Abstract
A limited number of transcription factors have been suggested to be regulated directly by Erks within the Ras/mitogen-activated protein kinase signaling pathway. In this paper we demonstrate that ERF, a ubiquitously expressed transcriptional repressor that belongs to the Ets family, is physically associated with and phosphorylated in vitro and in vivo by Erks. This phosphorylation determines the ERF subcellular localization. Upon mitogenic stimulation, ERF is immediately phosphorylated and exported to the cytoplasm. The export is blocked by specific Erk inhibitors and is abolished when residues undergoing phosphorylation are mutated to alanine. Upon growth factor deprivation, ERF is rapidly dephosphorylated and transported back into the nucleus. Phosphorylation-defective ERF mutations suppress Ras-induced tumorigenicity and arrest the cells at the G0/G1 phase of the cell cycle. Our findings strongly suggest that ERF may be important in the control of cellular proliferation during the G0/G1 transition and that it may be one of the effectors in the mammalian Ras signaling pathway.
Mitogen-activated protein kinase (MAPK) pathways are a central relay of many extracellular signals leading to change in gene expression. At least three MAPK pathways, which have high structural homology and identity in biochemical mechanisms of activation, have been identified in mammalian cells. The JNK (c-Jun amino-terminal kinase) and p38 pathways are involved primarily in the transduction of stress and cytokine stimuli. The Erk (extracellular signal-regulated kinase) pathway plays a major role in transduction of mitogenic and differentiation stimuli (for reviews, see references 41 and 47). Ras small GTPases have a pivotal role in regulation of proliferation from both receptor tyrosine kinases (RTK) and G protein-mediated receptors, (for reviews, see references 6 and 30). Notably, Ras plays an essential role in the activation of the Raf kinase, which directly phosphorylates and activates the Mek kinase, leading to the activation of Erk1 and Erk2 by phosphorylation on threonine and tyrosine residues. Phosphorylated Erks form homodimers (22) and translocate to the nucleus, where they phosphorylate proteins involved in gene regulation. Besides the Raf/Mek/Erk kinase cascade, other downstream Ras effectors are known to participate in the proliferative response (for a review, see reference 31). For example, phosphoinositide 3-OH kinase (PI3-K) (42) and members of the Rho family (for reviews, see references 17 and 23) have been shown to be responsible for morphological changes induced by Ras and are required for Ras-dependent transformation. Nevertheless, although the implication of Ras pathways, and in particular the Raf/Erk pathway, in the control of proliferation is well established, links with the control of the cell cycle machinery are not clear. Ras-dependent exit from G0 (45) and progression through G1 via the control of the retinoblastoma tumor suppressor protein (Rb) (34, 37) have been demonstrated. However, the transcription factors implicated in these responses and their target genes are poorly documented. Thus, identification of the nuclear targets of the MAPK pathways is of critical interest.
Several transcriptional factors have been proposed to be targeted by MAPKs, but the precise mechanism of their regulation by phosphorylation is not always known. For example, ATF-2 activity is regulated by both JNK and p38 kinase (16, 28, 48). The activities of MEF2C (18) and Chop (51) are enhanced through phosphorylation by p38 kinase. Binding and subsequent phosphorylation of the c-Jun transactivation domain by JNK leads to an increasing c-Jun activity (7, 25). JNK may also play a role in the phosphorylation of the transcription factor NFAT4 that leads to NFAT4 nuclear exclusion (3, 59). The Erks seem to regulate transcriptional activities of several members of the Ets family. The pointed domain of Ets2 (by analogy with Ets-domain protein pointed-P2 [PntP2]) is required for transcriptional activity and is phosphorylated by Erks in vitro (32, 56). The ternary complex factor (TCF) Elk1 is a target for all three MAPK pathways but through different determinants, within the same region, for Erk, JNK, and p38 (57, 58). Elk1 phosphorylation in its carboxyl-terminal transactivation domain by MAPK leads to enhanced DNA binding and TCF transcriptional activities (38, 55). Sap-1, another TCF family member, is preferentially targeted by Erk and p38 (38, 54, 55). ER81 (19) and ERM (20) also appear to be targets of the Ras/Raf/Mek/Erk signaling cascade, whereas Spi-B is phosphorylated by both Erks and JNK (29). Ets1 and Ets2 transcriptional activities are positively regulated by Ras (39, 56). The Drosophila gene product Yan is an Ets transcriptional repressor that is negatively regulated by Erk phosphorylation. The phosphorylation affects the subcellular localization of the protein (40) and also the stability of the protein. Another Drosophila Ets-domain protein, PntP2, is activated after phosphorylation by Erk (2). Thus, the Ras/Erk pathway controls the development of R7 fate in the Drosophila eye through phosphorylation of two antagonizing transcription factors of the Ets family, Yan and PntP2 (36). The involvement of the ets genes in the proliferation processes is further supported by their oncogenic potential and by the identification of ets gene rearrangements in human tumors (for a review, see reference 8).
ERF (Ets2 repressor factor) is a ubiquitously expressed transcriptional repressor and member of the Ets family, as defined by its DNA binding domain. ERF has no homology outside the DNA-binding domain with other ets genes, except for PE-1 (24), with which it forms a new subclass of ets genes. We have previously shown that ERF could act as a tumor suppressor gene able to revert ets- and fos-induced tumorigenicity and that the protein is probably regulated by phosphorylation during the cell cycle and after mitogenic stimulation (44). In this study, we show that Erks directly bind ERF and phosphorylate it at multiple sites. Erk-dependent phosphorylation of ERF governs its subcellular localization and thus its activity as a transcriptional repressor. Consistent with an implication of the Ras/Raf/Mek/Erk pathway, ERF acts as an immediate response factor to both mitogenic stimuli and arrest signals. Upon serum stimulation, ERF is immediately phosphorylated and exported to the cytoplasm. Conversely, upon serum withdrawal, ERF is rapidly dephosphorylated and imported into the nucleus. To investigate ERF function in response to MAPK phosphorylation, we used phosphorylation-deficient ERF mutants. Mutated ERF proteins exhibit predominantly nuclear localization and arrest cells in the G0/G1 phase of the cell cycle. Moreover, the constitutively nucleus-localized ERF mutants, in contrast to the wild-type (wt) ERF, can suppress cellular transformation induced by oncogenic Ras. Taken together, these results suggest that regulation of ERF transcriptional repressor activity is critical for the control of cellular proliferation.
MATERIALS AND METHODS
Plasmids.
ERF mutations were generated by PCR with the following mutant primers: T526A, CAGAGCTCACCCGCCTTGGGGcGAGGG; T357A, GCCCATGGCACCCGAGgCCCCACC; T271A, GGCCAGGTGGGTGGGCGcCATGGGC; SS246-251AA, AGGGATCCAGGACCGGCCAGAGGCGcCACAGGGAAGGGGcgAGG; S161A, GAAGATGAAGAGCAGGCTGGTGGTGcGCGG; and T148A, GGGGTCCTCGGTGGGGGACAGCACCTCGGAGGGCGcTGAG (lowercase letters indicate the mutated nucleotides). The PCR products were cleaved with the appropriate restriction endonucleases (T526A with SacI and NheI, T357A with BstXI, SS246-251AA with EaeI and XmnI, S161A with SapI and XmnI, and T148A with AvaII and EcoRI). The mutated DNA fragments were used to replace the corresponding fragment of the wt ERF in the pSG5 vector (44). The amplified regions were sequenced in their entirety to confirm the mutation and the absence of any other PCR-generated mutations. Multiple mutations were generated by swapping of the appropriate restriction fragments (EcoRI-AvaII for T148A, EcoRI-SapI for S161A, BamHI for SS246-251AA, BstXI for T271A, KpnI-NheI for T357A, and NheI-SstI for T527A). Clones were verified to contain the correct mutation and orientation by sequencing. The green fluorescence protein (GFP)-ERF fusion plasmid was generated by inserting the 1.9-kb SmaI-BstEII fragment of ERF in the XhoI site of pEGFP-C1 (Clontech). The GFP-ERF mutant plasmids were generated by replacing the 1.63-kb EcoRI-DraI fragment from the corresponding pSG5/ERF mutants. The in-frame fusion was verified by the reactivity to the ERF carboxyl-terminal antibody S17S. The promoter reporter plasmid (pTK-GATA.CAT), the plasmid expressing the kinase domain of c-Raf-1 (pBXB), and the plasmid containing the activated form of the Ha-ras gene (pT24) have been previously described (44).
Cell lines, transfection, and transformation.
HeLa cells were maintained in Dulbecco’s modified minimal essential medium (DMEM) supplemented with 10% bovine serum, NIH 3T3 cells were maintained in DMEM with 8% bovine serum, and Ref-1 cells were maintained in DMEM with 10% fetal bovine serum. The three cell lines have a progressively increased ability to suppress MAPK activity in the absence of serum. Cell lines were transfected with calcium phosphate or Lipofectamine (Life Technologies) by the company protocol and analyzed 24 to 48 h after transfection. Cell lines expressing ERF were generated by their ability to express the cotransfected neomycin resistance gene (neo) and proliferate in the presence of 400 μg of gentamicin sulfate (Life Technologies) per ml. Individual cell lines were tested for their ability to express elevated levels of ERF mRNA and protein. Low-serum growth was tested in DMEM–20 mM HEPES–1 or 0.2% bovine serum for 7 to 14 days. Serum starvation of Ref-1 cells was performed in DMEM–20 mM HEPES–0.04% bovine serum albumin (BSA) for the indicated times (usually 1 or 20 h). Serum stimulation was performed by the addition of fetal bovine serum to the starvation media to a 10 to 20% final concentration. Tumorigenic potential was assessed by subcutaneous injection of 3 × 105 to 1 × 106 cells in athymic mice; the animals were monitored twice a week for tumor development and general health.
Protein detection and phosphorylation.
In vivo and in vitro ERF labeling, detection, and phosphopeptide analysis were performed as previously described (44) (see Fig. 3). Immunoprecipitation of Erk complexes was performed in 10 mM Tris-HCl–100 mM NaCl–0.1% Triton X-100 (TNT buffer) with 1 μg of a rabbit polyclonal antibody against Erk1 and -2 that can specifically recognize the native Erk proteins (Zymed) for every 100 μg of extract. In vitro association of active or inactive glutathione transferase (GST)-Erk2 fusions (Upstate Biotechnology Inc.) with in vitro-translated ERF was tested under the following conditions. ERF proteins were produced by the TnT in vitro translation system (Promega), and at the end of the translation reaction, 20 ng of GST-Erk2 per ml was added and left for 15 min at 30°C. The reaction mixtures were diluted fivefold with TNT buffer, and the complexes were precipitated with glutathione (GSH)-Sepharose (Pharmacia) for 1 h at room temperature, washed five times with TNT buffer, and analyzed. The subcellular localization of GFP and GFP-ERF fusions was detected by GFP autofluorescence. Transfected cells were fixed with 4% paraformaldehyde and analyzed by microscopy. The subcellular localization of ERF and ERF mutant proteins was determined by immunofluorescence. The ERF protein was detected in methanol-acetone-fixed cells by using either S17S or M15C at a 1:30 dilution in 50 mM Tris-HCl–138 mM NaCl–2.7 mM KCl (TBS) plus 3% BSA and visualized with a biotin-labeled goat antirabbit antibody at a 1:100 dilution and streptavidin-fluorescein isothiocyanate (Jackson ImmunoResearch) at a 1:500 dilution in the same buffer by fluorescence microscopy. Replacement of the ERF-specific antibody with normal rabbit serum or addition of the immunizing peptide at 10 μg/ml resulted in the loss of the signal. Nuclear and cytosolic fractions for immunoblotting were obtained by lysing the cells with 35 strokes in a glass Dounce homogenizer (pestle B) in 10 mM Tris-HCl (pH 7.5)–300 mM sucrose–1 mM EDTA. Nuclei were pelleted at 2,000 × g for 5 min and washed with 10 mM HEPES–10 mM NaCl–1 mM KH2PO4–5 mM NaHCO3–1 mM CaCl2–0.5 mM MgCl2–0.1% Nonidet P-40. All buffers were supplemented with 1 mM orthovanadate, 1 μg of microcystin per ml, 1 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin per ml, 10 μg of aprotinin per ml, and 10 μg of leupeptin per ml. In immunoblotting, ERF was detected with either the S17S or the M15C rabbit polyclonal antibody at a 1:1,000 dilution in TBS plus 0.05% Tween 20 (TBST). Erks were detected by immunoblotting with a rabbit polyclonal antibody (Zymed) at a 1:1,000 dilution in TBST and an antibody against the phosphorylated forms of the enzymes (New England Biolabs) at a 1:600 dilution in TBST. JNK was detected with a rabbit polyclonal antibody (Santa Cruz) at a 1:100 dilution in TBST. In all cases a goat antirabbit antibody conjugated with horseradish peroxidase (Caltag) was used at a 1:3,000 dilution in TBST as the secondary antibody in immunoblotting detection. The results were visualized by exposure to either X-ray or Polaroid films. For in-gel MAPK assay, Erks were immunoprecipitated with the anti-Erk rabbit polyclonal antibody (Zymed) and analyzed in a 10% polyacrylamide gel containing 0.5 mg of myelin basic protein per ml. At the end of the electrophoresis, the gel was washed at room temperature for 1 h with 20% propanol–50 mM Tris (pH 8), for 1 h with 50 mM Tris (pH 8)–5 mM 2-mercaptoethanol, for 1 h with 6 M guanidine-HCl, and for 16 h at 4°C with 50 mM Tris (pH 8)–5 mM 2-mercaptoethanol–0.04% Tween 20. The kinase reaction was performed for 60 min at 25°C in 40 mM HEPES (pH 8)–2 mM dithiothreitol–5 mM MgCl–0.1 mM EGTA–10 μCi of [γ-32P]ATP per ml. The gel was washed with 5% trichloroacetic acid–1% sodium pyrophosphate, dried, and exposed to X-ray film. The same method was utilized to determine the JNK activity by use of the anti-JNK antibody (Santa Cruz) for the immunoprecipitation and GST–c-Jun protein as the in-gel kinase substrate.
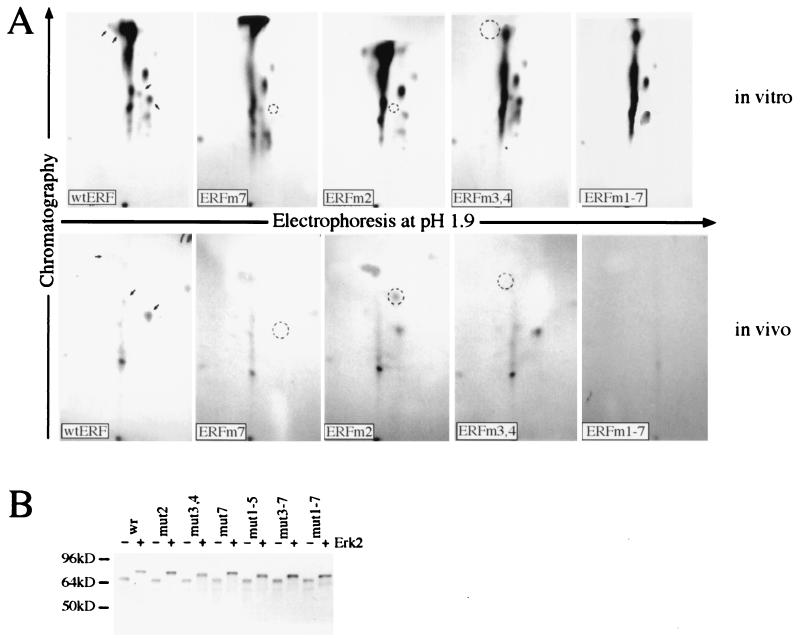
FIG. 3.
Identification of MAPK phosphorylation sites. (A) Upper panels, the indicated in vitro-produced proteins were purified by immunoprecipitation and labeled with [γ-32P]ATP and recombinant Erk2 kinase. Lower panels, for the in vivo labeling, plasmids encoding the indicated proteins were transfected into NIH 3T3 cells. Twenty hours after transfection, the cells were serum deprived, labeled for 4 h with 32P, and stimulated for 10 min with serum, and the ERF proteins were immunoprecipitated with the S17S antibody. The labeled proteins were isolated after SDS gel electrophoresis and digested with trypsin, and the tryptic peptides were analyzed by chromatography and electrophoresis. The arrows in the wt ERF panels indicate the phosphopeptides eliminated or modified by the subsequent mutations. The circles indicate the positions of the eliminated or modified peptides. (B) The indicated ERF proteins were synthesized in vitro and labeled with [35S]methionine. Half of each sample (lanes +) was phosphorylated with active GST-Erk2 and ATP. The effect of the mutations on ERF mobility was determined after SDS gel electrophoresis.
Analysis of cell DNA content.
Analysis of the DNA content of live NIH 3T3 cells by flow cytometry was performed after transfection of 5 × 105 cells with 8 μg of plasmid DNAs (total). For cotransfections, 4 μg of each plasmid was used. One hour before harvest, 10 μg of Hoechst 33342 dye per ml was added to the medium. At 24 or 48 h after the transfection, the cells were trypsinized, placed in complete medium, and kept on ice until the analysis, which took place within 1 h. The live cells were analyzed by using a two-laser flow cytometer (Innova 70 [488 nm] and Innova 305 [MLUV] argon lasers). The cell cycle profile of GFP-positive cells was obtained by using the ModFit LT 2.0 program. Monitoring of the newly synthesized DNA in Ref-1 cells was performed after transfection with a CD4-expressing plasmid as a marker of the transfected cells and the appropriate ERF-expressing plasmid. At 8 h before harvest, 50 μM bromodeoxyuridine (BrdU) was added to the culture medium. At 24 and 48 h after transfection, the cells were fixed and stained with an anti-BrdU mouse monoclonal antibody (Sigma) at a 1:100 dilution in TBS plus 3% BSA, visualized with a biotin-labeled goat antimouse antibody at a 1:100 dilution in TBS plus 3% BSA and streptavidin-lissamine rhodamine at a 1:500 dilution in TBS plus 3% BSA (Jackson ImmunoResearch), and analyzed by fluorescence microscopy. CD4-positive cells were detected by using a fluorescein isothiocyanate-conjugated anti-CD4 mouse monoclonal antibody (Immunotech) at a 1:10 dilution in 10 mM sodium phosphate (pH 7.5)–138 mM NaCl–2.7 mM KCl–1 mM CaCl2.
RESULTS
Physical association and phosphorylation of ERF by Erks.
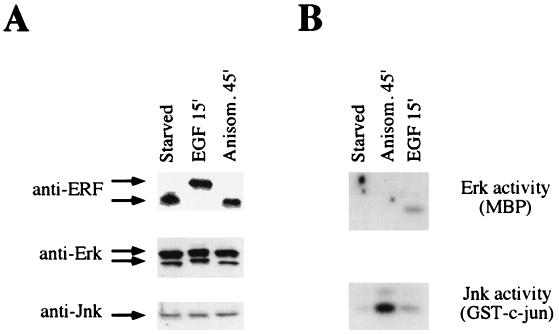
Our previous work suggested that ERF may be a substrate for MAPKs. The members of this family of serine/threonine kinases phosphorylate similar sites, but they are distinguishable both in their response to extracellular stimuli and in the repertoire of proteins that they phosphorylate. MAPKs and Erks are activated in response to mitogenic stimuli (47) as downstream effectors of the Ras signaling pathway, whereas stress-activated protein kinases (JNKs and p38) are activated in response to stress factors in the cdc42/rac pathway (5, 33). To determine if ERF was phosphorylated as a result of the activation of either pathway, we analyzed its in vivo phosphorylation following cellular stimulation by either EGF (mitogen) or noninhibitory levels of anisomycin (stress). The activation of MAPKs and stress-activated protein kinases, respectively, by these agents has been shown to be preferential for each class (25, 52). As shown by the in-gel kinase assay, the observed activation of Erks and JNKs by the respective stimuli was specific (Fig. 1). Although no quantitative changes were evident at the protein level, ERF as well as Erk p42 and p44 proteins exhibited the characteristic mobility shift associated with their phosphorylation. In contrast, ERF is not phosphorylated as a result of stress-activated kinases. Similar results were obtained when the cells were induced with either serum or phorbol 12-myristate 13-acetate (Erk induction) or were irradiated by UV light (JNK and p38 induction) (data not shown). Furthermore, incubation with the specific kinase inhibitors rapamycin and wortmannin 30 min prior to the induction with serum or phorbol 12-myristate 13-acetate did not affect ERF phosphorylation, indicating that neither PI3-K nor S6 kinase was involved in ERF phosphorylation. The ability of Erk2 to readily phosphorylate ERF in vitro and the good in vivo correlation between the activation of Erks and the phosphorylation of ERF are indications that ERF may be phosphorylated in vivo by Erks.
FIG. 1.
ERF is phosphorylated in response to mitogens but not in response to stress. (A) Ref-1 cells expressing the ERF gene were arrested by serum starvation for 20 h and then induced for the indicated times with either 100 ng of epidermal growth factor (EGF) per ml or 25 ng of anisomycin (Anisom.) per ml. Thirty micrograms of cellular extract was analyzed by SDS gel electrophoresis, and proteins were detected by immunoblotting with the indicated antibodies. (B) Three hundred micrograms of the same cellular extracts was immunoprecipitated with anti-Erk or anti-JNK antibodies, as indicated. The kinase activity was detected by in-gel kinase assay with, as substrates, myelin basic protein (MBP) for Erks and the amino terminus of c-Jun for JNKs.
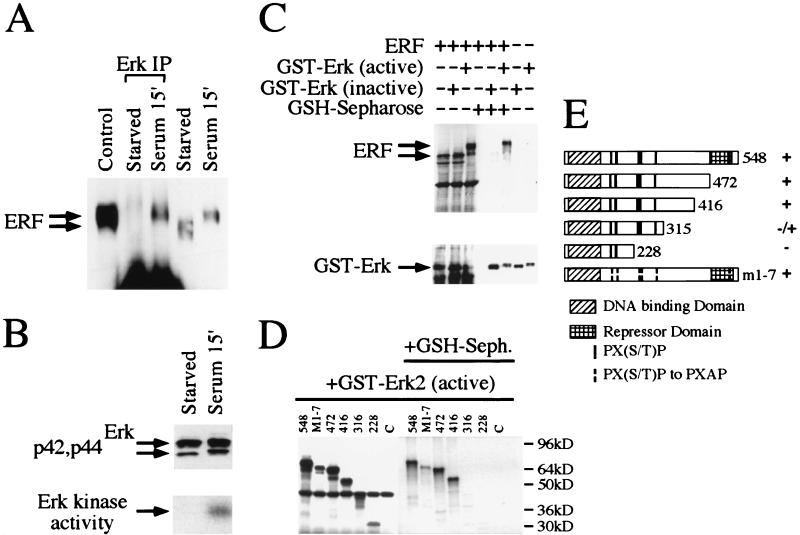
A strong indication of the in vivo phosphorylation of a substrate by a specific kinase is their physical association. We performed an immunoprecipitation under nondenaturing conditions with a rabbit anti-Erk polyclonal antibody (Zymed) that recognizes nondenatured Erks (Fig. 2A, lanes 2 and 3), and the immunoprecipitated complexes were examined for the presence of ERF with the S17S anti-ERF specific antibody. ERF can be found in the complexes immunoprecipitated by the anti-Erk antibody, but only under conditions in which the Erks are activated (Fig. 2A, lane 3, and B, lane 2). Under these conditions, ERF is phosphorylated, as shown by the mobility shift of the protein (Fig. 2A, lanes 4 and 5). The Erk-ERF interaction could also be detected in vitro by using the active or the inactive forms of bacterially expressed GST-Erk2 fusion proteins in combination with ERF synthesized in a cell-free system (Fig. 2C). The active form of GST-Erk2 could phosphorylate ERF, and their physical association was evident by the detection of ERF in GSH-Sepharose-associated complexes (Fig. 2C, upper panel, lane 6). In contrast, the interaction between inactive GST-Erk2 and ERF was at least an order of magnitude weaker (Fig. 2C, upper panel, lane 5). To determine the region of ERF mediating the interaction with Erk2, we analyzed the interaction between ERF deletion mutants and activated GST-Erk2 (Fig. 2D). Our data suggest that the region between amino acids 316 and 416 of the ERF protein is required for strong interaction with GST-Erk2. The ERF mutant with all seven putative MAPK sites mutated to alanine (see below) (Fig. 2D, M1-7) could still effectively associate with Erk2, indicating that phosphorylation of ERF at these sites did not affect the interaction.
FIG. 2.
Physical interaction of ERF and Erk in vivo and in vitro. (A) Ref-1 cells were grown in the absence of serum for 20 h (starved) and then were induced for 15 min with 20% fetal calf serum (serum). Five hundred micrograms of cellular extracts from Ref-1 cells expressing ERF was immunoprecipitated with an anti-Erk rabbit polyclonal antibody under nondenaturing conditions (Erk IP) and analyzed by SDS gel electrophoresis. Twenty micrograms of cellular extracts before the addition of the anti-Erk antibody and of extracts from cells growing in complete medium (control) was also analyzed on the same gel. The presence of the ERF protein was detected by immunoblotting with the S17S anti-ERF specific antibody and is indicated by arrows. (B) Erk presence and activity in the same extracts were detected by immunoblotting (upper panel) and in-gel kinase assay (lower panel) as described for Fig. 1. (C) In vitro-translated ERF was mixed with active or inactive GST-Erk2 (Upstate Biotechnology Inc.), and the complexes were precipitated with GSH-Sepharose and analyzed by SDS gel electrophoresis. The presence or absence of ERF was detected by autoradiography (upper panel), and that of GST-Erk2 was detected by immunoblotting with an anti-Erk specific antibody (lower panel). (D) The presence or absence of truncated ERF proteins in complex with active GST-Erk2 was detected as described for panel C. The numbers indicate the carboxy-terminal amino acid of each deletion. M1-7 is a full-length ERF with all seven putative MAPK sites mutated to alanine. (E) Diagrammatic representation of the deletions and mutation used for panel D. The Ets DNA-binding domain, the previously identified repression domain, and the putative MAPK phosphorylation sites are indicated. Plus and minus indicate the interaction of each protein with active GST-Erk2 kinase.
We have previously shown that threonine 526 is a site of in vivo and in vitro phosphorylation by Erks (44). In order to determine other possible sites of phosphorylation by Erks, we employed the same approach of site-directed mutagenesis and two-dimensional phosphopeptide mapping for the remaining six optimal MAPK sites, at positions T148 (position 1), S161 (position 2), S246 (position 3), S251 (position 4), T271 (position 5), and T357 (position 6). For the in vivo phosphorylation we used activated GST-Erk2 kinase, and for the in vivo phosphorylation we used transfection and mitogenic stimulation. We were not able to obtain phosphopeptide maps by using either nonactivated GST-Erk2 or unstimulated cells because of the negligible 32P incorporation. Our analysis indicates that in addition to T526 (position 7), S161 (position 2), S246 (position 3), and S251 (position 4) are also phosphorylated in vitro by Erk2 and in vivo after mitogenic stimulation (Fig. 3A). The phosphorylation on these four sites (indicated by arrows in the wt panels in Fig. 3A) was determined either from the elimination of specific phosphopeptides or from the shift of a phosphopeptide to a more hydrophobic and less charged position (Fig. 3A, ERFm2, in vivo) and is suggestive of the elimination of one phospho residue from a peptide phosphorylated at multiple sites. No changes in the phosphopeptide maps could be detected with the other three ERF mutations (at T148, T271, and T357), either in vitro or in vivo. As an additional indication of phosphorylation, we determined the effects of these mutations on the electrophoretic mobilities of the phosphorylated and nonphosphorylated ERF proteins. We have previously shown that phosphorylation has a significant effect on the electrophoretic mobility of the ERF protein. Alanine mutations do not affect the mobility of the nonphosphorylated form of the protein but have a clear effect on the mobility of ERF after in vitro phosphorylation by Erk2. Phosphorylation at positions 3 and 4 appears to have a major effect on the mobility of the protein (Fig. 3B). In contrast, mutations at other sites have a minimal effect. These data support the phosphopeptide mapping findings and our previously published observations that the ERF mobility shift is due to protein phosphorylation. Our analysis also indicates that, at least under in vitro conditions, additional sites can be phosphorylated by Erk2, generating characteristic phosphopeptides (Fig. 3A, upper panels, ERFm1-7) that contribute to the mobility shift of the protein (Fig. 3B, mut1-7). However, we were not able to detect specific phosphopeptides with the 1-7 ERF mutation after mitogenic stimulation (Fig. 3A, lower panels, ERFm1-7), although the electrophoretic mobility of the 1-7 ERF mutant was affected by serum stimulation. ERF has eight additional suboptimal putative MAPK sites (44), and it is not clear at this point if any of these sites are actually utilized in vivo. The sites identified so far appear to be relevant and important for ERF regulation (see below).
Subcellular localization of ERF.
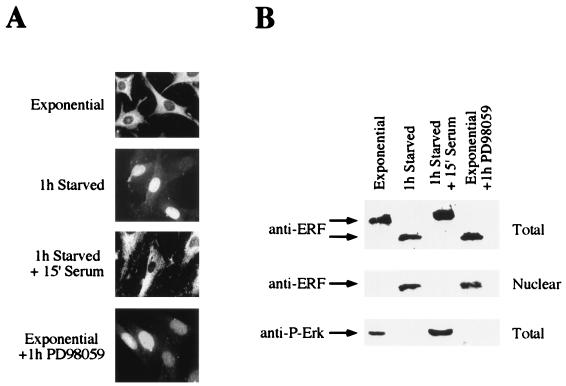
Our previous work suggested that phosphorylation of ERF did not affect its ability to bind to the specific Ets-binding site on DNA (44). In addition, we found no evidence that phosphorylation may affect protein stability in a way similar to Yan. Subcellular fractionation experiments suggested that ERF could be found mostly in the cytoplasmic fraction of proliferating cells. Therefore, we determined the subcellular localization of ERF in relation to its phosphorylation state. We employed direct immunofluorescence with two ERF-specific antibodies (S17S and M15C) to analyze the ERF subcellular localization. In exponentially growing cells, ERF could be detected almost exclusively in the cytoplasm (Fig. 4). However, upon serum starvation and exit from the cell cycle, ERF could be found only in the nucleus. This process was rapid; nuclear ERF could be detected within 15 min after serum withdrawal, and its nuclear relocalization was complete within 30 to 45 min. The entire process was reversed upon mitogenic stimulation, and ERF could be detected in the cytoplasm within 5 min following stimulation. This translocation was complete within 10 to 15 min. The nuclear localization of ERF was also induced in the presence of serum by the addition of the specific inhibitor PD98059, which specifically blocks Erk activation by MEK1 (9) (Fig. 4). The same inhibitor blocked the exit of ERF from the nuclei of serum-arrested cells after serum addition, suggesting that the subcellular localization of ERF is totally dependent on MAPK activation and phosphorylation.
FIG. 4.
Phosphorylation-dependent subcellular localization of ERF. (A) Ref-1 cells growing under the indicated conditions were fixed and stained with the S17S anti-ERF specific antibody and visualized by fluorescence microscopy (magnification, ×70). (B) Total and nuclear protein extracts from the same cells were analyzed by immunoblotting with the same anti-ERF specific antibody. Activated Erks were detected with an antibody directed against their phosphorylated form.
In order to determine whether this process was associated with the phosphorylation of ERF, we examined its phosphorylation state by immunoblotting (Fig. 4B). Nuclear and cytoplasmic extracts were prepared from stimulated or quiescent cells in the absence or presence of the MAPK activation inhibitor PD98059. Care was taken to minimize cross contamination between the nuclear and cytoplasmic fraction. Consistent with the immunofluorescence data, ERF could be found in the nuclear fraction either in quiescent cells or in the presence of the MEK1 inhibitor PD98059. Under these conditions, ERF was not phosphorylated as indicated by its faster mobility in sodium dodecyl sulfate (SDS) gel electrophoresis. Under the same conditions, we were unable to detect the phosphorylated form of MAPK with an anti-phospho-Erk specific antibody (New England Biolabs), indicating that the kinases are inactive (Fig. 4B).
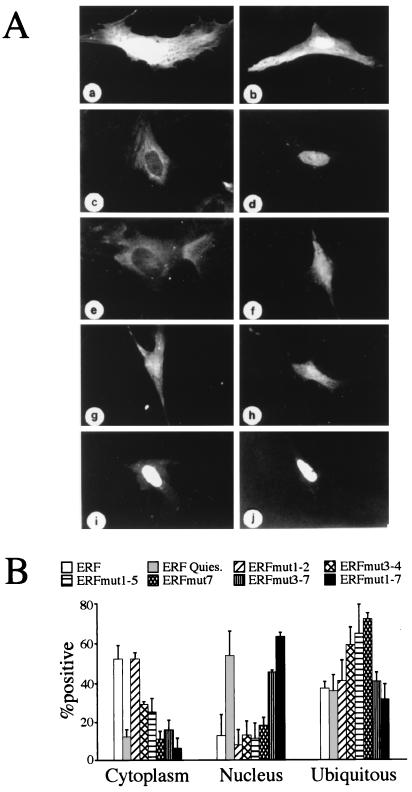
In order to determine if any of the MAPK phosphorylation sites that we have identified on ERF may be responsible for its regulated subcellular localization, we generated fusions of ERF mutants with GFP. The fusion of the wt ERF with GFP exhibited the same subcellular localization in response to growth arrest as the ERF protein (Fig. 5A), and it was active as a transcriptional repressor (not shown). Upon introduction of mutations that eliminate MAPK phosphorylation sites on ERF, an increased incidence of nuclear localization of ERF in proliferating cells was observed (Fig. 5B). Mutations at amino acids 246 (position 3), 251 (position 4), and 526 (position 7), in particular, contributed to an increased nuclear localization. However, none of the three mutations alone or in combination was sufficient for the exclusive nuclear localization of ERF. In contrast, mutation of all seven putative MAPK sites resulted in the predominantly nuclear localization of ERF, suggesting that although position 161 by itself does not appear to affect the localization of the protein, it is necessary for this process (Fig. 5B, ERFmut3-7 versus ERFmut 1-7). Mutations to glutamic acid at positions 3, 4, and 7, alone or in combination, caused a subcellular distribution similar to the wt, suggesting either that the negative charge of the glutamic acid may not be sufficient to mimic phosphorylation or that other positions are also required. Alternatively, phosphorylation may not be important for nuclear import but may be required for nuclear export. None of the mutations inhibited the ability of ERF to translocate to the nucleus upon serum withdrawal. These data further indicate that the direct phosphorylation of ERF by MAPK is responsible for the regulation of its subcellular localization.
FIG. 5.
Site-specific phosphorylation determines ERF localization. (A) Ref-1 cells were transfected with a GFP-expressing plasmid (a and b) or with plasmids expressing GFP-ERF fusion proteins (c to j). The localization of the proteins was determined by the GFP fluorescence in exponentially growing cells (a, c, and e to j) or 1 h after serum withdrawal (b and d). (a and b) GFP; (c and d) GFP-ERF; (e) GFP-ERFmut1-2; (f) GFP-ERFmut3-4; (g) GFP-ERFmut7; (h) GFP-ERFmut1-5; (i) GFP-ERFmut3-7; (j) GFP-ERFmut1-7. (B) The localization of hybrid proteins expressed from the same plasmids as for panel A was scored in at least three different experiments. The values are the averages from at least 100 positive cells. Cells with proteins localized exclusively in the nucleus or in the cytoplasm were scored in the respective category. Ubiquitous distribution includes cells with detectable fluorescence in both compartments even when the distribution was not even.
Growth arrest by nuclear ERF.
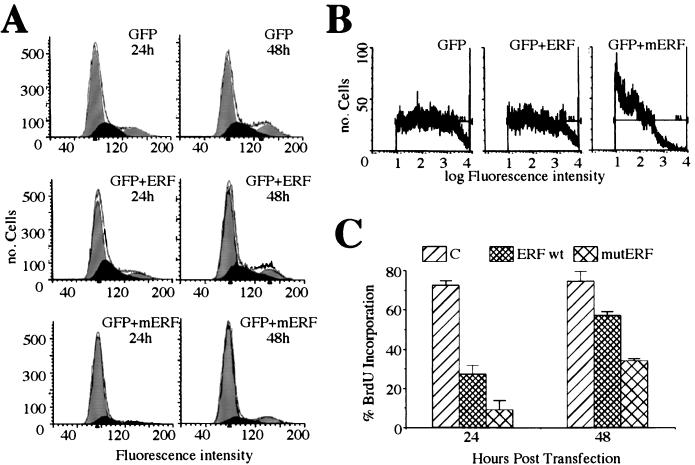
The rapid translocation of ERF into the nucleus upon growth factor deprivation and its subsequent immediate export upon mitogenic induction suggested that ERF may contribute to proliferation control. Such a hypothesis is consistent with our difficulties in obtaining cell lines expressing high levels of ERF and with the inability to obtain cell lines expressing nucleus-localized ERF mutants. In order to determine the fate of cells expressing an ERF mutant that is localized predominantly in the nucleus (ERFm1-7), we analyzed the DNA content of transiently transfected cells. NIH 3T3 cells were transfected with a GFP-expressing plasmid and either an empty vector or a vector encoding the wt ERF or the ERF mutated in all seven sites (Fig. 6A, mERF). At 24 and 48 h posttransfection, live cells were analyzed by flow cytometry. The fluorescence of the GFP was used to score the transfected-cell population. The DNA content of these cells was determined by the fluorescence intensity of Hoechst 32224 dye, which stains DNA in live cells (Fig. 6A, horizontal axis). In our experiment G0/G1 and G2/M cells have average fluorescence intensities of 80 and 160, respectively. The area in each segment defines the number of cells in each stage of the cell cycle. Our data (Fig. 6A) indicate that about 83% of the cells expressing the nucleus-specific ERF mutant had a DNA content corresponding to G0/G1 cells, in contrast to 70% of cells expressing the wt ERF and 63% of cells expressing GFP alone. Similar data were obtained when GFP-ERF fusions were used instead of GFP and ERF cotransfections. However, the level of expression of the mutated ERF, either when GFP-ERF fusions were used or when GFP- and ERF-expressing plasmids were cotransfected, was more than 1 order of magnitude lower than that of wt ERF or GFP alone. This could be estimated by the intensity of the GFP fluorescence of the transfected cells, shown on the horizontal axis of Fig. 6B in logarithmic scale. When cell populations with comparable levels of GFP or GFP-ERF expression were used for this analysis, with a GFP intensity 102 to 103, more than 95% of the cells expressing mutated ERF were in the G0/G1 state, in contrast to 73 and 65%, respectively, for wt ERF- and GFP-expressing cells. We also tested the ability of cells transfected with ERF to progress in the cell cycle by examining their competence in incorporating BrdU in newly synthesized DNA. Ref-1 cells were transfected with a plasmid encoding CD4, to allow scoring of the transfected cells, and either an empty vector or a vector encoding for wt ERF or ERF mutated in all seven sites (Fig. 6C, mutERF). At 16 and 40 h posttransfection, the cells were labeled with BrdU for 8 h. CD4-positive cells were scored for BrdU incorporation under fluorescence microscopy. Figure 6C shows that fewer than 10% of the cells expressing the ERFm1-7 mutant could incorporate BrdU 24 h after the transfection, in contrast to 30% of the wt ERF-expressing cells and more than 70% of those with the CD4 marker plasmid alone. These numbers were proportionally higher 48 h after the transfection. Both the flow cytometry (Fig. 6A) and the nuclear morphology (Fig. 4A and 5A) data provided no indication of apoptosis with either the wt or the mutated ERF. These data suggest that entry of ERF into the nucleus results in the inhibition of cellular proliferation by cell cycle arrest, consistent with its possible role as a growth-controlling protein.
FIG. 6.
Cell cycle arrest by ERF. (A) NIH 3T3 cells were cotransfected with a GFP-expressing plasmid and an empty vector plasmid, a plasmid expressing wt ERF (ERF), or a plasmid expressing the ERFm1-7 mutant (mERF). At 24 and 48 h after transfection, the cells were harvested and the DNA content of GFP-positive cells was determined by flow cytometry from the fluorescence of Hoechst 33342 dye. The data were analyzed with the ModFit LT 2.0 program. The black area represents S phase cells. (B) The protein expression level in the transfected cells was determined by the intensity of the GFP autofluorescence. (C) Ref-1 cells were cotransfected with a CD4-expressing plasmid and an empty vector plasmid (C), a plasmid expressing wt ERF (ERF), or a plasmid expressing the ERFm1-7 mutant (mutERF). Cells were scored for CD4 expression and BrdU incorporation by immunofluorescence. The values are the averages for at least 300 CD4+ cells from three independent experiments. The bars indicate statistical error.
Suppression of ras-induced tumorigenicity by ERF.
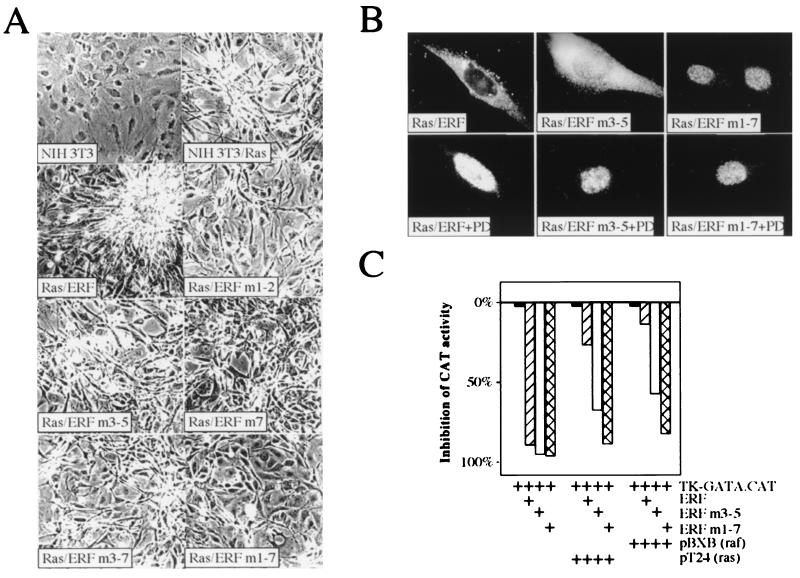
The phosphorylation and regulation of ERF by MAPKs indicated that it could be an effector in the branch of the Ras signal transduction pathway mediated by Erks. We have previously shown that ERF, in contrast to v-ets and v-fos, does not effect ras-induced tumorigenicity and that addition of activated ras or raf in transient-transfection assays inhibits the ERF transcription repressor activity (44). In order to determine if the phosphorylation-dependent subcellular localization of ERF correlated with its ability to inhibit cellular transformation, we introduced phosphorylation-deficient ERF mutations in Ha-ras-transformed NIH 3T3 cells. ERF-transfected cell lines were obtained at a frequency inversely proportional to the number of mutations carried by the ERF gene. Only two cell lines carrying the ERFm1-7 mutation were obtained, and the level of expression of the mutated protein, as determined by immunoblotting, was 10- to 20-fold lower than that of the other ERF mutants and 3- to 4-fold higher than that of the endogenous ERF protein. Established cell lines, in which expression of ERF, retention of the activated Ha-ras gene, and activation of MAPKs could be confirmed, were analyzed for growth rate, morphological changes, ability to grow in low-serum and suspension media, and tumorigenic potential. The growth rates of clones obtained with all ERF mutations were comparable, with no statistically significant difference for any of the mutations (Table 1). However, differences were evident in the other tests. wt ERF had no detectable effect on ras-transformed cells. Their morphology, ability to grow in low-serum medium and in soft agar, and ability to cause tumors after injection into athymic mice were identical to those of the parental Ha-ras-transformed NIH 3T3 cells (Fig. 7A and Table 1). In contrast, ras-transformed cells transfected with ERF mutants that had alanine mutations at the sites of phosphorylation by MAPK exhibited distinct phenotypes dependent on the specific mutations. Single mutations had no detectable effect, except for mutation 7, which affected tumor development but not morphology or soft-agar colony formation (Fig. 7A, Table 1, and unpublished data). Threonine 526 is the major serum-inducible phosphorylation site, and we have previously shown that phosphorylation at this site decreased the repressor activity of ERF (44). Multiple mutations that abolished more than one phosphorylation site caused altered morphology, decreased ability to grow in soft agar, and decreased ability to cause tumors following injection into nude mice (Fig. 7A, Table 1, and unpublished data). Mutation 1-7 caused a morphology reminiscent of that of nontransformed NIH 3T3 cells and exhibited comparable tumorigenic potential. We were not able to detect expression of the ERFm1-7 mutant protein in the tumors that arose from these cells, although the presence of the transgene was detectable. Similar results were obtained when pools instead of individual clones were used for the tumor assays. However, tumors developed 1 week earlier on average, possibly due to the variable expression of the mutated ERF transgene introduced by cotransfection with the selectable marker or even to a total lack of transgene expression. We were not able to use pools for the ERFm1-7 mutant due to the lack of clones. However, the results for all the other mutations were consistent, minimizing the possibility of clonal variation effects in our results. In addition, independent clones, with comparable levels of expression of a given mutant, gave similar phenotypes regarding morphology, growth rate, and soft-agar growth. In contrast, low-level expression of mutated ERF proteins (other than the ERFm1-7 mutant) gave a much weaker suppression of the ras-induced transformation in these assays.
TABLE 1.
Suppression of ras tumorigenicity by ERF mutationsa
| Cell line | Doubling time (h)b | No. of mice with tumors/total at wk:
|
|||||
|---|---|---|---|---|---|---|---|
| 3 | 4 | 5 | 6 | 7 | 8 | ||
| 3T3 | 26 | 1/5 | 1/5 | ||||
| Ras | 24 | 5/5 | |||||
| Ras/ERF | 23 | 5/5 | |||||
| Ras/ERFm1-2 | 23 | 1/5 | 2/5 | 3/5 | 5/5 | ||
| Ras/ERFm3-5 | 24 | 2/5 | 4/5 | 5/5 | |||
| Ras/ERFm7 | 22 | 1/5 | 3/5 | 5/5 | |||
| Ras/ERFm3-7 | 24 | 1/5 | 3/5 | 4/5 | 5/5 | ||
| Ras/ERFm1-7 | 24 | 1/5 | 2/5 | ||||
One-quarter million of G418-selected NIH 3T3 cells generated after transfection with the indicated plasmids were injected subcutaneously into athymic mice. The animals were monitored twice a week for tumor development and general health. The mutations are as in Fig. 7.
In complete growth medium.
FIG. 7.
ERF mutants can suppress the ras-transformed phenotype. (A) NIH 3T3 cells transformed with pT24 Ha-ras were transfected with wt or phosphorylation-deficient ERF mutants. Transformed cells were selected with G418 to establish colonies and cell lines. Cells from established cell lines were grown in the presence of 1% serum for 10 days to determine effects on cell morphology and photographed with a 10× phase-contrast lens. The numbers indicate the mutation(s) for each plasmid: 1, T148A; 2, S161A; 3, S246A; 4, S251A; 5, T271A; 6, T357A, and 7, T526A. (B) The subcellular localization of ERF mutants in ras-transformed NIH 3T3 cells was determined by immunofluorescence in exponentially growing cells in the presence or absence of the specific MEK1 inhibitor PD98059 (PD). (C) One microgram of the pTK-GATA.CAT reporter plasmid was cotransfected into HeLa cells with 0.5 μg of the indicated ERF plasmid and 1 μg of the indicated Ha-ras and c-raf-1 plasmids. The bars indicate inhibition of chloramphenicol acetyltransferase (CAT) activity relative to that in the samples in the absence of exogenous ERF.
The abilities of the various ERF mutants to suppress ras-induced transformation were consistent with their subcellular localization. wt ERF was cytoplasmic; ERF mutated at positions 246, 251, and 273 (ERFm3-5) could be detected in both compartments; and the protein mutated in all seven sites (ERFm1-7) was nuclear (Fig. 7B). The localization of ERF in ras-transformed cells was insensitive to serum withdrawal but sensitive to the MEK1 inhibitor PD98059 (Fig. 7B and unpublished data). These data suggest that subcellular localization may be the major mode of ERF regulation. The transformation data were also consistent with the transcriptional inhibition of a reporter gene by ERF. wt and mutated ERFs were tested for their ability to repress a reporter gene in the presence of MAPK activators. We utilized the artificial promoter reporter plasmid carrying two copies of the Ets-binding site of the GATA-1 gene. This reporter has been previously used successfully by us and others (43, 44) and has the advantage of having detectable activity in the presence of ERF, so the level of derepression can be estimated accurately. Similar derepression was also observed by using reporter genes with other Ets-dependent native or artificial promoters. In these transient-transactivation assays, ERF mutants that exhibited nuclear localization were insensitive to inactivation by MAPK activators such as ras and raf, in contrast to the wt ERF (Fig. 7C), suggesting again that nuclear localization may be the major mode of regulation of the ERF repressor activity.
Collectively our findings suggest that the phosphorylation of ERF by MAPK and its subsequent inactivation by export to the cytoplasm may be required steps for cellular proliferation and contributing factors to ras transforming ability.
DISCUSSION
The RTK/Ras/Erk signaling pathway is probably the most extensively studied pathway and has been associated with a plethora of physiological and malignant conditions. Several transcription factors that mediate this signal transduction pathway and result in the modification of the cellular transcription program have been identified. However, it is not always clear how these possible effectors are regulated by the RTK/Ras/Erk pathway. In this work, we provide evidence that ERF, a ubiquitously expressed transcriptional repressor encoded by a gene belonging to the ets family of genes, is directly phosphorylated by Erks and is exported from the nucleus as a result of this phosphorylation. Furthermore, we provide evidence suggesting that ERF may control cellular proliferation and that its inactivation by phosphorylation may be a required step in the development of ras-induced transformation. Thus, ERF appears to be one effector of a major signaling pathway in mammalian cells.
ERF is a MAPK target.
Erks have been shown to partially translocate into the nucleus upon activation (11, 12), where they phosphorylate transcription factors and alter their function. However, the repertoire of putative targets of Erks is rather limited (e.g., c-Myc [15]) and includes mostly members of the Ets family of transcription factors (Elk1 [58], Yan [40], Lin1 [46], and Ets2 [32]). Our data demonstrate that ERF is a specific target for Erks and is phosphorylated in vivo and in vitro at multiple sites. Four such sites have been so far identified, and additional sites cannot be excluded (Fig. 3). Additional sites on ERF can be phosphorylated by Erk2 in vitro, and in vivo phosphorylation at other sites by Erks or other kinases cannot be excluded. Indeed, the ERFm1-7 mutant exhibits a slower mobility than the ERF protein isolated from resting cells, indicating either that additional phosphorylation sites are utilized or that the protein undergoes additional modification that may be phosphorylation dependent. However, in all of the cases that we tested, we were not able to identify any condition where ERF activity, phosphorylation, and subcellular localization changes were independent of Erk activity. Our attempts included the use of stress factors; protein kinase A and C inducers; calcium ionophore; inhibitors of PI3-K, S6 kinase, and p38 kinase; and phosphatase inhibitors (unpublished data). Erk phosphorylation sites appear to be readily involved in the regulation of the ERF protein. In contrast to most other transcription factors that may be Erk substrates, it appears that ERF is phosphorylated in vivo predominantly, if not exclusively, by members of the Erk subclass (Fig. 1). This was also the case in vitro, where phosphorylation of ERF by MAPKs other than Erks was minimal or absent (unpublished data). We cannot exclude the possibility that other members of the MAPK family of kinases may be able to phosphorylate ERF in vivo. However, the total dependence of ERF regulation on the specific MEK1 inhibitor PD98059 strongly argues that Erks are the predominant regulators of its function. The specificity of this phosphorylation is further supported by the physical association of the two proteins in vivo (Fig. 2). This association is fairly strong, and almost 10% of the total ERF can be found in complexes with Erks after mitogenic stimulation. A similar percentage of the total Erk protein could be found associated with GST-ERF when pull-down experiments were performed (unpublished data). It is of interest that the activated but not the inactive form of Erk preferentially associates with ERF. This preference, observed in the in vitro experiments (Fig. 2), may be related to structural changes associated with the activation of Erk, including its dimerization (22). In contrast, phosphorylation of ERF does not appear to affect the association with Erk (Fig. 2 and unpublished data). In vivo, the apparent specificity of the activated Erks towards ERF is further enhanced due to the different localization of the two proteins. ERF is nuclear in resting cells, whereas Erks are cytoplasmic; thus, any physical interaction would not be possible.
It has been shown (58) that a specific region of the Elk1 protein, called the D domain, mediates the interaction of Elk1 with Erks. We were not able to identify any region within the ERF protein that has sequence similarity with the Elk1 D domain. Similar tertiary structures can be formed by two proteins even when no homology in the primary sequence is detected. Such a possibility cannot be excluded for the Erk interaction domains of ERF and Elk1. However, Elk1 and ERF are members of the same family; thus, if their respective Erk interaction domains are functionally similar, some sequence similarity would be expected, as in the case of their DNA-binding domains. An alternative hypothesis is that the two Erk interaction domains are distinct. This hypothesis is further supported by the fact that ERF does not appear to be a substrate for JNK or p38, in contrast to Elk1, which interacts with JNK and p38 through the same D domain. Thus, it appears that the Elk1 D domain is a structural determinant for binding by all three MAPK subclasses, whereas the Erk association domain of ERF seems to be specific for Erk1 and Erk2. Further analysis is necessary to identify the minimal structural determinants of ERF required for its interaction with Erks.
Nuclear-cytoplasmic shuttling of ERF.
Subcellular distribution of growth-regulating proteins (Rb, p53, Myc, Fos, and NF-κB [for a review, see reference 49]) as a function of the cell cycle stage has been observed and has been suggested as a mode of their regulation. Controlled nuclear import upon stimulation appears to be the most common process for these proteins. In contrast, stimulation-induced nuclear export of transcription factors has been recently described for NFAT4, which is exported from the nucleus upon phosphorylation by JNK (3) or casein kinase II (59). In both cases, however, phosphorylation has been implicated directly or indirectly as one of the possible regulatory mechanisms (21). The experiments presented here indicate that ERF is regulated during nuclear import and/or export and that this process depends on its phosphorylation by Erks (Fig. 4 and 5). In resting cells, ERF is located in the nucleus and upon mitogenic stimulation is phosphorylated by Erks and exported immediately into the cytoplasm, where it stays with a half-life of more than 4 h. This process is blocked by leptomycin B, suggesting an active nuclear export (unpublished data). Conversely, upon growth factor withdrawal, ERF is rapidly dephosphorylated and imported into the nucleus. The process is dependent primarily on Erks, and it can take place in the presence of growth factors when MAPKs are inhibited by the specific inhibitor PD98059, which inhibits the phosphorylation of Erks by MEK1 (9). In contrast, the p38 inhibitor SB203580 does not affect ERF localization when Erks are active (unpublished data). However, preliminary observations suggest that stress-induced pathways may control dephosphorylation of ERF and contribute to the mechanism of its nuclear import.
Our data demonstrate that ERF is found in its unphosphorylated form when isolated from nuclear extracts (Fig. 4B) and that phosphorylation of ERF at multiple sites is required in order for it to be effectively exported from the nucleus (Fig. 5A). Although ERF has a nuclear localization signal (NLS) within the DNA-binding domain, like other members of the Ets family, it does not contain a recognizable nuclear export signal with any similarity to the previously identified ones (for reviews, see references 4 and 14), and thus it may represent a new class of exported proteins. The extensive mobility shift of the protein after phosphorylation suggests a drastic change in the protein structure upon phosphorylation. Most of this change can be attributed to phosphorylation in the middle section of the protein (Fig. 2B). However, phosphorylation at only these sites is not sufficient to drive the protein into the cytoplasm (Fig. 5A), suggesting that further phosphorylation at the other sites (sites 2 and 7) may be involved in the substrate recognition of the export machinery. Another effect of the phosphorylation at these sites may be to expose recognition determinants for proteins responsible for sequestering a transiently exported ERF protein in the cytoplasm. Masking of the NLS, in contrast to the recently reported case of NFAT4 (59), does not appear to be a likely mechanism of ERF regulation. Replacement of the entire ERF DNA-binding domain with other NLS-carrying domains results in hybrid proteins that exhibit a subcellular distribution similar to that of the wt ERF (unpublished data). In addition, we have previously shown that phosphorylation does not affect the DNA binding ability of ERF in vitro, which argues against extensive structural changes within the NLS-containing DNA-binding domain of ERF. The localization of the ERF glutamic acid mutants indicates that a negative charge at positions 3, 4, and 7 is not sufficient to inhibit nuclear import, suggesting that dephosphorylation might not be critical for the import process per se. It is conceivable that the rate of the export process is dependent on phosphorylation. However, under quiescent conditions, cofactors required for ERF recognition and relay to the export machinery may not be available or active; thus, the glutamic acid mutants would stay in the nucleus. Overall, the extensive phosphorylation throughout ERF, required for its proper subcellular localization, suggests multiple controlling events (e.g., conditional recognition by the export and/or import machinery or conditional sequestration in either compartment) and supports the hypothesis that the immediate response of ERF to growth stimulation or arrest is part of the mechanism that controls cellular proliferation.
The control of the ERF subcellular localization is similar to the one identified for Yan (36, 40), where upon induction of the Sevenless pathway, Yan is phosphorylated, exported into the cytoplasm and degraded after ubiquitination. ERF is also phosphorylated and exported into the cytoplasm as a result of the Ras signaling pathway, but in contrast to the case for Yan, we were not able to detect any effects on the protein stability as a result of phosphorylation. ERF remains in the cytoplasm of proliferating cells, and upon growth arrest it rapidly translocates back into the nucleus. This immediate response suggests that ERF may have a distinct role in the regulation of the G0/G1 transition. Although ERF and Yan have only limited homology within the Ets DNA-binding domain, the conservation of two Ets-domain transcriptional repressors regulated within the same pathway and in a similar manner is suggestive of their fundamental role.
Growth inhibition by ERF.
The Ras signaling pathway is one of the major and better-characterized pathways of mitogenic response, and it results in the activation of transcription factors that alter the cell transcription program and, ultimately, proliferation. Noticeably, the branch of the Ras pathway mediated by Erks modifies the activities of Ets transcription factors (Elk1 [57], PEA-3 [35] and Ets2 [32, 56]) that have been shown to be oncogenic themselves and have been suggested to contribute to ras transformation. ERF is a member of the Ets family that, in contrast to most other members, is a transcriptional inhibitor. Our data demonstrate that activation of the Ras/Erk pathway leads to ERF inactivation by nuclear export. In addition, forced nuclear localization of ERF is able to suppress ras-induced transformation (Fig. 7 and Table 1), suggesting that its export from the nucleus is required for the development of the transforming phenotype. Finally, nucleus-localized ERF is capable of arresting cells at the G0/G1 stage of the cell cycle (Fig. 6). These data suggest that ERF is a growth-regulating protein within the Ras/MAPK signaling pathway.
Overexpression of Ets-domain proteins has been shown to inhibit ras transformation, probably by occupying Ets-binding sites with a nonactive transcription factor and thus inhibiting normal Ets function (53). Overexpression of normal or VP16-fused Ets2 also has an inhibitory effect on ras transformation (10), indicating that overexpression experiments may interfere with the function of other Ets-domain proteins and contribute to the observed phenotype. For the same reasons, overexpression-related effects in our system cannot be excluded. However, there is evidence suggesting that ERF is indeed a transcription factor related to cellular proliferation and the Ras signaling pathway. ERF nuclear localization is consistent with cellular proliferation; thus, arrest by either serum deprivation or contact inhibition results in the nuclear localization of this transcriptional repressor. ERF physically interacts with Erks and is regulated via phosphorylation by an established Ras effector. Furthermore, we have previously shown that an overexpressed ERF mutant with a mutation that eliminates its repressor domain, although nuclear, cannot suppress E26-induced transformation, arguing against the competition hypothesis (44). Replacement of the ERF DNA-binding domain with the Fli1 DNA-binding domains results in a protein similarly regulated but with a distinct phenotype, arguing for the recognition of specific target genes related to the suppression effect (unpublished data). We have not been able to establish cell clones that express high levels of wt ERF in nontransformed cells, in contrast to transformed cells (44). Neither have we been able to establish cell lines expressing high levels of ERF mutants that localize in the nucleus (more than 10-fold over that of the endogenous ERF, as determined by Western analysis), even in transformed cells. Only lines that express low levels of ERF mutants (four- to eightfold over the endogenous level) could be obtained, with a very low frequency. The case of Yan, the Drosophila Ets-domain protein with similar function and regulation (36, 40), suggests that a functionally similar Ets protein conceivably exists in mammalian cells. The ability of ERF to suppress v-fos-induced tumorigenicity (44) argues that ERF may be within the pathway of mitogenic response. Finally, the hypothesis that ERF is a growth-regulating gene within the Ras signaling pathway is consistent with the notion that a fundamental cellular process is likely to be at least partly mediated by ubiquitous effectors. ERF, in contrast to many other ets genes suggested to be regulated by the Ras/MAPK pathway, is expressed in all cells, tissues, and developmental stages tested and at fairly constant levels (27).
It is unclear at this point which genes are controlled by ERF and mediate its effect on cell proliferation. D cyclin genes, which respond to Ras activation (26) and have been suggested to be regulated by Ets-domain proteins (1), are possible candidate genes. In addition the p21 (13) and p53 (50) genes are also cell-cycle-regulated genes reported to be affected by ets genes. However, a plethora of ets targets have been reported, including transcription factors (Ets2, Ets1, JunB, c-Fos, NF-κB, Myc, and GATA-1), matrix receptors and ligands (integrins αV, α2, and β2 and osteopontin), proteinases (stromelysine-1, collagenase, gelatinase, and urokinase-type plasminogen activator), and keratins. Several of these genes can be repressed by ERF in transient-transactivation assays (unpublished data). However, in this type of assay, ERF can repress all of the tested reporter genes that contain an Ets-binding site, suggesting that such an approach may not be informative and that gene elimination studies may be necessary to identify ERF target genes. The combination of a multitude of ets genes with similar DNA-binding specificities and the plethora of possible ets targets makes the identification of a specific gene as a target of a single ets gene fairly uncertain. It appears, though, that the possible targets of ERF are ubiquitously expressed cell-cycle-regulated genes, absent in the G0 state, that are required for cell cycle entry and/or progression. To that extent, components of the cell cycle machinery and immediate-early mitogenic response genes are valid candidates.
Concluding remarks.
The proliferation-dependent subcellular localization of ERF, its ubiquitous cell type distribution, and its regulation by a major mitogenic pathway suggest that ERF may be involved in the control of cellular proliferation and may be an effector in the Ras signaling pathway in mammalian cells. Furthermore, ERF may represent a class of proteins exported from the nucleus and may provide a novel domain for substrate recognition and specific interaction with Erks. Finally, its possible role in cellular proliferation, its physical association with Erks, and the control of its activity by Erks provide a new interface for targeting efforts to control cellular growth.
There are many questions regarding ERF function and regulation that will need to be addressed, including the mechanism of nuclear import and export, the mechanism of transcriptional repression, possible mediation of other signaling pathways in its control, and the identification of genes regulated by ERF. However, the elucidation of critical steps in its regulation and function presented here should facilitate these efforts.
ACKNOWLEDGMENTS
We thank D. Blair and M. Athanasiou for comments and support; A. Moustakas, L. Virgilio, S. Doucet-Brutin, and B. Popko for helpful comments and discussions; B. Biteau for help and support; D. Liu and K. Borboudaki for technical support; and K. Noer for the flow cytometry analysis.
This work was partly supported by EU grants FMRX-CT96-0041 and BMH4-CT96-1355.
REFERENCES
- 1.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 2.Brunner D, Ducker K, Oellers N, Hafen E, Scholz H, Klambt C. The ETS domain protein pointed-P2 is a target of MAP kinase in the sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- 3.Chow C W, Rincon M, Cavanagh J, Dickens M, Davis R J. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 4.Corbett A H, Silver P A. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 6.Denhardt D T. Signal-transducing protein phosphorylation cascades mediated by Ras/Rho proteins in the mammalian cell: the potential for multiplex signalling. Biochem J. 1996;318:729–747. doi: 10.1042/bj3180729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 8.Dittmer J, Nordheim A. Ets transcription factors and human disease. Biochim Biophys Acta. 1998;1377:F1–F11. doi: 10.1016/s0304-419x(97)00039-5. [DOI] [PubMed] [Google Scholar]
- 9.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foos G, Garcia-Ramirez J J, Galang C K, Hauser C A. Elevated expression of Ets2 or distinct portions of Ets2 can reverse Ras-mediated cellular transformation. J Biol Chem. 1998;273:18871–18880. doi: 10.1074/jbc.273.30.18871. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 12.Fukuda M, Gotoh Y, Nishida E. Interaction of MAP kinase with MAP kinase kinase: its possible role in the control of nucleocytoplasmic transport of MAP kinase. EMBO J. 1997;16:1901–1908. doi: 10.1093/emboj/16.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funaoka K, Shindoh M, Yoshida K, Hanzawa M, Hida K, Nishikata S, Totsuka Y, Fujinaga K. Activation of the p21(Waf1/Cip1) promoter by the ets oncogene family transcription factor E1AF. Biochim Biophys Res Commun. 1997;236:79–82. doi: 10.1006/bbrc.1997.6909. [DOI] [PubMed] [Google Scholar]
- 14.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1538. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 15.Gupta S, Davis R J. MAP kinase binds to the NH2-terminal activation domain of c-Myc. FEBS Lett. 1994;353:281–285. doi: 10.1016/0014-5793(94)01052-8. [DOI] [PubMed] [Google Scholar]
- 16.Gupta S, Campbell D, Derijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 17.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 18.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 19.Janknecht R. Analysis of the ERK-stimulated ETS transcription factor ER81. Mol Cell Biol. 1996;16:1550–1556. doi: 10.1128/mcb.16.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janknecht R, Monte D, Baert J L, de Launoit Y. The ETS-related transcription factor ERM is a nuclear target of signaling cascades involving MAPK and PKA. Oncogene. 1996;13:1745–1754. [PubMed] [Google Scholar]
- 21.Jans D A. The regulation of protein transport to the nucleus by phosphorylation. Biochem J. 1995;311:705–716. doi: 10.1042/bj3110705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khoklatchev A V, Carnagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb M H. Phosphorylation of MAP kinase ERK2 promotes its homodimerisation and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 23.Khosravi-Far R, Campbell S, Rossman K L, Der C J. Increasing complexity of Ras signal transduction: involvement of Rho family proteins. Adv Cancer Res. 1998;72:57–107. doi: 10.1016/s0065-230x(08)60700-9. [DOI] [PubMed] [Google Scholar]
- 24.Klemsz M, Hromas R, Raskind W, Bruno E, Hoffman R. PE-1, a novel ETS oncogene family member, localizes to chromosome 1q21-q23. Genomics. 1994;20:291–294. doi: 10.1006/geno.1994.1169. [DOI] [PubMed] [Google Scholar]
- 25.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 26.Lavoie J N, L’Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 27.Liu D, Pavlopoulos E, Modi W, Moschonas N, Mavrothalassitis G. ERF: genomic organization, chromosomal localization and promoter analysis of the human and mouse genes. Oncogene. 1997;14:1445–1451. doi: 10.1038/sj.onc.1200965. [DOI] [PubMed] [Google Scholar]
- 28.Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao C, Ray-Gallet D, Tavitian A, Moreau-Gachelin F. Differential phosphorylations of Spi-B and Spi-1 transcription factors. Oncogene. 1996;12:863–873. [PubMed] [Google Scholar]
- 30.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 31.Marshall C J. Ras effectors. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 32.McCarthy S A, Chen D, Yang B S, Garcia Ramirez J J, Cherwinski H, Chen X R, Klagsbrun M, Hauser C A, Ostrowski M C, McMahon M. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol Cell Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minden A, Lin A, Claret F X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 34.Mittnacht S, Paterson H, Olson M F, Marshall C J. Ras signalling is required for inactivation of the tumour suppressor pRb cell-cycle control protein. Curr Biol. 1997;7:219–221. doi: 10.1016/s0960-9822(97)70094-0. [DOI] [PubMed] [Google Scholar]
- 35.O’Hagan R C, Tozer R G, Symons M, McCormick F, Hassell J A. The activity of the Ets transcription factor PEA3 is regulated by two distinct MAPK cascades. Oncogene. 1996;13:1323–1333. [PubMed] [Google Scholar]
- 36.O’Neill E M, Rebay I, Tjian R, Rubin G M. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 37.Peeper D S, Upton T M, Ladha M H, Neuman E, Zalvide J, Bernards R, DeCaprio J A, Ewen M E. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- 38.Price M A, Cruzalegui F H, Treisman R. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 1996;15:6552–6563. [PMC free article] [PubMed] [Google Scholar]
- 39.Rabault B, Roussel M F, Quang C T, Ghysdael J. Phosphorylation of Ets1 regulates the complementation of a CSF-1 receptor impaired in mitogenesis. Oncogene. 1996;13:877–881. [PubMed] [Google Scholar]
- 40.Rebay I, Rubin G M. Yan functions as a general inhibitor of differentiation and is negatively regulated by activation of the Ras1/MAPK pathway. Cell. 1995;81:857–866. doi: 10.1016/0092-8674(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 41.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 43.Seth A, Robinson L, Thompson D M, Watson D K, Papas T S. Transactivation of GATA-1 promoter with ETS1, ETS2 and ERGB/Hu-FLI-1 proteins: stabilization of the ETS1 protein binding on GATA-1 promoter sequences by monoclonal antibody. Oncogene. 1993;8:1783–1790. [PubMed] [Google Scholar]
- 44.Sgouras D N, Athanasiou M A, Beal G J, Jr, Fisher R J, Blair D G, Mavrothalassitis G J. ERF, an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J. 1995;14:4781–4793. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stacey D W, Kung H F. Transformation of NIH 3T3 cells by microinjection of Ha-ras p21 protein. Nature. 1984;310:508–511. doi: 10.1038/310508a0. [DOI] [PubMed] [Google Scholar]
- 46.Tan P B, Lackner M R, Kim S K. MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulva induction. Cell. 1998;93:569–580. doi: 10.1016/s0092-8674(00)81186-1. [DOI] [PubMed] [Google Scholar]
- 47.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–615. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 48.van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vandromme M, Gauthier-Rouviere C, Lamb N, Fernandez A. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- 50.Venanzoni M C, Robinson L R, Hodge D R, Kola I, Seth A. ETS1 and ETS2 in p53 regulation: spatial separation of ETS binding sites (EBS) modulate protein: DNA interaction. Oncogene. 1996;12:1199–1204. [PubMed] [Google Scholar]
- 51.Wang X Z, Ron D. Stress-induced phosphorylation and activation of the transcription factor CHOP (GADD153) by p38 MAP kinase. Science. 1996;272:1347–1349. doi: 10.1126/science.272.5266.1347. [DOI] [PubMed] [Google Scholar]
- 52.Waskiewicz A J, Cooper J A. Mitogen and stress response pathways: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 53.Wasylyk C, Maira S M, Sobieszczuk P, Wasylyk B. Reversion of Ras transformed cells by Ets transdominant mutants. Oncogene. 1994;9:3665–3673. [PubMed] [Google Scholar]
- 54.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 55.Whitmarsh A J, Yang S H, Su M S, Sharrocks A D, Davis R J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang B S, Hauser C A, Henkel G, Colman M S, Van Beveren C, Stacey K J, Hume D A, Maki R A, Ostrowski M C. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang S H, Whitmarsh A J, Davis R J, Sharrocks A D. Differential targeting of MAP kinases to the ETS-domain transcription factor Elk-1. EMBO J. 1998;17:1740–1749. doi: 10.1093/emboj/17.6.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang S H, Yates P R, Whitmarsh A J, Davis R J, Sharrocks A D. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu J, Shibasaki F, Price R, Guillemot J, Yano T, Dötsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]