ABSTRACT
Background and Objectives
Several types of needles are available for EUS–guided tissue sampling of pancreatic lesions. Whereas fine-needle aspiration (FNA) needles typically provide cytological samples, fine-needle biopsy (FNB) needles are designed to obtain microcores with preserved tissue architecture. The aim of this study was to compare tissue amount and diagnostic yield between a modified Franseen-type FNB needle (TopGain; Medi-Globe GmbH, Grassau, Germany) and a standard FNA needle.
Methods
We performed a prospective, multicenter randomized controlled study between June 2020 and September 2021, including patients with a solid pancreatic lesion referred for EUS-guided tissue sampling at 3 centers in Denmark. The patients were randomized 1:1 to either FNA needle or the novel FNB needle. Primary outcomes included the number of obtained tissue microcores and total and diagnostic tissue area.
Results
Sixty-four patients were included. The median number of tissue microcores procured per pass was significantly higher in the FNB group compared with FNA (3 vs. 2, P < 0.001). Similarly, the mean total tissue area (2.74 vs. 0.44 mm2, P < 0.001) and mean diagnostic tissue area (1.74 vs. 0.28 mm2, P < 0.001) were more than 6-fold larger in the FNB samples compared with FNA. The median number of passes needed for a diagnostic sample was 1 for the FNB needle and 2 for FNA needle (P = 0.12). The novel FNB needle provided a higher percentage of samples of excellent quality (P = 0.002).
Conclusions
The novel Franseen-type FNB needle seems to be significantly superior to a conventional FNA needle. The results of this study underline excellent performance of crown-cut needles.
Keywords: Pancreatic neoplasms; EUS-FNA; Biopsy, fine-needle; Biopsy, large-core needle; Image-guided biopsy
INTRODUCTION
Since its introduction in the early 1990s, EUS–guided tissue sampling has undergone major changes, and the development has mainly focused on improvement of the EUS needle design.[1] Although conventional fine-needle aspiration (FNA) needles have shown a very good performance, an increasing need for preserved tissue architecture in the form of tissue microcores led to development of fine-needle biopsy (FNB) needles.[2] This is especially true in case of pancreatic lesions where diagnosis of pancreatic cancer may prove difficult due to a substantial amount of tumor-induced desmoplastic fibrosis, but also in cases where specific diagnosis requires additional immunohistochemical staining and/or genetic analyses.
Current third-generation FNB needles are characterized by modifications of the needle tip, which is based on either 3 symmetrically distributed bevels around the needle lumen (Franseen design) or 2 bevels of unequal length on the opposite side of the needle lumen, resulting in 6 asymmetric cutting surfaces (fork-tip design).[3] In the last couple of years, several large randomized controlled studies (RCTs) have compared FNA and FNB needles, showing that FNB outperforms FNA mainly in the amount of procured histology-grade tissue and suitability of samples for ancillary analyses.[4,5] In a meta-analysis including 18 RCTs, FNB had a slightly higher pooled diagnostic accuracy compared with FNA (87% vs. 80%, P = 0.02).[6] Similarly, tissue microcores were more often obtained with an FNB needle (80% vs. 62%, P = 0.002), which overall required fewer passes to establish a diagnosis (P = 0.03).[6]
However, much of the current literature is focused on the older type of FNB needles with a side bevel, and there are only a few high-quality studies on newer crown-cut FNB needles (Franseen and fork-tip). In one of these, FNB was found to be the needle of choice when determining the presence of microsatellite instability in pancreatic tumors; successful evaluation was observed more often with FNB compared with FNA (89% vs. 36%, P = 0.03).[7] In another large multicenter RCT, Franseen-type FNB was superior to FNA with respect to the diagnostic accuracy per pass.[8] However, when the clinical question is to rule out pancreatic malignancy, FNA needle still has a strong foothold at many endoscopy units around the world. A recent modified Franseen FNB needle (SonoTip, TopGain; Medi-Globe GmbH, Grassau, Germany) uses a slightly modified tip geometry, where the bevel angle of the spikes was decreased from 15 degrees as seen in the original Franseen-type needle to 12 degrees. The choice of angle was based on bench studies showing that this change in angle resulted in less penetration force without sacrificing the tissue integrity and specimen size. Utility of this needle has so far only been mentioned in a single retrospective study of several needle and lesion types combined, and there are currently no prospective data.[9] The primary aim of this study was to compare the tissue amount and diagnostic yield of a novel FNB needle (TopGain; Medi-Globe GmbH) and a standard FNA needle [Figure 1].
Figure 1.
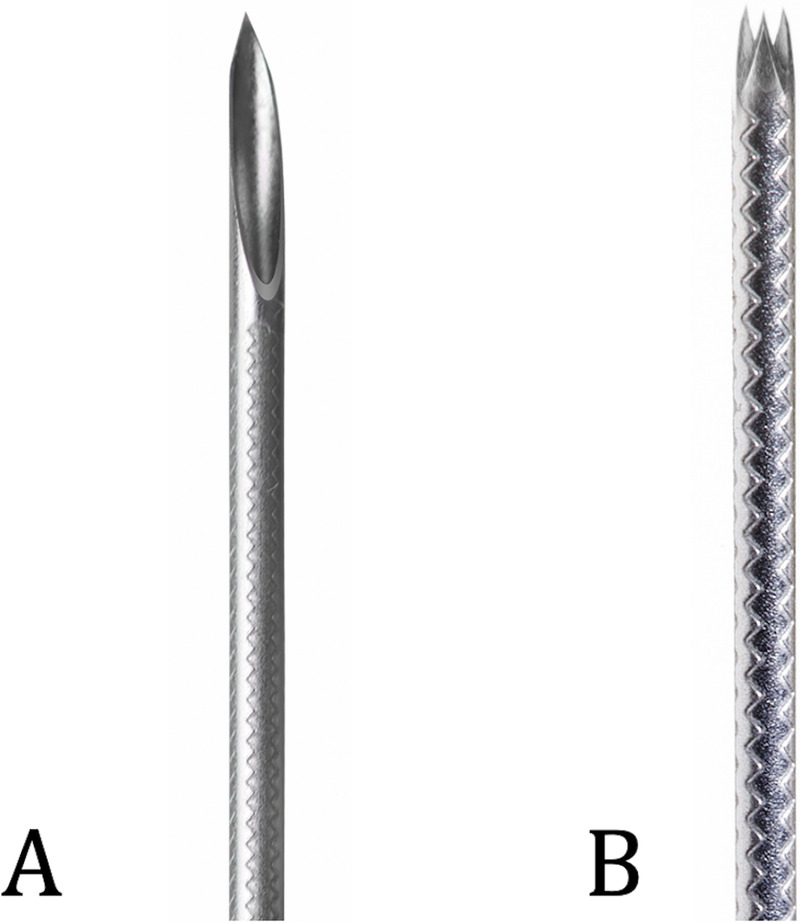
The 2 EUS-guided needles used in this study: standard FNA needle (A) and the novel modified Franseen needle (B). FNA, fine-needle aspiration.
PATIENTS AND METHODS
Patients
This was a prospective, multicenter, and randomized controlled trial from 3 centers in Denmark (Herlev, Hvidovre, and Odense university hospitals). Adult patients with a solid pancreatic lesion referred for EUS-guided tissue sampling were eligible for inclusion, given that they provided a written informed consent form. Exclusion criteria were uncorrected coagulopathies (international normalized ratio >1.5 or platelet count <50 000/mL) or anticoagulant therapy that could not be discontinued, pregnant or lactating females, and patients with interposed, large vessels between the transducer and the target lesion. The study was approved by the Regional Ethics Committee (no. H-20006798) and registered as a clinical trial (ClinicalTrials.gov NCT04687410).
Randomization and blinding
Following initial EUS examination, patients were randomized 1:1 to a 22-gauge FNA (SonoTip, ProControl; Medi-Globe, GmbH Grassau, Germany) or a 22-gauge FNB needle (SonoTip, TopGain; Medi-Globe GmbH). A computer-generated stratified block randomization method with 2 strata (size of the lesion ≤3 cm and presumed transgastric or transduodenal position for biopsy) was constructed by an author not participating in patient inclusion, randomization, or any following analyses. The concealed allocation table was uploaded to a web-based research database (REDCap), which was used for data collection and randomization.[10,11] Sample evaluation and quantitative real-time polymerase chain reaction (qRT-PCR) runs were performed in a blinded manner.
Study outcomes and sample size
Primary outcome was the amount of tissue (median number of tissue microcores and diagnostic and total tissue areas in millimeters squared). Secondary outcomes included diagnostic performance (diagnostic accuracy, sensitivity, and specificity), adverse event (AE) rate, and suitability for molecular analyses (mean DNA concentration and successful mutational analysis). Regarding diagnostic performance, final diagnosis was established by either subsequent histopathological confirmation or other evidence (clinical, radiological) of malignant disease at a minimum of 12 months of follow-up. Adverse events were defined according to the American Society of Gastrointestinal Endoscopy lexicon.[12] Sample size calculation was based on the expected difference in the mean number of tissue microfragments of 1.5, with an SD of 1.6 and 90% power.[13] This yielded 48 patients in total, but because of assumption of non-Gaussian distribution of the data, 15% was added according to Lehmann and D'Abrera.[14] This, together with an expected dropout rate of 10%, yielded a total of 64 patients to ensure even group distribution.
Sampling technique
All procedures were performed in an outpatient setting by experienced endosonographers using a linear array echoendoscope (EG-3870 UTK; Pentax [Tokyo, Japan]; HI Vision Preirus, Hitachi Medical Systems [Tokyo, Japan]). The procedures were performed in propofol sedation, administered by either an anesthesiologist or a specially trained nurse. Following initial EUS examination and randomization, EUS-guided tissue acquisition was performed in a standardized manner in all 3 centers. After the target lesion was accessed, samples were obtained using fanning technique and stylet slow-pull method. Ideally, 3 passes were performed, but the final choice was at the discretion of the endosonographer. Rapid on-site evaluation was not utilized because it is not standard of care in Denmark.
Sample preparation and evaluation
Samples were collected each in a separate vial following reinsertion of the needle stylet. Any remaining tissue was recovered by flushing the channel with an air-filled syringe. Samples were immediately fixated in formalin and subsequently embedded in paraffin. Remaining fluid was centrifuged and processed as cell blocks. Two dedicated expert cytopathologists (A.T. and S.D.) were responsible for the diagnostic evaluation of the specimens. Histopathological measurements were performed on digital slides by the same 2 pathologists who were blinded to the allocation group. Area was calculated using a computer software (NDP View; Hamamatsu Photonics K.K., Hamamatsu, Japan) through manual marking of the corresponding tissue fragments on digital slides [Figure 2], either as diagnostic (containing adequate tissue to evaluate or define the lesion) or nondiagnostic (providing no useful diagnostic information about the solid lesion sampled). Specimen quality was evaluated using the following criteria: insufficient, limited cytology, adequate cytology, low-quality histology (microcores <550 μm), good-quality histology (1–5 microcores, each >550 μm), high-quality histology (6–10 microcores, each >550 μm), and excellent-quality histology (>10 microcores or total tissue length >5550 μm). Cellularity was classified as fair (<100 cells), good (100–1000 cells), and excellent (>1000 cells), whereas blood and gastrointestinal (GI) tract contamination were classified as absent, minimal (<25%), moderate (25%–50%), and significant (>50%) for each pass.
Figure 2.
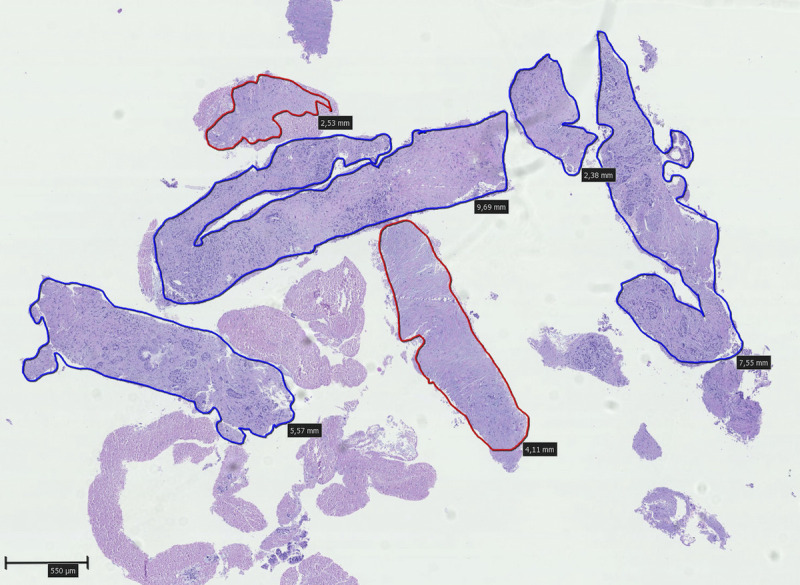
Digital pathology slide from one of the samples. Diagnostic histocores with a diameter of at least 550 μm are marked with blue color, whereas the remaining nondiagnostic histocores with a diameter of at least 550 μm are marked red (hematoxylin-eosin stain, original magnification ×2.5).
Quantitative real-time polymerase chain reaction
Samples were subjected to KRAS mutational analysis with qRT-PCR. Full description of genetic analyses is available in the Supplementary Material, http://links.lww.com/ENUS/A325. Briefly, DNA was extracted from predetermined amount of formalin-fixed paraffin-embedded tissue slides using QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). Subsequently, mutational status was assessed using the KRAS mutation analysis kit (Entrogen, Woodland Hills, California), which relies on amplification of mutated DNA using allele-specific primers and fluorescent hydrolysis probes and allows detection of 12 mutations in codons 12, 13, and 61 of the KRAS gene, which are the codons most often found mutated in pancreatic ductal adenocarcinoma. Based on detection of an endogenous gene marked with a fluorescent probe and acting as an internal control, the PCR reaction was described as successful (sufficient DNA loading), partially successful (insufficient DNA loading, but detectable mutation), or failed.
Statistical analyses
Continuous data were summarized by means with SD or medians with interquartile range, when appropriate. Area comparisons were performed using Student t test, whereas nonparametric Mann-Whitney U test was used for comparison of the median numbers of tissue microcores. Similarly, tissue quality, cellularity, and blood and GI tract contamination were compared using Mann-Whitney U test. Benjamini-Hochberg correction for multiple testing was utilized. Statistical significance for corrected P values was set at .05, and R version 4.2.0 (R studio build 492) was used for all statistical calculations. All authors had access to the study data and reviewed and approved the final manuscript.
RESULTS
Patient inclusion was conducted between June 2020 and December 2021. After initial screening of 84 patients, 64 were included in the study and constituted the study cohort [Figure 3]. Causes of exclusion were as follows: no visible lesion in the pancreas (n = 10), patient declined participation (n = 4), cystic pancreatic lesions (n = 3), and procedure aborted because of insufficient sedation (n = 3). No patients were lost to follow-up.
Figure 3.
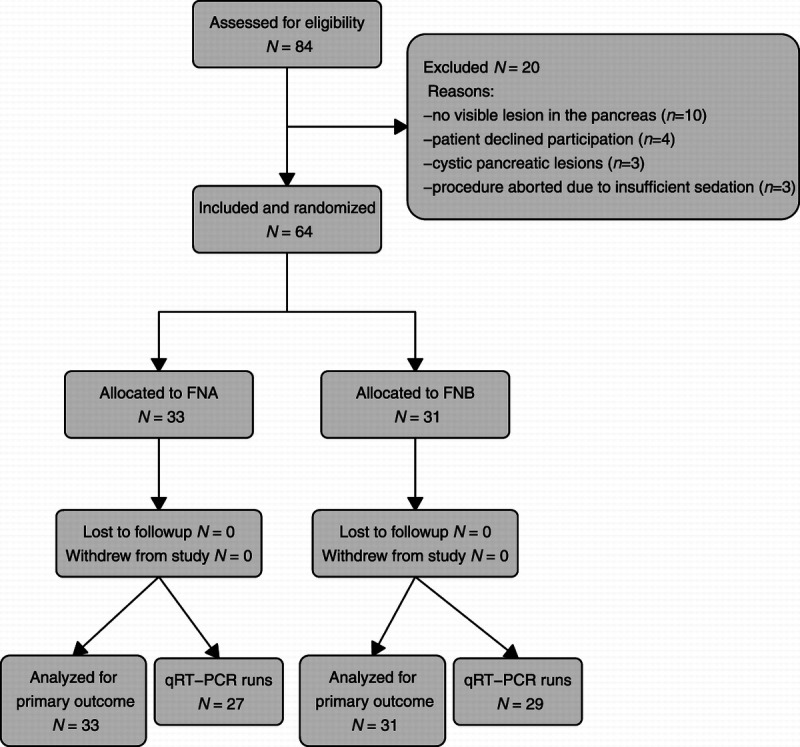
CONSORT flow diagram. CONSORT, Consolidated Standards of Reporting Trials; qRT-PCR, quantitative real-time polymerase chain reaction.
Patient characteristics
Mean age of the study cohort was 69 years, and 53.1% were male [Table 1]. Observed lesions measured in average 29.6 mm and were in most cases located in the head of the pancreas (42.2%). Final diagnosis consisted of pancreatic adenocarcinoma in 45 cases (70.3%), pancreatic neuroendocrine neoplasm (n = 8 [12.5%]), metastasis from other tumor (n = 5 [7.8%]), and chronic pancreatitis/benign tissue in the remaining 6 patients (9.4%).
Table 1.
Demographic and lesion data.
| FNA (n = 33) | FNB (n = 31) | Overall (n = 64) | |
|---|---|---|---|
| Age, year | |||
| Mean (SD) | 68.8 (9.89) | 69.3 (11.0) | 69.0 (10.4) |
| Median [min, max] | 68.0 [52.0, 88.0] | 74.0 [40.0, 89.0] | 68.5 [40.0, 89.0] |
| Gender, n (%) | |||
| Male | 20 (60.6) | 14 (45.2) | 34 (53.1) |
| Female | 13 (39.4) | 17 (54.8) | 30 (46.9) |
| Body mass index, kg/m2 | |||
| Mean (SD) | 24.6 (3.99) | 24.3 (4.00) | 24.4 (3.97) |
| Median [min, max] | 25.3 [17.7, 31.2] | 23.9 [15.8, 33.3] | 24.2 [15.8, 33.3] |
| ASA score, n (%) | |||
| I | 10 (30.3) | 8 (25.8) | 18 (28.1) |
| II | 15 (45.5) | 17 (54.8) | 32 (50.0) |
| III | 8 (24.2) | 6 (19.4) | 14 (21.9) |
| Location of the lesion, n (%) | |||
| Head | 15 (45.5) | 12 (38.7) | 27 (42.2) |
| Neck | 4 (12.1) | 2 (6.5) | 6 (9.4) |
| Body | 6 (18.2) | 9 (29.0) | 15 (23.4) |
| Tail | 3 (9.1) | 3 (9.7) | 6 (9.4) |
| Uncinate | 4 (12.1) | 5 (16.1) | 9 (14.1) |
| Diffuse involvement | 1 (3.0) | 0 (0) | 1 (1.6) |
| Size of the lesion, mm | |||
| Mean (SD) | 29.7 (13.1) | 29.5 (14.2) | 29.6 (13.5) |
| Median [min, max] | 30.0 [5.00, 60.0] | 30.0 [8.00, 72.0] | 30.0 [5.00, 72.0] |
| Final diagnosis, n (%) | |||
| Adenocarcinoma | 25 (75.8) | 20 (64.5) | 45 (70.3) |
| Neuroendocrine neoplasm | 3 (9.1) | 5 (16.1) | 8 (12.5) |
| Chronic pancreatitis | 1 (3.0) | 1 (3.2) | 2 (3.1) |
| Metastasis from other tumor | 1 (3.0) | 4 (12.9) | 5 (7.8) |
| Normal pancreatic tissue | 2 (6.1) | 1 (3.2) | 3 (4.7) |
| Adenoma | 1 (3.0) | 0 (0) | 1 (1.6) |
ASA, American Society of Anesthesiologists; FNA, fine-needle aspiration; FNB, fine-needle biopsy.
Tissue amount
All procedures were performed with the allocated needle for all 3 passes; no technical difficulties were observed. The median number of tissue microcores was significantly higher in the FNB group (3 vs. 2, P < 0.001). This difference was observed in all 3 passes, as well as in the pooled results [Table 2]. Similarly, the mean total tissue area, as well as the diagnostic tissue area, was larger in the FNB group compared with FNA (mean total tissue area, 2.74 vs. 0.44 mm2 [P < 0.001]; mean diagnostic tissue area, 1.74 vs. 0.28 mm2 [P < 0.001]). The observed effects persisted in all 3 passes [Table 2].
Table 2.
Amount of procured tissue and number of microcores in the 2 groups.
| FNA | FNB | Overall | P | |
|---|---|---|---|---|
| Median no. of microcores (IQR) | ||||
| Pass 1 | 0 (0–1) | 4 (1.2–10.2) | 1 (0–4) | |
| Pass 2 | 0.5 (0–1) | 2 (1–9.8) | 1 (0–4) | |
| Pass 3 | 1 (0–2) | 3 (1–6.5) | 1.5 (0.2–4) | |
| Overall | 2 (1–2.5) | 3 (2.1–7) | 2.2 (1.5–3.5) | <.001 |
| Total tissue area, mean (SD), mm2 | ||||
| Pass 1 | 0.6 (1.4) | 3 (3.3) | 1.7 (2.7) | |
| Pass 2 | 0.3 (0.5) | 2.5 (2.7) | 1.4 (2.2) | |
| Pass 3 | 0.5 (0.7) | 2.5 (2.8) | 1.5 (2.3) | |
| Overall | 0.4 (0.6) | 2.7 (2.4) | 1.6 (2.1) | <.001 |
| Diagnostic tissue area, mean (SD), mm2 | ||||
| Pass 1 | 0.4 (0.9) | 1.6 (2.1) | 0.9 (1.7) | |
| Pass 2 | 0.2 (0.4) | 2 (2.4) | 1.1 (1.9) | |
| Pass 3 | 0.4 (0.7) | 1.7 (2.3) | 1 (1.8) | |
| Overall | 0.3 (0.5) | 1.7 (1.6) | 1 (1.4) | <.001 |
FNA, fine-needle aspiration; FNB, fine-needle biopsy; IQR, interquartile range.
Diagnostic performance and sample quality
Overall accuracy for malignancy/neoplasia was 69.7% (95% confidence interval, 51.3%–84.4%) in the FNA group and 90.3% (95% confidence interval, 74.2%–98%) in the FNB group. Sensitivity for malignancy/neoplasia showed similar trends and was 65.5% in the FNA group and 89.7% in the FNB group. Hence, the number of false-negative results was 3 in the FNB group and 10 in the FNA group. As there were no false-positive results, observed specificity was 100% in both groups. The proportion of diagnostic samples was higher in the FNB group in the first (61.3% vs. 24.2%, P = 0.016) and second passes (66.7% vs. 29%, P = 0.016), but not in the third pass (70.4% vs. 44.4%, P = 0.154). Similarly, the median number of passes required to achieve diagnosis was 2 in the FNA compared with 1 in the FNB group (P = 0.120).
The FNB needle provided a higher proportion of samples with histology grade tissue with a higher median tissue quality score (5: good-quality histology vs. 4: low-quality histology P = 0.002). There were 2 samples insufficient for diagnosis in each group; whereas FNA samples were mostly cytological or with low-quality histology grade tissue, FNB provided an overweight of good- and/or excellent-quality histology grade tissue [Figure 4]. Furthermore, median cellularity score was higher in FNB samples where samples of excellent cellularity were observed more often compared with FNA samples (P = 0.017). There was no difference in the amount of blood contamination (P = 0.092) and GI tract contamination (P = 0.430).
Figure 4.
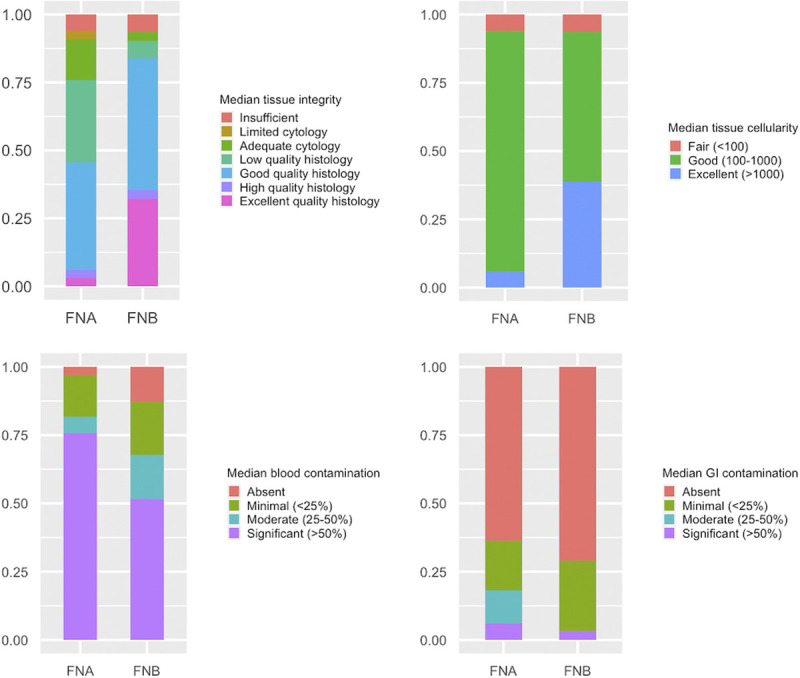
Sample quality and contamination in the FNA and FNB groups. FNA, fine-needle aspiration; FNB, fine-needle biopsy.
Suitability for qRT-PCR
After excluding samples with no apparent pancreatic tissue, qRT-PCR was performed in a total of 56 samples (FNA: n = 27, FNB: n = 29). The analysis was successful in 25 samples (44.6%), partially successful in 7 cases (12.5%), and failed in remaining 24 cases (42.9%). Successful or partially successful qRT-PCT runs were seen in 15 of 27 FNA samples (55.6%) and 17 of 29 FNB samples (58.6%). There was no association between successful qRT-PCR runs and the type of needle (P = 0.670), nor did the mean DNA concentration differ between the 2 groups (FNA: 0.73 ng/μL, FNB: 1.11 ng/μL; P = 0.166) (Supplementary Table S1, http://links.lww.com/ENUS/A325). For the samples with successful or partially successful (n = 32) runs, the majority of identified mutations was in codon 12 (90.6%, n = 29). Two patients harbored a mutation in codon 61 (Gln61His), and 1 sample was KRAS wild type.
Adverse events
Four AEs were observed, all of which were mild. Three of those occurred in the FNA group: 1 case of mild hemorrhage in the lesion, 1 case of postprocedural pain and vomiting, and 1 case of fever that was treated with antibiotics. The last AE was in the FNB group and was due to respiratory complications during the sedation, necessitating subsequent hospitalization. All 4 patients were discharged within 24 hours after the procedure.
DISCUSSION
This study was the first to prospectively evaluate a novel FNB needle with modified Franseen design (SonoTip, TopGain; Medi-Globe GmbH). Our results show that this needle on average obtained twice the number of tissue microcores, yielding 6 times as much tissue as the standard FNA needle well in line with currently published literature.
In an RCT by Bang et al[15] comparing fork-tip and Franseen needles, the mean diagnostic (tumor) tissue area was almost identical to what has been observed in the present study. Similar results have been reported in another smaller RCT comparing the original Franseen design with an FNA needle.[16] Our study also demonstrated an overall high accuracy and sensitivity for malignancy of FNB of approximately 90%. Although the study does not have sufficient power to show a difference in diagnostic performance, the results seem consistent with earlier studies showing superiority of crown-cut needles in EUS-guided biopsy of solid pancreatic lesions. The question remains whether there is a place for standard FNA needles in the current era of precision and personal medicine. Market competition has led to a lowered price of next-generation needles in the last couple of years, leading to a current price difference of some 20 euros. This fact might provide FNB needles with an additional advantage over FNA because they obliterate the need for rapid on-site evaluation. The design of this FNB needle is very similar to the original Franseen needle; why any differences in performance between these 2 needles are probably inconsequential. Therefore, any head-to-head comparison would demand a large number of participants to show any difference.
Strengths of this study are the sufficient power for comparison of the primary outcomes, a multicenter parallel design, randomization, and blinding of the expert pathologists evaluating the samples. More often comparative studies of EUS needles implement crossover design rather than parallel groups. Whereas a crossover design has several advantages, herein the lower-sample-size, parallel design offers more reliable estimates of variables on a group level. Prior needle insertion might well affect not only variables such as sample bloodiness, but also cellularity and the number of microcores, which is why a parallel design was chosen in this study. Furthermore, we chose needles of the same size and predefined the amount of needle passes and sampling technique to minimize possible bias. The 2 needles were compared on several different parameters not limited to diagnostic yield. We examined in detail the amount of blood and GI tract contamination, as well as cellularity and tissue area. Nonetheless, our study has some limitations. The study was not powered to detect differences in the amount of DNA procured, and mutational analyses were limited to qRT-PCR of KRAS alone rather than targeted next-generation sequencing of several genes. Whereas next-generation sequencing is not limited to several known mutational spots, commercial sequencing kits and analyses are more sensitive and may be feasible in the actual clinical situation. However, parallel reactions combined with result validation demand a larger amount of tumor tissue and may explain a relatively high amount of failed/inconclusive qRT-PCR runs in this study. Furthermore, sole use of KRAS gene for support of pathological diagnosis may be inappropriate, as some nonmalignant lesions such as pancreatic intraepithelial neoplasia are also known to harbor mutations in the KRAS gene. Therefore, in routine clinical setting, a gene panel is usually selected. In a meta-analysis by Fuccio et al[17] examining the performance of EUS-FNA and KRAS mutational status, a false-positive rate of up to 10% was observed. Therefore, other oncogenes should be included in a mutational panel when using ancillary genetic analyses. Our study did not show any significant difference in the number of samples suitable for qRT-PCR or in the mean DNA concentration and was probably underpowered to answer these questions. Lastly, this study focused solely on solid pancreatic lesions, and our results cannot be extrapolated to other types of lesions of the GI tract.
CONCLUSIONS
In conclusion, This study demonstrated a clear superiority of the novel modified Franseen-type FNB needle in regard to the amount of tissue procured and sample quality in pancreatic solid lesions suggestive of malignancy compared with the standard FNA needle. Both needles seem equally suitable for ancillary molecular analyses, in this case qRT-PCR of the KRAS gene.
Funding
Medi-Globe GmbH has provided needles and genetic analyses used in this study free of charge. The company did not have any influence on the design and execution of the study, interpretation of the data, or the decision to publish.
Conflicts of Interest
P.V. is a consultant to Medi-Globe GmbH. The other authors declare that they have no financial conflict of interest with regard to the content of this report.
Clinical Registry Name and Registration Number
Author Contributions
Bojan Kovacevic: conceptualization of the project, sample size calculation, protocol draft and finalization, institutional review board approvals, randomization table, interpretation of the results, statistical calculations, manuscript draft and finalization, and lead author. Anders Toxværd: conceptualization of the project, critical review of the protocol, histopathological evaluation of the samples, interpretation of the results, and manuscript draft and finalization. Pia Klausen: conceptualization of the project, sample size calculation, protocol draft and finalization, institutional review board approvals, and critical review of the manuscript and finalization. Michael H. Larsen: critical review of the protocol, patient inclusion and randomization, collection of demographical and procedure-related data, performed EUS-FNA/FNB procedures, and critical review of the manuscript and finalization. Simon Grützmeier: critical review of the protocol, patient inclusion and randomization, collection of demographical and procedure-related data, statistical calculations, and critical review of the manuscript and finalization. Sönke Detlefsen: conceptualization of the project, critical review of the protocol, histopathological evaluation of the samples, interpretation of the results, and manuscript draft and finalization. John Gásdal Karstensen: critical review of the protocol, patient inclusion and randomization, collection of demographical and procedure-related data, performed EUS-FNA/FNB procedures, and critical review of the manuscript and finalization. Lene Brink: critical review of the protocol, patient inclusion, collection of demographical and procedure-related data, performed EUS-FNA/FNB, and critical review of the manuscript and finalization. Hazem Hassan: critical review of the protocol, patient inclusion, collection of demographical and procedure-related data, performed EUS-FNA/FNB, and critical review of the manuscript and finalization. Estrid Høgdall: conceptualization of the project, critical review of the protocol, performed and interpreted DNA sequencing analyses, interpretation of the results, and critical review of the manuscript and finalization. Peter Vilmann: conceptualization of the project, critical review of the protocol, patient inclusion, collection of demographical and procedure-related data, performed EUS-FNA/FNB procedures, critical review of the manuscript and finalization, and senior author. All authors read and approved the final manuscript.
Footnotes
Supplemental digital content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal's Web site (www.eusjournal.com).
Published online: 25 July 2023
Contributor Information
Anders Toxværd, Email: anders.toxvaerd.01@regionh.dk.
Pia Klausen, Email: pia.helene.klausen@regionh.dk.
Michael H. Larsen, Email: Michael.Hareskov.Larsen@rsyd.dk.
Simon Grützmeier, Email: simon.ezban.gruetzmeier@regionh.dk.
Sönke Detlefsen, Email: Sonke.Detlefsen@rsyd.dk.
John Gásdal Karstensen, Email: john.gasdal.karstensen@regionh.dk.
Lene Brink, Email: lene.brink@regionh.dk.
Hazem Hassan, Email: hazem.hassan.al-hashemi@regionh.dk.
Estrid Høgdall, Email: Estrid.Hoegdall@regionh.dk.
Peter Vilmann, Email: peter.vilmann@regionh.dk.
References
- 1.Vilmann P, Hancke S. A new biopsy handle instrument for endoscopic ultrasound–guided fine-needle aspiration biopsy. Gastrointest Endosc 1996;43:238–242. [DOI] [PubMed] [Google Scholar]
- 2.Levy MJ Jondal ML Clain J, et al. Preliminary experience with an EUS-guided Trucut biopsy needle compared with EUS-guided FNA. Gastrointest Endosc 2003;57:101–106. [DOI] [PubMed] [Google Scholar]
- 3.Kovacevic B, Vilmann P. EUS tissue acquisition: from A to B. Endosc Ultrasound 2020;9:225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng B Zhang Y Chen Q, et al. Analysis of fine-needle biopsy vs fine-needle aspiration in diagnosis of pancreatic and abdominal masses: a prospective, multicenter, randomized controlled trial. Clin Gastroenterol Hepatol 2018;16:1314–221. [DOI] [PubMed] [Google Scholar]
- 5.van Riet PA Larghi A Attili F, et al. A multicenter randomized trial comparing a 25-gauge EUS fine-needle aspiration device with a 20-gauge EUS fine-needle biopsy device. Gastrointest Endosc 2019;89:329–339. [DOI] [PubMed] [Google Scholar]
- 6.van Riet PA Erler NS Bruno MJ, et al. Comparison of fine-needle aspiration and fine-needle biopsy devices for endoscopic ultrasound–guided sampling of solid lesions: a systemic review and meta-analysis. Endoscopy 2021;53:411–223. [DOI] [PubMed] [Google Scholar]
- 7.Sugimoto M Irie H Takagi T, et al. Efficacy of EUS-guided FNB using a Franseen needle for tissue acquisition and microsatellite instability evaluation in unresectable pancreatic lesions. BMC Cancer 2020;20:1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itonaga M Yasukawa S Fukutake N, et al. Comparison of 22-gauge standard and Franseen needles in EUS-guided tissue acquisition for diagnosing solid pancreatic lesions: a multicenter randomized controlled trial. Gastrointest Endosc 2022;96:57–66.e2. [DOI] [PubMed] [Google Scholar]
- 9.Kuraoka N Hashimoto S Matsui S, et al. Effectiveness of EUS-guided fine-needle biopsy versus EUS-guided fine-needle aspiration: a retrospective analysis. Diagnostics (Basel) 2021;11:965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris PA Taylor R Thielke R, et al. Research electronic data capture (REDCap) – A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris PA Taylor R Minor BL, et al. REDCap Consortium, The REDCap consortium: building an international community of software partners. J Biomed Inform 2019. doi: 10.1016/j.jbi.2019.103208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotton PB Eisen GM Aabakken L, et al. A lexicon for endoscopic adverse events: report of an ASGE workshop. Gastrointest Endosc 2010;71:446–454. [DOI] [PubMed] [Google Scholar]
- 13.Alatawi A Beuvon F Grabar S, et al. Comparison of 22G reverse-beveled versus standard needle for endoscopic ultrasound–guided sampling of solid pancreatic lesions. United Eur Gastroenterol J 2015;3:343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehmann E, D'Abrera H. Nonparametrics: statistical methods based on ranks. 1975
- 15.Bang JY Hebert-Magee S Navaneethan U, et al. Randomized trial comparing the Franseen and fork-tip needles for EUS-guided fine-needle biopsy sampling of solid pancreatic mass lesions. Gastrointest Endosc 2018;87:1432–1438. [DOI] [PubMed] [Google Scholar]
- 16.Asokkumar R Yung Ka C Loh T, et al. Comparison of tissue and molecular yield between fine-needle biopsy (FNB) and fine-needle aspiration (FNA): a randomized study. Endosc Int Open 2019;7:E955–E963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuccio L Hassan C Laterza L, et al. The role of K-ras gene mutation analysis in EUS-guided FNA cytology specimens for the differential diagnosis of pancreatic solid masses: a meta-analysis of prospective studies. Gastrointest Endosc 2013;78:596–608. [DOI] [PubMed] [Google Scholar]


