Abstract
Neddylation is necessary for activation of Cullin-RING ligases (CRLs), which degrade various immune regulatory proteins. Our recent study showed that while depletion of neddylation E2–E3 pair Ube2f-Sag in regulatory T (Treg) cells had no obvious phenotype, the same depletion of either Ube2m or Rbx1 caused inflammation disorders with different severity. Whether these E2s or E3s compensate each other in functional regulations of Treg cells is, however, previously unknown. In this report, we generated Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl or Foxp3Cre;Rbx1fl/fl;Sagfl/fl double-null mice by simultaneous deletion of both neddylation E2s or E3s in Treg cells, respectively. Remarkably, Ube2m&Ube2f double-null mice developed much severe autoimmune phenotypes than did Ube2m-null mice, indicating that Ube2m markedly compensates Ube2f in Treg cells. The minor worsened autoimmune phenotypes seen at the very early stage in Rbx1&Sag double-null than Rbx1-null mice is likely due to already severe phenotypes of the later, indicating a minor compensation of Rbx1 for Sag. The RNA profiling-based analyses revealed that up- and down-regulations of few signaling pathways in Treg cells are associated with the severity of autoimmune phenotypes. Finally, severer inflammation phenotypes seen in mice with double E3-null than with double E2-null Treg cells indicate a neddylation-independent mechanism of 2 E3s, also known to serve as the RING component of CRLs in regulation of Treg cell fitness.
Introduction
Neddylation, a ubiquitination-like process, is catalyzed by an enzyme cascade, consisting of NEDD8 E1 activation enzyme, NEDD8 E2 conjugating enzyme, and NEDD8 E3 ligases. In mammalian cells, the heterodimer of NEDD8 activating enzyme E1 subunit 1 (NAE1/APPBP1) and ubiquitin like modifier activating enzyme 3 (UBA3/NAEβ) is the only one E1 found; ubiquitin conjugating enzyme E2 M (UBE2M, also known as UBC12) and ubiquitin conjugating enzyme E2 F (UBE2F) are 2 known E2s. Among over a dozen of E3s, Ring-box 1 (RBX1) and Ring-box 2 (RBX2, also known as SAG/RNF7/ROC2) are most well studied for cullin neddylation [1,2].
Cullin-RING ligases (CRLs) are the largest E3 ubiquitin ligase family, consisting of 4 subunits: (a) scaffold cullins, (b) adaptor proteins, (c) RING component (RBX1/2), and (d) substrate-recognizing receptor. There are 8 members of cullin family, including cullin-1 to cullin-3, cullin-4A and cullin-4B, cullin-5, cullin-7, and cullin-9, with cullin-1 to cullin-5 being most well studied, which are physiological substrates of neddylation. Cullin neddylation triggers the conformation change, leading to activation of CRLs [3]. The UBE2M-RBX1 E2–E3 pair catalyzes neddylation of cullin-1 to cullin-4, while the UBE2F-RBX2/SAG axis catalyzes cullin-5 neddylation [4]. In addition, RBX1 and RBX2/SAG also act as the RING component of cullin-1 to cullin-4 and cullin-5, respectively, for targeted ubiquitylation and subsequent proteasome degradation of numerous substrates [5–7]. Thus, RBX1 and RBX2/SAG are dual E3s for both neddylation and ubiquitylation mediated by CRLs.
Our previous studies revealed that during mouse embryonic development, Rbx1 and Sag/Rbx2 are functional nonredundant, since in mice total knockout of Rbx1 or Sag leads to embryonic lethality at different stage with different mechanism [8,9]. Although whether Ube2m or Ube2f is also functionally redundant during development is currently unknown, the fact that neddylation of cullin-1 to cullin-4 versus cullin-5 by the Ube2m-Rbx1 pair versus the Ube2f-Sag pair [4] strongly suggests their functional independency. On the other hand, we recently showed a cross-talk between UBE2M and UBE2F, 2 neddylation E2s, in which UBE2M serves as a ubiquitylation E2 to promote ubiquitylation of UBE2F for proteasome degradation [10]. Whether there is a cross-talk between RBX1 and SAG remains unknown.
Regulatory T (Treg) cells are a suppressive subpopulation of CD4+ T lymphocytes, and transcription factor Foxp3 is the master marker of Treg cells [11–13]. Treg cells are essential for immune homeostasis, which is illustrated by the early-onset fatal inflammation caused by depletion of Treg cells [14]. We recently investigated the role of neddylation-CRL system in Treg cells using Foxp3Cre-LoxP system by generating 4 conditional knockout mouse models with deletion in Treg cells of 2 neddylation E2s, Ube2f or Ube2m, or 2 dual E3s, Sag or Rbx1, individually [15]. Interestingly, mice with the deletion of Ube2f or Sag in Treg cells had no visible phenotype, indicating that the Ube2f-Sag axis plays little, if any, role in regulation of Treg cells under physiological condition. Meanwhile, mice with the deletion of Ube2m or Rbx1 in Treg cells suffered from severe autoimmune inflammatory phenotypes, indicating the functional requirement of the Ube2m-Rbx1 axis in Treg cells [15]. The fact that much severer phenotypes are observed in Rbx1-null mice than in Ube2m-null mice indicates a neddylation-independent role of Rbx1 in Treg cells. However, it is still unknown whether 2 neddylation E2s or E3s are redundant in functional regulation of Treg cells.
In this study, we addressed this functionally redundant question by generating Treg double knockout mouse models of 2 E2s, Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl, or 2 E3s, Foxp3Cre;Rbx1fl/fl;Sagfl/fl, for simultaneous depletion of Ube2m and Ube2f or Rbx1 and Sag in Treg cells, respectively. Both Treg cell double knockout mice developed severe autoimmune disorders with an early-onset fatality. Given that Ube2f deletion in Treg cells had no phenotype, much severe autoimmune phenotypes in Ube2m&Ube2f double deletion than in Ube2m single deletion in Treg cells indicate that Ube2m compensates the function of Ube2f in Treg cells. On the other hand, minor increased severity in Treg Rbx1&Sag double deletion than Rbx1 single deletion suggests a major role of Rbx1 in regulation of Treg cell function. Furthermore, a greater severity in autoimmune phenotypes of mice with Rbx1&Sag deficiency than Ube2m&Ube2f deficiency in Treg cells suggests a neddylation-independent function of Rbx1/Sag. The comparison of RNA profiling in Treg cells with paired genotypes revealed a positive correlation of up- or down-regulation of few key signaling pathways with the severity of autoimmune phenotypes of Treg cell knockout mice.
Results
Fatal inflammation in Foxp3CreUbe2mfl/fl;Ube2ffl/fl mice
We have previously reported the phenotypical changes caused by individual deletion of Ube2m or Ube2f in Treg cells, and found that while the Foxp3Cre;Ube2ffl/fl mice had no visible phenotypic changes, the Foxp3Cre;Ube2mfl/fl mice developed severe inflammation disorders, where ~50% of mice died at about 4 months [15]. To study possible functional redundancy of 2 neddylation E2s in Treg cells, we intercrossed the single-null mice [15] and Foxp3YFP-Cre (Foxp3Cre) mice [16] to generate conditional knockout mice with deletion of both Ube2m and Ube2f in Treg cells simultaneously (designated as “Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl”). Strikingly, the Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice suffered from an early-onset alterations of appearance, including reduced body size, collapsed ears, festered skin (Fig. 1, A and B), and a significantly shortened life span with 100% of death rate at p55 (Fig. 1C). In Foxp3Cre;Ube2mfl/fl;Ube2f fl/fl mice at ~p20, the organ hematoxylin and eosin (H&E) staining revealed lymphocyte infiltration in liver, lung, kidney, stomach, colon, and skin (Fig. 1D). The autopsy revealed swollen peripheral immune organs, including lymph nodes and spleens, although the cellularity did not reach the statistically significant level (Fig. 1E and Fig. S1A). A more detailed characterization revealed a robust activation of immune cells from Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (~p20), as evidenced by a decreased ratio of CD4+/CD8+ T cells (Fig. S1B) and increased proportion of effector/memory T cells (CD44hiCD62Llo, Teff/mem cells) among conventional T cells (CD4+Foxp3−, Tcon cells) (Fig. 1F and Fig. S1C). Such fatal inflammatory disorders recaptured the phenotypes observed in mice with depleted Treg cells [14] in severity, indicating a pivotal role of neddylation E2s in the Treg cells.
Fig. 1.
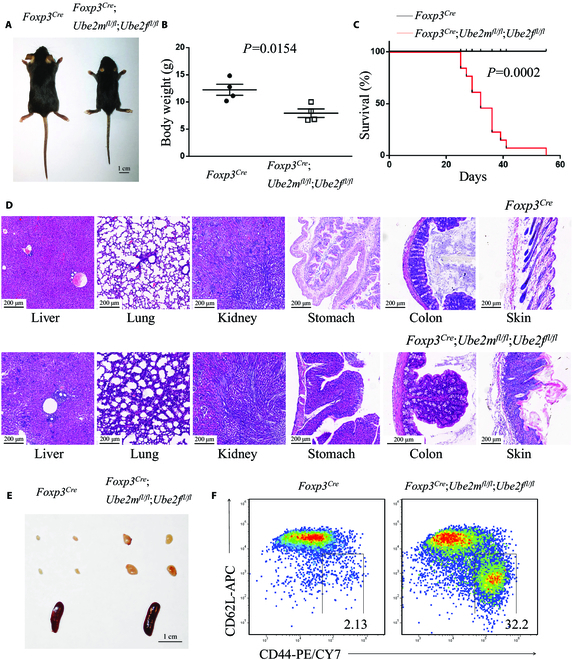
Deletion of Ube2m and Ube2f in Treg cells leads to an early-onset fatal inflammatory disorder. (A) Representative images of Foxp3Cre and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (p20, scale bar =1 cm). (B) Gross body weight of Foxp3Cre and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (n = 4). (C) Survival curves of Foxp3Cre and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (n = 13). (D) H&E staining of multiple organs from Foxp3Cre and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (p19 to p20, scale bar = 200 μm). (E) Representative images of the peripheral lymph nodes (top) and spleen (bottom) from Foxp3Cre and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (p22, scale bar = 1 cm). (F) Expression of CD44 and CD62L in Tcon cells from peripheral lymph nodes of Foxp3Cre and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (p21).
Impaired suppressive functions and altered transcriptome of Ube2m&Ube2f-deficient Treg cells
Given that both the Treg/CD4+ ratios (Fig. 2, A and B) and absolute numbers of Treg cells (Fig. 2C) are largely similar in peripheral lymph nodes derived from either Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl or Foxp3Cre control mice around p20, we hypothesized that the severe inflammation disorders observed in Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice at the same age is most likely attributable to the loss of the suppressive function of Treg cells. To test this hypothesis, we performed an in vivo suppression assay in immunodeficient Rag1−/− mice. Transfer of naive T (Tnai) cells (CD4+Foxp3−CD44loCD62Lhi) into Rag1−/− recipients led to a wasting disease with colitis within 4 weeks. We found that such colitis was prevented by simultaneous transfer of Treg cells from wild-type mice, but not from Ube2m&Ube2f-deficient mice (Fig. 2D), demonstrating that double deletion of neddylation E2s impairs the suppressive function of Treg cells.
Fig. 2.
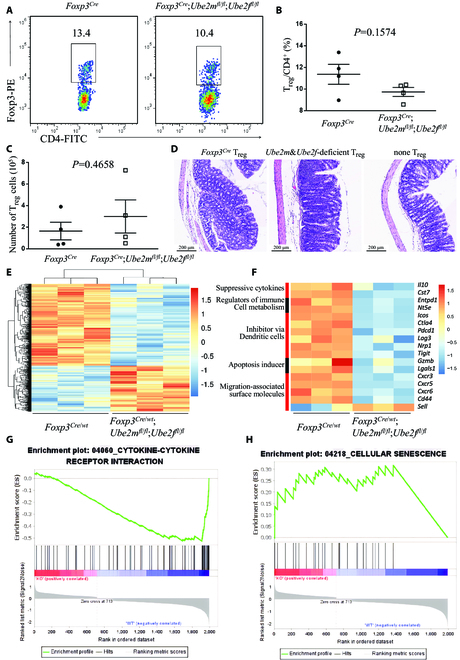
Impaired suppressive function of Ube2m&Ube2f-deficient Treg cells. (A) Expression of Foxp3 in CD4+ T cells in peripheral lymph nodes from Foxp3Cre and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice at p20. (B) Treg/CD4+ ratios in peripheral lymph nodes from Foxp3Cre and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (p19 to p21, n = 4). (C) Treg cell numbers in peripheral lymph nodes from Foxp3Cre and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (p19 to p21, n = 4). (D) Representative images of distal colon after H&E staining (scale bar = 200 μm). (E) Unsupervised cluster analysis of the transcriptional alterations in Ube2m&Ube2f-deficient Treg cells compared to the Foxp3Cre control Treg cells with Fc > 1.5 and P < 0.05. (F) Differentially expressed genes related to Treg cell function in CD4+YFP+ Treg cells from Foxp3Cre/wt and Foxp3Cre/wt;Ube2mfl/fl;Ube2ffl/fl mice, determined by transcriptional profiling with Fc > 1.5 and P < 0.05. (G) Gene set enrichment analysis (GSEA) of cytokine–cytokine receptor interaction genes in CD4+YFP+ Treg cells from Foxp3Cre/wt and Foxp3Cre/wt;Ube2mfl/fl;Ube2ffl/fl mice, determined by transcriptional profiling with Fc > 1.5 and P < 0.05. (H) GSEA of cellular senescence genes in CD4+YFP+ Treg cells from Foxp3Cre/wt and Foxp3Cre/wt;Ube2mfl/fl;Ube2ffl/fl mice, determined by transcriptional profiling with Fc > 1.5 and P < 0.05.
To define the underlying mechanism, we performed RNA profiling and analyzed the transcriptome alterations caused by Ube2m&Ube2f deletion in Treg cells. Considering that the inflammation in Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice would affect the transcript signature of Treg cells, we generated the inflammation-free female mice, in which one X chromosome expresses wild-type Foxp3 allele (Foxp3wt) and the other expresses Foxp3Cre mutant (designated as Foxp3Cre/wt). The random inactivation of one X chromosome in each somatic cell caused about half of Treg cells to express Foxp3Cre mutant, so Ube2m and Ube2f genes were deleted in these Treg cells; meanwhile, the other Treg cells expressed normal Foxp3wt allele and were kept absolutely normal. Due to the protection of the normal Treg cells, the Foxp3Cre/wt;Ube2mfl/fl;Ube2ffl/fl female mice are inflammation-free and healthy, so the CD4+YFP+ Treg cells in Foxp3Cre/wt;Ube2mfl/fl;Ube2ffl/fl mice were deficient of Ube2m&Ube2f and avoided the suffering from inflammation at the same time.
We then sorted the CD4+YFP+ Treg cells from Foxp3Cre/wt; Ube2mfl/fl;Ube2ffl/fl and Foxp3Cre/wt mice, respectively, followed by transcriptome analysis. The Ube2m&Ube2f-deficient Treg cells (Fig. S2A) showed significant alterations in transcriptome, with 714 genes up-regulated and 1,289 genes down-regulated with fold change (Fc) > 1.5 and P value < 0.05 (Fig 2E and Fig. S2, B and C). Among them, many Treg cell function-related genes were down-regulated, including Il10 [16] and Cst7 [17] (suppressive cytokines); Entpd1, Nt5e [18], and Il2r [19] (regulators of immune cell metabolism); Icos [20], Ctla4 [21], Pdcd1 [22], Lag3 [23], Nrp1 [24], and Tigit [25] (inhibitor via dendritic cells); and Gzmb [26] and Lgals3 [27] (apoptosis inducer) (Fig. 2F). Altered expression in migration-associated surface molecules was also observed in Ube2m&Ube2f-deficient Treg cells, such as down-regulation of Cxcr3, Cxcr5, Cxcr6, and Cd44 [28] and up-regulation of Sell [29] (Fig. 2F). On the other hand, among the top 20 genes up-regulated in Ube2m&Ube2f-deficienct Treg cells (Fig. S2D), none of them are previously known to negatively regulate Treg cells upon induction, which is certainly an interesting subject for future investigation.
Gene set enrichment analysis (GSEA) of the altered genes revealed dramatic changes in multiple pathways upon Ube2m&Ube2f deficiency (Fig. S2E), specially down-regulation of Treg-related pathways, such as cytokine–cytokine receptor interaction [30] (Fig. 2G) and T helper 17 (TH17) cell differentiation [31] (Fig. S2F). Some up-regulated pathways were also seen in Ube2m&Ubef-deficienct Treg cells, including DNA replication and repair, cell cycle, and metabolisms of amino acids tyrosine, phenylalanine, and tryptophan, as well as cellular senescence (Fig. S2E and Fig. 2H). How these pathways, upon double knockout of neddylation E2s, are up-regulated and how they functionally regulate Treg cells are the open and interesting questions for future investigation, particularly for possible involvement of senescence pathway.
Phenotype and transcriptome comparison between Foxp3Cre;Ube2mfl/fl and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice
A direct life-span comparison between Foxp3Cre;Ube2mfl/fl [15] and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice revealed a much shortened life span in double E2-null mice (Fig. 3A), indicating a significant contribution of Ube2f in the maintenance of Treg cells to the overall survival of mice. On the other hand, nondetectable phenotypic change in Ube2f Treg-depleted mice [15] indicates that the function of Ube2f in Treg cells is fully compensated by Ube2m. Thus, 2 neddylation E2s are functionally redundant in the regulation of Treg cells. While the main functions of Ube2f in Treg cells are compensated by Ube2m, Ube2f fails to functionally compensate Ube2m.
Fig. 3.
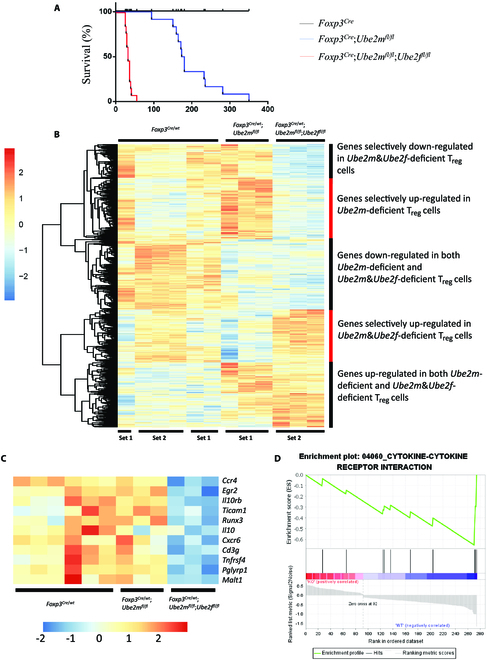
Enhanced inflammation phenotype and transcription alteration caused by Ube2m&Ube2f deficiency compared to Ube2m deficiency in Treg cells. (A) Survival curves of Foxp3Cre, Foxp3Cre;Ube2mfl/fl [15], and Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice. (B) Unsupervised cluster analysis of the transcriptional alterations in Ube2m&Ube2f- and Ube2m-deficient Treg cells compared to the Foxp3Cre control Treg cells. (C) Genes related with Treg cell function or regulation, and also down-regulated in Ube2m&Ube2f-deficient, but not Ube2m-deficient, Treg cells. (D) GSEA of cytokine–cytokine receptor interaction genes altered in Ube2m&Ube2f-deficient, but not Ube2m-deficient, Treg cells.
We next compared the transcriptome alterations between Ube2m&Ube2f-deficient and Ube2m-deficient Treg cells [15], along with the Foxp3Cre control. We normalized 2 sets of data (each from 3 individual mice, run at 2 different time periods), followed by unsupervised cluster analysis. The results showed that the transcriptional profile patterns were largely comparable in Foxp3Cre control Treg cells from 2 batches of experiments (Fig. 3B), suggesting a reproducibility of the experiments. The profiling comparison revealed 5 clusters (groups) of genes with altered expressions among 3 groups with an overall greater change seen in Ube2m&Ube2f double-deficient Treg cells (Fig. 3B).
Among the genes selectively down-regulated in double E2-deficient Treg cells (group 1), many genes are known to be related with functional regulation of Treg cells, including Ccr4 [32], Egr2 [33], Il10rb [34], Tccam1 [35], Runx3 [36], IL10 [16], Cd3g [37], Tnfrsf4 [38], Pglyrp1 [39], and Malt1 [40] (Fig. 3C). Among top 20 genes up-regulated in double E2-deficient Treg cells (group 4) (Fig. S3A), Smad7 is the only one known to be involved in regulation of Treg cells, whose up-regulation by the EZH2-FOXP3-RUNX1 axis inhibited Treg cell differentiation in rheumatoid arthritis [41].
GSEA analysis of the genes specially altered in double-null Treg cells revealed the changes in multiple pathways (Fig. S3B), and some down-regulated pathways are known to be involved in Treg cell regulation, such as cytokine–cytokine receptor interaction [30] (Fig. 3D) and TH17 cell differentiation [31] (Fig. S3C), whereas lysosome pathway, which is negatively correlated with function of Treg cells [42], along with cell cycle and phosphatidylinositol 3-kinase/Akt pathways, is up-regulated in Ube2m&Ube2f double-deficient Treg cells (Fig. S3, B and D). Thus, these greater alterations in gene expression are associated with and likely contributed to severer inflammatory disorders in Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice than in Foxp3Cre;Ube2mfl/fl mice.
Early-onset fatal inflammatory disorders in Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice
We next determined potential functional redundancy in Treg cells between 2 E3s, Rbx1 and Sag/Rbx2, dual for both neddylation and ubiquitylation by CRLs. We crossed the Rbx1-flox, Sag-flox, and Foxp3YFP-Cre (Foxp3Cre) mice [16] to generate the Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice, with simultaneous deletion of both Rbx1 and Sag, the only 2 known catalytic subunits of CRLs, in Treg cells. The Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice phenocopied the mice ablated of Treg cells in vivo [14], with smaller body size (Fig. 4, A and B), much shortened life span (Fig. 4C), inflammatory changes in multiple organs (Fig. 4D), swollen peripheral immune organs (Fig. 4E and Fig. S4A), and robust activation of immune cells (Fig. 4F and Fig. S7, B and C). Thus, Rbx1 and Sag are absolutely essential for the maintenance of Treg cell fitness.
Fig. 4.
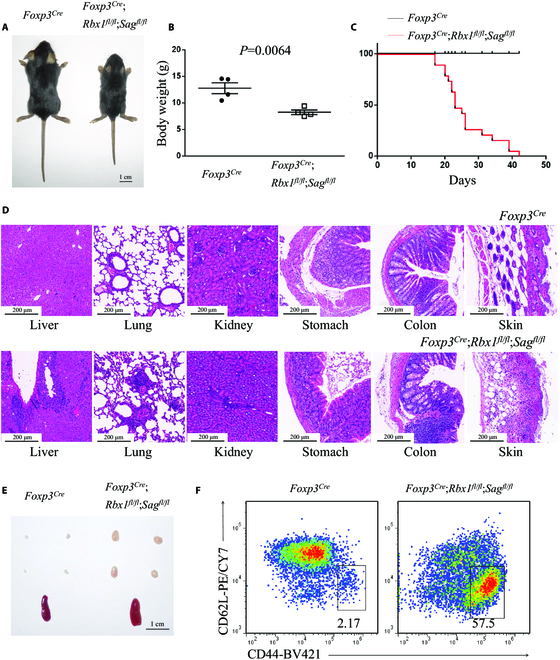
Deletion of Rbx1 and Sag in Treg cells leads to an early-onset fatal inflammatory disorder. (A) Representative images of Foxp3Cre and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (p19, scale bar =1 cm). (B) Gross body weight of Foxp3Cre and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (n = 4). (C) Survival curves of Foxp3Cre and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (n = 19). (D) H&E staining of multiple organs from Foxp3Cre and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (p20 to p21, scale bar = 200 μm). (E) Representative images of the peripheral lymph nodes (top) and spleen (bottom) from Foxp3Cre and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (p19, scale bar = 1 cm). (F) Expression of CD44 and CD62L in Tcon cells from peripheral lymph nodes of Foxp3Cre and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (p20).
We then performed the in vivo suppression assay in Rag1−/− immunodeficient mice to confirm the role of Rbx1&Sag deficiency in the suppressive function of Treg cells and found that the Rbx1&Sag-deficient Treg cells failed to prevent the colitis induced by Tnai cells (Fig. 5A), indicating impaired suppressive function. Thus, despite that the ratios and number of Rbx1&Sag-deficient Treg cells were dropped dramatically around p20 in vivo (Fig. 5, B to D), the severe inflammation disorder in Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice was not the simple consequence caused by the decreased number of Treg cells.
Fig. 5.
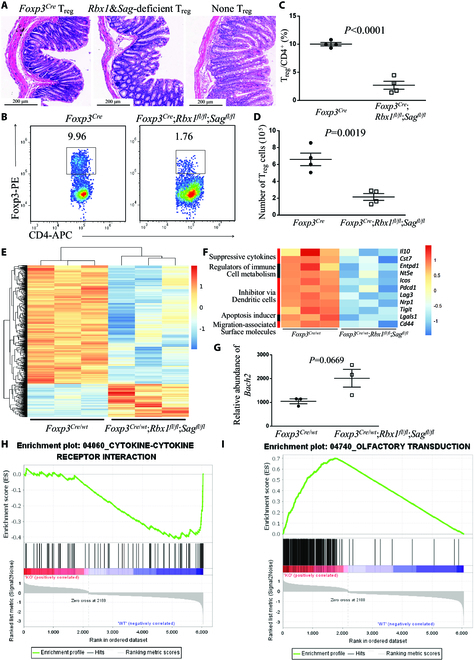
Impaired suppressive function of Rbx1&Sag-deficient Treg cells. (A) Representative images of distal colon after H&E staining (scale bar = 200 μm). (B) Expression of Foxp3 in CD4+ T cells in peripheral lymph nodes from Foxp3Cre and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice at p20. (C) Treg/CD4+ ratios in peripheral lymph nodes from Foxp3Cre and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (p19 to p20, n = 4). (D) Treg cell numbers in peripheral lymph nodes from Foxp3Cre and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (p19 to p20, n = 4). (E) Unsupervised cluster analysis of the transcriptional alterations in Rbx1&Sag-deficient Treg cells compared to the Foxp3Cre control Treg cells with Fc > 1.5 and P < 0.05. (F) Differentially expressed genes related to Treg cell function in CD4+YFP+ Treg cells from Foxp3Cre/wt and Foxp3Cre/wt;Rbx1fl/fl;Sagfl/fl mice, determined by transcriptional profiling with Fc > 1.5 and P < 0.05. (G) Relative abundance of Bach2 mRNA in CD4+YFP+ Treg cells from Foxp3Cre/wt and Foxp3Cre/wt;Rbx1fl/fl;Sagfl/fl mice, determined by transcriptional profiling. (H) GSEA of cytokine–cytokine receptor interaction genes in CD4+YFP+ Treg cells from Foxp3Cre/wt and Foxp3Cre/wt;Rbx1fl/fl;Sagfl/fl mice, determined by transcriptional profiling with Fc > 1.5 and P < 0.05. (I) GSEA of olfactory transduction genes in CD4+YFP+ Treg cells from Foxp3Cre/wt and Foxp3Cre/wt;Rbx1fl/fl;Sagfl/fl mice, determined by transcriptional profiling with Fc > 1.5 and P < 0.05.
To explore possible underlying mechanism(s) that mediated the function of Rbx1&Sag in Treg cells, we sorted the CD4+YFP+ Treg cells from inflammation-free female Foxp3Cre/wt;Rbx1fl/fl;Sagfl/fl and Foxp3Cre/wt control mice and performed transcription profiling. Deletion of Rbx1&Sag (Fig. S5A) led to a comprehensive change of the levels of multiple genes in Treg cells, with 641 genes up-regulated and 2,296 genes down-regulated with Fc > 2 and P < 0.05 (Fig. 5E and Fig. S5, B and C). Among down-regulated genes, many are reported to be related to the function of Treg cells, including Il10 [16] and Cst7 [17] (suppressive cytokines); Entpd1 and Nt5e [18] (regulators of immune cell metabolism); Icos [20], Pdcd1 [22], Lag3 [23], Nrp1 [24], and Tigit [25] (inhibitor via dendritic cells); and Lgals3 [27] (apoptosis inducer), and impairment of migration-associated surface molecules, including down-regulation of Cxcr3, Cxcr5, and Cd44 [28] (Fig. 5F). Although none of the top 20 up-regulated genes, including Plk2, Sik1, Nr4a2, and Dusp10 (Fig. S5D), are known to negatively regulate Treg cell functions upon induction, one of the up-regulated genes, Bach2 (up-regulated with Fc = 1.866 and P = 0.013) (Fig. 5G), is known as a negative regulator of Treg cell function [43].
The GSEA pathway analysis of the altered genes (Fc > 1.5 and P < 0.05) also revealed the changes of many pathways (Fig. S5E), including down-regulation of Treg-related pathways, such as cytokine–cytokine receptor interaction [30] (Fig. 5H), TH17 cell differentiation [31] (Fig. S5F), and up-regulation of pathways involving olfactory transduction (Fig. 5I) and metabolisms of retinol and arachidonic acid (Fig. S5E).
Earlier occurrence of inflammatory disorder in Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice than in Foxp3Cre;Rbx1fl/fl mice
We next addressed the question of functional redundancy between 2 E3s in Treg cells. Since mice with Rbx1-deficient Treg cells already had an early-onset fatal phenotype, whereas mice with Sag-deficient Treg cells had no phenotype [15], we first compared the survival of mice with 2 genotypes, Rbx1&Sag double deletion versus Rbx1 single deletion [15], in Treg cells and found no difference, both having early-onset fatal death (Fig. S6A). Moreover, no difference was found in the activation of immune response when mice were near moribund at age of ~20 days (Fig. S6B). We then focused on potential difference in inflammatory responses occurring at the very early stage of 8 days after the birth between 2 types of mice. Compared to the Foxp3Cre control, the ratio of Teff/mem cells (CD4+Foxp3−CD44hiCD62Llo) among Tcon cells (CD4+Foxp3−) is elevated dramatically in Rbx1-null or Rbx1&Sag double-null mice (Fig. 6A), indicating a robust immune activation. A substantially higher level was seen in double-null mice (Fig.A and B), demonstrating a more severe immune overactivation in vivo. Thus, like the Ube2m-Ube2f E2 pair, the Rbx1-Sag E3 pair also showed a functional redundancy in Treg cells, although it is minor due to the dominant effect of Rbx1 to mask the Sag effect. Collectively, the Sag function is being fully compensated by Rbx1, whereas the Rbx1 function cannot be compensated by Sag in Treg cells.
Fig. 6.
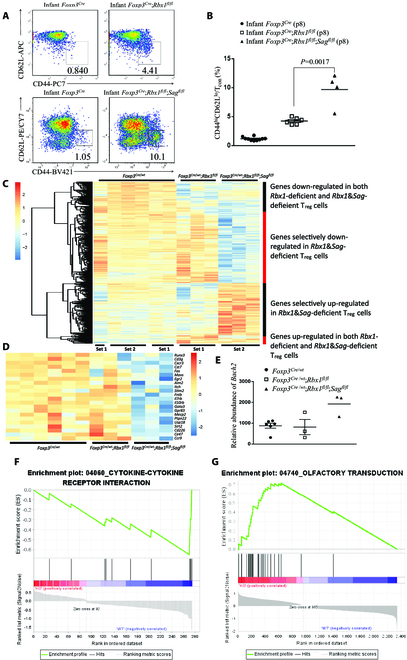
Enhanced inflammation phenotype and transcription alteration caused by Rbx1&Sag deficiency compared to Rbx1 deficiency in Treg cells. (A) Expression of CD44 and CD62L in Tcon cells from peripheral lymph nodes of infant Foxp3Cre, Foxp3Cre;Rbx1fl/fl, and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (p8). The upper panel was cited from our previous study [15] for comparison purpose. (B) Proportion of CD44hiCD62lo effector/memory cells among Tcon cells in peripheral lymph nodes from infant Foxp3Cre, Foxp3Cre;Rbx1fl/fl, and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (p8). (C) Unsupervised cluster analysis of the transcriptional alterations in Rbx1&Sag- and Rbx1-deficient Treg cells compared to the Foxp3Cre control Treg cells. (D) Genes related with Treg cell function or regulation, and also down-regulated in Rbx1&Sag-deficient, but not Rbx1-deficient, Treg cells. (E) Relative abundance of Bach2 mRNA in CD4+YFP+ Treg cells from Foxp3Cre/wt, Foxp3Cre/wt;Rbx1fl/fl, and Foxp3Cre/wt;Rbx1fl/fl;Sagfl/fl mice, determined by transcriptional profiling. (F) GSEA of cytokine–cytokine receptor interaction genes altered in Rbx1&Sag-deficient, but not Rbx1-deficient, Treg cells. (G) GSEA of olfactory transduction genes altered in Rbx1&Sag-deficient, but not Rbx1-deficient, Treg cells.
Enhanced transcription alterations caused by Rbx1&Sag double deficiency in comparison to Rbx1 single deficiency in Treg cells
We further compared the transcriptome alterations between Rbx1&Sag- and Rbx1-deficient Treg cells, along with Foxp3Cre control Treg cells, generated previously [15] and in this study, which showed large reproducibility (Fig. 6C, first 6 columns). Unsupervised cluster analysis of the transcriptome data revealed 4 groups of alterations: (a) genes down-regulated in both Rbx1&Sag double-deficient and Rbx1 single-deficient Treg cells; (b) genes selectively down-regulated in Rbx1&Sag double-deficient Treg cells; (c) genes selectively up-regulated in Rbx1&Sag double-deficient Treg cells; and (d) genes up-regulated in both Rbx1&Sag double-deficient and Rbx1 single-deficient Treg cells (Fig. 6C). Many group 2 genes are related with Treg cell regulation, including Cxcr3 [44], Itch [45], Il7rb [46], and Il10rb [34] (Fig. 6D). Among the top 20 up-regulated genes in group 3 (Fig. S7A), none of them are known to negatively regulate Treg cell function upon induction, and Bach2 in group 3 (not in the list of top 20 genes) appears to be the only gene known as a negative regulator of Treg cells [43] (Fig. 6E).
GSEA analysis of the genes specially altered in Rbx1&Sag-deficient Treg cells revealed changes of multiple pathways (Fig. S7B), where cytokine–cytokine receptor interaction was down-regulated (Fig. 6F), which are involved in Treg cell regulation [30] and up-regulation of multiple pathways, such as the olfactory transduction pathway (Fig. 6G). Taken together, it appears that many genes are associated with and may contribute to severer inflammatory disorder seen in Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice.
Phenotypes and transcriptome comparisons of mice and Treg cells with double E2 versus double E3 deficiency
It is well established that cullin neddylation is essential for activation of CRLs, and Rbx1 and Sag serve as dual E3 for both neddylation and CRL-medicated ubiquitylation [47,48]. To verify the functional relationship between neddylation and CRLs in Treg cells, we compare the degree of autoimmune disorders and transcriptional alterations caused by Ube2m&Ube2f versus Rbx1&Sag deficiency in Treg cells.
Although both Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice and Foxp3Cre; Rbx1fl/fl;Sagfl/fl mice suffered from severe autoimmune disorders with much shortened life span, the degree of severity still differs. In fact, the survival of Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice is significantly shorter than that of Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice, with P = 0.0350 (Fig. 7A). In the peripheral lymph nodes from mice at p19 to p21, the ratios of Teff/mem cells (CD4+Foxp3−CD44hiCD62Llo) among Tcon cells (CD4+Foxp3−) are higher in Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice than in Foxp3Cre; Ube2mfl/fl; Ube2ffl/fl mice, although the difference is not statistically significant due to variations among mice (Fig. 7B). Thus, double E3-null mice had more dramatic immune overactivation than double E2-null mice.
Fig. 7.
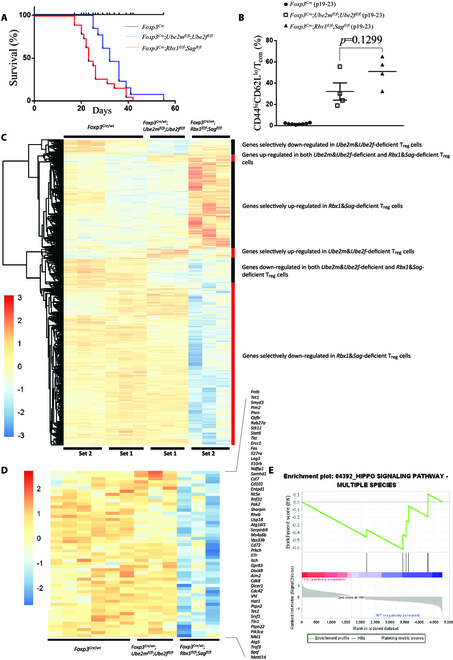
Enhanced inflammation phenotype and transcription alteration caused by Rbx1&Sag deficiency compared to Ube2m&Ube2f deficiency in Treg cells. (A) Survival curves of Foxp3Cre, Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl, and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice; P = 0.0350 between the survivals of Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice. (B) Proportion of CD44hiCD62lo effector/memory cells among Tcon cells in peripheral lymph nodes from Foxp3Cre, Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl, and Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice (p19 to p23). (C) Unsupervised cluster analysis of the transcriptional alterations in Ube2m&Ube2f- and Rbx1&Sag-deficient Treg cells compared to the Foxp3Cre control Treg cells. (D) Genes related with Treg cell function or regulation, and also down-regulated in Rbx1&Sag-deficient, but not in Ube2m&Ube2f-deficient, Treg cells. (E) GSEA of Hippo signaling pathway altered in Rbx1&Sag-deficient, but not Ube2m&Ube2f-deficient, Treg cells.
The comparison of transcriptome data of CD4+YFP+ Treg cells from Foxp3Cre/wt, Foxp3Cre/wt;Ube2mfl/fl;Ube2ffl/fl, and Foxp3Cre/wt;Rbx1fl/fl;Sagfl/fl mice revealed 6 groups of altered genes (up or down), unique to either double E2-deficient or double E3-deficient or both deficient Treg cells (Fig. 7C). We mainly focused on genes uniquely altered in double E3-null or double E2-null Treg cells. Among selectively down-regulated genes in double E3-null Treg cells, many are related to function or regulation of Treg cells (Fig. 7D), whereas among up-regulated genes (Fig. S8A), none of them are known to negatively regulate Treg cells upon induction. Similarly, among selectively altered genes (down and up) in double E2-null Treg cells (Fig. S9, A and B), few down-regulated, but not up-regulated, genes were known to regulate Treg cell functions. Nevertheless, the changes unique in double E3-null Treg cells likely represent CRL-dependent mechanism, whereas the changes unique in double E2-null Treg cells likely represent CRL-independent, but neddylation-dependent, mechanisms, consistent with our recent studies [49–51].
Similar GSEA pathway analysis of 2 groups of genes unique to double E3-deficient Treg cells revealed remarkable alterations in many pathways (Fig. S8B), including down-regulation of Hippo signaling pathway (Fig. 7E), which plays an important role in the maintenance of Treg cell function [52], and of pathways related to lysosome, as well as up-regulation of olfactory transduction and metabolisms of arachidonic acid and retinol (Fig. S8, B and C), which are previously unknown in regulation of the Treg cell functions.
GSEA pathway analysis of 2 groups of genes unique to double E2-deficient Treg cells also revealed alterations in a number of pathways (Fig. S9C), including down-regulation of ubiquitin-mediated proteolysis and up-regulation of DNA replication pathways (Fig. S9, D and E). Notably, the number of genes unique to double E2-deficient Treg cells is relatively smaller than in double E3-deficient Treg cells, which is consistent with the observation that Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice suffer more dramatic autoimmune disorders than do the Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice.
Collectively, the early-onset fatal disorders seen in both double E2- and double E3-deficient mice indicate functional similarity of neddylation and CRLs in regulation of Treg cells. More severe inflammatory disorders in double E3-deficient mice suggest a neddylation-independent function of CRLs in Treg cells.
Discussion
In this study, we investigated the role of the neddylation-CRL axis in functional regulation of Treg cells using several in vivo conditional knockout mouse models. Double deletion of neddylation E2s Ube2m&Ube2f or E3s Rbx1&Sag in Treg cells leads to the impairment of suppressive function of Treg cells and results in severe autoimmune disorders, fully demonstrating the pivotal role of the neddylation-CRL axis in the maintenance of Treg cell fitness. Thus, the neddylation-CRL axis is essential for Treg cell functions as the new key regulators of Treg cells beyond Foxp3 [11–13].
The functional independence of the Ube2m-Rbx1 and Ube2f-Sag axes was previously demonstrated both biochemically [4,53] and biologically [8,9]. Specifically, our recent study revealed that the Ube2m-Rbx1 axis plays a pivotal role in functional regulation of Treg cells, while the Ube2f-Sag axis is dispensable in Treg cells at the steady status [15]. This Treg-based functional study also supports the notion of functional independence between the Ube2m-Rbx1 and Ube2f-Sag axes.
Here, we used double knockout of either neddylation E2s or E3s in Treg cells and showed that both E2s and E3s are functionally redundant in Treg cells, as evidenced by the following: (a) The phenotypes of autoimmune disorders in Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl (double E2 knockout in Treg cells) mice are much more severe than in Foxp3Cre;Ube2mfl/fl (single E2 knockout in Treg cells) mice with much shortened life span (Fig. 3A). (b) The same autoimmune phenotypes are severer in Foxp3Cre;Rbx1fl/fl;Sagfl/fl (double E3s knockout in Treg cells) mice than in Foxp3Cre;Rbx1fl/fl (single E3 knockout in Treg cells) mice at the very early age only (Fig. 6A and B). We further found that the autoimmune phenotypes were much severer in Foxp3Cre;Rbx1fl/fl;Sagfl/fl mice than in Foxp3Cre;Ube2mfl/fl;Ube2ffl/fl mice (Fig. 7A).
We conclude from these results that (a) the Ube2f-Sag axis plays a role in Treg cells, which is, however, compensated by the Ube2m-Rbx1 axis. More specifically, the autoimmune phenotypes by Ube2f depletion were largely compensated by Ube2m, whereas the same autoimmune phenotypes by Sag depletion were minimally compensated by Rbx1, due to the dominant role of Rbx1 in Treg cells. (b) The functional redundancy in Treg cells is in one-way direction from the Ube2m-Rbx1 axis to the Ube2f-Sag axis, indicating much more significant role of CRL1 to CRL4 than of CRL5. (c) Rbx1 and Sag play neddylation-dependent and neddylation-independent roles in regulating Treg cell fitness.
To pursue the possible underlying mechanism, we performed the comprehensive transcriptome profiling analyses of Treg cells derived from the wild-type control or double knockout mice, and compared between single-null versus double-null Treg cells for both E2s or E3s, respectively. Compared to the wild-type control Treg cells, Treg cell deficiency of Ube2m&Ube2f caused dramatic alterations in mRNA transcriptome. Many genes (such as Il10, Cst7, and Cxcr3 to Cxcr6) and pathways (e.g., cytokine–cytokine receptor interaction and TH17 cell differentiation), previously known to regulate Treg cells, were down-regulated (Fig. 2 and Fig. S2), supporting the lost-of-function phenotypes. On the other hand, interestingly, many up-regulated genes (none of the top 20 genes) (Fig. S2D), with the exception of Sell (Fig. 2F) [29], or pathways (Fig. S2E) are previously unknown to negatively regulate Treg cell function upon activation. Nevertheless, some up-regulated pathways, such as cellular senescence (Fig. 2H), could contribute to the loss-of-function phenotype and deserve detailed future investigation.
Likewise, compared to the wild-type control Treg cells, Treg cell deficiency of Rbx1&Sag also caused dramatic alterations in mRNA transcriptome. Many genes (such as Il10, Cst7, Entpd1, and Nt5e) and pathways (again cytokine–cytokine receptor interaction and TH17 cell differentiation), previously known to regulate Treg cells, were down-regulated (Fig. 5 and Fig. S5), further supporting the lost-of-function phenotypes. On the other hand, interestingly, many up-regulated genes (again none of the top 20 genes) or pathways are previously unknown to regulate Treg cell function upon activation. One exception is Bach2, a known negative regulator of Treg cell functions [43], which is up-regulated in Rbx1&Sag-deficienct Treg cells (Fig. 5G). However, it is completely unknown how some up-regulated pathways, such as those involving olfactory transduction (Fig. 5I and Fig. S5E), negatively regulate Treg cell functions.
Transcriptome comparison between Treg cell deficiency of Ube2m&Ube2f versus Ube2m also showed many alterations. A list of 11 genes (e.g., Ccr4, Egr2, and Il10), uniquely down-regulated in double E2 null (Fig. 3C), are all known to regulate Treg cell functions. Again, the pathways associated with cytokine–cytokine receptor interaction and TH17 cell differentiation were also down-regulated uniquely to double E2 null Treg cells (Fig. 3D and Fig. S3C). Among top 20 up-regulated genes (Fig. S3A), Smad7 is the only one reported to inhibit Treg cell differentiation in rheumatoid arthritis upon up-regulation by the EZH2-FOXP3-RUNX1 axis [41]. The other up-regulated genes, along with up-regulated pathways (Fig. S3B), unknown to negatively regulate Treg cell functions, open a new window to study neddylation involvement in Treg cell functions, for example, how up-regulation of lysosome pathway (Fig. S3B and D) contributes to the loss-of-function phenotype of Treg cells.
Transcriptome comparison between Treg cell deficiency of Rbx1&Sag versus Rbx1 also showed many alterations, but to a lesser extent (Fig. 6C and Fig. S7). Down-regulation of Cxcr3, Fas, Egr2, and Itch and of cytokine–cytokine receptor interaction pathway appeared to be unique to double E3-null Treg cells (Fig. 6, D and F). Among up-regulated genes and pathways (Fig. S7), Bach2 and olfactory transduction (Fig. 6, E and G) appeared to be unique. Given a minor phenotypic difference between Rbx1/Sag double-null and Rbx1 single-null mice, these alterations are unlikely to play a major role in the loss-of-function phenotypes.
Finally, we compared the phenotypes of Foxp3Cre;Ube2mfl/fl; Ube2ffl/fl (double E2-null) mice with Foxp3Cre;Rbx1fl/fl;Sagfl/fl (double E3-null) mice and found that double E3-null mice suffer more severe autoimmune disorders with a shorter life span (Fig. 7A). Consistently, the transcriptional alterations caused by Rbx1&Sag deficiency in Treg cells are more dramatic than those in Ube2m&Ube2f-deficient Treg cells, with a more remarkable decrease of Treg cell regulatory genes and down-regulation of Hippo signaling pathway (Fig. 7, D and E), which was known to play an important role in the maintenance of Treg cell function [52]. Among up-regulated genes and pathways, such as olfactory transduction and metabolisms of retinol and arachidonic acid, which are unique to double E3-null (Fig. S8B and C), it is completely unknown whether and how they negatively regulate Treg cell function, nor their contribution to severe phenotypes. In addition, we also identified genes and pathways unique to double E2 knockout Treg cells (Fig. 7C and Fig. S9). These alterations are likely independent of inactivation of Rbx1/Sag-CRLs but involve neddylation modification of non-cullin substrates via other dual E3s for both neddylation and ubiquitylation [49–51].
It is not surprising that many changes in gene expression occur in these Treg conditional knockout models, given the fact that neddylation E2/E3s are required for cullin neddylation to activate CRLs, which are responsible for ubiquitylation and degradation of ~20% cellular proteins doomed for proteasome degradation [54], thus regulating many biochemical and biological processes. Mechanistically, it is anticipated that inactivation of CRLs, resulting from depletion of neddylation E2/E3, will cause the accumulation of many cellular substrates, including (a) transcription factors and repressors, which would directly affect gene expression, and (b) other signal molecules and cell cycle regulators, which would indirectly affect gene expression. Unfortunately, given that Treg cells are a rare population in vivo and no in vitro cell lines were established, it is technically challenging to define the detailed underlying mechanisms by these altered genes and pathways in our Treg cell conditional knockout setting. Nevertheless, our study defined genes and pathways responsible for both neddylation-dependent and neddylation-independent roles for neddylation E2s and dual E3s.
In summary, in this study, we used mouse models of Treg cell double knockout of neddylation E2s or E3s and made 3 major findings: (a) Neddylation enzymes play an absolute essential role in the maintenance of Treg cell fitness and their dual disruption causes severer autoimmune phenotypes; (b) there is previously unknown functional redundancy between Ube2m and Ube2f, and to a lesser extent between Rbx1 and Sag; and (c) dual E3s Rbx1 and Sag have neddylation-dependent and neddylation-independent role. Finally, our study has sound physiological and pathological relevance to autoimmune diseases, given the fact that a variety of human autoimmune diseases, including systemic lupus erythematosus, inflammatory bowel disease, and rheumatoid arthritis, are subjected to fine regulation by CRLs [55], whose activation requires neddylation E2, UBE2M/UBE2F, and E3 RBX1/RBX2.
Materials and Methods
Mice
The Foxp3YFP-Cre mice (The Jackson Laboratory, no. 016959) were provided by X. Feng (Peking Union Medical College, China) [56]. The Ube2m-flox, Ube2f-flox, Rbx1-flox, and Sag-flox mice were generated previously [15,57]. The Rag1−/− mice were obtained from GemPharmatech Co. Ltd., China (strain no. T004753).
Mice were fed in specific pathogen-free (SPF) conditions. All animal experiments were approved by the Animal Ethics Committee of Zhejiang University; animal care was provided in accordance with the principles and procedures by the regulatory standards at Zhejiang University Laboratory Animal Center.
Flow cytometry
Peripheral lymph nodes and spleens from indicated mice were grinded into single cells. Cells were washed in phosphate-buffered saline containing 2% (w/v) fetal bovine serum and then stained with indicated antibodies for analysis of surface proteins. For intracellular proteins, cells were fixed and permeabilized with Pharmingen Transcription Factor Buffer Set (BD Pharmingen, 562574). Flow cytometry was performed on CytoFLEX LX (Beckman).
Antibodies used were listed as follows: anti-CD4 (GK1.5, eBioscience), anti-CD8α (53-6.7, BD Pharmingen), anti-CD44 (IM7, BioLegend), anti-CD62L (MEL-14, BioLegend), and anti-Foxp3 (150D, BioLegend).
Cell sorting
The CD4+ T cells were isolated from the single-cell suspension of peripheral lymph nodes and spleens by Mouse CD4 T Lymphocyte Enrichment Set-DM (BD Biosciences, 558131), followed by fluorescence-activated cell sorting to purify Treg cells (CD4+YFP+) and/or Tnai cells (CD4+YFP−CD44loCD62Lhi) with purities >99%, performed on SONY Cell Sorter (SH800S).
In vivo suppression assay
Tnai cells (CD4+YFP−CD44loCD62Lhi, 4 × 105) and Treg cells (CD4+YFP+, 2 × 105), sorted from the peripheral lymph nodes and spleens of indicated mice (8 to 12 weeks old), were administrated into Rag1−/− mice via intraperitoneal injection as described [58]. Four weeks after the intraperitoneal injection, mouse colons were harvested and fixed by formalin, followed by H&E staining.
Transcriptome profiling
CD4+YFP+ Treg cells were sorted from the peripheral lymph nodes and spleens of indicated mice (8 to 12 weeks old). To generate enough materials, 2 to 3 mice were pooled for one sample. RNA was purified from the sorted cells with the miRNeasy Mini Kit (Qiagen, 21704). The RNAs were reverse-transcribed, amplified, and labeled (Affymetrix GeneChip Pico Kit, 703308) to achieve enough cDNAs. Then, the cDNA samples were hybridized to Clariom S Arrays, mouse (902931). The Applied Biosystems Expression Console Software 1.4 was employed to analyze the microarray datasets.
Statistical analysis
The P values were calculated by Mann–Whitney test, 2-tailed unpaired Student’s t test, using GraphPad Prism software. Mouse survival and respective P values were analyzed by the log-rank test. The statistical significance was evaluated by P < 0.05. All error bars represent SEM (n ≥ 3).
Acknowledgments
We thank X. Feng for providing the Foxp3YFP-Cre mice. We also thank C. Bi and Y. Xing from Core Facilities, Zhejiang University School of Medicine for their technical support. Funding: The project was funded by the National Key R&D Program of China (2021YFA1101000 and 2022YFC3401500 to Y.S.), National Natural Science Foundation of China (82172699 and 81801567 to D.W. and U22A20317 and 92253203 to Y.S.), and Zhejiang Provincial Natural Science Foundation of China (LY21H100005 to D.W. and LD22H300003 to Y.S.). Author contributions: D.W. designed and performed experiments, analyzed data, and wrote the manuscript. Y.S. conceived project, designed experiments, analyzed data, wrote and finalized the manuscript, and oversaw the project. Competing interests: The authors declare that they have no competing interests.
Data Availability
The microarray data generated in this study have been deposited in Gene Expression Omnibus under accession code GSE237499.
Supplementary Materials
Figs. S1 to S9
References
- 1.Zhou L, Zhang W, Sun Y, Jia L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell. Signal. 2018;44:92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao Y, Morgan MA, Sun Y. Targeting neddylation pathways to inactivate Cullin-RING ligases for anti-cancer therapy. Antioxid. Redox Signal. 2014;21(17):2383–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Y, Sun Y. Cullin-RING ligases as attractive anti-cancer targets. Curr. Pharm. Des. 2013;19(18):3215–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell. 2009;33(4):483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakayama KI, Nakayama K. Ubiquitin ligases: Cell-cycle control and cancer. Nat. Rev. Cancer. 2006;6(5):369–381. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, Xiong X, Sun Y. Cullin-RING ligase 5: Functional characterization and its role in human cancers. Semin. Cancer Biol. 2020. [DOI] [PubMed] [Google Scholar]
- 7.Zhou W, Wei W, Sun Y. Genetically engineered mouse models for functional studies of SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligases. Cell Res. 2013;23(5):599–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan M, Davis SW, Saunders TL, Zhu Y, Sun Y. RBX1/ROC1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of p27. Proc. Natl. Acad. Sci. U.S.A. 2009;106(15):6203–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan M, Zhao Y, Kim SJ, Liu M, Jia L, Saunders TL, Zhu Y, Sun Y. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev. Cell. 2011;21(6):1062–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou W, Xu J, Tan M, Li H, Li H, Wei W, Sun Y. UBE2M is a stress-inducible dual E2 for neddylation and ubiquitylation that promotes targeted degradation of UBE2F. Mol. Cell. 2018;70(6):1008–1024.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4(4):330–336. [DOI] [PubMed] [Google Scholar]
- 12.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299(5609):1057–1061. [DOI] [PubMed] [Google Scholar]
- 13.Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003;4(4):337–342. [DOI] [PubMed] [Google Scholar]
- 14.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007;8(2):191–197. [DOI] [PubMed] [Google Scholar]
- 15.Wu D, Li H, Liu M, Qin J, Sun Y. The Ube2m-Rbx1 neddylation-Cullin-RING-ligase proteins are essential for the maintenance of regulatory T cell fitness. Nat. Commun. 2022;13(1):3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubtsov YP, Rasmussen JP, Chi EY, Fontenot J, Castelli L, Ye X, Treuting P, Siewe L, Roers A, Henderson WR Jr, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28(4):546–558. [DOI] [PubMed] [Google Scholar]
- 17.Zacchigna S, Martinelli V, Moimas S, Colliva A, Anzini M, Nordio A, Costa A, Rehman M, Vodret S, Pierro C, et al. Paracrine effect of regulatory T cells promotes cardiomyocyte proliferation during pregnancy and after myocardial infarction. Nat. Commun. 2018;9(1):2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, Chen JF, Enjyoji K, Linden J, Oukka M, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 2007;204(6):1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, Gasteiger G, Feng Y, Fontenot JD, Rudensky AY. An essential role for the IL-2 receptor in T(reg) cell function. Nat. Immunol. 2016;17(11):1322–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman AE, Freeman GJ, Mathis D, Benoist C. CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J. Exp. Med. 2004;199(11):1479–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte–associated antigen 4 plays an essential role in the function of CD25+CD4+ regulatory cells that control intestinal inflammation. J. Exp. Med. 2000;192(2):295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs JF, Idema AJ, Bol KF, Nierkens S, Grauer OM, Wesseling P, Grotenhuis JÁ, Hoogerbrugge PM, de Vries IJM, Adema GJ. Regulatory T cells and the PD-L1/PD-1 pathway mediate immune suppression in malignant human brain tumors. Neuro Oncol. 2009;11(4):394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang CT, Workman CJ, Flies D, Pan X, Marson AL, Zhou G, Hipkiss EL, Ravi S, Kowalski J, Levitsky HI, et al. Role of LAG-3 in regulatory T cells. Immunity. 2004;21(4):503–513. [DOI] [PubMed] [Google Scholar]
- 24.Delgoffe GM, Woo SR, Turnis ME, Gravano DM, Guy C, Overacre AE, Bettini ML, Vogel P, Finkelstein D, Bonnevier J, et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013;501(7466):252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joller N, Lozano E, Burkett PR, Patel B, Xiao S, Zhu C, Xia J, Tan TG, Sefik E, Yajnik V, et al. Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity. 2014;40(4):569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao X, Cai SF, Fehniger TA, Song J, Collins LI, Piwnica-Worms DR, Ley TJ. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity. 2007;27(4):635–646. [DOI] [PubMed] [Google Scholar]
- 27.Garin MI, Chu C-C, Golshayan D, Cernuda-Morollón E, Wait R, Lechler RI. Galectin-1: A key effector of regulation mediated by CD4+CD25+ T cells. Blood. 2007;109(5):2058–2065. [DOI] [PubMed] [Google Scholar]
- 28.Firan M, Dhillon S, Estess P, Siegelman MH. Suppressor activity and potency among regulatory T cells is discriminated by functionally active CD44. Blood. 2006;107(2):619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor PA, Panoskaltsis-Mortari A, Swedin JM, Lucas PJ, Gress RE, Levine BL, June CH, Serody JS, Blazar BR. L-Selectinhi but not the L-selectinlo CD4+25+ T-regulatory cells are potent inhibitors of GVHD and BM graft rejection. Blood. 2004;104(12):3804–3812. [DOI] [PubMed] [Google Scholar]
- 30.Toomer KH, Malek TR. Cytokine signaling in the development and homeostasis of regulatory T cells. Cold Spring Harb. Perspect. Biol. 2018;10(3):Article a028597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee GR. The balance of Th17 versus Treg cells in autoimmunity. Int. J. Mol. Sci. 2018;19(3):730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y, You M, Fu J, Tian M, Zhong X, du C, Hong Z, Zhu Z, Liu J, Markowitz GJ, et al. Intratumoral stem-like CCR4+ regulatory T cells orchestrate the immunosuppressive microenvironment in HCC associated with hepatitis B. J. Hepatol. 2022;76(1):148–159. [DOI] [PubMed] [Google Scholar]
- 33.Zhang M, Wang Y, Wang JS, Liu J, Liu MM, Yang HB. The roles of Egr-2 in autoimmune diseases. Inflammation. 2015;38(3):972–977. [DOI] [PubMed] [Google Scholar]
- 34.Murai M, Turovskaya O, Kim G, Madan R, Karp CL, Cheroutre H, Kronenberg M. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat. Immunol. 2009;10(11):1178–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Luca A, Montagnoli C, Zelante T, Bonifazi P, Bozza S, Moretti S, D’Angelo C, Vacca C, Boon L, Bistoni F, et al. Functional yet balanced reactivity to Candida albicans requires TRIF, MyD88, and IDO-dependent inhibition of Rorc. J. Immunol. 2007;179(9):5999–6008. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Patsoukis N, Petkova V, Boussiotis VA. Runx1 and Runx3 are involved in the generation and function of highly suppressive IL-17-producing T regulatory cells. PLOS ONE. 2012;7(9):Article e45115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rowe JH, Delmonte OM, Keles S, Stadinski BD, Dobbs AK, Henderson LA, Yamazaki Y, Allende LM, Bonilla FA, Gonzalez-Granado LI, et al. Patients with CD3G mutations reveal a role for human CD3γ in Treg diversity and suppressive function. Blood. 2018;131(21):2335–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinterbrandner M, Rubino V, Stoll C, Forster S, Schnüriger N, Radpour R, Baerlocher GM, Ochsenbein AF, Riether C. Tnfrsf4-expressing regulatory T cells promote immune escape of chronic myeloid leukemia stem cells. JCI Insight. 2021;6(23):Article e151797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park SY, Jing X, Gupta D, Dziarski R. Peptidoglycan recognition protein 1 enhances experimental asthma by promoting Th2 and Th17 and limiting regulatory T cell and plasmacytoid dendritic cell responses. J. Immunol. 2013;190(7):3480–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng L, Deng N, Yang N, Zhao X, Lin X. Malt1 protease is critical in maintaining function of regulatory T cells and may be a therapeutic target for antitumor immunity. J. Immunol. 2019;202(10):3008–3019. [DOI] [PubMed] [Google Scholar]
- 41.Xiao XY, Li YT, Jiang X, Ji X, Lu X, Yang B, Wu LJ, Wang XH, Guo JB, Zhao LD, et al. EZH2 deficiency attenuates Treg differentiation in rheumatoid arthritis. J. Autoimmun. 2020;108:Article 102404. [DOI] [PubMed] [Google Scholar]
- 42.Du T, Nagai Y, Xiao Y, Greene MI, Zhang H. Lysosome-dependent p300/FOXP3 degradation and limits Treg cell functions and enhances targeted therapy against cancers. Exp. Mol. Pathol. 2013;95(1):38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sidwell T, Liao Y, Garnham AL, Vasanthakumar A, Gloury R, Blume J, Teh PP, Chisanga D, Thelemann C, de Labastida Rivera F, et al. Attenuation of TCR-induced transcription by Bach2 controls regulatory T cell differentiation and homeostasis. Nat. Commun. 2020;11(1):252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kornete M, Mason ES, Girouard J, Lafferty EI, Qureshi S, Piccirillo CA. Th1-like ICOS+ Foxp3+ Treg cells preferentially express CXCR3 and home to β-islets during pre-diabetes in BDC2.5 NOD mice. PLOS ONE. 2015;10(5):Article e0126311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin HS, Park Y, Elly C, Liu YC. Itch expression by Treg cells controls Th2 inflammatory responses. J. Clin. Invest. 2013;123(11):4923–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmaler M, Broggi MAS, Lagarde N, Stöcklin BF, King CG, Finke D, Rossi SW. IL-7R signaling in regulatory T cells maintains peripheral and allograft tolerance in mice. Proc. Natl. Acad. Sci. U.S.A. 2015;112(43):13330–13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 1999;15:435–467. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Y, Morgan MA, Sun Y. Targeting neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid. Redox Signal. 2014;21(17):2383–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li C, Zhang L, Qian D, Cheng M, Hu H, Hong Z, Cui Y, Yu H, Wang Q, Zhu J, et al. RNF111-facilitated neddylation potentiates cGAS-mediated antiviral innate immune response. PLOS Pathog. 2021;17(3):Article e1009401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fei X, Li Z, Yang D, Kong X, Lu X, Shen Y, Li X, Xie S, Wang J, Zhao Y, et al. Neddylation of Coro1a determines the fate of multivesicular bodies and biogenesis of extracellular vesicles. J Extracell Vesicles. 2021;10(12):Article e12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu X, Kong X, Wu H, Hao J, Li S, Gu Z, Zeng X, Shen Y, Wang S, Chen J, et al. UBE2M-mediated neddylation of TRIM21 regulates obesity-induced inflammation and metabolic disorders. Cell Metab. 2023. [DOI] [PubMed] [Google Scholar]
- 52.Shi H, Liu C, Tan H, Li Y, Nguyen TLM, Dhungana Y, Guy C, Vogel P, Neale G, Rankin S, et al. Hippo kinases Mst1 and Mst2 sense and amplify IL-2R-STAT5 signaling in regulatory T cells to establish stable regulatory activity. Immunity. 2018;49(5):899–914.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuang P, Tan M, Zhou W, Zhang Q, Sun Y. SAG/RBX2 E3 ligase complexes with UBCH10 and UBE2S E2s to ubiquitylate β-TrCP1 via K11-linkage for degradation. Sci. Rep. 2016;6:37441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458(7239):732–736. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X, Liu Y, Zhang T, Tan Y, Dai X, Yang YG, Zhang X. Advances in the potential roles of Cullin-RING ligases in regulating autoimmune diseases. Front. Immunol. 2023;14:Article 1125224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu D, Luo Y, Guo W, Niu Q, Xue T, Yang F, Sun X, Chen S, Liu Y, Liu J, et al. Lkb1 maintains T(reg) cell lineage identity. Nat. Commun. 2017;8:15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Tan M, Jia L, Wei D, Zhao Y, Chen G, Xu J, Zhao L, Thomas D, Beer DG, et al. Inactivation of SAG/RBX2 E3 ubiquitin ligase suppresses KrasG12D-driven lung tumorigenesis. J. Clin. Invest. 2014;124(2):835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouyang W, Liao W, Luo CT, Yin N, Huse M, Kim MV, Peng M, Chan P, Ma Q, Mo Y, et al. Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature. 2012;491(7425):554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Data Availability Statement
The microarray data generated in this study have been deposited in Gene Expression Omnibus under accession code GSE237499.


