Abstract
Mitogen-activated protein (MAP) kinase-mediated signalling to the nucleus is an important event in the conversion of extracellular signals into a cellular response. However, the existence of multiple MAP kinases which phosphorylate similar phosphoacceptor motifs poses a problem in maintaining substrate specificity and hence the correct biological response. Both the extracellular signal-regulated kinase (ERK) and c-Jun NH2-terminal kinase (JNK) subfamilies of MAP kinases use a second specificity determinant and require docking to their transcription factor substrates to achieve maximal substrate activation. In this study, we demonstrate that among the different MAP kinases, the MADS-box transcription factors MEF2A and MEF2C are preferentially phosphorylated and activated by the p38 subfamily members p38α and p38β2. The efficiency of phosphorylation in vitro and transcriptional activation in vivo of MEF2A and MEF2C by these p38 subtypes requires the presence of a kinase docking domain (D-domain). Furthermore, the D-domain from MEF2A is sufficient to confer p38 responsiveness on different transcription factors, and reciprocal effects are observed upon the introduction of alternative D-domains into MEF2A. These results therefore contribute to our understanding of signalling to MEF2 transcription factors and demonstrate that the requirement for substrate binding by MAP kinases is an important facet of three different subclasses of MAP kinases (ERK, JNK, and p38).
The mitogen-activated protein (MAP) kinase pathways transduce extracellular signals into distinct nuclear responses (reviewed in references 29 and 33). These pathways are conserved in a diverse set of organisms, ranging from Saccharomyces cerevisiae to humans, and have been implicated in regulating multiple cellular processes (reviewed in reference 29). In humans, there are at least four separate subclasses of MAP kinases (extracellular signal-regulated kinase [ERK], c-Jun NH2-terminal kinase [JNK], p38, and ERK5/BMK), with members of each subclass having significant sequence similarity and apparently, in most cases, sharing several upstream components. Furthermore, the same signals often activate overlapping subsets of MAP kinases, although the speed and efficiency of activation often differ (reviewed in reference 1).
An increasing number of nuclear targets for the different MAP kinase cascades are being identified, and in most cases, it appears that each transcription factor acts as a target for one or a limited subset of MAP kinases (reviewed in reference 29). Several recent advances have contributed to our understanding of how these kinases achieve their substrate specificity and elicit distinct biological responses (reviewed in references 17 and 27). For example, in yeast, the binding of the inactive Kss1p MAP kinase to its nuclear targets (Ste12p-Tec1p) inhibits the activation of this transcription factor complex by the parallel MAP kinase cascade containing Fus3p. Similarly, Fus3p appears to have a reciprocal inhibitory action, in this case by blocking the upstream pheromone-responsive module from spuriously activating Kss1p (2, 16; reviewed in references 17 and 27). In mammals, MAP kinases have also been shown to bind to their nuclear targets, and it has been proposed that this might contribute to their substrate specificity (reviewed in reference 29). In the case of c-Jun, the binding of MAP kinases to a domain which is distinct from the phosphoacceptor motifs, the δ-domain, has been shown to be an important part of the kinase specificity-determining mechanism and is required for its efficient phosphorylation by the JNK MAP kinases (3, 4, 7, 12, 13). Similar docking domains have been identified in activating transcription factor 2 (ATF-2) (for JNK MAP kinases) (6, 7, 15) and in Elk-1 (for JNK and ERK MAP kinases) (37, 38). In Elk-1, the docking domain appears to act to allow the convergence of two diverse MAP kinase pathways on a single transcription factor, whereas the δ-domain in c-Jun permits phosphorylation by a single type of cascade. It is currently unclear how p38 MAP kinases are targeted to transcription factors, although the docking domain in ATF-2 is thought to permit targeting by p38α in addition to the JNK MAP kinases (11a).
It has recently been demonstrated that p38α associates with the MADS-box family transcription factor MEF2C. Subsequent phosphorylation of MEF2C stimulates its ability to activate transcription (8). In mammals, there are four MEF2C-related genes, MEF2A, MEF2B, MEF2C, and MEF2D, that encode proteins which exhibit significant amino acid sequence similarity within their DNA binding domains and to a lesser extent throughout the rest of the proteins. One of the major roles of these proteins is to regulate muscle-specific gene expression (reviewed in references 19 and 24), although they have recently been shown to also contribute to the activation of c-jun transcription (8, 9, 14). Until recently, little was known about MAP kinase signalling to MEF2 family proteins other than MEF2C, which, in addition to being a p38α target, can also be regulated by ERK5/BMK (14, 36). In this study, we have investigated the phosphorylation of MEF2C and the related family member MEF2A by the p38 subclass of MAP kinases. We demonstrate p38α- and p38β2-specific activation of MEF2A and MEF2C and demonstrate the existence of a MAP kinase docking domain which is specific for these two MAP kinase subtypes. Furthermore, we demonstrate the interchangeability of MAP kinase docking domains and that these domains play a major role in determining the specificity of MAP kinase signalling to nuclear transcription factors.
MATERIALS AND METHODS
Plasmid constructs.
The following plasmids were used for expressing glutathione S-transferase (GST) fusion proteins in Escherichia coli. pAS860 (encoding GST-MEF2A; MEF2A amino acids 266 to 413), pAS861 (encoding GST-MEF2AΔD; MEF2A amino acids 283 to 413), pAS865 (encoding GST-MEF2C; MEF2C amino acids 249 to 378), pAS866 (encoding GST-MEF2CΔD; MEF2C amino acids 265 to 378), and pAS869 (encoding GST-MEF2B; MEF2B amino acids 201 to 300) were constructed by inserting BamHI/EcoRI-cleaved PCR-derived fragments into the same sites of pGEX-3X (Pharmacia). pAS406 (encoding GST–Elk-1ΔD; Elk-1 amino acids 330 to 428), pAS545 (encoding GST–Elk-1; Elk-1 amino acids 310 to 428) (37), and a plasmid encoding GST-Jun (c-Jun amino acids 1 to 79) (4) have been described previously. pAS862, pAS863, and pAS864 (encoding GST-MEF2A mutants) are derivatives of pAS860 with the site-directed mutations R269A and K270A (GST-MEF2A[M1]), L273A and V275A (GST-MEF2A[M2]), and I277A and P278A (GST-MEF2A[M3]), respectively. pAS865 (encoding GST–MEF2A–Elk-1; MEF2A amino acids 266 to 292; Elk-1 amino acids 330 to 428), pAS866 (encoding GST–MEF2A–c-Jun; MEF2A amino acids 266 to 292; c-Jun amino acids 55 to 79), pAS870 (encoding GST-MEF2B-MEF2A; MEF2B amino acids 201 to 224; MEF2A amino acids 292 to 413), pAS871 (encoding GST-cJunδ-MEF2A; c-Jun amino acids 1 to 55; MEF2A amino acids 283 to 413), pAS872 (encoding GST-SAPD-MEF2A; SRF accessory protein 1 [SAP-1] amino acids 290 to 334; MEF2A amino acids 283 to 413), and pAS873 (encoding GST-ElkD-MEF2A; Elk-1 amino acids 310 to 327; MEF2A amino acids 283 to 413) were constructed by inserting a BamHI/EcoRI-cleaved PCR-derived fragment into the same sites of pGEX-3X. PCR fragments were generated by a two-step PCR protocol (31) with a mutagenic primer consisting of nucleotides homologous to each half of the chimeric product, two flanking primers, and the following pairs of templates in each step: pAS860 and pAS407, pAS860 and GST-Jun, pAS860 and pAS869, GST-Jun and pAS860, GST-SAPC (32) and pAS860, and pAS407 and pAS860, respectively.
The following plasmids were constructed for use in mammalian cell transfections. pG5E1b contains five GAL4 DNA binding sites cloned upstream of a minimal promoter element and the firefly luciferase gene (23). pSG424 encodes the GAL4 DNA binding domain (22), and the vectors pCMV5-HA-ERK2 (18), pcDNA3-F-JNK2 (26), pCMV5-F-p38α (21), pcDNA3-F-p38β2 (5), pcDNA3-F-p38γ (5), and pcDNA3-F-p38δ (30) encoding hemagglutinin (HA)- or Flag (F)-tagged MAP kinases, pCMV5-HA-MEK1 (ΔNS218E-S222D), pcDNA3-F-MKK6(E), and pcDNA3-F-MKK7β, encoding HA- or Flag-tagged wild-type (MKK7β) or constitutively active (MEK1 and MKK6) MAP kinase kinases (MKKs) have been described previously (35). pAS874 (GAL4-MEF2A), pAS875 (GAL4-MEF2AΔD), pAS876 (GAL4-MEF2A[M1]), pAS877 (GAL4-MEF2A[M2]), pAS878 (GAL4-MEF2A[M3]), pAS879 (GAL4–MEF2A–Elk-1), pAS880 (GAL4-MEF2A-cJun), pAS881 (GAL4-MEF2C), and pAS882 (GAL4-MEF2CΔD) were constructed by ligating BamHI/XbaI fragments from pAS860 to pAS868, respectively, into the same sites of pSG424. pAS897 (GAL4-Elk330 [GAL4–Elk-1ΔD]) and pAS898 (GAL4-Elk310 [GAL4–Elk-1]) were constructed by ligating BamHI/XbaI fragments from pAS406 and pAS545 (37), respectively, into the same sites of pSG424. GAL–c-Jun has previously been described (10). pAS883 and pAS891 encode the same series of GAL4 fusion proteins under the control of the cytomegalovirus (CMV) promoter and were constructed by ligating the HindIII/XbaI fragments from pAS874 to pAS882 into the same sites of pCMV5. pAS900 and pAS1351 encode the GAL4 fusion proteins GAL4–Elk-1 and GAL4–Elk-1ΔD, respectively, under the control of the CMV promoter and were constructed by ligating the HindIII/XbaI fragments from pAS897 and pAS898 into the same sites of pCMV5. pAS572 (encoding CMV promoter-regulated GAL4–Elk-205) has been described previously (37). Details of PCR primers can be supplied on request. All plasmid constructs made by PCR were verified by automated dideoxy sequencing.
The following plasmids were constructed for the production of proteins by in vitro transcription and translation. pAS522 (encoding full-length MEF2A; amino acids 1 to 486) was constructed by inserting an NcoI fragment from pT7C4 (20) into the NcoI site in pAS37. pAS1201 is identical to pAS522 except that an XhoI site was introduced into the 3′ end of the coding region (by using the primer ADS575) and the 3′ untranslated region was also lost from the clone. pAS1203 (encoding C-terminally Flag- or His-tagged full-length MEF2A under the control of a T7 promoter) was constructed by inserting an NcoI/XhoI fragment from pAS1201 into the same sites in pETnefPFH. pAS1209 (encoding C-terminally Flag- and His-tagged full-length MEF2A under the control of a T3 promoter) was constructed by inserting an NcoI/EcoRI fragment from pAS1203 into the same sites in pAS798 (31). pAS1397 (encoding full-length MEF2A with the point mutations I277A and P278A) was constructed by replacing the BglII/XhoI fragment in pAS1209 with a PCR-derived fragment incorporating the two point mutations.
Protein expression and purification.
GST fusion proteins were expressed in E. coli JM101 and purified as described previously (25, 37). In vitro-translated proteins were produced by sequential transcription and translation with rabbit reticulocyte lysate (Promega). Tagged proteins were immunoprecipitated prior to use in protein kinase assays, by using 10 μl of anti-Flag antibody conjugated to agarose beads (50% slurry; Sigma). Proteins were bound to the beads for 1 h at room temperature, washed five times with Triton lysis buffer (TLB) (37), and resuspended in an equal volume of TLB.
Tissue culture, cell transfection, and reporter gene assays.
COS-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco BRL). CHO cells were maintained in F12 medium supplemented with 5% FBS. 293 cells were maintained in DMEM supplemented with 10% FBS. HeLa cells were maintained in DMEM supplemented with 10% FBS. Transfection experiments were carried out with Superfect transfection reagent (Qiagen) as described previously (37, 38).
For reporter gene assays, a luciferase reporter construct controlled by a GAL4-driven promoter was cotransfected with CMV promoter-driven vectors encoding various GAL4 fusion proteins. The activities of the GAL4 fusion proteins (usually 50 ng of plasmid DNA but 5 ng with GAL4-MEF2C derivatives) were measured in cotransfection assays in all cell lines by using 1 μg of the reporter plasmid pG5E1bLuc and, where indicated, 250 ng of vectors encoding a MAP kinase and constitutively activated MKK. Transfection efficiencies were normalized by measuring the activity from a cotransfected plasmid (1 μg) which expresses β-galactosidase (pCH110; Pharmacia KB Biotechnology Inc.). To stimulate the endogenous MAP kinase pathways, COS-7 cells were treated with 50 nM epidermal growth factor (EGF) (Sigma), and CHO and HeLa cells were treated with 10 ng of mouse interleukin 1α (IL-1α) (Genzyme Corp.) and left for 12 h. Cell extracts were prepared and luciferase and β-galactosidase assays were carried out as described previously (37, 38).
Protein kinase assays.
In order to prepare recombinant JNK2 and p38 MAP kinases (α, β2, γ, and δ), COS-7 cells were transfected with constructs encoding Flag epitope-tagged MAP kinases. Kinases were activated by stimulation with UV light for 30 min (for JNK2) or were cotransfected with a constitutively activated form of MKK6 [MKK6(E)] (for p38α, β2, γ, and δ), and the active kinases were purified by immunoprecipitation as described previously (37). Purified kinases were eluted from beads by competing with 0.1 mg of Flag peptide per ml. Recombinant active ERK2 was obtained from New England Biolabs. The kinase assays were carried out in 20-μl reaction volumes containing in vitro-translated proteins bound to anti-Flag antibody conjugated to agarose beads or with 5 pmol of GST fusion proteins as a substrate as described previously (37, 38). The phosphorylation of substrate proteins was examined by autoradiography following sodium dodecyl phosphate-polyacrylamide gel electrophoresis and quantified by phosphorimaging (Fuji BAS1500; TINA 2.08e software), and the data were presented graphically after curve fitting with the appropriate equation by using BIOSOFT Fig.P or Microsoft Excel software. Peptide competition experiments were carried out essentially as described above, except for the preincubation of the p38 MAP kinases with 50 to 5,000 pmol of the peptide competitors before the kinase reactions. Final peptide concentrations were 2.5 to 25 μM (a 10- to 1,000-fold excess over GST fusion protein substrates).
Western blot analysis.
GAL4 fusion proteins were detected in total 293 cell extracts by using the anti-GAL4 antibody directed against the amino-terminal DNA-binding domain (Santa Cruz). Elk-1 phosphorylated on Ser383 was detected with an anti-phospho-Elk-1(Ser383) antibody (New England Biolabs). Immunocomplexes were detected by using horseradish peroxidase-conjugated secondary antibody followed by enhanced chemiluminescence (Amersham).
Figure generation and data quantification.
All figures were generated electronically from scans of autoradiographic images by using Picture Publisher (Micrografix) and Powerpoint version 7.0 (Microsoft) software. Final images are representative of the original autoradiographic images. Phosphorimager data were quantified by using Tina software (version 2.08e).
RESULTS
Phosphorylation and activation of MEF2 transcription factors by p38 MAP kinases.
The transcription factor MEF2C is phosphorylated and activated by the p38α MAP kinase in response to inflammation (8), thereby providing a link between the p38 pathway and a member of the MEF2 group of transcription factors. However, several distinct members of the p38 MAP kinases have been identified, including p38α, -β, -γ, and -δ (reviewed in references 1 and 30). The abilities of these different p38 MAP kinases to phosphorylate MEF2 proteins in vitro and stimulate their transcriptional activation properties in vivo were tested (Fig. 1). Firstly, the phosphorylation of a GST fusion protein containing the transcriptional activation domain (TAD) of MEF2A and MEF2C by the different p38 MAP kinases was tested (Fig. 1B). These fusion proteins contain the minimal TAD, which in the case of MEF2C, has been shown to be sufficient for maximal phosphorylation-inducible transcriptional activation (8). This region contains the major p38α phosphorylation sites T293 and T300 in MEF2C (8) and the corresponding amino acid residues T304 and T311 in MEF2A (Fig. 1A). An independent study has confirmed that these two residues in MEF2A are the major p38α phosphorylation sites (39; T312 and T319 in the MEF2A isoform were used in that study). GST-MEF2A and GST-MEF2C are both phosphorylated by p38α and p38β2 (Fig. 1B, lanes 1 and 2). However, in comparison, neither p38γ nor p38δ can efficiently phosphorylate GST-MEF2A and GST-MEF2C (Fig. 1B, lanes 3 and 4). Moreover, GST-MEF2A and GST-MEF2C appear to be phosphorylated selectively by a subgroup of p38 MAP kinases but not by other classes of MAP kinases, as ERK and JNK family members only poorly phosphorylate these substrates in vitro (data not shown).
FIG. 1.
Phosphorylation and activation of MEF2A or MEF2C by different p38 MAP kinases. (A) Diagram illustrating the domain structure of full-length MEF2C and truncated MEF2A and MEF2C proteins fused to either GST or the GAL4 DNA binding domain (open boxes). The location of the DNA binding domain (DBD), minimal TAD, and p38α (T293 and T300) and ERK5 (S387) phosphorylation sites in MEF2C are indicated. The black box represents the putative kinase docking domain (D-domain) of MEF2A/C, and the numbers of the N- and C-terminal amino acids in the MEF2A or MEF2C moiety are indicated. The sequences of MEF2A and MEF2C around the major phosphoacceptor motifs for p38α MAP kinase are shown. (B) The phosphorylation of GST-MEF2A, GST-MEF2C, and GST–Elk-310 by the p38 MAP kinase subtypes p38α (lane 1), p38β2 (lane 2), p38γ (lane 3), and p38δ (lane 4) was examined by a protein kinase assay. The activity of each protein kinase was standardized towards the substrate GST–Elk-310 (bottom panel). Kinase assays were performed for 15 min at 30°C with 5 pmol of GST-MEF2A and GST-MEF2C as substrates. (C) COS-7 cells were cotransfected with either GAL4-MEF2A or GAL4-MEF2C expression vectors, a constitutively activated form of MKK6 (MKK6[E]), the indicated p38 MAP kinases, and a GAL4-driven luciferase reporter plasmid. (D) COS-7 cells were cotransfected with expression vectors encoding GAL4-MEF2A, an overexpressed (MKK7β) or constitutively activated form of MKK (MEK and MKK6), a MAP kinase (ERK2, JNK2, and p38β2), and a GAL4-driven luciferase reporter plasmid. (E) HeLa cells were cotransfected with vectors encoding GAL4-MEF2A and a GAL4-driven luciferase reporter plasmid. Cells either were left unstimulated or were stimulated with IL-1 for 18 h in the absence or presence of the indicated JNK pathway (cotransfected dominant negative form of MKK4 [DN-MKK4]) or p38 pathway (SB202190) inhibitors. (F) COS-7 (for EGF stimulation of ERKs), CHO (for IL-1 stimulation of JNKs), and HeLa (for IL-1 stimulation of p38s) cells were cotransfected with vectors encoding GAL4-MEF2A or GAL4–Elk-1 and a GAL4-driven luciferase reporter plasmid. The cells either were left unstimulated (white bars) or were stimulated (black bars for Elk-1 and grey bars for MEF2A) with EGF or IL-1 as in panel E. Transfection efficiencies were monitored by using the β-galactosidase expression vector pCH110. The normalized luciferase activities (means ± standard errors; n, 2 or 3) are presented.
In order to determine whether this differential phosphorylation by p38 MAP kinases reflects a difference in their response to these pathways in vivo, GAL4 fusion proteins containing the TADs of MEF2A and MEF2C were constructed (Fig. 1A) and were tested for their ability to activate a GAL4-driven luciferase reporter gene. The activation of distinct MAP kinase cascades was achieved by cotransfecting a constitutively activated form of MKK6, together with either p38α, p38β2, p38γ, or p38δ. In the cell line used in this study (COS-7), the expression of MKK6 alone had little effect on the activity of the GAL4 fusion proteins (Fig. 1C), although the activation of MEF2A can be detected by using MKK6 alone in other cell lines (e.g., 293 cells [39]). MEF2A and MEF2C were efficiently activated by p38α and p38β2 but not by p38γ and p38δ (Fig. 1C). To test the ability of the ERK2 and JNK2 pathways to activate MEF2A, similar experiments were performed with the cotransfection of their respective upstream kinases (MAP kinase [or Erk] kinase [MEK] and MKK7β, respectively). However, in comparison to p38β2 (∼50-fold induction), both ERK2 and JNK2 had little effect on the activity of MEF2A (Fig. 1D). Treatment with MEK/ERK2 and MKK7β/JNK2 is sufficient to activate other nuclear substrates (see Fig. 6) (37, 39).
FIG. 6.
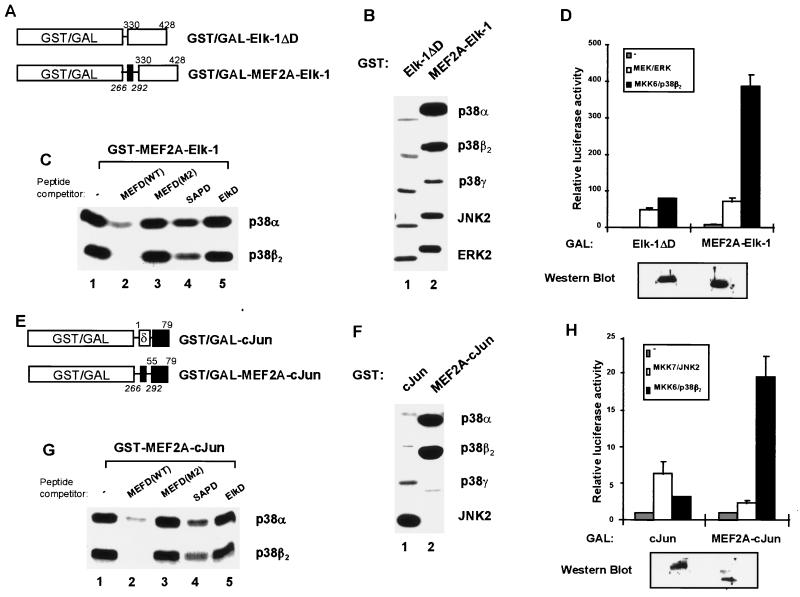
The p38-binding domain of MEF2A (D-domain) functions in a heterologous context. The phosphorylation and activation of GST and GAL4 fusions to MEF2A–Elk-1 (A to D) and MEF2A-cJun (E to H) chimeras by MAP kinases were analyzed. (A and E) Diagrams illustrating fusions of GST to Elk-1ΔD, MEF2A–Elk-1, cJun, and MEF2A-cJun. The numbers of the N- and C-terminal Elk-1/cJun (roman type) and MEF2A (italics) amino acids included in these constructs are indicated above and below each construct, respectively. The MEF2A D-domain and cJun δ-domain are indicated by solid and open boxes, respectively. The Elk-1 and cJun transcriptional activation domains are shown by open and black boxes, respectively. (B and F) Kinase assays of GST fusion proteins (5 pmol of each substrate) for the indicated MAP kinases were carried out for 15 min as described in the legend to Fig. 1. (C and G) Phosphorylation of the GST–MEF2A–Elk-1 and GST-MEF2A-cJun chimeras by MAP kinases was analyzed in the presence of competitor peptides as described in the legend to Fig. 4. Five picomoles of each chimeric protein was used in the reactions. The indicated p38 MAP kinases were preincubated in the absence (lane 1) or presence of 500 pmol of the following peptides: MEF2A WT D-domain (lane 2; MEFD[WT]), MEF2A mutant D-domain peptide (lane 3; MEFD[M2]), SAP-1 D-domain peptide (lane 4; SAPD), and Elk-1 D-domain peptide (lane 5; ElkD). (D and H) COS-7 cells were cotransfected with vectors encoding GAL4–Elk-1ΔD, GAL4–MEF2A–Elk-1, GAL4-cJun, and GAL4-MEF2A-cJun fusions, a GAL4-driven luciferase reporter plasmid, and vectors encoding MAP kinases (ERK2, JNK2, and p38β2) and an overexpressed (MKK7β) or constitutively activated form (MEKΔN and MKK6[E]) of the upstream MAP kinase kinases. The expression levels of the GAL4 fusion proteins in unstimulated cells were examined by Western blotting with an anti-GAL4 antibody (bottom panel).
The response of MEF2A to the activation of endogenous MAP kinase pathways following stimulation by mitogens and cytokines in the absence of overexpressed pathway components was subsequently investigated. Initially, HeLa cells were stimulated with IL-1. This treatment results in the stimulation of GAL4-MEF2A via the p38 pathway, as the JNK pathway inhibitor (DN-MKK4) has little effect, while the p38 pathway inhibitor (SB202190) almost completely blocks this stimulation (Fig. 1E). To directly compare the effect of stimulating individual MAP kinase pathways on MEF2A activation, COS-7, CHO, and HeLa cells were transfected with GAL4-MEF2A and treated with either EGF (to activate the ERK pathway in COS-7 cells [37]) or IL-1 (to activate the JNK pathway in CHO cells [34] or the p38 pathway in HeLa cells [Fig. 1E]). While the treatment of COS-7 cells with EGF and that of CHO cells with IL-1 lead to the activation of GAL4-Elk-1 via the ERK and JNK pathways, neither of these treatments activates GAL4-MEF2A (Fig. 1F). However, in comparison, the stimulation of the endogenous p38 pathway by IL-1 treatment of HeLa cells results in a comparable activation of GAL4–Elk-1 and GAL4-MEF2A (Fig. 1F).
Taken together, these results demonstrate that like MEF2C, MEF2A is phosphorylated and activated by p38α. This is in agreement with the findings of two independent studies (18a, 39). Moreover, while both MEF2A and MEF2C are also targeted by p38β2, neither appears to be a target of p38γ and p38δ. Thus, these MEF2 family proteins appear to be substrates of a subset of p38 MAP kinases in vitro and in vivo.
Requirement of the MEF2 D-domain for phosphorylation by p38α and p38β2 MAP kinases in vitro and in vivo.
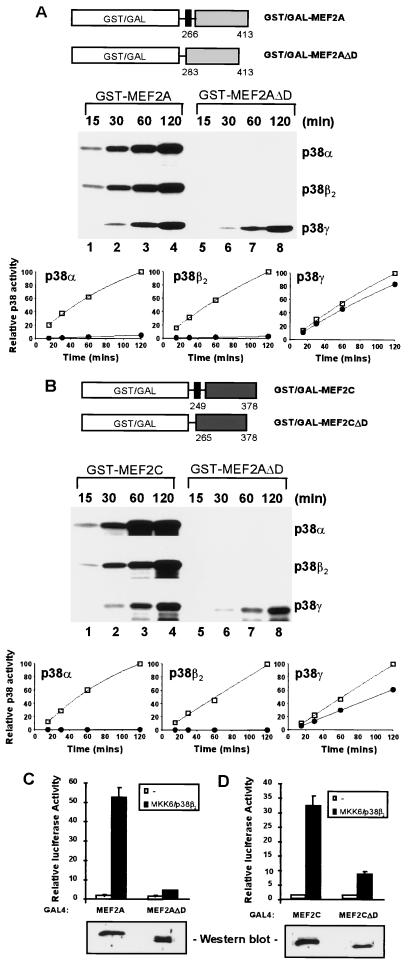
We have previously identified within the transcription factor Elk-1 a kinase docking domain, the D-domain, that contains specificity determinants and therefore enhances the phosphorylation of Elk-1 by ERK and JNK MAP kinases. However, this domain does not appear to affect the phosphorylation of Elk-1 by the p38 MAP kinases (37, 38). Inspection of the sequence of MEF2A and MEF2C indicated the presence of a motif which exhibits limited similarity with the Elk-1 D-domain (see Fig. 5A). To investigate whether the efficiency of MEF2A and MEF2C phosphorylation by p38α and p38β2 is enhanced by the presence of a kinase docking domain, the GST fusion proteins GST-MEF2AΔD and GST-MEF2CΔD, which contain the TAD and associated phosphoacceptor motifs but lack the region which resembles the Elk-1 D-domain (Fig. 2A and B), were created. These GST fusion proteins were tested as in vitro MAP kinase substrates, in comparison to analogous proteins which contain the putative MAP kinase docking site (Fig. 2A and B). The activity of the kinases towards GST–Elk-1 was initially standardized, and equivalent activities were used in the kinase assays. The kinetics of phosphorylation of GST-MEF2A (Fig. 2A, lanes 1 to 4) and GST-MEF2C (Fig. 2B, lanes 1 to 4) by p38α, p38β2, and p38γ were virtually indistinguishable. In contrast, the phosphorylation of GST-MEF2AΔD (Fig. 2A, lanes 5 to 8) and GST-MEF2CΔD (Fig. 2B, lanes 5 to 8) by p38α and p38β2 was greatly reduced over the same time period. However, the phosphorylation of these two substrates by p38γ was virtually indistinguishable from that when the putative docking site was present (compare the graphs and lanes 1 to 4 and 5 to 8 in the bottom panels of Fig. 2A and B).
FIG. 5.
The MEF2A D-domain acts as a binding site for the p38α and p38β2 MAP kinases. (A) The sequences of the competitor peptides corresponding to the MAP kinase docking domains from Elk-1, SAP-1, and MEF2A. Sequences are aligned to give maximal similarities, with identical and highly conserved residues highlighted. Residues altered in the mutant MEF2A peptide (MEFD[M2]) are shown in bold. (B) Phosphorylation of GST-MEF2A (5 pmol) by p38 MAP kinases in the presence of indicated competitor peptides. The peptide competition assay was based on the kinase assays described in the legend to Fig. 1, except that the p38 MAP kinases were preincubated in the absence (lanes 1 and 8) or presence of competitor peptides (a 10- to 1,000-fold excess over MEF2A substrate) at 50 pmol (lanes 2 and 5), 500 pmol (lanes 3 and 6), and 5 nmol (lanes 4, 7, 9, and 10), respectively. Increases in the concentration of added competitor peptides are indicated schematically above each set of lanes. The activity of each kinase was standardized relative to the phosphorylation of GST-MEF2A(WT). (C) Specificity of action of inhibitory peptides and phosphorylation of transcription factor substrates (5 pmol) by MAP kinases (combinations indicated above each panel) in the presence of the MEFD(WT) peptide. Assays were carried out as in panel B in the presence of 0 pmol (lanes 1, 5, and 9), 5 pmol (lanes 2, 6, and 10), 50 pmol (lanes 3, 7, and 11), and 500 pmol (lanes 4, 8, and 12) of the competitor peptide. The quantification of the data is shown graphically below each panel, relative to the phosphorylation of each substrate in the absence of added competitor peptide.
FIG. 2.
Identification of a domain required for efficient p38-mediated activation of MEF2A and MEF2C. (A and B) Requirement of the D-domain for phosphorylation of MEF2A and MEF2C in vitro. Diagrammatic illustrations of truncated MEF2A or MEF2C and MEF2AΔD or MEF2CΔD proteins fused to either GST or the GAL4 DNA binding domain (open boxes) are shown above each gel. The black box represents the putative kinase docking domain (D-domain) of MEF2A, and the numbers of the N- and C-terminal amino acids in MEF2A moiety are shown. (A) Kinetic analysis of GST-MEF2A phosphorylation by p38α, p38β2, and p38γ in vitro. GST-MEF2A (lanes 1 to 4) and GST-MEF2AΔD (lanes 5 to 8) were phosphorylated by p38α, p38β2, and p38γ MAP kinases for the times indicated above each lane. Due to the lower levels of phosphorylation by p38γ, the bottom panels were exposed for longer times than the other panels. The results are presented graphically below these panels with data obtained with GST-MEF2A (squares) and GST-MEF2AΔD (circles) as p38 substrates. Data are presented relative to the phosphorylation of GST-MEF2A after 120 min (taken as 100). (B) As described for panel A, except that the GST-MEF2C derivatives were used as substrates. (C and D) The D-domain is essential for efficient p38β2-inducible transcriptional activation by MEF2A and MEF2C in vivo. (C) COS-7 cells were cotransfected with expression vectors encoding GAL4-MEF2A derivatives, a constitutively activated form of MKK6 (MKK6[E]), p38β2 MAP kinase, and a GAL4-driven luciferase reporter plasmid. The expression levels of the GAL4 fusion proteins in the unstimulated cells were examined by Western blot analysis with an anti-GAL4 antibody (bottom panel). (D) Assays were carried out as described for panel C, except that GAL4-MEF2C was examined. Transfection efficiencies were monitored by using the β-galactosidase expression vector pCH110. The normalized luciferase activities (means ± standard errors; n = 3) are presented.
To examine the requirement of the D-domain for the activation of MEF2 transcription factors by MAP kinases in vivo, GAL4 fusion proteins to MEF2A and MEF2C which either contain or lack this domain were constructed and tested for their ability to activate a GAL4-driven luciferase reporter gene in response to MAP kinase activation (Fig. 2C and D). In the absence of cotransfected MKK6(E) and p38β2 (Fig. 2C and D), all the fusion proteins activated the reporter gene to low levels. However, upon cotransfection of MKK6(E) together with p38β2 and either GAL4-MEF2A (Fig. 2C) or GAL4-MEF2C (Fig. 2D), greatly enhanced transcriptional activation was observed. In contrast, the deletion of the D-domain in both MEF2A and MEF2C caused a large reduction in the GAL4-MEF2A- and GAL4-MEF2C-mediated transcriptional activation (Fig. 2C and D). Western blotting indicates that the deletion of the D-domain had little effect on the levels of the GAL4 fusion proteins (Fig. 2C and D, bottom panels). Virtually identical results were observed for the activation of these chimeric proteins by p38α (data not shown).
These data therefore indicate that both MEF2A and MEF2C contain a domain, the D-domain, which is distinct from their phosphoacceptor motifs and is required for efficient phosphorylation in vitro and stimulation of their transcriptional activation potential in vivo by the p38α and p38β2 MAP kinases.
Identification of important residues in the D-domain required for efficient phosphorylation by p38 MAP kinases.
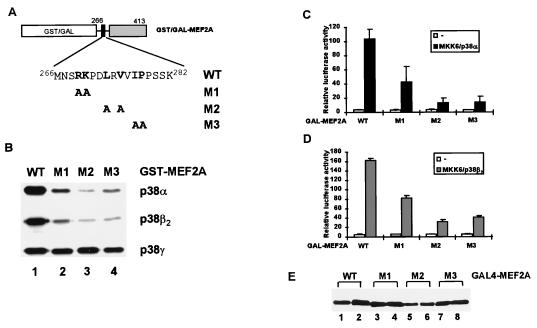
The D-domain plays an important role in enhancing phosphorylation and stimulation of MEF2A- and MEF2C-mediated transcriptional activation by the p38α and p38β2 MAP kinases. In order to investigate the contribution of residues within this domain of MEF2A towards this function, pairs of amino acids conserved between MEF2A and MEF2C (see Fig. 8A) were mutated to alanine residues (Fig. 3A). Such mutations should preserve any structural motifs which are present but remove side chains which are available for intermolecular interactions. The mutant proteins were examined as substrates for p38 MAP kinases (Fig. 3B). All three of the mutants tested (M1, M2, and M3) exhibited a reduction in the efficiency of their phosphorylation by p38α and p38β2 (Fig. 3B). In contrast, none of the mutations within the D-domain resulted in a decrease in the efficiency of MEF2A phosphorylation by p38γ (Fig. 3B). GAL4 fusion proteins were also constructed with each of the mutant MEF2A derivatives to investigate their activation by p38α and p38β2 in vivo. In comparison to the wild-type (WT) protein, the M1, M2, and M3 mutant GAL4-MEF2A fusion proteins exhibit reduced activation of transcription in response to MKK6(E) and p38α (Fig. 3C) or MKK6(E) and p38β2 (Fig. 3D) in vivo. Western blotting indicates that all the mutant proteins were expressed to equivalent levels in the presence and absence of activated upstream cascades (Fig. 3E). Collectively, these data demonstrate that the conserved residues within the MEF2A D-domain play key roles in determining its efficient phosphorylation and transcriptional activation by p38α and p38β2. Furthermore, the critical residues for activation by p38α and p38β2 appear to be indistinguishable.
FIG. 8.
The identity of the kinase docking domain determines the specificity of MAP kinases towards MEF2A. (A) Diagrammatic illustration of GST fusions to MEF2B, the MEF2B-MEF2A chimera, and MEF2A. The numbers of the N- and C-terminal MEF2A (italics) and MEF2B (roman type) amino acids are indicated above and below each construct. The p38-binding motifs in MEF2A and MEF2B are indicated by black and grey boxes, respectively, the amino acid sequences of these regions in MEF2A, MEF2B, MEF2C and MEF2D are shown, and identical residues are highlighted. (B) Kinase assays with the indicated GST fusion proteins and p38 MAP kinases were carried out as described in the legend to Fig. 1. (C) Diagram illustrating fusions of GST to MEF2A and MEF2AΔD and the chimeric proteins cJunδ-MEF2A, SAPD-MEF2A, and ElkD-MEF2A. These constructs contain the transcriptional activation domain of MEF2A and the kinase docking domain of cJun (δ), SAP-1 (D), Elk-1 (white box), MEF2A (black box), or no docking site, respectively. The numbers of the N- and C-terminal c-Jun/SAP-1/Elk-1 (italics) and MEF2A (roman type) amino acids are indicated above and below each construct, respectively. (D) Kinase assays with the indicated chimeric GST fusion proteins as substrates for MAP kinases (as indicated on the right) were carried out with 5 pmol of each protein for 15-min reactions as described in the legend to Fig. 1.
FIG. 3.
Mapping the residues in the D-domain of MEF2A required for targeting by p38α and p38β2. (A) Amino acid sequences of the WT and D-domain mutants R269A/K270A (M1), L273A/V275A (M2), and I277A/P278A (M3) are shown. Numbers above the sequences represent the N- and C-terminal residues in the D-domain. (B) Kinase assays of WT and mutant GST-MEF2A fusion proteins by p38 MAP kinases were carried out as described in the legend to Fig. 1. The activity of each kinase was standardized relative to the phosphorylation of GST-MEF2A(WT). (C) p38α-inducible transcriptional activation by WT and mutant GAL4-MEF2A fusion proteins. COS-7 cells were cotransfected with expression vectors encoding GAL4 fusions to either WT or D-domain mutant (M1 to M3) MEF2A derivatives, vectors encoding MKK6(E) and p38α, and a GAL4-driven luciferase reporter plasmid. The luciferase activities relative to GAL4-MEF2A-mediated reporter activation in the absence of cotransfected kinases are presented (means ± standard errors; n = 3). (D) Assays were carried out as in panel C, except that vectors encoding p38β2 were cotransfected where indicated. (E) Expression levels of the GAL4 fusion proteins, in the absence (lanes 1, 3, 5, and 7) and presence (lanes 2, 4, 6, and 8) of cotransfected MKK6(E) and p38α, were examined by Western blotting with an anti-GAL4 antibody.
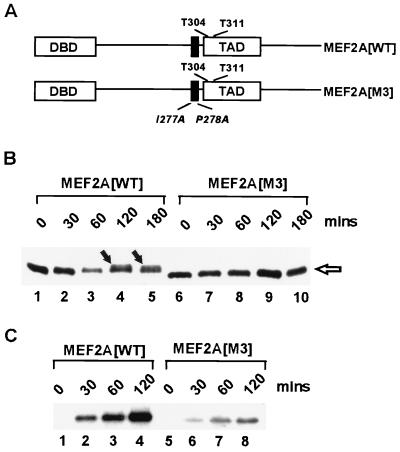
In order to demonstrate the importance of the D-domain in targeting p38 MAP kinases to MEF2A in the context of a full-length protein rather than truncated fusion proteins, we created a mutant version of MEF2A with two point mutations in the D-domain (MEF2A[M3]; Fig. 4A), which reduce the phosphorylation and activation of truncated MEF2A derivatives by p38α and p38β2 (Fig. 3). Full-length MEF2A was initially tested as a substrate for the different p38 isoforms (data not shown) with results virtually identical to those obtained with truncated GST fusion proteins (Fig. 1B). We then compared the ability of p38β2 to phosphorylate MEF2A(WT) and the mutant MEF2A(M3) in vitro. Phosphorylation of MEF2A(WT) was observed by the presence of a slower migrating band (Fig. 4B, lanes 4 and 5) and the incorporation of 32P-labelled ATP (Fig. 4C, lanes 2 to 4). In contrast, over the same time period, much-reduced MEF2A(M3) phosphorylation was detected by these assays (Fig. 4B, lanes 6 to 10, and C, lanes 5 to 8). Thus, the D-domain plays an important role in directing the efficiency of MEF2A phosphorylation by p38β2 in the context of the full-length protein.
FIG. 4.
The D-domain is required for the efficient phosphorylation of full-length MEF2A by p38β2. (A) Diagrammatic representation of the domain structure of full-length MEF2A. The locations of the MADS-MEF DNA-binding domain (DBD), the MAP kinase docking domain (black box), and the key phosphoacceptor motifs (T304 and T311) in the TAD are shown. The locations of the mutations in MEF2A(M3) are indicated in italics. (B and C) Phosphorylation of MEF2A(WT) and MEF2A(M3) was carried out in vitro with p38β2. Equal amounts of in vitro-translated WT and mutant MEF2A proteins were phosphorylated by p38β2 for the indicated times in the absence (B) or presence (C) of 32P-labelled ATP. Phosphorylated MEF2A was detected by the reduced mobility of the proteins (arrows in panel B) or by the incorporation of 32P-labelled ATP (C).
The MEF2A D-domain acts as a binding motif for the p38 MAP kinases.
The ability of ERK2 to phosphorylate Elk-1 and that of JNK to phosphorylate c-Jun correlate with their ability to bind to these substrates via a docking domain (13, 37). However, while stable interactions are readily detectable in these cases, such stable interactions are not observed between MAP kinases and other substrates (e.g., JNK and Elk-1; 38). Similarly, we were unable to demonstrate a physical interaction between MEF2A and p38 MAP kinases under a variety of experimental conditions (data not shown), although others have previously demonstrated such an interaction (8). We therefore adopted a peptide competition assay to investigate the binding of the kinases to MEF2A. This approach has previously been used to allow the comparison of ERK and JNK binding to Elk-1 under identical experimental conditions and relies on the fact that peptides which bind to the kinase will act as competitors for the binding of the kinase to docking sites in the transcription factor targets (38). Peptides were synthesized which correspond to MEF2A amino acids 266 to 283 (encompassing the D-domain) (MEFD[WT]) and the same region with two amino acid substitutions (MEFD[M2]) (Fig. 5A). Increasing amounts of these peptides were included in kinase assays to compete for binding of the p38 MAP kinases to MEF2A via the D-domain (Fig. 5B). The MEFD(WT) peptide acted as an inhibitor of p38α- and p38β2-mediated phosphorylation of MEF2A in a concentration-dependent manner (Fig. 5B, lanes 1 to 4). However, little effect was seen on the efficiency of phosphorylation by p38γ except at the highest concentrations of the wild-type and mutant peptides used (Fig. 5B, lanes 1 to 4, bottom panel). In contrast, the mutant MEFD(M2) peptide did not act as a competitor (Fig. 5B, lanes 5 to 7). This is consistent with the observation that the M2 mutant GST- and GAL-MEF2A fusion proteins are poor targets for p38α and p38β2 (Fig. 3). Peptides corresponding to the kinase docking domains from Elk-1 (ElkD) and SAP-1 (SAPD) (Fig. 5A) were also tested (Fig. 5B). The SAPD peptide also acted as an efficient inhibitor (Fig. 5B, lane 9), whereas the ElkD peptide did not act as an inhibitor of the phosphorylation of MEF2A by p38α and p38β2 (Fig. 5B, lane 10). These results are consistent with the observation that the SAP-1 D-domain acts as a docking site for both the p38α and p38β2 MAP kinases (5a), whereas the Elk-1 D-domain does not act as a p38 MAP kinase docking site (38).
In order to investigate the specificity of action of the MEF2A D-domain peptide as a p38α and p38β2 MAP kinase inhibitor, the ability of the D-domain peptide to inhibit MEF2A phosphorylation by p38β2 was compared to its inhibitory properties on other MAP kinase-substrate combinations (Fig. 5C). Of the three kinases tested, the D-domain peptide acted as a more efficient competitor of p38β2 than ERK and JNK. This is most apparent at the highest concentrations of peptide used (Fig. 5C, lanes 4, 8, and 12), although the inhibition of all three MAP kinases can be observed to some degree. This might reflect some conservation of the peptide binding site on the MAP kinases, as these proteins are all related and the relative order of peptide inhibition efficiency is consistent with the observation that the highest similarity is between the p38 and ERK MAP kinases.
These data therefore demonstrate that the D-domain of MEF2A preferentially acts as a binding site for p38α and p38β2 MAP kinases in vitro.
The MAP kinase targeting domain of MEF2A is sufficient to confer p38 responsiveness to Elk-1 and c-Jun.
In order to investigate whether the MEF2A D-domain can act in a heterologous context to allow p38 targeting and hence transduce signals via the p38 pathway to different substrates, chimeric proteins were created. In these proteins, the p38 targeting domain of MEF2A was fused to the minimal TAD of Elk-1 (which responds to both the ERK and JNK pathways) and c-Jun (which responds to the JNK pathway) and either GST or the GAL4-DNA-binding domain (Fig. 6A and E). These minimal TADs lack their natural kinase binding domains. Firstly, the efficiencies of phosphorylation of the GST fusion proteins by different MAP kinases were compared (Fig. 6B and F). GST–Elk-1ΔD, which lacks its own kinase targeting domain, represents a relatively poor MAP kinase substrate (Fig. 6B, lane 1). However in comparison, GST–MEF2A–Elk-1 was efficiently phosphorylated by p38α and p38β2 (Fig. 6B, lane 2). In contrast, little enhancement of Elk-1 phosphorylation by ERK2, JNK2, and p38γ was observed by the inclusion of the MAP kinase docking site from MEF2A (Fig. 6B, lane 2). Moreover, the fusion of the MEF2A D-domain to c-Jun in the GST-MEF2A-cJun chimera converts c-Jun into a better substrate for p38α and p38β2 but not for p38γ (Fig. 6F, lane 2). c-Jun can usually be efficiently phosphorylated only by JNK and not p38 MAP kinases (Fig. 6F, lane 1), and the propensity of c-Jun as a JNK substrate is severely reduced in the presence of the MEF2A MAP kinase docking site (Fig. 6F, lane 2, bottom panel).
In order to demonstrate that these changes in substrate specificity towards MAP kinases, which are dictated by the MEF2A D-domain, reflect a difference in the ability of these domains to bind to the MAP kinases, peptide competition experiments were performed with the chimeric proteins MEF2A–Elk-1 (Fig. 6C) and MEF2A-cJun (Fig. 6G). The MEFD(WT) and, to a lesser extent, SAPD peptides acted as competitors of phosphorylation of these chimeras by p38α and p38β2 (Fig. 6C and G, lanes 2 and 4), whereas the MEFD(M2) and ElkD peptides were ineffectual competitors (Fig. 6C and G, lanes 3 and 5). These results are essentially the same as those observed with wild-type GST-MEF2A (Fig. 6B), thereby demonstrating that the MEF2A D-domain acts in a similar manner to permit p38α and p38β2 binding in a heterologous context.
The analogous GAL4 fusion proteins were subsequently analyzed for their activation by p38α (data not shown) and p38β2 (Fig. 6D and H) in vivo. All these GAL4 derivatives are expressed to similar levels (Fig. 6D and H, bottom panels). GAL4–Elk-1ΔD, which lacks the kinase docking domain, is moderately activated by the ERK and p38 pathways (Fig. 6D) (37, 38). In comparison, GAL4–MEF2A–Elk-1 exhibits greatly enhanced transcriptional activation in response to MKK6/p38β2-mediated stimulation in vivo (Fig. 6D), whereas the response to MEK/ERK-mediated stimulation barely changes (Fig. 6D). Similarly, c-Jun is usually activated by the JNK pathway but only poorly by the p38β2 pathway in vivo. However, reciprocal effects are seen with the MEF2A-cJun chimera, which is efficiently activated by the p38β2 pathway but in comparison is poorly activated by the JNK pathway (Fig. 6H).
Taken together, the results clearly demonstrate that the D-domain of MEF2A is sufficient to confer responsiveness to the p38α and p38β2 signalling pathways in heterologous contexts both in vitro and in vivo.
The p38-binding domain of MEF2A directs phosphorylation of key phosphoacceptor motifs.
The D-domain of MEF2A is sufficient to permit the targeting of p38α and p38β2 MAP kinases to heterologous substrates. In order to demonstrate that physiologically relevant residues are targeted for phosphorylation by the D-domain, we investigated the phosphorylation of Ser383 in MEF2A–Elk-1 chimeras, which has been shown to be one of the key residues which must be phosphorylated to trigger the DNA binding and transcriptional activation properties of Elk-1 (reviewed in reference 33). The phosphorylation of Ser383 was assessed by Western blotting with a phospho–Elk-1(Ser383) antibody.
Firstly, GST–MEF2A–Elk-1 was phosphorylated by p38β2 in vitro. In comparison to GST–Elk-1ΔD, which lacks a p38 docking site, GST–MEF2A–Elk-1 was efficiently phosphorylated on Ser383 (Fig. 7A). In order to analyze whether the same effect could be observed in vivo, the phosphorylation of GAL4–MEF2A–Elk-1 was compared to the phosphorylation of GAL4–Elk-1 in the presence of cotransfected MKK6/p38β2 (Fig. 7B). The overall phosphorylation of Elk-1 was greater in the presence of the D-domain from MEF2A (Fig. 7B; compare lanes 2 and 4, bottom panel). Moreover, the phosphorylation of Ser383 was greatly enhanced in the GAL4–MEF2A–Elk-1 chimera (Fig. 7B; compare lanes 2 and 4, top panel).
FIG. 7.
The p38-binding domain of MEF2A directs the phosphorylation of the key phosphoacceptor motifs in MEF2A–Elk-1 chimeras. (A) In vitro kinase assays of GST fusion proteins (5 pmol of each substrate) by p38β2 MAP kinase were carried out for 15 min. Phosphorylation of Ser383 was detected by Western blotting with an anti-phospho–Elk-1(Ser383) antibody. (B) In vivo phosphorylation of MEF2A–Elk-1 chimeras. 293 cells were transfected with vectors encoding GAL–Elk-1 or GAL–MEF2A–Elk-1 and a GAL4-driven luciferase reporter plasmid and, where indicated, p38β2 and MKK6(E). The phosphorylation of the Elk-1 moiety at Ser383 in cell extracts was detected by Western blotting as in panel A (top panel), and the total levels of each GAL4 fusion protein were detected with an anti-GAL4 antibody (bottom panel). The locations of the bands corresponding to nonphosphorylated and phosphorylated GAL4 fusion proteins are indicated.
Furthermore, in the MEF2A-cJun chimeras analyzed in Fig. 6, there are only two potential phosphoacceptor motifs in the c-Jun moiety which correspond to the physiologically relevant sites. Together these results therefore demonstrate that in addition to enhancing the overall efficiency of substrate phosphorylation by p38α and p38β2, the MEF2A D-domain also directs the phosphorylation of physiologically relevant sites in vitro and in vivo.
The kinase docking domain determines the specificity of MAP kinases towards MEF2A.
The kinase docking domain of MEF2A is sufficient to change the specificity of heterologous proteins as substrates for phosphorylation and activation by different MAP kinases. Reciprocal constructs were made in which either known or putative kinase docking domains from other transcription factors were fused to the transcription activation domain of MEF2A (Fig. 8A and C). Firstly, the homologous region of MEF2B was substituted for the kinase docking domain of MEF2A (Fig. 8A), and the resulting chimeric protein was tested as a substrate for p38 MAP kinases. In contrast to MEF2A, MEF2B appears not to be phosphorylated by p38 MAP kinases (Fig. 8B; compare lanes 1 and 3). Moreover, the putative kinase docking domain in MEF2B is unable to permit the efficient phosphorylation of MEF2A in the GST-MEF2B-MEF2A chimera (Fig. 8B, lane 2). Thus, the amino acid sequence differences in this region of MEF2B inactivate its ability to act as a kinase binding motif.
The MAP kinase docking domains from c-Jun (for JNK MAP kinases) (4, 12, 13), SAP-1 (for ERK, p38α, and p38β2 MAP kinases) (5a), and Elk-1 (for JNK and ERK MAP kinases) (37, 38) were fused with the transcriptional activation domain of MEF2A (Fig. 8C) and tested as substrates for different MAP kinases (Fig. 8D). MEF2A is phosphorylated efficiently by p38α and p38β2 but is a poor substrate for the ERK and JNK MAP kinases (Fig. 8D, lane 4, and Fig. 1). However, in the absence of the D-domain, MEF2A is a poor substrate for all of these MAP kinases (Fig. 8D, lane 5, and Fig. 2). Significantly, the introduction of the c-Jun δ-domain converts MEF2A into a good JNK substrate (Fig. 8D, lane 1). Similarly, the ElkD-MEF2A chimera is a good JNK substrate (Fig. 8D, lane 3). In contrast, the fusion of the SAP-1 D-domain to MEF2A restores its ability to act as a substrate for p38α and p38β2 (Fig. 8D, lane 2). None of the chimeric proteins are good ERK2 substrates (Fig. 8D, top panel), indicating that the docking site alone is insufficient to permit the targeting of ERK MAP kinases (see Discussion).
Together these results demonstrate that the docking domains from several different transcription factors can direct the targeting of specific subsets of stress-activated MAP kinases to MEF2A. Thus, MAP kinase docking sites appear to be common, conserved elements in transcription factors which can act as independent domains and contribute significantly to the specificity of action of the diverse MAP kinase cascades.
DISCUSSION
The MAP kinase cascades play important roles in regulating multiple diverse cellular processes. However, as different MAP kinases all phosphorylate similar sites containing the minimal consensus motif (Ser/Thr)-Pro, it is unclear how substrate specificity is achieved. It has recently been shown that one component of this specificity-determining mechanism for the JNK and ERK MAP kinases involves binding of the MAP kinases to a docking domain on their nuclear targets (3, 4, 6, 7, 12, 13, 26, 37, 38). In this study, we demonstrate that members of the p38 subclass of MAP kinases are targeted to the transcription factors MEF2A and MEF2C by a docking domain which is distinct from the phosphoacceptor motifs. This domain is sufficient to confer p38 responsiveness on heterologous substrates and can be functionally replaced by alternative kinase docking motifs from other transcription factors. Similarly, a docking site in ATF-2 is required for efficient activation by p38α (11a). Our results therefore demonstrate the generality of the phenomenon of MAP kinase docking with transcription factors as a major part of the mechanism of substrate specificity determination.
MAP kinase docking domains.
The sequence of the MAP kinase docking domain in MEF2A and MEF2C is highly conserved between these two proteins (Fig. 8A) and is also evolutionarily conserved in the Drosophila MEF2 homologue (28). This sequence conservation is reflected by the functional conservation of these domains as docking sites for both p38α and p38β2 (Fig. 2). However, neither domain appears to be a docking site for p38γ (Fig. 2A and B) and p38δ (data not shown), as their deletion does not affect the efficiency of phosphorylation of MEF2A and MEF2C. It is unclear whether p38γ uses a different docking domain or alternatively, as suggested by in vivo experiments (Fig. 1) (39), MEF2A and MEF2C do not represent true substrates for this kinase. A similar phenomenon has been noted for Elk-1, for which it was proposed that the lack of a targeting domain for the p38 MAP kinases is reflected in its poor response to the p38 pathways in vivo (38).
Residues throughout the docking domain are important in targeting p38α and p38β2 to MEF2A (Fig. 3 and 4), and in common with other kinase docking domains (37, 38), hydrophobic residues play the most important roles. Moreover, an excellent correlation exists between the ability of the docking site to permit efficient phosphorylation in vitro and transcriptional activation in vivo (compare Fig. 2A and B, Fig. 2C and D, and Fig. 3B and C and D), indicating the functional importance of these sites. The specificity of action of the D-domain of MEF2A towards p38α and p38β2 subtypes in vitro and in vivo was demonstrated by using chimeric transcription factors which contain the MEF2A docking domain and transcriptional activation domains of different proteins (Fig. 6 and 7). Neither the JNK nor the ERK MAP kinases can be targeted to transcription factors by the MAP kinase docking domain from MEF2A. However, by using reciprocal chimeric proteins with the MEF2A transcriptional activation domain and the kinase docking domains of either c-Jun, Elk-1, or SAP-1, the propensity of MEF2A as a substrate for the JNK and p38 MAP kinases can be altered in a manner which is predictable from the known characteristics of each domain (Fig. 8). Interestingly, MEF2A could not be converted into an ERK substrate by the inclusion of the ERK binding domain from Elk-1 (Fig. 8D), indicating that the docking domain itself is not sufficient to direct efficient phosphorylation. A similar chimeric protein with the c-Jun transcriptional activation domain is also not responsive to ERKs (38). This might reflect the requirement for additional docking components such as that observed for ERK targeting to the ETS domain transcription factors Elk-1 and LIN-1, which uses both a docking domain located N-terminally from the phosphoacceptor residues and an FXFP motif located C-terminally from these residues to target ERKs to these proteins (11). Moreover, in addition to these docking domains, the local context of the phosphoacceptor motifs may also play a major role in determining substrate specificity towards MAP kinases as suggested previously for c-Jun (13). In MEF2A and c-Jun, the phosphoacceptor motifs presumably represent good JNK, p38α, and p38β2 sites, and it is the presence of the correct docking site which ultimately determines which MAP kinase phosphorylates these transcription factors (Fig. 6F to H).
Recently, the existence of cytoplasmic MAP kinase modules has been proposed in yeast (reviewed in references 17 and 27) and mammals (35), in which the MAP kinases are stably associated with the upstream components in the cascade by specific docking proteins. The identification of MAP kinase docking sites in nuclear transcription factors suggests that nuclear complexes might also exist. However, in general, MAP kinases do not appear to stably associate with transcription factors in their active state, with the exception of ERK-2 and Elk-1 (37). Furthermore, the binding of inactive kinases appears to be weak, as we are unable to detect significant stable binding of p38α to MEF2C or that of JNKs to c-Jun under standard binding conditions, although others have detected these interactions (3, 4, 7, 9, 12, 13). However, binding can be directly inferred from peptide competition experiments (Fig. 4A and 5C and G) (38).
To date, MAP kinase targeting domains have been identified in c-Jun (3, 4, 7, 12, 13), Elk-1 (37, 38), SAP-1 (5a), MEF2A (this study), and ATF-2 (6, 7, 11a, 15). In all cases, the docking site is located immediately N-terminally to the phosphoacceptor motifs within the transcriptional activation domain. It remains unclear whether this positioning is functionally significant or reflects the evolutionary conservation of a MAP kinase recognition module. In c-Jun, a spacer can be inserted between the docking domain and its transcriptional activation domain (13), and in Elk-1 deletions can be made between these modules (5a) without changing their proficiency as MAP kinase substrates. Recently, an additional docking domain has been identified in several substrates, which is located C-terminally from the phosphoacceptor motifs (11). Further systematic studies, however, are required to examine the requirement for specific spatial alignment of the docking domains and phosphoacceptor motifs.
Activation of MEF2 family members by the p38 MAP kinases.
The MEF2 subfamily of MADS-box transcription factors is composed of four members: MEF2A, MEF2B, MEF2C, and MEF2D (reviewed in references 19 and 24). Of these, MEF2A and MEF2C exhibit significant sequence conservation throughout their lengths (≈56% identity), including that within their transcription activation domains and the MAP kinase phosphorylation sites, which have been shown to be functionally important in MEF2C. Both p38α (8) and ERK5/BMK (14, 36) have been shown to phosphorylate and enhance the transcriptional activation potential of MEF2C. MEF2D contains the sites for p38α but lacks the site for ERK5/BMK, while MEF2B appears to contain only one of the p38α phosphoacceptor motifs. Here we demonstrate that p38α phosphorylates and activates the transcription activation domains of both MEF2A and MEF2C and requires targeting by a docking domain. Similarly, p38β2 activates both these transcription factors and requires docking via the same domain. Neither p38γ nor p38δ efficiently phosphorylates and activates MEF2A and MEF2C. Thus, only a subset of p38 MAP kinase subtypes activate the transcriptional activation domain of MEF2A and MEF2C. Similarly, neither the ERK nor JNK MAP kinases significantly activate MEF2A and MEF2C (Fig. 1D and E).
Our data are consistent with the findings of two independent studies, which demonstrate that MEF2A is a p38α substrate (18a, 39), although this study is different in that we clearly demonstrate that p38β2 phosphorylates and activates MEF2A, whereas p38β1 apparently does not phosphorylate MEF2A (39). This discrepancy might be attributable to the differences observed between p38β1 and p38β2 (identical apart from an 8-amino-acid deletion in p38β1), which confer on p38β2 the ability to phosphorylate other substrates more efficiently than p38β1 (5). MEF2B lacks one of the p38α phosphorylation sites found in MEF2A and MEF2C but still exhibits some sequence similarity with the D-domain of MEF2A/C (Fig. 8A). However, MEF2B is a poor substrate for the p38 MAP kinases (Fig. 8B). Moreover, the putative MAP kinase docking site is unable to act as a p38 docking motif in a heterologous substrate (Fig. 8B). Thus, MEF2B differs from MEF2A and MEF2C, as it is unable to either recruit p38 MAP kinases or act as a p38 substrate. It is currently unknown whether MEF2D is regulated by p38 MAP kinase subtypes, although from sequence conservation, it might represent a p38α and p38β2 substrate, as both the phosphoacceptor motifs and the docking domain are conserved. However, in the case of the docking domain, sufficient differences exist (especially in the C-terminal half) to suggest that its MAP kinase targeting properties might differ from those of MEF2A and MEF2C (Fig. 8A). MEF2D, however, lacks the C-terminal ERK5/BMK phosphoacceptor motif. Interestingly, MEF2A and MEF2C have been shown to physically associate with ERK5/BMK (36) via a motif contained within the minimal DNA-binding domain, which is distinct from the docking sites described here. However, the functional consequences of this interaction have not been investigated. As MEF2 proteins act as dimers and can associate to form different combinations of heterodimers, the resulting complexes will respond differently to different MAP kinase cascades, depending on the docking domains and phosphoacceptor motifs present in each subset. Such diversity in response to p38 MAP kinase cascades might contribute to different responses by alternative MEF2-containing heterodimers. The activation of MEF2C by p38α, it has been proposed, is important in inflammatory responses in the immune system (8). In addition, the activation of MEF2C by ERK5/BMK has been shown to be an important part of the mechanism of serum induction of the c-jun promoter. As the MEF2 proteins play a major role in regulating muscle-specific gene expression (19), it will be interesting to discover whether p38α and/or p38β2-mediated signalling to MEF2 proteins plays a role in muscle differentiation and/or proliferation during development or in response to damage.
Conclusion.
It is becoming clear that the binding of MAP kinases to transcription factors is a critical determinant of their specificity. Here we demonstrate the selective targeting of p38α and p38β2 MAP kinases to MEF2A and MEF2C by binding to a distinct docking domain and that this binding is an important determinant of the efficiency of p38α- and p38β2-mediated phosphorylation of MEF2A and MEF2C in vitro and transcriptional activation in vivo. Members of three classes of MAP kinases in mammals (ERK, JNK, and p38) have now been shown to require docking sites on transcription factors for their correct function. It is likely that such interactions will prove to be essential specificity determinants in other MAP kinase-transcription factor signalling modules.
ACKNOWLEDGMENTS
We thank Margaret Bell and Katherine Stewart for excellent technical and secretarial assistance and Bob Liddell for DNA sequencing. We are grateful to John McDermott, Alan Whitmarsh, and members of our laboratories for comments on the manuscript and for stimulating discussions and to Nic Jones and John McDermott for communicating data prior to publication. We also thank Adam West for help with the inception of this project and Fei-Ling Lim for constructing some of the intermediate plasmids. We are grateful to Stuart Lipton, Richard Treisman, Alan Whitmarsh, and Roger Davis for providing reagents and to Simon Ridley for helpful advice about p38 activation.
This work was supported by the North of England Cancer Research Campaign, the Cancer Research Campaign (CRC), the Wellcome Trust, and a Jeffcock Ph.D. studentship from the University of Newcastle Upon Tyne to A.G. A.D.S. is a Research Fellow of the Lister Institute of Preventative Medicine.
REFERENCES
- 1.Cohen P. The search for physiological substrates of MAP and SAP kinases in mammalian cells. Trends Cell Biol. 1997;7:353–361. doi: 10.1016/S0962-8924(97)01105-7. [DOI] [PubMed] [Google Scholar]
- 2.Cook J G, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 3.Dai T, Rubie E, Franklin C C, Kraft A, Gillespie D A F, Avruch J, Kyriakis J M, Woodgett J R. Stress-activated protein kinases bind directly to the δ domain of c-Jun in resting cells: implications for repression of c-jun function. Oncogene. 1995;10:849–855. [PubMed] [Google Scholar]
- 4.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 5.Enslen H, Raingeaud J, Davis R J. Selective activation of p38 MAP kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J Biol Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 5a.Galanis, A., and A. D. Sharrocks. Unpublished data.
- 6.Gupta S, Campbell D, Dérijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 7.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Dérijard B, Davis R J. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 8.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 9.Han T-H, Prywes R. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol Cell Biol. 1995;15:2907–2915. doi: 10.1128/mcb.15.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs D, Glossip D, Xing H, Muslin A J, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 11a.Jones, N. Personal communication.
- 12.Kallunki T, Su B, Tsigelny I, Sluss H K, Dérijard B, Moore G, Davis R J, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 13.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 14.Kato Y, Kravchenko V V, Tapping R I, Han J, Ulevitch R J, Lee J-D. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–674. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 17.Madhani H D, Fink G R. The riddle of MAP kinase signalling. Trends Genet. 1998;14:151–155. doi: 10.1016/s0168-9525(98)01425-5. [DOI] [PubMed] [Google Scholar]
- 18.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande-Woude G F, Ahn N G. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 18a.McDermott, J. Personal communication.
- 19.Molkentin J D, Olson E N. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 21.Raingeaud J, Gupta S, Rogers J S, Dickens M, Han J, Ulevitch R J, Davis R J. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 22.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seth A, Gonzalez F A, Gupta S, Raden D L, Davis R J. Signal transduction within the nucleus by mitogen-activated protein kinase. J Biol Chem. 1992;267:24796–24804. [PubMed] [Google Scholar]
- 24.Shore P, Sharrocks A D. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 25.Shore P, Bisset L, Lakey J, Waltho J P, Virden R, Sharrocks A D. Characterization of the Elk-1 ETS DNA-binding domain. J Biol Chem. 1995;270:5805–5811. doi: 10.1074/jbc.270.11.5805. [DOI] [PubMed] [Google Scholar]
- 26.Sluss H K, Barrett T, Dérijard B, Davis R J. Signal transduction by tumor necrosis factor mediated by JNK protein kinases. Mol Cell Biol. 1994;14:8376–8384. doi: 10.1128/mcb.14.12.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sprague G F. Control of MAP kinase signalling specificity or how not to go HOG wild. Genes Dev. 1998;12:2817–2820. doi: 10.1101/gad.12.18.2817. [DOI] [PubMed] [Google Scholar]
- 28.Taylor M V, Beatty K E, Hunter H K, Baylies M K. Drosophila MEF2 is regulated by twist and is expressed in both the primordia and differentiated cells of the embryonic somatic, visceral and heart musculature. Mech Dev. 1995;50:29–41. doi: 10.1016/0925-4773(94)00323-f. [DOI] [PubMed] [Google Scholar]
- 29.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 30.Wang X S, Diener K, Manthey C L, Wang S-W, Rosenzweig B, Bray J, Delaney J, Cole C N, Chan-Hui P-Y, Mantlo N, Lichenstein H S, Zukowski M, Yao Z. Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase. J Biol Chem. 1997;272:23668–23674. doi: 10.1074/jbc.272.38.23668. [DOI] [PubMed] [Google Scholar]
- 31.West A G, Causier B, Davies B, Sharrocks A D. DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res. 1998;26:5277–5287. doi: 10.1093/nar/26.23.5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 33.Whitmarsh A J, Davis R J. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J Mol Med. 1996;74:589–607. doi: 10.1007/s001090050063. [DOI] [PubMed] [Google Scholar]
- 34.Whitmarsh A J, Yang S-H, Su M S-S, Sharrocks A D, Davis R J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;17:2360–2371. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 36.Yang C-C, Ornatsky O I, McDermott J C, Cruz T F, Prody C A. Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen-activated protein kinase, ERK5/BMK1. Nucleic Acids Res. 1998;26:4771–4777. doi: 10.1093/nar/26.20.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang S-H, Yates P R, Whitmarsh A J, Davis R J, Sharrocks A D. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang S-H, Whitmarsh A J, Davis R J, Sharrocks A D. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998;17:1740–1749. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao M, New L, Kravchenko V, Kato Y, Gram H, Di Padova F, Olson E, Ulevitch R J, Han J. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]