Abstract
Homologous recombination is an important DNA repair mechanism in vegetative cells. During the repair of double-strand breaks, genetic information is transferred between the interacting DNA sequences (gene conversion). This event is often accompanied by a reciprocal exchange between the homologous molecules, resulting in crossing over. The repair of DNA damage by homologous recombination with repeated sequences dispersed throughout the genome might result in chromosomal aberrations or in the inactivation of genes. It is therefore important to understand how the suitable homologous partner for recombination is chosen. We have developed a system in the yeast Saccharomyces cerevisiae that can monitor the fate of a chromosomal double-strand break without the need to select for recombinants. The broken chromosome is efficiently repaired by recombination with one of two potential partners located elsewhere in the genome. One of the partners has homology to the broken ends of the chromosome, whereas the other is homologous to sequences distant from the break. Surprisingly, a large proportion of the repair is carried out by recombination involving the sequences distant from the broken ends. This repair is very efficient, despite the fact that it requires the processing of a large chromosomal region flanking the break. Our results imply that the homology search involves extensive regions of the broken chromosome and is not carried out exclusively by sequences adjacent to the double-strand break. We show that the mechanism that governs the choice of homologous partners is affected by the length and sequence divergence of the interacting partners, as well as by mutations in the mismatch repair genes. We present a model to explain how the suitable homologous partner is chosen during recombinational repair. The model provides a mechanism that may guard the integrity of the genome by preventing recombination between dispersed repeated sequences.
Homologous recombination is a universal process; it plays a role in generating diversity during meiosis and is an important DNA repair mechanism in vegetative cells. Recombination results in the transfer of genetic information from one DNA molecule to a homologous one (gene conversion) and in the reciprocal exchange of DNA fragments between chromosomes (crossing over). Reciprocal and nonreciprocal recombination show a nonrandom association. For meiotic cells, tetrad analysis has shown that when a heterozygous marker has converted, there is often (18 to 66% of the time) an associated crossover of flanking markers (reviewed in reference 29). This coupling holds true for mitotic recombination, although the level of association seems to be lower (13, 17). These results have led to the assumption that gene conversion and crossing over are mechanistically related.
Double-strand breaks (DSBs) in the DNA of living organisms occur as a consequence of the natural cell metabolism or can be created by exogenous sources, such as chemical agents or radiation. In addition, DSBs are generated during certain developmental processes, such as V(D)J recombination in lymphoid cells (33), meiosis (47), and mating-type switch in yeast (46). If left unrepaired, DSBs result in broken chromosomes and cell death (20, 24). On the other hand, repair of the DSBs by a mechanism that alters the original chromosomal structure or sequence may lead to carcinogenesis or inborn defects. Therefore, the accurate repair of this damage by mitotic recombination is important for cell survival and for genome stability (29).
A basic model for DSB-initiated recombination was proposed by Szostak et al. (44); since then, many of its basic features have been proven to be true for yeast cells undergoing meiosis (reviewed in reference 39). Briefly, single-strand degradation at the broken DNA molecule creates protruding 3′-OH ends that can invade the homologous regions in the other chromosome, forming two cross-stranded structures (Holliday junctions) surrounded by regions of heteroduplex DNA. Mismatch repair (MMR) in the heteroduplex DNA may lead to gene conversion, and depending on the resolution of the intermediates, this conversion may or may not be associated with a reciprocal exchange (44). An alternative model, termed synthesis-dependent strand annealing, has recently been supported by findings from several laboratories. In this model, following single-stranded DNA resection, one of the broken arms invades the homologous regions in the other chromosome, allowing DNA synthesis to take place. The newly synthesized DNA can then reanneal with the other broken end, resulting in a gene conversion event that is never accompanied by a crossover (5, 11, 12, 25, 28, 45).
The MMR system is composed of proteins that are able to identify mismatched DNA created during replication or recombination and to ensure their correction. The MMR proteins of prokaryotes and eukaryotes have been extensively analyzed. Homologs of the bacterial MutS protein (which binds to mismatched base pairs) and of the MutL protein (which interacts with MutS) have been identified in many eukaryotic organisms (19, 23). In eukaryotes the MMR proteins form several alternative complexes that usually include two different MutS homologs and two different MutL homologs (19). The MMR proteins that have been implicated in mitotic recombination include the MutS homologs Msh2p, Msh3p, and Msh6p (9, 10, 35). Some of the MMR proteins have been shown to prevent recombination between divergent DNA sequences in prokaryotes and eukaryotes (4, 8, 14). This antirecombinational effect is presumably aimed at preventing recombination between divergent sequences and may play a central role in the creation of new species (8, 31).
Recently, evidence for a direct role of Msh2p and Msh3p in recombination has accumulated. Msh2p binds in vitro to Holliday junction structures; its binding affinity for such structures is even higher than that seen for its interactions with mismatched DNA (3). In addition, mutations in the MSH2 and MSH3 genes cause an increase in recombination between identical sequences, which cannot generate mismatches during recombination (10). Mutations in the MSH2 and MSH3 genes reduce the efficiency of recombination of a broken plasmid, which requires the removal of nonhomologous ends for its repair (27, 41). Hence, the MMR proteins may directly participate in the process of recombination, playing a role different from MMR per se.
Recombination between homologous DNA sequences located at different places in the genome takes place at frequencies similar to that seen between alleles located at the same position along homologous chromosomes. This is true for both vegetative cells and cells undergoing meiosis, although in the latter case the homologous chromosomes are paired along their entire length (17, 18, 21). We have previously shown that a DSB can be efficiently repaired by gene conversion when a 500-bp homologous sequence located on a different chromosome is provided as a template (15). The repair of such a chromosomal break involved the scanning of the whole genome in order to recognize the small stretch of sequence homology. Nothing is known about the mechanism that executes the homology search and the choice of partner during the process of recombination.
In the present study we have analyzed the repair of a chromosomal DSB in haploid yeast cells. We have studied the mechanism by which homologous partners are chosen during mitotic recombination.
MATERIALS AND METHODS
Media and growth conditions.
S. cerevisiae strains were grown at 30°C. Standard YEP medium (1% yeast extract, 2% Bacto Peptone) supplemented with 3% glycerol (YEPGly), 2% galactose (YEPGal), or 2% dextrose (YEPD) was used for nonselective growth; 1.8% Bacto Agar was added for solid media (36).
Yeast strains.
All of the strains used in the present work are isogenic derivatives of OI27 (MATa-inc ura3-HOcs-inc ade3::GALHO ade2-1 leu2-3,112 his3-11,15 trp1-1 can1-100). Strain OI29 carries at the LYS2 locus on chromosome II a 1.2-kb HindIII fragment containing the ura3-HOcs allele. This allele was created by inserting a 39-bp oligonucleotide at the NcoI site of the URA3 gene (43). The ura3-HOcs-inc allele was similarly created and differs from the ura3-HOcs allele at three positions: it carries a G-to-A change at the HOcs, which prevents the recognition by the HO endonuclease (26), and substitutions to the right and to the left of the HOcs, which introduce BamHI and EcoRI sites, respectively. In strain OI70 a 5.6-kb BamHI fragment containing the ura3-HOcs allele was inserted into the LYS2 locus. The different ura3-HOcs alleles used were inserted at the HpaI site within LYS2 sequences by means of the integrative plasmid pOI5 (TRP1 LYS2 URA3). They were integrated into the yeast chromosome II by a two-step replacement method (1). Strains MK181 and OI78 were created by transforming strains OI29 and OI70, respectively, with plasmid pM141. This plasmid enables the integration of a LYS2-containing 4.9-kb XbaI-HindIII DNA fragment close to the HIS3 locus on chromosome XV. Strain OI76 was constructed by deleting the MMR genes of strain MK181 by one- or two-step transplacement with plasmid pSR395 for MSH2, pRK366 for MSH3, and pRK465 for MSH6 (4, 9). Strain MK182 is identical to strain MK181 except that it bears, at the LYS2 locus on chromosome II, a 1.2-kb HindIII fragment containing the ura3-HOcs allele, into which changes were introduced by site-specific mutagenesis at ∼100-bp intervals (43). Strain OI77 was created by deleting the MSH2, MSH3, and MSH6 genes of strain MK182 as explained above.
Recombination assay.
Single colonies were resuspended in rich YEPGly medium, grown to logarithmic phase, centrifuged, and resuspended in YEPGal or YEPD medium. DNA was extracted from samples at various times and subjected to Southern blot analysis with LYS2 sequences as probes. The blots were quantified with a Fujix BAS1000 phosphorimager. For each strain tested, the experiment was repeated at least three times.
The statistical differences were examined by a two-tailed Mann-Whitney two-sample test (38).
RESULTS
In order to study the mechanism of recombination, we have developed an assay for DSB-initiated interchromosomal recombination (Fig. 1). A haploid yeast strain (OI29) bears two copies of the URA3 gene; one allele carries the recognition site for the yeast HO site-specific endonuclease (26, 46) inserted as a short oligonucleotide (ura3-HOcs). The second copy, located on another chromosome, carries a similar site containing a single-base-pair mutation that prevents recognition by the endonuclease (ura3-HOcs-inc) (43). In addition, the two ura3 alleles differ at two restriction sites, located to the right (BamHI) and to the left (EcoRI) of the HOcs-inc insertion; these polymorphisms are used to monitor the transfer of genetic information between the chromosomes (15, 16). In these strains the HO gene is under the transcriptional control of the GAL1 promoter. Upon transfer of the cells to galactose-containing medium, the HO endonuclease is produced at elevated levels. The enzyme creates a chromosomal DSB; if left unrepaired, such a lesion leads to cell death (references 20 and 24 and data not shown). The presence of homologous information in the genome can be used to repair the DSB by a recombinational repair mechanism. Cell survival is very high: approximately 90% of the cells plated on galactose medium are able to recover and form colonies. It is possible to monitor the kinetics of recombination in real time, by extracting DNA from aliquots taken at intervals and subjecting them to Southern blot analysis. This analysis has shown that a DSB is produced in essentially every cell of the population and is repaired efficiently by a gene conversion event that transfers genetic information from chromosome V to chromosome II (15, 16).
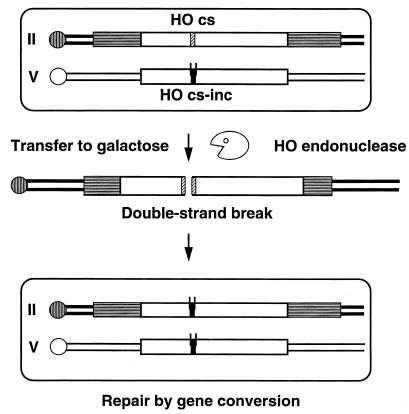
FIG. 1.
Schematic representation of the experimental system (strain OI29). Open rectangles represent the ura3 alleles on chromosomes II and V. Striped rectangles represent LYS2 sequences on chromosome II. A hatched box represents the HO cut site (HOcs); black boxes represent the inactive HOcs-inc, with the two lines representing polymorphic sites recognized by the EcoRI and BamHI restriction enzymes. Transfer of the cells to galactose-containing medium induces the production of the HO endonuclease, which recognizes the HOcs and creates a DSB. If this DSB is not repaired, it leads to cell death. We have found that the break is repaired in all of the cells of the population by a gene conversion event that transfers the HOcs-inc information to chromosome II, thus preventing further recognition by the HO endonuclease (16).
A system to study the choice of homologous partner during recombination.
Once a DSB is created in a chromosome, an efficient search is carried out throughout the genome to localize homologous sequences that can serve as templates during the repair by gene conversion. In order to study this still-uncharacterized mechanism which governs the homology search and choice of partner, we created strain MK181. This strain carries two alleles of ura3, and in addition, it bears a 4.9-kb fragment containing the LYS2 gene integrated on chromosome XV (Fig. 2). Upon creation of a DSB at the ura3-HOcs allele, the break can be repaired by recombination with either the ura3 genetic information on chromosome V or the LYS2 sequences on chromosome XV. In the first case, recombination takes place with sequences homologous to the very ends of the broken chromosomes, whereas in the second case, recombination takes place with genetic information homologous to internal sequences which are not adjacent to the broken ends (Fig. 2). We can ask whether the search for homology is restricted to a short region flanking the DSB or whether it is carried out along a larger chromosomal domain. It is important to note that the repair of the DSB by a gene conversion event involving the distant LYS2 sequences requires the processing of the broken arms in order to remove nonhomologous information, represented in this case by the 1.2-kb ura3-HOcs allele.
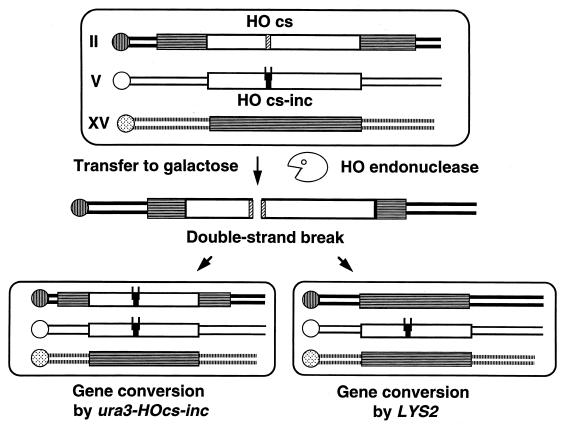
FIG. 2.
Schematic representation of the homologous-partner choice experimental system. Open rectangles represent the ura3 alleles on chromosomes II and V. Striped rectangles represent LYS2 sequences on chromosomes II and XV. A hatched box represents the HOcs; black boxes represent the inactive HOcs-inc, with the two lines representing polymorphic sites recognized by the EcoRI and BamHI restriction enzymes. Transfer of the cells to galactose-containing medium induces the production of HO endonuclease, which recognizes the HOcs and creates a DSB. This break is repaired by gene conversion with one of two alternative partners: the ura3 allele on chromosome V and the LYS2 allele on chromosome XV. In the latter case, the ura3-HOcs allele has to be removed.
The kinetics of DSB repair of strain MK181 were analyzed in real time following the transfer of a population of cells to galactose-containing medium, in which the HO endonuclease is induced. Aliquots were taken at various times, and the DNA was extracted and subjected to Southern blot analysis, which allows monitoring of the fate of the broken chromosome. The results of such an experiment are shown in Fig. 3. Within 30 min after transfer, two new bands were seen, representing the broken chromosome arms. These bands accumulated, reaching about 80% of the cells 90 min after transfer to galactose. Most of the cell population remained with broken chromosomes for another 2 h, and then the percentage of cells bearing a DSB diminished. At the same time as the bands representing the DSB started to decrease, new bands were seen, representing gene conversion events that involved LYS2 information. Seven hours after transfer to galactose, when no broken chromosomes could be detected, 91% of the cells had completed a gene conversion event involving LYS2 sequences. Only a minority of the cells (9%) repaired the DSB by a gene conversion event involving the ura3 sequences, which have homology with the very ends of the broken chromosome (Fig. 3B). Essentially the whole cell population of strain MK181 (more than 99% of the cells) repaired the DSB by gene conversion with either the ura3 or the LYS2 alleles.
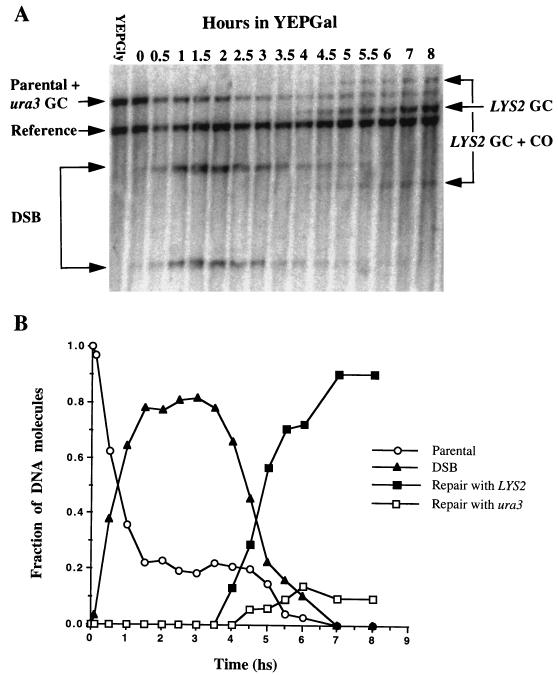
FIG. 3.
(A) Southern blot analysis of DNA extracted from strain MK181 at various times after transfer to galactose-containing medium. The DNA was digested with ClaI and probed with a fragment of chromosome II carrying the LYS2 gene. The reference band represents the LYS2 copy on chromosome XV, which is used to quantitate the amount of DNA in each lane. The band designated parental + ura3 GC represents the parental ura3 allele on chromosome II and the product of gene conversion with its allele on chromosome V. The parental band can be separated from the gene conversion product by digesting the DNA with EcoRI or BamHI, since these sites are transferred to the broken chromosome during the ura3 gene conversion event (data not shown). The band designated LYS2 GC represents the product of gene conversion events with LYS2, whereas the bands designated LYS2 GC + CO represent events in which the gene conversion with LYS2 was accompanied by a crossing over. (B) Kinetics of DSB repair. The Southern blot shown in panel A was scanned; the relative intensity of each band after normalization with the reference band is presented. The apparent delay in the appearance of ura3 recombinants compared to LYS2 recombinants is probably due to a technical reason: for the sample taken 4 h after transfer to galactose, the band representing gene conversion with ura3 cannot be distinguished from the background.
Gene conversion events are often associated with reciprocal crossing over (13, 17, 18, 21, 29). In the strains analyzed in the present study, if the repair of the DSB by a gene conversion event involving LYS2 sequences is coupled to a crossing-over event, it results in a translocation between chromosomes II and XV. Bands representing the translocation accumulated together with those resulting from the gene conversion event, reaching a maximal level of 13% of this cell population (Fig. 3). This level of association between gene conversion and crossing over is in agreement with that seen in previous studies (13, 15, 17, 29). In addition, no associated crossing over was seen among the cells that repaired the DSB by gene conversion with ura3 sequences, as observed before for such limited lengths of homology (15).
In strain OI29 (which has only one potential partner), every cell of the population repaired the DSB by a gene conversion event with ura3 sequences. The same high efficiency of repair was observed in strain MK181, where two alternative repair possibilities existed. In the competition between the two homologous partners, ura3 and LYS2 sequences, more than 90% of the cells chose to repair the DSB by transferring LYS2 genetic information, which is not adjacent to the broken chromosomal ends. During this repair, 1.2 kb of nonhomologous sequences was efficiently removed from the broken ends. These results show that sequences distant from the DSB can successfully participate in the search for homology and imply that the search is carried out by a large chromosomal region.
Effect of the length of the ura3 allele on the choice of homologous partner.
The DSB in strain MK181 is created within ura3-HOcs sequences, which have 1.2 kb of homology to the ura3-HOcs-inc allele on chromosome V. Most of the recombination events, however, were carried out with the 4.9-kb LYS2 sequences despite the fact that the DNA ends were not homologous and had to be removed to complete the repair. In order to study the effect of homology length on the competition between the two substrates, we created a strain in which the ura3 allele was fivefold longer: strain OI78 carries a 5.6-kb ura3 allele on chromosome II and a 4.9-kb copy of LYS2 on chromosome XV, and the two alleles can compete to serve as templates for the repair of the DSB.
When cells of strain OI78 were transferred to galactose-containing medium, 59% of them repaired the DSB by a gene conversion event with the ura3 allele (Table 1). This value is sixfold higher than that obtained with MK181 (1.2 kb of ura3 homology), where only 9% of the cell population repaired the DSB by gene conversion with the ura3 allele. Similar to the results obtained for strain MK181, 12% of the cells of OI78 that repaired the DSB by gene conversion with LYS2 exhibited an associated crossover that created a translocation between chromosomes II and XV (Table 1).
TABLE 1.
Influence of sequence length and divergence and mutations in the MMR genes on choice of homologous partner and on coupling of crossing over to gene conversion
| Strain | ura3 allele (kb) | MMR function | % of gene conversion with the ura3/LYS2 alleles | % of LYS2 gene conversion events associated with crossing over |
|---|---|---|---|---|
| OI78 | 5.6 | MMR+ | 59/41 | 12 |
| MK181 | 1.2 | MMR+ | 9/91 | 13 |
| OI76 | 1.2 | MMR− | 26/74 | 3.2 |
| MK182 | 1.2 (1% divergence) | MMR+ | 6/94 | 19 |
| OI77 | 1.2 (1% divergence) | MMR− | 4/96 | 2.9 |
We conclude that the length of the homologous sequences adjacent to the DSB affects the choice of homologous partner: when these sequences are longer, a higher proportion of the cells use them to repair the DSB. It is important to note, however, that 40% of the cells of strain OI78 still repair the break by recombination with a region homologous to the distant sequences; this repair requires processing of 5.6 kb of nonhomologous DNA from the broken ends.
Effect of mutation in the MMR genes on the choice of homologous partner.
Recently it has been shown that some of the MMR proteins are needed for the removal of terminal heterologous regions in DSB repair of plasmids (27, 41) and in the recombination between chromosomal direct repeats (34, 41). In order to test the involvement of MMR proteins in a chromosomal system, we created strain OI76. This strain is identical to strain MK181 (1.2-kb ura3 alleles and LYS2 sequences on chromosome XV) except that it has three of the MutS homologous genes, MSH2, MSH3, and MSH6, deleted. We will refer to the triple-mutant strains as MMR−, since it is unlikely that they have any functional mitotic MMR activity that may affect recombination (9, 10, 19, 23, 35, 41). When cells of strain OI76 were transferred to galactose-containing medium, the DSB created in each cell was still efficiently repaired by recombination. In this strain 26% of the cells repaired the DSB by a gene conversion event involving the ura3 allele (Table 1). This proportion is almost threefold higher than that obtained with strain MK181 (9%); the difference between the two strains is statistically significant (P < 0.024). Among the cells that repaired the DSB by gene conversion with LYS2 sequences, only 3% exhibited an associated crossover that resulted in a translocation between chromosomes II and XV. This level of association is fourfold lower than the one seen for strain MK181 (Table 1).
Three conclusions can be drawn from these results. (i) The MMR proteins play a role in the choice of homologous partner. Their presence in the cells channels the repair of the DSB towards gene conversion with LYS2 sequences. (ii) Although deletion of the MMR genes influences the choice between the competing partners, the overall efficiency of repair remained unchanged; 100% of the population still is able to repair the break by gene conversion. Thus, mitotic gene conversion can take place without an active MMR system (16, 30). (iii) In several systems in which crossing over can be monitored, mutations in the MMR genes lead to an increase in crossing-over levels (4, 8, 14, 35). This has led to the proposal that the MMR system plays an antirecombinational role, presumably in order to prevent recombination between divergent sequences (4, 8, 14, 31). We show that in our system the MMR proteins play a positive role in coupling crossing over to gene conversion: mutations in the MMR genes drastically reduce the level of associated crossovers among LYS2 recombinants.
Effect of sequence divergence of the ura3 alleles on the choice of homologous partner.
Mutations in the MMR system increase the proportion of cells that repair the DSB by gene conversion with the ura3 allele, implying that the activity of the MMR proteins results in a channeling of the repair process towards recombination with LYS2 sequences. This could be due to two alternative roles played by the MMR system: (i) the MMR proteins may be involved in the processing of the nonhomologous DNA ends (e.g., removing the nonhomologous ura3 sequences from the broken chromosomal ends to allow recombination with the LYS2 allele), or (ii) the MMR system may cause a higher proportion of recombination with LYS2 by recognizing mismatches created during gene conversion events involving the ura3 alleles. In all of the strains examined, the ura3 alleles differ at three positions within the 1.2 kb of homology; thus, during gene conversion, only three potential mismatches may be created. In order to test whether the competition between the two potential partners is affected by heterology between the interacting sequences, we created strain MK182. This strain bears a 1.2-kb ura3-HOcs allele inserted at chromosome II that has been modified by introducing a sequence change every ∼100 bp (43). Thus, the two ura3 alleles show 1% sequence divergence.
When cells of strain MK182 were transferred to galactose-containing medium, only 6% of the cells repaired the DSB by gene conversion with the divergent ura3 allele (Table 1). This result is significantly different from that obtained for MK181, which bears near-identical ura3 alleles (P < 0.001). Southern blot analysis indicates that 19% of the gene conversion events with LYS2 sequences were associated with a reciprocal crossing over, a result which is not significantly different from the one seen with strain MK181 (Table 1). We conclude that the 1% sequence divergence between the ura3 alleles affects the choice between the two potential recombination partners, causing most of the DSBs to be repaired by gene conversion with the LYS2 sequences. In contrast, the ura3 sequence divergence played no role in determining the level of association between crossing over and gene conversion by the LYS2 allele.
Combined effect of sequence divergence and the MMR system on the choice of homologous partner.
We have shown that mutations in the MMR genes increased the proportion of cells that chose to repair the DSB by gene conversion with the ura3 allele, whereas sequence divergence decreased this proportion. In order to study the interaction between these two factors, we created strain OI77. This strain is identical to MK182 (1% divergent ura3 alleles) but, like OI76, has the MSH2, MSH3, and MSH6 MutS homologs deleted. When cells of strain OI77 were transferred to galactose-containing medium, only 4% of them repaired the DSB by gene conversion with the divergent ura3 sequences (Table 1). Thus, this strain does not show the increase in ura3 gene conversion seen in the MMR− strain OI76. Moreover, the low ura3 gene conversion of strain OI77 is similar to that seen in strain MK182 (1% divergence, MMR+) (Table 1). We therefore conclude that the effect of sequence divergence overcomes the effect of mutations in the MMR system in the competition between the potential recombination partners.
Southern blot analysis of strain OI77 (1% divergent ura3 alleles, MMR−) showed that only 3% of the cells that repaired the break by gene conversion with LYS2 exhibited reciprocal crossovers (Table 1). This result is different from the one seen for strain MK182 (1% divergent ura3 alleles, MMR+) but similar to that seen for strain OI76 (near-identical ura3 alleles, MMR−). Thus, the reduction in coupling of crossing over to gene conversion with LYS2 seen in MMR− strains is independent of the presence of sequence divergence in the ura3 allele flanking the DSB.
DISCUSSION
When a chromosome is broken at more than one site, random joining of the broken fragments can create deleterious chromosomal aberrations. By using homologous sequences as templates during repair, recombination is able to restore the original chromosomal structure. In addition, during the recombinational repair of DSBs, the transfer of genetic information prevents mutagenesis by restoring information that may have been eliminated at the site of the DNA damage. Recombinational repair of DNA damage by using repetitive sequences as templates may result in chromosomal aberrations. The genomic instability resulting from incorrectly repaired DSBs may lead to carcinogenesis or inborn defects (13, 15–18, 21, 29). Hence, it is important to understand how the search for the suitable homologous partner and the subsequent repair processing take place.
The study of the mechanism by which the homology is searched throughout the genome is hampered by the fact that monitoring of recombination usually requires the selection of rare genetic products. This introduces biases in the type of events studied and precludes an understanding of the processes that take place in the majority of the cells. In order to overcome these problems, we have developed a recombination system that allows monitoring of the fate of a cell population that has suffered a chromosomal break, without the need to select for recombinants. The repair of the broken chromosome can be carried out by a gene conversion event involving either sequences immediately adjacent to the DSB (the ura3 allele) or sequences distant from the DSB (the LYS2 allele). We have shown that there is a competition between the two potential homologous partners. From the analysis of factors that influence this competition, we can learn about the mechanism that searches the genome for homologous sequences suitable to serve as templates for recombinational repair.
Our results show that the creation of a DSB initiates an efficient search for homologous sequences which encompasses the whole genome. This homology search involves extensive regions of the broken chromosome and is not carried out exclusively by sequences adjacent to the DSB. Previous evidence for such an activation of a large chromosomal region has been reported (32). We have shown that when the DSB was created within 1.2 kb of ura3 sequences (strain MK181), only a minority of the cells (9%) repaired the break by recombination with the ura3 homologous allele. When the terminal ura3 sequences were enlarged to 5.6 kb, 60% of the cells used this allele to repair the break (strain OI78). However, 40% of the cells still repaired the DSB by a gene conversion event with the LYS2 sequences. These cells managed to find a homologous partner to regions distant from the DSB, and they successfully repaired the break by gene conversion with these sequences. Although in this strain recombination with the LYS2 allele required the removal of a large 5.6-kb fragment of nonhomologous DNA, the repair was very accurate: the whole ura3 allele was removed from the LYS2 locus during this process.
In this report we show a feature of the recombinational repair mechanism that helps to maintain the stability of the genome. In order to avoid the deleterious consequences of using repetitive sequences as templates for the repair of DSBs, the search for a homologous partner takes place along a wide region of the chromosome. This allows confirmation that the partner involved has homology along a large region, and thus it ensures that the broken chromosome is repaired with the suitable homologous partner (such as the homolog or the sister chromatid).
Figure 4 shows a possible model that explains how the choice of homologous partner takes place during recombinational repair. This model is based on the current version of the DSB-initiated model of recombination (39, 44) but incorporates several features present in the synthesis-depended strand annealing model (5, 11, 12, 25, 28, 45). The DSB creation activates a large chromosomal region (Fig. 4A). The activation initiates a search for homology which encompasses the whole genome. At this stage the DNA at the broken ends is subjected to single-stranded DNA resection. Evidence for the 5′-to-3′ degradation of the broken DNA ends in meiosis (42) and in vegetative cells (7, 40) has been obtained. The search for the homologous partner by sequences distant from the DNA ends may be carried out by single-stranded DNA and thus require extensive chromosomal resection. If, during the resection, homologous sequences flanking the DSB are exposed, the break can be repaired by a single-strand annealing mechanism (22, 40).
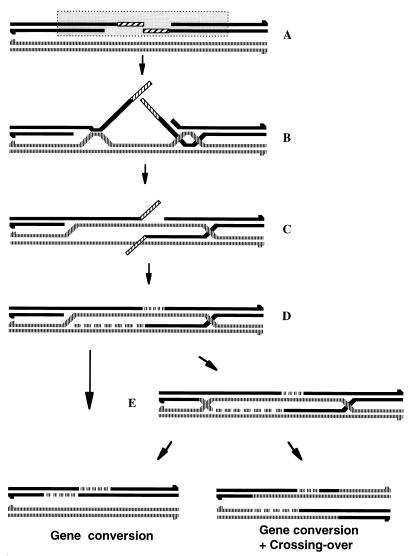
FIG. 4.
A model for recombination initiated by sequences distant from the chromosomal DSB. (A) The creation of a DSB activates a large chromosomal region, which initiates a search for suitable homologous partners. Single-stranded DNA resection of the broken ends takes place at this stage. (B) Weak interactions occur between the broken chromosome and homologous sequences in the genome. These interactions may involve single-stranded or double-stranded DNA. (C) The interactions are stabilized and extended in both directions, until the border of homology is reached. (D) The nonhomologous terminal sequences are removed by a process in which the MMR system participates. The exposed DNA ends can now serve as primers for DNA synthesis. (E) An intermediate consisting of two Holliday junctions is created by ligation. Such an intermediate can be resolved to give gene conversion events or gene conversion events accompanied by a crossover. Alternatively, the intermediate depicted in panel D can disassemble; reannealing between the newly synthesized strands gives rise to gene conversion events (15).
The search for a homologous partner leads to interactions between short stretches of DNA (Fig. 4B). The searching mechanism may consist of the probing of weak paranemic interactions by the activated chromosomal region. Once homologous contacts are made, they may be converted into more stable plectonemic interactions that can be extended in both directions (6). This extension of the homologous interaction allows testing of whether the recombination partner is suitable (homolog or sister chromatid). If the interactions take place between short repeated sequences, the extended contact will soon reach the border of the homologous region and will lead to disengagement, allowing a second round of homology search. Once the borders of homology are defined, the nonhomologous tails are removed by a mechanism that is partially dependent on the MMR system (Fig. 4C). This removal may involve other proteins, such as the Rad1/Rad10 endonuclease and the Srs2p helicase (27, 41). If the DSB is repaired by gene conversion with LYS2 sequences, the fate of the ura3 allele is similar to that of a natural repetitive sequence in the genome: it does not serve as template for repair and is precisely removed. We have never detected ura3 genetic information left when the repair took place by gene conversion with the LYS2 allele. This implies that the mechanism of repair always involves genetic information on both sides of the DSB. Our results thus do not support one-handed models of recombination in which each broken arm independently searches for homology (5, 28, 37, 39).
Following the removal of nonhomologous sequences, the newly exposed DNA ends may now serve as primers for DNA synthesis in accordance with the current models of recombination (25, 28, 39, 44) (Fig. 4D). As we have previously suggested, recombination can then continue by two different pathways (15). In one of them, the invading DNA strands complete DNA synthesis and undergo ligation, resulting in the formation of a double Holliday junction (Fig. 4E) which can then be resolved to give either a gene conversion event or a gene conversion event accompanied by a crossover (44). In the second pathway, the intermediate shown in Fig. 4D is disassembled, and the newly synthesized DNA molecules reanneal to each other, resulting in a gene conversion event that is never accompanied by a crossover. Thus, the stability of this intermediate determines whether there is a coupling between gene conversion and crossing over (15).
One feature of this model is that it does not require the presence of homologous sequences at the broken DNA ends. Since according to the model a large chromosomal segment is activated and participates in the search for homology, in many cases the partner chosen could be the LYS2 allele. When the homologous partner found is the ura3 allele, the repair takes place similarly, except that no heterologous DNA tails have to be removed from the broken ends.
We have shown that different factors affect the choice of homologous partner: length of homology, sequence divergence, and activity of the MMR system. A low level of sequence divergence (1%) of the region close to the break causes a shift in partner choice: more cells repair their broken chromosome with the LYS2 allele (compare strains MK181 and MK182 in Table 1). We envision two alternative ways in which the sequence heterology can influence this choice: (i) the creation of mismatches during recombination can elicit the action of the MMR system, or (ii) the mismatches can affect recombination directly, e.g., by reducing the homologous recognition or DNA branch migration, etc. We prefer the second hypothesis, since mutations in the MMR genes did not affect the distribution of events between the two homologous partners in strains with many potential mismatches (compare strains MK182 and OI77 in Table 1). On the other hand, mutations in the MMR genes did affect the partner choice in strains bearing near-identical ura3 alleles (compare strains MK181 and OI76 in Table 1). When functional, the MMR system contributes to the repair by gene conversion with the LYS2 allele, probably through the removal of the nonhomologous DNA ends (27, 41).
The mechanism of recombination is often regarded as a unidirectional process in which once two homologous sequences make contact, they are committed to a certain pathway. If this interaction is not productive, it may lead to cell death. An alternative view is that partner choice during recombination is a dynamic process: if a difficulty is encountered during a homologous interaction, the molecules can disengage and a second round of homology search can take place. The effect of sequence divergence and mutations in MMR genes on the partner choice supports this dynamic view of the homology search mechanism.
An association between gene conversion events and reciprocal crossing over is often observed (13, 15, 17, 18, 21, 29). We have shown that the association was seen in about 14% of the cells that repaired the DSB by gene conversion with LYS2 sequences. The coupling was independent of the length of the nonhomologous DNA ends or of the divergence level between the sequences close to the break. In contrast, we found that mutations in the MMR genes reduced the coupling: in two MMR− strains more than 99% of the cells were able to repair the DSB, but only 3% of these events were associated with a reciprocal crossing over. This reduction in the association was independent of the level of divergence, suggesting that the effect of the MMR system on crossing over has no relation to the information defined as nonhomologous.
The reduction in crossing over seen in MMR− mutants is in contradiction with results in the literature which show that mutations in MMR genes cause an increase in crossing over (4, 8, 14, 15, 35). We have thus uncovered a positive role for the MMR proteins in promoting crossing over. A positive role of the MutS homologs in recombination between chromosomal direct repeats has been also reported (34, 41). It has been shown that the Msh2 protein, alone or in complexes with Msh3p and Msh6p, is able to recognize looped mispairs and branched DNA structures such as Holliday junctions (2, 3). The presence of a nonhomologous tail may be recognized by Msh2p, which may then act as a recruitment factor to direct other proteins to the recombination intermediates, allowing its resolution as a crossover. The activity of the MMR proteins in the removal of the nonhomologous DNA ends may cause the stabilization of the intermediate depicted in Fig. 4D, allowing the formation of the intermediate seen in Fig. 4E, which can then be resolved as a gene conversion associated with a crossover. The positive role of the MMR system we have shown may therefore be related to the function of these proteins in the processing of the nonhomologous ends of the chromosome.
It is important to note that consistent with our previous results (16), mitotic gene conversion with both LYS2 and ura3 sequences can take place in vegetative cells in the absence of a functional MMR system. This is different from what the current models for recombination predict and from what is observed in MMR− cells undergoing meiosis, in which gene conversion is greatly reduced (4, 8, 14, 19, 39). We have proposed a model for MMR-independent mitotic recombination (15) that can explain this phenomenon.
To summarize, we have shown that the search for a homologous partner takes place by a mechanism that encompasses a large chromosomal domain. Regions distant from the broken ends participate in the search for a suitable homologous partner. The homology search mechanism is dynamic, as seen by the influence on the partner choice of the length and sequence divergence of the region adjacent to the break, as well as by the effect of mutations in the MMR genes. The presence of many repeated sequences in the genome requires that the appropriate partners be found in order to allow the repair of a chromosome without causing karyotypic rearrangements. We have presented a model that explains how the homologous partner is chosen; this model provides a mechanism that may guard the integrity of the genome by preventing recombination between dispersed repeated sequences.
ACKNOWLEDGMENTS
We thank J. Nickoloff, S. Jinks-Robertson, and R. Kolodner for the generous gift of plasmids.
This work was supported by a grant from the Israel Science Foundation to M.K. O.I. was partially supported by a scholarship from the Constantiner Institute for Molecular Genetics.
REFERENCES
- 1.Alani E, Cao I, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E, Chi N W, Kolodner R D. The Saccharomyces cerevisiae MSH2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base-pairs and loops insertions. Genes Dev. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- 3.Alani E, Lee S, Kane M F, Griffith J, Kolodner R D. Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J Mol Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 4.Alani E, Reenan R A G, Kolodner R D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belmaaza A, Chartrad P. One-side invasion events in homologous recombination at double-strand-breaks. Mutat Res. 1994;314:199–208. doi: 10.1016/0921-8777(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi M, DasGupta C, Radding C M. Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell. 1983;34:931–939. doi: 10.1016/0092-8674(83)90550-0. [DOI] [PubMed] [Google Scholar]
- 7.Bitan, G., O. Inbar, and M. Kupiec. Unpublished data.
- 8.Chambers S R, Hunter N, Louis E J, Borts R H. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol Cell Biol. 1996;16:6110–6120. doi: 10.1128/mcb.16.11.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datta A, Adjiri A, New L, Crouse G F, Jinks-Robertson S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. Dual role for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson D O, Holloman W K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Formosa T, Alberts B M. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 13.Harris S, Rudnicki K S, Haber J E. Gene conversions and crossing-over during homologous and homeologous ectopic recombination in Saccharomyces cerevisiae. Genetics. 1993;135:5–16. doi: 10.1093/genetics/135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter N, Borts R H. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 1997;11:1573–1582. doi: 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- 15.Inbar, O., G. Bitan, and M. Kupiec. The mechanism that prevents genomic rearrangements during recombinational DNA repair. Submitted for publication.
- 16.Inbar, O., and M. Kupiec. Recombination between divergent sequences leads to cell death in a mismatch-repair-independent manner. Submitted for publication. [DOI] [PubMed]
- 17.Jinks-Robertson S, Michelitch M, Ramcharan R. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinks-Robertson S, Petes T D. Chromosomal translocations generated by high frequency meiotic recombination between repeated yeast genes. Genetics. 1986;114:731–752. doi: 10.1093/genetics/114.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 20.Kramer K M, Brock J A, Bloom K, Moore J K, Haber J E. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichten M, Borts R H, Haber J E. Meiotic gene conversion and crossing over between dispersed homologous sequences occur frequently in Saccharomyces cerevisiae. Genetics. 1987;115:233–246. doi: 10.1093/genetics/115.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin F, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 24.Moore J K, Haber J E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nassif N, Penney J, Pal S, Engels E R, Gloor G B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nickoloff J A, Singer J D, Heffron F. In vivo analysis of the Saccharomyces cerevisiae HO nuclease recognition site by site-directed mutagenesis. Mol Cell Biol. 1990;10:1174–1179. doi: 10.1128/mcb.10.3.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paques F, Leung W Y, Haber J E. Expansions and contractions in tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- 30.Porter G, Westmoreland J, Priebe S, Resnick M. Homologous and homeologous intermolecular gene conversion are not differentially affected by mutations in DNA damage or mismatch repair genes RAD1, RAD50, RAD51, RAD52, RAD54, PMS1 and MSH2. Genetics. 1996;143:755–767. doi: 10.1093/genetics/143.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Radman M. Avoidance of inter-repeat recombination by sequence divergence and a mechanism of neutral evolution. Biochimie. 1991;73:357–361. doi: 10.1016/0300-9084(91)90101-6. [DOI] [PubMed] [Google Scholar]
- 32.Ray A, Machin N, Stahl F W. A DNA double chain break stimulates triparental recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:6225–6229. doi: 10.1073/pnas.86.16.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth D B, Craig N L. VDJ recombination: a transposase goes to work. Cell. 1998;94:411–414. doi: 10.1016/s0092-8674(00)81580-9. [DOI] [PubMed] [Google Scholar]
- 34.Saparbaev M, Prakash L, Prakash S. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selva E M, New L, Crouse G F, Lahue R S. Mismatch correction acts as barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 37.Silberman R, Kupiec M. Plasmid-mediated induction of recombination in yeast. Genetics. 1994;134:41–48. doi: 10.1093/genetics/137.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokal R R, Rolf F J. Biometry. 2nd ed. San Francisco, Calif: W. H. Freeman; 1981. [Google Scholar]
- 39.Stahl F W. Meiotic recombination in yeast: coronation of the double-strand repair model for recombination. Cell. 1996;87:965–968. doi: 10.1016/s0092-8674(00)81791-2. [DOI] [PubMed] [Google Scholar]
- 40.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-strand DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugawara N, Paques F, Colaiacovo M, Haber J E. Role of Msh2 and Msh3 repair proteins in double-strand break induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun H, Treco D, Szostak J W. Extensive 3′-overhanging single-stranded DNA associated with meiosis-specific double-strand breaks induced at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 43.Sweetser D B, Hough H, Whelden J F, Arbuckle M, Nickoloff J A. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol Cell Biol. 1994;14:3863–3875. doi: 10.1128/mcb.14.6.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szostak J W, Orr-Weaver T L, Rothstein R, Stahl F W. The double-strand break repair model for recombination. Cell. 1983;3:725–738. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 45.Weng Y, Nickoloff J A. Evidence for independent mismatch repair processing on opposite sides of a double-strand break in Saccharomyces cerevisiae. Genetics. 1998;148:59–70. doi: 10.1093/genetics/148.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–674. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zenvirth D, Arbel T, Sherman A, Goldway M, Klein S, Simchen G. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 1992;11:3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]