Abstract
The A+U-rich RNA-binding factor AUF1 exhibits characteristics of a trans-acting factor contributing to the rapid turnover of many cellular mRNAs. Structural mapping of the AUF1 gene and its transcribed mRNA has revealed alternative splicing events within the 3′ untranslated region (3′-UTR). In K562 erythroleukemia cells, we have identified four alternatively spliced AUF1 3′-UTR variants, including a population of AUF1 mRNA containing a highly conserved 107-nucleotide (nt) 3′-UTR exon (exon 9) and the adjacent downstream intron (intron 9). Functional analyses using luciferase–AUF1 3′-UTR chimeric transcripts demonstrated that the presence of either a spliceable or an unspliceable intron 9 in the 3′-UTR repressed luciferase expression in cis, indicating that intron 9 sequences may down-regulate gene expression by two distinct mechanisms. In the case of the unspliceable intron, repression of luciferase expression likely involved two AUF1-binding sequences, since luciferase expression was increased by deletion of these sites. However, inclusion of the spliceable intron in the luciferase 3′-UTR down-regulated expression independent of the AUF1-binding sequences. This is likely due to nonsense-mediated mRNA decay (NMD) owing to the generation of exon-exon junctions more than 50 nt downstream of the luciferase termination codon. AUF1 mRNA splice variants generated by selective excision of intron 9 are thus also likely to be subject to NMD since intron 9 is always positioned >137 nt downstream of the stop codon. The distribution of alternatively spliced AUF1 transcripts in K562 cells is consistent with this model of regulated AUF1 expression.
The cytoplasmic steady-state level of any given mRNA, and hence its potential for translation, is a cumulative function of its rates of synthesis (transcription), nuclear pre-mRNA processing, nucleocytoplasmic transport, and cytoplasmic decay. Thus, the potential exists for cis-acting elements to regulate the abundance of individual mRNAs at many stages (reviewed in reference 6). Genetic determinants regulating the rate of mRNA turnover are frequently localized to the 3′ untranslated region (3′-UTR) (reviewed in references 4, 36, and 38).
A+U-rich elements (AREs) are cis-acting determinants of rapid cytoplasmic mRNA turnover found in the 3′-UTRs of many constitutively labile transcripts, including some encoding oncoproteins, inflammatory mediators, cytokines, and G-protein-coupled receptors (reviewed in references 10 and 36). AREs are variable in length but frequently consist of a number of overlapping AUUUA pentamers located within or adjacent to a U-rich region. While sequence and functional heterogeneity is observed among these destabilizing elements from different mRNAs, ARE-directed mRNA turnover is generally characterized by rapid deadenylation followed by decay of the mRNA body. Since many ARE-containing mRNAs encode essential components of pathways regulating cell growth, inflammation, and immune responses, the control of mRNA decay through these sequences influences many biological processes.
AUF1 is an ARE-binding protein originally identified as an activity capable of accelerating the turnover of c-myc mRNA in cell-free mRNA decay assays (5). Several additional lines of evidence indicate that association of AUF1 with ARE-containing mRNAs targets rapid turnover of these transcripts (47). First, the binding affinity of recombinant AUF1 for AREs closely correlates with their ability to destabilize mRNA in cis (14). Second, accelerated turnover of β-adrenergic receptor (β-AR) mRNA in failing human heart and isoproterenol-treated DDT1-MF2 smooth muscle cells is accompanied by an increase in intracellular AUF1 concentrations (31). Third, GRO and interleukin-1β mRNAs are stabilized during monocyte adherence concomitant with loss of an ARE-binding activity containing AUF1 (42). Moreover, treatment of monocytes with inhibitors of either tyrosine kinases or mitogen-activated protein kinases is accompanied by both retention of this ARE-binding activity in adhered monocytes and rapid turnover of interleukin-1β and GRO mRNAs. Thus, the magnitude of AUF1-dependent ARE-binding activity is a major determinant of mRNA turnover rates in cells.
AUF1 is expressed as a family of four protein isoforms designated by their apparent molecular masses as p37AUF1, p40AUF1, p42AUF1, and p45AUF1 (9, 17, 51). Recent mapping of the AUF1 gene demonstrated that the various protein isoforms are generated by alternative splicing of a common pre-mRNA (44). The AUF1 gene is a single-copy locus mapping to human chromosome 4 region q21 (44, 45) and consists of 10 exons. The translational termination codon lies within exon 8, indicating that exons 9 and 10 encode exclusively 3′-UTR sequences. This is an unusual gene structure, since in most genes the termination codon lies within the 3′-terminal exon (27). In any event, since exons 8 to 10 encode potential 3′-UTR sequences, alternative pre-mRNA splicing events involving these exons could alter the sequence composition of the 3′-UTR. This could have significant regulatory consequences, since 3′-UTR elements can control gene expression at several levels, including translation, mRNA stability, and subcellular mRNA localization (reviewed in reference 13).
For this reason, the focus of this study was twofold: first, to test the hypothesis that alternative pre-mRNA splicing generates AUF1 mRNAs with a variety of 3′-UTR structures; and second, to examine the effects of specific AUF1 3′-UTR structures on gene expression. We demonstrate that alternative pre-mRNA splicing does control the expression of highly conserved 3′-UTR elements (exon 9 and intron 9) in AUF1 mRNA. In addition, highly conserved intron sequences specifically associate with AUF1 protein in vitro and can participate in cis repression of gene expression in vivo. Further functional studies suggest that expression of a subset of AUF1 3′-UTR splice variants may also be regulated by the nonsense-mediated mRNA decay (NMD) pathway. Finally, we discuss potential roles for regulation of AUF1 expression involving elements in the 3′-UTR.
MATERIALS AND METHODS
PCR and reverse transcription-PCR (RT-PCR).
DNA fragments were amplified from plasmid or cosmid templates by PCR using Pfu DNA polymerase (Stratagene) according to the manufacturer’s instructions. In cases where restriction sites were incorporated into fragment ends, amplification cycles were as follows: 96°C for 2 min; 10 cycles of 96°C for 40 s, 52°C for 1 min, 72°C for 3 min; 20 cycles of 96°C for 40 s, 60°C for 1 min, 72°C for 3 min; and 72°C for 10 min. In all other cases, amplification was performed as follows: 96°C for 2 min, 30 cycles of (96°C for 40 s, 52°C for 1 min, 72°C for 3 min), 72°C for 10 min. Oligonucleotide primers (Operon) used for each amplification are indicated where necessary. Amplified fragments were purified using a High Pure PCR product purification kit (Boehringer Mannheim) prior to subcloning.
RT-PCR was performed with an Access RT-PCR kit (Promega) with the buffer provided by the manufacturer. Reaction mixtures were assembled in a final volume of 50 μl containing sense and antisense primers (50 pmol each), deoxynucleoside triphosphates (0.2 mM), MgSO4 (1 mM), K562 cell total RNA (0.5 μg), avian myeloblastosis virus reverse transcriptase (5 U), and Tfl DNA polymerase (5 U). The reverse transcription reaction was performed at 48°C for 45 min, and then the reverse transcriptase was inactivated by heating to 94°C for 2 min. Triton X-100 (Sigma) was added to a final concentration of 1% (vol/vol), and cDNA fragments were amplified as follows: 94°C for 2 min; 30 cycles of 94°C for 40 s, 53°C for 1 min, 68°C for 2 min; and 68°C for 8 min. For subcloning procedures, fragments generated by RT-PCR were purified as described above.
Southern blotting.
RT-PCR products were fractionated on 1.2% agarose gels and transferred to nylon membranes by capillary blotting in 10× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium acetate [pH 7.0]) as described previously (39). Blots were prehybridized at 47°C for 4 h in 6× SSC containing polyvinylpyrrolidone (0.2%), Ficoll (0.2%), pyrophosphate (0.1%), and sodium dodecyl sulfate (0.2%). Oligonucleotide probes were radiolabeled with [γ-32P]ATP (4,500 Ci/mmol; ICN) by using T4 polynucleotide kinase, injected directly into the hybridization solution, and incubated for 16 to 24 h at 47°C. Blots were then washed four times for 10 min each with 6× SSC–0.2% sodium dodecyl sulfate at 47°C. Hybridized fragments were visualized by autoradiography.
Construction of plasmids.
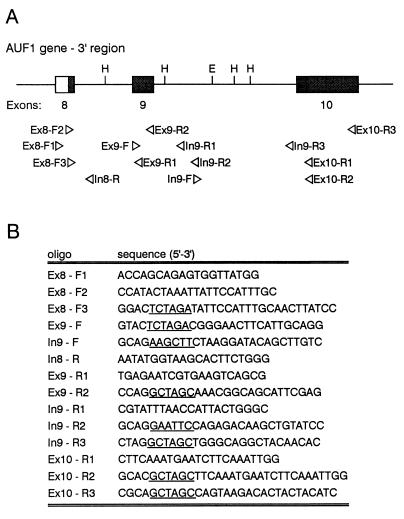
Standard subcloning procedures were used to generate all recombinant plasmids (39). Automated sequencing was performed on all PCR-generated inserts to verify fidelity. A 1.2-kb fragment spanning most of the 3′-UTR of the AUF1 gene was amplified from cosmid 10A (44) by PCR from primers Ex8-F2 and Ex10-R1 (Fig. 1). This fragment was subcloned into the SmaI site of pGEM7Z(+) (Promega) to generate pG7(+)In8-10. This plasmid was digested with HindIII, and a 302-nucleotide (nt) fragment containing exon 9 and some flanking intron sequence was purified and subcloned into the HindIII site of pGEM7Z(+). Linearization of this construct, termed pG7(+)RPA-A, with BamHI produced the template for riboprobe A. The template for riboprobe B was amplified by RT-PCR from K562 cell total RNA by using primers Ex8-F1 and Ex10-R1. The primary amplified product of 139 bp contained only sequences from exons 8 and 10 and was subcloned into the SmaI site of pGEM7Z(+). This plasmid was linearized by digestion with HindIII to generate the riboprobe B template. A 320-bp fragment was amplified from cosmid 10A by using primers Ex9-F and In9-R2. After digestion with HindIII plus EcoRI, a 151-bp fragment was subcloned into the HindIII-EcoRI sites of pGEM7Z(+). The riboprobe In9-C template was generated by digestion of this plasmid with EcoRI. A 570-bp fragment was amplified from cosmid 10A by using primers In9-F and Ex10-R1. Digestion with EcoRI plus HindIII generated a 130-bp fragment that was subcloned into the HindIII-EcoRI sites of pGEM7Z(+). The template for riboprobe In9-D was prepared by digesting this plasmid with EcoRI. The construction of templates used for generation of the fos ARE and β-globin riboprobes was described earlier (51).
FIG. 1.
Oligonucleotides used in this study. (A) Schematic of the 3′ end of the AUF1 gene. Untranslated exon sequences are shaded; restriction endonuclease cleavage sites used in subsequent plasmid constructions (H, HindIII; E, EcoRI) are shown. Below, the location and orientation of each oligonucleotide is noted. (B) Sequence of each oligonucleotide. Incorporated restriction sites are underlined.
All AUF1 3′-UTR fragments linked to the coding region of luciferase cDNA were subcloned into the XbaI site of pGL3-Promoter (Promega). The insert designated Ex8:Ex10 was generated by RT-PCR from K562 cell total RNA by using primers Ex8-F3 and Ex10-R3. This construct lacks any insertions between exons 8 and 10. The Ex9 insert was amplified by PCR from pG7(+)In8-10 by using primers Ex9-F and Ex9-R2. Inserts Ex9:In9:Ex10 and Ex9:In9 were similarly amplified by using primer sets Ex9-F, Ex10-R2 and Ex9-F, In9-R3, respectively. The AUF1-binding site (ABS) insert was generated as a 248-bp HindIII-EcoRI fragment from pG7(+)In8-10. Fragment ends were blunted with Klenow enzyme followed by ligation of XbaI linkers (Promega) and subcloning into pGL3-Promoter. To generate an AUF1 3′-UTR template lacking the ABS, a 490-bp fragment containing the 3′ half of intron 9 and the 5′ 40 nt of exon 10 was isolated from pG7(+)In8-10 with EcoRI and was subcloned into the EcoRI site of pG7(+)RPA-A, generating pG7(+)Ex9-10ΔABS. This plasmid served as the template for amplification of inserts Ex9:In9:Ex10ΔABS and Ex9:In9ΔABS from primer sets Ex9-F, Ex10-R2 and Ex9-F, In9-R3, respectively.
Cell culture, transfections, and luciferase assays.
K562 erythroleukemia cells were maintained in RPMI 1640 medium containing 10% newborn calf serum (Gibco/BRL). HeLa cells were grown in Dulbecco modified Eagle medium containing 10% fetal calf serum (HyClone). For transient transfections of HeLa cells, 1 × 105 to 2 × 105 cells were seeded into each well of a six-well tissue culture plate 24 h prior to transfection. Plasmids used in transfection experiments were prepared by using a Qiagen Plasmid Maxi kit and were judged to be >95% supercoiled by agarose gel electrophoresis. Expression vectors containing luciferase–AUF1 3′-UTR chimeras (1 μg/well) were introduced along with control vector pRL-SV40 (Promega) (10 ng/well) into cells, using Lipofectin reagent (Gibco/BRL) according to the manufacturer’s instructions and a 5-h incubation time. Cells were then washed briefly with OPTI-MEM before addition of Dulbecco modified Eagle medium–10% fetal calf serum. After 24 h, cells were harvested and assayed for both firefly and Renilla luciferase activities, using a Dual-Luciferase assay kit (Promega) as described by the manufacturer and measured with a model TD-20/20 luminometer (Turner Designs). All transfections were performed in triplicate, and data were analyzed by normalizing firefly luciferase activity to Renilla luciferase activity for each sample. Data sets were compared by the unpaired Student’s t test, and significant differences were considered to be those with a P of <0.05.
Synthesis of riboprobes.
Riboprobes used in RNase protection assays (RPAs) and gel mobility shift assays were generated by in vitro runoff transcription using T3, T7, or SP6 RNA polymerase (Promega) incorporating [α-32P]UTP (ICN). Following digestion of linearized DNA templates with RQ1 DNase (Promega), radiolabeled probes were purified by duplicate extractions with phenol-chloroform. Unincorporated nucleotides were removed from the preparation by spin column chromatography through RNase-free G-50 Quick Spin columns (Boehringer Mannheim). Riboprobe yields were determined by liquid scintillation counting, and probe integrity was monitored by denaturing polyacrylamide gel electrophoresis.
Preparation of cellular RNA and RPAs.
Total RNA was purified from K562 cells by using TRIzol reagent (Gibco/BRL) according to the manufacturer’s instructions. Cytoplasmic and nuclear fractions were obtained by resuspension of cell pellets in buffer A (10 mM Tris-HCl [pH 7.5], 1 mM potassium acetate, 1.5 mM magnesium acetate, 2 mM dithiothreitol)–0.5% Nonidet P-40 (NP-40) at 5 × 107 cells/ml. Lysis was performed with five strokes of a loose-fitting Dounce homogenizer and verified by phase-contrast microscopy. Nuclei were pelleted by centrifugation at 1,000 × g for 5 min and were lysed directly in TRIzol reagent. Cytoplasmic RNA was recovered by precipitation with 1 volume (vol/vol) of isopropanol, then solubilized in TRIzol, and purified according to the manufacturer’s instructions. All TRIzol lysates were sheared 5 to 10 times with a 25-gauge needle prior to chloroform extraction.
For purification of K562 polyribosomal and cytosolic RNA fractions, postnuclear supernatants were prepared as described above in buffer A lacking NP-40 and then divided into two equal portions. To one aliquot, EDTA (20 mM) was added to disrupt polysomes. Samples were then layered on 30% (wt/vol) sucrose pads (with or without EDTA) and centrifuged at 130,000 × g in an SW50.1 rotor (Beckman) for 2.5 h at 0°C. Pelleted material (polyribosomal fraction) was solubilized directly in TRIzol reagent. Material remaining above the sucrose pad (cytosol) was precipitated with 1 volume (vol/vol) of isopropanol, followed by solubilization in TRIzol. RNA was then purified from each fraction as described above.
Quantitation and mapping of cellular mRNAs was achieved by using RPAs with RNases P1 and T1 as described previously (7). 32P-labeled antisense riboprobes were synthesized as described above to specific activities of 1 × 104 to 2 × 104 cpm/fmol. Protected RNA fragments were fractionated by denaturing gel electrophoresis and were visualized with a PhosphorImager (Molecular Dynamics).
Detection and quantitation of luciferase and luciferase–AUF1 3′-UTR chimeric mRNAs in transfected HeLa cells.
HeLa cells (106) seeded in 60-mm-diameter dishes were transfected as described above with expression vectors containing luciferase–AUF1 3′-UTR chimeras (5 μg/plate) along with the vector pRL-SV40 (2 μg/plate) as an internal control for transfection efficiency. For mock transfections, heat-denatured calf thymus DNA was substituted for plasmid DNA. After 24 h, cells were washed twice with phosphate-buffered saline and scraped from plates in 0.5 ml of buffer A–0.5% NP-40. Cytoplasmic RNA was purified as described above, fractionated on 1.2% formaldehyde agarose gels (5 μg/lane), and transferred to nylon membranes by capillary blotting in 10× SSC and UV cross-linking (Stratalinker; Stratagene). RNA blots were probed with the 130-bp XbaI-HpaII fragment of pGL3-Promoter containing sequences immediately upstream of the simian virus 40 late polyadenylation/termination signal common to both the firefly (pGL3 series) and Renilla (pRL-SV40) expression cassettes. The gel-purified DNA fragment was radiolabeled by random priming incorporating [α-32P]dCTP, using a Prime-It random priming kit (Stratagene). Hybridization was performed as described elsewhere (48) with luciferase and luciferase–AUF1 3′-UTR chimeric mRNAs detected by PhosphorImager scan.
Gel mobility shift assays.
The prokaryotic expression vector pTrcHisB-p37AUF1[1-257] encodes an N-terminal His6-tagged deletion mutant of p37AUF1 lacking 29 amino acid residues from the C terminus. Construction of this plasmid was described previously (15). The recombinant His6-p37AUF1[1-257] polypeptide exhibits ARE-binding activity similar to that of wild-type p37AUF1 (15), but it is more stable and is produced with greater yields than the full-length protein (46). Recombinant His6-p37AUF1[1-257] was purified from isopropyl-β-d-thiogalactopyranoside (1 mM)-induced Escherichia coli TOP10 cells by Ni2+ affinity chromatography using the Xpress system (InVitrogen) under native conditions as described previously (31). RNA-protein binding reactions were assembled with various amounts of purified His6-p37AUF1[1-257] in a 10-μl final volume with 32P-labeled RNA (1 fmol ≈ 1 × 104 to 2 × 104 cpm), Tris-HCl (pH 7.5) (10 mM), magnesium acetate (5 mM), potassium acetate (100 mM), dithiothreitol (2 mM), spermine (0.1 mM), acetylated bovine serum albumin (0.1 μg/μl), RNasin (8 U), and a mixture of nonspecific competitors [yeast tRNA (0.2 μg/μl), heparin (5 μg/μl), and poly(C) (0.1 μg/μl)]. Reactions were incubated on ice for 10 min before fractionation on 4% (40:1 acrylamide-bis acrylamide) gels in 0.5× TBE (1× TBE is 90 mM Tris-borate [pH 8.3] plus 2 mM EDTA) at 13 V/cm for 2.5 h. Gels were prerun for 30 min prior to loading. After electrophoresis, the gels were dried and visualized with a PhosphorImager (Molecular Dynamics).
RESULTS
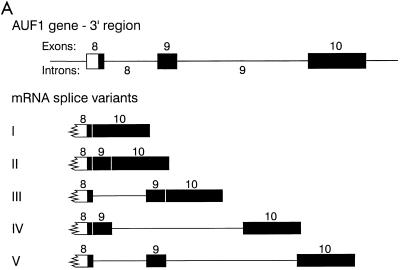
Four splice variants of the AUF1 3′-UTR are detectable in K562 cells.
Five potential splice variants of the AUF1 3′-UTR are predicted based on the organization of the 3′ region of the AUF1 gene (Fig. 2A). Variant V represents the unspliced 3′-UTR, while generation of variant I requires that intron 8, exon 9, and intron 9 be removed in a single splicing event. The other variants (II, III, and IV) are predicted based on the selective loss of intron 8 and/or intron 9.
FIG. 2.
Splicing variants of the AUF1 3′-UTR. (A) Potential 3′-UTR splice variants based on exon-intron organization of the AUF1 gene. (B) RT-PCR was performed from K562 cell total RNA as described in Materials and Methods. Oligonucleotide Ex8-F1 was used as the forward primer for each reaction, with reverse primers shown. Southern blots of RT-PCR products were probed with 32P-labeled oligonucleotides specific for exon 8 (Ex8-F2) or exon 9 (Ex9-F). A longer exposure of lane 8 yielded the signal depicted in lane 9. Potential AUF1 3′-UTR splice variants encoding hybridized fragments are indicated to the right of each band.
Using total RNA isolated from K562 cells, we performed RT-PCR and Southern blotting to detect AUF1 3′-UTR splice variants (Fig. 2B). Lane 1 shows the fragment amplified from primers in exon 8 (forward) and intron 8 (reverse). As expected, a single product of 217 bp which hybridized to an internal oligonucleotide probe from exon 8 (lane 1) but not to an exon 9 probe (lane 5) was generated. cDNAs amplified from a reverse primer in exon 9 identified AUF1 transcripts containing or lacking intron 8 (lane 2; cf. 448- and 146-bp products). These results are consistent with amplification from splice variants III or V and splice variants II or IV. Similar results were observed when a reverse primer from intron 9 (lane 3; cf. 654- and 352-bp products) was used. Retention of exon 9 sequence in each amplified fragment was verified by Southern hybridization (cf. lanes 2 and 6 and lanes 3 and 7). The 654-bp product primed from intron 9 (lane 3) thus corresponds to the AUF1 pre-mRNA (variant V), while the 352-bp fragment identifies variant IV. This observation was further confirmed by the selective hybridization of an intron 8 probe to the 654-bp fragment (data not shown).
Reverse transcription from exon 10 resulted in the predominant amplification of a 139-bp product lacking exon 9 sequences (cf. lanes 4 and lane 8), consistent with variant I. However, hybridization of an exon 9 probe to these products also identified a 246-bp product corresponding to variant II (lane 9). Although all AUF1 3′-UTR splice variants are targets for amplification using the exon 10 reverse primer, only variants I and II were observed. Two factors likely contributed to this observation. First, levels of AUF1 mRNA variant I are significantly higher in K562 cells than are levels of variants II to V (Fig. 3B). Second, cDNA fragments containing intron 9 sequence are amplified with relatively poor efficiency (Fig. 2B; cf. lanes 2 and 6 with lanes 3 and 7). As a result, fragments corresponding to variants IV and V coamplifying with variant I were not detected. However, taken together with amplification reactions containing intron 9-specific reverse primers, these experiments unambiguously identified AUF1 3′-UTR splice variants I, II, IV, and V in K562 cells.
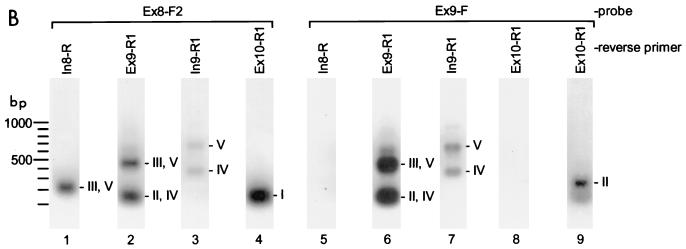
FIG. 3.
Detection of AUF1 3′-UTR splicing variants by RPA. (A) 32P-labeled riboprobe A was generated as described in Materials and Methods. The position of the riboprobe is shown along with the predicted sizes of RNA fragments protected by hybridization with each AUF1 3′-UTR splice variant (left). Riboprobe A (5 fmol) was used to program RPA reactions (right) containing 20 μg of total RNA from K562 cells (whole cell) or an equal mass of yeast tRNA. In experiments with subcellular fractionated RNA, 20 μg of cytoplasmic RNA was assayed pairwise with equal cellular equivalents of nuclear RNA (≈15 μg). A lane containing undigested probe (0.1 fmol) was also included to verify probe excess and to ensure that sample digests were complete. (B) Similar analyses performed with 32P-labeled riboprobe B to discriminate AUF1 mRNAs containing fully spliced 3′-UTRs (variant I) from those containing inserts in this region. An estimate of the relative levels of variant I versus variants II to V was calculated by PhosphorImager analysis of the 140- and 100-nt bands, respectively, and normalization to the number of uridylate residues protected in each fragment (see text).
A population of AUF1 mRNA containing exon 9 and intron 9 accumulates and may be translated.
RPAs were used to further examine the distribution of AUF1 3′-UTR splice variants in K562 cells. First, an antisense riboprobe spanning exon 9 with flanking sequences from introns 8 and 9 was used to identify variants II, III, IV, and V in K562 total RNA or RNA purified from K562 nuclei or cytoplasm (Fig. 3A). Protected fragments corresponding to splice variants IV and V were detected in assays with K562 cell RNA, while variants II and III were not detectable above background signals by this technique. Variant IV was most abundantly detected in these assays, with >95% localized to the nucleus. However, variant IV was also readily detected in the cytoplasm. Similar experiments using poly(A)-selected RNA demonstrated that splice variants IV and V were polyadenylated in these cells (data not shown).
RPAs were also performed with a second riboprobe spanning exon 8 and the 5′ end of exon 10, allowing discrimination of AUF1 mRNAs containing or lacking inserts in the 3′-UTR (Fig. 3B). The relative levels of AUF1 transcripts containing or lacking 3′-UTR inserts were then determined by quantitation of the 100- and 140-nt protected fragments, respectively, using a PhosphorImager (Molecular Dynamics). Measured signals were corrected for background and normalized to the number of radiolabeled (uridylate) residues retained in each fragment. By this analysis, approximately 25% of AUF1 mRNA in K562 cell nuclei was observed to contain some inserted 3′-UTR sequence (corresponding to variants II to V), while variant I represented >95% of the AUF1 mRNA detected in cytoplasm. These data indicate that while the predominant form of AUF1 mRNA is fully spliced (i.e., lacks exon 9) in K562 cells, a population of polyadenylated AUF1 mRNA containing exon 9 and intron 9 (variant IV) does accumulate, although largely in the nucleus.
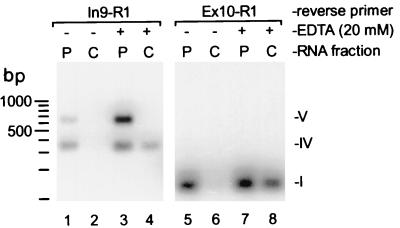
To rule out the possibility that variant IV mRNA recovered in K562 cytoplasm was the result of nuclear leakage during subcellular fractionation, we examined whether this variant was localized to polysomes. This was achieved by separating polysomes from cytosol by ultracentrifugation of cytoplasm through a 30% sucrose cushion at 130,000 × g. RNA was purified and analyzed by RT-PCR and Southern blotting. Variant IV and V mRNAs were largely detected in the polyribosomal pellet of fractionated K562 cytoplasm (Fig. 4; cf. lanes 1 and 2). Treatment of crude cytoplasm with EDTA (20 mM) prior to ultracentrifugation released a portion of variant IV mRNA from polysomes; variant V was not released (cf. lanes 3 and 4). Since release of mRNPs from polysomes in the presence of EDTA is consistent with their active translation (2, 40), these results suggest that cytoplasmic variant IV mRNA is exported to the cytoplasm, associates with polysomes, and may be translated. While variant V mRNA was also detected in the cytoplasmic 130,000 × g pellet, its retention in the presence of EDTA raises the possibility that it is contained within a translationally inactive complex (28) or that it may localize to an EDTA-insensitive structure such as the cytoskeleton (20, 22) or membranous components (29). As a control, parallel reactions amplifying a fragment of the fully spliced AUF1 3′-UTR (variant I) were also assembled. As expected, cytoplasmic variant I mRNA was most abundantly detected in the polysomal RNA pellet and was partially released into the postpolysomal cytosol by treatment with EDTA, consistent with a portion being translated (Fig. 4, lanes 5 to 8). These results indicate that variant IV may be a biologically active molecule in the cytoplasm (i.e., is translated) and that it is not solely a pre-mRNA processing intermediate.
FIG. 4.
Detection of AUF1 3′-UTR splice variants in polysomes. Fragments of AUF1 3′-UTR splice variants were amplified by RT-PCR from polysomal (P) or postpolysomal cytosolic (C) RNA fractions prepared with or without EDTA (20 mM) from K562 cell cytoplasm as described in Materials and Methods. Amplification reactions were programmed with equal cellular equivalents of RNA from each fraction. Oligonucleotide Ex8-F1 was used as the forward primer for each reaction, with reverse primers shown. Products from one-fifth of each reaction were fractionated by agarose gel electrophoresis and Southern blotted. Amplified fragments were detected by probing with 32P-labeled oligonucleotide Ex8-F2. The locations of fragments corresponding to selected 3′-UTR splice variants are indicated.
Exon 9 and the 5′ half of intron 9 are highly conserved.
Since variant IV is likely translated, we examined the sequence of the AUF1 3′-UTR inserts for determinants of possible function and to establish their likely importance by comparison to the corresponding murine sequences. Sequencing of the human AUF1 locus between exons 8 and 10 revealed a remarkable degree of conservation between the human and murine sequences (Fig. 5). In particular, exon 9 is 100% conserved between these species, while the 5′ half of intron 9 is also highly conserved (99%). In the AUF1 coding sequence, only exon 3, encoding the N-terminal RNA recognition motif, shows comparable sequence identity (99%); the remaining exons of the AUF1 locus are 88 to 97% conserved between humans and mice (references 17 and 44 and data not shown).
FIG. 5.
Sequence identity between human and murine AUF1 3′-UTR inserts. A fragment of the human AUF1 gene spanning intron 8, exon 9, and intron 9 was amplified from cosmid 10A (44) by using oligonucleotide primers Ex8-F2 and Ex10-R1 to generate plasmid pG7(+)In8-10 as described in Materials and Methods. Sequence was obtained on both strands of this insert by automated sequencing and showed only minor variations from an archived sequence (16) (GenBank accession no. AF026126). This sequence (HS [Homo sapiens]) was then compared by using NALIGN (PC/GENE; Intelligenetics) to the murine (MM [Mus musculus]) 3′-UTR insert (24), with sequence modifications based on murine EST submissions (25, 26) identified by BLAST homology search (3). Identical nucleotides are indicated with vertical lines. Exon 9 sequences are in boldface. Conserved intron 9 sequences similar to AREs are boxed. The 5′ and 3′ limits of riboprobes In9-C and In9-D (Fig. 6) are indicated by arrowheads above the corresponding human sequence.
This extraordinary conservation of homology between mouse and human AUF1 sequences downstream of the translational termination codon suggests that exon 9 and intron 9 may contribute some essential regulatory function. For example, conserved A+U-rich sequences in the 5′ half of intron 9 display significant similarity to the AREs present in some labile mRNAs (Fig. 5, boxed regions). Since AREs are well-characterized determinants of rapid cytoplasmic mRNA decay, their presence in intron 9 may contribute to the low steady-state levels of variant IV and V mRNAs observed in K562 cytoplasm (Fig. 3). Furthermore, the high binding affinity of AUF1 proteins for AREs (14, 44) raises the possibility that AUF1 associates with these elements in its own pre-mRNA, leading to the potential for autoregulated expression of this protein.
AUF1 binds intron 9 A+U-rich sequences in vitro.
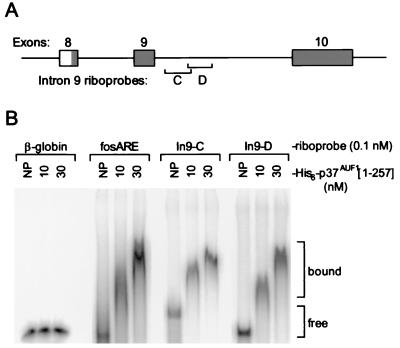
To test whether AUF1 protein could bind sequences near the 5′ end of intron 9, two riboprobes which spanned this region were synthesized (Fig. 6A, In9-C and D; see also Fig. 5). In gel mobility shift assays, specific binding events between recombinant AUF1 and both of these riboprobes were observed (Fig. 6B). Similar binding to the ARE from c-fos mRNA, a high-affinity ABS (14), was observed, while no binding to a sequence from β-globin mRNA was detected. In addition, minimal binding activity was observed with riboprobes spanning other regions of the AUF1 3′-UTR (data not shown).
FIG. 6.
Association of recombinant AUF1 with A+U-rich sequences in intron 9. (A) Schematic showing the relative positions of intron 9 sense riboprobes In9-C and In9-D. Riboprobe In9-C spans 151 nt from bases 464 to 614 (Fig. 5, human sequence). Riboprobe In9-D is 130 nt in length and corresponds to the sequence from bases 588 to 717 (Fig. 5, human sequence). (B) Riboprobe binding to recombinant AUF1 was monitored by gel mobility shift assay either without added AUF1 (NP) or in the presence of 10 or 30 nM His6-p37AUF1[1-257] as described in Materials and Methods. Identical reactions using fos ARE and β-globin riboprobes as positive and negative controls, respectively, were assembled. The positions of free and bound riboprobes are indicated.
In a previous study, binding of His6-p37AUF1[1-257] to the fos ARE with a Kd of 5.3 nM was observed (15). Thus, complete association of the fos ARE, In9-C, and In9-D riboprobes with 10 nM His6-p37AUF1[1-257] (Fig. 6B) indicates that AUF1 binds the intron 9 riboprobes with affinities comparable to those of potent A+U-rich mRNA-destabilizing sequences, which typically exhibit AUF1 binding with a Kd of <50 nM (14). Since AUF1 may participate in the rapid turnover of many labile mRNAs, these data suggest that the expression of AUF1 may also be modulated by autoregulatory mRNA decay events. Accordingly, functional analyses of these alternatively spliced 3′-UTR sequences were performed to establish their potential for regulating gene expression in cis.
Two mechanisms involving the AUF1 3′-UTR repress gene expression in cis.
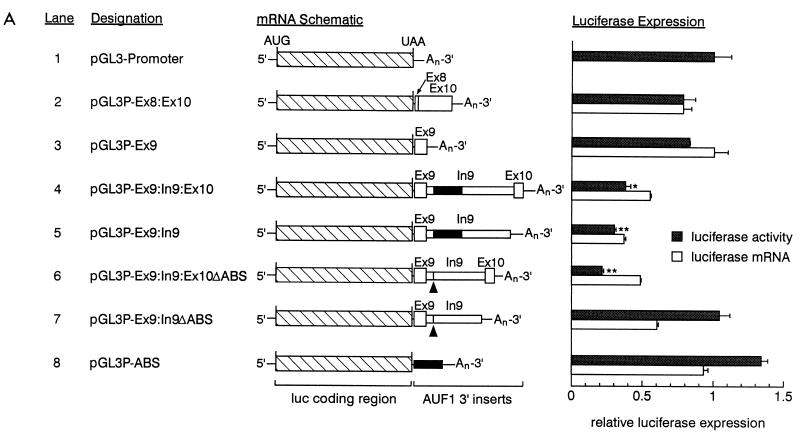
A series of expression constructs encoding chimeric mRNAs was constructed such that the coding sequence of firefly luciferase was linked to various regions of the AUF1 3′-UTR (Fig. 7A). The relative firefly luciferase activity and mRNA expressed from each construct was determined by transient transfection into HeLa cells and was normalized to expression from the internal control plasmid pRL-SV40, encoding Renilla luciferase. The effects of inserted 3′ sequences on reporter expression were evaluated in comparison to expression from pGL3-Promoter, which lacks any AUF1 3′-UTR inserts (lane 1).
FIG. 7.
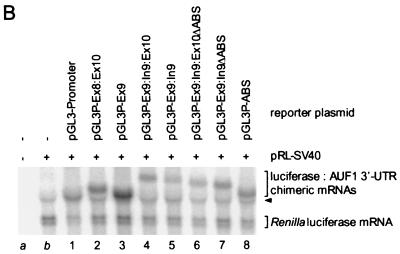
Identification of cis-acting regulatory elements in AUF1 3′-UTR splice variants. (A) A series of luciferase–AUF1 3′-UTR chimeric constructs was assembled as described in Materials and Methods. A schematic of each chimeric mRNA is shown along with a lane number for text reference (left). Exon and intron sequences are labeled, and the ABSs in intron 9 are denoted by black boxes. In lanes 6 and 7, the locations of the deleted ABSs are indicated with arrowheads. Transfection into HeLa cells and analyses of luciferase activities and mRNA levels were performed for each plasmid as described in Materials and Methods. Normalized firefly luciferase activity from each construct (shaded bars) is presented relative to the expression from pGL3-Promoter (lane 1) and represents the mean + standard deviation of triplicate transfections. (∗, P < 0.01; ∗∗, P < 0.005 relative to pGL3-Promoter). Levels of cytoplasmic firefly luciferase and luciferase–AUF1 3′-UTR chimeric mRNAs normalized to Renilla luciferase mRNA (open bars) represent the mean + spread of duplicate samples. Since firefly luciferase mRNA (lane 1) comigrated with an extended transcript generated from the pRL-SV40 expression cassette, accurate quantitation of this mRNA was not possible. Accordingly, all mRNA levels are expressed relative to the mRNA encoded by pGL3P-Ex8:Ex10. (B) Representative blot of cytoplasmic RNA from transiently transfected HeLa cells performed as described in Materials and Methods. RNA was purified from the cytoplasm of mock-transfected cells (lane a), cells transfected with the control Renilla vector pRL-SV40 alone (lane b), or cells cotransfected with pRL-SV40 and a luciferase–AUF1 3′-UTR chimera (lanes 1 to 8). Positions of luciferase–AUF1 3′-UTR chimeric transcripts and the Renilla luciferase mRNA are bracketed; the extended transcript generated from pRL-SV40 is indicated by the arrowhead.
The Ex8:Ex10 insert consists of the fully spliced AUF1 3′-UTR sequence (variant I) between the translational termination codon and the polyadenylation signal. Neither this sequence nor the completely conserved exon 9 fragment (Ex9) contributed to any significant change in reporter gene expression at the level of luciferase activity or mRNA (cf. lanes 1, 2, and 3). Inclusion of intron 9 sequences, however, either with (lane 4) or without (lane 5) a fragment of exon 10, resulted in a 60 to 70% decrease in luciferase activity, indicating the presence of elements contributing to repression of gene expression in cis. The decreases in mRNA levels for these luciferase–AUF1 3′-UTR chimeras were consistent with the changes in luciferase activity. Addition of the 5′ 40 nt of exon 10 in Ex9:In9:Ex10 (lane 4) allowed the intron 9 sequence to be contained within a spliceable unit. By contrast, removal of the 3′ 30 nt of intron 9 in Ex9:In9 (lane 5) prevented excision of this sequence by the pre-mRNA processing machinery. While similar effects on luciferase expression were observed with each construct, the potential for splicing of intron 9 from the Ex9:In9:Ex10 insert suggested that elements in addition to intron 9 may exert a negative influence on reporter gene expression (see Discussion).
Since the ABS are localized to the 5′ half of intron 9, additional plasmids were constructed in which the ABS were deleted from the expression cassettes. In the case where intron 9 could not be excised, removal of the ABS completely abrogated cis repression of luciferase activity (cf. lanes 7 and 5). However, mRNA levels were only partially restored by deletion of the ABS from this construct, raising the possibility that translational effects may also be involved. Nevertheless, these data demonstrated that sequences binding AUF1 are required for down-regulation of expression involving intron 9. However, fusion of the ABS alone to the luciferase coding region was ineffective in repressing reporter expression (cf. lanes 8 and 1). This result indicated that while the ABSs are necessary for cis repression when intron 9 is present, they are not sufficient when removed from the context of intron 9.
In the case where intron 9 could be removed by a splicing event, deletion of the ABS did not alleviate the repression of reporter expression at the level of either mRNA or luciferase activity (cf. lanes 6 and 4). These data suggest that a second regulatory mechanism, perhaps independent of AUF1 binding, also functions to repress gene expression in this system. In this case, retention of intron 9 was not required, yet sequences from exon 9 (lane 3) and exon 10 (lane 2) showed no independent cis-acting effects on reporter activity. Based on these observations, we conclude that the down-regulation of luciferase expression observed with the Ex9:In9:Ex10 (lane 4) and Ex9:In9:Ex10ΔABS (lane 6) constructs is likely linked to the splicing of intron 9. Taken together, these data suggest a model for controlling the levels of AUF1 mRNA 3′-UTR splice variants based on the parallel actions of two regulatory pathways (see Discussion).
DISCUSSION
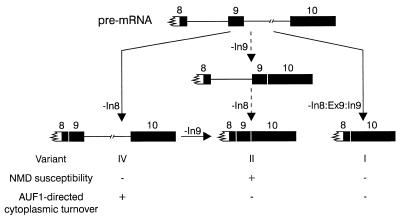
The organization of the 3′ region of the AUF1 locus presents several possible regulatory events leading to the accumulation of splice variants either containing or lacking exon 9 (Fig. 8). The initial splicing event within the 3′-UTR serves to commit the transcript to either (i) removal of exon 9 by excision of intron 8, exon 9, and intron 9 as a single unit or (ii) retention of exon 9 by splicing intron 8 and/or intron 9 individually. In K562 cells, the majority of AUF1 mRNA contained a fully spliced 3′-UTR (variant I), indicating that significant quantities of AUF1 mRNA containing inserted 3′-UTR sequences were not required in these cells. However, a splicing variant containing exon 9 and intron 9 (variant IV) was also observed, suggesting that the processing pathway leading to formation of an exon 9-containing transcript remained active. Since this variant is incapable of splicing exon 9, it cannot serve as a pre-mRNA splicing intermediate in the generation of the fully spliced 3′-UTR (variant I). However, poor detection of the subsequent intron 9-spliced transcript retaining exon 9 (variant II) suggests that its accumulation is repressed either by inhibition of intron 9 removal or by rapid turnover of the product mRNA.
FIG. 8.
Putative modulation of AUF1 3′-UTR splice variants through regulated splicing or RNA turnover rates. A schematic of the 3′ end of the AUF1 pre-mRNA is shown (top) with the splicing pathways necessary to generate each possible splicing variant. Exon sequences downstream of the translation termination codon in exon 8 are shaded. Potential regulatory events modulating the levels of splice variants I, II, and IV are indicated and further described in the text. The arrows flanking the intron 9-excised mRNA intermediate are dashed (right) because this splice variant (III) was not observed in K562 cells.
The functional analyses of luciferase–AUF1 3′-UTR chimeric transcripts in HeLa cells indicated that elements within the AUF1 3′-UTR appear to repress gene expression by at least two distinct mechanisms. In the absence of intron 9 splicing, inhibition of luciferase expression was dependent on the presence of ABS in intron 9. The presence of high-affinity ABS coupled with active translation of the mRNA is characteristic of ARE-directed cytoplasmic mRNA turnover (1, 12, 40). This is also consistent with the low steady-state levels of variant IV mRNA detected in K562 cytoplasm relative to nuclear RNA fractions. By contrast, expression from constructs capable of intron 9 splicing was repressed regardless of the presence of AUF1-binding sequences. These observations raise the possibility that some negative regulatory event may also occur concomitant with or as a result of intron 9 excision.
Recent data indicate that in mammalian cells an exon-exon junction located more than 50 nt downstream of a translational termination codon may function to promote mRNA turnover by the NMD pathway (27, 43). NMD, which has been extensively investigated in the yeast Saccharomyces cerevisiae, is characterized by the presence of a downstream element located 3′ of a nonsense codon (30, 37) interacting with several essential trans-acting factors (11, 19, 30). In higher eukaryotes, untranslated exon-exon junctions located more than 50 nt downstream of termination codons may also be recognized as downstream elements (43, 49, 50), possibly through retention of selected components of the splicing machinery. In the case of AUF1, splicing of any 3′-UTR sequence from the pre-mRNA generates an exon-exon junction downstream of the termination codon (Fig. 8). However, in the two splice variants predominantly detected in K562 cells (I and IV), this junction is located only 30 nt downstream of the termination codon and would therefore not be a likely target for NMD by this model. Conversely, variants lacking intron 8 (II and III) contain exon-exon junctions well beyond the 50-nt threshold. Consistent with the NMD hypothesis, variant II is barely detectable in K562 cells, while variant III has not been observed. Furthermore, the exon-exon junctions generated by splicing of intron 9 from luciferase–AUF1 fusion mRNAs Ex9:In9:Ex10 and Ex9:In9:Ex10ΔABS were located 120 nt downstream of the stop codon and were also accompanied by repression of reporter expression. Further experimentation will be necessary, however, to verify whether NMD is involved in the regulated expression of endogenous AUF1 transcripts.
The strong sequence identity between murine and human AUF1 in exon 9 and intron 9 implies that these sequences contribute some important function. In K562 cells, the expression of AUF1 transcripts containing exon 9 is repressed, possibly involving the mechanisms mentioned above. Under some circumstances, however, it seems likely that inclusion of exon 9 would be required in vivo. Several observations indicate that exon 9 may be retained for some developmental function. First, BLAST homology searches (3) identified expressed sequence tag (EST) submissions analogous to AUF1 variant II transcripts from both human (21) and murine (26) fetal sources. Comparable sequences were not found among ESTs derived from adult sources. Second, studies of two other genes capable of generating exon-exon junctions more than 50 nt downstream of the translational termination codon indicate rigid temporal and tissue-specific regulation of their expression. The chicken homeobox gene Gnot1 is expressed in a position- and cell fate-dependent manner early during chick development and encodes a transcript containing a single 3′-UTR intron 58 nt downstream of the termination codon (33). Accumulation of Gnot1 mRNA coincides with establishment of the embryonic body axis and is largely (≈80%) accompanied by retention of the 3′-UTR intron (23). As for AUF1 variant IV mRNA, retention of the most 3′ intron sequence during Gnot1 mRNA accumulation would likely exempt this transcript from NMD by the above model. The second example of a gene encoding exon-exon junctions >50 nt 3′ of the termination codon is human HLA 6.0, which contains two introns in its 3′-UTR (18). Expression of HLA 6.0 mRNA is atypical among the HLA genes in that it is restricted to tissues such as fetal eye and thymus; it is not detected in adult tissues (41). Mechanisms contributing to the developmental expression of HLA 6.0 mRNA are unknown. However, the expression of Gnot1 and HLA 6.0 provide examples where mRNAs capable of generating distal 3′-UTR exon-exon junctions may selectively accumulate during development.
The data presented in this work indicate that posttranscriptional regulatory events involving alternatively spliced elements in the AUF1 3′-UTR may contribute to the regulation of its expression. While a developmental role for modulation of AUF1 expression may be postulated based on parallels with other genes containing similar 3′-UTR features, as described above, restriction of AUF1 expression at multiple levels of regulatory control may also serve as a general protective mechanism for cell growth and function. Current evidence indicates that AUF1 proteins contribute to the turnover of many mRNAs, including several whose encoded products are responsible for control of cell growth and division. Examples of such transcripts are those encoding the oncoproteins c-Fos and c-Myc, whose overexpression may contribute to cell transformation (32, 34). As such, it is likely that the expression and/or activity of AUF1 is also tightly regulated, since decreases in AUF1 protein levels or activity may contribute to stabilization of ARE-containing transcripts. This has been observed in 5637 bladder carcinoma cells, where inhibition of ARE-directed mRNA turnover (8, 35) occurs concomitant with reduced ARE-binding activity of AUF1 (9). However, overexpression of AUF1 may also present deleterious effects. For example, in patients with congestive heart failure, a twofold increase in AUF1 mRNA and protein levels, relative to normal heart, leads to a twofold reduction in cardiac β1-AR mRNA and protein levels (31). The net result of this decrease in β-AR expression is desensitization of the β-AR/G protein/adenylyl cyclase signalling pathway necessary for maintenance of normal cardiac output. The pathological implications of dysregulated AUF1 expression thus raises the possibility that the repression of AUF1 expression involving 3′-UTR elements described in this work is indicative of some global mechanism restricting cellular AUF1 levels.
ACKNOWLEDGMENTS
We thank Doug Lyles for critical reading of the manuscript.
This work was supported by grant CA 52443 (National Institutes of Health) to G.B. Automated DNA sequencing was performed by Elyse Jung at the DNA Sequence and Gene Analysis Core Laboratory, Comprehensive Cancer Center, Wake Forest University, supported in part by grant P30 CA 12197 (National Institutes of Health).
REFERENCES
- 1.Aharon T, Schneider R J. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol Cell Biol. 1993;13:1971–1980. doi: 10.1128/mcb.13.3.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfonso-Pizzaro A, Carlson D P, Ross J. Subcellular localization of RNAs in transfected cells: role of sequences at the 5′ terminus. Nucleic Acids Res. 1984;12:8363–8380. doi: 10.1093/nar/12.22.8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atwater J A, Wisdom R, Verma I M. Regulated mRNA stability. Annu Rev Genet. 1990;24:519–541. doi: 10.1146/annurev.ge.24.120190.002511. [DOI] [PubMed] [Google Scholar]
- 5.Brewer G. An A+U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewer G. Post-transcriptional considerations of gene expression: translation, mRNA stability, and poly(A) processing. In: Wolff J A, editor. Gene therapeutics. Cambridge, Mass: Birkhäuser Boston; 1994. pp. 40–59. [Google Scholar]
- 7.Brewer G, Ross J. Messenger RNA turnover in cell-free extracts. Methods Enzymol. 1990;181:202–209. doi: 10.1016/0076-6879(90)81122-b. [DOI] [PubMed] [Google Scholar]
- 8.Brown C Y, Lagnado C A, Goodall G J. A cytokine mRNA-destabilizing element that is structurally and functionally distinct from A+U-rich elements. Proc Natl Acad Sci USA. 1996;93:13721–13725. doi: 10.1073/pnas.93.24.13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buzby J S, Lee S, van Winkle P, DeMaria C T, Brewer G, Cairo M S. Increased granulocyte-macrophage colony-stimulating factor mRNA instability in cord versus adult mononuclear cells is translation-dependent and associated with increased levels of A+U-rich element binding factor. Blood. 1996;88:2889–2897. [PubMed] [Google Scholar]
- 10.Chen C-Y A, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 11.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 12.Curatola A M, Nadal M S, Schneider R J. Rapid degradation of AU-rich element (ARE) mRNAs is activated by ribosome transit and blocked by secondary structure at any position 5′ to the ARE. Mol Cell Biol. 1995;15:6331–6340. doi: 10.1128/mcb.15.11.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decker C J, Parker R. Diversity of cytoplasmic functions for the 3′ region of eukaryotic transcripts. Curr Opin Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 14.DeMaria C T, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 15.DeMaria C T, Sun Y, Long L, Wagner B J, Brewer G. Structural determinants in AUF1 required for high affinity binding to A+U-rich elements. J Biol Chem. 1997;272:27635–27643. doi: 10.1074/jbc.272.44.27635. [DOI] [PubMed] [Google Scholar]
- 16.Dempsey L A, Li M, DePace A, Bray-Ward P, Maizels N. The human HNRPD locus maps to 4q21 and encodes a highly conserved protein. Genomics. 1998;49:378–384. doi: 10.1006/geno.1998.5237. [DOI] [PubMed] [Google Scholar]
- 17.Ehrenman K, Long L, Wagner B J, Brewer G. Characterization of cDNAs encoding the murine A+U-rich RNA-binding protein AUF1. Gene. 1994;149:315–319. doi: 10.1016/0378-1119(94)90168-6. [DOI] [PubMed] [Google Scholar]
- 18.Geraghty D E, Koller B H, Orr H T. A human major histocompatibility complex class I gene that encodes a protein with a shortened cytoplasmic segment. Proc Natl Acad Sci USA. 1987;84:9145–9149. doi: 10.1073/pnas.84.24.9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 20.Hesketh J E, Pryme I F. Interaction between mRNA, ribosomes and the cytoskeleton. Biochem J. 1991;277:1–10. doi: 10.1042/bj2770001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hillier, L., N. Clark, T. Dubuque, K. Elliston, M. Hawkins, M. Holman, M. Hultman, T. Kucaba, M. Le, G. Lennon, M. Marra, J. Parsons, L. Rifkin, T. Rohlfing, F. Tan, E. Trevaskis, R. Waterston, A. Williamson, P. Wohldmann, and R. Wilson. 1995. GenBank accession no. H49797.
- 22.Howe J G, Hershey J W B. Translational initiation factor and ribosome association with the cytoskeletal framework fraction from HeLa cells. Cell. 1984;37:85–93. doi: 10.1016/0092-8674(84)90303-9. [DOI] [PubMed] [Google Scholar]
- 23.Knezevic V, Ranson M, Mackem S. The organizer-associated chick homeobox gene, Gnot1, is expressed before gastrulation and regulated synergistically by activin and retinoic acid. Dev Biol. 1995;171:458–470. doi: 10.1006/dbio.1995.1296. [DOI] [PubMed] [Google Scholar]
- 24.Lafon I, Carballes F, Brewer G, Poiret M, Morello D. Developmental expression of AUF1 and HuR, two c-myc mRNA binding proteins. Oncogene. 1998;16:3413–3421. doi: 10.1038/sj.onc.1201895. [DOI] [PubMed] [Google Scholar]
- 25.Marra, M., L. Hillier, M. Allen, M. Bowles, N. Dietrich, T. Dubuque, S. Geisel, T. Kucaba, M. Lacy, M. Le, J. Martin, M. Morris, K. Schellenberg, M. Steptoe, F. Tan, K. Underwood, B. Moore, B. Theising, T. Wylie, G. Lennon, B. Soares, R. Wilson, and R. Waterston. 1996. GenBank accession no. AA250225.
- 26.Marra, M., L. Hillier, M. Allen, M. Bowles, N. Dietrich, T. Dubuque, S. Geisel, T. Kucaba, M. Lacy, M. Le, J. Martin, M. Morris, K. Schellenberg, M. Steptoe, F. Tan, K. Underwood, B. Moore, B. Theising, T. Wylie, G. Lennon, B. Soares, R. Wilson, and R. Waterston. 1996. GenBank accession no. AA645815.
- 27.Nagy E, Maquat L E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen F C, Gammeltoft S, Christiansen J. Translational discrimination of mRNAs coding for insulin-like growth factor II. J Biol Chem. 1990;265:13431–13434. [PubMed] [Google Scholar]
- 29.Ouelette A J, Silverberg M B, Malt R A. Association of poly(adenylate)-deficient messenger ribonucleic acid with membranes in mouse kidney. Biochemistry. 1981;20:3561–3567. doi: 10.1021/bi00515a040. [DOI] [PubMed] [Google Scholar]
- 30.Peltz S W, Brown A H, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 31.Pende A, Tremmel K D, DeMaria C T, Blaxall B C, Minobe W A, Sherman J A, Bisognano J D, Bristow M R, Brewer G, Port J D. Regulation of the mRNA-binding protein AUF1 by activation of the β-adrenergic receptor signal transduction pathway. J Biol Chem. 1996;271:8493–8501. doi: 10.1074/jbc.271.14.8493. [DOI] [PubMed] [Google Scholar]
- 32.Piechaczyk M, Yang J, Blanchard J, Jeanteur P, Marcu K B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985;42:589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- 33.Ranson M, Tickle C, Mahon K A, Mackem S. Gnot1, a member of a new homeobox gene subfamily, is expressed in a dynamic, region-specific domain along the proximodistal axis of the developing limb. Mech Dev. 1995;51:17–30. doi: 10.1016/0925-4773(94)00344-m. [DOI] [PubMed] [Google Scholar]
- 34.Raymond V, Atwater J A, Verma I M. Removal of an mRNA destabilizing element correlates with increased oncogenicity of proto-oncogene c-fos. Oncogene Res. 1989;4:861–865. [PubMed] [Google Scholar]
- 35.Ross H, Sato N, Ueyama Y, Koeffler H. Cytokine messenger RNA stability is enhanced in tumor cells. Blood. 1991;77:1787–1795. [PubMed] [Google Scholar]
- 36.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Echevarria M J, González C I, Peltz S W. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sachs A B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 40.Savant-Bhonsale S, Cleveland D W. Evidence for instability of mRNAs containing AUUUA motifs mediated through translation-dependent assembly of a >20S degradation complex. Genes Dev. 1992;6:1927–1939. doi: 10.1101/gad.6.10.1927. [DOI] [PubMed] [Google Scholar]
- 41.Shukla H, Swaroop A, Srivastava R, Weissman S M. The mRNA of a human class I gene HLA G/HLA 6.0 exhibits a restricted pattern of expression. Nucleic Acids Res. 1990;18:2189. doi: 10.1093/nar/18.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sirenko O I, Lofquist A K, DeMaria C T, Morris J S, Brewer G, Haskill J S. Adhesion-dependent regulation of an A+U-rich element-binding activity associated with AUF1. Mol Cell Biol. 1997;17:3898–3906. doi: 10.1128/mcb.17.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze M W, Kulozik A E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner B J, DeMaria C T, Sun Y, Wilson G M, Brewer G. Structure and genomic organization of the human AUF1 gene: alternative pre-RNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 45.Wagner B J, Long L, Rao P N, Pettenati M J, Brewer G. Localization and physical mapping of genes encoding the A+U-rich element RNA-binding protein AUF1 to human chromosomes 4 and X. Genomics. 1996;34:219–222. doi: 10.1006/geno.1996.0269. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, G. M., and G. Brewer. Unpublished observations.
- 47.Wilson G M, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acid Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 48.Wilson G M, Roberts E A, Deeley R G. Modulation of LDL receptor mRNA stability by phorbol esters in human liver cell culture models. J Lipid Res. 1997;38:437–446. [PubMed] [Google Scholar]
- 49.Zhang J, Sun X, Qian Y, LaDuca J P, Maquat L E. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, Sun X, Qian Y, Maquat L E. Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]