Abstract
Peptide sequences obtained from the accessory subunit of Xenopus laevis mitochondrial DNA (mtDNA) polymerase γ (pol γ) were used to clone the cDNA encoding this protein. Amino-terminal sequencing of the mitochondrial protein indicated the presence of a 44-amino-acid mitochondrial targeting sequence, leaving a predicted mature protein with 419 amino acids and a molecular mass of 47.3 kDa. This protein is associated with the larger, catalytic subunit in preparations of active mtDNA polymerase. The small subunit exhibits homology to its human, mouse, and Drosophila counterparts. Interestingly, significant homology to glycyl-tRNA synthetases from prokaryotic organisms reveals a likely evolutionary relationship. Since attempts to produce an enzymatically active recombinant catalytic subunit of Xenopus DNA pol γ have not been successful, we tested the effects of adding the small subunit of the Xenopus enzyme to the catalytic subunit of human DNA pol γ purified from baculovirus-infected insect cells. These experiments provide the first functional evidence that the small subunit of DNA pol γ stimulates processive DNA synthesis by the human catalytic subunit under physiological salt conditions.
Mitochondrial DNA (mtDNA) is replicated by a DNA polymerase, DNA polymerase γ (pol γ), that is distinct from nuclear DNA polymerases α, β, δ, ɛ, and ζ. Since DNA pol γ represents only a small fraction of total cellular DNA polymerase, purification and characterization of the subunit composition of this enzyme have been difficult. Molecular cloning has contributed greatly to understanding the structure of DNA pol γ in different organisms. The catalytic subunits of DNA pol γ have been cloned for several organisms (6, 16, 17, 27, 34) and have been found to resemble family A of DNA polymerases, related to Escherichia coli DNA pol I. In Saccharomyces cerevisiae DNA pol γ is composed of a single polypeptide, while in Drosophila melanogaster DNA pol γ is comprised of two different polypeptides, a catalytic subunit of 125 kDa and an accessory subunit of 41 kDa (24, 33). The Drosophila subunits copurify and have been shown to interact, but the recombinant proteins have not yet been shown to be functional. The function of the small subunit, which we refer to as pol γB, is unknown. It has been proposed to influence the processivity of the catalytic subunit. Putative mammalian homologs of the Drosophila accessory subunit have been identified in sequence databases. One published purification scheme for human DNA pol γ suggested the existence of a small subunit (11), but the potential relationship between this polypeptide and Drosophila pol γB has not been established. Recently, the catalytic subunit of human DNA pol γ was expressed in an active form (10, 19). Surprisingly, the recombinant catalytic subunit alone displayed most of the characteristics of the enzyme purified from human cells, which did not appear to contain a stoichiometric amount of a small subunit. Thus, it is not clear what role, if any, is played by putative human DNA pol γB.
We have previously described the cloning of the large subunit of Xenopus laevis mtDNA polymerase (34). Here we describe the cloning and initial characterization of the accessory subunit of Xenopus mtDNA polymerase, which is homologous to its Drosophila and human counterparts. The cDNA encodes a protein of 463 amino acids with a mitochondrial targeting sequence of 44 amino acids. Both the catalytic subunit and the accessory subunit copurify through several steps of chromatography, and coimmunoprecipitation assays indicated that they are associated in a holoenzyme. We were not able to study the effect of the small subunit on the catalytic subunit of Xenopus pol γ, since the recombinant Xenopus catalytic subunit was inactive following expression in bacteria or insect cells. Therefore, we attempted to determine whether the Xenopus small subunit would stimulate the recombinant human catalytic subunit. Our data indicates that the Xenopus accessory subunit confers processivity on the human catalytic subunit. A striking similarity found between the accessory subunit and some class II aminoacyl-tRNA synthetases suggests a common evolutionary origin.
MATERIALS AND METHODS
Purification of Xenopus pol γ and sequencing of the small subunit.
Purification of X. laevis DNA pol γ has been described elsewhere (15, 23). Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) separated two polypeptides of 140 and 45 kDa. The smaller polypeptide was gel purified and subjected to digestion with endoprotease LysC, and peptides were sequenced by Edman degradation as described previously (32). The intact small subunit was also subjected to direct N-terminal sequencing after blotting to a polyvinylidene difluoride (PVDF) membrane (Immobilon P; Millipore).
Cloning.
Degenerate primers were synthesized from peptide sequences, and two rounds of nested PCR were carried out. Primer XLGB1 (GARTAYGTICCIWSIATGGA; sense strand, EYVPSME; R is A or G, Y is C or T, W is A or T, and S is C or G), primer XLGB2 (TTISWIACRTGIATIGTYTC; antisense strand, ETIHVSK), primer XLGB3 (AARGARACIATICAYGT; sense strand, KETIHV), primer XLGB4 (ACRTGIATIGTYTCYTT; antisense strand, KETIHV), and primer XLGB5 (CAYCCIACIYTIGCICC; sense strand, HPTLAP) were used in “touchdown” PCR (5). Nested PCR with primers XLGB1 and XLGB4 followed by primers XLGB5 and XLGB4 yielded a 281-bp PCR product that was cloned by TA cloning (Invitrogen) and sequenced.
A probe made from this clone was used to screen 500,000 clones from a lambda gt11 Xenopus cDNA library made with oligo(dT) and random primers (Clontech). Eight positive clones were identified and analyzed by PCR. All of them contained inserts of the same size, and one of them, designated pJAC3, was subcloned in pBluescript KS (Stratagene). This 1,491-bp cDNA clone was completely sequenced on both strands and found to contain an open reading frame coding for a protein of 463 amino acids and including a short 5′ untranslated region (UTR) and a 3′ UTR with a putative polyadenylation signal located 25 bp upstream from the beginning of a poly(A) tail.
Protein expression.
A PCR primer was designed to contain an NdeI site positioned to permit translation initiation at an ATG codon immediately preceding the first residue of the mature mitochondrial protein (without the 44-amino-acid mitochondrial presequence). Another primer was made with a NotI site at the position of the stop codon for the protein to permit in-frame linkage to a C-terminal His tag in the vector. PCR with these primers and pJAC3 DNA as a template was used to amplify a fragment of DNA that was subsequently digested with NdeI and NotI and cloned into pET22b(+) (Novagen) to generate clone pJAC8. Induction of expression of the protein from this clone in E. coli BL21(DE3) produced a C-terminally His-tagged version of the mitochondrial protein containing an extra methionine in the amino terminus. The majority of this protein was insoluble under native conditions. When the protein was solubilized with urea, it did not bind well to an Ni-nitrilotriacetic acid (NTA) column (Qiagen). Electrophoretic purification with a Prep-Cell 491 apparatus (Bio-Rad) was used to prepare protein to inoculate a rabbit (13).
For baculovirus expression, the pFastBac I vector (Life Technologies) was modified to contain a His tag derived from pET22b(+) in such a way that a C-terminally His-tagged protein could be produced. Briefly, pET22b(+) was digested with BlpI, an isoschizomer of Bpu1102I, and the ends were made blunt by filling in with Pfu polymerase and then digested with NotI to generate a NotI-blunted fragment coding for a His tag. In a similar way, pFastBac I was digested with HindIII, and the ends were made blunt and then cut with NotI. The NotI-blunted fragment from pET22b(+) was then cloned in pFastBac I, regenerating the HindIII site but not the BlpI site. An oligonucleotide was used to insert an NdeI restriction site following the BamHI site in pFastBac I. This step allowed cloning of the NdeI-NotI insert directly from pJAC8 into the modified baculovirus expression vector. The resulting clone was named pJAC30. Expression in Spodoptera frugiperda (Sf9) cells was achieved by following the procedures recommended by the manufacturer of the Bac-to-Bac baculovirus expression system (Life Technologies). The C-terminally His-tagged protein was purified by affinity chromatography on Ni-NTA superflow columns (Qiagen) with phosphate buffer under native conditions as recommended by the supplier, followed by chromatography on Mono S. The protein was concentrated by a second round of chromatography on Ni-NTA resin. The His-tagged recombinant catalytic subunit of human DNA pol γ was expressed and purified as described by Longley et al. (19). To quantify the recombinant proteins, various quantities of the catalytic and accessory subunits were subjected to SDS-PAGE along with various amounts of quantitative protein standards (glutamate dehydrogenase and phosphorylase b; Boehringer Mannheim). The protein concentrations were determined by densitometry of the Coomassie blue-stained gel.
Antibody methods.
Proteins were separated by SDS-PAGE with 10% gels, electroblotted onto Immobilon P membranes, and incubated overnight at room temperature with a 1:20,000 dilution of antiserum in phosphate-buffered saline containing 0.5% Tween 20. In one experiment (see Fig. 3), polyclonal sera directed against Xenopus pol γA and pol γB were used. In another experiment (see Fig. 4), an antipeptide serum directed against the sequence TRRAVEPTWLTASN(C) was used to detect either Xenopus or human DNA pol γA (34). Proteins were detected by successive incubation of the blots with a commercial alkaline phosphatase-conjugated goat anti-rabbit secondary antibody and 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium (Kirkegaard and Perry Laboratories). Affinity-purified antibodies against Xenopus DNA pol γB were prepared by adsorption of antibodies to a column containing a recombinant protein cross-linked to an Affiprep 10 matrix (Bio-Rad) and elution with 50 mM glycine (pH 2.3)–0.5 M NaCl–0.02% Triton X-100. The solution was quickly neutralized by the addition of one-fourth volume of 0.2 M Na2HPO4.
FIG. 3.
Copurification of large and small subunits of pol γ. Protein fractions collected during different steps of the purification of DNA pol γ were separated by SDS-PAGE and electroblotted onto PVDF membranes. The membranes were cut into upper and lower halves by use of prestained protein molecular weight markers as a guide. The upper part of each membrane was probed with an antibody directed against the DNA pol γ large subunit (L). The lower part of each membrane was probed with an antibody directed against the small subunit (S). Fractions are indicated by numbers, and FT denotes the flowthrough fraction. (A) Cation exchange (S Sepharose). (B) Gel filtration (Superdex 200 HiLoad). (C) Hydrophobic interaction (Poros PH).
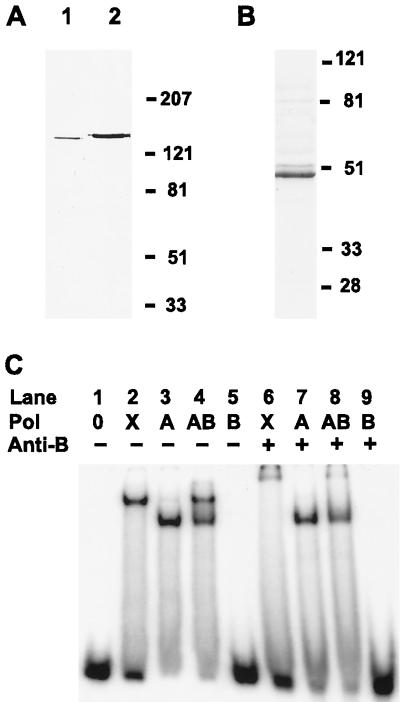
FIG. 4.
Characterization of recombinant pol γA and pol γB and analysis of primer-template binding by electrophoretic mobility shift assays (EMSA). (A) Xenopus pol γ (1,500 U) and recombinant human pol γA (180 U) were run in lanes 1 and 2, respectively, of an SDS–6% polyacrylamide gel, blotted to a PVDF membrane, and detected with a 1:20,000 dilution of antipeptide antibodies directed against the sequence TRRAVEPTWLTASN(C), contained in both human and Xenopus pol γA. (B) Coomassie blue-stain SDS-PAGE analysis of Xenopus pol γB purified from baculovirus-infected cells as described in Materials and Methods. Numbers at the right in panels A and B indicate the mobilities of protein mass standards in kilodaltons. (C) Autoradiogram of EMSA results. A 5′-32P-labeled 44-mer hook oligonucleotide was incubated either alone (lane 1) or in the presence of either pol γ purified from X. laevis ovaries (lanes 2 and 6; labeled X) or recombinant subunits of pol γ, either human pol γA (lanes 3 and 7; labeled A), human pol γA plus Xenopus pol γB (lanes 4 and 8; labeled AB), or Xenopus pol γB alone (lanes 5 and 9; labeled B). Following incubation for 10 min at 25°C, affinity-purified antibodies raised against Xenopus DNA pol γB were added to reaction mixtures in lanes 6 through 9. Incubation was continued for an additional 10 min. Samples were subjected to electrophoresis under nondenaturing conditions, and 32P radioactivity in the dried gel was detected by phosphorimager analysis.
DNA polymerase assays.
DNA pol γ activity was calibrated in reactions with poly(rA)-oligo(dT) as described previously (15). One unit corresponds to the incorporation of 1 pmol of deoxynucleoside monophosphate into acid-insoluble products in a 30-min reaction. All polymerase assays with DNA templates were performed at 30°C with a buffer consisting of 10 mM Tris (pH 8); 50 mM KCl; 8 mM MgCl2; 2 mM dithiothreitol; 25 μM each TTP, dATP, dGTP, and dCTP; and 100 μg of bovine serum albumin per ml (the KCl and MgCl2 concentrations were varied as noted in the figures). For primer extension reactions, oligonucleotide dT24 or dT16 and the M13 −20 sequencing primer (GTAAAACGACGGCCAGT) were phosphorylated with [γ-32P]ATP and polynucleotide kinase and gel purified by standard methods (28). Oligo(dT) was annealed to poly(dA) (estimated chain length, ∼1 to 1.2 kb), and the −20 primer was annealed to M13mp9 DNA in a buffer containing 100 mM NaCl, 10 mM Tris (pH 7.5), and 1 mM EDTA. Primer extension reactions included a final concentration of 20 nM primer-template. Samples were withdrawn at various times, mixed with 2 volumes of formamide DNA loading solution, boiled, and analyzed by electrophoresis on 20% polyacrylamide gels containing 8 M urea.
DNA binding assays were conducted by an electrophoretic mobility shift assay as described by Mikhailov and Bogenhagen (23) with 40 fmol of a 44-mer oligonucleotide (5′-32P-CCATCTAAGCAGACTCACGAATTCACCTAGTTGTTCTAGGTGAA) labeled with [γ-32P]ATP and polynucleotide kinase. This oligonucleotide sequence is expected to fold into a partial hairpin with a 9-bp duplex and a 19-nucleotide single-stranded tail to provide a primer-template structure. The quantities of polymerase used in binding reactions are specified in the figure legends.
RESULTS
Cloning of the X. laevis mtDNA polymerase accessory subunit.
DNA pol γ was purified from X. laevis ovary mitochondria as previously described (15, 23). SDS-PAGE analysis of the purified enzyme revealed the presence of two polypeptides, of approximately 140 and 45 kDa (Fig. 1). The larger polypeptide has been previously identified as the catalytic subunit of DNA pol γ (15, 34). The smaller polypeptide was assumed to be the accessory subunit of X. laevis DNA pol γ by analogy to the situation for Drosophila (33). The purified small subunit was subjected to amino-terminal and internal sequencing. For internal protein sequencing, the gel-purified protein was digested with endoprotease LysC, and peptides were separated by high-pressure liquid chromatography and sequenced. PCR was carried out on first-strand cDNA (synthesized from Xenopus ovary mRNA) by use of degenerate primers derived from those peptides. A 281-bp product was amplified, cloned, and sequenced. A putative open reading frame encoding 93 amino acids spanned the whole cDNA fragment and showed homology to part of the human and Drosophila homologs. This fragment was used to screen an X. laevis ovary cDNA library. A positive clone was completely sequenced. The 1,491-bp long cDNA clone encodes a putative protein of 463 amino acids and contains short 5′ and 3′ UTRs. Amino-terminal sequencing of the purified protein revealed a mitochondrial targeting sequence of 44 amino acids, leaving a mature protein of 419 amino acids with a predicted molecular mass of 47.3 kDa, which agrees with the size of 45 kDa estimated by electrophoresis. An mRNA of approximately 1,600 nucleotides was identified on a Northern blot (data not shown). We believe that the cDNA clone is a full-length cDNA clone, since it corresponds well with the size of the mRNA and includes the N terminus of the mature protein with an apparently complete mitochondrial signal sequence. Furthermore, a stop codon is located in frame six codons before the putative start codon. We refer to the product of this cDNA clone as the B subunit of DNA pol γ, or pol γB, while the larger catalytic subunit is referred to as pol γA.
FIG. 1.
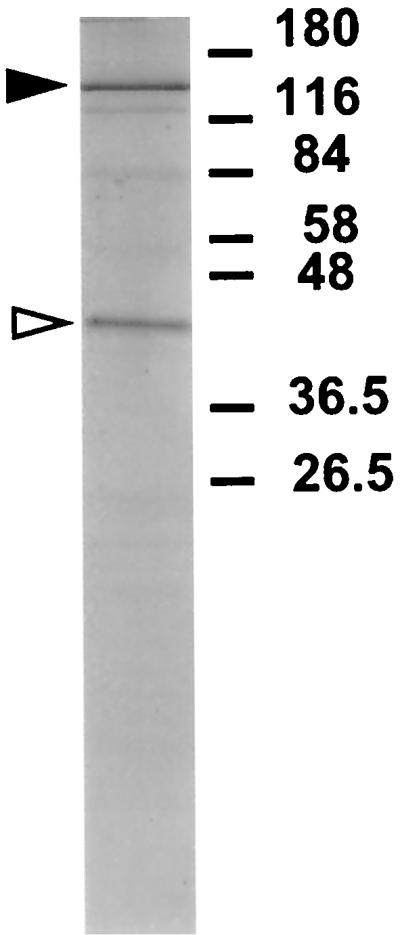
Polypeptide composition of X. laevis pol γ. Purified DNA pol γ was analyzed by SDS-PAGE and Coomassie blue staining. The solid arrowhead indicates the large subunit, and the open arrowhead indicates the small subunit. Numbers on the right indicate the molecular masses in kilodaltons of prestained protein mobility markers.
Vertebrate DNA pol γB shows primary sequence similarity to glycyl-tRNA synthetases.
The protein sequence of mature X. laevis DNA pol γB, excluding the putative mitochondrial targeting sequence, was used to search the GenBank database. This search revealed 21.05% identity and 29.53% similarity (including identical residues) with Drosophila DNA pol γB and 52.69% identity and 66.13% similarity with a human cDNA product identified as a likely homolog of Drosophila DNA pol γB (33). Surprisingly, this search also revealed a highly significant similarity between Xenopus DNA pol γB and certain class II aminoacyl-tRNA synthetases. An alignment of the DNA pol γB sequences and one tRNA synthetase sequence is shown in Fig. 2. X. laevis DNA pol γB has 25.54% identity with Thermus thermophilus glycyl-tRNA synthetase, an identity higher than that reported above with Drosophila DNA pol γB. The significance of these homologies can be interpreted with reference to the E values or expectation values reported in a Blast search for matches to X. laevis pol γB, which indicate the probability that the match could occur by chance. The E values for the match to human pol γB, T. thermophilus glycyl-tRNA synthetase, and Drosophila pol γB are e−109, 4e−24, and 2e−10, respectively. It is interesting that Drosophila DNA pol γB shows significantly less similarity to tRNA synthetases, a finding which may account for the fact that no homology between Drosophila DNA pol γB and tRNA synthetases was noted in the original report by Wang et al. (33).
FIG. 2.
X. laevis, Drosophila, and putative human pol γ small subunits (XLGB, GenBank accession no. AF124606; DMGB, accession no. U94702; and HSGB, accession no. U94703, respectively) were aligned with the glycyl-tRNA synthetase from T. thermophilus (TTGRS; accession no. P56206) by use of the program CLUSTAL from PCGene. Amino acids conserved among all four proteins are indicated by asterisks; similar amino acids are indicated by dots. Amino acids conserved between XLGB and TTGRS are indicated in bold. The three conserved motifs identified in class II glycyl-tRNA synthetases are also labeled under the sequences. Note that the X. laevis pol γB sequence begins with the N terminus of the mature protein and does not include the signal sequence.
Xenopus DNA pol γ is a stable heterodimer.
Bacterially expressed recombinant protein was used to generate a rabbit antiserum against DNA pol γB. The antibodies recognize specifically both the recombinant protein and the protein purified from ovary mitochondria. To determine whether the two subunits of DNA pol γ copurified, column fractions obtained at several steps of the purification of the polymerase from Xenopus ovaries were analyzed by Western blotting with antibodies directed against pol γA and pol γB. Both proteins were found to copurify on cation-exchange, gel filtration, and hydrophobic interaction chromatography (Fig. 3). Antibodies against the small subunit were able to coimmunoprecipitate the large subunit from a partially purified preparation of active DNA pol γ (data not shown). This result indicates that the two subunits of X. laevis DNA pol γ are associated in a holoenzyme. The molecular mass predicted for a heterodimer containing one molecule each of X. laevis DNA pol γA and pol γB is 181.5 kDa, consistent with the molecular mass of 185 kDa determined for native X. laevis pol γ by sedimentation and gel filtration (15).
We expressed both subunits of X. laevis DNA pol γ in insect cells by using baculovirus vectors in an attempt to characterize the potential effects of the small subunit on the catalytic subunit. We were unable to recover polymerase activity from preparations of the recombinant catalytic subunit, so we were not able to assess the effect of the small subunit on polymerase activity. We also did not observe enzyme activity when insect cells were coinfected with baculoviruses carrying the genes encoding both the catalytic and the accessory subunits. A deleterious mutation in the recombinant catalytic subunit could account for these results. During the course of experiments carried out to characterize the DNA pol γ gene, we identified several discrepancies in the amino-terminal portion of the sequence of the catalytic subunit of X. laevis pol γ initially reported by Ye et al. (34), who had cloned the cDNA by using 5′ rapid amplification of cDNA ends. To correct these errors, most of the cDNA was recloned with a Marathon cDNA kit (Clontech; synthesized from Xenopus mRNA by use of Pfu polymerase). The revised cDNA sequence was found to agree with the genomic DNA sequence at each site at which the original cDNA sequence differed from the genomic DNA sequence. Nevertheless, the proteins expressed from clones constructed from the revised sequence exhibited no polymerase activity. We currently suspect that recombinant protein misfolding or the absence of required posttranslational modifications may be responsible for the lack of activity.
The Xenopus small subunit stimulates the human catalytic subunit at physiological salt concentrations.
As all our attempts to recover activity from the Xenopus recombinant catalytic subunit failed, we decided to determine whether the Xenopus small subunit would alter the properties of the active recombinant human catalytic subunit purified from insect cells (19). Figure 4A shows an immunoblot analysis of samples of Xenopus pol γ [lane 1; 1,500 U of enzyme, as assayed by TMP incorporation on poly(rA)-oligo(dT)] and recombinant human pol γA (lane 2; 180 U of enzyme, 14 ng of polypeptide). Both proteins were detected with an antibody raised against a peptide sequence that is identical in the two proteins. We estimate that on a per-molecule basis, recombinant human pol γA is 10- to 20-fold less active than the native Xenopus protein in the poly(rA)-oligo(dT) assay at low salt concentrations. It is possible that all of the recombinant protein is not correctly folded or that posttranslational modifications of the authentic mitochondrial protein increase its activity. Nevertheless, we judged that the quality of the recombinant human DNA pol γA preparation was sufficient for some mechanistic studies. Figure 4B shows a Coomassie blue-stained SDS-polyacrylamide gel used for analysis of the recombinant Xenopus small subunit.
Electrophoretic mobility shift assays were conducted with these proteins to examine their ability to bind a 44-mer hook oligonucleotide primer-template. We have previously shown that authentic Xenopus DNA pol γ binds to oligonucleotide substrates and retains polymerase activity following native gel electrophoresis (23). Binding of the human catalytic subunit alone produced a shifted complex with a more rapid gel mobility than that observed for the Xenopus pol γ holoenzyme (compare lanes 2 and 3 in Fig. 4C). When the Xenopus accessory subunit was included in the binding reactions, a complex with the same electrophoretic mobility as that of the authentic Xenopus holoenzyme was observed (compare lanes 2 and 4 in Fig. 4C). However, we were not able to convert all of the complexes containing pol γA to this more slowly migrating form. X. laevis pol γB was not able to bind to DNA in the absence of the catalytic subunit (lane 5). These results suggest that Xenopus DNA pol γB does not have appreciable DNA binding ability on its own but is able to associate with the human catalytic subunit in a complex with a primer-template. To test this hypothesis, parallel binding reactions in which affinity-purified antibodies directed against Xenopus pol γB were added after 10 min were performed. The addition of antibodies resulted in a clear supershift of the complex containing Xenopus pol γ (lane 6), suggesting that the small subunit is present in this complex. The addition of anti-DNA pol γB antibodies did not affect the complex with recombinant human pol γA (lane 7). The addition of antibodies to a reaction mixture containing both human pol γA and Xenopus pol γB (Fig. 4C, lane 8) provided mixed results consistent with the possibility that some complexes were disrupted by the antibodies while a minor fraction was supershifted. As expected, the anti-pol γB antibodies did not affect the complexes in lane 8 which did not appear to contain DNA pol γB.
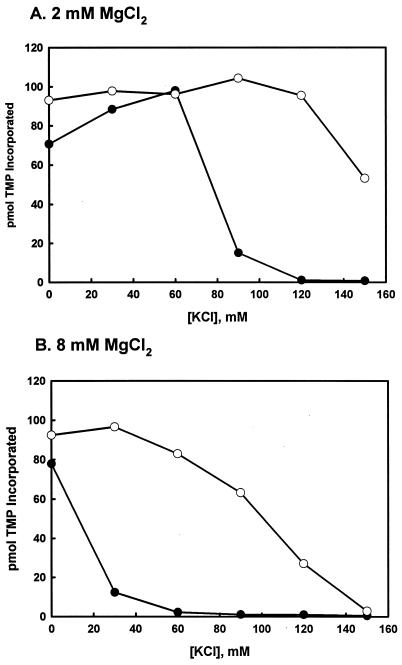
A variety of primer-template constructs were used to test whether X. laevis DNA pol γB influences the catalytic activity of human DNA pol γA. Figure 5 shows the results of an experiment in which recombinant human DNA pol γA was assayed on a poly(dA)-oligo(dT) substrate with and without Xenopus DNA pol γB. In this experiment, we varied the monovalent salt concentration in reactions performed with either 2 or 8 mM MgCl2 to encompass the range of physiological salt concentrations. Human pol γA was found to be very sensitive to increasing ionic strength when assayed at either 2 or 8 mM MgCl2, as reported previously for reactions performed with 1 mM MgCl2 (19). The addition of the Xenopus small subunit markedly stimulated the activity of the human catalytic subunit at higher, more physiological KCl concentrations, but not under low-salt conditions. Heating the factor to 80°C for 10 min completely abolished this stimulation (data not shown).
FIG. 5.
Stimulation of human DNA pol γA by Xenopus DNA pol γB. Polymerase activity of human DNA pol γA (●) and an equimolar mixture of human pol γA plus Xenopus pol γB (○) on oligo(dT)-poly(dA) was measured with reaction mixtures containing 2 (A) or 8 (B) mM MgCl2. All reaction mixtures contained 0.2 pmol of the indicated polymerase subunits.
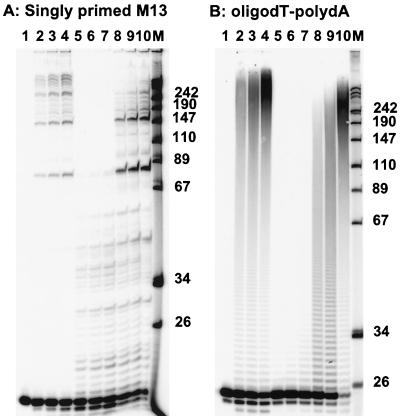
To determine whether Xenopus pol γB increased the processivity of DNA synthesis by human pol γA, primer extension assays were conducted under conditions of excess primer-template with two different kinds of templates, singly primed M13 DNA and poly(dA)-oligo(dT). The homopolymeric template was included in this analysis since we have previously shown that this is a preferred template for DNA pol γ because it does not present the enzyme with blocks to replication that require a mitochondrial single-stranded DNA binding protein (SSB) as an additional processivity factor (23). In each case, the primers were 32P end labeled for the primer extension assays. In order to compare the activities of the authentic Xenopus and recombinant human enzymes in these experiments, it was necessary to consider that the polypeptide concentration of the Xenopus enzyme could not be determined accurately due to the small amount of material available and that its specific activity was certainly higher than that of the recombinant human enzyme, as noted in Fig. 4. Conducting these reactions with excess primer-template tends to make the precise polymerase concentration a secondary concern. Xenopus pol γ generated products as long as 500 nucleotides in the first minute of incubation, with a progressive accumulation of products as the reaction continued. Recombinant human pol γA alone synthesized only short products under these relatively high-salt conditions. The addition of the Xenopus small subunit significantly increased the processivity of the human polymerase on both templates, permitting the synthesis of DNA chains nearly as long as those synthesized by the Xenopus DNA pol γ holoenzyme (Fig. 6). These results strongly suggest a role for the small subunit as a processivity factor for pol γ in both Xenopus and humans.
FIG. 6.
Xenopus DNA pol γB confers increased processivity on human pol γA. Primer extension assays with singly primed M13 single-stranded DNA (A) or oligo(dT)-poly(dA) (B) were carried out as described in Materials and Methods. Each reaction included 200 fmol of annealed primer. In both panels, the reaction mixture in lane 1 contained no enzyme; those in lanes 2, 3, and 4 contained 75 U of Xenopus pol γ and were incubated for 1, 3, and 10 min, respectively. Reaction mixtures in lanes 5, 6, and 7 contained 9 U of recombinant human pol γA, equivalent to 50 fmol of polypeptide, and were incubated for the same periods of time; reactions in lanes 8, 9, and 10 were carried out with 50 fmol of recombinant human pol γA plus 50 fmol of recombinant Xenopus pol γB, incubated as in the previous reactions. Lane M shows DNA molecular weight markers of 32P-labeled MspI fragments of plasmid pUC18, with the sizes, in nucleotides, indicated to the right of each panel. Due to space constraints, the larger marker fragments, of 501, 489, 404, and 353 nucleotides, are not individually labeled.
DISCUSSION
mtDNA pol γ has a variable subunit structure in different organisms, with or without a processivity factor.
Our results indicate that X. laevis DNA pol γ is a heterodimer. Both polypeptides copurify through several steps of chromatography and can be coimmunoprecipitated. The mass predicted for a heterodimer containing one molecule each of pol γA and pol γB agrees very well with the native size determined by sedimentation and gel filtration. This situation is similar to what has been described for Drosophila mtDNA pol, in which both subunits appear to be tightly associated (33). It appears that not all DNA pol γ enzymes are stable heterodimers. No small subunit has been reported for DNA pol γ in the yeast S. cerevisiae (6). The complete S. cerevisiae genome sequence does not appear to contain a homolog for Xenopus or Drosophila DNA pol γB. The sequences of the DNA pol γ catalytic subunits have been reported for other yeasts (26, 34), but it is currently not known whether these organisms use a small subunit of DNA pol γ. The understanding of the machinery involved in the maintenance of mtDNA in yeasts is still incomplete, so other accessory factors that assist yeast DNA pol γ may remain to be identified. In light of our observation that vertebrate DNA pol γB is related to tRNA synthetases, it is interesting that tyrosyl-tRNA synthetase (a class I aminoacyl-tRNA synthetase) has been found to rescue a defect in mtDNA maintenance caused by a mutation in another uncharacterized gene, MGM104 (12). It has been proposed that tyrosyl-tRNA synthetase may have a biochemical function that enhances the activity of the MGM104 gene product.
It is not clear whether human DNA pol γ exists in vivo as a monomer or a heterodimer. The evidence bearing on this point has been discussed above. The possibility that the human proteins are not tightly associated as a heterodimer would not preclude the association of these two proteins during DNA replication. We do not yet know whether pol γB influences any of the template binding properties of DNA pol γ in a way that would contribute to the regulation of mtDNA replication. In Drosophila and Xenopus, the two subunits in the pol γ heterodimer contain complete or nearly complete leucine zipper domains that may mediate their dimerization. The human pol γ subunits lack complete leucine zipper motifs. Experiments are under way to define the sequences in pol γ subunits responsible for their interaction.
Our observation that Xenopus pol γB can act as a processivity factor for the human catalytic subunit suggests that it is likely to play the same role for its homologous partner. It has been suggested that the accessory subunit of Drosophila pol γ is a processivity factor, but this suggestion could not be demonstrated because the recombinant enzyme was inactive (33). We have also been unable to recover activity from the recombinant Xenopus catalytic subunit. Xenopus pol γ resembles its Drosophila counterpart in that both enzymes seem to be composed of tightly associated A and B subunits. It is interesting that the only pol γ enzymes that have been expressed in an active form to date, the human and yeast enzymes, can also be purified from mitochondria without a tightly associated small subunit. It is tempting to suggest that the Xenopus and Drosophila enzymes may require the coordinate assembly of the large and small subunits in a mitochondrial environment.
The current view of DNA pol γ is reminiscent of the situation for the prokaryotic DNA polymerases E. coli DNA polymerase I and T7 DNA polymerase. The latter polymerase is well known to use host thioredoxin as an accessory processivity factor (30). E. coli DNA polymerase I acts as a rather distributive polymerase without an accessory factor and functions mainly as a repair polymerase. Interestingly, grafting the thioredoxin interaction domain of T7 DNA polymerase onto the Klenow fragment of E. coli DNA polymerase I permits the engineered polymerase to interact with thioredoxin and improves its processivity (1). Clearly, further experiments are required to explore the relationship between the two subunits of pol γ in a variety of organisms to arrive at a similarly sophisticated model for their interaction.
DNA pol γB is related to aminoacyl-tRNA synthetases.
Several key enzymes involved in mtDNA maintenance are closely related to their prokaryotic counterparts; these include mitochondrial RNA polymerase (3, 21), SSB protein (4, 9, 20, 31), the high-mobility-group-box DNA binding protein mitochondrial transcription factor A (25), and the catalytic subunit of DNA pol γ (17, 26, 27, 34). Nevertheless, the observation that the small subunit of DNA pol γ is related to prokaryotic aminoacyl-tRNA synthetases is surprising, since the latter represent a different class of enzyme involved in nucleic acid metabolism. Other systems have provided some evidence for structural similarities between components of the replication machinery and tRNA synthetases. Bochkarev et al. (2) have shown that the structure of the single-stranded DNA binding domain of human replication protein A is related to that of yeast aspartyl-tRNA synthetase bound to tRNA. However, the similarity that we describe is unusual in that it is apparent at the level of primary sequence homology.
The closest homolog that we have identified for X. laevis DNA pol γB (other than human DNA pol γB) is the glycyl-tRNA synthetase from T. thermophilus and Mycobacterium tuberculosis. This glycyl-tRNA synthetase is similar to the archaeal and eukaryotic enzymes, which are dimeric, but is highly divergent from other prokaryotic enzymes, such as that of E. coli, which are tetrameric (22). E. coli glycyl-tRNA synthetase can charge only prokaryotic tRNAGly, whereas the enzyme from T. thermophilus is also able to charge eukaryotic tRNAGly. We have not tested whether X. laevis DNA pol γB has tRNA synthetase activity. In the experiment shown in Fig. 3, we did not detect a pool of the small subunit free of the large subunit, a result which may have been expected if the small subunit served a secondary role as a tRNA synthetase.
The tRNA synthetases closely related to the pol γ accessory subunit belong to the class II family of aminoacyl-tRNA synthetases, which differ from the class I enzymes in the aminoacylation site in the tRNA (3′ OH versus 2′ OH of the terminal ribose, with the exception of phenylalanyl-tRNA synthetase) (7, 14, 29) as well as in the structure of the active site of the enzyme. Class II enzymes contain a seven-stranded antiparallel β-sheet motif (18). Three sequence motifs have been identified in class II aminoacyl-tRNA synthetases. These are believed to be involved in multimerization, recognition of the tRNA, and catalysis of the aminoacylation reaction (for a review, see reference 8). As shown in Fig. 2, the sequence conservation between X. laevis DNA pol γB and T. thermophilus glycyl-tRNA synthetase is relatively high near the three motifs that form the active site of class II aminoacyl-tRNA synthetases, as well as in the C-terminal portion of each protein, which has been implicated in tRNA recognition. The fact that the regions of aminoacyl-tRNA synthetases that are conserved as functional domains show the greatest similarity to pol γB is striking and may suggest a functional relationship between both types of proteins. It will be interesting to determine whether the structure of X. laevis DNA pol γB is similar to the known structure of T. thermophilus glycyl-tRNA synthetase (18).
ACKNOWLEDGMENTS
This work was supported by NIH grant GM29681 and NIEHS grant P01-04068 to D.F.B.
We thank Kevin Pinz for technical assistance.
REFERENCES
- 1.Bedford E, Tabor S, Richardson C C. The thioredoxin binding domain of bacteriophage T7 DNA polymerase confers processivity on Escherichia coli DNA polymerase I. Proc Natl Acad Sci USA. 1997;94:479–484. doi: 10.1073/pnas.94.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochkarev A, Pfuetzner R A, Edwards A M, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 3.Cermakian N, Ikeda T M, Cedergren R, Gray M W. Sequences homologous to yeast mitochondrial and bacteriophage T3 and T7 RNA polymerases are widespread throughout the eukaryotic lineage. Nucleic Acids Res. 1996;24:648–654. doi: 10.1093/nar/24.4.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curth U, Urbanke C, Greipel J, Gerberding H, Tiranti V, Zeviani M. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur J Biochem. 1994;221:435–445. doi: 10.1111/j.1432-1033.1994.tb18756.x. [DOI] [PubMed] [Google Scholar]
- 5.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ’Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:9007–9049. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foury F. Cloning and sequencing of the nuclear gene MIP 1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem. 1989;264:20552–20560. [PubMed] [Google Scholar]
- 7.Fraser T H, Rich A. Amino acids are not all initially attached to the same position on transfer RNA molecules. Proc Natl Acad Sci USA. 1975;72:3044–3048. doi: 10.1073/pnas.72.8.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freist W, Logan D T, Gauss D H. Glycyl-tRNA synthetase. Biol Chem Hoppe-Seyler. 1996;377:343–356. [PubMed] [Google Scholar]
- 9.Ghrir R, Lecaer J P, Dufresne C, Gueride M. Primary structure of the two variants of Xenopus laevis mtSSB, a mitochondrial DNA binding protein. Arch Biochem Biophys. 1991;291:395–400. doi: 10.1016/0003-9861(91)90152-9. [DOI] [PubMed] [Google Scholar]
- 10.Graves S W, Johnson A A, Johnson K A. Expression, purification, and initial kinetic characterization of the large subunit of the human mitochondrial DNA polymerase. Biochemistry. 1998;37:6050–6058. doi: 10.1021/bi972685u. [DOI] [PubMed] [Google Scholar]
- 11.Gray H, Wong T W. Purification and identification of subunit structure of the human mitochondrial DNA polymerase. J Biol Chem. 1992;267:5835–5841. [PubMed] [Google Scholar]
- 12.Guan M-X. Cytoplasmic tyrosyl-tRNA synthetase rescues the defect in mitochondrial genome maintenance caused by the nuclear mutation mgm 104-1 in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1997;255:525–532. doi: 10.1007/s004380050525. [DOI] [PubMed] [Google Scholar]
- 13.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 14.Hecht S M. Transfer RNA: structure, properties and recognition. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1979. [Google Scholar]
- 15.Insdorf N F, Bogenhagen D F. DNA polymerase gamma from Xenopus laevis. I. The identification of a high molecular weight catalytic subunit by a novel DNA polymerase photolabeling procedure. J Biol Chem. 1989;264:21491–21497. [PubMed] [Google Scholar]
- 16.Lecrenier N, Van Der Bruggen P, Foury F. Mitochondrial DNA polymerases from yeast to man: a new family of polymerases. Gene. 1997;185:147–152. doi: 10.1016/s0378-1119(96)00663-4. [DOI] [PubMed] [Google Scholar]
- 17.Lewis D L, Farr C L, Wang Y, Lagina A T, Kaguni L S. Catalytic subunit of mitochondrial DNA polymerase from Drosophila embryos. Cloning, bacterial overexpression, and biochemical characterization. J Biol Chem. 1996;271:23389–23394. doi: 10.1074/jbc.271.38.23389. [DOI] [PubMed] [Google Scholar]
- 18.Logan D T, Mazauric M H, Kern D, Moras D. Crystal structure of glycyl-tRNA synthetase from Thermus thermophilus. EMBO J. 1995;14:4156–4167. doi: 10.1002/j.1460-2075.1995.tb00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longley M J, Ropp P A, Lim S E, Copeland W C. Characterization of the native and recombinant catalytic subunit of human DNA polymerase γ: identification of residues critical for exonuclease activity and dideoxynucleotide sensitivity. Biochemistry. 1998;37:10529–10539. doi: 10.1021/bi980772w. [DOI] [PubMed] [Google Scholar]
- 20.Mahoungou C, Ghrir R, Lecaer J P, Mignotte B, Barat-Gueride M. The amino-terminal sequence of the Xenopus laevis mitochondrial SSB is homologous to that of the Escherichia coli protein. FEBS Lett. 1988;235:267–270. doi: 10.1016/0014-5793(88)81276-6. [DOI] [PubMed] [Google Scholar]
- 21.Masters B S, Stohl L L, Clayton D A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 22.Mazauric M H, Keith G, Logan D, Kreutzer R, Giege R, Kern D. Glycyl-tRNA synthetase from Thermus thermophilus—wide structural divergence with other prokaryotic glycyl-tRNA synthetases and functional inter-relation with prokaryotic and eukaryotic glycylation systems. Eur J Biochem. 1998;251:744–757. doi: 10.1046/j.1432-1327.1998.2510744.x. [DOI] [PubMed] [Google Scholar]
- 23.Mikhailov V S, Bogenhagen D F. Effects of Xenopus laevis mitochondrial single-stranded DNA binding protein on primer-template binding and 3′→5′ exonuclease activity of DNA polymerase γ. J Biol Chem. 1996;271:18939–18946. doi: 10.1074/jbc.271.31.18939. [DOI] [PubMed] [Google Scholar]
- 24.Olson M W, Wang Y, Elder R H, Kaguni L S. Subunit structure of mitochondrial DNA polymerase from Drosophila embryos. Physical and immunological studies. J Biol Chem. 1995;270:28932–28937. doi: 10.1074/jbc.270.48.28932. [DOI] [PubMed] [Google Scholar]
- 25.Parisi M A, Clayton D A. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 1991;252:965–969. doi: 10.1126/science.2035027. [DOI] [PubMed] [Google Scholar]
- 26.Ropp P A, Copeland W C. Characterization of a new DNA polymerase from Schizosaccharomyces pombe: a probable homologue of the Saccharomyces cerevisiae DNA polymerase gamma. Gene. 1995;165:103–107. doi: 10.1016/0378-1119(95)00412-y. [DOI] [PubMed] [Google Scholar]
- 27.Ropp P A, Copeland W C. Cloning and characterization of the human mitochondrial DNA polymerase, DNA polymerase γ. Genomics. 1996;36:449–458. doi: 10.1006/geno.1996.0490. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sprinzl M, Cramer F. Site of aminoacylation of tRNAs from Escherichia coli with respect to the 2′- or 3′-hydroxyl group of the terminal adenosine. Proc Natl Acad Sci USA. 1975;72:3049–3053. doi: 10.1073/pnas.72.8.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabor S, Huber H E, Richardson C C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J Biol Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 31.Van Dyck E, Foury F, Stillman B, Brill S J. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 1992;11:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Kobayashi R, Bishop J. Cellular adherence elicits ligand-independent activation of the Met cell-surface receptor. Proc Natl Acad Sci USA. 1996;93:8425–8430. doi: 10.1073/pnas.93.16.8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Farr C L, Kaguni L S. Accessory subunit of mitochondrial DNA polymerase from Drosophila embryos. J Biol Chem. 1997;272:13640–13646. doi: 10.1074/jbc.272.21.13640. [DOI] [PubMed] [Google Scholar]
- 34.Ye F, Carrodeguas J A, Bogenhagen D F. The γ subfamily of DNA polymerases: cloning of a developmentally regulated cDNA encoding Xenopus laevis mitochondrial DNA polymerase γ. Nucleic Acids Res. 1996;24:1481–1488. doi: 10.1093/nar/24.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]