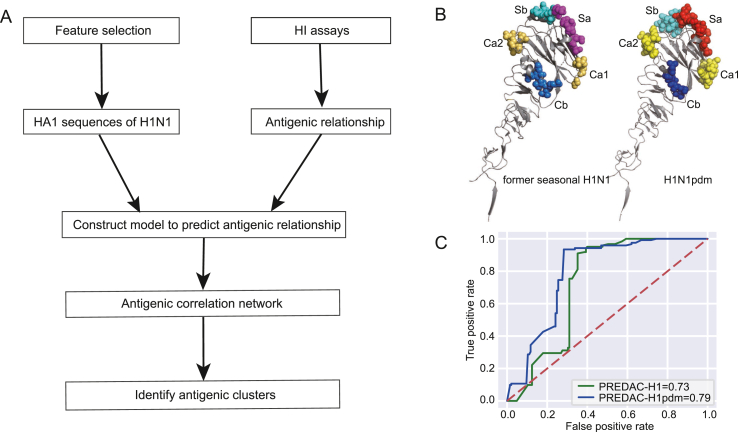
The antigenic relationships between viruses were predicted using a machine-learning approach, based on our previously developed model for former seasonal influenza H1N1(Liu et al., 2015). This model took into account the structural and physicochemical features that underlie antigen-antibody interactions, including five known H1N1 virus HA epitopes, five physicochemical properties of amino acids (hydrophobicity, volume, charge, polarity and accessible surface area), receptor binding and glycosylation. Based on the training dataset, we calculated a threshold cut-off for each feature, and then a Naïve Bayes classifier was built based on these features to infer the antigenic relationship. To improve the model performance for H1N1pdm viruses, we mainly made the following two refinements. The first refinement was to collect a more reliable training dataset, as we described in the HI data collection section. The second refinement was to use different definitions of epitopes for the former seasonal H1N1 virus and H1N1pdm virus. As epitopes were important features in our model, the epitope definition was crucial. Xu et al. (2010) determined the crystal structure of the HA from the H1N1 A/California/04/2009 virus, and revealed that the 2009 H1 HA shared conserved antigenic epitopes with human and swine H1 viruses from the early 20th century, while the seasonal human H1 HAs gradually diverged from the swine lineage (Smith et al., 2009). We used the epitope definition in this work for H1N1pdm, and the definition in Caton et al.‘s work (Caton et al., 1982) for the former seasonal H1N1, as shown in Fig. 1B, the detailed epitope sites were listed in Supplementary Table S3. With these refinements, we built a new model for H1N1pdm viruses.
Fig. 1.
Development of PREDAC-H1pdm. A Workflow of the PREDAC-H1pdm method. B Epitope definitions on HA1 protein for former seasonal H1N1 (left, 1RU7) and H1N1pdm (right, 3LZG). C The Receiver Operating Characteristic (ROC) curve of initial and modified antigenic relationship prediction models.
To validate whether our method could perform well on the dataset after 2009, we used the antigenic relationship dataset of former seasonal H1N1 viruses we described above as a training dataset and tested it on the HI dataset after 2009. The model performance was evaluated by the following parameters:
| (2) |
| (3) |
| (4) |
where, TP, TN, FP and FN represent true positive samples, true negative samples, false positive samples and false negative samples, respectively.
2.4. Development of PREDAC-H1pdm
PREDAC-H1pdm is an updated version of PREDAC-H1, which was used to model antigenic clusters of H1N1 viruses. After predicting the antigenic relationship between each pair of viruses in a group of H1N1pdm viruses, an antigenic correlation network was constructed by connecting pairs of viruses inferred to be antigenically similar. Then, groups of viruses with similar antigenicity, denoted as antigenic clusters, were identified from the network using the MCL program (Enright et al., 2002). The workflow was shown in Fig. 1A.
3. Results
3.1. Predicting the antigenic relationship between H1N1pdm strains
Since we have developed a model to predict the antigenic relationship between former seasonal influenza H1N1 viruses, we first tested the performance of this model on the HI dataset of H1N1pdm virus. With former seasonal H1N1 dataset as training dataset and H1N1pdm dataset as test dataset, the prediction accuracy of this initial model was 71.5%, and the sensitivity and specificity were 74.6% and 69.0%, respectively. After the refinement as we described in the method section, the accuracy, sensitivity and specificity of the retrospective test of our refined model were 80.9%, 91.8%, and 71.7%, respectively. The Receiver Operating Characteristic (ROC) curves of the initial model and our refined model were shown in Fig. 1C, with area under curve (AUC) of 0.73 and 0.79, respectively.
We noticed that the specificity of our model was somewhat low on this testing dataset, and we further analyzed the false-positive pairs of viruses. Among these false-positive pairs, 15 out of 41 pairs were A/California/7/2009 with strains after 2018, and they were antigenically similar in ferret HI tests, but some viruses have already showed poor inhibition by some post-vaccination human serum pools since 2016. We further listed the predicted antigenic relationships and mutations between vaccine strains in Supplementary Table S4. Pairs A/California/7/2009-A/Brisbane/02/2018, A/California/7/2009-A/Guangdong-Maonan/SWL1536/2019 and A/Brisbane/02/2018-A/Guangdong-Maonan/SWL1536/2019 were predicted to be antigenically variant, while the ferret HI assays showed that they were antigenically similar. In WHO reports, 6B.1A viruses with the HA1 amino acid substitution of S183P caused HI titer reduction in some post-vaccination human sera (World Health Organization, 2019a). The A/California/7/2009-A/Brisbane/02/2018 and A/California/7/2009-A/Guangdong-Maonan/SWL1536/2019 pairs all had S183P substitutions, and the prediction results were rational considering that they were antigenic variants. Additionally, for the A/Brisbane/02/2018-A/Guangdong-Maonan/SWL1536/2019 pair, the ferret HI results showed that they were antigenically similar, but in the WHO report, human HI titers against viruses in the 5A clade with substitutions D187A and Q189E, as in GD19, showed reductions compared to HI titers against A/Brisbane/02/2018-like viruses (World Health Organization, 2020). All false-positive pairs of viruses with predicted scores were listed in Supplementary Table S5, and the pairs with substitution S183P or substitutions D187A and Q189E were highlighted. Our model showed fairly good performance in predicting antigenic variants, with better sensitivity than the ferret HI assays.
3.2. Mapping the antigenic clusters of H1N1pdm strains
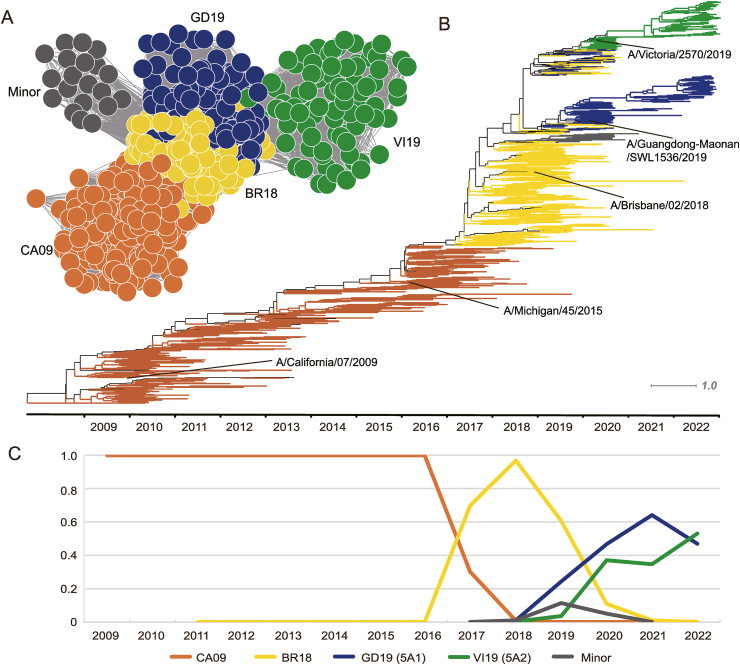
After constructing the antigenic relationship prediction model for H1N1pdm, we further developed PREDAC-H1pdm to model the antigenic evolution of H1N1pdm viruses (available at http://www.computationalbiology.cn/H1N1pdm/html/index.php). Since 2009, the WHO has recommended several vaccine strains: A/California/7/2009, A/Michigan/45/2015, A/Brisbane/02/2018, A/Guangdong-Maonan/SWL1536/2019, and A/Victoria/2570/2019. In the most recent 2023 southern hemisphere influenza season, A/Sydney/5/2021 (H1N1) pdm09-like virus was recommended as the vaccine strain. With the PREDAC-H1pdm method, we identified five antigenic clusters for H1N1pdm viruses after 2009: CA09, BR18, GD19, VI19 and Minor, named after the vaccine strain abbreviations they contained. A/Michigan/45/2015 was included in the CA09 cluster, which was reasonable since they were antigenically similar. The Minor cluster contained a small number of virus strains that mainly circulated in 2019 and 2020, and in the phylogenetic tree, it mainly belonged to the 6B.1A.5b clade.
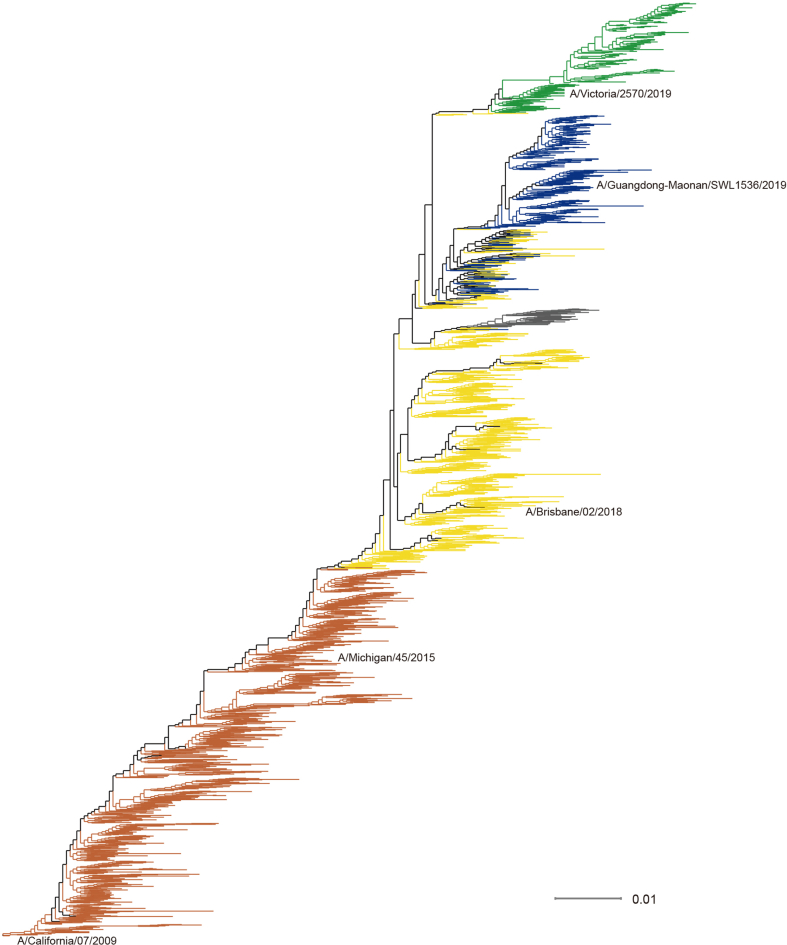
The antigenic correlation network is shown in Fig. 2A, and a time-scaled tree was built to show the genetic relationships between those clusters in Fig. 2B, and the genetic distance tree was shown in Supplementary Figure S1. We then mapped the dynamic changes in the percentage of antigenic clusters in Fig. 2C, the percentage of one antigenic cluster in a year was calculated by the percentage of viral isolates belonging to this antigenic cluster this year. Cluster CA09 mainly circulated from 2009 to 2016, while BR18 arose in 2017 and dominated from 2017 to 2019. After 2019, three clusters emerged and cocirculated: the Minor cluster mainly circulated in 2019 and disappeared soon after, while clusters GD19 and VI19 cocirculated from 2019 and re-emerged in 2020 and 2021. In 2022, the VI19 cluster arose and GD19 declined. Those results were corroborated in the WHO reports. All H1N1pdm viruses were antigenically indistinguishable from A/California/7/2009 and A/Michigan/45/2015 since 2009, until 2017, when human serum from vaccinated adults (A/Michigan/45/2015-like) showed reduced protection to viruses circulated in 2017 (World Health Organization, 2017). According to WHO report in 2019, most circulating viruses were antigenically similar to A/Brisbane/02/2018, except for a small number of viruses with HA1 amino acid substitutions at residues 155 or 156 (World Health Organization, 2019b), which was consistent with the emergence of GD19 cluster in Fig. 2C. Also, in our results, the GD19 and VI19 clusters have co-circulated since 2020, which was reported in WHO report in 2021 (World Health Organization, 2021).
Fig. 2.
Antigenic and genetic evolution of human influenza A (H1N1) pdm09 viruses. A Inferred antigenic correlation network of H1N1pdm viruses. B Mapping of antigenic clusters on time-scaled tree of the HA1 region of the H1N1 HA nucleotide sequences. C Distribution of antigenic clusters by year.
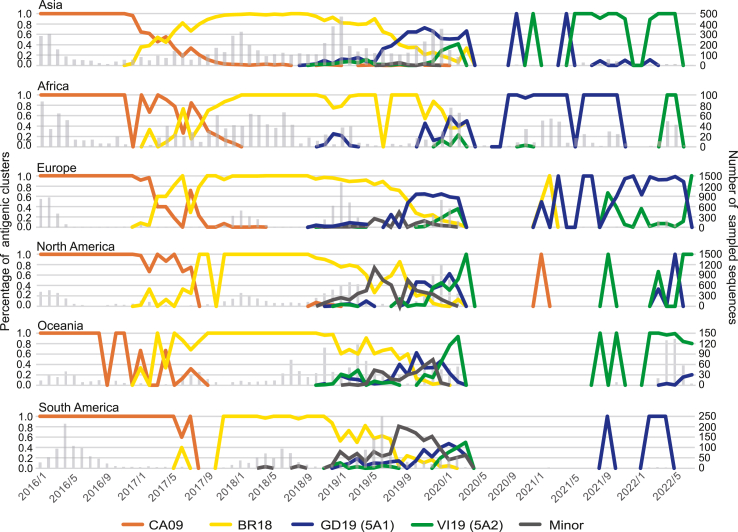
We further analyzed the circulation of antigenic clusters in different districts, as illustrated in Fig. 3. After the long-term circulation of the CA09 cluster, the BR18 cluster emerged in most continents in late 2016 or early 2017. However, in South America, the first emergence of BR18 was in June 2017. Since 2019, three clusters (GD19, VI19 and Minor) have co-circulated, gradually replacing the BR18 cluster. The circulation of H1N1pdm antigenic clusters was inconsistent in those continents, especially in the last four years. The Minor cluster dominated in North America from the end of 2018 to the end of 2019, and in South America from the end of 2018 to the beginning of 2020, and it didn't reach dominance at any time in other regions. In Asia, the GD19 cluster circulated since early 2019, dominated since August 2019 and was replaced by the VI19 cluster in the middle of 2021. In Europe, GD19 was the main circulating cluster during the 2019–2020 season, and the VI19 cluster emerged in June 2021. However, the VI19 cluster did not become dominant until July 2022. In Africa, the GD19 cluster dominated from the 2019–2020 season to late 2021, and VI19 became the main circulating cluster in 2022. Meanwhile, the epidemic patterns in North America, Oceania and South America seemed more complex. From mid-2019 to mid-2020, three clusters (GD19, VI19 and Minor) co-circulated for almost one year, and few strains were detected from mid-2020 to mid-2021 based on sequence data. Since March 2020, the VI19 cluster has dominated in North America and Oceania. In South America, few strains were sequenced since May 2020, and the small number of strains present from September 2021 to July 2022 belonged to the GD19 cluster. These results reveal a more complex antigenic evolutionary route of influenza H1N1pdm in recent years, with the cocirculation of different antigenic clusters in some regions.
Fig. 3.
Antigenic evolution in different regions. Dynamic changes in the percentage of antigenic clusters from Jan. 2016 to July. 2022 were recorded monthly. Different antigenic clusters were colored as in Fig. 2. The number of sampled sequences was shown in gray bars.
3.3. Indentifying cluster-specific amino acid substitutions in H1N1pdm viruses
As we determined the antigenic clusters for H1N1pdm viruses, we were able to find the cluster-specific amino acid substitutions which may serve as the determinants of antigenic variants. Here, we define the cluster-specific amino acid substitutions as follows: if more than 70 percent of amino acids in position n were X in one cluster, and Y in another cluster, then the cluster-specific amino acid substitution was XnY. We calculated the cluster-specific amino acid substitutions between CA09-BR18, BR18-GD19, BR18-VI19, and BR18-Minor, as shown in Table 1, and the substitutions for former seasonal H1N1 are also shown in Supplementary Table S6, and the ratio of strains with the specific amino acids were also marked. We found that most substitutions were on epitope Sa, or within or around the receptor-binding domain (RBD). CA09-BR18 had four specific substitutions, located on epitopes Sa and Cb and one on RBD. The two major clusters, GD19 and VI19, which circulated predominantly in the last three years, had different mutation routes. GD19 mainly mutated on epitope Sb, while VI19 mainly mutated on epitope Sa. Additionally, both clusters had an N129D substitution located on the 130-loop of the RBD. Interestingly, the 160 position was quite conserved in former seasonal H1N1 viruses, and the K160 M substitution between BR18 and Minor only existed in the Minor cluster after 2009. This site was located in the Sa epitope and was conserved in other H1N1 strains. The Minor cluster also contained substitutions on the 130-loop and 220-loop of the receptor-binding area, which may influence receptor binding properties. Although the Minor cluster did not circulate all over the world and existed for only a short time, these specific mutations are worthy of constant concern. Additionally, we observed a small clade on the phylogenetic tree that seemed to have evolved from early VI19 viruses, which mainly consist of viruses in 2021 and 2022. We compared the amino acids between VI19 and this small clade and found several specific substitutions, which contain A186T and Q189E on epitope Sb, and the Q189E substitution was also a specific substitution between BR18 and GD19. In the latest WHO report (World Health Organization, 2022), A/Sydney/5/2021 from this clade was included in the recommended composition of vaccines for use in the 2023 southern hemisphere influenza season.
Table 1.
The antigenic-specific substitutions of H1N1pdm antigenic clusters.
| Sa | Sb | Ca1/Ca2 | Cb | RBD | other | |
|---|---|---|---|---|---|---|
| CA09-BR18 | (99.6%)S164T(88.8%) | (99.3%)S74R (97.0%) | (96.8%)S183Pa(70.1%) | (98.7%)I295V(99.8%) | ||
| BR18-GD19 | (99.3%)D187A(79.4%)(99.9%)Q189E(82.8%) | (70.6%)N129D(99.3%) | ||||
| BR18-VI19 | (99.3%)N156K(99.1%)(94.0%)L161I(91.8%) | (70.6%)N129D(97.5%) (99.8%)K130N(96.9%) | (98.3%)V250A(92.2%) | |||
| BR18-Minor | (99.3%)K160M(99.6%) | (95.3%)E235D (99.9%) | (99.8%)K130N(92.8%)(99.5%)T216K(99.7%) | (99.9%)H296N(89.3%) |
The sites in the receptor-binding domain were in bold.
We also compared the specific substitutions of H1N1pdm viruses with former H1N1 seasonal viruses. Most substitutions of former seasonal H1N1 were located on the Sb epitope and RBD, while most of the specific substitutions for H1N1pdm were on the Sa epitope and RBD, which may suggest different variant preferences. Interestingly, the S183P substitution was not only the main cause of antigenic variant for CA09-BR18 but also appeared in the BE95-NE99 cluster transition. These results showed different preferences in antigenic evolution of former seasonal H1N1 and H1N1pdm, meanwhile, the same substitution might reappear, thus the substitutions from the former seasonal H1N1 also deserve special attention.
3.4. Prediction of antigenic variants helps to estimate vaccine effectiveness
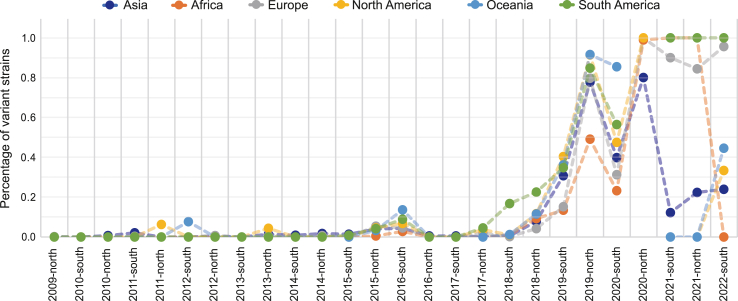
As our model had a good performance in predicting antigenic variants, we calculated the percentage of variant strains in each continent by influenza season, as shown in Fig. 4. Each node represents the percentage of variant strains in one season, compared with the vaccine strain recommended in the last influenza season (the vaccine strains recommended each season are listed in Supplementary Table S7). Taking North America as an example, the variant strain percentage experienced a rapid increase from the 2018-north season to the 2019-north season, which was consistent with the gradually reduced vaccine efficacy data in the 2018–2019 season and 2019–2020 season in the United States, where the vaccine effectiveness dropped from 62% in 2017–2018 season (Rolfes et al., 2019) to 44% in 2018–2019 season (Chung et al., 2020) and 30% in 2019–2020 season (Tenforde et al., 2021) against H1N1pdm. Similar situations occurred in other districts, due to the emergence of the BR18 cluster and the subsequent GD19 and VI19 clusters, and the mismatch between circulating strains and vaccine strains. We also observed different variant patterns in different districts since 2021. From 2021 to 2022, the mismatch of vaccine strains with circulation strains was observed in Europe, North America and South America but not in other districts. With the rapid evaluation of antigenicity by our model, the percentage of variant strains could be used as an initial estimate of vaccine effectiveness, which may further assist vaccine recommendations.
Fig. 4.
Percentage of antigenic variant strains. The percentage of antigenic variant strains in each continent was mapped by influenza seasons. The percentage was calculated as the percentage of antigenic variant strains in a given season, compared to the last season's recommended vaccine strain.
4. Discussion
The influenza A/(H1N1) pdm09 viruses have circulated seasonally since 2009, and continued to cause epidemics. As the continuous genetic evolution of this virus, high-throughput measurements for antigenic variants are crucial for influenza surveillance. In this study, we developed PREDAC-H1pdm to model the antigenic evolution of human influenza H1N1pdm viruses. Our antigenic prediction model demonstrated strong performance in retrospective tests and exhibited greater sensitivity in predicting antigenic variant strains compared to ferret HI assays. Since our antigenic prediction model only relies on the sequences of viruses and is faster and easier than the cost-worthy human serological analyses, it could further assist in vaccine recommendations for H1N1pdm viruses. Also, since our model was trained on a dataset of the former seasonal influenza H1N1, this provided a possible way to predict the antigenic relationship of newly emergent strains like H1N1pdm, when insufficient HI data is available. In recent years, deep learning models have been developed to predict antigenic relationships between influenza viruses and had very good performance. We tested the performance of a deep learning method IAV-CNN (Yin et al., 2019) on the H1N1pdm dataset, the accuracy of 5-fold cross-validation testing was 64.8%; while our feature-based model performed an accuracy of 81.3%. With sufficient HI data, we could build a deep learning model for H1N1pdm to achieve better performance in the future.
With this method, we also generated antigenic clusters for H1N1pdm. Compared with the former seasonal influenza A (H1N1) viruses, antigenic variant determined sites of the H1N1pdm viruses were somewhat different. Except for the sites located in RBD regions, substitutions on the Sb epitope were usually important for former seasonal H1N1, while the antigenic evolution of H1N1pdm tended to exhibit more substitutions on the Sa epitope, which suggest potential difference in immuno-dominance between the two virus types. Since H1N1pdm has not experienced long-term evolution, this observation may have some biases. The preference of substitutions on the Sa epitope of H1N1pdm may be influenced by human population immunity, or the inherent characteristics of H1N1pdm, which need further investigation with more experimental evidence in the future.
For the antigenic relationship prediction model, we used HI assay data to generate training and testing datasets, as most computational methods that predict antigenic relationships or distances between influenza viruses did. However, we noticed in the WHO reports that some antigenically similar strains in ferret HI assay showed poorly protection in human post-vaccination serum. This inconsistency might result from differences in immunodominance patterns between human and ferret serum (Liu et al., 2018), or might be influenced by previous influenza infections and vaccinations in human (Hensley, 2014; Li et al., 2013). With the growing number of HI assays by human post-vaccination sera, these results could be collected to refine our datasets to generate prediction models with more sensitivity in predicting antigenic variant strains.
As demonstrated in our previous work (Liu et al., 2015), the co-circulation of different antigenic clusters was quite normal in former seasonal H1N1 evolution, and this phenomenon is even more remarkable in H1N1pdm. The co-circulation of GD19, VI19, and Minor clusters was observed in late 2019 and early 2020 in North America, Oceania and South America. A localized epidemic pattern was also observed in the last two years, as the main circulating cluster in a given season varied across different continents. Since WHO recommends only one vaccine strain for the H1N1 subtype each season, a mismatch between the vaccine strain and the actual circulating strains exists in some regions. This suggests the necessity of region-specific vaccine recommendations for H1N1pdm. This local-persistent pattern could be a consequence of the SARS-CoV-2 pandemic, which brought reduction in international travel and caused less frequent transmission between different districts. As the SARS-CoV-2 pandemic continues, new transmission patterns of influenza worth constant concern.
Overall, we developed a computational method that effectively predicts antigenic variants for H1N1pdm viruses, which may further assist vaccine recommendations. Additionally, mapping the antigenic evolution of H1N1pdm viruses deepens our understanding of the evolution and epidemic pattern of this new virus, contributing to better surveillance and vaccination strategies. Although H1N1pdm virus has circulated for over 13 years and experienced certain antigenic drift, its history is still short compared to that of former seasonal influenza H1N1 and seasonal influenza H3N2. Consequently, the antigenic characteristics and epidemic patterns of H1N1pdm viruses need sustained attention.
Data availability
We developed a user-friendly web server PREDAV-H1pdm for predicting antigenic variants of influenza H1N1pdm viruses (available at http://www.computationalbiology.cn/H1N1pdm/html/index.php).
Ethics statement
No human or animal subjects were involved in this study.
Author contributions
Mi Liu: conceptualization, formal analysis, investigation, writing-original draft, writing-review and editing. Jingze Liu: data curation and methodology. Wenjun Song: methodology. Yousong Peng: data curation and methodology. Xiao Ding: methodology. Lizong Deng: methodology and funding acquisition. Taijiao Jiang: conceptualization, funding acquisition, supervision, writing-review and editing.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgement
This research was funded by the National Natural Science Foundation of China (32070678); the National Key Research and Development Program of China (2021YFC2302001).
Footnotes
Appendix A
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2023.05.008.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
References
- Adabor E.S. A statistical analysis of antigenic similarity among influenza A (H3N2) viruses. Heliyon. 2021;7 doi: 10.1016/j.heliyon.2021.e08384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.S., McCall P.R., Stern H.A., Yang H., Topham D.J. Antigenic cartography of H1N1 influenza viruses using sequence-based antigenic distance calculation. BMC Bioinf. 2018;19:1–11. doi: 10.1186/s12859-018-2042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archetti I., Horsfall F.L.J. Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. J. Exp. Med. 1950;92:441–462. doi: 10.1084/jem.92.5.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., Heled J., Jones G., Kühnert D., De Maio N., Matschiner M., Mendes F.K., Müller N.F., Ogilvie H.A., Du Plessis L., Popinga A., Rambaut A., Rasmussen D., Siveroni I., Suchard M.A., Wu C.H., Xie D., Zhang C., Stadler T., Drummond A.J. Beast 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:1–28. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton A.J., Brownlee G.G., Yewdell J.W., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Chung J.R., Rolfes M.A., Flannery B., Prasad P., O'Halloran A., Garg S., Fry A.M., Singleton J.A., Patel M., Reed C., Kim S.S., Martin E.T., Monto A.S., Jackson M.L., Jackson L.A., McLean H.Q., Belongia E.A., King J.P., Zimmerman R.K., Nowalk M.P., Balasubramani G.K., Bear T.M., Hickey R., Raviotta J.M., Suyama J., Weissman A.J., Williams J.V., Gaglani M., Raiyani C., Smith M., Murthy K., Clipper L., Reis M., Rao A., Walker K., Volz M., Mutnal M., Cummings C.N., Yousey-Hindes K., McMullen C., Chai S.J., Anderson E.J., Monroe M.L., Risk I., Herlihy R., Kim S., Spina N., Billing L., Schaffner W., Keipp Talbot H., Thomas A., McMahon M. Effects of influenza vaccination in the United States during the 2018-2019 influenza season. Clin. Infect. Dis. 2020;71:E368–E376. doi: 10.1093/cid/ciz1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Dong L., Lan Y., Peng Y., Wu A., Zhang Y., Huang W., Wang D., Wang M., Guo Y., Shu Y., Jiang T. Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nat. Commun. 2012;3:709. doi: 10.1038/ncomms1710. [DOI] [PubMed] [Google Scholar]
- Enright A.J., Van Dongen S., Ouzounis C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost W.H. Statistics of influenza morbidity: with special reference to certain factors in case incidence and case fatality. Publ. Health Rep. 1920;35:584–597. [Google Scholar]
- Harvey W.T., Benton D.J., Gregory V., Hall J.P.J., Daniels R.S., Bedford T., Haydon D.T., Hay A.J., McCauley J.W., Reeve R. Identification of low- and high-impact hemagglutinin amino acid substitutions that drive antigenic drift of influenza A(H1N1) viruses. PLoS Pathog. 2016;12:1–23. doi: 10.1371/journal.ppat.1005526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley S.E. ScienceDirect Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Curr. Opin. Virol. 2014;8:85–89. doi: 10.1016/j.coviro.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Richter D.C., Rausch C., Dezulian T., Franz M., Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinf. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbourne E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006;12:9–14. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingen T.R., Reimering S., Guzmán C.A., McHardy A.C. Silico vaccine strain prediction for human influenza viruses. Trends microbiol. 2018;26:119–131. doi: 10.1016/j.tim.2017.09.001. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., Mcgettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 20. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee E.K., Tian H., Nakaya H.I. Antigenicity prediction and vaccine recommendation of human influenza virus A (H3N2) using convolutional neural networks. Hum. Vaccines Immunother. 2020;16:2690–2708. doi: 10.1080/21645515.2020.1734397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Chang D., Han L., Zhang X., Zaia J., Wan X.F. Multi-task learning sparse group lasso: a method for quantifying antigenicity of influenza A(H1N1) virus using mutations and variations in glycosylation of Hemagglutinin. BMC Bioinf. 2020;21:1–22. doi: 10.1186/s12859-020-3527-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li Y., Myers J.L., Bostick D.L., Sullivan C.B., Madara J., Linderman S.L., Liu Q., Carter D.M., Wrammert J., Esposito S., Principi N., Plotkin J.B., Ross T.M., Ahmed R., Wilson P.C., Hensley S.E. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J. Exp. Med. 2013;210:1493–1500. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.-C., Lee M.-S., Ko C.-Y., Hsiung C.a. Bioinformatics models for predicting antigenic variants of influenza A/H3N2 virus. Bioinformatics. 2008;24:505–512. doi: 10.1093/bioinformatics/btm638. [DOI] [PubMed] [Google Scholar]
- Liu M., Zhao X., Hua S., Du X., Peng Y., Li X., Lan Y., Wang D., Wu A., Shu Y., Jiang T. Antigenic patterns and evolution of the human influenza A (H1N1) Virus. Sci. Rep. 2015;5:1–8. doi: 10.1038/srep14171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.T.H., Behzadi M.A., Sun W., Freyn A.W., Liu W.C., Broecker F., Albrecht R.A., Bouvier N.M., Simon V., Nachbagauer R., Krammer F., Palese P. Antigenic sites in influenza H1 hemagglutinin display species-specific immunodominance. J. Clin. Invest. 2018;128:4992–4996. doi: 10.1172/JCI122895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monto A.S. Reflections on the global influenza surveillance and Response System (GISRS) at 65 Years: an expanding framework for influenza detection, prevention and control. Influenza other respi. Viruses. 2018;12:10–12. doi: 10.1111/irv.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Wang D., Wang J., Li K., Tan Z., Shu Y., Jiang T. A universal computational model for predicting antigenic variants of influenza A virus based on conserved antigenic structures. Sci. Rep. 2017;7:1–7. doi: 10.1038/srep42051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. Fasttree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Tian X., Liu Y., Lu T., Wang H., Shi Z., Lu S., Xu D., Qiu T. Univ-flu: a structure-based model of influenza A virus hemagglutinin for universal antigenic prediction. Comput. Struct. Biotechnol. J. 2022;20:4656–4666. doi: 10.1016/j.csbj.2022.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfes M.A., Flannery B., Chung J.R., O'Halloran A., Garg S., Belongia E.A., Gaglani M., Zimmerman R.K., Jackson M.L., Monto A.S., Alden N.B., Anderson E., Bennett N.M., Billing L., Eckel S., Kirley P.D., Lynfield R., Monroe M.L., Spencer M., Spina N., Talbot H.K., Thomas A., Torres S.M., Yousey-Hindes K., Singleton J.A., Patel M., Reed C., Fry A.M., McLean H.Q., King J.P., Nowalk M.P., Balasubramani G.K., Bear T.M., Hickey R., Williams J.V., Reis E.C., Moehling K.K., Eng H., Jackson L.A., Smith Michael, Raiyani C., Clipper L., Murthy K., Chen W., Reis M., Petrie J.G., Malosh R.E., McSpadden E.J., Segaloff H.E., Cheng C.K., Truscon R., Johnson E., Lamerato L.E., Rosenblum B., Ford S., Johnson M., Raviotta J.M., Sax T., Steele J., Susick M., Chabra R., Garofolo E., Iozzi P., Kevish B., Middleton D.B., Urbanski L., Ponder T., Crumbaker T., Iosefo I., Sleeth P., Gandy V., Bounds K., Kylberg M., Rao A., Fader R., Walker K., Volz M., Ray J., Price D., Thomas J., Wehbe-Janek H., Beeram M., Boyd J., Walkowiak J., Probe R., Couchman G., Motakef S., Arroliga A., Kaniclides A., Bouldin E., Baker C., Berke K., Smith Mackenzie, Rajesh N., Alleman E., Bauer S., Groesbeck M., Brundidge K., Hafeez N., Jackson J., Anastasia I., Kadoo G., Petnic S., Ryan A., Maslar A., Meek J., Chen R., Stephens S., Thomas S., Segler S., Openo K., Fawcett E., Farley M., Martin A., Ryan P., Sunkel R., Lutich T., Perlmutter R., Grace B., Blood T., Zerrlaut C., McMahon M., Strain A., Christensen J., Angeles K., Butler L., Khanlian S., Mansmann R., McMullen C., Pradhan E., Manzi K., Felsen C., Gaitan M., Long K., Fisher N., Hawley E., O'Shaughnessy R., Scott M., Crawford C., Schaffner W., Markus T., Leib K., Dyer K., Santibanez T., Zhai Y., Lu P., Srivastav A., Hung M.C. Effects of influenza vaccination in the United States during the 2017-2018 influenza season. Clin. Infect. Dis. 2019;69:1845–1853. doi: 10.1093/cid/ciz075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandbulte M.R., Westgeest K.B., Gao J., Xu X., Klimov A.I., Russell C., Burke a, Smith D.F., Fouchier D.J., R. a M., Eichelberger M.C. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., Mccauley J. Gisaid : global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22:2–4. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.J.D., Bahl J., Vijaykrishna D., Zhang J., Poon L.L.M., Chen H., Webster R.G., Peiris J.S.M., Guan Y. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11709–11712. doi: 10.1073/pnas.0904991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenforde M.W., Kondor R.J.G., Chung J.R., Zimmerman R.K., Nowalk M.P., Jackson M.L., Jackson L.A., Monto A.S., Martin E.T., Belongia E.A., McLean H.Q., Gaglani M., Rao A., Kim S.S., Stark T.J., Barnes J.R., Wentworth D.E., Patel M.M., Flannery B. Effect of antigenic drift on influenza vaccine effectiveness in the United States-2019-2020. Clin. Infect. Dis. 2021;73:e4244–e4250. doi: 10.1093/cid/ciaa1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Recommended composition of influenza virus vaccines for use in the 2023 southern hemisphere influenza season. Wkly. Epidemiol. Rec. 2022;43:537–566. [Google Scholar]
- World Health Organization Recommended composition of influenza virus vaccines for use in the 2021–2022 northern hemisphere influenza season. Wkly. Epidemiol. Rec. 2021;11:77–88. [Google Scholar]
- World Health Organization Recommended composition of influenza virus vaccines for use in the 2020–2021 northern hemisphere influenza season. Wkly. Epidemiol. Rec. 2020;12:105–116. [Google Scholar]
- World Health Organization Recommended composition of influenza virus vaccines for use in the 2019–2020 northern hemisphere influenza season. 2019;12:141–150. [Google Scholar]
- World Health Organization Recommended composition of influenza virus vaccines for use in the 2020 southern hemisphere influenza season September. Wkly. Epidemiol. Rec. 2019;42:473–484. [Google Scholar]
- World Health Organization Recommended composition of influenza virus vaccines for use in the 2018 southern hemisphere influenza season. Wkly. Epidemiol. Rec. 2017;42:625–633. [PubMed] [Google Scholar]
- World Health Organization Recommended viruses for influenza vaccines for use in the 2010-2011 northern hemisphere influenza season. Wkly. Epidemiol. Rec. 2010;85:81–92. [PubMed] [Google Scholar]
- World Health Organization Influenza. Wkly. Epidemiol. Rec. 1978;53:21–24. [Google Scholar]
- Xia Y.L., Li W., Li Y., Ji X.L., Fu Y.X., Liu S.Q. A deep learning approach for predicting antigenic variation of influenza A H3N2. Comput. Math. Methods Med. 2021;2021:1–10. doi: 10.1155/2021/9997669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Ekiert D.C., Krause J.C., Hai R., Crowe J.E., Wilson I.A. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R., Thwin N.N., Zhuang P., Lin Z., Kwoh C.K. IAV-CNN: a 2D convolutional neural network model to predict antigenic variants of influenza A virus. IEEE ACM Trans. Comput. Biol. Bioinf. 2019;19:3497–3506. doi: 10.1109/TCBB.2021.3108971. [DOI] [PubMed] [Google Scholar]
- Yin R., Tran V.H., Zhou X., Zheng J., Kwoh C.K. Predicting antigenic variants of H1N1 influenza virus based on epidemics and pandemics using a stacking model. PLoS One. 2018;13:1–16. doi: 10.1371/journal.pone.0207777. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We developed a user-friendly web server PREDAV-H1pdm for predicting antigenic variants of influenza H1N1pdm viruses (available at http://www.computationalbiology.cn/H1N1pdm/html/index.php).