Abstract
Objective
Heatlhcare institutions are establishing frameworks to govern and promote the implementation of accurate, actionable, and reliable machine learning models that integrate with clinical workflow. Such governance frameworks require an accompanying technical framework to deploy models in a resource efficient, safe and high-quality manner. Here we present DEPLOYR, a technical framework for enabling real-time deployment and monitoring of researcher-created models into a widely used electronic medical record system.
Materials and Methods
We discuss core functionality and design decisions, including mechanisms to trigger inference based on actions within electronic medical record software, modules that collect real-time data to make inferences, mechanisms that close-the-loop by displaying inferences back to end-users within their workflow, monitoring modules that track performance of deployed models over time, silent deployment capabilities, and mechanisms to prospectively evaluate a deployed model’s impact.
Results
We demonstrate the use of DEPLOYR by silently deploying and prospectively evaluating 12 machine learning models trained using electronic medical record data that predict laboratory diagnostic results, triggered by clinician button-clicks in Stanford Health Care’s electronic medical record.
Discussion
Our study highlights the need and feasibility for such silent deployment, because prospectively measured performance varies from retrospective estimates. When possible, we recommend using prospectively estimated performance measures during silent trials to make final go decisions for model deployment.
Conclusion
Machine learning applications in healthcare are extensively researched, but successful translations to the bedside are rare. By describing DEPLOYR, we aim to inform machine learning deployment best practices and help bridge the model implementation gap.
Keywords: machine learning, artificial intelligence, organizational readiness, computational infrastructure, healthcare organizations, clinical decision support
BACKGROUND AND SIGNIFICANCE
Access to real-world data streams like electronic medical records (EMRs) accelerates the promise of machine learning (ML) in healthcare. Over 250 000 studies exist related to risk-stratification models alone, many published in the past 10 years.1,2 Despite the hype, a sizeable gap separates ML models in research articles from those impacting clinical care.3,4 This implementation gap leaves most published clinical ML applications lost in the “model graveyard”.5
The gap between research and implementation demonstrates that strong predictive performance alone is not sufficient for feasible and worthwhile translation of clinical ML models to the bedside. Beyond demonstrating predictive accuracy, model champions must articulate actions that can be taken as a result of inferences (model outputs).6 Actions must incur utility, some measurably positive effect on a clinical outcome of interest benefiting the deployment population.7–9 Positive impact on average, however, does not imply positive impact for all. ML applications to healthcare must satisfy fairness principles, ensuring performance and accrued utility are not unfavorable across certain patient populations, particularly those traditionally under-served.10–12 Institutions leading the charge in the translation of clinical ML applications are establishing governance frameworks ensuring models are safe, reliable and useful throughout their deployment life-cycles.13–17
Even if the above are addressed, ML applications must overcome technical feasibility hurdles related to their deployment. Traditionally, if such technical implementations were possible at all within the context of existing vendor and legacy infrastructure, massive overhead was required to establish and maintain computational frameworks enabling ML deployment and integration into the EMR.18–20 Approaches of early-adopting institutions can be broken down into 2 broad categories.21 Some institutions rely on platforms native to their EMR vendor (eg, Epic Nebula) to deploy ML applications.22,23 These frameworks reduce maintenance overhead and facilitate model sharing across hospitals that use the same EMR.24,25 But deployment of researcher-developed models trained using an institution’s clinical data warehouse remains difficult, leading other institutions to develop custom frameworks. These solutions often tap into daily refreshes of EMR data (eg, from Epic Clarity) at inference time that do not support real-time use-cases.21,26 Successful solutions largely spawn from collaborations of data scientists who build and validate models and hospital information technology (IT) personnel who know the ins-and-outs of their institution’s EMR. These combinations of expertise are valuable but rare. Custom implementations allow flexibility out-of-the-box EMR vendor solutions do not, but design details largely remain internal knowledge, leaving other institutions to either re-invent the wheel or be left behind. This has led to a sparse and eclectic set of solutions with limited academic discourse of best practices.
OBJECTIVE
Through a collaboration of data scientists at the Stanford School of Medicine, and IT persona at both Stanford Health Care and Stanford Children’s Health, our objective was to develop and describe DEPLOYR—a technical framework for deploying researcher created clinical ML applications directly into the EMR. The DEPLOYR framework does not rely on EMR vendor solutions like Epic Nebula, and supports real-time ML applications by tapping into an up-to-date data stream. In this article, we detail the mechanisms DEPLOYR enables and design decisions made during its formation to provide a blueprint for development and best practices for execution. Functions reviewed here include real-time closed-loop model trigger, data retrieval, inference, user-interface integration, and continuous monitoring. DEPLOYR enables enhanced prospective evaluations of ML models that extend beyond traditional retrospective evaluations. We demonstrate this capability and importance by silently deploying 12 previously retrospectively validated ML models triggered by clinician button-clicks in the EMR.
MATERIALS AND METHODS
Core functions of the DEPLOYR framework include data sourcing, inference triggers, and EMR integration. We detail our implementation of a monitoring module that tracks performance of deployed models over time, mechanisms that enable silent trial deployments, and prospective evaluations of model impact. DEPLOYR enables these mechanisms by leveraging integration capabilities native to Stanford Health Care’s EMR vendor (Epic Systems) in conjunction with 3 software applications: DEPLOYR-dev (a python package used for model development and validation), DEPLOYR-serve (a python Azure Function application to expose trained models as APIs), and DEPLOYR-dash (a dashboard implemented using the streamlit python package).27,28 Figure 1 provides a system level overview of DEPLOYR.
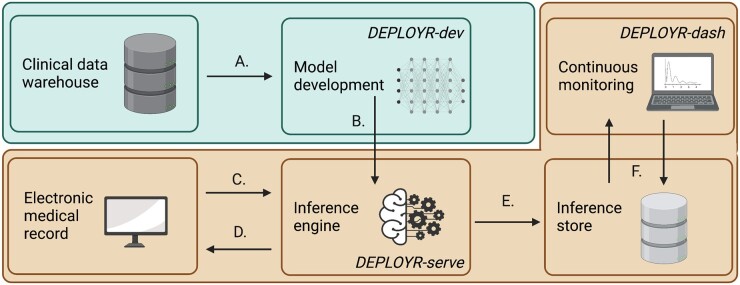
Figure 1.
Summary of a DEPLOYR enabled model deployment. Blue shading indicates infrastructure operated by the Stanford School of Medicine (academic research). Orange shading indicates infrastructure operated by Stanford Health Care (clinical operations). (A) De-identified EMR data are sourced from Stanford’s clinical data warehouse (STARR), a model is developed and retrospectively validated using the DEPLOYR-dev python package. (B) The model is deployed to the inference engine, and exposed as a REST API using DEPLOYR-serve. (C) Inference is triggered, spawning an HTTPS request from the EMR directed at the exposed model. (D) The request results in the collection of a feature vector from the EMR’s transactional database using REST (both FHIR and EMR specific) APIs maintained by the EMR vendor. Inference is performed on the real-time retrieved data, and routed back to the EMR closing the loop with end-users and integrating into workflow. (E) Inferences and relevant metadata are additionally saved to the inference store, (F) and consumed by monitoring software (DEPLOYR-dash) that continuously tracks model performance via dashboard. REST: representation state transfer; HTTPS: hypertext transfer protocol secure; FHIR: fast healthcare interoperability resources.
Data sources
The design of ML deployment frameworks requires data sourcing decisions. To effectively translate research models, training data should be sourced from research-grade clinical data warehouses. At inference time, many ML use-cases require real-time and up-to-date data streams.16,29–31 Since clinical data warehouses are typically several transformations removed from these data streams, mappings must be implemented to ensure models receive the same data elements in training and deployment environments.
Training data source
DEPLOYR uses data from Stanford’s clinical data warehouse (STARR) for training.32 STARR contains deidentified EMR data from over 2.4 million unique patients spanning 2009–2021 who have visited Stanford Hospital (academic medical center in Palo Alto, CA), ValleyCare hospital (community hospital in Pleasanton, CA) and Stanford University Healthcare Alliance affiliated ambulatory clinics. STARR data are derived from the EMR through a series of ETLs (exchange transform load) developed and maintained by a research IT team within the school of medicine. Quality checks are executed after each transformation to mitigate error propagation and persistence.32 Use of STARR data was approved by the institutional review board of the Stanford University School of Medicine.
Inference data source
In production, DEPLOYR sources data from the EMR’s transactional database, Epic Chronicles, which contains real-time patient data.33 We access this data stream using vendor specific and Fast Healthcare Interoperability Resources (FHIR) representational state transfer (REST) application programming interfaces (APIs) documented by our EMR vendor.34,35 Credentials enabling authentication to these APIs were provisioned upon registration of a back-end application to our EMR vendor’s application marketplace, Epic’s App Orchard. Use of this data source falls under the umbrella of hospital operational work specifically scoped to quality improvement. Data sources and mappings are depicted in Figure 2.
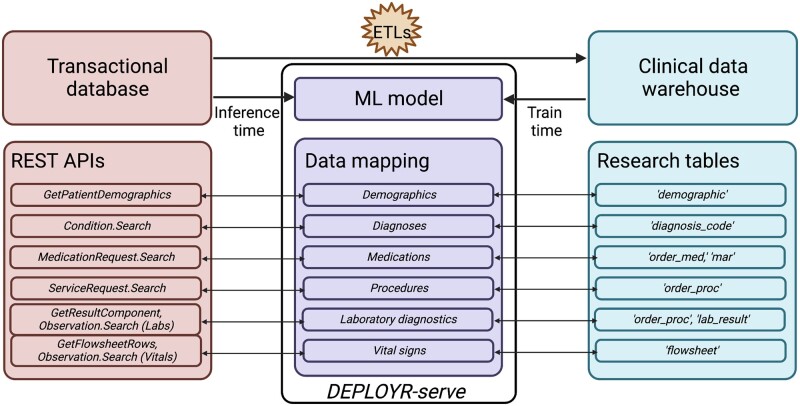
Figure 2.
ML models are trained using data sourced from Stanford’s clinical data warehouse (STARR). In production, real-time data are sourced from the EMR’s transactional database (Epic Chronicles) through Epic and FHIR REST APIs. STARR data are several ETLs (extract, transform, loads) removed from the transactional database. Data mapping is necessary at inference time to ensure features seen during training match features seen in production. Mappings and inferences are invoked in DEPLOYR-serve.
Model inference triggers
Inference triggering mechanisms dictate how model’s can integrate into workflow. Additionally, because triggering logic specifies a model’s deployment population, it should be considered during cohort development to ensure the population in a researcher’s retrospective test set matches what is seen in production. DEPLOYR supports 2 classes of inference mechanisms—event- and time-based triggers.
Event-based triggers
Event-based triggers execute as a direct result of a clinical action. Examples include order entry for a laboratory diagnostic test, signature of a progress note, inpatient admission, or discharge. We enable event-based triggers by exposing models as REST APIs that listen for inbound HTTPS requests from the EMR. Models are wrapped in custom python functions using DEPLOYR-serve, an Azure Function application deployed to Stanford Health Care’s instance of Azure.27 Requests are spawned from the EMR through use of EMR alerts (eg, Epic Best Practice Advisories), rules, and programming points configured to execute upon button-clicks in the EMR’s graphical-user-interface, Epic Hyperspace.36,37 Requests from the EMR transmit patient identifiers to DEPLOYR-serve functions enabling patient specific feature vector collection.
Time-based triggers
Time-based triggers initiate inference periodically on batches of patients. Example applications include models that periodically monitor patients for deterioration, sepsis, or acute kidney injury.38–41 DEPLOYR-serve supports time-based triggering through use of Azure Function timer triggers configured using cron logic.27,42 Because time-based triggers do not originate from the EMR, DEPLOYR needs to determine which patients to perform inference on. DEPLOYR-serve functions select patients using vendor maintained APIs that collect patient identifiers, for example, within specific hospital units. Event- and time-based triggers are summarized in Figure 3.
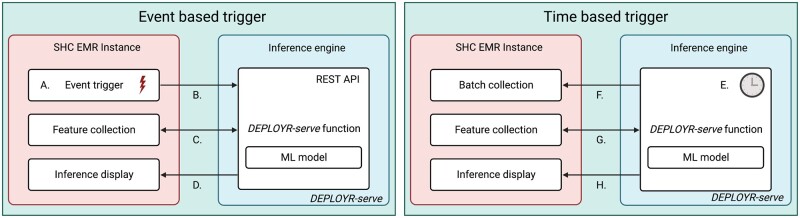
Figure 3.
DEPLOYR triggering mechanisms. Models deployed with event-based triggering logic are exposed as REST APIs on the inference engine using a python Azure Function application (DEPLOYR-serve). An event (A) in the EMR (eg, clinician button-click initiating a laboratory order) transmits an HTTPS request (B) directed at the exposed DEPLOYR-serve function, which wraps an ML model. The function transmits HTTPS requests (C) to REST APIs documented in Epic’s App Orchard to collect a feature vector, performs model inference, and directs the inference and resulting clinical decision support via HTTPS request (D) back into the EMR to interface with end-users. Models deployed with time-based triggering logic perform inference at set intervals (E) through use of Azure Function timer triggers. Every time interval (eg, 15 min), a DEPLOYR-serve function transmits HTTPS requests (F) to REST APIs to retrieve a batch of patient identifiers for whom inference should be made. Feature vectors are collected for the batch of patients (G), and inferences are transmitted back into the EMR (H). SHC: Stanford Health Care.
Directing model outputs
Deployment frameworks require mechanisms to direct model inferences and recommendations back to end-users, closing the loop. Although possible to communicate inferences via third-party web or mobile application, DEPLOYR integrates inferences into the EMR, reducing technical overhead and best fitting clinical workflow. Integration with the EMR is possible through several mechanisms, implemented in DEPLOYR-serve.
Passive integration
Inferences can be written to the EMR without interrupting clinical workflow, for example to an external model score column visible in inpatient lists and outpatient schedules. In Epic, score columns can be configured to display probabilities, binary flags, feature values and their contributions to promote interpretability.43 Additionally, inferences can be written to flowsheet rows and dynamic data elements, for example Epic Smart Data Values, which can be used to trigger downstream clinical decision support.44 Inferences can also be directed at clinicians using the EMR’s internal messaging system, for example Epic in-basket messages. Inferences are written to the EMR using APIs exposed and documented by our EMR vendor.
Active integration
Some applications better integrate with workflow through interruptive alerts. Consider the ML use-case of flagging low-yield laboratory diagnostic testing to reduce wasteful ordering behavior.31 In production, inference could be triggered upon order entry of the diagnostic and interrupt the ordering process. Such active integration is possible through use of typical EMR alerts. Specifically, we use Epic Best Practice Advisory web-services. Epic supports 2 styles of web-services: classic clinical document architecture (CDA) and CDS web-hooks.45 DEPLOYR currently uses classic CDA web-services, which direct HTTPS requests at DEPLOYR-serve functions and await XML (Extensible Markup Language) responses.46 We show mock-ups of EMR integration capabilities in Figure 4.
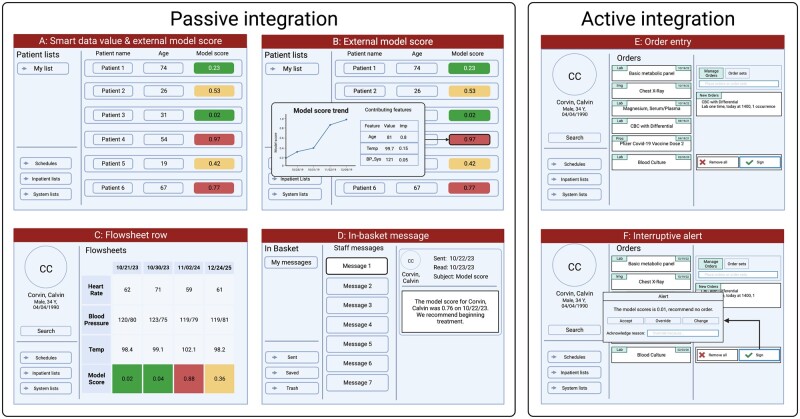
Figure 4.
Mock-up frames depicting the EMR user-interface and mechanisms in which inferences (model outputs) can be displayed to end-users. Inferences can be directed back into the EMR passively, without interrupting clinical workflow. They can be written as smart data values or as external model score columns displayed in inpatient lists and outpatient schedules (A). Hovering the mouse over an external model score column value in a patient list displays a pop-up window (B) depicting the trend of the model score over time, feature values, and contribution of those features to the resulting inference. Model inferences can be written to flowsheet rows (C) and visualized over time in conjunction with other vital sign data (eg, heart rate, blood pressure, temperature). Inferences and suggested interventions can be directed as in-basket messages (D) to specific providers. Additionally, inferences can be integrated with the EMR actively through use of interruptive alerts (E, F) that trigger as a result of button-clicks (eg, signature of a laboratory order) in the user-interface. Inference integration is implemented in DEPLOYR-serve.
Continuous performance monitoring
Deployed models must be monitored to ensure continued reliability in production.47–49 All models are subject to potential performance decay over time due to distribution shift. Examples include covariate shift (changes over features), label shift (changes over labels), and concept shift (changes to the relationship between features and labels).2,50
Extracting labels in production
DEPLOYR uses LabelExtactors to collect labels in production. LabelExtractors, implemented in DEPLOYR-serve, are specific to a deployed model. They execute periodically using cron logic. At inference time, in addition to integrating model outputs to the EMR, we package inferences with relevant metadata (identifiers, timestamps, features) in inference packets and save them to the inference store, an Azure Cosmos database maintained by Stanford Health Care. LabelExtactors consume inference packets and pair to them their corresponding labels (once observable). A model tasked with predicting unplanned (30-day readmission) might produce inference packets that include the patient’s FHIR identifier, discharge time, and inference. The corresponding LabelExtractor would use the FHIR identifier and discharge time to make requests directed at relevant APIs, determining whether the patient was readmitted to the hospital within 30 days of the produced inference.
Tracking model performance
After a LabelExtractor has collected labels, performance metrics can be estimated. When monitoring binary classifiers, metrics may include threshold dependent measures like accuracy, sensitivity, specificity, precision along with threshold independent metrics like area under the receiver operator characteristics curve (AUROC). Plots that track discrimination ability (ROC and precision-recall curves) and calibration (calibration curves) can be constructed. In addition to population estimates, metrics can be monitored over patient subgroups, including protected demographic classes. Beyond classic ML metrics, measures of model usefulness such as net benefit and expected utility can be tracked to ensure model use is yielding more good than harm.7,51
Tracking distribution shifts in features and labels
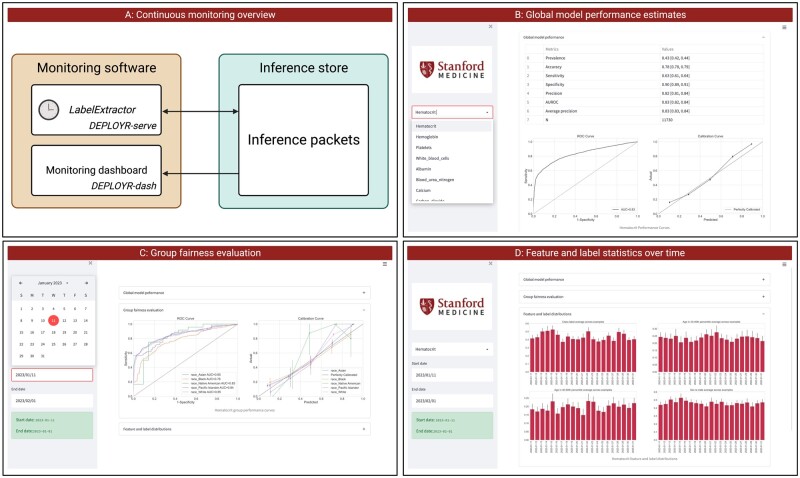
Performance decay over time is often attributable to distributional shift. Beyond calculating prospective performance metrics, DEPLOYR monitoring infrastructure tracks statistics that describe the distributions of features, labels, and predictions over time. Both model performance and distributional statistics are displayed to a dashboard created using the streamlit python package, which we show in Figure 5.28
Figure 5.
DEPLOYR performance monitoring. Panel A depicts the flow of data from monitoring software to the inference store. A LabelExtactor is implemented in DEPLOYR-serve as an Azure Function timer trigger. When executed it collects inference packets and pairs them with their corresponding labels. Inference packets paired with labels are consumed by a monitoring dashboard, implemented in DEPLOYR-dash. Panel B shows a screenshot of the dashboard displaying global model performance for the user-specified model. Panel C shows a group fairness evaluation across protected demographic classes for the user-specified time window. Panel D shows feature and label distribution statistics collected and tracked over time.
Enabling silent deployment
Deployment infrastructure requires the ability to silently test models prospectively.14,52,53 Silent trials allow data scientists to ensure data pipes are appropriately linked, and provide more robust appraisals of model performance than typical retrospective analyses because they occur directly in the intended production environment.54 Additionally, silent deployment can uncover faulty cohort design, for instance when a retrospective cohort was generated using exclusion criteria observable only after inference time, common when generating ML cohorts using case–control study design.55,56 DEPLOYR event- and time-based triggers can be configured to execute in the background, enabling silent trials.
Enabling prospective evaluation of impact
The ultimate evaluation of healthcare ML impact is not a prospective ROC curve, but rather estimation of the causal effect of the model’s implementation on a clinical or operational outcome. In some use-cases, a model’s inference and corresponding recommendation will only be displayed to the end-user if predicted risk exceeds some threshold.26 Here, regression discontinuity designs may be suitable to estimate the local treatment effect of the model’s implementation.57 In other cases when an average treatment effect on the deployment population at large is desired, tracking end-user adherence to model suggestions (eg, whether an alert was accepted) can enable observational estimates of the intervention’s effect.58 These analyses require traditional assumptions from causal inference literature, for example no unobserved confounding, positivity, and consistency to produce unbiased effect estimates. Under equipoise, randomized study designs may be appropriate to achieve unbiased effect estimates.59,60 DEPLOYR supports functionality to inject randomization into the inference-directing mechanism to enable prospective determination of a model’s impact.
Silent trial deployment of laboratory prediction models
To evaluate DEPLOYR, we silently deployed 12 binary classifiers that predict laboratory diagnostic results, building off of previous retrospective analyses of similar models designed to reduce wasteful laboratory utilization.31,61–63 Models were trained using STARR data, and exposed as REST APIs using DEPLOYR-serve. EMR alerts were configured to trigger the deployed models upon signature of the diagnostic test whose result each model aimed to predict.
Cohort and prediction task definitions
Three retrospective cohorts specific to a laboratory diagnostic exam were constructed. The unit of observation was an order for the exam. One cohort included orders for complete blood count (CBC) with differential diagnostics. Another included orders of metabolic panels. A third included orders for magnesium diagnostics. CBC and metabolic panel diagnostics result in several components, whereas magnesium exams yield a single result. Consistent with prior work, we trained binary classifiers per component to, at order time, predict whether the test result would fall outside the clinical laboratory defined normal reference range.31,62 Four binary classifiers were trained for the CBC cohort that separately predicted hematocrit, hemogloblin, white blood cell, and platelet results. Seven binary classifiers were trained for the metabolic panel cohort to predict albumin, blood urea nitrogen, calcium, carbon dioxide, creatinine, potassium, and sodium results. One classifier was trained to predict magnesium results. Retrospective cohorts spanned from 2015 to 2021, with 2000 diagnostic tests sampled randomly per year for a total of 14 000 orders per task. Corresponding prospective cohorts were collected in real-time during our silent deployment trial—spanning the dates January 11 to February 15, 2023.
Model training and evaluation
We trained and deployed random forest classifiers, as tree-based models are strong baselines for EMR-based ML tasks.64,65 A description of random forests as well as other model classes considered are detailed in Supplementary Note S1. Features included patient demographics, diagnosis codes mentioned on the problem list, medication orders, and prior lab results. Features were represented as counts, as detailed in Supplementary Note S2.66 All 12 classifiers were evaluated using retrospective and prospective test sets. We measured model discrimination ability by estimating the AUROC. The 95% confidence intervals were estimated by bootstrapping the corresponding test set 1000 times.
RESULTS
Here we present the results of our retrospective and prospective silent trial evaluations. In Table 1, we summarize demographic characteristics of patients in both retrospective and prospective cohorts. In Table 2, we summarize model performance estimated using retrospective and prospective test sets. We compute prevalence of the positive class (an abnormal diagnostic result), as well as AUROC. Additionally, we constructed retrospective and prospective receiver operating characteristics (ROC) curves, precision recall (PR) curves, and calibration plots—shown in the Supplementary Figures S1–S3. We estimated model performance across patient subgroups stratified by protected demographic classes, which we show in Supplementary Tables S1–S12. Additionally, we show retrospective and prospective AUROC stratified by the number of available features in Supplementary Figure S4.
Table 1.
Demographic breakdown of retrospective and prospective cohorts
| Demographic breakdown |
Retrospective cohorts |
Prospective cohorts |
|||||
|---|---|---|---|---|---|---|---|
| Diagnostic | CBC | Magnesium | Metabolic | CBC | Magnesium | Metabolic | |
| N unique patients | 13 362 | 11 771 | 13 410 | 18 982 | 5234 | 16 441 | |
| Age, mean (SD) | 51.4 (23.7) | 53.1 (24.2) | 54.6 (21.4) | 55.7 (21.4) | 62.1 (17.8) | 55.4 (21.1) | |
| Sex, n (%) | Female | 7103 (53.2) | 5432 (46.1) | 6923 (51.6) | 10 092 (53.2) | 2425 (46.3) | 8711 (53.0) |
| Male | 6259 (46.8) | 6338 (53.8) | 6485 (48.4) | 8884 (46.8) | 2808 (53.6) | 7724 (47.0) | |
| Unknown | 0 (0.0) | 1 (0.0) | 2 (0.0) | 6 (0.0) | 1 (0.0) | 6 (0.0) | |
| Race, n (%) | White | 6888 (51.5) | 6038 (51.3) | 6915 (51.6) | 9009 (47.5) | 2726 (52.1) | 7636 (46.4) |
| Other | 2870 (21.5) | 2899 (24.6) | 2680 (20.0) | 4545 (23.9) | 1067 (20.4) | 4103 (25.0) | |
| Asian | 2444 (18.3) | 1739 (14.8) | 2600 (19.4) | 3612 (19.0) | 928 (17.7) | 3125 (19.0) | |
| Black | 557 (4.2) | 587 (5.0) | 583 (4.3) | 1060 (5.6) | 327 (6.2) | 884 (5.4) | |
| Unknown | 391 (2.9) | 238 (2.0) | 420 (3.1) | 382 (2.0) | 56 (1.1) | 353 (2.1) | |
| Pacific Islander | 168 (1.3) | 212 (1.8) | 171 (1.3) | 294 (1.5) | 98 (1.9) | 261 (1.6) | |
| Native American | 44 (0.3) | 58 (0.5) | 41 (0.3) | 80 (0.4) | 32 (0.6) | 79 (0.5) | |
Note: Retrospective cohorts were sourced from Stanford’s clinical data warehouse (STARR). Prospective cohorts were collected in real-time through the EMR transactional database (Epic Chronicles) as diagnostic orders triggered model inferences between January 11 and February 15, 2023.
CBC: complete blood count.
Table 2.
Model performance estimates on retrospective and prospectively collected test sets for all twelve models
| Model performance |
Positive (abnormal) prevalence |
AUROC |
|||
|---|---|---|---|---|---|
| Diagnostic | Component | Retrospective | Prospective | Retrospective | Prospective |
| CBC with differential | Hematocrit | 0.47 [0.45, 0.49] | 0.43 [0.42, 0.44] | 0.86 [0.85, 0.88] | 0.83 [0.83, 0.84] |
| Hemoglobin | 0.50 [0.48, 0.52] | 0.47 [0.46, 0.47] | 0.88 [0.86, 0.89] | 0.83 [0.83, 0.84] | |
| Platelets | 0.26 [0.24, 0.28] | 0.21 [0.21, 0.22] | 0.79 [0.77, 0.82] | 0.77 [0.76, 0.78] | |
| White blood cell | 0.29 [0.27, 0.31] | 0.27 [0.27, 0.28] | 0.76 [0.74, 0.79] | 0.69 [0.68, 0.70] | |
| Metabolic panel | Albumin | 0.20 [0.18, 0.21] | 0.21 [0.20, 0.21] | 0.88 [0.86, 0.91] | 0.85 [0.84, 0.86] |
| Blood urea nitrogen | 0.22 [0.20, 0.24] | 0.24 [0.23, 0.25] | 0.85 [0.83, 0.87] | 0.80 [0.79, 0.81] | |
| Calcium | 0.11 [0.10, 0.13] | 0.11 [0.11, 0.12] | 0.80 [0.76, 0.83] | 0.79 [0.78, 0.81] | |
| Carbon dioxide | 0.16 [0.14, 0.17] | 0.20 [0.20, 0.21] | 0.69 [0.66, 0.72] | 0.62 [0.61, 0.63] | |
| Creatinine | 0.31 [0.29, 0.33] | 0.32 [0.31, 0.33] | 0.78 [0.75, 0.80] | 0.75 [0.74, 0.76] | |
| Potassium | 0.06 [0.05, 0.08] | 0.08 [0.08, 0.09] | 0.67 [0.61, 0.72] | 0.60 [0.59, 0.62] | |
| Sodium | 0.12 [0.10, 0.13] | 0.17 [0.16, 0.17] | 0.79 [0.75, 0.82] | 0.71 [0.70, 0.72] | |
| Magnesium | Magnesium | 0.15 [0.14, 0.17] | 0.14 [0.13, 0.15] | 0.70 [0.67, 0.73] | 0.65 [0.63, 0.67] |
AUROC: area under the receiver operating characteristics curve; CBC: complete blood count.
DISCUSSION
Our silent trial results underscore the necessity of prospective evaluations before ML applications are integrated into clinical work-streams.14 Across our 12 laboratory predictions tasks, AUROC in our prospective test sets were generally several percentage points lower than what is seen retrospectively. Drops in performance were similarly apparent in our ROC and PR curves, though the shapes of the curves remained similar across settings. Poorer performance could be attributable to data drift, data-elements appearing to be accessible at inference time in the clinical data warehouse not actually being accessible in production, and imperfect mappings between training and inference data sources. In Supplementary Figure S4 we show model performance stratified by number of available features at inference time, which differs between retrospective and prospective settings. Due to these deviations, we recommend using performance measures estimated from prospectively collected test sets during silent trials to make final go decisions when deploying clinical ML models.
In our evaluation we silently deploy 12 random forest classifiers, but DEPLOYR allows integration of arbitrary model classes ranging from simple regressions to complicated neural networks.67,68 This feature is attributable to DEPLOYR’s use of server-less compute, which is separate from the EMR and allows users to expose arbitrary code as REST endpoints.22,25 While DEPLOYR is model class agnostic, latency will increase with increasing model complexity. If latency is an issue, data scientists should consider whether the more complicated model is necessary to achieve desired model performance, and if so serve the model using a graphics processing unit (GPU), which server-less functions support.69
Continuous monitoring tools are essential to any deployment framework.47 Statistically significant and clinically meaningful variations in performance can indicate models need re-training or decommissioning.70–72 Careful attention to feedback mechanisms from interventions administered as a result of model integration must be considered. Feedback mechanisms occur when predictions lead to interventions that alter the distribution from which prospective data are generated. When monitoring risk-stratification models, interventions assigned to high risk patients intending to reduce risk can make models appear as “victims of their own success”.73 Models intending to discourage diagnostics test orders whose results are highly predictable, for example those we silently evaluate in this study, will induce censoring of the most predictable labels. Feedback mechanisms can cause drastic deviations between observed and actual performance.73–75 Naively updating these models do more harm than good.76 DEPLOYR supports injecting randomization into inference integration, which can be used in tandem with traditional weighting estimators to recover true performance in the presence of feedback mechanisms.77 The ability to randomize is similarly critical for deliberate randomized controlled trial evaluations of ML model impact on clinical outcomes.59,60
DEPLOYR can serve and evaluate a broad class of models for a range of clinical applications. Though we deploy binary classifiers in this study, the framework naturally extends to multi-class, multi-label and regression settings, which may be more suitable for alternative clinical applications including emergency department triage, recommender systems, and insulin dose optimization.78–80 Performance metrics a data scientist chooses to monitor will vary depending on the task. In a binary setting, a data scientist might monitor an ROC curve, in a multi-class setting a confusion matrix, and in a regression setting mean-squared-error. Irrespective of the exact metric set, the overarching goals of continuously monitoring remain the same. Because DEPLOYR leverages server-less compute that allows execution of arbitrary software, data scientists have control over postprocessing of inferences and the metrics they choose monitor in deployment.
The DEPLOYR framework can be extended to institutions that (1) have access to a clinical data warehouse for model development and (2) maintain 21st Century Cures Act compliant FHIR APIs.81,82 Though the DEPLOYR code base does also utilize Epic specific APIs, making extensions to institutions that vendor with Epic Systems easier, similar FHIR-based APIs exist that serve as suitable alternatives.83 Additionally, while our installation of DEPLOYR uses an EMR alerting mechanism native to Epic Systems to trigger model inferences (Epic Best Practice Advisories), similar alerting mechanisms are maintained by alternative EMRs that would enable equivalent functionality.84,85
Successful installation and maintenance of DEPLOYR requires dedicated financial resources and personnel. DEPLOYR’s computing infrastructure is resource efficient. The total cloud computing cost for our silent deployment which lasted between January 11 and February 15, 2023 (Azure Function executions and data storage in Azure Cosmos) was less than $300.86,87 To assign dedicated personnel, Stanford Health Care’s IT department has formed a data science team. Expertise in engineering is required to adequately connect data pipes and manage cloud infrastructure. Data scientists are required to develop, monitor and evaluate deployed models. EMR integration expertise is required to set up EMR alerts, ensure the EMR can communicate with the server-less function application, and maintain integrations in the advent of EMR upgrades. Networking and security personnel are required to ensure data are appropriately routed between the EMR and the institution’s cloud infrastructure. At Stanford Health Care, data never leaves our internal network.
Limitations to this study include that, while the DEPLOYR framework is general and can be extended to additional institutions that use alternative EMRs, expertise and support from an institution’s IT department would be required to translate the Epic specific integrations we leverage (eg, Best Practice Advisories) and data mappings to alternative EMR solutions and clinical data warehouse structures. The design decisions and considerations we have outlined in this article remain relevant to any institution attempting to install a deployment framework of their own. We note that EMR data are notoriously messy and frequently incomplete. Missing data can influence performance of clinical ML models, which we demonstrate in Supplementary Figure S4. We show that performance of our ML models increases with the number of available features at inference time. Data scientists tasked with deploying ML models may benefit from specifying a tolerance threshold on the sparsity of a feature vector used for inference, which may boost performance in production by restricting inferences to examples a model is more likely to correctly classify. While DEPLOYR can even include unstructured EMR data represented in clinical notes, future upgrades would be necessary to support a broader array of clinical ML applications that use multiple data modalities.88–90
CONCLUSION
There exist frameworks to govern and promote the implementation of accurate, actionable and reliable models that integrate with clinical workflow.14,15 The creation of such frameworks has created the need for accompanying technical solutions that enable executing best practices laid out as desiderata.53 To enable adherence to such guidance, we have developed and demonstrated DEPLOYR, a technical framework for rapidly deploying as well as prospectively monitoring custom, researcher developed ML models trained using clinical data warehouses. The framework’s design elements and capabilities are demonstrated via silent deployment of multiple ML models triggered by button-clicks in Stanford Healthcare’s production EMR instance, enabling prospective evaluation of the models’ performance on unseen data directly in the deployment environment. The ability to perform enhanced prospective evaluations of researcher created ML models that extend beyond typical retrospective analyses supports the adoption of best practices that can close the ML implementation gap.
Supplementary Material
ACKNOWLEDGMENTS
We used data provided by STARR, “STAnford medicine Research data Repository”, a clinical data warehouse containing de-identified Epic data from Stanford Health Care (SHC), the University Healthcare Alliance (UHA) and Packard Children’s Health Alliance (PCHA) clinics and other auxiliary data from Hospital applications such as radiology PACS. The STARR platform is developed and operated by the Stanford Medicine Research IT team and is made possible by Stanford School of Medicine Research Office. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or Stanford Health Care. Figures 1–5 were created with BioRender.com.
Contributor Information
Conor K Corbin, Department of Biomedical Data Science, Stanford, California, USA.
Rob Maclay, Stanford Children’s Health, Palo Alto, California, USA.
Aakash Acharya, Stanford Health Care, Palo Alto, California, USA.
Sreedevi Mony, Stanford Health Care, Palo Alto, California, USA.
Soumya Punnathanam, Stanford Health Care, Palo Alto, California, USA.
Rahul Thapa, Stanford Health Care, Palo Alto, California, USA.
Nikesh Kotecha, Stanford Health Care, Palo Alto, California, USA.
Nigam H Shah, Center for Biomedical Informatics Research, Division of Hospital Medicine, Department of Medicine, Stanford University, School of Medicine, Stanford, California, USA.
Jonathan H Chen, Center for Biomedical Informatics Research, Division of Hospital Medicine, Department of Medicine, Stanford University, School of Medicine, Stanford, California, USA.
FUNDING
This work was funded in part from the NIH/National Institute on Drug Abuse Clinical Trials Network (UG1DA015815–CTN-0136), Stanford Artificial Intelligence in Medicine and Imaging– Human-Centered Artificial Intelligence Partnership Grant, Doris Duke Charitable Foundation—Covid-19 Fund to Retain Clinical Scientists (20211260), Google Inc (in a research collaboration to leverage health data to predict clinical outcomes), and the American Heart Association—Strategically Focused Research Network—Diversity in Clinical Trials.
AUTHOR CONTRIBUTIONS
CKC and RM are co-first authors indicating equal contribution. CKC contributed to conceptualization, designed the experiments, co-developed the deployment framework, and drafted the manuscript. RM contributed to conceptualization, co-developed the deployment framework and contributed to manuscript drafting and editing. AA, SM, and SP contributed to conceptualization and execution of experiments. RT contributed to design of framework and analysis. NK, NS, and JHC contributed to design of experiments, manuscript draft, and supplied supervision. All authors contributed significantly to editing and finalizing the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
JHC reported receiving consulting fees from Sutton Pierce and Younker Hyde MacFarlane PLLC and being a co-founder of Reaction Explorer LLC, a company that develops and licenses organic chemistry education software using rule-based artificial intelligence technology. CKC reported receiving consultation fees from Fountain Therapeutics, Inc.
CODE AVAILABILITY
The DEPLOYR framework uses 3 distinct software repositories, DEPLOYR-dev to train and validate models, DEPLOYR-serve to expose models to the inference engine and DEPLOYR-dash to monitor and visualize prospective model performance characteristics. While this software is specifically designed to interact with Stanford data models and systems, we make DEPLOYR-dev, DEPLOYR-dash and an example of DEPLOYR-serve publicly available for reference.
DEPLOYR-dev: https://github.com/HealthRex/deployr-dev
DEPLOYR-serve: https://github.com/HealthRex/deployr-serve
DEPLOYR-dash: https://github.com/HealthRex/deployr-dash
DATA AVAILABILITY
De-identified electronic medical record data used here for model training is made available through STARR, STAnford medicine Research data Repository.32 The data can be accessed for research purposes after Institutional Review Board approval via the Stanford Research Informatics Center.
REFERENCES
- 1. Challener DW, Prokop LJ, Abu-Saleh O. The proliferation of reports on clinical scoring systems: issues about uptake and clinical utility. JAMA 2019; 321 (24): 2405–6. [DOI] [PubMed] [Google Scholar]
- 2. Guo LL, Pfohl SR, Fries J, et al. Systematic review of approaches to preserve machine learning performance in the presence of temporal dataset shift in clinical medicine. Appl Clin Inform 2021; 12 (4): 808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen JH, Asch SM. Machine learning and prediction in medicine—beyond the peak of inflated expectations. N Engl J Med 2017; 376 (26): 2507–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matheny M, Israni ST, Ahmed M, Whicher D. Artificial Intelligence in Health Care: The Hope, the Hype, the Promise, the Peril. Washington, DC: National Academy of Medicine; 2019. [PubMed] [Google Scholar]
- 5. Seneviratne MG, Shah NH, Chu L. Bridging the implementation gap of machine learning in healthcare. BMJ Innov 2020; 6 (2): 45–7. [Google Scholar]
- 6. Callahan A, Shah NH. Machine learning in healthcare. In: Sheikh A, Cresswell K, Wright A, Bates D, eds. Key Advances in Clinical Informatics. Amsterdam: Elsevier; 2017: 279–91. [Google Scholar]
- 7. Shah NH, Milstein A, Bagley SC. Making machine learning models clinically useful. JAMA 2019; 322 (14): 1351–2. [DOI] [PubMed] [Google Scholar]
- 8. Ko M, Chen E, Agrawal A, et al. Improving hospital readmission prediction using individualized utility analysis. J Biomed Inform 2021; 119: 103826. [DOI] [PubMed] [Google Scholar]
- 9. Jung K, Kashyap S, Avati A, et al. A framework for making predictive models useful in practice. J Am Med Inform Assoc 2021; 28 (6): 1149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hernandez-Boussard T, Bozkurt S, Ioannidis JP, Shah NH. MINIMAR (MINimum Information for Medical AI Reporting): developing reporting standards for artificial intelligence in health care. J Am Med Inform Assoc 2020; 27 (12): 2011–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu J, Sattler A, Wang S, et al. Considerations in the reliability and fairness audits of predictive models for advance care planning. Front Digit Health 2022; 4: 943768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Char DS, Shah NH, Magnus D. Implementing machine learning in health care—addressing ethical challenges. N Engl J Med 2018; 378 (11): 981–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Armitage H. Researchers create guide for fair and equitable AI in health care. Logo Left ContentLogo Right Content 10,000+ Posts Scope Stanford University School of Medicine Blog; 2022. https://scopeblog.stanford.edu/2022/10/03/researchers-create-guide-for-fair-and-equitable-ai-in-health-care/. Accessed January 12, 2023. [Google Scholar]
- 14. Bedoya AD, Economou-Zavlanos NJ, Goldstein BA, et al. A framework for the oversight and local deployment of safe and high-quality prediction models. J Am Med Inform Assoc 2022; 29 (9): 1631–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reddy S, Allan S, Coghlan S, Cooper P. A governance model for the application of AI in health care. J Am Med Inform Assoc 2020; 27 (3): 491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wiens J, Saria S, Sendak M, et al. Do no harm: a roadmap for responsible machine learning for health care. Nat Med 2019; 25 (9): 1337–40. [DOI] [PubMed] [Google Scholar]
- 17. Kim JY, Boag W, Gulamali F, et al. Organizational governance of emerging technologies: AI adoption in healthcare. In: proceedings of the 2023 ACM conference on fairness, accountability, and transparency; 2023, p. 1396–417. [Google Scholar]
- 18. Sendak MP, Balu S, Schulman KA. Barriers to achieving economies of scale in analysis of EHR data. Appl Clin Inform 2017; 8 (3): 826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sculley D, Holt G, Golovin D, et al. Hidden technical debt in machine learning systems. Adv Neural Inform Process Syst 2015; 28. [Google Scholar]
- 20. Morse KE, Bagley SC, Shah NH. Estimate the hidden deployment cost of predictive models to improve patient care. Nat Med 2020; 26 (1): 18–9. [DOI] [PubMed] [Google Scholar]
- 21. Kashyap S, Morse KE, Patel B, Shah NH. A survey of extant organizational and computational setups for deploying predictive models in health systems. J Am Med Inform Assoc 2021; 28 (11): 2445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siwicki B. An epic cognitive computing platform primer; 2021. https://www.healthcareitnews.com/news/epic-cognitive-computing-platform-primer. Accessed January 14, 2023.
- 23. Major VJ, Jones SA, Razavian N, et al. Evaluating the effect of a COVID-19 predictive model to facilitate discharge: a randomized controlled trial. Appl Clin Inform 2022; 13 (3): 632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang E, Major VJ, Adler N, et al. Supporting acute advance care planning with precise, timely mortality risk predictions. NEJM Catal Innov Care Deliv 2021; 2 (3). 10.1056/CAT.20.0655. [DOI] [Google Scholar]
- 25. Afshar M, Adelaine S, Resnik F, et al. Deployment of real-time natural language processing and deep learning clinical decision support in the electronic health record: pipeline implementation for an opioid misuse screener in hospitalized adults. JMIR Med Inform 2023; 11: e44977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li RC, Smith M, Lu J, et al. Using AI to empower collaborative team workflows: two implementations for advance care planning and care escalation. NEJM Catal Innov Care Deliv 2022; 3 (4): CAT–21. [Google Scholar]
- 27.Microsoft. Azure Functions—Serverless Functions in Computing—Microsoft Azure. 2023. azure.microsoft.com. https://azure.microsoft.com/en-us/products/functions/. Accessed January 19, 2023.
- 28. Streamlit. Streamlit: a faster way to build and share data apps; 2023. https://streamlit.io/. Accessed May 15, 2023.
- 29. Dash S, Shakyawar SK, Sharma M, Kaushik S. Big data in healthcare: management, analysis and future prospects. J Big Data 2019; 6 (1): 1–25. [Google Scholar]
- 30. Corbin CK, Sung L, Chattopadhyay A, et al. Personalized antibiograms for machine learning driven antibiotic selection. Commun Med 2022; 2 (1): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu S, Hom J, Balasubramanian S, et al. Prevalence and predictability of low-yield inpatient laboratory diagnostic tests. JAMA Netw Open 2019; 2 (9): e1910967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Datta S, Posada J, Olson G, et al. A new paradigm for accelerating clinical data science at Stanford Medicine. arXiv preprint arXiv:200310534. 2020.
- 33. Krall M. Acceptance and performance by clinicians using an ambulatory electronic medical record in an HMO. In: proceedings of the Annual Symposium on Computer Application in Medical Care; American Medical Informatics Association; October 28–November 1; 1995, p. 708. New Orleans, Louisiana. [PMC free article] [PubMed]
- 34. Bender D, Sartipi K. HL7 FHIR: an agile and RESTful approach to healthcare information exchange. In: proceedings of the 26th IEEE international symposium on computer-based medical systems. IEEE; June 20–June 22; 2013, pp. 326–31. Porto, Portugal.
- 35. Barker W, Johnson C. The ecosystem of apps and software integrated with certified health information technology. J Am Med Inform Assoc 2021; 28 (11): 2379–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klatt TE, Hopp E. Effect of a best-practice alert on the rate of influenza vaccination of pregnant women. Obstet Gynecol 2012; 119 (2 Pt 1): 301–5. [DOI] [PubMed] [Google Scholar]
- 37. Ahmed AM, Bose S, Breeden M, Smeds M. Interacting with best practice advisory (BPA) notifications in epic significantly improves screening rates for abdominal aortic aneurysms. J Vasc Surg 2022; 76 (4): e75–6. [Google Scholar]
- 38. Ye C, Wang O, Liu M, et al. A real-time early warning system for monitoring inpatient mortality risk: prospective study using electronic medical record data. J Med Internet Res 2019; 21 (7): e13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nemati S, Holder A, Razmi F, Stanley MD, Clifford GD, Buchman TG. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit Care Med 2018; 46 (4): 547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saqib M, Sha Y, Wang MD. Early prediction of sepsis in EMR records using traditional ML techniques and deep learning LSTM networks. In: 2018 40th annual international conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE; July 17–July 21; 2018, pp. 4038–41. Honolulu, HI. [DOI] [PubMed]
- 41. Tomašev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 2019; 572 (7767): 116–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Keller MS. Take command: cron: job scheduler. Linux J 1999; 1999 (65es): 15-es. [Google Scholar]
- 43. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: advances in neural information processing systems; December 4–December 9; 2017;30. Long Beach, CA. [Google Scholar]
- 44. Flynn MS, Jiang SW, Stauffer T, Kwock JT, Nicholas MW. Tracking procedure outcomes using Epic SmartText and SmartData Elements following minor dermatologic procedures in the ambulatory setting. J Am Acad Dermatol 2023; 88 (3): 659–61. [DOI] [PubMed] [Google Scholar]
- 45. Goldberg HS, Paterno MD, Grundmeier RW, et al. Use of a remote clinical decision support service for a multicenter trial to implement prediction rules for children with minor blunt head trauma. Int J Med Inform 2016; 87: 101–10. [DOI] [PubMed] [Google Scholar]
- 46. Bloomfield RA Jr, Polo-Wood F, Mandel JC, Mandl KD. Opening the Duke electronic health record to apps: implementing SMART on FHIR. Int J Med Inform 2017; 99: 1–10. [DOI] [PubMed] [Google Scholar]
- 47. Feng J, Phillips RV, Malenica I, et al. Clinical artificial intelligence quality improvement: towards continual monitoring and updating of AI algorithms in healthcare. NPJ Digit Med 2022; 5 (1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schröder T, Schulz M. Monitoring machine learning models: a categorization of challenges and methods. Data Sci Manage 2022; 5 (3): 105–16. [Google Scholar]
- 49. Klaise J, Van Looveren A, Cox C, Vacanti G, Coca A. Monitoring and explainability of models in production. arXiv preprint arXiv:200706299. 2020.
- 50. Jung K, Shah NH. Implications of non-stationarity on predictive modeling using EHRs. J Biomed Inform 2015; 58: 168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vickers AJ, Van Calster B, Steyerberg EW. Net benefit approaches to the evaluation of prediction models, molecular markers, and diagnostic tests. BMJ 2016; 352: i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tonekaboni S, Morgenshtern G, Assadi A, et al. How to validate machine learning models prior to deployment: silent trial protocol for evaluation of real-time models at ICU. In: conference on health, inference, and learning. PMLR; April 7–April 8; 2022, pp. 169–82. Virtual Event.
- 53.Blueprint for trustworthy AI implementation guidance and assurance for healthcare. https://tinyurl.com/CHAI-paper. Accessed March &, 2023.
- 54. Otles E, Oh J, Li B, et al. Mind the performance gap: examining dataset shift during prospective validation. In: Machine learning for healthcare conference. PMLR; August 6–August 7; 2021, pp. 506–34. Virtual Event.
- 55. Krautenbacher N, Theis FJ, Fuchs C. Correcting classifiers for sample selection bias in two-phase case-control studies. Comput Math Methods Med 2017; 2017: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reps JM, Ryan PB, Rijnbeek PR, Schuemie MJ. Design matters in patient-level prediction: evaluation of a cohort vs. case-control design when developing predictive models in observational healthcare datasets. J Big Data 2021; 8 (1): 1–18.33425651 [Google Scholar]
- 57. David G, Smith-McLallen A, Ukert B. The effect of predictive analytics-driven interventions on healthcare utilization. J Health Econ 2019; 64: 68–79. [DOI] [PubMed] [Google Scholar]
- 58. Adams R, Henry KE, Sridharan A, et al. Prospective, multi-site study of patient outcomes after implementation of the TREWS machine learning-based early warning system for sepsis. Nat Med 2022; 28 (7): 1455–60. [DOI] [PubMed] [Google Scholar]
- 59. Nemati S, Shashikumar SP, Holder AL, Wardi G, Owens RL. Randomized clinical trials or convenient controls: TREWS or FALSE? medRxiv. 2022.
- 60. Escobar GJ, Liu VX, Schuler A, Lawson B, Greene JD, Kipnis P. Automated identification of adults at risk for in-hospital clinical deterioration. N Engl J Med 2020; 383 (20): 1951–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Aikens RC, Balasubramanian S, Chen JH. A machine learning approach to predicting the stability of inpatient lab test results. AMIA Summits Transl Sci Proc 2019; 2019: 515. [PMC free article] [PubMed] [Google Scholar]
- 62. Rabbani N, Ma SP, Li RC, et al. Targeting repetitive laboratory testing with electronic health records-embedded predictive decision support: a pre-implementation study. Clin Biochem 2023; 113: 70–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kim GY, Noshad M, Stehr H, et al. Machine learning predictability of clinical next generation sequencing for hematologic malignancies to guide high-value precision medicine. In: AMIA annual symposium proceedings. vol. 2021. American Medical Informatics Association; October 30–November 3; 2021, p. 641. San Diego, California. [PMC free article] [PubMed]
- 64. Steinberg E, Jung K, Fries JA, Corbin CK, Pfohl SR, Shah NH. Language models are an effective representation learning technique for electronic health record data. J Biomed Inform 2021; 113: 103637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Corbin CK, Medford RJ, Osei K, Chen JH. Personalized antibiograms: machine learning for precision selection of empiric antibiotics. AMIA Summits Transl Sci Proc 2020; 2020: 108. [PMC free article] [PubMed] [Google Scholar]
- 66. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018; 1 (1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Grinsztajn L, Oyallon E, Varoquaux G. Why do tree-based models still outperform deep learning on typical tabular data? In: thirty-sixth conference on neural information processing systems datasets and benchmarks track; 2022.
- 68. Hastie T, Tibshirani R, Friedman JH, Friedman JH. The Elements of Statistical Learning: data Mining, Inference, and Prediction. vol. 2. New York, NY: Springer; 2009. [Google Scholar]
- 69. Garg A. Why use azure functions for ML inference? 2020. https://techcommunity.microsoft.com/t5/apps-on-azure-blog/why-use-azure-functions-for-ml-inference/ba-p/1416728. Accessed January 9, 2023.
- 70. Rabanser S, Günnemann S, Lipton Z. Failing loudly: an empirical study of methods for detecting dataset shift. Adv Neural Inf Process Syst 2019; 32. [Google Scholar]
- 71. Davis SE, Greevy RA Jr, Lasko TA, Walsh CG, Matheny ME. Detection of calibration drift in clinical prediction models to inform model updating. J Biomed Inform 2020; 112: 103611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Davis SE, Greevy RA Jr, Fonnesbeck C, Lasko TA, Walsh CG, Matheny ME. A nonparametric updating method to correct clinical prediction model drift. J Am Med Inform Assoc 2019; 26 (12): 1448–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lenert MC, Matheny ME, Walsh CG. Prognostic models will be victims of their own success, unless. J Am Med Inform Assoc 2019; 26 (12): 1645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Perdomo J, Zrnic T, Mendler-Dünner C, Hardt M. Performative prediction. In: International conference on machine learning. PMLR; July 12–July 18; 2020, pp. 7599–609. Virtual Event.
- 75. Adam GA, Chang CHK, Haibe-Kains B, Goldenberg A. Hidden risks of machine learning applied to healthcare: unintended feedback loops between models and future data causing model degradation. In: Machine learning for healthcare conference. PMLR; 2020, pp. 710–31.
- 76. Adam GA, Chang CHK, Haibe-Kains B, Goldenberg A. Error amplification when updating deployed machine learning models. In: Proceedings of the Machine Learning for Healthcare Conference, Durham, NC, USA; 2022, pp. 5–6.
- 77. Corbin CK, Baiocchi M, Chen JH. Avoiding biased clinical machine learning model performance estimates in the presence of label selection. arXiv preprint arXiv:220909188. 2022. [PMC free article] [PubMed]
- 78. Nguyen M, Corbin CK, Eulalio T, et al. Developing machine learning models to personalize care levels among emergency room patients for hospital admission. J Am Med Inform Assoc 2021; 28 (11): 2423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chen JH, Podchiyska T, Altman RB. OrderRex: clinical order decision support and outcome predictions by data-mining electronic medical records. J Am Med Inform Assoc 2016; 23 (2): 339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Nguyen M, Jankovic I, Kalesinskas L, Baiocchi M, Chen JH. Machine learning for initial insulin estimation in hospitalized patients. J Am Med Inform Assoc 2021; 28 (10): 2212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gordon WJ, Mandl KD. The 21st century cures act: a competitive apps market and the risk of innovation blocking. J Med Internet Res 2020; 22 (12): e24824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Centers for Medicare & Medicaid Services (CMS). Interoperability and patient access. 2023. https://www.cms.gov/regulations-and-guidance/guidance/interoperability/index. Accessed January 17, 2023.
- 83. FHIR (Fast Healthcare Interoperability Resources). HTTP—FHIR v4.0.1; 2023. https://build.fhir.org/http.html. Accessed May 15, 2023.
- 84. Rico PM, Pérez AV, Martínez JML. Electronic alerting and decision support for early sepsis detection and management: impact on clinical outcomes. Eur J Clin Pharm: Farm 2017; 19 (1): 33–40. [Google Scholar]
- 85. Cerner. Health Systems Solutions; 2023. https://www.cerner.com/solutions/health-systems. Accessed May 9, 2023.
- 86. Pricing—functions: Microsoft Azure. https://azure.microsoft.com/en-us/pricing/details/functions/. Accessed January 9, 2023.
- 87. Microsoft. Azure cosmos DB autoscale provisioned throughput; 2023. https://azure.microsoft.com/en-us/pricing/details/cosmos-db/autoscale-provisioned/. Accessed May 10, 2023.
- 88. Rakha EA, Toss M, Shiino S, et al. Current and future applications of artificial intelligence in pathology: a clinical perspective. J Clin Pathol 2021; 74 (7): 409–14. [DOI] [PubMed] [Google Scholar]
- 89. Rezazade Mehrizi MH, van Ooijen P, Homan M. Applications of artificial intelligence (AI) in diagnostic radiology: a technography study. Eur Radiol 2021; 31 (4): 1805–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Eapen BR. Artificial intelligence in dermatology: a practical introduction to a paradigm shift. Indian Dermatol Online J 2020; 11 (6): 881–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified electronic medical record data used here for model training is made available through STARR, STAnford medicine Research data Repository.32 The data can be accessed for research purposes after Institutional Review Board approval via the Stanford Research Informatics Center.