SUMMARY
Argonaute proteins are at the core of the microRNA-mediated gene silencing pathway essential for animals. In C. elegans, the microRNA-specific Argonautes ALG-1 and ALG-2 regulate multiple processes required for proper animal developmental timing and viability. Here, we identified a phosphorylation site on ALG-1 that modulates microRNA association. Mutating ALG-1 serine 642 into a phospho-mimicking residue impairs microRNA binding and causes embryonic lethality and post-embryonic phenotypes that are consistent with alteration of microRNA functions. Monitoring microRNA levels in alg-1 phosphorylation mutant animals reveal that microRNA passenger strands increase in abundance but are not preferentially loaded into ALG-1, indicating that the miRNA binding defects could lead to microRNA duplex accumulation. Our genetic and biochemical experiments support the protein kinase A (PKA) KIN-1 as the putative kinase that phosphorylates ALG-1 serine 642. Altogether, our data indicate that PKA triggers the ALG-1 phosphorylation to regulate its microRNAs association during C. elegans development.
Keywords: miRISC, ALG-1, Protein Kinase A, PKA, KIN-1, post-translational modification, gene regulation
INTRODUCTION
MicroRNAs (miRNAs) were identified in C. elegans as potent regulators of developmental timing1-3 and since then, have been shown to regulate a wide range of cellular processes in animals and plants4-6. MiRNAs are ~22 nucleotide (nt) long RNAs, which are typically produced by RNA polymerase II and a successive processing by Drosha and Dicer7,8. The resulting mature miRNA duplex associates with an Argonaute (AGO) protein, which is held in an open conformation, and the closing of the structure triggers the unwinding of the duplex followed by the ejection of the passenger strand, also called miR*9-11. The discarded strand is then subjected to degradation by exonucleases12-14. In many cases, there is a preference for a specific arm of the duplex to be loaded into AGO (originating from either the 5′ or the 3′ of the precursor miRNA), leading to a disproportionate number of copies for one of the two strands in cells. Although less abundant, specific miR* can be loaded into AGO and repress mRNA containing target sites, suggesting that some of them might be functional15.
The association between the miRNA and the AGO protein form the core of the miRNA-induced silencing complex (miRISC). Typically, the miRNA guides the miRISC to the 3′ untranslated region (UTR) of mRNAs and binds sequences that are partially complementary, which leads to translational repression and destabilization of the target16,17. The two miRNA-specific AGOs in C. elegans are ALG-1 and ALG-218,19; orthologous to the human AGO1-4. A third C. elegans AGO, ALG-5, also binds a small subset of miRNAs and is primarily expressed in the germline20. AGOs are bilobed proteins composed of six domains: the N, L1, PAZ, L2, MID and PIWI21. The N domain serves as a wedge to unwind the miRNA duplex, during the loading step, while the PAZ and the MID domains bind the 3′ and the 5′ ends of the miRNA, respectively. The L1 linker between the N and PAZ domains and the L2 linker between the PAZ and MID domains, contribute to the structural stability of the RISC. The C-terminal PIWI domain resembles RNase H proteins 22 allow the cleavage (or slicing) of target RNA that have an extended complementarity. In humans, AGO2 and AGO3 are slicer competent23-25. In C. elegans, the slicing activity of ALG-1 and ALG-2 was shown to be implicated in the production of functional miRISC26. The PIWI domain also contains two tryptophan (W) binding pockets27 which bind GW182 through its Glycine (G) Tyrptophan (W) GW/WG repeats28-31 that contribute to the removal of the poly(A) tail of targeted mRNA16.
AGO function is modulated by the addition and removal of post-translational modifications. In human cells, hypoxia increases the AGO2 hydroxylation and stabilizes it32. Upon hypoxic stresses, EGFR causes the AGO2 phosphorylation at tyrosine 393 (Y393) leading to defective maturation of specific miRNAs33. The same residue can be dephosphorylated by PTPB1 and the inactivation of this phosphatase affects the function of H-RASv12-induced oncogenic miRNAs34. Under cell stress, AGO2 localizes to stress granules where it is poly ADP-ribosylated to relieve miRNA-mediated repression of translation35. Beside these stress-induced modifications, the phosphorylation of AGO specific residues has been observed in normoxic conditions. The p38 MAPK and AKT3 pathways converge to phosphorylate the serine 387 (S387) to regulate AGO localization to processing bodies and drive translational repression36,37. This specific modification was shown to regulate the association between AGO2 and LIMD1 and facilitate the AGO2 binding to GW182 protein TNRC638. Interestingly, the biological significance of S387 phosphorylation was highlighted by its implication in the control of dendritic spine growth and maturation39. In addition to the regulation of localization and protein interactions, phosphorylation of specific AGO residues was also shown to affect the binding to miRNA and miRNA targets. The phosphorylation of tyrosine 529 (Y529) in the MID domain prevents the binding to small RNAs40. Our recent systematic analysis of AGO phosphorylation identified a conserved serine/threonine phosphorylation cluster in the PIWI domain that is essential for miRNA-mediated gene silencing in vivo and showed that its hyper-phosphorylation impairs binding to miRNA targets41. The phosphorylation and de-phosphorylation of this cluster were shown to be mediated by the kinase CSNKA1 and the phosphatase PPP6C in human cells42. It was also reported that its phosphorylation impairs miRISC binding to miRNA targets and the lack of AGO2 phosphorylation on these residues leads to an expansion of the miRNA target repertoire42.
Here we report the phosphorylation of serine 642, a residue located in the MID domain of C. elegans ALG-1, which drastically reduces its ability to bind miRNAs. Mutation of this serine (S) into a phospho-mimicking negatively charged glutamate (E) leads to phenotypes reminiscent of animals completely depleted of alg-1. Developmental delays are observed in both the non-phosphorylatable alanine (A) and the phospho-mimicking glutamate mutants, indicating that this phosphorylation regulates key developmental events during animal growth. Our sequencing analysis further shows an miR* accumulation that are not bound to the ALG-1S642E mutant suggesting that the lack of miRNA-binding by ALG-1 leads to an accumulation of miRNA duplexes. Last, we show that the Protein Kinase A (PKA) kin-1 interacts genetically with alg-1, and this was strongly suppressed by a non-phosphorylatable alg-1(S642A). This data, along with the in vitro phosphorylation of serine 642, suggest that PKA regulates the miRNA-mediated gene silencing in C. elegans through the phosphorylation of ALG-1.
RESULTS
Assessment of a novel AGO phosphorylation site
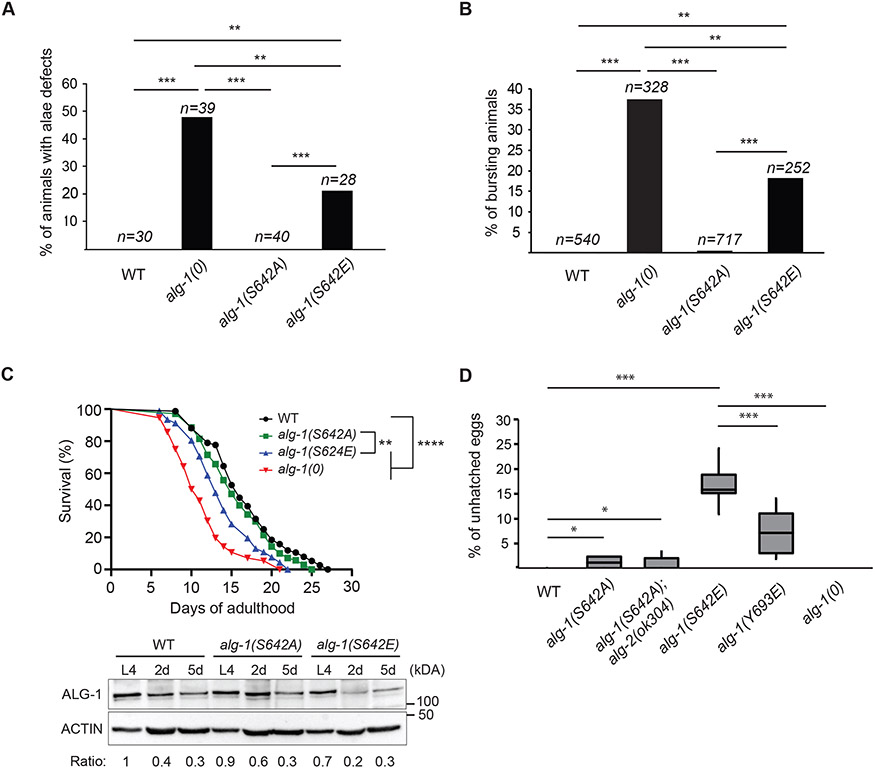
Previously, we characterized in vivo effects of ALG-1 phosphorylation on a highly conserved serine/threonine cluster located on the surface of the PIWI domain41. Mass spectrometry analyses of immunopurified ALG-1 from C. elegans extracts identified an additional phosphorylation site on ALG-1 MID domain that had not yet been identified and characterized on AGOs (Figure S1A). To determine whether this phosphorylation event can affect the function of ALG-1 in vivo, we expressed transgenes carrying a non-phosphorylatable (alanine: A) or a phospho-mimicking (glutamate: E) mutation in an alg-1 knockout strain alg-1(gk214) (hereafter called alg-1(0)). To determine whether these mutant transgenes would rescue the loss of alg-1, we monitored the alae structure on young adult animals. During the transition from the fourth and final larval stage of C. elegans (L4) to young adult, seam cells exit the cell cycle, terminally differentiate and fuse in a syncytium. Differentiated seam cells secrete a cuticular structure known as the alae and the loss of alg-1 function leads to an abnormal number of seam cells and defective cell fusion producing incomplete alae (gapped) or breaks along the structure18,19. Rescue experiments showed that phospho-mimicking alg-1(S642E) transgenes were unable to rescue alae defects in alg-1(0) (Figure S1B) and, unlike the non-phosphorylatable alg-1(S642A) transgenes, they were unable to suppress larval arrest and sterility upon depletion of alg-2 with RNA interference (RNAi) (Figure S1C). To determine whether non-phosphorylatable or phospho-mimicking variants of ALG-1 would impair alg-1 function, when expressed from the endogenous alg-1 loci, we produced these mutations in a wild-type background using CRISPR-Cas9 gene editing method. Mutation of this residue, on the endogenous loci of alg-1, revealed that alg-1(S642E) but not alg-1(S642A) displayed alae defects (Figure 1A). Likewise, a significant fraction of alg-1(S642E) mutant animals died at the larval to adult transition from vulva rupturing (Figure 1B); a phenotype that can be attributed to the impairment of the let-7 miRNA family3,18,43,44. These results support that the phosphorylation of serine 642 leads to the loss of alg-1 function as the phospho-mimicking alg-1(S642E) mutation phenocopies alg-1(0).
Figure 1. Phospho-mimicking ALG-1 S642E impairs C. elegans development.
(A) Edited ALG-1 of serine 642 (S642), into non-phosphorylatable alanine (A) and phospho-mimicking glutamate (E) mutant animals were monitored with Nomarski DIC microscopy to evaluate the incidence of alae formation defects at young adult stage. The graph indicates the percentage of animals affected by alae defects. P-values were measured with a two-tailed Fisher's exact test, ** indicates p<0.01 and *** p<0.001. The sample size (n=) used for quantification is indicated. (B) Lethality through vulva bursting. Percentage of alg-1(0), alg-1(S642A) and alg-1(S642E) animal populations that burst through the vulva when reaching adulthood. P-values were measured with a two-tailed Fisher's exact test, ** indicates p<0.01 and *** p<0.001. The sample size (n=) used for quantification is indicated. (C) Top: Lifespan assay. Survival curves of wild-type (WT) animals (black), alg-1(S642A) (green), alg-1(S642E) (blue) and alg-1(0) (red). P-values were calculated using the Mantel-Cox log-rank test, ****: p<0.0001 and **: p<0.01. Lifespan assays were performed with a population of n > 70 animals for each genotype. Bottom: Western blot analysis of ALG-1 at L4 larval stage, two days into adulthood (2d) and five days into adulthood (5d). Exactly 75 worms were used for each genotype and time point. Actin was used as a loading control. The ratios of ALG-1 levels relative to wild-type animals at L4 stage after normalization on Actin levels are shown. This is representative of three biological replicates. (D) Embryonic lethality. Embryos that die during development (unhatched) are counted and reported on the total progeny as a percentage. For each genotype, 8 P0 animals were allowed to lay eggs for 24h before removing them from their respective plate. After 48 hours, unhatched eggs and the total progeny (embryos and larvae) were counted. The boxplot reports the percentage of unhatched eggs relative to the total progeny for the indicated genotypes. P-values were measured by a two-tailed Fisher's exact test accounting for the total number of unhatched eggs and progeny of each P0 animal for each genotype. *: p<0.05 and ***: p<0.001.
During adulthood, ALG-1 and specific miRNAs are involved in the aging process and regulate C. elegans lifespan. Loss of alg-145,46 and miRNAs such as lin-4, miR-71, miR-228, miR-238 and miR-24647-51 leads to a shortened lifespan. We found that, as reported for alg-1(0)46, alg-1(S642E) mutant animals have an average lifespan that is significantly shorter than that of wild-type animals (Figure 1C). Moreover, upon entering adulthood, ALG-1 protein levels were shown to drastically decrease46. We observed a similar decrease for wild-type ALG-1, ALG-1 S642A and ALG-1 S642E proteins after animals had reached adulthood (Figure 1C), suggesting that a decrease in ALG-1 expression levels in alg-1(S642E) mutants is not sufficient to explain the shorter lifespan (Figure 1C) and instead hinting at a defect in ALG-1 activity.
In contrast to alg-1(0), the expression of the phospho-mimicking mutant caused defects in embryonic development (Figure 1D). Specifically, phospho-mimicking alg-1(S642E) adult animals laid eggs that remained unhatched, indicating a defective development leading to embryonic arrest. In animals completely depleted of alg-1, the activity of ALG-2 is sufficient to maintain the miRNA activity required for viability, but the simultaneous depletion of alg-1 and -2 leads to embryonic arrest19. In agreement with this, we were unable to isolate homozygous mutants of alg-1(S642E) and alg-2(0) but the double mutant alg-1(S642A); alg-2(0) were viable (Figure 1D). The embryonic lethality observed in alg-1(S642E) suggests that this mutation is more deleterious than the loss of alg-1. Concurrently, incomplete penetrance of this phenotype indicates that the miRNA pathway and ALG-2 retain at least partial function in the mutant animals. It also reveals that the defects in alg-1(S642E) cannot be explained by a decreased expression of ALG-1 alone, since embryonic lethality does not occur upon loss of alg-1.
The phosphorylation site serine 642 is located on the AGO MID domain, which is responsible for the 5′ nucleotide recognition and binding of the miRNA52,53. To determine whether the embryonic lethality could occur in animals in which ALG-1 is defective for miRNA binding, we produced a mutant strain, alg-1(Y693E). The mutation of this conserved tyrosine to a glutamate located within the 5′ nucleotide binding pocket of the AGO was shown to disrupt miRNA loading of the human AGO2 (Y529E)40. As for the phospho-mimicking alg-1(S642E) mutants, alg-1(Y693E) animals also showed a noticeable number of unhatched eggs (Figure 1D). Taken together, these results show that phospho-mimicking alg-1(S642E) mutation leads to a loss of function of alg-1 and impairs C. elegans development during embryogenesis as well as post-embryonically. Furthermore, dead embryos were observed in animals expressing a miRNA-binding mutant of alg-1, alg-1(Y693E) as well as in alg-1(S642E) mutants but not in alg-1(0), which suggests that S642E and Y693E could affect ALG-1 function in a similar way.
Delayed larval development in phospho-mimicking ALG-1 S642E mutants
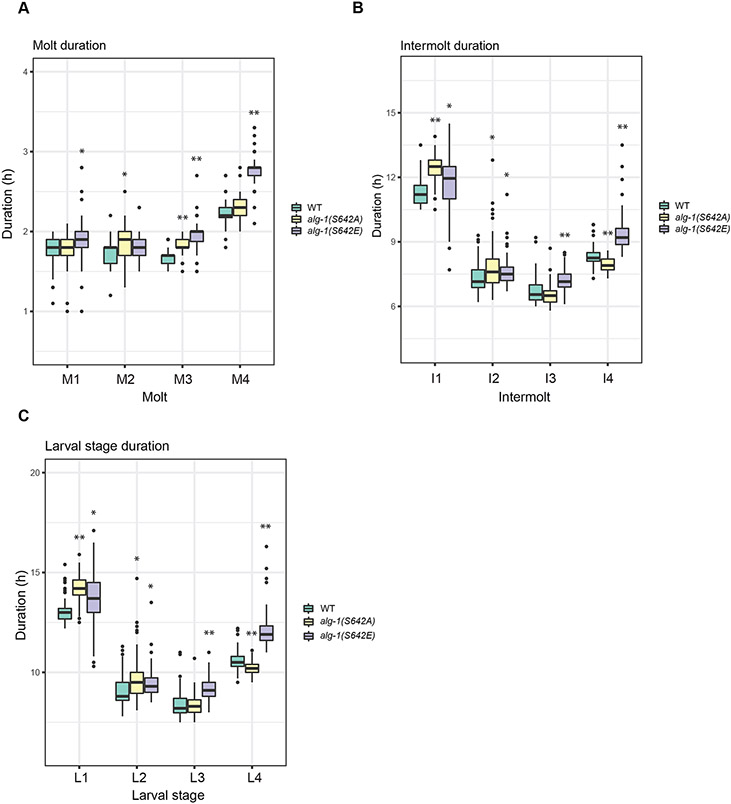
Since our results indicate that constitutive phospho-mimicking of ALG-1 (S642E) leads to a loss of function of the miRNA-specific AGO, we were interested to know whether this phosphorylation event could be important at any point during C. elegans development, where several processes are controlled by miRNAs54. We noticed that alg-1(S642E) mutants reached adulthood a few hours after wild-type animals. To determine whether those delays were caused at a specific stage during development and help us understand the biological relevance of this phosphorylation site, we used a luminescence-based assay55,56 to quantify developmental tempo. This assay detects lack of food uptake in animals during lethargus (molt) through a drop in luminescence signal. Specifically, we cultured animals expressing luciferase from a single copy integrated transgene in the presence of the luciferase substrate D-luciferin. Luminescence emission requires the ingestion of luciferin, which only occurs outside molts, i.e., in intermolts. Hence, by tracking animals individually in the wells of a multiwell plate, we can identify molt entry through a sudden drop in luminescence signal, and molt exit through a steep increase in signal. We can thus quantify the duration of molts (time between drop and increase) and intermolts (between increase and drop). A larval stage is the sum of duration of molts and intermolts.
We found that for the phospho-mimicking alg-1(S642E) mutant animals, all four larval stages were lengthened due to an increase in the durations of all intermolts and molts except for molt 2 (Figure 2). Moreover, although alg-1(S642A) mutant animals appeared wild-type in our other assays, they displayed some alterations in developmental tempo. Thus, the durations of the first and second larval stages were increased relative to wild-type animals due to a lengthening of intermolts, most prominently the first intermolt (Figure 2A and 2B). Additionally, the durations of molts 2 and 3 were modestly increased and that of intermolt 4 decreased. We conclude that both mutations alter developmental timing with a stronger effect observed for alg-1(S642E).
Figure 2. ALG-1 phosphorylation mutants affect larval developmental timing.
Quantification of developmental durations of single animal molts expressing the xeSi296 transgene (A), intermolts (B) and larval stages (C) in WT (n=64), alg-1(S642A) (n=68) and alg-1(S642E) (n=68) as determined by a luciferase assay. The boxplot represents median (thick black line within the box), interquartile range (box), 1.5 times the interquartile range (whiskers); data falling outside this range are plotted as outliers (circles). P-values were measured by Welch two-sample and two-sided t-test. Asterisks represent statistically significant (* p<0.05, ** p<0.01, *** p<0.001).
The ALG-1 S642E phospho-mimicking mutant does not affect its interaction with AIN-1 nor its ability to silence mRNAs
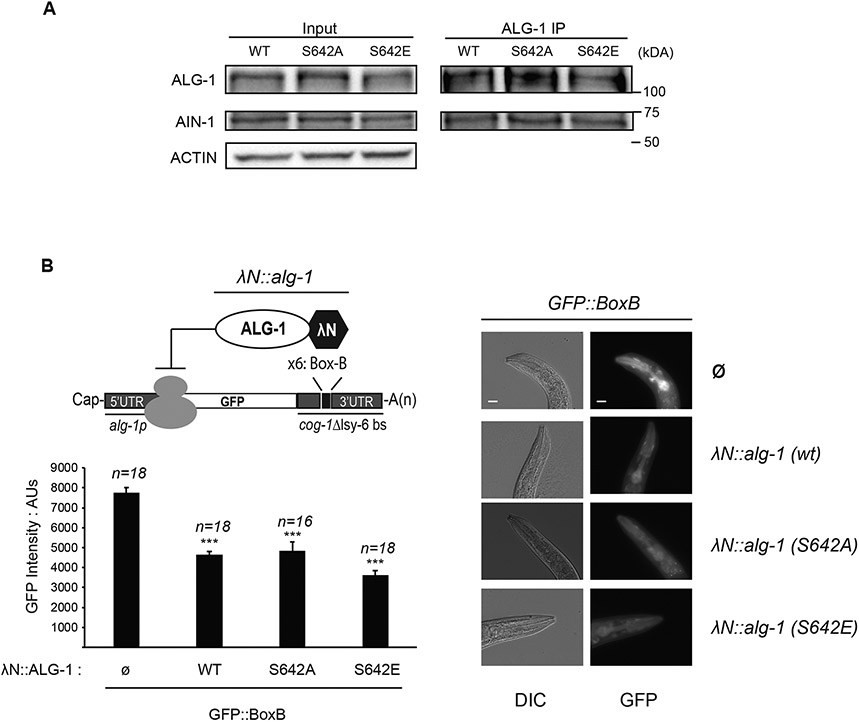
Protein phosphorylation can have a broad range of effects on its conformation, stability, localization or interacting partners. An important component of the miRISC is the scaffold protein GW182 that directly interacts with AGO to recruit deadenylation enzymes such as PAN2/3 or the CCR4/NOT complex that removes the poly(A) tail of the mRNA16. Co-immunoprecipitation experiments of ALG-1 and the GW182 protein AIN-1 showed that wild-type ALG-1, ALG-1 S642A and ALG-1 S642E mutants interacted comparably with AIN-1 (Figure 3A). This suggests that the phosphorylation status of serine 642 does not affect the interaction between AGO and GW182. To determine if the phosphorylation of serine 642 might affect the ability of ALG-1 to reduce protein synthesis once bound to mRNAs, we used a λN/Box-B tethering reporter that enables gene silencing without the requirement for miRNA:mRNA interaction41,57. Endogenously tagged λN::ALG-1 variants were co-expressed with a single copy integrated transgene of GFP with Box-B sequences in the 3′ UTR. Expressing a λN tagged ALG-1 S642A or ALG-1 S642E was as efficient as the λN tagged wild-type ALG-1 to silence the expression of GFP (Figure 3B). Taken together, these results indicate that the phosphorylation status of serine 642 does not affect the silencing efficiency of ALG-1 artificially bound to the 3' UTR of a target mRNA.
Figure 3. Phosphorylation of ALG-1 serine 642 does not affect its interaction with GW182 homolog AIN-1 nor its ability to silence a mRNA.
(A) ALG-1 co-immunoprecipitation with GW182 homolog AIN-1. ALG-1 was immunoprecipitated from adult extract of wild type, alg-1(S642A) or alg-1(S642E) and polyclonal antibodies for AIN-1 and ALG-1 were used for western blotting. The inputs are 10% of the total protein extracts used for immunoprecipitations. Actin served as loading control. The blots are representative of three biological replicates. (B) ALG-1 S642 phosphorylation does not impair mRNA silencing when tethered to a mRNA 3′ UTR. Top left: Schematic representation of AGO tethering system. A GFP reporter under the control of an alg-1 promoter fused with the sequence of cog-1 3′UTR where the lsy-6 miRNA binding sites (delta lsy-6 bs) are replaced by six copies of the Box-B element (x6: Box-B). The high affinity between the Box-B RNA secondary structure and the λN peptide fused to ALG-1 leads to its recruitment. A strain with a single integrated copy of alg-1p::GFP::Box-B reporter carrying endogenous alg-1 alleles tagged with a λN sequence at the 5′ end of the coding sequence was used to edit λN::alg-1 into non-phosphorylatable λN::alg-1 (S642A) and phospho-mimicking λN::alg-1 (S642E). The expression level of the GFP reporter was measured in the pharynx. Bottom left: The GFP level expressed in the pharynx of young adult worms was quantified using arbitrary units (AU). The error bars represent the 95% confidence interval, and the P-values indicated were measured by a two-tailed Student’s t-test; *** p<0.001. The number of animals scored (n=) is indicated and the graph is representative of two independent measurements. Right: Representative images of animals expressing only the GFP reporter (Ø) or GFP reporter and different versions of λN-tagged alg-1 (λN::alg-1) gene are shown. The scale bar indicates 20 μm. Images were obtained at the same time of exposure, on the same slide, and with the same area of measure for each animal strain.
The ALG-1 S642E mutation affects the abundance of both guide and passenger miRNA strands
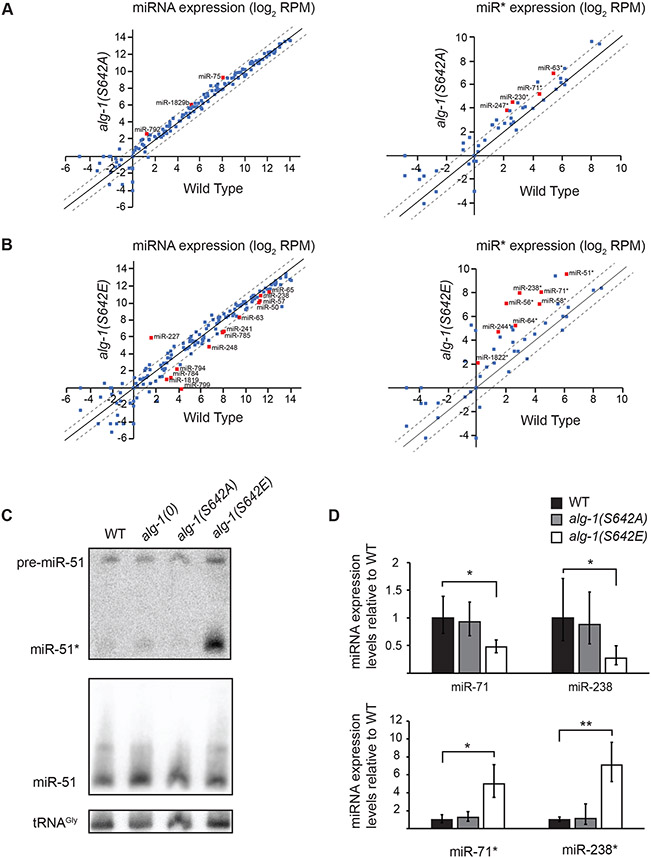
AGOs are at the core of the miRISC and, as the carrier of miRNAs, they play a major role in promoting their maturation from the pre-miRNA to their mature form10,58-60 and protect the mature miRNA from degradation by exonucleases. Hence, variations in the ability of AGO to bind miRNAs is expected to have repercussions on the miRNA levels. To assess whether serine 642 phosphorylation could have such an effect, we measured the abundance of miRNAs by high-throughput sequencing. We found that, while miRNA levels in alg-1(S642A) mutants were overall indistinguishable from wild-type animals (Figure 4A), several miRNAs were significantly decreased in adult alg-1(S642E) mutant animals (Figure 4B and 4D) and throughout the larval development (Figure S2 and S3). alg-1(S642E) mutants showed alae formation defects and vulva rupturing (Figure 1A and 1B) which are typically caused by misregulation of the let-7 miRNA family. While miR-48-5p and miR-84-5p were expressed at similar levels compared to wild type at various time points, the expression of let-7-5p was delayed by a few hours and miR-241-5p was decreased at all time points (Data S1). These data indicate that misregulation of the let-7 miRNA family during development could indeed contribute to the larval phenotypes in alg-1(S642E) (Figure 1A and 1B). We performed miRNA quantification in embryo by RT-qPCR for miR-35-42 and miR-51-56 miRNA families because these two miRNA families are required for embryogenesis61-63. We observed a modest but significant decrease for miR-35-42 miRNA family that occurred in both alg-1(S642A) and alg-1(S642E), while miR-51-56 miRNA family was not affected (Figure S4A), suggesting a potential contribution of miR-35-42 miRNA family in the embryonic phenotypes observed in alg-1(S642E) mutants (Figure 1D).
Figure 4. microRNA abundance is altered in phospho-mimicking ALG-1 S642E.
(A and B) Normalized miRNA reads expressed as log2 of reads per million (RPM) sequenced from small RNA cloning total RNA extracts of (A) alg-1(S642A) mutant animals and (B) alg-1(S642E) mutant animals compared to wild-type animals at adult stage. The scatterplot reports the guide miRNA (Left) and passenger strand miRNA (miR*) (Right) abundance in mutant vs wild-type animals. The abscissa measures the abundance in wild-type animals and the ordinate measures the abundance in alg-1(S642A) or alg-1(S642E). Each point represents the values for a specific miRNA averaged on three biological replicates. Red squares are miRNAs for which the difference in number of normalized reads compared to wild type was significant as evaluated with an unpaired Student’s t-test; p<0.05. The dashed diagonals indicate the two-fold change, and the middle diagonal (black) represents the x=y slope. (C) Detection of miR-51 by Northern Blotting in wild-type, alg-1(0), alg-1(S642A) and alg-1(S642E) adult animals. The mature guide miRNA (miR-51) or passenger strand miRNA (miR-51*) are indicated as well as the precursor molecule (pre-miR-51). The detection of tRNA glycine (tRNAgly) was used as a loading control. Representative images of two biological replicates. (D) Guide and passenger miRNA strands quantification by RT-qPCR. The levels of the guide and passenger strands of miR-71 and miR-238 in alg-1(S642A) and alg-1(S642E) gravid adult animals were measured by RT-qPCR and normalized to the levels of wild-type animals. Small nucleolar RNA sn2841 was used as an internal control gene. The error bars represent the 95% confidence interval from three biological replicates and the P-values were calculated using a two-tailed Student’s t-test; * p<0.05, ** p<0.01.
Interestingly, this sequencing data showed that several passenger strands (miR*) were more abundant in the alg-1(S642E) mutants than those measured in wild-type animals (Figure 4B right panel). Although many passenger strands were more abundant in alg-1(S642E) mutant animals, their reads never exceeded those of the guide strands. Specifically, passenger strands that showed a significant increase (p<0.05) in alg-1(S642E) compared to wild-type animals are miR-51-3p (10 fold), miR-56-5p (6 fold), miR-58-5p (7 fold), miR-64-3p (6 fold), miR-71-3p (12 fold), miR-238-5p (32 fold), miR-244-3p (9 fold), and miR-1822-5p (4 fold). We confirmed the increase of miR-51-3p by Northern blot (Figure 4C) and miR-71-3p and miR-238-5p by RT-qPCR (Figure 4D). Taken together, these results show that phosphorylation of ALG-1 serine 642 affects the abundance of both the guide and passenger strands during C. elegans development.
Phosphorylation of ALG-1 serine 642 affects miRNA binding
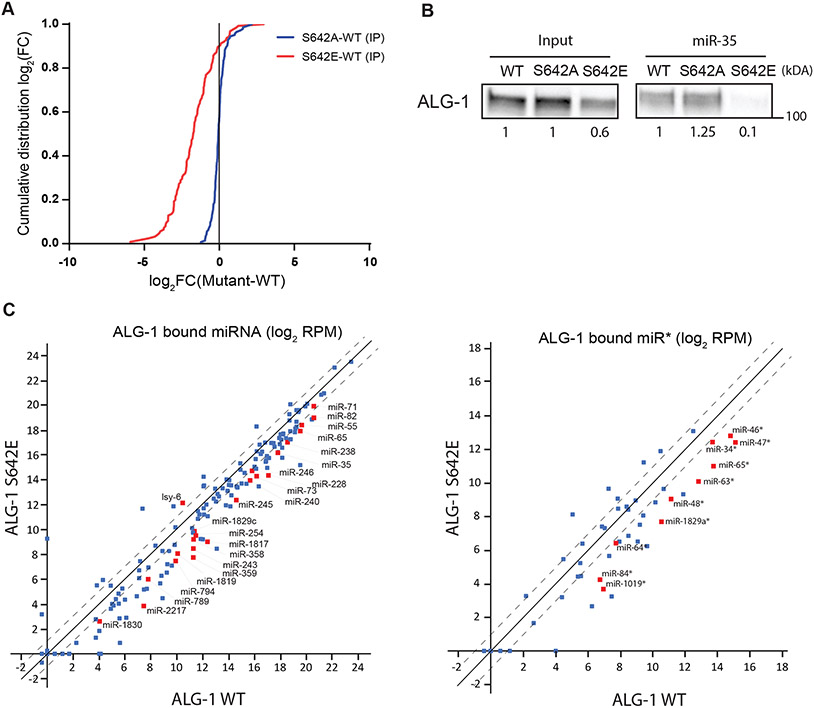
Alteration of miRNA levels can be attributed to different mechanisms mediated by AGO. First, the processing of some precursor miRNA into mature miRNA is decreased in the absence of ALG-118,26, which did not occur for miR-51, as the levels of the precursor form was not affected in alg-1(0) nor alg-1(S642E) (Figure 4C). Second, AGO stabilizes miRNAs through binding and thus protects them from nuclease-mediated degradation, a mechanism that can be regulated in cells by exposing the 3′ end of miRNA to modifications or by degradation of AGO by the ubiquitin-proteasome64-67. To investigate if ALG-1 phosphorylation at serine 642 could prevent miRNA binding and thus lead to the differences in miRNA levels observed in the phospho-mimicking alg-1(S642E) mutant, we immunopurified wild-type ALG-1, ALG-1 S642A and ALG-1 S642E and quantified the associated miRNAs with high-throughput sequencing. We observed a global decrease in miRNA population bound to the phospho-mimicking ALG-1 S642E compared to wild type (Figure 5A). In contrast, the abundance of miRNAs associated with the non-phosphorylatable ALG-1 S642A mutant was indistinguishable from wild-type ALG-1 (Figure 5A). The decrease in miRNA association of ALG-1 S642E was also observed when we pulled down miR-35 miRISC (Figure 5B). These results reveal that phospho-mimicking ALG-1 S642E impairs the function of ALG-1 by decreasing its ability to bind miRNAs.
Figure 5. Phospho-mimicking ALG-1 S642E impairs binding to miRNAs.
(A) ALG-1 immunoprecipitation (IP) and small RNA sequencing experiments using wild-type (WT) and phosphorylation mutants animal populations. The plot shows the cumulative distribution of the log2-fold changes (log2(FC)) for miRNA reads in ALG-1 mutants IP vs ALG-1 WT IP averaged over three biological replicates. ALG-1 S642A mutant bind miRNAs similarly to ALG-1 WT (Blue). The ALG-1 S642E mutant binds far less miRNAs compared to WT (Red). The vertical line at 0 represents a log2FC of zero compared to WT. (B) miR-35 miRISC pulldown of ALG-1. Proteins bound to miR-35 miRNA in gravid adult extracts were pulled-down using a 2′-O-methylated and 5′ biotinylated RNA fully complementary oligonucleotide. The levels of ALG-1 pulled down in wild-type (WT) animal extracts or in phosphorylation mutants S642A and S642E were evaluated by western blotting. The ALG-1 levels in the input and in the pulldown relative to the signal in WT are shown. Representative image of three biological replicates. (C) Scatterplot of miRNA bound to ALG-1 averaged on three biological replicates and expressed as Log2 of reads per million (RPM). The guide strands (Left) and passenger strands (miR*) (Right) associated to ALG-1 are plotted comparing ALG-1 S642E and ALG-1 WT. The dashed lines indicate the two-fold change, and the middle diagonal represents the x=y slope. Red squares indicate miRNAs for which the number of reads were significantly different between WT and S642E IP, as determined with an unpaired Student’s t-test (p<0.05). miRNA reads obtained from ALG-1 IP were normalized on the number of miR-48-5p reads in each replicate as miR-48 binding to ALG-1 WT, ALG-1 S642A and ALG-1 S642E is robustly identical (Figure S5A).
Surprisingly, although passenger strands accumulated in the total RNA of alg-1(S642E) mutant animals (Figure 4B-4D), this did not coincide with an increased passenger strand binding to ALG-1 S642E (Figure 5C). This suggests that the affected passenger strands accumulate without binding to ALG-1. Indeed, most matching guide strands did not exhibit a substantial decrease in levels (Figure S4B), suggesting that the alg-1(S642E) mutant phenotypes do not generally derive from a switch in miRNA strand loading onto ALG-1. Instead, these data may be parsimoniously explained by a defect in loading of the miRNA guide:passenger strand duplex onto ALG-1. Duplexes may thus accumulate unbound to ALG-1 in the cytoplasm, and binding to the guide strand may render the passenger strand refractory to single-strand nucleases that would normally degrade evicted passenger strands after loading of the guide strands into ALG-1. Concurrently, this would diminish the amounts of functional, ALG-1-loaded guide strands and thus explain why miRNA activity is decreased despite little changes in overall cellular miRNA guide strand levels.
The protein kinase A phosphorylates serine 642 in vitro and genetically interacts with alg-1
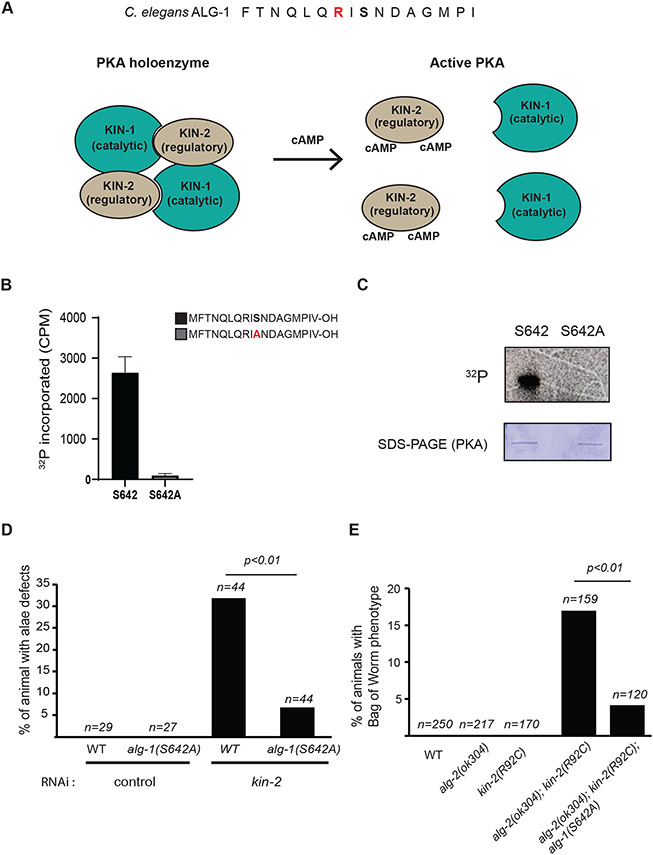
To determine which pathway(s) can regulate ALG-1 through phosphorylation of serine 642 residue, we first used Netphos 3.168 to predict kinases that can use this specific amino acid as substrate. The protein kinase A (PKA) was the only kinase with a positive score (0.62). PKA is a 3′,5′-cyclic adenosine monophosphate (cAMP)-dependent serine/threonine kinase complex composed of a catalytic and a regulatory subunit known as KIN-1 and KIN-2 in C. elegans, respectively69-71. In the absence of cAMP, PKA forms an inactive tetramer (holoenzyme) of two regulatory and two catalytic subunits. Upon stimulation, adenylyl cyclase produces cAMP from ATP. The regulatory subunit binds to the cAMP and triggers a conformational change that reduces the affinity for the catalytic subunit and allows it to phosphorylate its substrates (Figure 6A). To determine whether ALG-1 serine 642 can be phosphorylated by PKA, we performed an in vitro kinase assay using different peptides of ALG-1 spanning 18 amino acids and incubated them with recombinant PKA (Figure 6B-C). Incorporation of radioactive phosphate was reproducibly detected, and the in vitro phosphorylation was specific to serine 642 as the signal was lost when we used a peptide in which the serine had been replaced by an alanine (Figure 6B-C). The consensus sequence along with the in vitro phosphorylation of ALG-1 peptides suggest that PKA could target ALG-1 to regulate its activity.
Figure 6. Protein Kinase A (PKA) genetically interacts with alg-1 and alg-1(S642A).
(A) Schematic of PKA activation. The consensus sequences for PKA substrates are shown on ALG-1 sequence for serine 642 (Top). The basic side chain of arginine (R) (Red) is commonly found at position -2 and -3 of PKA substrates. PKA is a holoenzyme complex (Bottom) formed by two regulatory subunits and two catalytic subunits: KIN-1 and KIN-2 respectively in C. elegans. Activation of GPCRs and downstream synthesis of cyclic adenosine monophosphate (cAMP) by adenylate cyclase leads to the dissociation of the complex which allows the catalytic subunit to phosphorylate its substrates. (B) ALG-1 peptides are phosphorylated in vitro by PKA at serine 642. The graph indicates the incorporation of 32P as measured with a liquid scintillation counter following the incubation of peptides with recombinant PKA. 18 amino-acid peptides of ALG-1 spanning the S642 phosphorylation site or a peptide in which the serine (S) is replaced with an alanine (A) were used. The error bars show the 95% confidence interval from three independent experiments (n=3). (C) Autoradiogram and Coomassie blue stained gels of in vitro PKA kinase reaction. (D) Alae defects caused by kin-2 RNAi are suppressed by the non-phosphorylatable mutant alg-1(S642A). First stage larvae L1 were fed with bacteria expressing double-stranded RNA targeting kin-2 mRNA or a control double-stranded RNA. Young adults staged wild-type animals or alg-1(S642A) mutants were monitored for alae defects. The p-value was measured by Fisher's exact test. The graph is representative of three biological replicates. (E) alg-1(S642A) suppresses kin-2 loss of function mutant Bag of worms phenotype in alg-2(0) animals. The hypomorphic point mutation kin-2(R92C) was edited with CRISPR-Cas9 in alg-2(0) mutant and in the double mutant alg-2(0);alg-1(S642A). The double mutant alg-2(0); kin-2(R92C) animals die at adult stage caused by a defect in egg laying (Egl) leading to embryos hatching inside the worm (Bag of worms). This phenotype was significantly suppressed in animals that cannot be phosphorylated on ALG-1 serine 642. The sample size (n=) used for quantification for each genotype is indicated and the p-value was calculated with Fisher’s exact test.
To investigate whether PKA contributes to the regulation of ALG-1 in vivo, we first determined whether PKA genetically interacts with alg-1 and the miRNA pathway by depleting the regulatory subunit, kin-2, with RNAi. A decrease in KIN-2 levels will stimulate KIN-1 activity and could thus cause similar phenotypes as those observed in alg-1(S642E) if serine 642 is a bona fide substrate of PKA. We observed that, as for alg-1(S642E) mutants (Figure 1A), kin-2 RNAi treated animals showed alae defects. More importantly, those defects were suppressed in the non-phosphorylatable alg-1(S642A) mutant exposed to kin-2 RNAi (Figure 6D). These data indicate that the alae defects occurring upon the kin-2 knockdown in animals require the phospho-acceptor residue serine 642, suggesting that PKA might regulate ALG-1 phosphorylation in vivo. Since kin-2 RNAi produced severe developmental defects and early adult lethality, we decided to use a hypomorphic kin-2(ce179) allele to gain further evidence of the involvement of PKA in this process. This kin-2 allele contains a point mutation on a conserved residue (R92C) in the auto-inhibitory domain that interacts with the catalytic subunit leading to an increased PKA activity72,73. kin-2(R92C) larvae mutant animals are viable but take a longer time to reach adulthood, produce fewer progeny than wild-type animals and have an egg laying phenotype (Egl)73. To strengthen the evidence that PKA antagonizes alg-1, we tested whether the depletion of alg-2 in the kin-2 mutant would cause a synthetic lethality phenotype as it is the case for the loss of both alg-1 and alg-219. To do so, we generated a double mutant with kin-2(R92C) and a knockout strain of alg-2 (alg-2(ok304)). We observed that the loss of alg-2 in the hyperactive PKA strain led to adult lethality caused by egg laying defects (Bag of worms phenotype) along with a few larvally arrested worms. These phenotypes were suppressed in a triple mutant including the non-phosphorylatable alg-1(S642A) mutant (Figure 6E). This genetic assessment shows that activated PKA antagonizes ALG-1 in vivo, in a serine 642 dependent manner. The in vitro phosphorylation assays strongly suggests that PKA could regulate ALG-1 function through its phosphorylation on serine 642. Overall, our results demonstrate a regulatory mechanism for AGO protein in C. elegans, in which ALG-1 phosphorylation on serine 642 reduces its binding to miRNAs and impairs the formation of functional miRISC, and present evidence that the PKA signalling pathway is likely responsible for this phosphorylation in vivo.
DISCUSSION
In this study, we report the phosphorylation of a specific serine residue on the MID domain of the C. elegans AGO ALG-1 that modulates its ability to bind miRNAs. Our in vivo analysis of the phosphorylation mutants of ALG-1 serine 642 reveals that the constitutive phosphorylation of this residue (mimicked by S642E) produces loss-of-function phenotypes during embryonic and larval development. Since embryonic lethality from the loss of miRNA function arises from the lack of both AGOs ALG-1 and ALG-2 activity in embryos, this phenotype in alg-1(S642E) and alg-1(Y693E) mutants suggests that miRNA-binding deficient ALG-1 would interfere with ALG-2 function by reducing the availability of important interactors for miRNA maturation and/or gene silencing. The evidence for such misregulation is the interaction between ALG-1 S642E and the GW182 protein AIN-1 (Figure 3A). We have previously reported that the interaction between AGO and GW182 is not strictly required for embryonic viability57, suggesting the possibility that other factors at play produce this phenotype. Therefore, we looked at the interaction between ALG-1 S642E and DCR-1 that cleaves the precursor miRNAs to produce mature miRNA duplexes. ALG-1 S642E maintained its interaction with DCR-1 in adults (Figure S5B) and embryos (Figure S5C) despite that ALG-1 S642E mutant binds miRNA less efficiently. Thus, we speculate that, in addition to the reduced miRNAs binding (Figure 5), ALG-1 S642E mutants could impair the ALG-2 function through its association with DCR-1 and the RISC loading complex.
The C. elegans ALG-1 serine 642 was found to be phosphorylated in vivo (Figure S1A) which is a conserved residue on all four human AGOs (Figure S6A). Since this serine is conserved on the C. elegans ALG-2 (Figure S6A) and located in the vicinity of a basic amino acid (Arginine (R)) at position −2, which is typically found in PKA substrates, it will be interesting to determine whether ALG-2 can be phosphorylated. Furthermore, differences in the phosphorylation statuses between ALG-1 and ALG-2 during development could potentially explain the previously reported preferential loading of specific miRNAs19,20. The abovementioned key residues are not found in ALG-5, another C. elegans AGO capable of binding to a specific subset of miRNAs in the germline (Figure S6A). Therefore, it is difficult to speculate if a specific post-translational modification on ALG-5 could share a similar function. Despite the sequence conservation around the residue corresponding the ALG-1 serine 642 on human AGOs (Figure S6A), we could not detect phosphorylation on the human AGO1-4 purified from cell cultures. It is possible that the phosphorylation on human AGOs only occurs in specific cell types or that the differences in their structural features prevent this phosphorylation. When we looked at the previously determined structures of the guide- and guide-target duplex-bound AGO1-4, most of them occludes the hydroxyl group on the side chain of the corresponding serine (Table S1)25,27,65,74-79. In contrast, the hydroxyl group on the side chain of serine 642 is solvent-exposed in the model of apo-ALG-1 (RNA-free form of ALG-1) available from the AlphaFold Protein Structure Database EMBL server (https://alphafold.ebi.ac.uk) (Figure S6B-C). These observations suggest that the phosphorylation of this serine could either be C. elegans specific or that residue can only be phosphorylated before the miRISC formation. We performed experiments in HEK 293T to determine whether mutations of AGO2 serine 478 (the serine corresponding to ALG-1 serine 642) impair miRNA function, and observed that although miRNA levels were not affected upon overexpression of Flag/HA AGO2 S478E (Figure S6D), both Flag/HA AGO2 S478E and the miRNA binding mutant Flag/HA AGO2 Y529E decreased binding to mRNAs (Figure S6E). Although the conservation of this phosphorylation event in human cells is not yet clear or under which biological conditions it would occur, these results indicate that serine 478 is an important residue for AGO2 function. The addition of a negative charge at this position, by either phosphorylation or a somatic mutation, would impair the miRNA function and impact human cell homeostasis.
As opposed to the human AGO2 phosphorylation site Y529 forming the miRNA binding pocket40, ALG-1 serine 642 is located on a helix at the surface of the MID domain (Figure S6C). While both phosphorylation impair binding to miRNA, the modification of Y529 is thought to prevent miRNA binding by sterically hindering the 5′ phosphate of the miRNA or by the proximity of the negative charges carried by both the phospho-tyrosine and the miRNA 5′ phosphate. Conversely, ALG-1 serine 642 is located on the surface of the MID domain and thus does not seem to interact with the miRNA 5′ end or its sugar-phosphate backbone. Therefore, this phospho-serine would decrease the binding to miRNAs by a different mechanism, such as locking the AGO in an open conformation, and preventing the transition toward the closed conformation after miRNA binding. Although the bulk of miRNAs are strongly decreased in ALG-1 S642E IP compared to wild-type ALG-1, the binding to specific miRNAs remains reproducibly efficient (Figure 5C and S5A), which could deny the possibility that the phosphomimetic mutant ALG-1 S642E remains in open conformation. We speculate that there could be a sequence bias or that the stability of the miRNA duplex end could allow specific miRNAs to be sorted into ALG-1 S642E as efficiently as for the wild-type protein and form of a functional miRISC.
Mutations in conserved residues of ALG-1 MID domain that result in accumulation of passenger strands, like in alg-1(S642E), have been reported in C. elegans80,81. Genetically, the antimorphic mutant allele alg-1(ma202) contains an amino acid substitution G553R and has phenotypes that are more penetrant than for alg-1(0) mutant animals. In comparison, alg-1(S642E) mutants have less severe phenotypes compared to alg-1(0) during larval development (Figure 1A and 1B) but more severe defects during embryonic development (Figure 1D). The difference in phenotypes and their severity indicate that alg-1(S642E) and alg-1(G553R) have distinct molecular effects. As observed for ALG-1 S642E (Figure S5B-C), ALG-1 G533R proteins associate with DCR-1 but unlike ALG-1 S642E, ALG-1 G553R poorly associates with the miRISC effector AIN-1, suggesting that the latter sequester miRNAs in ineffective complexes. ALG-1 S642E associate with AIN-1 (Figure 3A) and silence GFP protein expression as effectively as wild-type ALG-1 when tethered to the 3′ UTR of the mRNA (Figure 3B), suggesting that ALG-1 S642E can form an effective miRISC despite the decrease in miRNA binding. Both mutants show a strong increase in passenger strands but apparently for different reasons. In alg-1(G553R) mutants, passenger strands are inappropriately loaded in ALG-1, which in turn, protects them from degradation. In contrast, ALG-1 S642E does not selectively bind passenger strands when compared to wild-type ALG-1 (Figure 5C). This indicates that passenger strands in alg-1(S642E) might not be dissociated from the guide strands and accumulate as duplexes which could also explain the modest decrease for many guide miRNAs (Figure 4A, S4A and S4B) despite their inefficient binding to ALG-1 S642E (Figure 5A-C).
In the alg-1(S642E) mutant animals, while we observed a robust decrease in miRNAs associated to ALG-1, we also noticed a decrease in ALG-1 levels at L4 stage and in adults (Figure 1C and S1D). Although a decrease in ALG-1 protein levels could contribute to the phenotypes observed in alg-1(S642E) mutants, the stronger phenotypes compared to alg-1(0) animals (Figure 1D) shows that this decrease in ALG-1 S642E levels is not sufficient to explain them. Various reports show that apo AGOs are selectively degraded by either the proteasome or through the lysosome82-85. Blocking the proteasome using MG132 proteasome inhibitor was not sufficient to restore ALG-1 S642E levels (Figure S1D). It will be interesting to test whether ALG-1 S642E mutant is sorted into autophagy as recently found in fly Ago1 miRNA-binding mutants85,86.
PKA is implicated in several biological processes, including lipid metabolism, rhythmic behavior, locomotion, immunity and stress response87-92. Beside its important role to regulate the transcriptional activation of target genes through the phosphorylation of CREB family proteins93, PKA was shown to regulate mRNA translation94-96 and mRNA decay97. The regulation of the AGO phosphorylation by the cAMP signalling provides an additional mechanism for post-transcriptional regulation of gene expression. Our phenotypical analyses in the non-phosphorylatable alg-1(S642A) mutants indicates that ALG-1 serine 642 phosphorylation is not required for the synthesis of the alae, the formation of the vulva and during embryogenesis (Figure 1A-B and 1D). Therefore, we do not foresee a sustained regulation of ALG-1 by PKA in those tissues throughout development. In the context of PKA signalling, the activity of the catalytic subunit KIN-1 is under tight regulation and inhibited by the regulatory subunit KIN-2 in cells until its activation. In this signalling pathway and upstream of the second messenger cAMP that activates KIN-1 by releasing KIN-2, are found the G-protein coupled receptors (GPCRs). There are approximately 1,300 predicted or curated genes encoding GPCRs in C. elegans98, that respond to different extracellular stimuli and ligands. The model that we envision for the role of ALG-1 inactivation by PKA is as follows: upon activation of PKA, several genes are upregulated or downregulated by transcriptional and post transcriptional means. Among the transcripts that are positively regulated by PKA, some are also putative miRISC targets which hinders their expression. The phosphorylation of ALG-1 serine 642 by PKA decreases ALG-1 binding to miRNA and thereby, promotes the expression of PKA induced genes that contain miRNA binding sites. In C. elegans, PKA have been shown to upregulate a specific set of antimicrobial genes in the neurons, in response to Salmonella enterica infection91. It will be an interesting way to test our model by exposing alg-1(S642A) mutants to S. enterica infection to determine if 1) ALG-1 is involved, through its modification by PKA, in innate immunity response and 2) whether serine 642 phosphorylation promotes the expression of the same set of genes as PKA, upon PKA activation in the neurons.
Altogether our study indicates miRNA-mediated gene regulation pathway interacts with the cAMP signalling pathway. Specifically, phosphorylation of ALG-1 on serine 642 by PKA decreases AGO ability to bind miRNAs. As only specific cells might undergo activation of PKA at a given time during normal growth conditions and might be restricted by compartmentalization in others, the identification of the processes that incurs the miRISC inactivation will be crucial to understand its biological function. For this purpose, it will be important in the future to survey different conditions and environmental stresses if we want to uncover how this important gene regulation pathway is controlled.
Limitations of the study
There are potential caveats in our approach that needs to be considered when interpreting the data. Phospho-mimicking ALG-1 mutations were used to characterize the effect of serine 642 phosphorylation. While those variants are broadly used to study protein phosphorylation, they do not always fully recapitulate the phospho-substrate (for example99,100), hence the data reported here only describes the molecular and biological effect of ALG-1 phospho-mimicking substitution. The contribution of serine 642 phosphorylation in vivo could differ from what is observed with ALG-1 S642E mutant.
We showed that kin-2 interacts genetically with alg-1. The genetic suppression of kin-2 phenotypes by alg-1(S642A) indicates that ALG-1 phosphorylation ablation counteracts the effects of KIN-1 activation. While this could mean that alg-1 and kin-1/kin-2 are part of the same pathway, this data alone is not sufficient to conclude that ALG-1 is a direct substrate of KIN-1. KIN-1 activation could affect different kinases and phosphatases that regulate ALG-1 and hence, stimulate serine 642 phosphorylation indirectly. In vitro phosphorylation of serine 642 were conducted with recombinant PKA enzyme and ALG-1 peptides. This data provides evidence that PKA can recognize and phosphorylate serine 642 site, but does not recapitulate the context of protein folding, structure and protein-protein interactions in vivo.
This study focuses on ALG-1 serine 642 phosphorylation; the phosphorylation of ALG-2 and its biological relevance remains to be investigated. Our phenotypical analyses of the non-phosphorylatable alg-1(S642A) mutant animals showed developmental delays during the first two larval stages, indicating that serine 642 phosphorylation positively regulates early larval development. Quantitative mass spectrometry analyses of ALG-1 phosphorylation at different time points during development will be required to determine whether there is a prominent phosphorylation at those stages.
Last, this study did not address the localization of ALG-1 in cells. The mislocalization of ALG-1 S642E could affect the loading process and the miRISC turnover dynamic. Similarly, in vitro analysis of phospho-mimicking ALG-1 S642E miRNA binding and duplex unwinding will be a matter of future study to address the biochemical effect of serine 642 phosphorylation on AGO.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Martin J. Simard (martin.simard@crchudequebec.ulaval.ca).
Materials availability
All unique and stable reagents and strains generated in this study are available from the lead contact without restriction.
Data and Code availability
Raw and processed small RNA-seq datasets have been deposited at NCBI’s Gene Expression Omnibus (GEO) repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-beta Actin Monoclonal Antibody, HRP Conjugated, Clone AC-15 | Abcam | Cat# ab49900, RRID:AB_867494 |
| Peroxidase-AffiniPure Goat Anti-Rabbit IgG (H+L) (min X Hu,Ms,Rat Sr Prot) antibody | Jackson Immunoresearch Labs. | Cat# 111-035-144, RRID:AB_2307391 |
| Rabbit Anti-ALG-1 polyclonal antibody | (Vasquez-Rifo et al., 2012) | N/A |
| Rabbit Anti-AIN-1 polyclonal antibody | (Jannot et al., 2016) | N/A |
| Rabbit Anti-DCR-1 polyclonal antibody | (Duchaine et al., 2006) | N/A |
| Mouse Anti-Ubiquitin Antibody (P4D1) monoclonal antibody | Santa Cruz | Cat# sc-8017, RRID:AB_628423 |
| Mouse Anti-HA.11 Monoclonal Antibody, Unconjugated, Clone 16B12 | Covance | Cat# MMS-101P-500, RRID:AB_291261 |
| IRDye 800CW Goat anti-Mouse IgG | Li-Cor Biosciences | Cat# 925-32210, RRID: AB_2687825 |
| Bacterial and virus strains | ||
| E. coli strain: OP50 | Caenorhabditis Genetics Center | CGC: OP50, RRID: WB-STRAIN:WBStrain00041969 |
| E. coli strain: HT115(DE3) | Caenorhabditis Genetics Center | CGC: HT115(DE3), RRID: WB-STRAIN:WBStrain00041080 |
| Chemicals, peptides, and recombinant proteins | ||
| Biotin-MFTNQLQRISNDAGMPIV-OH | This study | N/A |
| Biotin-MFTNQLQRIANDAGMPIV-OH | This study | N/A |
| Firefly D-Luciferin | PJK GmbH | Cat# 102111 |
| MG-132 | Sigma-Aldrich | Cat# 474790 |
| Tri Reagent | Sigma-Aldrich | Cat# T9424 |
| Critical commercial assays | ||
| TaqMan Universal PCR Master Mix, no AmpErase UNG | Thermo Fisher Scientific | Cat# 4324018 |
| Taqman MicroRNA Assay | Thermo Fisher Scientific | Cat# 4440886 |
| Q5 Site-Directed Mutagenesis Kit | New England Biolabs | Cat# E0554S |
| Dynabeads M-280 Streptavidin | Thermo Fisher Scientific | Cat# 11205D |
| Dynabeads Protein G for Immunoprecipitation | Thermo Fisher Scientific | Cat# 10004D |
| Anti-FLAG M2 Magnetic Beads | Sigma Aldrich | Cat# M8823 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat# 4368814 |
| Edit-R tracrRNA | Horizon Discovery | Cat# U-002005 |
| Edit-R crRNA | Horizon Discovery | Cat# Custom0280 |
| QIAseq miRNA Library Kit | Qiagen | Cat# 331505 |
| Takyon No Rox SYBR MasterMix dTTP Blue | Eurogentec | Cat# UF-NSMT-B0701 |
| Deposited data | ||
| Small RNA sequencing reads from Caenorhabditis elegans adult worms (total RNA and ALG-1 IP) | This study | GEO: GSE198352 |
| Small RNA sequencing reads at different time points during larval development | This study | GEO: GSE174368 |
| Experimental models: Cell lines | ||
| Human: HEK293T cells | ATCC | RRID:CVCL_0063 |
| Experimental models: Organisms/strains | ||
| Strain N2 Bristol (WT) | Caenorhabditis Genetics Center | CGC: N2, RRID:WB-STRAIN:WBStrain00000001 |
| Strain VC446: alg-1(gk214) X | Caenorhabditis Genetics Center | CGC: VC446, RRID: WB-STRAIN: WBStrain00035775 |
| Strain KG532: kin-2(ce179) X | Caenorhabditis Genetics Center | CGC: KG532, RRID:WB-STRAIN:WBStrain00023482 |
| Strain MJS218: alg-1(gk214) Ex[alg-1p::lambdaN::mCherry::alg-1::alg-1 3'UTR; prf4(rol-6(su1006))] | (Quévillon Huberdeau et al., 2017) | N/A |
| Strain MJS258: alg-1(gk214) Ex[λN::mcherry::alg-1(S642E); prf4(rol-6(su1006))] | This paper | N/A |
| Strain MJS259: alg-1(gk214) Ex[λN::mcherry::alg-1(S642A); prf4(rol-6(su1006))] | This paper | N/A |
| Strain MJS275: alg-1(qbc23[S642A]) X | This paper | N/A |
| Strain MJS306: alg-1(qbc46[S642E]) X | This paper | N/A |
| Strain HW1939: xeSi296[Peft-3::luc::gfp::unc-54 3'UTR, unc-119(+)] II | (Meeuse et al., 2020) | N/A |
| Strain HW3060: xeSi296[Peft-3::luc::gfp::unc-54 3'UTR, unc-119(+)] II; alg-1(qbc23[S642A]) X | This paper | N/A |
| Strain HW3061: xeSi296[Peft-3::luc::gfp::unc-54 3'UTR, unc-119(+)] II; alg-1(qbc46[S642E]) X | This paper | N/A |
| Strain MJS237: qbcSi03[Palg-1::GFP::cog-1-boxb-cb-unc-19(+)] IV; alg-1(qbc18[lambdaN::alg-1]) X | (Quévillon Huberdeau et al., 2017) | N/A |
| Strain MJS345: qbcSi03[alg-1p::GFP::cog-1-boxb;cb-unc-119(+)] IV; alg-1(qbc66[λN::alg-1(S642E)] X | This paper | N/A |
| Strain MJS349: qbcSi03[alg-1p::GFP::cog-1-boxb;cb-unc-119(+)] IV; alg-1(qbc67[λN::alg-1(S642A)] X | This paper | N/A |
| Strain MJS446: alg-2(ok304) II; kin-2(qbc92[R92C]) X | This paper | N/A |
| Strain MJS 447: alg-2(ok304) II; kin-2(qbc92[R92C]) alg-1(qbc67[S642A]) X | This paper | N/A |
| Oligonucleotides | ||
| Primers used for cloning are listed in Table S2. | This study | N/A |
| CRISPR RNA and repair templates to generate alg-1(S642A), alg-1(S642E), alg-1(Y693E) and kin-2(R92C) are listed in Table S2. | This study | N/A |
| Primers for RT-qPCR are listed in Table S2. | This study | N/A |
| Recombinant DNA | ||
| Plasmid L4440: untargeted RNAi vector | Addgene | Cat# 1654 |
| Plasmid MSP163: RNAi vector targeting alg-2 | (Bouasker and Simard, 2012) | N/A |
| Plasmid MSP437: RNAi vector targeting kin-2 | This paper | N/A |
| Plasmid MSP186: alg-1p::lambdaN::mCherry::alg-1::alg-1 3'UTR | Jannot et al. 2016 | N/A |
| Plasmid MSP438: alg-1p::lambdaN::mCherry::alg-1(S642A)::alg-1 3'UTR | This study | N/A |
| Plasmid MSP439: alg-1p::lambdaN::mCherry::alg-1(S642E)::alg-1 3'UTR | This study | N/A |
| Software and algorithms | ||
| Bowtie (version 0.12.7) | Langmead, B. et al, 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| Bowtie (version 1.2.2) | Langmead, B. et al, 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| Cutadapt (version 2.3) | Martin,M. et al., 2011 | https://cutadapt.readthedocs.io/en/v2.3/ |
| Samtools (version 1.9) | Li, H. et al, 2009 | https://sourceforge.net/projects/samtools/files/samtools/1.9/ |
| HTSeq (version 0.11.2) | Anders, S. et al, 2015 | https://htseq.readthedocs.io/en/master/ |
| Image Lab | Bio-Rad | https://www.bio-rad.com/en-ca/product/image-lab-software?ID=KRE6P5E8Z |
| Other | ||
| Breathable sealing membrane (Breathe-EASIER) | Diversified Biotech | Cat# BERM-2000 |
| Luminometer | Berthold Technologies | Centro XS3 LB 960 |
| Gel Imaging System | Bio-Rad | ChemiDoc XRS+ System |
This paper does not report original code.
Additional information required to reanalyze the data in this study is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans model and methods
All C. elegans strains were cultured on nematode growth medium (NGM) agar, fed with E. coli OP50 and handled using standard methods101 unless indicated otherwise. Hermaphrodite animals were used for all C. elegans experiments. Developmentally staged embryos, larvae and young adult animals were used. The animal stage used for each experiment can be found in the respective figure’s legend. The transgenic strains were obtained by micro-injection in young adults to produce progeny carrying extrachromosomal non-integrated transgene arrays102. The plasmid alg-1p::lambdaN::mCherry::alg-1g::alg-1 3'UTR plasmid (MSp0186) was generated in57. MSp0186 alg-1 serine 642 codon was mutated into alanine or glutamate with the oligonucleotides listed in Table S2, using Q5 Site-Directed Mutagenesis Kit (NEB). Genome editing of C. elegans with CRISPR-Cas9 methods was carried out by micro-injection in young adult animals with reconstituted Cas9 RNP mix [Cas9 protein (2.5 μg/μL), tracrRNA (1 μg/μL), CRISPR guide RNA (crRNA) (0.4 μg/μL) and repair templates with short homology arms (ssODN; 1.625μM)]103 (Table S2). F1 heterozygotes and F2 homozygotes were determined by PCR genotyping and Sanger sequencing. Missense mutations of alg-1 loci were carried out in a wild-type N2 (Bristol) strain. Missense mutations of alg-1 were obtained additionally in a strain where alg-1 is endogenously tagged in N-terminus with λN in a genetic background containing a single-copy insertion of GFP::cog-1-boxb reporter41,57. kin-2(R92C) was produced in alg-2(ok304) and in the double mutant alg-1(ok304); alg-1(S642A) background using CRISPR-Cas9, as described above. All strains that were used for this study are listed in the key resources table and the oligonucleotides used for genome editing can be found in Table S2.
Cultivation of HEK293 T cells
HEK 293T cells were cultivated under standard conditions (37 °C, 5 % CO2) using Dulbecco’s modified Eagle Medium (DMEM, Gibco) supplemented with 10 % FBS (Sigma-Aldrich) and 1% penicillin-streptomycin (Sigma-Aldrich). For cultivation, cells were passaged to a new dish every two to three days. Authentication was not performed. HEK 293T cells originate from a female embryo.
METHOD DETAILS
RNA interference
Knockdown of alg-2 and kin-2 were carried by feeding104. cDNA fragments of alg-2 and kin-2 were cloned into L4440 plasmid and transformed in the inducible IPTG HT115 (DE3) bacterial strain. L1 staged worms were grown at 20°C on IPTG Agar plates seeded with the respective bacterial strains or with bacteria transformed with L4440 control plasmid.
Lifespan analyses
All strains were cultured under standard conditions and synchronized by alkaline hypochlorite solution treatment. Lifespan assays46 were conducted at 20°C. Embryos were plated on NGM plates containing OP50. At the L4 stage, 20 animals were transferred to one plate, and 5 plates were counted for each strain. First day after the L4 stage was noted as day 1. Adult worms were transferred every 2 days during active reproduction and scored for viability. Animals were scored as dead when they stopped responding to gentle prodding with a platinum wire pick. Dead animals were removed from the plates. Animals that died by internal hatching, vulval bursting, or crawling on the side of the plates were censored from the lifespan analysis. P-values were calculated using the Mantel-Cox log-rank test.
Preparation of protein extracts, immunoprecipitation and Western Blotting analysis
Synchronized worm populations were obtained by alkaline hypochlorite solution treatment and plated onto NGM agar plates seeded with Escherichia coli OP50 bacteria. Animals were cultured at 20°C until adult stage then washed in M9 buffer, resuspended in ice-cold lysis buffer solution (100 mM potassium acetate, 30 mM Hepes-KOH pH 7, 2 mM magnesium acetate, 1 mM DTT, 1.5% [v/v] Triton X-100, 1 tablet/10 ml Complete Mini Protease Inhibitor without EDTA (Roche)) and lysed using a Dounce homogenizer. For immunoprecipitation, 12.5 μL of Dynabeads protein G (Thermo Fisher Scientific) were washed three times with lysis buffer and then incubated with ALG-1 antibody in 200 μL PBST for 1 hour with rotation. Beads were washed three times with lysis buffer and incubated with 1mg of worm extract (500 μL), for 3 hours at 4°C with rotation. Beads were resuspended in 20 μL 2X SDS loading buffer and eluted by heating at 95°C for 10 min before loading on gel for SDS-PAGE. For ALG-1 Western blotting, primary rabbit polyclonal ALG-1 antibodies were used at 1:1000 dilution in PBST supplemented with 1% [v/v] bovine serum albumin, AIN-1 and DCR-1 antibodies105 were used at 1:5000 in PBST; 5% [w/v] dried milk and beta-ACTIN (abcam, ab49900) was diluted 1:10000 in PBST 5% [w/v] dried milk with overnight incubation at 4°C. For ALG-1 expression at L4 and adult stage in Figure 1C, exactly 75 worms were picked and directly boiled in SDS Sample loading buffer (1 mM Tris-HCl [pH 6.8], 2% [w/v] SDS, 100mM DDT and 10% [v/v] glycerol) for western blot analysis.
Assessment of ALG-1 expression and proteasomal degradation
Hand-picked animals were harvested at the young adult stage. The worms were treated by rotating them in suspension for two hours with vehicle only (DMSO) or with 50μM of MG132 (Sigma-Aldrich, 474790) in M9. Harvested animals were collected and washed in M9 before being lyzed in SDS loading buffer (1 mM Tris-HCl [pH 6.8], 2% [w/v] SDS, 100mM DDT and 10% [v/v] glycerol). The homogenized extract was clarified by centrifugation at 17,000× g for 5 min at 4°C. To detect ALG-1or ACTIN, the total protein extract was boiled for 10 minutes in SDS loading buffer and proteins were resolved on 8% acrylamide gel and transferred to Protran Premium NC membranes (GE Healthcare). Membranes were incubated overnight at 4°C with either antibody: (i) Rabbit polyclonal against ALG-1 diluted 1:1,000 or (ii) Mouse monoclonal against beta-ACTIN (Abcam, ab49900) diluted 1:20,000; Ubiquitin (Santa Cruz, sc-8017) diluted 1:400. Antibodies were diluted in PBST-1% bovine serum albumin solution (137mM NaCl, 10 mM Phosphate, 2.7mM KCl [pH 7.4], 0.05% [v/v] Tween-20 and 1% [w/v] bovine serum albumin). The membrane was incubated for 1 hour at room temperature with HRP-conjugated secondary antibody in PBST and then revealed using Western Lightening ECL Kit (Perkin Elmer) and visualized using Chemidoc imaging system (BioRad).
Luciferase assays
To perform Luciferase assays56, C. elegans expressing the xeSi296 transgene [eft-3p::luc::gfp::unc-54 3'UTR, unc-119(+)] II were grown until they became gravid adults. The embryos were extracted using an alkaline hypochlorite solution treatment and single eggs were transferred by pipetting into a well of a flat-bottom, white 384-well plate (Berthold Technologies, 32505). The embryos hatched and developed in 90 μl S-Basal medium containing food (E. coli OP50 at OD600 = 0.9) and substrate for luciferase (100 μM Firefly D-Luciferin) (p.j.k., 102111). Plates were sealed with a breathable sealing membrane (Breathe Easier, Diversified Biotech, BERM-2000). Luminescence signal was measured using a Luminometer (Berthold Technologies, Centro XS3 LB 960) for 72 hours of development at 20°C in an incubator for 0.5 seconds every 10 minutes in a temperature-controlled incubator. Analysis of luminescence data was done in MATLAB using an automated algorithm to detect the hatch and the molts56.
Time-course small RNA sequencing and processing
N2 and alg-1 mutant animals were grown until gravid adults. Synchronized L1s were prepared by alkaline hypochlorite solution treatment and hatching them in absence of food in M9 buffer for 15h. The worms were plated on 2% NGM agar plates with Escherichia coli OP50 bacteria and placed at 25°C. From 18h to 30h of development, worms were collected hourly by washing them off the plates with M9. Worms were subjected to five cycles of freeze thawing in liquid nitrogen and a 42°C heat block, respectively, in TRI Reagent (LucernaChem, TR-118). After the lysis of worms, RNA isolation was performed with phenol-chloroform extraction (adapted from106). Subsequently RNA was treated with DNase (Life Technologies) and used for preparing small-RNA libraries.
For the time course experiments, the libraries were prepared using a QIAseq miRNA Library Kit (Qiagen) according to manufacturer’s protocol. This was followed by sequencing using the HiSeq 50 Cycle Single end reads protocol on HiSeq 2500. 3' adapters (AACTGTAGGCACCATCAAT) were trimmed with cutadapt107 with the following options (--error-rate 0.1, --minimum-length 15, --overlap 3). Reads were mapped to the genome with bowtie108 (version 1.2.2) with the following options (-v 1, -m 100,--best, --strata, --fr). Alignments were sorted and indexed with samtools109 (version 1.9). Reads mapping to rRNA, rRNA_pseudogene, tRNA, tRNA_pseudogene or from the mitochondrial chromosome were excluded (based on the Wormbase 4 WS270 annotation110). The mature miRNAs from miRBase111 (version 22) and reads were counted with HTSeq112 (version 0.11.2) with the following options (--stranded yes, --type miRNA, --idattr Name, --mode union).
RNA isolation of adult animals, Northern blot and RT-qPCR
Total RNA was purified by resuspending worm pellet in TRI Reagent (Sigma) and lysed by flash-freezing in liquid nitrogen three times. 30 micrograms of total RNAs were used for northern blot. Samples were mixed with an equal volume of 2x Loading Dye (8 M urea, 25 mM EDTA, 0.025% [w/v] xylene cyanol (XC), 0.025% [w/v] bromophenol blue (BB)) and heated at 80°C. 15% Urea Gel PAGE (Sequagel solutions) was pre-ran for 20 minutes before loading the samples. The gel was transferred onto Amersham Hybond-ECL nitrocellulose membrane (GE Healthcare). The RNA was cross-linked to the membrane with an EDC solution [0.373g (1-ethyl-3-(3-dimethylaminoprophy) carbodiimide and 1x methylimidazole (127.5 mM 1-methylimidazole-HCl [pH 8]) at 60°C for 1 hour. The membrane was then washed with water several times and baked at 80°C for 10 minutes. The membrane was pre-hybridized in a hybridization bottle with 50 mL (5X SSC, 20 mM Na2HPO4 pH 7.2, 7% [w/v] SDS, 2X Denhardt’s Solution and 1 mg of freshly denatured sheared salmon sperm DNA) at 50°C for 2 hours with rotation. Probes were radiolabelled with IDT StarFire reagents (discontinued), heated at 85°C for 5 minutes, directly added to the pre-hybridization solution, along with the membrane, and incubated overnight at 50°C with rotation. The membrane was washed three times in non-stringent wash solution (3X SSC, 5% [w/v] SDS) and once with stringent wash solution (1X SSC, 1% [w/v] SDS), at 50°C for 20 minutes with rotation. Phosphorimager screen was exposed with the membrane (overnight for miR-51 probes and 1 hour for tRNA glycine probe) and revealed by autoradiography. Membranes were stripped by adding 100 mL of boiled 0.1% [w/v] SDS solution and incubating at 50°C for 20 minutes with rotation.
RT-qPCR were performed with TaqMan miRNA Assay reagents (Life Technologies). ΔΔCT values were obtained using snoRNA sn2841 as the endogenous control. To quantify the level of miRNA bound to ALG-1, 1 mg of total protein extract was used to immunoprecipitate ALG-1. 10% of the beads was suspended in 2x Laemmli denaturing buffer, heated at 95°C and loaded on 8 % SDS-PAGE to assess the efficiency of the immunoprecipitation by western blotting. 90 % of the remaining beads were suspended in 2X PK buffer (100 mM Tris-Cl [pH 7.5], 200 mM NaCl, 1% [w/v] SDS) and digested with proteinase K (20μg) at 50°C for 20 minutes. RNA was extracted from the solution using TriReagent (Sigma). Samples were spiked-in and normalized with synthetic human miR-20a (50fmol).
Small RNA cloning, sequencing and analysis of adult worms
Recombinant enzymes and oligonucleotides used for small RNA cloning were synthesized and provided by Dr. Weifeng Gu Laboratory113,114. 1μg of total RNA or RNA purified from ALG-1 IP (1mg IP) was ligated in 3' with a 5' adenylated DNA oligonucleotide (AppAGATCGGAAGAGCACACGTCTGAACTCCAGTCA/3ddC/) and triphosphorylated small RNAs were dephosphorylated with recombinant C. elegans PIR-1 in a 10 μL reaction [50 mM Tris (pH 7.5), 10 mM DTT, and 10 mM MgCl2, PEG-8000 25% [v/v], 0.25 μM oligonucleotide, 0.25 μM truncated T4 RNA ligase 2, 0.25 μM PIR-1] for 2 hours at room temperature. The reaction was heat inactivated for 10 minutes at 65°C and then annealed in 3' with 5 μM oligonucleotide GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT starting at 65°C, decreasing at a rate of 0.1°C/second to 20°C over 5 minutes.
5' ligation was performed by adding 9.5 μL of ligation reaction [0.5 mM ATP, 0.125 μM T4 RNA ligase 1, and 0.2 μM RNA oligonucleotide (rArCrArCrUrCrUrUrUrCrCrCrUrArCrArCrGrArCrGrCrUrCrUrUrCrCrGrArUrCrU)] to the above reaction samples and incubated at room temperature for 2 hours. Reverse transcription was performed by adding 4.125 μL of [0.5 mM dNTP, 25 mM Tris, pH 8.8, 75 mM KCl, 5mM DTT and 0.125 μM Superscript II] to the reaction, incubated at 42°C for 30 minutes and heat inactivated at 85°C for 5 minutes.
PCR amplifications were carried out in a 50 μL PCR reaction of [1× PFU buffer (20mM Tris-HCl (pH 8.8), 10mM KCl, 10mM (NH4)2SO4, 2mM MgSO4, 0.1% [v/v] TritonX-100, 0.1 mg/ml BSA), 15 mM tetramethylammonium chloride, 0.1 mM dNTPs, 0.1 μM 5' and 3' oligonucleotides, 5 μL reverse transcribed samples, and 1× PFU polymerase]. 5' primers (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGA) and the 3' primers containing index (CAAGCAGAAGACGGCATACGAGAT-index-GTGACTGGAGTTCAGACGTGT) were used. The PCR was first amplified for 5 cycles (94°C 20s; 53°C 20s; 68°C 30s) and then amplified for 11 cycles (94°C 20s; 68°C 40s). Additional 0.6 μM 5′ and 3′ primers were added to the mixture and the PCR was amplified for 2 more cycles (94°C 20s; 68°C 40s).
PCR products were compared on 8% native PAGE gel, pooled according to the measured ratio, purified by phenol-chloroform, precipitated and gel-purified.
Hiseq 4000 SR50 sequencing reads were mapped to the genome and cDNA using custom PERL (5.10.1) scripts and Bowtie 0.12.7108. Databases used include C. elegans genome (WormBase release WS215), Repbase 15.10115, and miRBase 16116. The Generic Genome Browser117 was used to visualize the alignments. Detailed PERL scripts and related database files and analyses in this study are available upon request.
For the analysis of the total RNA datasets, the samples were normalized to the total small RNAs including miRNAs, 22G-RNAs and 21U-RNAs. For ALG-1 bound miRNAs (IP), miRNA and miR* reads were normalized on miR-48-5p reads bound to ALG-1 as it binds ALG-1 WT, S642A and S642E equally (Figure S5A). Three replicates were used for statistical analysis.
Specific miRISC purification: 2'-O-methyl pull down
2'-O-methyl pull down118 were performed with adult animal lysates homogenized in 2 volumes of lysis buffer (100 mM potassium acetate, 30 mM Hepes-KOH pH 7, 2 mM magnesium acetate, 1 mM DTT, 1.5% [v/v] Triton X-100, 1 tablet/10 ml Complete Mini Protease Inhibitor without EDTA (Roche)) using a Dounce homogenizer. The extracts were clarified by centrifugation at 10,000x g. 4 mg of worm lysate was pre-cleared with M-280 streptavidin Dynabeads (Thermo Fisher Scientific) coupled with non-specific 2′-O-Me Luciferase oligonucleotides (10pmol) for 1h. The supernatant was incubated with biotinylated 2'-O-Me oligonucleotides (10pmol) bound to streptavidin beads for 1h at room temperature. Beads were washed three times using ice-cold lysis buffer. Beads were resuspended in 20 μL 2X SDS loading buffer and eluted by heating at 95°C for 10 min before loading on gel for western blotting. 2'-O-Me oligonucleotides for miR-35-3p pull-down and the non-specific oligonucleotides targeting luciferase are listed in Table S2.
In vitro phosphorylation assay
In vitro PKA kinase assays were performed using assay conditions adapted from the manufacturer’s recommendations (Recombinant PKA, P6000S, NEB). All reactions were performed in a 25 μl volume for 60 min at 30°C. Assay buffer was composed of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM EGTA, 0.1 mM EDTA, 2 mM DTT, 0.01% Brij 35, 2 units of recombinant CK2, and 200 μM [γ-32P] ATP [6000 Ci/mmol; Perkin Elmer]. Reactions were terminated on ice using 7.5 mM Guanidine hydrochloride and the biotinylated peptides were spotted on streptavidin-coated membranes. Samples were washed three times in 2M NaCl followed by four times in 2M NaCl with 1% H3PO4. Incorporated 32P was measured using a liquid scintillation counter in CPM (counts per minute). The data were also analyzed by Tricine-SDS-PAGE and autoradiography. Total protein levels of PKA were observed by Coomassie staining.
Cell culture and transfection
The vectors for Flag/HA-tagged human AGO2 as well as Flag/HA-tagged EGFP23 and the AGO2 Y529E mutant40 were used. S478 mutations were introduced by site-directed mutagenesis strategy using the primers in Table S2. Constructs were verified by Sanger sequencing.
HEK 293T cells were cultivated under standard conditions (37 °C, 5 % CO2) using Dulbecco’s modified Eagle Medium (DMEM, Gibco) supplemented with 10 % FBS (Sigma-Aldrich) and 1% penicillin-streptomycin (Sigma-Aldrich). Cells were grown on 15 cm dishes and calcium-phosphate transfected using 10 μg of plasmid DNA per dish. For transfection of the AGO2 S478E mutant, 20 μg of DNA were used, for transfection of EGFP 5 μg.
For analysis of AGO2 target interactions, cells were harvested 48 h after transfection, washed with PBS and lysed in 1 mL NET buffer (50 mM Tris/HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.5 % (v/v) NP-40 alternative, 10 % glycerol, 1 mM NaF, 1 mM DTT and 1 mM AEBSF) for 20 min on ice. Lysates were cleared by centrifugation at 15,000 g for 20 min at 4 °C and input samples were taken before performing immunoprecipitation.
Immunoprecipitation (IP) of Flag/HA AGO2
For IP of Flag-tagged proteins and subsequent analysis of AGO2 target interaction, 50 μl packed volume of ANTI-FLAG M2 agarose beads (Sigma-Aldrich) was used. Prior to use, beads were washed twice with cold PBS (1 min, 1,000 g, 4 °C). The beads were incubated with lysate for 2.5 h at 4 °C while rotating. Afterwards, the samples were washed with NET lysis buffer (50 mM Tris/HCl pH 7.5, 5 mM EDTA, 0.5 % NP-40 alternative, 10 % glycerol, 1 mM NaF, 1 mM DTT, 1 mM AEBSF) + 300 mM NaCl twice, once with lysis buffer + 450 mM NaCl, once with 600 mM NaCl, once with 450 mM NaCl, followed by one washing step with PBS. During the last washing step, samples were split into RNA (75 %) and Western blot (25 %) samples.
After adding 50 μl of 2.5 x Laemmli sample buffer to the Western blot samples, samples were incubated for 5 min at 95 °C. The denatured proteins were separated on a 10 % SDS gel and semidry blotted. Flag/HA-tagged AGO2 proteins were detected with anti-HA antibody (Covance 16B12, 1:1000) in combination with IRDye 800CW goat anti-mouse IgG secondary antibody (Li-Cor Biosciences, 1:10 000). Signals were detected and quantified with the Odyssey Imaging System (Li-Cor Biosciences).
mRNA quantification of and Flag/HA AGO2 IP transfected cells
RNA of input and IP samples was extracted using Trizol reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol, including a second extraction step with chloroform. 1 μg of input and the complete RNA of IP samples were digested with DNaseI (Thermo Fisher Scientific), and cDNA was synthesized using the First-Strand cDNA synthesis kit (Thermo Fisher Scientific) with random hexamer primers following the manufacturer’s protocol. RT-qPCR was performed with Takyon No Rox SYBR MasterMix dTTP Blue (Eurogentec) using the primers listed in Table S2. Data were analyzed using the ΔΔCt method and normalized to the AGO2 expression in the Western blot IP sample.
Northern blot analysis of miRNA expression in Flag/HA AGO2 transfected cells
RNA of Flag/HA AGO2 transfected HEK 293T cells input samples was extracted using Trizol (Thermo Fisher Scientific) according to the manufacturer’s protocol. Northern blots were performed with 1 μg of input RNA119. RNA was separated on 12 % denaturing polyacrylamide gels (Rotiphorese, Roth), semidry blotted, EDC cross-linked, and hybridized overnight at 50°C. Blots were washed twice with 5× SSC, 1 % [w/v] SDS, and once with 1× SSC, 1 % [w/v] SDS. Signals were detected by exposure to a screen and scanning with the PMI imaging system (BioRad).
QUANTIFICATION AND STATISTICAL ANALYSIS
Experiments with C. elegans
Statistical analysis for phenotypes (Figure 1A, B and D; Figure 6D and E; Figure S1B-C) is presented as percentage of population scored and statistical significance was determined with a two-tailed Fisher’s exact test in Excel. For the lifespan assay (Figure 1C), P-values were calculated with a Mantel-Cox log-rank test with GraphPad Prism. For developmental timing (Figure 2), P-values were calculated with a Welch two-sample and two-sided t-test. For Figure S2 and S3, read counts of each timepoint for each sample were divided by total number of reads and multiplied by the average library size. A pseudocount of 8 was added after log2 transformation. Pearson’s correlation was performed using Seaborn package in Python and PCA was performed using sklearn.decomposition package in python. Statistical analysis for miRNA quantification (Figure 4A, B and D; Figure 5A and C; Figure S4A; Figure S5A) was evaluated with a two-tailed Student’s t-test in Excel. λN/Box-B tethering reporter fluorescence intensity (Figure 3B) was quantified with Zen Software (Zeiss), presented as mean. P-values were calculated with a two-tailed Student’s t-test in Excel. Western blot bands intensity (Figure 1C; Figure 5B; Figure S1D; Figure S5A) were measured with ImageJ software and t-test were used to assess differences in protein levels where applicable, using biological triplicates. Statistical details, including the sample size (n=) and the number of biological replicates for each experiment, can be found in the respective figure and figure legend. Normality and equal variance assumptions were not tested.
Experiments with human AGO2
Northern blot signals were quantified with the PMI software Quantity One, Western blot signals with the Odyssey software. Significance was tested using a two-tailed Student’s t-test in Excel. All experiments were performed in three biological replicates. Data is shown as the mean and error bars represent the standard deviation.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Dr. Weifeng Gu for the help with small RNA cloning and sequencing data analysis, Jean-Christophe Simard for technical help and Dr. Thomas Duchaine for providing DCR-1 antibody. We thank the members of our laboratory for helpful comments. The Canadian Institutes of Health Research (CIHR) supported this work. M.Q.H. and V.N.S. are recipients of scholarship from the Fonds de Recherche du Québec-Santé (FRQ-S). S.N. received funding from the EU Horizon 2020 Research and Innovation Program through a Marie Sklodowska-Curie grant (842386-miRhythm). H.G. received funding from the SNF through the NCCR “RNA & Disease” and the Novartis Research Foundation through the FMI. K.N. was supported by the National Institutes of Health (NIH; R01GM124320). G.M. received funding from the Deutsche Forschungsgemeinschaft (SFB 960).
Footnotes
DECLARATION OF INTEREST
The authors declare no competing interests.
REFERENCES
- 1.Lee RC, Feinbaum RL, and Ambros V (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, and Ruvkun G (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, and Ruvkun G (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403, 901–906. 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Peng Y, and Croce CM (2016). The role of MicroRNAs in human cancer. Signal Transduct Target Ther 1, 15004. 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra S, Vimal D, Sharma D, Rai V, Gupta SC, and Chowdhuri DK (2017). Role of miRNAs in development and disease: Lessons learnt from small organisms. Life sciences 185, 8–14. 10.1016/j.lfs.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 6.Hajieghrari B, and Farrokhi N (2022). Plant RNA-mediated gene regulatory network. Genomics 114, 409–442. 10.1016/j.ygeno.2021.12.020. [DOI] [PubMed] [Google Scholar]
- 7.Kim VN, Han J, and Siomi MC (2009). Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10, 126–139. 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 8.Ha M, and Kim VN (2014). Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524. 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 9.Dueck A, and Meister G (2014). Assembly and function of small RNA - argonaute protein complexes. Biol Chem 395, 611–629. 10.1515/hsz-2014-0116. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi H, and Tomari Y (2016). RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochimica et biophysica acta 1859, 71–81. 10.1016/j.bbagrm.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Nakanishi K (2016). Anatomy of RISC: how do small RNAs and chaperones activate Argonaute proteins? Wiley Interdiscip Rev RNA 7, 637–660. 10.1002/wrna.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran V, and Chen X (2008). Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321, 1490–1492. 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee S, and Grosshans H (2009). Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461, 546–549. 10.1038/nature08349. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee S, Fasler M, Bussing I, and Grosshans H (2011). Target-mediated protection of endogenous microRNAs in C. elegans. Dev Cell 20, 388–396. 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Medley JC, Panzade G, and Zinovyeva AY (2021). microRNA strand selection: Unwinding the rules. Wiley Interdiscip Rev RNA 12, e1627. 10.1002/wrna.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonas S, and Izaurralde E (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16, 421–433. 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 17.Frederick PM, and Simard MJ (2021). Regulation and different functions of the animal microRNA-induced silencing complex. Wiley Interdiscip Rev RNA 13, e1701. 10.1002/wrna.1701. [DOI] [PubMed] [Google Scholar]
- 18.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, and Mello CC (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 19.Vasquez-Rifo A, Jannot G, Armisen J, Labouesse M, Bukhari SI, Rondeau EL, Miska EA, and Simard MJ (2012). Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS One 7, e33750. 10.1371/journal.pone.0033750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown KC, Svendsen JM, Tucci RM, Montgomery BE, and Montgomery TA (2017). ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline. Nucleic Acids Res 45, 9093–9107. 10.1093/nar/gkx536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakanishi K (2022). Anatomy of four human Argonaute proteins. Nucleic Acids Res 50, 6618–6638. 10.1093/nar/gkac519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakanishi K, Weinberg DE, Bartel DP, and Patel DJ (2012). Structure of yeast Argonaute with guide RNA. Nature 486, 368–374. 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, and Tuschl T (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15, 185–197. 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, and Hannon GJ (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441. 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 25.Park MS, Phan HD, Busch F, Hinckley SH, Brackbill JA, Wysocki VH, and Nakanishi K (2017). Human Argonaute3 has slicer activity. Nucleic Acids Res 45, 11867–11877. 10.1093/nar/gkx916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouasker S, and Simard MJ (2012). The slicing activity of miRNA-specific Argonautes is essential for the miRNA pathway in C. elegans. Nucleic Acids Res 40, 10452–10462. 10.1093/nar/gks748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schirle NT, and MacRae IJ (2012). The crystal structure of human Argonaute2. Science 336, 1037–1040. 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, and Ladurner AG (2007). A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol 14, 897–903. 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 29.Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, and Chan EK (2009). The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA 15, 804–813. 10.1261/rna.1229409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazzaretti D, Tournier I, and Izaurralde E (2009). The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 15, 1059–1066. 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takimoto K, Wakiyama M, and Yokoyama S (2009). Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA 15, 1078–1089. 10.1261/rna.1363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J, and Shi Y (2008). Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 455, 421–424. 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, Du Y, Wang Y, Chang WC, Chen CH, et al. (2013). EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 497, 383–387. 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang M, Haase AD, Huang FK, Coulis G, Rivera KD, Dickinson BC, Chang CJ, Pappin DJ, Neubert TA, Hannon GJ, et al. (2014). Dephosphorylation of tyrosine 393 in argonaute 2 by protein tyrosine phosphatase 1B regulates gene silencing in oncogenic RAS-induced senescence. Mol Cell 55, 782–790. 10.1016/j.molcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, and Chang P (2011). Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42, 489–499. 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horman SR, Janas MM, Litterst C, Wang B, MacRae IJ, Sever MJ, Morrissey DV, Graves P, Luo B, Umesalma S, et al. (2013). Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol Cell 50, 356–367. 10.1016/j.molcel.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng Y, Sankala H, Zhang X, and Graves PR (2008). Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J 413, 429–436. 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 38.Bridge KS, Shah KM, Li Y, Foxler DE, Wong SCK, Miller DC, Davidson KM, Foster JG, Rose R, Hodgkinson MR, et al. (2017). Argonaute Utilization for miRNA Silencing Is Determined by Phosphorylation-Dependent Recruitment of LIM-Domain-Containing Proteins. Cell Rep 20, 173–187. 10.1016/j.celrep.2017.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paradis-Isler N, and Boehm J (2018). NMDA receptor-dependent dephosphorylation of serine 387 in Argonaute 2 increases its degradation and affects dendritic spine density and maturation. The Journal of biological chemistry 293, 9311–9325. 10.1074/jbc.RA117.001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rüdel S, Wang Y, Lenobel R, Korner R, Hsiao HH, Urlaub H, Patel D, and Meister G (2011). Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res 39, 2330–2343. 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quévillon Huberdeau M, Zeitler DM, Hauptmann J, Bruckmann A, Fressigne L, Danner J, Piquet S, Strieder N, Engelmann JC, Jannot G, et al. (2017). Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo. EMBO J 36, 2088–2106. 10.15252/embj.201696386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golden RJ, Chen B, Li T, Braun J, Manjunath H, Chen X, Wu J, Schmid V, Chang TC, Kopp F, et al. (2017). An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 542, 197–202. 10.1038/nature21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, and Ruvkun G (2000). The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5, 659–669. 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 44.Ecsedi M, Rausch M, and Grosshans H (2015). The let-7 microRNA directs vulval development through a single target. Dev Cell 32, 335–344. 10.1016/j.devcel.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 45.Kato M, Chen X, Inukai S, Zhao H, and Slack FJ (2011). Age-associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans. RNA 17, 1804–1820. 10.1261/rna.2714411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aalto AP, Nicastro IA, Broughton JP, Chipman LB, Schreiner WP, Chen JS, and Pasquinelli AE (2018). Opposing roles of microRNA Argonautes during Caenorhabditis elegans aging. PLoS genetics 14, e1007379. 10.1371/journal.pgen.1007379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boehm M, and Slack F (2005). A developmental timing microRNA and its target regulate life span in C. elegans. Science 310, 1954–1957. 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 48.de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, and Slack FJ (2010). MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol 20, 2159–2168. 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pincus Z, Smith-Vikos T, and Slack FJ (2011). MicroRNA predictors of longevity in Caenorhabditis elegans. PLoS genetics 7, e1002306. 10.1371/journal.pgen.1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boulias K, and Horvitz HR (2012). The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab 15, 439–450. 10.1016/j.cmet.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith-Vikos T, de Lencastre A, Inukai S, Shlomchik M, Holtrup B, and Slack FJ (2014). MicroRNAs mediate dietary-restriction-induced longevity through PHA-4/FOXA and SKN-1/Nrf transcription factors. Curr Biol 24, 2238–2246. 10.1016/j.cub.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank F, Sonenberg N, and Nagar B (2010). Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822. 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 53.Boland A, Tritschler F, Heimstadt S, Izaurralde E, and Weichenrieder O (2010). Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep 11, 522–527. 10.1038/embor.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ketting RF, and Cochella L (2021). Concepts and functions of small RNA pathways in C. elegans. Curr Top Dev Biol 144, 45–89. 10.1016/bs.ctdb.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 55.Olmedo M, Geibel M, Artal-Sanz M, and Merrow M (2015). A High-Throughput Method for the Analysis of Larval Developmental Phenotypes in Caenorhabditis elegans. Genetics 201, 443–448. 10.1534/genetics.115.179242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meeuse MW, Hauser YP, Morales Moya LJ, Hendriks GJ, Eglinger J, Bogaarts G, Tsiairis C, and Grosshans H (2020). Developmental function and state transitions of a gene expression oscillator in Caenorhabditis elegans. Mol Syst Biol 16, e9975. 10.15252/msb.209975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jannot G, Michaud P, Quevillon Huberdeau M, Morel-Berryman L, Brackbill JA, Piquet S, McJunkin K, Nakanishi K, and Simard MJ (2016). GW182-Free microRNA Silencing Complex Controls Post-transcriptional Gene Expression during Caenorhabditis elegans Embryogenesis. PLoS genetics 12, e1006484. 10.1371/journal.pgen.1006484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gregory RI, Chendrimada TP, Cooch N, and Shiekhattar R (2005). Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123, 631–640. 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 59.Diederichs S, and Haber DA (2007). Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131, 1097–1108. 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 60.Park JH, and Shin C (2015). Slicer-independent mechanism drives small-RNA strand separation during human RISC assembly. Nucleic Acids Res 43, 9418–9433. 10.1093/nar/gkv937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alvarez-Saavedra E, and Horvitz HR (2010). Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20, 367–373. 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaw WR, Armisen J, Lehrbach NJ, and Miska EA (2010). The conserved miR-51 microRNA family is redundantly required for embryonic development and pharynx attachment in Caenorhabditis elegans. Genetics 185, 897–905. 10.1534/genetics.110.117515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McJunkin K, and Ambros V (2014). The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 (Bethesda) 4, 1747–1754. 10.1534/g3.114.011973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghini F, Rubolino C, Climent M, Simeone I, Marzi MJ, and Nicassio F (2018). Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nature Communications 9, 3119. 10.1038/s41467-018-05182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheu-Gruttadauria J, Pawlica P, Klum SM, Wang S, Yario TA, Schirle Oakdale NT, Steitz JA, and MacRae IJ (2019). Structural Basis for Target-Directed MicroRNA Degradation. Mol Cell 75, 1243–1255. 10.1016/j.molcel.2019.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han J, LaVigne CA, Jones BT, Zhang H, Gillett F, and Mendell JT (2020). A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science 370, eabc9546. 10.1126/science.abc9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi CY, Kingston ER, Kleaveland B, Lin DH, Stubna MW, and Bartel DP (2020). The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation. Science 370, eabc9359. 10.1126/science.abc9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blom N, Gammeltoft S, and Brunak S (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294, 1351–1362. 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 69.Gross RE, Bagchi S, Lu X, and Rubin CS (1990). Cloning, characterization, and expression of the gene for the catalytic subunit of cAMP-dependent protein kinase in Caenorhabditis elegans. Identification of highly conserved and unique isoforms generated by alternative splicing. The Journal of biological chemistry 265, 6896–6907. [PubMed] [Google Scholar]
- 70.Pastok MW, Prescott MC, Dart C, Murray P, Rees HH, and Fisher MJ (2013). Structural diversity of the cAMP-dependent protein kinase regulatory subunit in Caenorhabditis elegans. Cell Signal 25, 168–177. 10.1016/j.cellsig.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 71.Søberg K, and Skalhegg BS (2018). The Molecular Basis for Specificity at the Level of the Protein Kinase a Catalytic Subunit. Front Endocrinol (Lausanne) 9, 538. 10.3389/fendo.2018.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buechler YJ, Herberg FW, and Taylor SS (1993). Regulation-defective mutants of type I cAMP-dependent protein kinase. Consequences of replacing arginine 94 and arginine 95. The Journal of biological chemistry 268, 16495–16503. [PubMed] [Google Scholar]
- 73.Schade MA, Reynolds NK, Dollins CM, and Miller KG (2005). Mutations that rescue the paralysis of Caenorhabditis elegans ric-8 (synembryn) mutants activate the G alpha(s) pathway and define a third major branch of the synaptic signaling network. Genetics 169, 631–649. 10.1534/genetics.104.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, and Joshua-Tor L (2012). The structure of human argonaute-2 in complex with miR-20a. Cell 150, 100–110. 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Faehnle CR, Elkayam E, Haase AD, Hannon GJ, and Joshua-Tor L (2013). The making of a slicer: activation of human Argonaute-1. Cell Rep 3, 1901–1909. 10.1016/j.celrep.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakanishi K, Ascano M, Gogakos T, Ishibe-Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, and Patel DJ (2013). Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep 3, 1893–1900. 10.1016/j.celrep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schirle NT, Sheu-Gruttadauria J, and MacRae IJ (2014). Structural basis for microRNA targeting. Science 346, 608–613. 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sheu-Gruttadauria J, Xiao Y, Gebert LF, and MacRae IJ (2019). Beyond the seed: structural basis for supplementary microRNA targeting by human Argonaute2. EMBO J 38, e101153. 10.15252/embj.2018101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park MS, Araya-Secchi R, Brackbill JA, Phan HD, Kehling AC, Abd El-Wahab EW, Dayeh DM, Sotomayor M, and Nakanishi K (2019). Multidomain Convergence of Argonaute during RISC Assembly Correlates with the Formation of Internal Water Clusters. Mol Cell 75, 725–740 e726. 10.1016/j.molcel.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zinovyeva AY, Bouasker S, Simard MJ, Hammell CM, and Ambros V (2014). Mutations in conserved residues of the C. elegans microRNA Argonaute ALG-1 identify separable functions in ALG-1 miRISC loading and target repression. PLoS genetics 10, e1004286. 10.1371/journal.pgen.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zinovyeva AY, Veksler-Lublinsky I, Vashisht AA, Wohlschlegel JA, and Ambros VR (2015). Caenorhabditis elegans ALG-1 antimorphic mutations uncover functions for Argonaute in microRNA guide strand selection and passenger strand disposal. Proceedings of the National Academy of Sciences of the United States of America 112, E5271–5280. 10.1073/pnas.1506576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Johnston M, Geoffroy MC, Sobala A, Hay R, and Hutvagner G (2010). HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Molecular biology of the cell 21, 1462–1469. 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Derrien B, Baumberger N, Schepetilnikov M, Viotti C, De Cillia J, Ziegler-Graff V, Isono E, Schumacher K, and Genschik P (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proceedings of the National Academy of Sciences of the United States of America 109, 15942–15946. 10.1073/pnas.1209487109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smibert P, Yang JS, Azzam G, Liu JL, and Lai EC (2013). Homeostatic control of Argonaute stability by microRNA availability. Nat Struct Mol Biol 20, 789–795. 10.1038/nsmb.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kobayashi H, Shoji K, Kiyokawa K, Negishi L, and Tomari Y (2019). VCP Machinery Mediates Autophagic Degradation of Empty Argonaute. Cell Rep 28, 1144–1153 e1144. 10.1016/j.celrep.2019.07.003. [DOI] [PubMed] [Google Scholar]
- 86.Kobayashi H, Shoji K, Kiyokawa K, Negishi L, and Tomari Y (2019). Iruka Eliminates Dysfunctional Argonaute by Selective Ubiquitination of Its Empty State. Mol Cell 73, 119–129. 10.1016/j.molcel.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 87.Majercak J, Kalderon D, and Edery I (1997). Drosophila melanogaster deficient in protein kinase A manifests behavior-specific arrhythmia but normal clock function. Molecular and cellular biology 17, 5915–5922. 10.1128/MCB.17.10.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang H, and Sieburth D (2013). PKA controls calcium influx into motor neurons during a rhythmic behavior. PLoS genetics 9, e1003831. 10.1371/journal.pgen.1003831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu F, Xiao Y, Ji XL, Zhang KQ, and Zou CG (2017). The cAMP-PKA pathway-mediated fat mobilization is required for cold tolerance in C. elegans. Sci Rep 7, 638. 10.1038/s41598-017-00630-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gottschling DC, Doring F, and Luersen K (2017). Locomotion Behavior Is Affected by the GalphaS Pathway and the Two-Pore-Domain K(+) Channel TWK-7 Interacting in GABAergic Motor Neurons in Caenorhabditis elegans. Genetics 206, 283–297. 10.1534/genetics.116.195669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiao Y, Liu F, Zhao PJ, Zou CG, and Zhang KQ (2017). PKA/KIN-1 mediates innate immune responses to bacterial pathogens in Caenorhabditis elegans. Innate immunity 23, 656–666. 10.1177/1753425917732822. [DOI] [PubMed] [Google Scholar]
- 92.Castaneda PG, Cecchetelli AD, Pettit HN, and Cram EJ (2020). Galpha/GSA-1 works upstream of PKA/KIN-1 to regulate calcium signaling and contractility in the Caenorhabditis elegans spermatheca. PLoS genetics 16, e1008644. 10.1371/journal.pgen.1008644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang H, Kong Q, Wang J, Jiang Y, and Hua H (2020). Complex roles of cAMP-PKA-CREB signaling in cancer. Exp Hematol Oncol 9, 32. 10.1186/s40164-020-00191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ranganathan G, Pokrovskaya I, Ranganathan S, and Kern PA (2005). Role of A kinase anchor proteins in the tissue-specific regulation of lipoprotein lipase. Molecular endocrinology 19, 2527–2534. 10.1210/me.2005-0144. [DOI] [PubMed] [Google Scholar]
- 95.Tudisca V, Simpson C, Castelli L, Lui J, Hoyle N, Moreno S, Ashe M, and Portela P (2012). PKA isoforms coordinate mRNA fate during nutrient starvation. Journal of cell science 125, 5221–5232. 10.1242/jcs.111534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jewell JL, Fu V, Hong AW, Yu FX, Meng D, Melick CH, Wang H, Lam WM, Yuan HX, Taylor SS, and Guan KL (2019). GPCR signaling inhibits mTORC1 via PKA phosphorylation of Raptor. eLife 8, e43038. 10.7554/eLife.43038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rataj F, Planel S, Desroches-Castan A, Le Douce J, Lamribet K, Denis J, Feige JJ, and Cherradi N (2016). The cAMP pathway regulates mRNA decay through phosphorylation of the RNA-binding protein TIS11b/BRF1. Molecular biology of the cell 27, 3841–3854. 10.1091/mbc.E16-06-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Venkatesh SR, and Singh V (2021). G protein-coupled receptors: The choreographers of innate immunity in Caenorhabditis elegans. PLoS Pathog 17, e1009151. 10.1371/journal.ppat.1009151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Somale D, Di Nardo G, di Blasio L, Puliafito A, Vara-Messler M, Chiaverina G, Palmiero M, Monica V, Gilardi G, Primo L, and Gagliardi PA (2020). Activation of RSK by phosphomimetic substitution in the activation loop is prevented by structural constraints. Sci Rep 10, 591. 10.1038/s41598-019-56937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kozelekova A, Naplavova A, Brom T, Gasparik N, Simek J, Houser J, and Hritz J (2022). Phosphorylated and Phosphomimicking Variants May Differ-A Case Study of 14-3-3 Protein. Front Chem 10, 835733. 10.3389/fchem.2022.835733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mello CC, Kramer JM, Stinchcomb D, and Ambros V (1991). Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10, 3959–3970. 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paix A, Folkmann A, Rasoloson D, and Seydoux G (2015). High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics 201, 47–54. 10.1534/genetics.115.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Timmons L, Court DL, and Fire A (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112. 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 105.Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr., Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, et al. (2006). Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124, 343–354. 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 106.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, and Antebi A (2009). Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science 324, 95–98. 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011). 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 108.Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and Genome Project Data Processing, S. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Cho J, Davis P, Gao S, Grove CA, Kishore R, et al. (2020). WormBase: a modern Model Organism Information Resource. Nucleic Acids Res 48, D762–D767. 10.1093/nar/gkz920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, and Enright AJ (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34, D140–144. 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li L, Dai H, Nguyen AP, and Gu W (2020). A convenient strategy to clone small RNA and mRNA for high-throughput sequencing. RNA 26, 218–227. 10.1261/rna.071605.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chaves DA, Dai H, Li L, Moresco JJ, Oh ME, Conte D Jr., Yates JR 3rd, Mello CC, and Gu W (2021). The RNA phosphatase PIR-1 regulates endogenous small RNA pathways in C. elegans. Mol Cell 81, 546–557. 10.1016/j.molcel.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, and Walichiewicz J (2005). Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110, 462–467. 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 116.Kozomara A, and Griffiths-Jones S (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39, D152–157. 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, and Lewis S (2002). The generic genome browser: a building block for a model organism system database. Genome Res 12, 1599–1610. 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jannot G, Vasquez-Rifo A, and Simard MJ (2011). Argonaute pull-down and RISC analysis using 2'-O-methylated oligonucleotides affinity matrices. Methods Mol Biol 725, 233–249. 10.1007/978-1-61779-046-1_16. [DOI] [PubMed] [Google Scholar]
- 119.Pall GS, and Hamilton AJ (2008). Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3, 1077–1084. 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed small RNA-seq datasets have been deposited at NCBI’s Gene Expression Omnibus (GEO) repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-beta Actin Monoclonal Antibody, HRP Conjugated, Clone AC-15 | Abcam | Cat# ab49900, RRID:AB_867494 |
| Peroxidase-AffiniPure Goat Anti-Rabbit IgG (H+L) (min X Hu,Ms,Rat Sr Prot) antibody | Jackson Immunoresearch Labs. | Cat# 111-035-144, RRID:AB_2307391 |
| Rabbit Anti-ALG-1 polyclonal antibody | (Vasquez-Rifo et al., 2012) | N/A |
| Rabbit Anti-AIN-1 polyclonal antibody | (Jannot et al., 2016) | N/A |
| Rabbit Anti-DCR-1 polyclonal antibody | (Duchaine et al., 2006) | N/A |
| Mouse Anti-Ubiquitin Antibody (P4D1) monoclonal antibody | Santa Cruz | Cat# sc-8017, RRID:AB_628423 |
| Mouse Anti-HA.11 Monoclonal Antibody, Unconjugated, Clone 16B12 | Covance | Cat# MMS-101P-500, RRID:AB_291261 |
| IRDye 800CW Goat anti-Mouse IgG | Li-Cor Biosciences | Cat# 925-32210, RRID: AB_2687825 |
| Bacterial and virus strains | ||
| E. coli strain: OP50 | Caenorhabditis Genetics Center | CGC: OP50, RRID: WB-STRAIN:WBStrain00041969 |
| E. coli strain: HT115(DE3) | Caenorhabditis Genetics Center | CGC: HT115(DE3), RRID: WB-STRAIN:WBStrain00041080 |
| Chemicals, peptides, and recombinant proteins | ||
| Biotin-MFTNQLQRISNDAGMPIV-OH | This study | N/A |
| Biotin-MFTNQLQRIANDAGMPIV-OH | This study | N/A |
| Firefly D-Luciferin | PJK GmbH | Cat# 102111 |
| MG-132 | Sigma-Aldrich | Cat# 474790 |
| Tri Reagent | Sigma-Aldrich | Cat# T9424 |
| Critical commercial assays | ||
| TaqMan Universal PCR Master Mix, no AmpErase UNG | Thermo Fisher Scientific | Cat# 4324018 |
| Taqman MicroRNA Assay | Thermo Fisher Scientific | Cat# 4440886 |
| Q5 Site-Directed Mutagenesis Kit | New England Biolabs | Cat# E0554S |
| Dynabeads M-280 Streptavidin | Thermo Fisher Scientific | Cat# 11205D |
| Dynabeads Protein G for Immunoprecipitation | Thermo Fisher Scientific | Cat# 10004D |
| Anti-FLAG M2 Magnetic Beads | Sigma Aldrich | Cat# M8823 |
| High-Capacity cDNA Reverse Transcription Kit | Thermo Fisher Scientific | Cat# 4368814 |
| Edit-R tracrRNA | Horizon Discovery | Cat# U-002005 |
| Edit-R crRNA | Horizon Discovery | Cat# Custom0280 |
| QIAseq miRNA Library Kit | Qiagen | Cat# 331505 |
| Takyon No Rox SYBR MasterMix dTTP Blue | Eurogentec | Cat# UF-NSMT-B0701 |
| Deposited data | ||
| Small RNA sequencing reads from Caenorhabditis elegans adult worms (total RNA and ALG-1 IP) | This study | GEO: GSE198352 |
| Small RNA sequencing reads at different time points during larval development | This study | GEO: GSE174368 |
| Experimental models: Cell lines | ||
| Human: HEK293T cells | ATCC | RRID:CVCL_0063 |
| Experimental models: Organisms/strains | ||
| Strain N2 Bristol (WT) | Caenorhabditis Genetics Center | CGC: N2, RRID:WB-STRAIN:WBStrain00000001 |
| Strain VC446: alg-1(gk214) X | Caenorhabditis Genetics Center | CGC: VC446, RRID: WB-STRAIN: WBStrain00035775 |
| Strain KG532: kin-2(ce179) X | Caenorhabditis Genetics Center | CGC: KG532, RRID:WB-STRAIN:WBStrain00023482 |
| Strain MJS218: alg-1(gk214) Ex[alg-1p::lambdaN::mCherry::alg-1::alg-1 3'UTR; prf4(rol-6(su1006))] | (Quévillon Huberdeau et al., 2017) | N/A |
| Strain MJS258: alg-1(gk214) Ex[λN::mcherry::alg-1(S642E); prf4(rol-6(su1006))] | This paper | N/A |
| Strain MJS259: alg-1(gk214) Ex[λN::mcherry::alg-1(S642A); prf4(rol-6(su1006))] | This paper | N/A |
| Strain MJS275: alg-1(qbc23[S642A]) X | This paper | N/A |
| Strain MJS306: alg-1(qbc46[S642E]) X | This paper | N/A |
| Strain HW1939: xeSi296[Peft-3::luc::gfp::unc-54 3'UTR, unc-119(+)] II | (Meeuse et al., 2020) | N/A |
| Strain HW3060: xeSi296[Peft-3::luc::gfp::unc-54 3'UTR, unc-119(+)] II; alg-1(qbc23[S642A]) X | This paper | N/A |
| Strain HW3061: xeSi296[Peft-3::luc::gfp::unc-54 3'UTR, unc-119(+)] II; alg-1(qbc46[S642E]) X | This paper | N/A |
| Strain MJS237: qbcSi03[Palg-1::GFP::cog-1-boxb-cb-unc-19(+)] IV; alg-1(qbc18[lambdaN::alg-1]) X | (Quévillon Huberdeau et al., 2017) | N/A |
| Strain MJS345: qbcSi03[alg-1p::GFP::cog-1-boxb;cb-unc-119(+)] IV; alg-1(qbc66[λN::alg-1(S642E)] X | This paper | N/A |
| Strain MJS349: qbcSi03[alg-1p::GFP::cog-1-boxb;cb-unc-119(+)] IV; alg-1(qbc67[λN::alg-1(S642A)] X | This paper | N/A |
| Strain MJS446: alg-2(ok304) II; kin-2(qbc92[R92C]) X | This paper | N/A |
| Strain MJS 447: alg-2(ok304) II; kin-2(qbc92[R92C]) alg-1(qbc67[S642A]) X | This paper | N/A |
| Oligonucleotides | ||
| Primers used for cloning are listed in Table S2. | This study | N/A |
| CRISPR RNA and repair templates to generate alg-1(S642A), alg-1(S642E), alg-1(Y693E) and kin-2(R92C) are listed in Table S2. | This study | N/A |
| Primers for RT-qPCR are listed in Table S2. | This study | N/A |
| Recombinant DNA | ||
| Plasmid L4440: untargeted RNAi vector | Addgene | Cat# 1654 |
| Plasmid MSP163: RNAi vector targeting alg-2 | (Bouasker and Simard, 2012) | N/A |
| Plasmid MSP437: RNAi vector targeting kin-2 | This paper | N/A |
| Plasmid MSP186: alg-1p::lambdaN::mCherry::alg-1::alg-1 3'UTR | Jannot et al. 2016 | N/A |
| Plasmid MSP438: alg-1p::lambdaN::mCherry::alg-1(S642A)::alg-1 3'UTR | This study | N/A |
| Plasmid MSP439: alg-1p::lambdaN::mCherry::alg-1(S642E)::alg-1 3'UTR | This study | N/A |
| Software and algorithms | ||
| Bowtie (version 0.12.7) | Langmead, B. et al, 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| Bowtie (version 1.2.2) | Langmead, B. et al, 2009 | http://bowtie-bio.sourceforge.net/index.shtml |
| Cutadapt (version 2.3) | Martin,M. et al., 2011 | https://cutadapt.readthedocs.io/en/v2.3/ |
| Samtools (version 1.9) | Li, H. et al, 2009 | https://sourceforge.net/projects/samtools/files/samtools/1.9/ |
| HTSeq (version 0.11.2) | Anders, S. et al, 2015 | https://htseq.readthedocs.io/en/master/ |
| Image Lab | Bio-Rad | https://www.bio-rad.com/en-ca/product/image-lab-software?ID=KRE6P5E8Z |
| Other | ||
| Breathable sealing membrane (Breathe-EASIER) | Diversified Biotech | Cat# BERM-2000 |
| Luminometer | Berthold Technologies | Centro XS3 LB 960 |
| Gel Imaging System | Bio-Rad | ChemiDoc XRS+ System |
This paper does not report original code.
Additional information required to reanalyze the data in this study is available from the lead contact upon request.