Abstract
The lymphoid cell-specific proteins RAG1 and RAG2 initiate V(D)J recombination by cleaving DNA adjacent to recombination signals, generating blunt signal ends and covalently sealed, hairpin coding ends. A critical next step in the reaction is opening of the hairpins, but the factor(s) responsible has not been identified and had been thought to be a ubiquitous component(s) of the DNA repair machinery. Here we demonstrate that RAG1 and RAG2 possess an intrinsic single-stranded nuclease activity capable of nicking hairpin coding ends at or near the hairpin tip. In Mn2+, a synthetic hairpin is nicked 5 nucleotides (nt) 5′ of the hairpin tip, with more distant sites of nicking suppressed by HMG2. In Mg2+, hairpins generated by V(D)J cleavage are nicked whereas synthetic hairpins are not. Cleavage-generated hairpins are nicked at the tip and predominantly 1 to 2 nt 5′ of the tip. RAG1 and RAG2 may therefore be responsible for initiating the processing of coding ends and for the generation of P nucleotides during V(D)J recombination.
V(D)J recombination assembles the variable regions of antigen receptor genes during lymphocyte development by joining together V (variable), J (joining), and in some cases D (diversity), coding gene segments (28). Recombination is specifically directed to coding elements by recombination signal sequences (RSSs) which flank the segments to be joined. These RSSs consist of a conserved heptamer which is contiguous to the coding flank and an AT-rich nonamer. Heptamer and nonamer are separated by a nonconserved spacer of either 12 or 23 bp, yielding the 12-RSS or 23-RSS, respectively. In vivo, recombination primarily occurs between coding elements with RSS spacers of different lengths, thereby preventing the joining of inappropriate elements. This restriction is referred to as the 12/23 rule.
Mechanistically, the recombination reaction is envisioned to occur in two stages. In the first stage, initiation of recombination is mediated by the proteins encoded by the recombination-activating genes, RAG1 and RAG2, which bind directly to RSSs (10, 32, 47, 51). Binding by RAG proteins is followed by RSS synapsis and concerted cleavage at both signals. Cleavage involves hydrolytic nicking at the heptamer-coding flank border, and the the 3′-hydroxyl thus generated serves as a nucleophile to attack the phosphodiester bond on the other DNA strand opposite the nick in a direct transesterification reaction (32, 52). The coding ends generated by cleavage are covalently sealed DNA hairpins, while signal ends are blunt and 5′ phosphorylated. This reaction is stimulated in vitro, especially at the 23-RSS, by addition of DNA-bending proteins HMG1 and HMG2 (44, 50). Coordinate cleavage in accordance with the 12/23 rule requires Mg2+ rather than Mn2+ as divalent metal cofactor and is stimulated by HMG1 or HMG2 (12, 43, 44, 50, 53). In the second stage of the reaction, coding ends are processed, and coding joints and signal joints form in reactions with similarities to the repair of DNA double-strand breaks by nonhomologous end joining (NHEJ) (9).
A hallmark of V(D)J recombination in many species is generation of receptor diversity required for specific immune recognition. Many coding elements can be joined in different combinations, and joining is imprecise by virtue of nucleotide deletions and insertions. The source of nucleotide deletions is unknown, but two mechanisms are known to contribute to insertions. Terminal deoxynucleotidyltransferase, a lymphoid cell-specific polymerase, adds untemplated nucleotides to DNA 3′ termini (16, 25). Another source of nucleotide insertions is asymmetric nicking of coding-end hairpins to generate palindromic extensions termed P nucleotides (26, 30, 33).
The opening of covalently sealed coding ends is a prerequisite for coding-joint formation, and evidence exists for a ubiquitous hairpin opening activity (4, 29, 48). Mice deficient in general factors required for repair of DNA double-strand breaks by NHEJ, including XRCC4, DNA ligase IV, Ku70, Ku80, and the DNA-dependent protein kinase (DNAPK), are deficient in coding-joint formation and lymphocyte development (13–15, 17, 18, 41, 49, 55). Deficiencies in Ku80 and DNAPK also result in the accumulation of hairpin coding ends (15, 41, 55), indicating that Ku and DNAPK may regulate the hairpin opening reaction. Together, these data have supported the hypothesis that hairpin opening in V(D)J recombination is probably performed by a ubiquitous factor (29). Recent evidence, however, indicates that the RAG proteins have a postcleavage role in coding-joint formation (27, 39) and that they remain associated with coding ends after cleavage (20). In an effort to understand their role in coupling the two stages of V(D)J recombination, we examined the alternative possibility that coding-end hairpins are opened by RAG1 and RAG2.
While this report was in preparation, Besmer et al. reported concurrent work demonstrating DNA hairpin opening by RAG proteins (5). Our results are consistent with their basic findings and lead to a number of novel conclusions concerning the hairpin opening reaction.
MATERIALS AND METHODS
Synthetic hairpin nicking reactions.
RAG and HMG2 proteins have been described previously (1, 47). RAG proteins (expressed individually or coexpressed) contained either a C-terminal polyhistidine and Myc antibody epitope (MH) tag (MH-RAGs) or an N-terminal glutathione S-transferase (GST) tag (GST-RAGs, GST-RAG1, and GST-RAG2). MH-RAGs (30 ng of each; 0.3 to 0.6 pmol) or GST-RAGs (30 ng of each; 0.3 to 0.4 pmol) and, where appropriate, HMG2 (75 ng; 2.5 pmol) were incubated with 25 to 50 fmol of 32P-end-labeled, annealed, synthetic hairpins at 30°C for 3 h (unless otherwise indicated). Reaction mixtures (20 μl) contained 20 mM HEPES (pH 7.5), 16 mM sodium acetate (pH 7.0), 34 mM NaCl, 10 μM ZnSO4, 2 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 15% glycerol, and 1 mM MnCl2 (unless otherwise indicated) and were terminated essentially as described previously (1). DNA samples were resuspended in loading buffer containing 80% formamide, 10 mM NaOH, and 1 mM EDTA. After heating for 2 min at 95°C, DNA was analyzed at 40 to 45°C by denaturing (8% polyacrylamide, 7 M urea, and 40% formamide) polyacrylamide gel electrophoresis (PAGE).
Oligonucleotides used in synthetic hairpin nicking reactions.
The 126-nucleotide (nt) hairpin oligonucleotide was HPBSAI (5′-GCGAGCGTCGGTCTCG CCAATCGAGCCATGTCGTCGTCGATCCTCTCATCGATGAGAGGATC CGGATCCTCTCATCGATGAGAGGATCGACGACGACATGGCTCGATT GGCGAGACCGACGCTCGC). This is identical to the 23 coding-end hairpin of pJH299 except that 14 bp (including a BsaI site) have been added at a position 51 bp from the hairpin tip. Hairpin oligonucleotides were denatured at 95°C and quick cooled on ice to favor intramolecular hairpin annealing rather than intermolecular association. Linear DNA duplex substrate was made by annealing the oligonucleotides HPBSAI-T (5′-GCGAGCGTCGGTCTCGCCAATCGAGCCATGTCGTCGTCGATCCTCTCATCGATGAGAGGATCC) and HPBSAI-B (5′-GGATCCTCTCATCGATGAGAGGATCGACGACGACATGGCTCGATTGGCGAGACCGACGCTCGC).
Detection of coding ends after 12/23-regulated cleavage.
Reactions were performed essentially as described above, with 10 mM MgCl2 in place of 1 mM MnCl2. Reaction mixtures (40 μl) contained 100 ng of DNA substrate and 60 ng each of MH-RAG1 and MH-RAG2 (and 150 ng of HMG2 where appropriate) and were incubated at 30°C for 3 h. The DNA substrate was pJH299 (19) or pJ2VIS6 (45), which contains the endogenous Vκ21C and Jκ1 gene segments (and their flanking RSSs) separated by 670 bp in inversional orientation. Reactions were terminated by addition of 0.06% sodium dodecyl sulfate (SDS), 1.1 mM EDTA, and proteinase K (180 ng/μl) and incubation at 55°C for 30 min. After organic extraction and ethanol precipitation, DNA was digested with HinfI for analysis of the 23 coding end of pJH299 (or MseI and SalI for analysis of the 12 and 23 coding ends, respectively, of pJ2VIS6) and analyzed as described above by denaturing PAGE. After electroblotting to Gene Screen Plus and base (0.4 N NaOH, 10 min) treatment of the membrane, blots were hybridized at 37°C for 18 h in a mixture containing 4× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 10× Denhardt’s solution, 0.1% SDS, fish sperm DNA (100 μg/ml), yeast RNA (250 μg/ml), and 5′-end-labeled lower-strand-specific oligonucleotide probe PSHP1 (5′-TCGCAGCAACTTGTCGCGCCAATCGAGCCA; 7 × 106 cpm/ml) for pJH299. Blots were washed at 37°C in 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.5% SDS and at 55°C in 2× SSC–0.5% SDS.
Detection of coding ends by LMPCR.
Anchor oligonucleotides used in ligation-mediated PCR (LMPCR) (DR19 and DR20), described previously (42), contained a terminal EcoRI site for cloning. Forward primers, all of which contained a terminal XbaI site for cloning, were CBLMP1-XB (5′-GCCTCTAGACAAGAACAGCAAGCAGCATTGAG; specific for sequences upstream of the 23 coding flank of pJH299, generating a 249-bp PCR product with a full-length coding end), J2LM-XB12 (5′-GCCTCTAGACGAAGATTGGCTGTGTCTCTAGG; 12 coding flank of pJ2V1S6; 309-bp product with a full-length coding end), and J2LM-XB23 (5′-GCCTCTAGAGCAAGATTCCGAATACCGCAAGC; 23 coding flank of J2V1S6; 294-bp product with a full-length coding end). Coding flank refers to coding-end sequences immediately adjacent to the RSS heptamer. Five percent of the DNA recovered from cleavage reactions was treated for 30 min at 37°C (or mock treated [−T4 reactions]) in 20-μl mixtures containing 0.75 U of T4 DNA polymerase (T4 DNAP; New England Biolabs), 0.2 mM deoxynucleoside triphosphates, 100 μg of bovine serum albumin per ml, and 1× T4 DNAP buffer. Reactions were terminated by heating for 10 min at 75°C. Ten percent of the DNA from T4 DNAP reactions was ligated to annealed anchor oligonucleotides (2 μM) in a 15-μl reaction mixture containing 1.5 U of T4 DNA ligase (Gibco/BRL) at 16°C for 18 h. PCR was performed by standard methods with annealing at 64°C for 23 to 26 cycles, and products were analyzed by native PAGE on 7% gels.
Cloning and sequencing of coding ends.
DNA from LMPCR reactions was phenol-chloroform extracted, precipitated, and double digested with EcoRI and XbaI. Products were gel purified, cloned into pBluescript II KS+, and sequenced on an ABI 373 automated DNA sequencer (Perkin-Elmer).
RESULTS
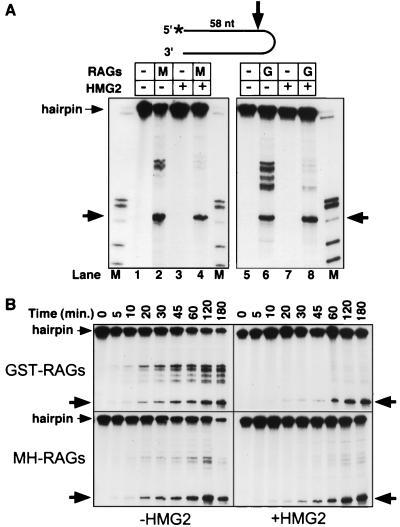
RAG proteins nick synthetic hairpins in Mn2+ but not Mg2+.
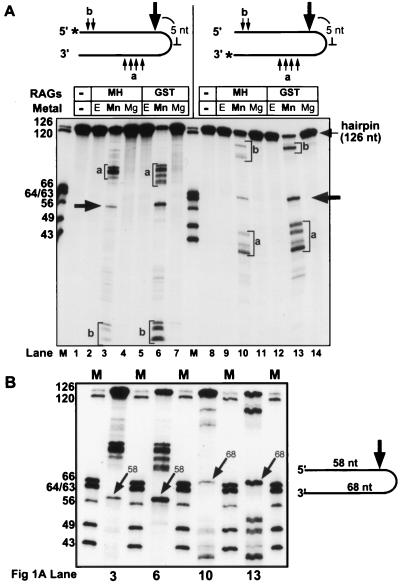
We first incubated the RAG proteins with a 126-nt synthetic hairpin DNA molecule and detected substrate cleavage by denaturing PAGE. With two different preparations of RAG proteins, cleavage was observed at multiple positions when reactions were performed in Mn2+, but not in Mg2+, EDTA (Fig. 1A, lanes 3, 6, 10, and 13), or Ca2+ (data not shown). Reciprocal-sized products were detected with substrates labeled at the 5′ and 3′ termini (compare lanes 3 and 6 with lanes 10 and 13), indicating that single-strand nicks, rather than double-strand breaks, were generated and that these fragments were the immediate products of nicking. One discrete product (Fig. 1A, large arrows) was shown to be the result of a nick 5 nt 5′ of the hairpin tip (generating 58- and 68-nt products with the 5′- and 3′-end-labeled substrates, respectively; higher-resolution mapping data is also shown (Fig. 1B). Other nicks occurred at a greater distance from the hairpin tip, with sites located predominantly in two clusters (Fig. 1A, small arrows labeled a and b).
FIG. 1.
Synthetic hairpin nicking mediated by the RAG proteins. (A) Synthetic hairpin nicking occurs in the presence of Mn2+ but not Mg2+. Annealed 5′- and 3′-end-labeled hairpin oligonucleotides were incubated with truncated, coexpressed MH- or GST-tagged RAG proteins. Sites of nicking distant from the hairpin tip (small arrows and lowercase letters) and 58-nt (5′ labeled) and 68-nt (3′ labeled) fragments corresponding to nicking at a position 5 nt 5′ of the hairpin tip (large arrows) are shown. Reactions included either 10 mM Mg2+ (Mg), 1 mM Mn2+ (Mn), or 10 mM EDTA (E). Positions of DNA molecular weight markers (M) are indicated in nucleotides. An asterisk is used to indicate the site of 32P end labeling. (B) High-resolution mapping of synthetic hairpin nicking products. Samples from lanes 3, 6, 10, and 13 of panel A were reanalyzed adjacent to marker fragments by denaturing PAGE. The 58- and 68-nt reciprocal products of hairpin nicking generated with 5′- and 3′-end-labeled substrates, respectively, are indicated with arrows.
Nicking of synthetic hairpins requires both RAG1 and RAG2 and is inhibited by anti-RAG1 antibody.
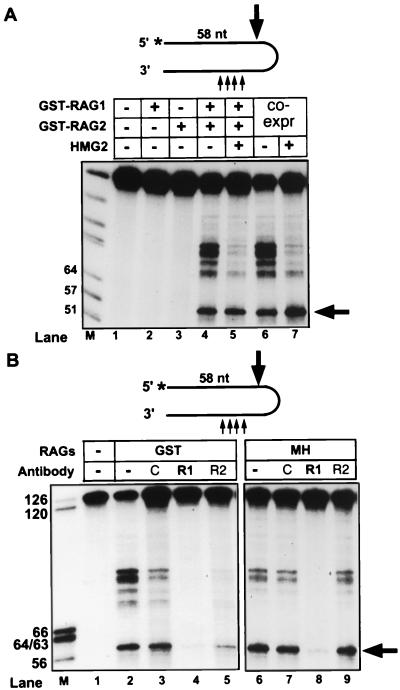
To confirm that the RAG proteins, and not a copurifying nuclease, were responsible for the nicking activity, we examined the activity of individually expressed and purified GST-RAG fusion proteins. No nicking could be detected with either protein alone, but the mixture of RAG1 and RAG2 displayed robust activity in Mn2+ (Fig. 2A). Identical results were obtained with a number of other highly purified, active preparations of recombinant RAG proteins, including RAG1 from bacteria and RAG2 purified from mammalian cells after infection with a vaccinia virus expression vector. In all cases, nicking was absolutely dependent on the presence of both RAG1 and RAG2 (data not shown). Furthermore, addition of anti-RAG1 antibodies to the reaction dramatically inhibited nicking activity, whereas anti-RAG2 antibodies did not have a reproducible effect (Fig. 2B). We conclude that the nicking activity is due to the action of RAG1 and RAG2 and cannot be attributed to contaminants in the preparations.
FIG. 2.
Nicking of synthetic hairpins requires both RAG1 and RAG2 and is inhibited by anti-RAG1 antibody. (A) 5′-end-labeled, annealed hairpin oligonucleotide was incubated with individually expressed (lanes 2 to 5) or coexpressed (co-expr) GST-RAG proteins, in the presence or absence of purified HMG2 (as indicated above the lanes). Reactions in both panels were performed in 1 mM Mn2+ and analyzed as for Fig. 1A; in each, the 58-nt fragment derived from nicking 5 nt 5′ of the hairpin tip is indicated with an arrow. (B) RAG proteins were preincubated for 25 min at 30°C with anti-RAG1 (R1) or anti-RAG2 (R2) antibodies or a rabbit immunoglobulin G specificity control (C) before addition of hairpin substrate. Antibodies specific for RAG1 (R1P8) and RAG2 have been described previously (2). Nicking reactions were carried out for 3 or 2 h with coexpressed GST- or MH-RAGs, respectively.
A hairpin terminus is not required for single-stranded nicking activity of the RAG proteins.
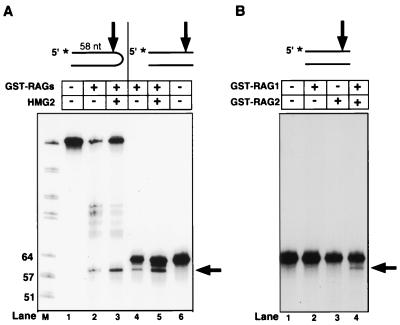
To investigate the role of the hairpin terminus in this reaction, we repeated the experiment with a linear duplex DNA substrate, identical in sequence to the hairpin substrate. A product that comigrated with that produced by nicking near the tip of the hairpin substrate was observed, indicating that the two substrates were nicked at identical positions 5 nt from the 3′ end, or tip, of the molecule (Fig. 3A). The linear duplex substrate was also nicked at a variety of sites on the bottom strand (data not shown), and all activity required the presence of both RAG1 and RAG2 (Fig. 3B). We conclude that a hairpin terminus is not required for the single-strand nuclease activity of the RAG proteins.
FIG. 3.
Single-stranded endonuclease activity of the RAG proteins is not specific for a hairpin terminus (A) and requires both RAG1 and RAG2 (B). Reactions were performed in 1 mM Mn2+ and analyzed as for Fig. 1A. The 58-nt fragment derived from nicking 5 nt 5′ of the hairpin tip and the corresponding fragment derived from nicking of the linear duplex DNA substrate are indicated with arrows. The 5′-end-labeled hairpin (lanes 1 to 3) and the corresponding linear duplex DNA substrates are indicated above the lanes.
HMG2 protein confines nicking to the vicinity of the hairpin tip.
Since the high-mobility group proteins HMG1 and HMG2 enhance DNA binding and cleavage by the RAG proteins (44, 50), we examined hairpin nicking in the presence of recombinant HMG2. With HMG2 added, nicking by the RAG proteins in Mn2+ was inhibited at all sites except that near the hairpin tip (Fig. 4A, lanes 4 and 8; Fig. 2A, lanes 5 and 7; Fig. 3, lane 3). To extend this observation and examine the kinetics of nicking, we performed time course experiments in the presence or absence of HMG2 (Fig. 4B). Nicked products could be observed by 10 min and accumulated for up to 3 h. HMG2 reduced the initial rate of nicking near the hairpin tip but did not decrease the final yield of this product, and it inhibited nicking at all other sites. HMG2 exhibited no nicking activity by itself on synthetic hairpins under any conditions tested, including those used in the coupled cleavage reactions described below (Fig. 4A, lanes 3 and 7, and data not shown). With the linear DNA duplex substrate, RAG-mediated nicking (as described above) was observed in the presence of HMG2 (Fig. 3A, lane 5). However, HMG2 did not consistently suppress nicking at other sites (data not shown). Therefore, while altered DNA structures at the DNA duplex end may be a preferential site for nicking, the effect of HMG2 in targeting the RAG proteins appears to be specific for the hairpin structure.
FIG. 4.
HMG2 inhibits nicking at sites distant from the hairpin tip. (A) Synthetic 5′-end-labeled hairpins were incubated with RAG proteins in the presence or absence of HMG2, or with HMG2 alone, as indicated above the lanes. Reactions were carried out for 3 or 2 h with coexpressed GST- or MH-RAGs, respectively. Reactions in both panels were performed in 1 mM Mn2+ and analyzed as for Fig. 1A; in each, the 58-nt fragment derived from nicking 5 nt 5′ of the hairpin tip is indicated with an arrow. (B) RAG proteins were preincubated with or without HMG2 for 15 min at 30°C before addition of hairpin substrate, and samples were analyzed at the indicated times.
RAG proteins mediate hairpin nicking in Mg2+ in the context of V(D)J cleavage.
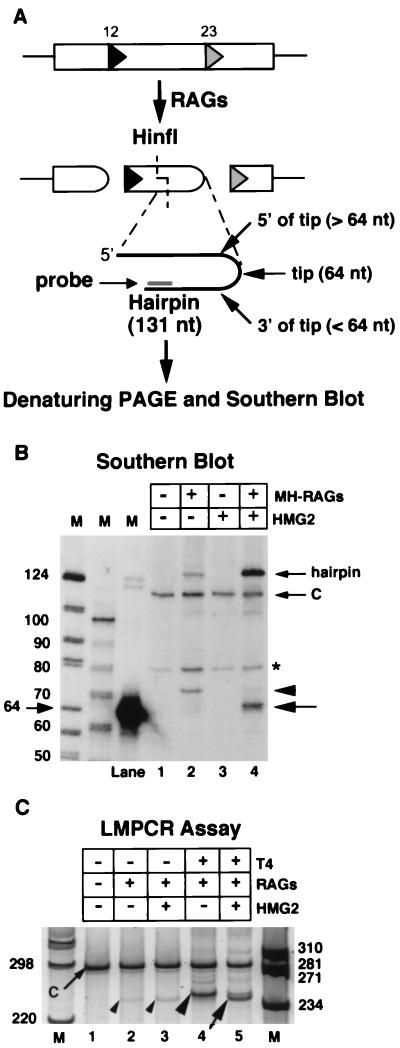
In Mg2+, synapsis of a 12-RSS and 23-RSS is required for efficient hairpin formation by the RAG proteins, whereas Mn2+ more efficiently supports disregulated cleavage at a single RSS (12, 32, 43, 53). By analogy, hairpin nicking in Mg2+ by the RAG proteins might require the formation of an appropriate postcleavage synaptic complex. To examine this possibility, cleavage reactions were performed in Mg2+ with a DNA substrate (pJH299) containing a 12-RSS and a 23-RSS, and the resulting hairpin coding ends (derived from cleavage at the 23-RSS) were analyzed for evidence of nicking by denaturing gel electrophoresis followed by Southern blotting with a strand-specific oligonucleotide probe (Fig. 5A; note that the synthetic hairpin substrate used in Fig. 1 to 4 has essentially the same nucleotide sequence as the 23-coding flank of this substrate [see Materials and Methods]). As expected, DNA cleavage, as measured by the production of the 131-nt hairpin product, was RAG dependent and stimulated by HMG2 (Fig. 5B). In addition, in the reaction containing the RAG and HMG2 proteins, we detected a broad band that migrated with approximately the same mobility as the 64-nt marker (Fig. 5B, lane 4, large arrow), consistent with nicking of the hairpin at or near its tip. In the reaction lacking HMG2, a somewhat slower migrating band was detected (lane 2, arrowhead).
FIG. 5.
RAG-mediated hairpin nicking in Mg2+ after V(D)J cleavage. (A) Diagram indicating possible coding-end products detectable after RAG-mediated cleavage of pJH299, digestion of DNA with HinfI, denaturing PAGE, Southern blotting, and hybridization with a lower-strand-specific oligonucleotide probe. Site of HinfI cleavage, location of probe, and 12- and 23-RSSs (triangles) are indicated. (B) Southern blot showing products detected by denaturing PAGE (as diagrammed in panel A) after cleavage of pJH299 with the RAG proteins in the absence (arrowhead) and presence (large arrow) of HMG2. A DNA fragment (SmaI-digested pJH299) added upon termination of the cleavage reaction to ensure equal DNA recovery during subsequent manipulations (C) and the intact hairpin (small arrows) are also indicated. The asterisk marks a background band present in all lanes. Control experiments indicate that the intact 131-nt hairpin is detected with approximately one-third the efficiency of a linear control oligonucleotide, presumably due to self-reannealing of the hairpin during or after transfer to the membrane. (C) Native PAGE showing pJH299 23 coding ends detected by LMPCR. Products resulting from T4 DNAP-treated coding ends from reactions containing RAG proteins alone (large arrowhead), or RAG proteins plus HMG2 (arrow) or coding ends not treated with T4 DNAP (small arrowheads), are visible. A slower-migrating product resulting from the control DNA fragment (C; small arrow) is also indicated. DNA products have been detected by staining with SYBR Green.
After cleavage, RAG proteins generate open coding ends predominantly with single-stranded extensions.
The structure of the putative open coding ends was examined further by LMPCR assay. In this assay, products of the cleavage reaction were treated with T4 DNAP to blunt 5′ or 3′ overhangs, or left untreated, then ligated to a blunt-ended, unphosphorylated linker, amplified with appropriate primers, and analyzed by native PAGE. This analysis revealed significantly greater amounts of product after T4 DNAP treatment (Fig. 5C; compare lanes 4 and 5 with lanes 2 and 3), indicating that a substantial majority of the ends in the cleavage reactions contained overhangs. In addition, with T4 DNAP-treated samples, the presence of HMG2 in the cleavage reaction resulted in a more rapidly migrating LMPCR product (compare lanes 4 and 5), consistent with the different-sized products seen in lanes 2 and 4 of Fig. 5B.
Coding-end LMPCR products are consistent with RAG proteins nicking precisely at or a few nucleotides 5′ to the hairpin tip.
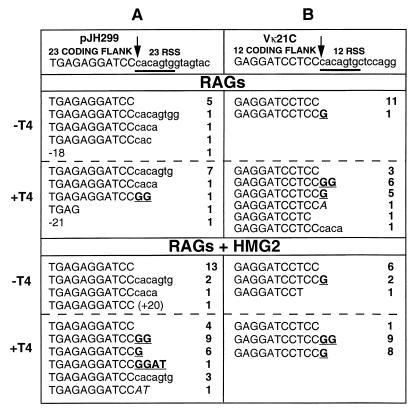
Cloning and sequencing of the LMPCR products revealed that the majority of blunt-ended molecules generated during the cleavage reaction terminated precisely at the last nucleotide of the coding flank (Fig. 6A, −T4 samples; Table 1). This structure is consistent with nicking at the tip of the coding-end hairpin. In T4 DNAP-treated samples, the presence of HMG2 in the cleavage reaction strongly influenced the structure of the products obtained (Fig. 6A, +T4 samples; Table 1). In the absence of HMG2, the dominant product contained the entire heptamer attached to the coding end, indicative of aberrant cleavage inside the 23-RSS. This explains the slower mobility of the products observed in the absence of HMG2 by Southern blotting (Fig. 5B, lane 2) and LMPCR (Fig. 5C, lane 4). Cleavage inside the RSS has been observed in previous studies, both in vivo and in vitro (27, 38, 46, 54). With HMG2 present, such events were much less frequent, and instead, ends with 1- or 2-bp palindromic extensions were the major products (Fig. 6A, bottom; Table 1). Such ends are likely the result of asymmetric nicking of the hairpin coding end to generate a 5′ overhang that was then filled in by T4 DNAP (3′ overhangs would be removed by T4 DNAP, resulting in deletions rather than additions). Thus, when coupled cleavage occurs under optimal conditions (RAG1, RAG2, HMG2, Mg2+), the RAG proteins open the resulting hairpin coding ends at a variety of sites at or close to the hairpin tip.
FIG. 6.
Sequences of LMPCR products derived from RAG-mediated cleavage of plasmid substrates in Mg2+. The top line shows the sequence of the coding flank (uppercase) and RSS (lowercase) from 5′ to 3′, with the heptamer underlined. The site of precise cleavage is indicated with an arrow. LMPCR was performed either before (−T4) or after (+T4) blunting of ends with T4 DNAP. Products of RAG (top half) or RAG-plus-HMG2 (bottom half) cleavage are displayed, with palindromic (boldface and underlined) and untemplated (italics) nucleotide additions indicated. The number of occurrences is indicated to the right of each sequence. The sizes of deletions and RSS extensions exceeding 10 bp are indicated with negative and positive numbers, respectively. (A) Data for the 23 coding end of pJH299, derived from LMPCR products from the experiment shown in Fig. 5. (B) Data for Vκ21C (which is flanked by a 12-RSS) of pJ2VIS6.
TABLE 1.
Summary of LMPCR coding-end sequences resulting from RAG-mediated hairpin opening after V(D)J cleavage of plasmid substrates
| Reaction | % of sequence obtaineda
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| pJH299 23 coding end
|
Vκ21C 12 coding end
|
Jκ1 23 coding endb
|
|||||||
| Tip | P | RSS | Tip | Pc | RSS | Tip | Pc | RSS | |
| RAGs | |||||||||
| −T4 | 56 | 0 | 33 | 92 | 8 | 0 | 18 | 9 | 64 |
| +T4 | 0 | 9 | 73 | 18 | 65 | 6 | 0 | 5 | 95 |
| RAGs + HMG2 | |||||||||
| −T4 | 76 | 0 | 24 | 67 | 22 | 0 | 64 | 0 | 9 |
| +T4 | 17 | 67 | 13 | 6 | 94 | 0 | 6 | 41 | 53 |
Values do not sum to 100% in some cases because ends with deletions or untemplated insertions are not included. Tip, full-length coding ends, indicative of nicking at the tip of the hairpin; P, coding ends containing palindromic extensions; RSS, products derived from cleavage within the RSS.
Numbers of sequences examined were 11 (−T4) and 20 (+T4) for RAGs and 11 (−T4) and 17 (+T4) for RAGs + HMG2 reactions.
The origin of the single (G) nucleotide extension amplified in the absence of T4 polishing is unknown.
Open coding ends are detected in the context of RAG-mediated, 12/23-regulated cleavage of Igκ locus coding elements and their flanking RSSs.
We then extended these analyses to coding ends derived from murine Vκ21C and Jκ1 gene segments. In Mn2+, synthetic hairpins representing these coding ends were nicked 5′ of the hairpin tip by the RAG proteins (data not shown). In Mg2+, hairpin nicking after coupled cleavage was assessed by Southern blotting and LMPCR using a plasmid containing the endogenous Jκ1 and Vκ21C gene segments and flanking RSSs. The results resembled those obtained for the pJH299 23-coding end: with HMG2, a significant proportion (10 to 30%) of hairpin coding ends were opened; opening appeared to occur at or near the hairpin tip; and the great majority of opened coding ends contained an overhang (Fig. 6, Table 1, and data not shown). Cloning and sequencing of the LMPCR products revealed that for Vκ21C (flanked by a 12-RSS), full-length coding ends were the predominant blunt-ended species (−T4 samples), while coding ends containing 1- or 2-nt palindromic extensions were most common after T4 DNAP treatment (+T4 samples; Fig. 6B and Table 1). HMG2 had little effect on the pattern of hairpin opening at this coding end, and cleavage inside the RSS was very rare. For Jκ1, cleavage inside its flanking 23-RSS was frequent in the absence of HMG2, while with HMG2 added, the previously observed pattern of blunt full-length coding ends (−T4) and an increase in the percentage of ends with short palindromic extensions (+T4) was again observed (Table 1 and data not shown).
DISCUSSION
Our data support a new model for V(D)J recombination in which the RAG proteins function as a regulated endonuclease that nicks hairpin coding ends in the context of a postcleavage complex and thereby serve to couple the cleavage and end-joining stages of the reaction. Since nicking of free hairpins by the RAG proteins does not occur in Mg2+, it is unlikely that the open coding ends that we observe in the context of coupled V(D)J cleavage derive from hairpin ends that have been released from this complex. While the complex in which Mg2+-based hairpin opening occurs has not been determined, it may well coincide with the recently identified cleaved signal complex thought to contain two coding ends and two signal ends (20). Several pieces of data support the idea that the RAG proteins are associated with a pair of signal ends during hairpin nicking. First, they bind tightly to pairs of, but not individual, signal ends (2, 20). Second, they bind more tightly to signal ends than coding ends (2, 20). Third, we and Besmer et al. (5) observe hairpin nicking in Mg2+ exclusively after cleavage at a pair of RSSs. The requirement for a postcleavage complex in RAG-mediated hairpin nicking provides a mechanism for regulating and targeting this nuclease activity. Further, we predict that in the postcleavage complex, nicking activity is directed to coding ends rather than signal ends for two reasons. First, signal ends may be protected by stably bound RAG proteins. Second, the active site may be oriented such that it predominantly contacts and cleaves DNA at the heptamer-coding flank border or nearby coding end hairpins, but not the signal ends to which these proteins are bound.
While our results of hairpin nicking by RAG proteins are consistent with those of Besmer et al. (5), some important differences are evident. In our experiments, nicking of synthetic hairpins occurs 5 nt 5′ of the hairpin tip, with other sites of nicking more distant from the tip. Nicking at the distant sites is suppressed in the presence of HMG2. In the experiments of Besmer et al., nicking by RAG proteins in Mn2+ occurs 2, 3, and 4 nt 5′ of the tip. A second difference is seen in experiments with nonhairpin, linear DNA substrates, in which we observe nicking 5 nt from the 3′ end whereas Besmer et al. observe a 2-nt 3′ end processing activity. Both of these differences may relate to substrate sequence differences, which could influence single-stranded DNA character at the hairpin tip or protein-DNA interactions. The nicking of hairpins by other single-strand-specific nucleases is influenced by the terminal 4 nt at the hairpin tip and loose nucleotide preferences exist for different nucleases (23). We note that nicking 2 nt from the 3′ end may also be occurring in our reactions with linear DNA substrates, but the product would not be well resolved from the input substrate (Fig. 3A, lanes 4 and 5).
A notable difference between our experiments and those of Besmer et al. (5) is that in their reactions involving 12/23-regulated cleavage with oligonucleotide substrates in Mg2+, the 23 coding end shows nicking exclusively at the hairpin tip (opening of the 12 coding end was not analyzed). In our reactions with plasmid substrates, both the 12 and 23 coding ends frequently exhibit 1- or 2-nt 5′ extensions, indicative of nicking 5′ of the hairpin tip. One explanation for the difference might be the use of different preparations of RAG proteins (MH-RAGs in our experiments; GST-RAGs in those of Besmer et al.). In support of this possibility, initial experiments reveal that in the context of coupled cleavage of our plasmid substrates, purified GST-RAG proteins, similar to those used by Besmer et al., preferentially nick coding-end hairpins at the tip (data not shown). It is possible that the GST moiety sterically interferes with the ability of the RAG proteins to nick the hairpin asymmetrically. Alternatively, since GST can dimerize, the postcleavage complex formed after GST-mediated dimerization of RAG proteins may differ structurally from that formed by RAG proteins lacking these tags. In vivo, the processing of coding ends is influenced by DNA sequence, and so sequence differences in the hairpin ends analyzed may also be relevant (reference 36 and references therein). The hairpins generated by our cleavage reactions contain 5′-ATCC, CTCC, and CCAC at their termini, compared to 5′-CCTA for the 23 coding end in the study of Besmer et al. Systematic analysis of a variety of hairpins generated by cleavage will help to elucidate possible sequence preferences for RAG-mediated hairpin nicking. Finally, the use of DNA substrates with different structures (plasmid versus oligonucleotide) might also contribute to differences observed in the two systems. The short palindromic extensions that we observe in the context of V(D)J cleavage (usually 1 to 2 nt in length) are consistent with the short stretches of P nucleotides (very often 1 to 2 bp in length) inserted in a significant percentage of V(D)J coding joints in vivo (28, 34). Thus, our data uniquely suggest that P nucleotides are generated by the RAG proteins.
The mechanism by which HMG2 constrains nicking of synthetic hairpins to a site near the hairpin tip is unclear, but it is unlikely to be due to nonspecific binding and protection of the DNA because at the concentration of HMG2 used (125 nM), it binds poorly to linear duplex DNA (7). Furthermore, addition of purified Ku protein, which can bind DNA ends and translocate to internal positions (11), inhibits nicking at all sites of the hairpin substrate equally (data not shown). Finally, nicking of DNA duplex substrates is not similarly influenced by HMG2. It is possible that HMG2 exerts its influence by virtue of its interaction with RAG1 (3, 40) and its higher affinity for distorted DNA structures (7), such as hairpins (6). Current evidence suggests that hairpin coding ends are usually opened within a few base pairs of the hairpin tip in vivo (28, 31, 36, 46), and hence this function of HMG2 may be significant during V(D)J recombination. The requirement for HMG2 in stimulating properly targeted RSS cleavage of plasmid substrates, and suppressing inappropriate cleavage at the 23-RSS but not the 12-RSS, is interesting in light of its selective enhancement of binding and cleavage at the 23-RSS (44, 50).
It is clear that vertebrate cells contain factors in addition to RAG1 and RAG2 that are capable of processing DNA hairpins (4, 29, 48). For example, the ubiquitous DNA double-strand break repair factor Mre11 can nick hairpin DNA and promote homology-mediated ligation of DNA ends in vitro (37). RAG1 and RAG2 appear to be in direct physical contact with hairpin coding ends after cleavage (20), and our data demonstrate that they can nick perfect hairpins. It is therefore plausible that they are responsible for some or all hairpin opening during V(D)J recombination. Processing of coding end hairpins by the RAG proteins might enhance subsequent recruitment and/or activation of general DNA repair factors, perhaps by altering the structure of the postcleavage complex. In addition, RAG protein-mediated coding-end processing raises the possibility that V(D)J end joining and general NHEJ involve somewhat distinct mechanisms.
These findings extend the mechanistic parallels noted between DNA cleavage in V(D)J recombination and bacterial transposition (1, 22, 35, 52). Tn10 transposase cleaves DNA by sequential hydrolysis (nicking) and strand transfer (hairpin formation) reactions, followed by nicking of the hairpin at its tip (24), and our data indicate that DNA cleavage by the RAG proteins proceeds via a similar series of chemical reactions. One principal difference in the reactions is that the RAG proteins can nick the hairpin at a variety of sites, which could contribute to the diversity of coding junctions and hence of the encoded antigen receptors. RAG-mediated hairpin nicking does not occur in Ca2+ and so in this respect more closely resembles RAG-mediated RSS nicking than RAG-mediated transposition (21, 22). A single bifunctional active site appears to mediate nicking and strand transfer by Tn10 transposase (8), and it is tempting to think that RSS nicking, hairpin formation, and hairpin nicking during V(D)J recombination similarly involve a single active site. This possibility is supported by the finding that mutations in RAG1 or RAG2 that interfere with cleavage also disrupt nicking of synthetic hairpins in Mn2+ (5). It will be important to determine whether, in the context of regulated cleavage in Mg2+, RSS nicking and hairpin nicking involve the same active site.
Recently it was shown that the RAG proteins are capable in vitro of mediating open-shut and hybrid joint formation using the 3′OH of cleaved signal ends as a nucleophile to attack the coding-end hairpin in a reaction which is the reverse of the V(D)J cleavage reaction (35). The hairpin nicking reactions that we observe may proceed by a similar mechanism but with water as the nucleophile to hydrolyze hairpin DNA. It remains to be proven that this mechanism operates in vivo to open hairpin coding ends or to form hybrid joints.
Two observations must be reconciled with our results and those of Besmer et al. and with the hypothesis that the RAG proteins open hairpin coding ends in vivo. First, a majority of the rare coding ends detected in normal lymphoid precursors have 3′ overhangs and show nucleotide deletion, in contrast to the 5′ overhangs that we detect in vitro (31, 46). However, the coding ends detected in vivo may have undergone additional processing events after initial hairpin nicking by RAG1 and RAG2. In addition, these coding ends may be products that do not go on to form coding joints, although a correlation was noted between the extent of deletion at coding ends and the structure of coding joints (46). Second, cells deficient in general DNA double-strand break repair proteins, such as DNAPK and Ku80, are deficient in coding-joint formation and accumulate hairpin coding-end intermediates, despite expression of RAG1 and RAG2 (15, 41, 55). Our results indicate that general repair proteins are not required for hairpin opening under the simplified conditions of our in vitro experiments. However, such factors may be necessary to regulate the extent and sites of RAG-mediated hairpin opening in vivo. They might do so directly, by posttranslational modification of RAG1, RAG2, or other proteins in the postcleavage complex or indirectly by regulating the structure of the postcleavage complex and association of the RAG proteins with coding ends.
ACKNOWLEDGMENTS
We thank Q. Eastman, I. Villey, D. Hesselein, A. Tevelev, E. Corbett, and K. K. Rodgers for work leading to and various preparations of purified proteins from mammalian cells and bacteria, L. Ptaszek for individually expressed GST-RAG1 and GST-RAG2, G. Chu and O. Hammarsten for purified Ku proteins, M. Bianchi for helpful discussions, A. Agrawal and A. Lee for helpful comments on the manuscript, and the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University for oligonucleotide synthesis and DNA sequencing.
This work was supported by a postdoctoral fellowship from the Arthritis Foundation to P.E.S. and by grant AI32524 from the National Institutes of Health to D.G.S. D.G.S. is an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Agrawal A, Eastman Q M, Schatz D G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal A, Schatz D G. RAG1 and RAG2 form a stable post-cleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 3.Aidinis, V., S. Santagata, and M. E. Bianchi. Personal communication.
- 4.Akgün E, Zahn J, Baumes S, Brown G, Liang F, Romanienko P, Lewis S, Jasin M. Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Besmer E, Mansilla-Soto J, Cassard S, Sawchuk D J, Brown G, Sadofsky M, Lewis S M, Nussenzweig M C, Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi, M. Personal communication.
- 7.Bianchi M E, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 8.Bolland S, Kleckner N. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell. 1996;84:223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- 9.Critchlow S E, Jackson S P. DNA end-joining: from yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 10.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 11.Dynan W S, Yoo S. Interaction of Ku protein and DNA-dependent protein kinase catalytic subunit with nucleic acids. Nucleic Acids Res. 1998;26:1551–1559. doi: 10.1093/nar/26.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 13.Frank K M, Sekiguchi J M, Seidl K J, Swat W, Rathbun G A, Cheng H L, Davidson L, Kangaloo L, Alt F W. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- 14.Gao Y, Sun Y, Frank K M, Dikkes P, Fujiwara Y, Seidl K J, Sekiguchi J M, Rathbun G A, Swat W, Wang J, Bronson R T, Malynn B A, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin S H, Greenberg M E, Alt F W. A critical role for DNA end joining proteins in both lymphomagenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y J, Chaudhuri J, Zhu C M, Davidson L, Weaver D T, Alt F W. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for Ku in V(D)J recombination. Immunity. 1998;9:367–376. doi: 10.1016/s1074-7613(00)80619-6. [DOI] [PubMed] [Google Scholar]
- 16.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 17.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber M R. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 18.Gu Y S, Seidl K J, Rathbun G A, Zhu C M, Manis J P, Vanderstoep N, Davidson L, Cheng H L, Sekiguchi J M, Frank K, Stanhopebaker P, Schlissel M S, Roth D B, Alt F W. Growth retardation and leaky scid phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 19.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 20.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 21.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 22.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 23.Kabotyanski E B, Zhu C M, Kallick D A, Roth D B. Hairpin opening by single-strand-specific nucleases. Nucleic Acids Res. 1995;23:3872–3881. doi: 10.1093/nar/23.19.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennedy A, Guhathakurta A, Kleckner N, Haniford D. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 25.Komori T, Okada A, Stewart V, Alt F W. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 26.Lafaille J J, DeCloux A, Bonneville M, Takagaki Y, Tonegawa S. Junctional sequences of T cell receptor gamma delta genes: implications for gamma delta T cell lineages and for a novel intermediate of V-(D)-J joining. Cell. 1989;59:859–870. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- 27.Leu T M J, Eastman Q M, Schatz D G. Coding joint formation in a cell free V(D)J recombination system. Immunity. 1997;7:303–314. doi: 10.1016/s1074-7613(00)80532-4. [DOI] [PubMed] [Google Scholar]
- 28.Lewis S M. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 29.Lewis S M. P nucleotide insertions and the resolution of hairpin DNA structures in mammalian cells. Proc Natl Acad Sci USA. 1994;91:1332–1336. doi: 10.1073/pnas.91.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieber M R. Site-specific recombination in the immune system. FASEB J. 1991;5:2934–2944. doi: 10.1096/fasebj.5.14.1752360. [DOI] [PubMed] [Google Scholar]
- 31.Livàk F, Schatz D G. Identification of V(D)J recombination coding end intermediates in normal thymocytes. J Mol Biol. 1997;267:1–9. doi: 10.1006/jmbi.1996.0834. [DOI] [PubMed] [Google Scholar]
- 32.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 33.McCormack W T, Tjoelker L W, Carlson L M, Petryniak B, Barth C F, Humphries E H, Thompson C B. Chicken IgL gene rearrangement involves deletion of a circular episome and addition of single nonrandom nucleotides to both coding segments. Cell. 1989;56:785–791. doi: 10.1016/0092-8674(89)90683-1. [DOI] [PubMed] [Google Scholar]
- 34.Meier J T, Lewis S M. P nucleotides in V(D)J recombination: a fine structure analysis. Mol Cell Biol. 1993;13:1078–1092. doi: 10.1128/mcb.13.2.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melek M, Gellert M, van Gent D C. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 1998;280:301–303. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- 36.Nadel B, Feeney A J. Nucleotide deletion and P addition in V(D)J recombination: a determinant role of the coding-end sequence. Mol Cell Biol. 1997;17:3768–3778. doi: 10.1128/mcb.17.7.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paull T T, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 38.Ramsden D A, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 39.Ramsden D A, Paull T T, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers, K. K., I. J. Villey, E. Corbett, D. G. Schatz, and J. E. Coleman. A dimer of RAG1 recognizes the recombination signal sequence and the complex stably incorporates HMG2. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 41.Roth D B, Menetski J P, Nakajima P B, Bosma M J, Gellert M. V(D)J recombination: broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 42.Roth D B, Zhu C M, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santagata S, Aidinis V, Spanopoulou E. The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J Biol Chem. 1998;273:16325–16331. doi: 10.1074/jbc.273.26.16325. [DOI] [PubMed] [Google Scholar]
- 44.Sawchuk D J, Weis-Garcia F, Malik S, Besmer E, Bustin M, Nussenzweig M C, Cortes P. V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA bending proteins. J Exp Med. 1997;185:2025–2032. doi: 10.1084/jem.185.11.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schatz D G, Baltimore D. Stable expression of immunoglobulin gene V(D)J recombinase activity by gene transfer into 3T3 fibroblasts. Cell. 1988;53:107–115. doi: 10.1016/0092-8674(88)90492-8. [DOI] [PubMed] [Google Scholar]
- 46.Schlissel M S. Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol Cell Biol. 1998;18:2029–2037. doi: 10.1128/mcb.18.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spanopoulou E, Zaitseva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. The homeodomain of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 48.Staunton J E, Weaver D T. Scid cells efficiently integrate hairpin and linear DNA substrates. Mol Cell Biol. 1994;14:3876–3883. doi: 10.1128/mcb.14.6.3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taccioli G E, Amatucci A G, Beamish H J, Gell D, Xiang X H, Arzayus M I T, Priestley A, Jackson S P, Rothstein A M, Jeggo P A, Herrera V L M. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity. 1998;9:355–366. doi: 10.1016/s1074-7613(00)80618-4. [DOI] [PubMed] [Google Scholar]
- 50.van Gent D C, Hiom K, Paull T T, Gellert M. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 1997;16:2665–2670. doi: 10.1093/emboj/16.10.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 52.van Gent D C, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 53.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 54.Weis-Garcia F, Besmer E, Sawchuk D J, Yu W, Hu Y, Cassard S, Nussenzweig M C, Cortes P. V(D)J recombination: in vitro coding joint formation. Mol Cell Biol. 1997;17:6379–6385. doi: 10.1128/mcb.17.11.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu C M, Bogue M A, Lim D S, Hasty P, Roth D B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]