Abstract
Background
Infections caused by avian pathogenic Escherichia coli (APEC) result in significant economic losses in poultry industry. APEC strains are known to form biofilms in various conditions allowing them to thrive even under harsh and nutrient-deficient conditions on different surfaces, and this ability enables them to evade chemical and biological eradication methods. Despite knowing the whole genome sequences of various APEC isolates, little has been reported regarding their biofilm-associated genes. A random transposon mutant library of the wild-type APEC IMT 5155 comprising 1,300 mutants was analyzed for biofilm formation under nutrient deprived conditions using Videoscan technology coupled with fluorescence microscopy. Seven transposon mutants were found to have reproducibly and significantly altered biofilm formation and their mutated genes were identified by arbitrary PCR and DNA sequencing. The intact genes were acquired from the wild-type strain, cloned in pACYC177 plasmid and transformed into the respective altered biofilm forming transposon mutants, and the biofilm formation was checked in comparison to the wild type and mutant strains under the same conditions.
Results
In this study, we report seven genes i.e., nhaA, fdeC, yjhB, lysU, ecpR, AJB35136 and fdtA of APEC with significant contribution to biofilm formation. Reintroduction of AJB35136 and fdtA, reversed the altered phenotype proving that a significant role being played by these two O-antigen related genes in APEC biofilm formation. Presence of these seven genes across nonpathogenic E. coli and APEC genomes was also analyzed showing that they are more prevalent in the latter.
Conclusions
The study has elucidated the role of these genes in APEC biofilm formation and compared them to adhesion expanding the knowledge and understanding of the economically significant pathogens.
Keywords: APEC, Biofilm, Gene complementation, Transposon mutant, VideoScan
Background
Avian pathogenic Escherichia coli (APEC), a pathotype belonging to extra-intestinal pathogenic E. coli (ExPEC), causes various localized or systemic infections called ‘colibacillosis’ in birds. It is characterized by fibrinous lesions surrounding different organs [1] resulting in colisepticemia, hemorrhagic septicemia, coligranuloma (Hjarre’s disease), air sac disease (chronic respiratory disease), swollen-head syndrome, venereal colibacillosis, coliform cellulitis (inflammatory or infectious process), peritonitis, salpingitis, orchitis, osteomyelitis/synovitis (including turkey osteomyelitis complex), panophthalmitis, omphalitis/yolk sac infection, and enteritis [2, 3]. APEC adversely affects all stages of production and all sectors of the poultry industry including broiler and egg laying hens commencing in high morbidity and mortality leading to significant economic losses [4]. Poultry serves as a host for APEC and consumption of infected undercooked meat can lead to zoonotic transmission in humans becoming potential reservoirs for the pathogen [5]. APEC are responsible for substantial economic losses possibly due to bacterial biofilms and associated resistance to antimicrobials making them highly resilient to the eradication and suppression strategies [1]. APEC share the common virulence factors with UPEC, thus linked to certain diseases in humans [3, 4].
Biofilms are well-defined as a population of bacterial cells that live in a matrix of self-produced exopolysaccharides (EPS), proteins and DNA known as extracellular polysaccharide matrix (EPM), contributing in their attachment and colonization to various inert and living surfaces [6–8]. Biofilm formation is a multi-step process beginning with the initial attachment of microbial community to the surface and then progressing on to detachment [9]. The biofilm EPM contributes not only in the cell to cell adhesions but also acts as a barrier to protect the bacterial community against the host’s defense systems [10]. Therefore, biofilms allow the bacterial cells to endure and persist against harsh environmental, chemical and antimicrobial pressures [11]. A number of acute and chronic infections are related to biofilm-associated drug resistance and tolerance [12–15].
Complete genomic sequencing of APEC strain (APEC O1:K1:H7) has revealed that the functions of a large number of genes are still unknown while the roles of certain genes in various steps of its pathogenesis are still hypothetical and speculative [16]. Biofilm formation in APEC and its role in infections and pathogenesis have not been well studied. Several qualitative and quantitative methods have been developed for the detection of biofilms in laboratory [17]. Qualitative test for the detection of biofilm formation includes Congo Red agar test and pellicle formation assay, whereas the quantitative analysis of biofilm includes microtiter plate assay with crystal violet (CV) staining [18]. As an alternative, the VideoScan technology promises a direct, rapid and automated detection and quantification of fluorescent objects [19], we have successfully used this technology for detection and quantification of biofilms formed by different E. coli pathotypes [20], P. aeruginosa [21] and Salmonella [22]. The current study applied this direct approach and screened and quantified biofilm formation of 1,300 transposon mutants of a previously reported library of a wild-type APEC strain, IMT 5155 [23]. The objective of this study was to identify the APEC genes involved in biofilm formation contributing to the understanding of mechanisms involved in this complex process of colonization and pathogenesis.
Results
Screening of transposon mutant library and identification of potential genes involved in biofilm formation
A VideoScan module already established and published by Schiebel et al. (2017) was used [20] with minor modifications to quantify the biofilm formation by bacteria (Fig. 1).
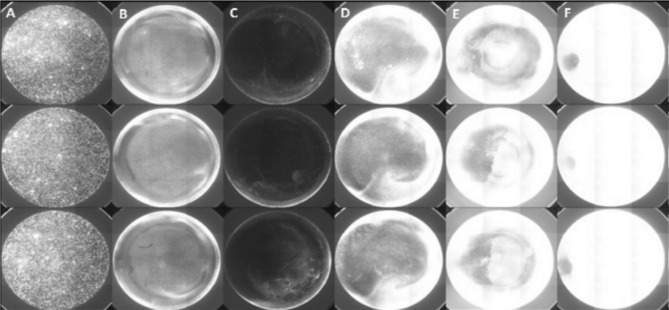
Fig. 1.
Overview of image of screening of transposon mutants with SYTO 9-stained biofilms. (A) plastic beads as internal reference, (B) wild-type strain IMT-5155 forming biofilm, (C) transposon mutant with down-regulated biofilm formation, (D) transposon mutant depicting slightly up-regulated biofilm formation, (E) transposon mutant exhibiting up-regulated biofilm formation and (F) transposon mutant with highly up-regulated biofilm
By applying this method, 1,300 transposon mutants were screened for biofilm formation in M9 minimal media at 37 °C after 72 h. Transposon mutants (n = 7) with reproducible (six wells with three independent experiments) increase or decrease in biofilm formation under the same conditions as the wild-type strain were branded as “biofilm-altered mutants”. These specific mutants were also subjected to growth assay which revealed that no impairment in growth rate. Sequencing of arbitrary PCR amplicons revealed the identity of the mutated genes in biofilm-altered mutants potentially contributing to biofilm formation (Table 1).
Table 1.
Transposon mutants with their respective disrupted genes in biofilm-altered mutants
| Mutant name | Gene name/ locus |
Complete name | (Possible) role/function | PSORT (location) |
Biofilm Formation |
|---|---|---|---|---|---|
| A10k | nhaA | Na+/H+ antiporter NhaA | Na+/H+ antiporter | Cytoplasmic membrane | Down |
| A7r | fdeC |
Intimin-like adhesin FdeC (factor adherence E. coli) |
Adhesin | Outer membrane | Down |
| C4s | yjhB | MFS transporter | Probably sialic acid transport | Cytoplasmic membrane | Up |
| D11r | lysU | lysine—tRNA ligase [multifunctional] | aminoacyl tRNA synthetases | Cytoplasmic | Up |
| D2q | ecpR | Helix-turn-helix transcriptional regulator/putative fimbrial transcriptional regulator | ECP transcriptional regulator | Unknown | Down |
| E4r | AJB35136 | Group 1 glycosyl transferase | Probably O-antigen synthesis | Cytoplasmic | Up |
| H6n | fdtA | TDP-4-oxo-6-deoxy-alpha-D-glucose-3,4-oxoisomerase | O-antigen synthesis | Cytoplasmic | Down |
Motility assays reveal contribution of selected biofilm-altered mutants to APEC motility
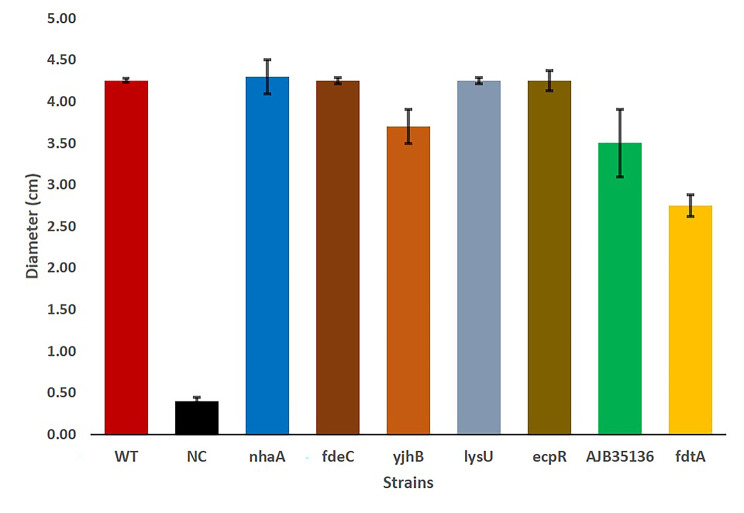
The results of the motility assay of biofilm-altered mutants in comparison to WT are shown in Fig. 2. One mutation which partially affected O-antigen synthesis (fdtA) showed more than 30% reduction in motility compared to the wild type. The yjhB (MFS transporter) and group 1 glycosyl transferase (GenBank accession no. AJB35136) mutants showed 4–18% lower motility in comparison to the wild type. Gene mutants such as nhaA, fdeC, lysU and ecpR had no significant difference in motility when compared to the wild-type strain.
Fig. 2.
Motility assay of APEC wild-type strain IMT5155 and selected transposon mutants. The data are shown as median values and standard deviation of three separate experiments. SAEC5148, non-motile strain used as a negative control
Complementation of intact genes from wild-type APEC IMT5155 confirms contribution of biofilm-altered mutants to APEC biofilm formation
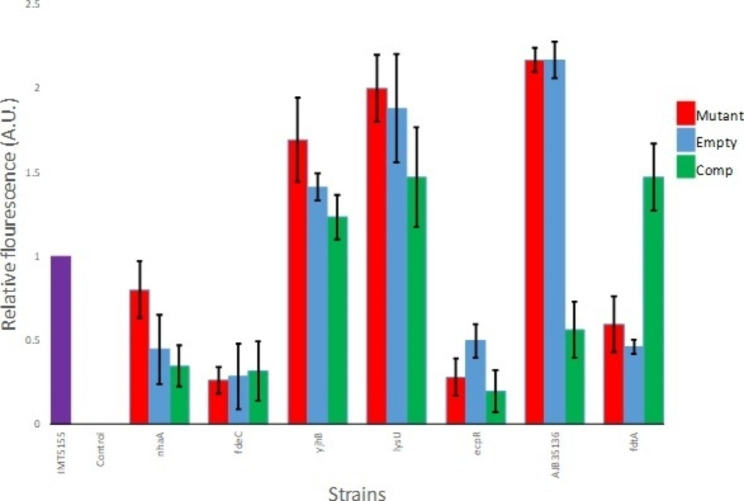
All biofilm-altered mutants when tested for susceptibility to ampicillin before complementation were found sensitive. The effect of complementation on bacterial growth was also analyzed in “complemented biofilm-altered mutants” via growth assays and no change in growth was detected. Biofilm assay for the screening of biofilm-altered mutants and complemented biofilm-altered mutants exhibited significant complementation effects for two of the genes identified in the original screen i.e., AJB35136 and fdtA (Fig. 3). Transformation of biofilm-altered mutants with only the empty vector plasmid alone exhibited no significant change for these mutants in biofilm formation. The complementation of fdtA resulted in biofilm formation comparable to that of the wild-type strain whereas its mutant formed really weak biofilm, proving its important role in this complex process. In case of complementation by AJB35136, drastic decrease of around 74% in biofilm formation was observed when compared to its mutant whose biofilms was twice as that of the wild-type strain. For Na+/H+ antiporter nhaA gene and MFS transporter yjhB gene, their complementation although decreased biofilm formation but this decrease was not significant as the mutant transformed with empty plasmid also affected biofilm formation. Both fdeC and ecpR, complementation with intact gene or empty plasmid did not cause any alteration in biofilm forming ability of these strains (Fig. 3).
Fig. 3.
Biofilm formation. APEC wild-type strain IMT5155 (purple bar), selected transposon mutants (Mutant, red bar), and transposon mutants transformed with either empty vector pACYC177 (empty, blue bar) or pACYC177 plasmids bearing wild-type versions of transposon mutated genes (Comp, green bar) in M9 minimal media at 37 °C after 72 h. The data shown are mean values and standard deviation of three separate experiments in six wells replicate for each experiment. Wells not incubated with bacteria were used as a negative control
Genes from biofilm-altered mutants have higher prevalence in APEC compared to nonpathogenic E. coli
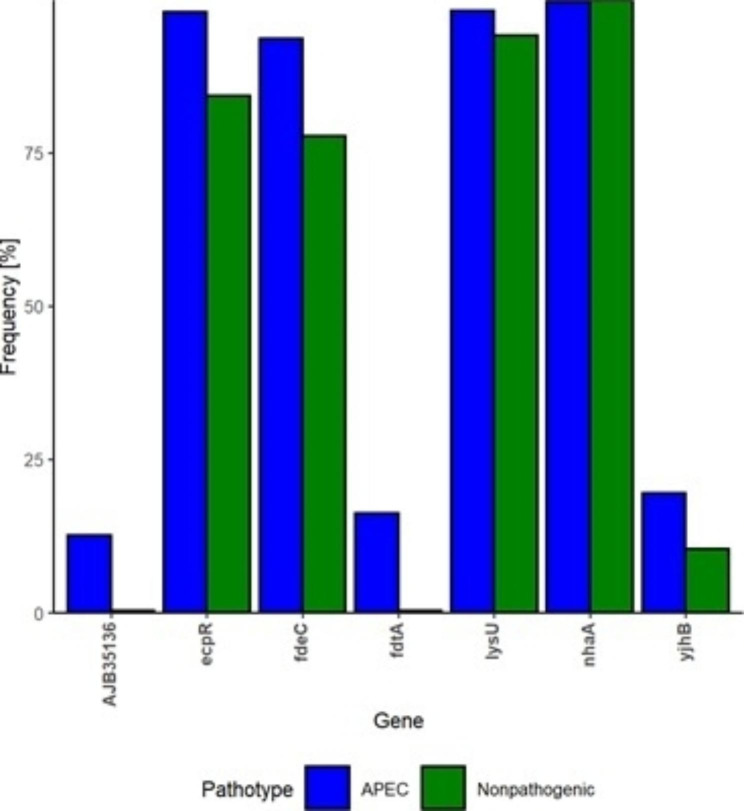
The prevalence of selected genes was carried among two groups to evaluate their frequency in E. coli present in nature as depicted in Fig. 4. Genes such as ecpR, fdeC and lysU were prevalent in higher number in both APEC and nonpathogenic E. coli but they were significantly more frequent in APEC genomes (p.BH < 0.0001). The genes which were less frequent in APEC (i.e., fdtA (16.23%), AJB35136 (12.57%) and yjhB (19.55%) were significantly (p.BH < 0.0001) scarcer in nonpathogenic E. coli (i.e., fdtA (0.36%), AJB35136 (0.33%) and yjhB (10.43%). Only nhaA gene was present across both APEC and nonpathogenic E. coli with similar prevalence (99%).
Fig. 4.
Prevalence of the selected genes in APEC and nonpathogenic E. coli. Gene prevalence/frequency percentage (y-axis) is shown for selected genes (x-axis) for the biofilm altered mutants. The analysis was carried out on 573 APEC and 3,069 nonpathogenic E. coli genomes
Discussion
Among pathogenic E. coli, APEC is a major bacterial pathogen of poultry with considerable zoonotic importance and can potentially serve as a virulence gene reservoir [24]. APEC strains generally form biofilms on different surfaces such as polystyrene, polypropylene, polyvinylchloride (PVC) under scarce nutrient conditions and different temperatures [1, 20] and these conditions can be easily encountered in commercial poultry facilities and farms in water systems, drinkers, feeding trays etc. [1]. APEC biofilm formation has been previously investigated but the focus has been on different biofilm formation types, suitable media [25, 26], characterization of strains from colibacillosis cases [27] along with other factors such as antibiotic resistance and virulence factors [28]. So far, the role of various genes in biofilm formation of APEC remains elusive and therefore a transposon mutant library of 1,300 APEC mutants was screened and selective intact genes were introduced or complemented in the mutants to evaluate their role in this complex process. Here, we used our already established method which is specific, rapid, and high-throughput to detect and quantify the biofilm formation [20].
Most of these genes have also been previously reported in our previous study where the same transposon library was analyzed for adhesion to CHIC-8E11 and Lovo cell lines and similar strategy was used whereby complementation of the mutated genes was carried out [29].
The gene fdtA encodes iTDP-4-oxo-6-deoxy-alpha-D-glucose-3,4-oxoisomerase involved in O-antigen synthesis and its corresponding transposon mutant (H6n) exhibited reduced adhesion previously, and reduced biofilm formation (almost half of that of wild type) in the current study. After complementation, a substantial increase in its adhesion property had been reported, now similarly its biofilm formation has also been found significantly increased and became at par with the wild type [29–31]. This showed that fdtA is not only involved in adhesion and surface to surface contact with eukaryotic cells but it is also potentially involved in initial attachment and colonization on non-living surfaces. In case of AJB35136 (encoding group 1 glycosyl transferase), it is involved in lipopolysaccharide biosynthesis and the corresponding transposon mutant had low adhesion to the cell lines [29]. It contrasts with our current findings of biofilm formation where the transposon mutant produced more than two-fold high biofilm under the tested conditions as compared to wild-type strain. The reintroduction of this gene decreased biofilm formation significantly and the biofilm became almost half of that of the wild type under same conditions. On biofilm formation, the opposite effect of complementation of both O-antigen synthesis related genes points towards the diversity in nature of genes, their expression and other complex underlying mechanisms determining their roles.
For other genes, complementation did not significantly affect biofilm formation such as yjhB nhaA, lysU, fdeC and ecpR when compared to wild type, although their transposon mutants differed both in adhesion and biofilm formation phenotypes. The transposon mutant of yjhB gene produced high biofilm and similarly the adhesion percentage on Lovo and CHIC-8E11 cell lines was also very high when compared to the wild-type strain [29]. In both cases, reintroduction of the same gene in the mutant did not have any significant effect on either of the phenotype. yjhB has been identified as putative sialic acid transporter and has been suggested to indirectly effect the regulation of expression of type 1 fimbriae [32, 33]. It can be concluded that it plays a role in both virulence attributes of adhesion and biofilm formation but the role has to be further confirmed by complementation via an appropriate expression system. A previous study showed that a mutation in nhaA gene encoding the Na/H antiporter in E. coli K-12 strain had increased motility and FliC protein expression [34], this may have translated into high adhesion [29] but did not significantly boost biofilm formation. Also the nhaA transposon mutant did not exhibit significantly high motility and this is in contrast to the previously reported study [34]. For motility assays, Smith et al., (2017) used YESCA agar and incubation temperature of 26 ºC, whereas LB agar and 37 ºC incubation were used in the current study. As it has been shown previously that media or temperature differences can effect gene expression of E. coli, the contradiction regarding the effect of nhaA mutation on motility could be due to methodological differences in design of motility experiments [35, 36].
Another interesting gene not identified in the previous study with its transposon mutant exhibiting really high biofilm formation under the above-mentioned conditions was lysU gene. The reintroduction of intact lysU gene in the transposon mutant D11r did not result in significant decrease in the biofilm formation. lysU has been identified as heat inducible lysyl-tRNA synthetase of E. coli and its deletion has been shown to grow adequately under normal conditions but the growth is hampered at higher temperatures (i.e., 44 °C) [37]. The partial effect of complementation of lysU in trans might be associated with the pACYC177 plasmid system used in this study. This system has been used successfully in multiple species in the past [36, 38]. The complemented mutant exhibited tendency to lower biofilm formation, but relatively high level of variation was observed indicating that other complementation systems e.g. inducible expression plasmids pBAD33 or pWRG30 could provide better results in further experiments for this gene.
The fdeC (encoding intimin-like adhesion) transposon mutant exhibited really low biofilm formation in contrast to the slightly high adhesion to the cell lines and complementation through pACYC177 did not result in any significant change in biofilm formation or adhesion on either of the cell lines. Contribution of FdeC to biofilm formation of STEC has been shown in study of Easton et al. [39] but the expression conditions were the crucial factor for FdeC contribution. Our gene frequency analysis revealed lower abundance of fdeC gene in nonpathogenic E. coli than in APEC. This result is in agreement with a previously reported study, where it showed the conservation of FdeC among strains of different E. coli pathotypes and elicit protection against urinary tract infections [40]. The ecpR transposon mutant i.e., D2q had very high adhesion rate and upon complementation decreased 3 folds and became similar to wild type only in case of CHIC-8E11 cell line. In case of biofilm formation, this mutant produced very low biofilm and complementation did not have any significant effect when the same system of complementation was used. The ecpR gene encodes the E. coli common pilus (ECP) regulator is a part of the ECP gene cluster [41].
In summary, we report the identification of 7 genes affecting biofilm formation of APEC under stressed conditions with low nutrition. This study also further analyzes the role of some of the previously identified genes effecting adhesion phenotype, evaluates their effect on biofilm formation and compares their influence on both phenotypes. Both are important in colonization and pathogenesis of infectious bacteria as adhesion deals with initial contact and colonization of bacteria to the living tissue and host cells and biofilm formation was studied as in this case it also involves initial attachment and colonization on a non-living surface. The study also shows that same transposon mutants show different trend in the two virulence mechanism depicting diverse role of these genes under different situations.
It is also important to note that when intact genes were cloned in pACYC177 plasmid and reintroduced in intact form into the transposon mutant, it did not result in complementation for all the cases. Therefore, the effect on phenotype holds true for only two genes i.e., fdtA and AJB35136 where the empty plasmid did not alter the biofilm formation and only the plasmid cloned with intact gene showed increase or decrease in the biofilm formation. For all the other genes, complementation did not significantly affect the phenotype under consideration. Similar issues were observed in study of Young et al., (2022), where complementation of APEC O18 deletion mutant using pBAD33 did not alter the biofilm formation [42]. This can be due to the nature of genes which needs to be expressed at a specific time point or under an inducible promoter instead of the constitutive promoter and points to a complex underlying mechanisms of biofilm formation which requires further research. This study contributes toward the understanding of APEC biofilm formation and compares the roles played by the same genes in adhesion.
Conclusion
APEC adversely affects all stages and sectors of poultry industry commencing in high morbidity and mortality leading to significant economic losses. This study identified seven genes that play a significant role in the biofilm formation of APEC under nutrient deprived conditions. Two O-antigen related genes, AJB35136 and fdtA, have a particularly important contribution to biofilm formation and regulation in APEC. This research provides valuable insights into the mechanisms of biofilm formation in APEC and may contribute to the development of effective strategies to control and prevent APEC persistence in the poultry industry. As phages provide useful alternative as surface decontaminants, targeting genes that also reduce biofilm formation with phages in the future might provide valuable resource in reduction of APEC burden in poultry production.
Materials and methods
Bacterial Strains
Wild-type clinical isolate APEC IMT 5155 along with its transposon mutant library of 1,300 random transposon mutants (coding Kanamycin resistance cassette) were acquired from Free University (FU) Berlin, Germany [23]. The mutants were revived on LB (Lysogeny Broth) agar plates with the appropriate antibiotic i.e., kanamycin (50 μg/ml) whereas the APEC IMT 5155 was revived on an antibiotic free CHROM agar plate (Mast Diagnostica GmbH, Reinfeld, Germany).
Screening of transposon mutant library for biofilm formation
Initially, wild-type APEC IMT 5155 was subjected to various biofilm forming conditions including four growth media i.e., brain heart infusion broth (BHI), LB broth, tryptic soya broth (TSB) and M9 minimal media; two incubation temperatures i.e., 28 and 37 °C, and three variable durations: 24, 48 and 72 h. The biofilms were quantified with our previously optimized protocol [20] with minor modifications. Since the wild-type APEC IMT 5155 formed adequate biofilm in M9 minimal media at 37 °C after 72 h, therefore the 1,300 transposon mutants were screened under these conditions and the up/down regulation of biofilm formation was evaluated in 96-well flat bottom polypropylene plates (Greiner Bio-One GmbH, Frickenhausen, Germany) to make biofilm [20]. E. coli strain K-12 MG1655 F’tet ΔtraD was used as biofilm forming positive control. The plates were covered with sealing films and incubated at 37 °C for 72 h. The non-adherent bacteria from the wells were aspirated and attached biofilms were washed once with 200 μl of sterile 0.9% NaCl followed by an incubation in the dark for 10 min with isotonic saline containing 5 μM SYTO 9 green fluorescent nucleic acid stain (Thermo Fisher Scientific GmbH, Dreieich, Germany). The plates were washed again with isotonic saline and analyzed using the automated VideoScan technology platform. Biofilm formation potential of each mutant was checked in triplicate wells with two independent experiments. The mutants suspected for up/down regulation of biofilm formation were further tested in six wells with three independent experiments and the mutants showing significantly up/down regulation in biofilm formation as compared to wild-type strain were designated as ‘biofilm-altered mutants’. The shake flask growth studies of the wild-type strain and biofilm-altered mutants were done at 37 °C to rule out any effect of transposon mutation on the bacterial growth [29].
Identification of transposon insertion sites in ‘biofilm-altered mutants’
The ‘biofilm-altered mutants’ were subjected to arbitrary polymerase chain reaction (PCR) using Ampli Taq Gold polymerase to locate the transposon cassette insertion site and identify the mutated genes [29]. The products of first round PCR were amplified again in another round/nested PCR using one arbitrary primer (Arbi2, as homologous to the 5/ sequence of Arbi5) and one transposon I terminus-specific primer (P6) as described earlier [23]. Sequences of all the primers used in the study are shown in Table 2. Genomic DNA of the wild-type APEC, and a reaction without template were used as templates for negative controls. The amplicons were purified with PCR purification kit (Thermo Scientific) and sent for commercial Sanger sequencing (LGC’s Agova Genomics, Berlin, Germany). The DNA sequences were analyzed in NCBI public databases using BLASTn and bacterial localization prediction tool (PSORTb 3.0) [43] and the genes mutated by transposon insertions and potentially involved in regulation of biofilm formation were selected for further experiments.
Table 2.
List of primers used in the study
| Gene name/Target Sequence | Primer Name | Sequence 5’to 3’ | |
|---|---|---|---|
| Primers for arbitrary PCR | |||
| Flanking sequence Tn5 | P9 | CGCAGGGCTTTATTGATT | |
| Arbitrary | Arbi5 | GGCCACGCGTCGACTAGTAC(N)10TACNG | |
| Flanking sequence Tn5 | P6 | CCTAGGCGGCCAGATCTGAT | |
| Arbitrary | Arbi2 | GGCCACGCGTCGACTAGTAC | |
| Primers for amplification of genes from wild-type APEC | |||
| nhaA | A10kFwd | ACATAAGCTTTTGACAATTAATCATCGGCTCG TATAATGTGTGGAGGAGGACAGCTATGAAAC ATCTGCATCGATTCTTTAGC | |
| A10kRev | ACATAAGCTTTCAAACTGATGGACGCAAACGA ACGCGTAACCAGC | ||
| fdeC | A7rFwd | ACATGGATCCTTGACAATTAATCATCGGCTCG TATAATGTGTGGAGGAGGACAGCTATGTCACA TTATAAAACAGGTC | |
| A7rRev | ACATGGATCCCTATTGCTGGGTAAGAGGC | ||
| yjhB | C4sFwd | ACATGGATCCTTGACAATTAATCATCGGCTCG TATAATGTGTGGAGGAGGACAGCTATGGCAAC AGCATGGTATAAAC | |
| C4sRev | ACATGGATCCTCATTTAGCCACGGATAG | ||
| lysU | D11rFwd | ACATGGATCCTTGACAATTAATCATCGGCTCGTATAATGTGTGGAGGAGGACAGCTATGTCTGAACAAGAAACACGGGG | |
| D11rRev | ACATGGATCCTTATTTCTGTGGGCGCATCG | ||
| ecpR | D2qFwd | ACATGGATCCTTGACAATTAATCATCGGCTCGTATAATGTGTGGAGGAGGACAGCTATGGAATG TCAAAACCGTTCTG | |
| D2qRev | ACATGGATCCTTACTGAACCAACTTATATATTTTTGAGTACAGC | ||
| AJB35136 | E4rFwd | ACATGGATCCTTGACAATTAATCATCGGCTCGTATAATGTGTGGAGGAGGACAGCTATGGAAG AAAATAATATGAAGACG | |
| E4rRev | ACATGGATCCTTAATAAATAGATTCATACATA GC | ||
| fdtA | H6nFwd | ACATGGATCCTTGACAATTAATCATCGGCTCG TATAATGTGTGGAGGAGGACAGCTATGGATAT TAAATTAATCTCTTTGC | |
| H6nRev | ACATGGATCCTTATGAATTCTCAATTGAATTTA CTCTTC | ||
| Insert sequencing after cloning | pACYC177Fwd | GTAGCGGTTCGGTTTATTGAC | |
| pACYC177Rev | TGTCCACGGTACGCCTGC | ||
Cloning of intact genes from wild-type APEC IMT 5155 into the pACYC177
The genes selected by arbitrary PCR genes were amplified in intact form from the wild-type APEC IMT 5155 strain by PCR using Phusion polymerase (Thermo Scientific, Germany) with primers designed for cloning in expression vector pACYC177. The PCR products were purified by PCR purification kit (Thermo Scientific, Germany) while the pACYC177 plasmid was isolated using the Midiprep Kit (Qiagen N.V., Venlo, NL) and DNA concentrations were measured with a Colibri Microvolume Spectrometer (Titertek-Berthold). The purified PCR products (1 μg) and plasmid DNA (4 μg) were digested with Fast Digest (FD) BamHI enzyme (Thermo Scientific) and purified by PCR purification kit (Thermo Scientific) and gel extraction kit respectively (Qiagen N.V., Venlo, NL). The digested and purified PCR products and plasmid were ligated by T4 DNA ligase (Thermo Scientific) following the manufacturer’s protocol for 30 min and transformed by heat shock at 42 °C for 45 s into already prepared E. coli XL1Blue chemocompetent cells [44]. The transformants were plated onto LB agar with Kan (50 μg/ml) for selection and incubated at 37 °C overnight and the colonies were tested with the rapid colony screening protocol [45]. The positive colonies were processed for overnight culture in 10 ml of LB broth with Kan (50 μg/mL) and plasmid was isolated by Miniprep plasmid extraction kit (Thermo Scientific) after 16–18 h growth and quantified. To confirm successful cloning, plasmids were purified and digested with BamHI and the plasmids, that showed two bands of desired sizes after restriction, were sent for Sanger sequencing commercially (LGC’s Agova Genomics, Berlin, Germany). The plasmid sequences were analysed and clones with accurate gene with promotor sequence and open reading frames were stored at -20 °C.
Complementation of the ‘biofilm-altered mutants’ with intact genes
The transposon mutants with altered biofilm forming abilities were complemented with intact genes taken from wild-type strain. For complementation, electrocompetent cells of each of the ‘biofilm-altered mutant’ were prepared according to the established protocol [46] with minor modifications. Briefly, LB medium (100 ml) was inoculated with 1 ml of overnight culture and incubated at 37 °C at 180 rpm until OD600 = 0.6 was achieved. The bacteria were then incubated on ice for 5 min, centrifuged at 6,000 x g for 10 min at 4 °C. The supernatant was discarded and the pellet was washed once with 50 ml of ice cold water and twice with 25 ml of ice cold 10% glycerol in water. Finally, washed bacterial cells were resuspended in 0.4 ml of 10% glycerol in water and aliquoted to 50 μl per tube. All the steps were carried out on ice. For transformation, plasmid (100 ng) was added to particular mutant’s electrocompetent cells, incubated on ice for 1 min, transferred to cuvette (4 mm wide) and electroporated at a voltage of 2.5 kV, with the capacity of 25 μF and resistance of 200 Ω. The bacteria were also transformed with empty plasmid (i.e., without insert) to observe effects of transformation by plasmid only. A volume of 900 μl of super optimal broth media (SOC) was added immediately after transformation and the bacteria were transferred to 2 ml tubes for incubation at 37 °C for 1 h with 180 rpm shaking. The bacteria were then centrifuged at 6,000 x g for 3 min, resuspended in 200 μl of medium, spread onto LB agar plates with ampicillin (100 μg/ml) and kept at 37 °C overnight. The bacterial colonies on agar plates were designated as ‘biofilm-altered mutants complemented with gene of interest’ and ‘biofilm-altered mutants complemented with empty plasmid only’.
To investigate the effect of gene complementation, the wild-type APEC IMT5155 and ‘biofilm-altered mutants’ with and without complementation were checked for their biofilm formation by automated VideoScan technology as described earlier in 6 well replicates and in three independent experiments. The average biofilm formation of wild-type bacteria in each of the 6 wells in three experiments was normalized to 1 and compared for statistical significance with similar treated values of bacteria of ‘biofilm-altered mutants’ with and without complementation with the genes.
Prevalence of potential biofilm regulating/affecting genes
Genes potentially involved in biofilm formation in the biofilm-altered mutants as selected by arbitrary PCR were checked for their prevalence in the reported APECs and nonpathogenic E. coli genomes. A collection of 573 APEC and 3,069 nonpathogenic E. coli genomes was obtained from RefSeq as reported earlier [47]. Briefly, when categorizing genomes as nonpathogenic E. coli, RefSeq collection of E. coli was blasted against virulence associated genes (VAGs) and pathotyped based on presence or absence of VAGs. Genomes encoding none of the VAGs or only one of virulence gene i.e., fimH, fyuA, iucC, neuC, sitA or yfcV were assigned as nonpathogenic E. coli. Moreover, genomes lacking information about coverage (except complete genome sequences), coverage less than 50 times, number of contigs more than 400 and 3rd generation sequencing platforms/technologies, i.e., “Oxford Nanopore” and “Pacific Biosciences”, were omitted [47]. APEC genomes were downloaded as assemblies or raw reads from BioProjects found in GenBank and reference APEC genomes. Genome assemblies were made with Shovill pipeline. The prevalence of genes listed in Table 1 in APEC and nonpathogenic E. coli, was tested with ABRicate. All results with values of 80% or above were considered positive. The prevalence of genes was compared with chi-squared test of independence [48].
Motility assays
Initial analysis of the genes mutated in transposon library and their associated literature pointed to their linkage with motility which is known to affect biofilm formation. Therefore, we verified selected strains with transposon mutations with altered biofilm formation and wild-type APEC IMT5155 for motility. The strain SAEC5148 was used as a negative control [29]. The bacteria were inoculated in 1 ml LB medium and incubated overnight at 37 °C with 180 rpm shaking. Each isolate was inoculated into the center of a motility agar plate (LB containing 0.25% agar), and the plates were sealed with paraffin film and kept at 37 °C for 16 h. The diameter of the diffusion zone from the point of inoculation was measured. The diameter of 5 mm or more was considered as motile, whereas less than 5 mm diameter was considered non-motile [20]. The experiments for each of the isolate were carried out three independent times.
Authors’ contributions
A.A., M.M.K., R.K. and P.S. conceived and designed the experiments. A.A., M.M.K., A.O. and G.L. collected the data and samples. M.M.K., A.A., R.K. and J.W. performed laboratory analysis. M.M.K., and A.A. analyzed the data and wrote the manuscript. All authors have read and approved the final draft of the manuscript.
Funding
The research was supported by Brandenburg Research Academy and International Network (BRAIN) program of the Brandenburg Ministry of Sciences, Research and Cultural Affairs, co-funded by the Marie Curie Program of the European Union and the Federal Ministry of Education and Research, Germany (BMBF InnoProfile-Transfer 03IPT611X and BMBF project 03PSZZF1A).
Open Access funding enabled and organized by Projekt DEAL.
Data Availability
All essential data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not Applicable.
Consent for publication -
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Oosterik LH, Tuntufye HN, Butaye P, Goddeeris BM. Effect of serogroup, surface material and disinfectant on biofilm formation by avian pathogenic Escherichia coli. Vet J. 2014;202(3):561–5. doi: 10.1016/j.tvjl.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Nolan LK, Barnes HJ, Vaillancourt JP, Abdul-Aziz T, Logue CM. Colibacillosis [Internet]. Wiley Online Library. John Wiley & Sons, Ltd; 2017 [cited 2022 Jan 29]. p. 751–805. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/9781119421481.ch18.
- 3.Vincent C, Boerlin P, Daignault D, Dozois CM, Dutil L, Galanakis C, et al. Food reservoir for Escherichia coli causing urinary tract infections. Emerg Infect Dis. 2010;16(1):88–95. doi: 10.3201/eid1601.091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordstrom L, Liu CM, Price LB. Foodborne urinary tract infections: a new paradigm for antimicrobial-resistant foodborne illness. Front Microbiol. 2013;4:29. doi: 10.3389/fmicb.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markland SM, LeStrange KJ, Sharma M, Kniel KE. Old friends in new places: exploring the role of extraintestinal E. coli in intestinal disease and foodborne illness. Zoonoses Public Health. 2015;62(7):491–6. doi: 10.1111/zph.12194. [DOI] [PubMed] [Google Scholar]
- 6.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–22. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 7.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, et al. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296(2):202–11. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Homøe P, Bjarnsholt T, Wessman M, Sørensen HCF, Johansen HK. Morphological evidence of biofilm formation in Greenlanders with chronic suppurative otitis media. Eur Arch Otorhinolaryngol. 2009;266(10):1533–8. doi: 10.1007/s00405-009-0940-9. [DOI] [PubMed] [Google Scholar]
- 9.Koo H, Allan RN, Howlin RP, Stoodley P, Hall-Stoodley L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat Rev Microbiol. 2017;15(12):740–55. doi: 10.1038/nrmicro.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hobley L, Harkins C, MacPhee CE, Stanley-Wall NR. Giving structure to the biofilm matrix: an overview of individual strategies and emerging common themes. FEMS Microbiol Rev. 2015;39(5):649–69. doi: 10.1093/femsre/fuv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35(4):322–32. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS. 2013;121:1–58. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 13.Bjarnsholt T, Jensen P, Jakobsen TH, Phipps R, Nielsen AK, Rybtke MT, et al. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE. 2010;5(4):e10115. doi: 10.1371/journal.pone.0010115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15(2):167–93. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis K. Multidrug tolerance of biofilms and persister cells. In: Romeo T, editor. Bacterial Biofilms [Internet]. Berlin, Heidelberg: Springer; 2008 [cited 2022 Mar 16]. p. 107–31. (Current Topics in Microbiology and Immunology). Available from: 10.1007/978-3-540-75418-3_6 [DOI] [PubMed]
- 16.Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, et al. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol. 2007;189(8):3228–36. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arciola CR, Campoccia D, Gamberini S, Cervellati M, Donati E, Montanaro L. Detection of slime production by means of an optimised Congo red agar plate test based on a colourimetric scale in Staphylococcus epidermidis clinical isolates genotyped for ica locus. Biomaterials. 2002;23(21):4233–9. doi: 10.1016/S0142-9612(02)00171-0. [DOI] [PubMed] [Google Scholar]
- 18.Stepanović S, Vuković D, Dakić I, Savić B, Švabić-Vlahović M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Methods. 2000;40(2):175–9. doi: 10.1016/S0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 19.Rödiger S, Schierack P, Böhm A, Nitschke J, Berger I, Frömmel U, et al. et al. A highly versatile microscope imaging technology platform for the multiplex real-time detection of biomolecules and autoimmune antibodies. In: Seitz H, Schumacher S, et al.et al., editors. Molecular Diagnostics. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. pp. 35–74. [DOI] [PubMed] [Google Scholar]
- 20.Schiebel J, Böhm A, Nitschke J, Burdukiewicz M, Weinreich J, Ali A et al. Genotypic and phenotypic characteristics associated with biofilm formation by human clinical Escherichia coli isolates of different pathotypes. Appl Environ Microbiol. 2017;83(24). [DOI] [PMC free article] [PubMed]
- 21.Awan AB, Schiebel J, Böhm A, Nitschke J, Sarwar Y, Schierack P, et al. Association of biofilm formation and cytotoxic potential with multidrug resistance in clinical isolates of Pseudomonas aeruginosa. EXCLI J. 2019;18:79–90. [PMC free article] [PubMed] [Google Scholar]
- 22.Nawaz S, Khan MM, Noack J, Awan AB, Schiebel J, Roggenbuck D et al. Rapid detection of biofilm formation by zoonotic serovars of Salmonella enterica and avian pathogenic E. coli isolates from poultry. Biofilms. 2020.
- 23.Li G, Laturnus C, Ewers C, Wieler LH. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect Immun. 2005;73(5):2818–27. doi: 10.1128/IAI.73.5.2818-2827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeStrange K, Markland SM, Hoover DG, Sharma M, Kniel KE. An evaluation of the virulence and adherence properties of avian pathogenic Escherichia coli. One Health. 2017;4:22–6. doi: 10.1016/j.onehlt.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen DW, Klimavicz JS, Cavender T, Wannemuehler Y, Barbieri NL, Nolan LK, et al. The impact of media, phylogenetic classification, and E. coli pathotypes on biofilm formation in extraintestinal and commensal E. coli from humans and animals. Front Microbiol. 2018;9:902–2. doi: 10.3389/fmicb.2018.00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skyberg JA, Siek KE, Doetkott C, Nolan LK. Biofilm formation by avian Escherichia coli in relation to media, source and phylogeny. J Appl Microbiol. 2007;102(2):548–54. doi: 10.1111/j.1365-2672.2006.03076.x. [DOI] [PubMed] [Google Scholar]
- 27.Newman DM, Barbieri NL, de Oliveira AL, Willis D, Nolan LK, Logue CM. Characterizing avian pathogenic Escherichia coli (APEC) from colibacillosis cases, 2018. PeerJ. 2021;9:e11025–5. doi: 10.7717/peerj.11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlickova S, Klancnik A, Dolezalova M, Mozina SS, Holko I. Antibiotic resistance, virulence factors and biofilm formation ability in Escherichia coli strains isolated from chicken meat and wildlife in the Czech Republic. J Environ Sci Health B. 2017;52(8):570–6. doi: 10.1080/03601234.2017.1318637. [DOI] [PubMed] [Google Scholar]
- 29.Ali A, Kolenda R, Khan MM, Weinreich J, Li G, Wieler LH et al. Novel avian pathogenic Escherichia coli genes responsible for adhesion to chicken and human cell lines. Björkroth J, editor. Appl Environ Microbiol. 2020;86(20):e01068-20. [DOI] [PMC free article] [PubMed]
- 30.Burns SM, Hull SI. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect Immun. 1998;66(9):4244–53. doi: 10.1128/IAI.66.9.4244-4253.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DebRoy C, Fratamico PM, Yan X, Baranzoni G, Liu Y, Needleman DS, et al. Comparison of O-antigen gene clusters of all O-serogroups of Escherichia coli and proposal for adopting a new nomenclature for O-typing. PLoS ONE. 2016;11(1):e0147434. doi: 10.1371/journal.pone.0147434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blomfield IC. Sialic acid and N-acetylglucosamine regulate type 1 fimbriae synthesis. Microbiol Spectr. 2015;3(3):3308. doi: 10.1128/microbiolspec.MBP-0015-2014. [DOI] [PubMed] [Google Scholar]
- 33.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62(1):1–34. doi: 10.1128/MMBR.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith DR, Price JE, Burby PE, Blanco LP, Chamberlain J, Chapman MR. The production of curli amyloid fibers is deeply integrated into the biology of Escherichia coli. Biomolecules. 2017;7(4):75. doi: 10.3390/biom7040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gadgil M, Kapur V, Hu WS. Transcriptional response of Escherichia coli to temperature shift. Biotechnol Prog. 2005;21(3):689–99. doi: 10.1021/bp049630l. [DOI] [PubMed] [Google Scholar]
- 36.Parker A, Cureoglu S, De Lay N, Majdalani N, Gottesman S. Alternative pathways for Escherichia coli Biofilm formation revealed by sRNA overproduction. Mol Microbiol. 2017;105(2):309–25. doi: 10.1111/mmi.13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark RL, Neidhardt FC. Roles of the two lysyl-tRNA synthetases of Escherichia coli: analysis of nucleotide sequences and mutant behavior. J Bacteriol. 1990;172(6):3237–43. doi: 10.1128/jb.172.6.3237-3243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolenda R, Burdukiewicz M, Schiebel J, Rödiger S, Sauer L, Szabo I, et al. Adhesion of Salmonella to pancreatic secretory granule membrane major glycoprotein GP2 of human and porcine origin depends on FimH sequence variation. Front Microbiol. 2018;9:1905–5. doi: 10.3389/fmicb.2018.01905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Easton DM, Allsopp LP, Phan MD, Moriel DG, Goh GK, Beatson SA, et al. The intimin-like protein FdeC is regulated by H-NS and temperature in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 2014;80(23):7337–47. doi: 10.1128/AEM.02114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nesta B, Spraggon G, Alteri C, Gomes Moriel D, Rosini R, Veggi D, et al. FdeC, a aovel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio. 2012;3(2):e00010–12. doi: 10.1128/mBio.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aleksandrowicz A, Khan MM, Sidorczuk K, Noszka M, Kolenda R. Whatever makes them stick – adhesins of avian pathogenic Escherichia coli. Vet Microbiol. 2021;257:109095. doi: 10.1016/j.vetmic.2021.109095. [DOI] [PubMed] [Google Scholar]
- 42.Young MM, de Oliveira AL, Nolan LK, Barbieri NL, Logue CM. Identification of novel genes involved in the biofilm formation process of avian pathogenic Escherichia coli (APEC) PLoS ONE. 2022;17(12):e0279206. doi: 10.1371/journal.pone.0279206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26(13):1608–15. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grzymajlo K, Ugorski M, Suchanski J, Kedzierska AE, Kolenda R, Jarzab A, et al. The novel type 1 fimbriae FimH receptor calreticulin plays a role in Salmonella host specificity. Front Cell Infect Microbiol. 2017;7:326–6. doi: 10.3389/fcimb.2017.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Casali N, Preston A. E. coli plasmid vectors: methods and applications. Volume 235. Springer Science & Business Media; 2003.
- 46.Sambrook J, Russell DW. Transformation of E. coli by Electroporation. CSH Protoc. 2006;2006(1):pdb.prot3933 [DOI] [PubMed]
- 47.Kolenda R, Sidorczuk K, Noszka M, Aleksandrowicz A, Khan MM, Burdukiewicz M, et al. Genome placement of alpha-hemolysin defines virulence-associated factor profile of Escherichia coli. Microb Genomics. 2021;7(12):000743. doi: 10.1099/mgen.0.000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidorczuk K. Aleksandrowicz, Adrianna, Burdukiewicz, Michał, Kingsley, Robert, Kolenda, Rafał. Genomic characterization of enterohemolysin-encoding hemolytic Escherichia coli of animal and human origin. Microb Genomics. 2023 Feb 27. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All essential data generated or analyzed during this study are included in this published article.