Abstract
A fungus-derived compound (OSI-2040) which induces fetal globin expression in the absence of erythroid cell differentiation was identified in a high-throughput drug discovery program. We utilized this compound to isolate γ-globin regulatory genes that are differentially expressed in OSI-2040-induced and uninduced cells in the human erythroleukemia cell line K562. Representational difference analysis (RDA) of cDNA revealed several genes that were significantly up- or down-regulated in OSI-2040-induced cells. One gene whose expression was markedly enhanced was the gene for the helix-loop-helix (HLH) transcription factor Id2. Southern analysis of RDA amplicons demonstrated progressive enrichment of Id2 with each successive subtraction of uninduced cDNA from induced cDNA. Northern analysis of OSI-2040-induced K562 cells confirmed that Id2 expression was directly up-regulated coordinately with γ-globin. Analysis of other inducers of fetal globin demonstrated up-regulation of Id2 with sodium butyrate but not with hemin. Retrovirus-mediated overexpression of Id2 in K562 cells reproduced the enhancement of endogenous globin expression observed with OSI-2040 induction. Functional assays demonstrated that an E-box element in hypersensitivity site 2 is required for Id2-dependent enhancement of γ-promoter activity. Protein binding studies suggest that alterations in E-box site occupancy by basic HLH proteins may influence this activity, thus expanding the potential role of these factors in globin gene regulation.
The genes of the human β-globin locus (ɛ, Gγ, Aγ, δ, β) are regulated in a developmental and tissue-specific manner. Inherent to this control are cis-acting promoter sequences which flank the genes themselves and distal regulatory sequences which reside 6 to 20 kb 5′ of the ɛ-gene, known as the locus control region (LCR). The coordinate action of the LCR and globin gene promoters results in high levels of expression of the individual genes at distinct developmental stages. The ɛ-gene is expressed during embryonic erythropoiesis and the γ-genes are expressed during fetal development, while expression of the δ- and β-genes is confined to adult erythroid cells (42). Studies of the globin promoters and the four erythroid-cell-specific DNase I hypersensitivity sites (HS1 through -4) which define the LCR have identified numerous protein-binding motifs for a variety of ubiquitous (Sp1, USF, YY1, and others) and erythroid-cell-specific (GATA-1, NF-E2, EKLF, and others) transcription factors (20, 35). Many of these proteins have been identified through recognition of their cognate binding sites in globin regulatory regions, although several have been defined through biochemical purification or molecular cloning techniques. These proteins are thought to act as transcriptional activators or modifiers of the chromatin structure of the locus. Despite these important functions, few of these factors have been shown to have discriminatory roles in the regulation of specific globin genes. EKLF is one exception to this, as mice nullizygous for the EKLF gene retain normal embryonic and fetal globin expression but die of a β-thalassemic illness at the onset of adult hematopoiesis (34, 37). Another exception appears to be the stage selector protein, which binds to the proximal γ-promoter and preferentially recruits the LCR during fetal erythropoiesis (21, 22).
The existence of other factors capable of altering the developmental profile of the β-globin cluster is suggested by studies of a diverse group of genetic mutations unified phenotypically by persistent fetal globin expression after birth. These mutations are known collectively as hereditary persistence of fetal hemoglobin (HPFH) and are largely due to point mutations in the γ-promoter that alter the binding of transcriptional activators or repressors (39). However, several kindred have been identified with HPFH that is not linked to the β-globin cluster, indicating the presence of as-yet-unknown trans-factor mutations that influence the developmental profile of γ-gene expression (12, 14). These factors may not be tissue-specific or developmentally restricted in their expression patterns but may exert their influence by augmenting the transcriptional activity of an already active γ-promoter. The importance of such factors lies in the observation that elevated levels of fetal hemoglobin have a protective effect on patients who inherit the devastating β-chain disorders β-thalassemia and sickle-cell disease (33). Hence, the identification of proteins capable of inducing fetal globin gene expression may provide treatment alternatives for these disorders through gene therapy or pharmacological strategies.
One approach to identifying these novel factors is the study of compounds that induce γ-gene expression in vitro or in vivo. To this end, we recently identified a fungus-derived compound (OSI-2040) which enhances γ-globin gene expression in the human erythroleukemia cell line K562 (29). This fungus-derived compound was found in a high-throughput robotic drug screen designed to identify agents which specifically transcriptionally activate target genes. The activity of this compound is comparable to that of the other potent γ-globin inducers, sodium butyrate and hydroxyurea. In this study, we employed representational difference analysis (RDA) of cDNA to identify genes differentially expressed coordinately with γ-gene induction by OSI-2040. Our results demonstrate that transcription factor Id2, a member of the helix-loop-helix (HLH) family of proteins, is up-regulated in OSI-2040-induced K562 cells. Retrovirus-mediated overexpression of Id2 in K562 cells reproduces the induction of γ-gene expression observed with OSI-2040, suggesting that this factor may play a direct role in the regulation of the β-globin cluster.
MATERIALS AND METHODS
Isolation of OSI-2040.
A library of fungal broth extracts (OSI Pharmaceuticals, Inc.) was screened against a stable human erythroleukemia cell line, K562, containing an HS2-γ–luciferase reporter gene. A total of 54,656 entities were screened. The fungal extract file yielded 460 hits from 378 different organisms. Compounds were prioritized on the basis of efficacy, potency, and cytotoxicity at 24- and 48-h exposure time points for bioassay-guided fractionation and purification by sequential organic extraction, reverse-phase high-pressure liquid chromatography (HPLC), and countercurrent liquid phase chromatography. The biological activity of pure compounds was further characterized by tissue culture (see Results), while the compound structures were determined by mass spectrometry and nuclear magnetic resonance imaging. OSI-2040 increased γ-globin transcription and γ-globin protein expression in three distinct assays: a hemoglobin-staining assay, a fetal hemoglobin (HbF) enzyme-linked immunosorbent assay, and a cation-exchange HPLC assay (16).
Cell lines.
The human erythroleukemia cell lines K562 and HEL were grown in RPMI medium supplemented with 10% fetal calf serum. Cells were induced with OSI-2040 at 0.8 mM, 25 mM hemin, or 1 mM sodium butyrate for 48 h prior to RNA extraction. The human embryonal kidney cell line 293T and the human hepatocellular carcinoma cell line Hep3B were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum.
RNA isolation and Northern analysis.
Two methods were used to isolate poly(A)+ RNA. For RDA, total RNA was isolated from 5 × 107 K562 cells by guanidinium isothiocyanate lysis (RNAzol; Biotecx Laboratories, Inc.) as described by the manufacturer. Polyadenylated RNA was then isolated with biotinylated oligo(dT) primers and streptavidin coupled to magnetic particles (PolyATract; Promega). Aliquots of 2 mg of poly(A)+ RNA were converted to cDNA by using an oligo(dT) primer and the RiboClone cDNA synthesis system (Promega).
For Northern analysis, poly(A)+ RNA was isolated from pools of 2.5 × 107 K562 cells with oligo(dT) cellulose (Boehringer). Northern analysis was performed as previously described (41).
RDA of cDNA.
Analyses of cDNAs prepared from drug-induced and uninduced K562 cells were performed as previously described with minor adjustments (19, 27). K562 cells were induced with OSI-2040 (0.8 mM) for 48 h, and RNA and cDNA were prepared as detailed above. Prior to the initial subtractive hybridization, DpnII-digested cDNA fragments were electrophoresed on a 1% agarose gel, transferred to a nitrocellulose filter, and probed with β-actin to normalize the pools. The tester and driver amplicons were hybridized to each other at molar ratios of 1:100, 1:400, and 1:80,000 for each successive round of RDA. The driver amplicons derived from uninduced cells were spiked with the γ-globin cDNA. After three rounds of hybridization and PCR, discrete DNA bands visualized on 1% agarose gels were excised, isolated, and cloned into the BamHI site of M13 mp19. They were then sequenced with an ABI dideoxy terminator cycle sequencer (Applied Biosystems) and compared to the GenBank and expressed sequence tag (EST) databases by using BLAST algorithms.
Reverse Northern blotting.
The M13 mp19 clones of the RDA fragments (100 ng) were blotted onto Genescreen Plus membranes in duplicate with a slot blot apparatus (Schleicher and Schuell). Fragments from several housekeeping genes (actin, S14, and ribosomal protein genes) were also included. The membranes were baked for 2 h at 80°C. Equivalent amounts of uninduced and drug-induced K562 cDNAs were radiolabelled by reducing the concentration of dCTP in the PCR to 42.5 mM and including 5 ml of [α-32P]dCTP in a 12-cycle PCR (95°C for 1 min and 72°C for 3 min). The labelled cDNAs were then purified through a Sephadex G-50 column (Pharmacia) and hybridized with filters as previously described (41). The signals from housekeeping genes were compared to ensure that cDNA probes with similar specific activity had been used.
Isolation of Id2 coding sequence.
The Id2 coding sequence was amplified from an λZAP K562 library with the primers 5′ CGGTCTCGAGTTCCTCGCGGTC and 3′ GAACCTCGAGTATTCAGCCACACAGTG, which correspond to nucleotides 72 to 93 and 488 to 514 (Genbank sequence HUMID2HC), respectively (44). The primers also incorporate an XhoI site for subsequent cloning. The first-round PCR included 200 ng of library DNA, 100 pmol of vector primer and 3′ Id2 primer, 0.4 mM deoxynucleoside triphosphates, 1 U of Taq polymerase, and 1× Taq buffer (Boehringer). PCR conditions were 95°C for 30 s, 55°C for 1 min, and 72°C for 1 min for a total of 33 cycles. Nested PCR was then performed with 1 ml of the first-round reaction product with both Id2-specific primers for 25 cycles. The integrity of the final PCR product was confirmed by sequencing as detailed above.
Generation of amphotropic retroviral supernatant and transduction of K562 cells.
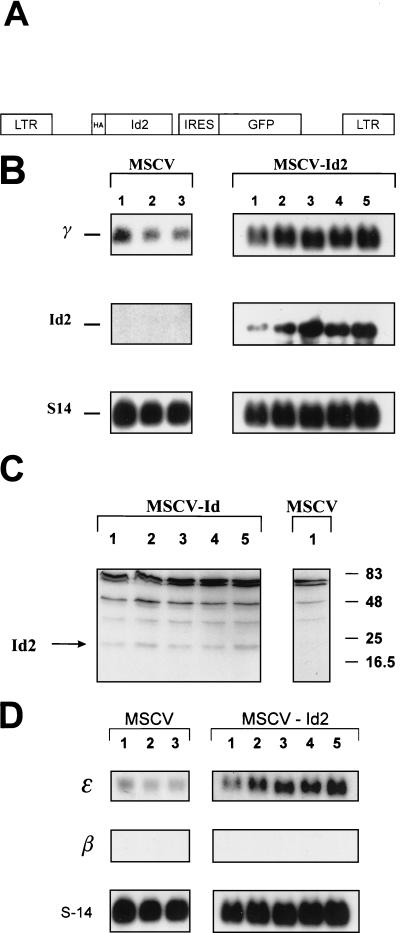
The Id2 coding region was cloned into the retroviral vector plasmid MSCV-HA at a unique XhoI site (17, 38). This bicistronic vector contains (i) the murine stem cell virus (MSCV) 5′ long terminal repeat, (ii) a hemagglutinin (HA) epitope tag with the Id2 coding sequence in frame, (iii) the encephalomyocarditis internal ribosomal entry site (IRES), (iv) the green fluorescent protein (GFP) cDNA, and (v) the MSCV 3′ long terminal repeat (see Fig. 5A). The plasmid was cotransfected with an amphotropic packaging plasmid into 293T cells by calcium phosphate precipitation. After 48 h, the supernatant containing the amphotropic particles was harvested, filtered, and added to either K562 or HEL cells every 12 h for 3 days. The K562 cells were allowed to recover for 72 h and then were analyzed for GFP expression by flow cytometry. The highest-expressing 10% of cells were sterilely sorted and subsequently expanded in pools. A biological titer of the supernatant on NIH 3T3 cells was equivalent to a concentration of 106 CFU/ml.
FIG. 5.
Overexpression of Id2 in K562 cells induces γ-gene expression. (A) Diagrammatic representation of the MSCV-Id2-IRES-GFP retroviral vector. The vector consists of the MSCV retroviral backbone, containing the Id2 coding sequence, followed by an IRES from the encephalomyocarditis virus linked to the GFP gene. (B) Northern analysis of K562 cell pools overexpressing Id2. Two milligrams of poly(A)+ RNA from K562 cells transduced with MSCV-Id2 or MSCV alone were analyzed with γ-gene and Id2 probes. S14 served as the control. (C) Western analysis of K562 cell pools overexpressing Id2. Whole-cell lysates from K562 cells transduced with MSCV-Id2 or MSCV alone were analyzed with anti-HA antisera. The specific HA-Id2 band is shown by an arrow. Numbers on the right represent the migration pattern of protein molecular weight standards (in thousands). (D) Northern analysis of K562 cell pools overexpressing Id2. Two milligrams of poly(A)+ RNA from K562 cells transduced with MSCV-Id2 or MSCV alone was analyzed with ɛ-gene and β-gene probes. S14 served as the control.
Oligonucleotides, nuclear extract, and EMSA.
The sequences of the top strand of the duplex oligonucleotides used for an electrophoretic mobility shift assay (EMSA) were the 8762 E-box sequence (CTAGAGGGCAGATGGCAA) and the USF consensus site (CACCCGGTCACGTGGCCTACACC). Nuclear extract of K562 cells was prepared by Dounce homogenization of isolated nuclei and utilized in EMSA as described previously (21).
DNA construction and transfection.
Constructs containing the HindIII-XbaI fragment of HS2 linked to the γ-promoter–luciferase gene or β-promoter–luciferase gene have been described previously (1). Constructs containing the mutant 8701 or 8762 E boxes were described in reference 15. Transient-transfection experiments were repeated six times with at least two independent preparations of DNA, as described previously (2). Reporter assays were performed with the luciferase assay kit from Promega according to the manufacturer’s instructions.
RESULTS
Identification and preliminary characterization of OSI-2040.
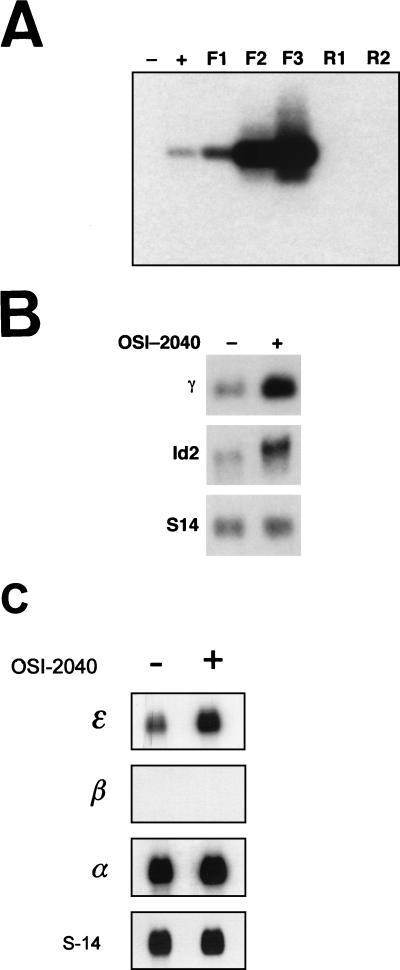
OSI-2040 is a fungus-derived compound identified as part of a proprietary high-throughput drug screening program designed to isolate novel agents which could induce fetal globin production (see Materials and Methods for details of isolation and purification). In the initial screening approach, a K562 cell line stably transfected with an artificial promoter construct containing the HS2 region of the LCR and a −260 γ-promoter fused to the luciferase gene was used (Fig. 1A). Transcriptional activity of the γ-promoter was increased in these cells in response to exposure to OSI-2040 for 24 h in a dose-dependent manner (Fig. 1B). No increase in the activity of a control bcr promoter-reporter in OSI-2040-induced K562 cells or of a control erythropoietin promoter-reporter in OSI-2040-induced Hep3B cells was observed (data not shown). To examine the effects of OSI-2040 in the context of the endogenous γ-globin genes, K562 cells were exposed to the drug for 96 h and stained with benzidine. Cells treated with hydroxyurea and sodium butyrate served as controls. As seen in Fig. 1C, globin synthesis increased with all three agents. These findings were confirmed by HbF enzyme-linked immunosorbent assay and combined HPLC-mass spectrometry studies (data not shown).
FIG. 1.
OSI-2040 increases γ-promoter activity and endogenous fetal globin expression. (A) Diagrammatic representation of the HS2-γ–luciferase construct used for screening compounds from fungus libraries. The open box represents the HindIII-BglII fragment of HS2, and the shaded box represents the −260 γ-promoter–luciferase reporter gene. (B) Dose-response curve of γ-promoter induction with OSI-2040. K562 cells containing the HS2-γ–luciferase construct were exposed to increasing concentrations of OSI-2040 as indicated. After 24 h, the cells were lysed and the reporter gene activity was measured and plotted as the induction compared with that of uninduced control K562 cells. The OSI-2040 dose axis is not to scale. (C) Benzidine staining in cells treated with γ-gene inducers. K562 cells were treated with OSI-2040 (0.8 mM), butyrate (1 mM), or hydroxyurea (500 mM) for 96 h. Untreated K562 cells served as a control.
RDA of cDNA of OSI-2040-induced and uninduced K562 cells.
Based on these findings, we proposed that OSI-2040 might be acting through a transcriptional mechanism involving a regulator of γ-gene expression. To identify a putative factor involved in this process, we performed RDA of cDNA, a PCR-based subtraction strategy on mRNA from OSI-2040-induced and uninduced K562 cells (see Materials and Methods). This procedure was performed with uninduced mRNA subtracted from induced mRNA to detect transcripts which were up-regulated by OSI-2040 (forward direction) and with induced RNA subtracted from uninduced RNA to detect suppressed transcripts (reverse direction). As seen in Fig. 2A, the complexity of the cDNA species decreased with successive rounds of subtraction and amplification in both directions. Discrete bands were then excised from agarose gels and cloned. To determine whether the isolated cDNAs were truly differentially expressed, a reverse Northern analysis was performed. As shown in Fig. 2B, the majority of clones isolated in the forward direction showed significant up-regulation (2- to 16-fold) in the induced cDNA pool. Clones isolated in the reverse direction were down-regulated in the induced pool (3- to 15-fold), indicating that both subtractions had identified differentially expressed genes. The identity of these genes was determined through sequence analysis and BLAST database searches. As seen in Table 1, a variety of clones was obtained, many with a role in cell-signaling pathways or cell cycle regulation. Of note was the isolation of an up-regulated transcription factor, Id2, in the induced pool.
FIG. 2.
RDA of cDNA of uninduced and OSI-2040-induced K562 cells. (A) Ethidium bromide-stained agarose gels showing RDA amplicons. Lanes − and +, cDNAs from uninduced and OSI-2040-induced K562 cells. Lanes F1 to F3 represent successive rounds of subtraction in the forward direction with induced K562 cDNA as the tester and uninduced K562 cDNA as the driver. Lanes R1 and R2 represent successive rounds of subtraction in the reverse direction with uninduced K562 cDNA as the tester and induced K562 cDNA as the driver. The migration of molecular weight standards is indicated. (B) Slot blot hybridization of 22 clones demonstrating differential representation between uninduced and induced amplicons. Actin, which is not differentially expressed, was used as a control.
TABLE 1.
Differential amplicons identified by RDA
| Amplicona | Fold change in hybridization signal |
|---|---|
| Forward direction (increase) | |
| Id2 | 9 |
| Purine nucleoside phosphorylase | 2 |
| CD24 | 12 |
| EST similar to CDC10 | 6 |
| 4-1BB ligand | 5 |
| IL-1-inducible EST | 16 |
| Hep27 | 7 |
| Clusterin | 7 |
| Reverse direction (decrease) | |
| Stat 5a | 12 |
| MBP-1 | 6 |
| Polo-like kinase | 10 |
| EST similar to cyclophilin B | 4 |
| Sorcin | 15 |
| Lamin B receptor | 4 |
| CSA-19 | 3 |
IL-1, interleukin-1.
Id2 is up-regulated in OSI-2040-induced K562 cells.
To validate that Id2 was differentially enriched during RDA, cDNA amplicons obtained at each step of the forward and reverse procedures were analyzed by Southern blotting with a full-length Id2 cDNA probe. As seen in Fig. 3A, no signal was observed in uninduced (lane −) K562 cells or in amplicons derived from one or two rounds of reverse subtraction (lanes R1 and R2). In contrast, the Id2 signal was apparent after drug treatment alone (lane +), with dramatic enhancement observed with sequential subtraction in the forward direction (lanes F1 to F3). To directly assess the effects of OSI-2040 treatment on Id2 expression, K562 cells were cultured in the presence of the drug for 48 h, and mRNA was prepared. As seen in Fig. 3B, Northern analysis revealed a significant up-regulation of Id2 in this setting. Concomitant up-regulation of endogenous γ-gene expression was also evident with OSI-2040 treatment. In contrast, other markers of erythroid-cell differentiation, including GATA-1 and glycophorin A, were not increased (data not shown). To assess the effects of OSI-2040 on other globin genes, we performed Northern analysis on induced K562 cells with ɛ-, β-, and α-globin probes. Interestingly, a significant induction of ɛ-gene expression was observed. No induction of either β- or α-globin expression was seen (Fig. 3C). We also examined the effects of OSI-2040 on another human erythroleukemia cell line, HEL, which constitutively expresses fetal globin genes. No induction of Id2 or γ-gene expression was observed with Northern analysis of this line. Reverse transcription-PCR analysis of uninduced and induced HEL mRNA revealed that Id2 was not expressed in this line (data not shown).
FIG. 3.
Id2 and γ-globin are up-regulated in OSI-2040-induced K562 cells. (A) Southern blot of the cDNA pools and amplicons shown in Fig. 2A obtained with an Id2 probe. Lanes − and +, cDNA from uninduced and OSI-2040-induced K562 cells. Lanes F1 to F3 represent successive rounds of subtraction in the forward direction with induced K562 cDNA as the tester and uninduced K562 cDNA as the driver. Lanes R1 and R2 represent successive rounds of subtraction in the reverse direction with uninduced K562 cDNA as the tester and induced K562 cDNA as the driver. (B) Northern analysis of OSI-2040-induced K562 cells. Two milligrams of poly(A)+ RNA from uninduced (−) or OSI-2040-induced (+) K562 cells was analyzed with γ-gene and Id2 probes. S14 served as the control. (C) Northern analysis of OSI-2040-induced K562 cells. Two milligrams of poly(A)+ RNA from uninduced (−) or OSI-2040-induced (+) K562 cells was analyzed with ɛ-gene, β-gene, and α-gene probes. S14 served as the control.
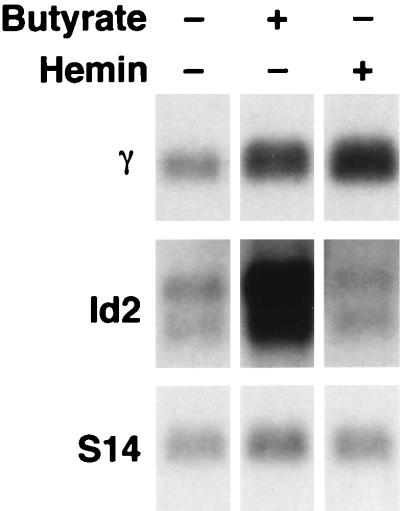
To determine whether other compounds capable of inducing fetal globin expression also induced Id2, we performed Northern analysis on K562 cells treated with sodium butyrate or hemin. As seen in Fig. 4, butyrate induced a significant increase in Id2 expression with a concomitant increase in γ-globin message (lane 2). In contrast, hemin failed to induce Id2 expression, despite effectively increasing the levels of γ-gene transcripts (lane 3). Quantitation of these results revealed that OSI-2040 induced γ-globin 4.9-fold and Id2 11-fold, whereas butyrate induced γ-globin 2.9-fold and Id2 7.9-fold when corrected for expression of the housekeeping gene S14. These results further indicated that a significant correlation existed between Id2 and γ-gene induction.
FIG. 4.
Id2 is induced by sodium butyrate but not hemin. Northern analysis of butyrate- or hemin-induced K562 cells is shown. Two milligrams of poly(A)+ RNA from uninduced K562 cells or K562 cells induced with either butyrate or hemin (as indicated) was analyzed with γ-gene and Id2 probes. S14 served as the control.
Overexpression of Id2 in K562 cells induces γ-gene expression.
To directly test whether Id2 alone could induce γ-gene expression, we constructed an MSCV-based retroviral vector containing Id2 cDNA. This bicistronic vector contains the GFP cDNA linked by the encephalomyocarditis IRES to the Id2 cDNA tagged with an HA epitope (Fig. 5A). The K562 cells were transduced with an amphotropic retrovirus derived from this vector, and positive cells were selected by repeated fluorescence-activated cell sorter sorting for green fluorescence (Id2+ K562 cells). Cells transduced with the MSCV vector lacking the Id2 cDNA served as the control (Id2− K562 cells). Multiple pools of Id2+ and Id2− K562 cells were obtained and analyzed by Northern analysis. As seen in Fig. 5B, a significant increase in Id2 expression was observed in all pools transduced with the Id2-containing vector (mean, 15-fold). Reprobing of the Northern blot with a γ-gene-specific probe demonstrated a concomitant increase in expression of the γ-gene in all Id2+ K562 cell pools, compared with controls (mean, fourfold). No increase in SCL, GATA-1, or E12 expression was observed (data not shown). Similar results were also obtained in a second γ-globin-expressing cell line, HEL, with a threefold induction of γ-gene expression observed in Id2+ lines compared with controls (data not shown). Western analysis of the Id2+ K562 cell pools demonstrated Id2 protein expression in all pools (Fig. 5C). Northern blotting analyses to examine the direct effect of Id2 on ɛ- and β-gene expression were also performed. As seen in Fig. 5D, ɛ-gene expression was induced to a level similar to that of the γ-gene. No induction of β-gene expression was observed.
An E-box motif in HS2 is essential for Id2-dependent enhancement of γ-promoter activity.
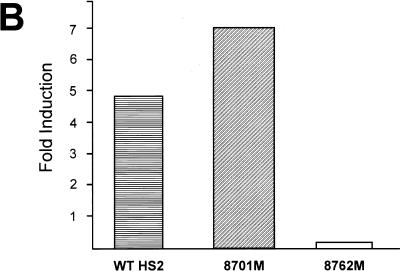
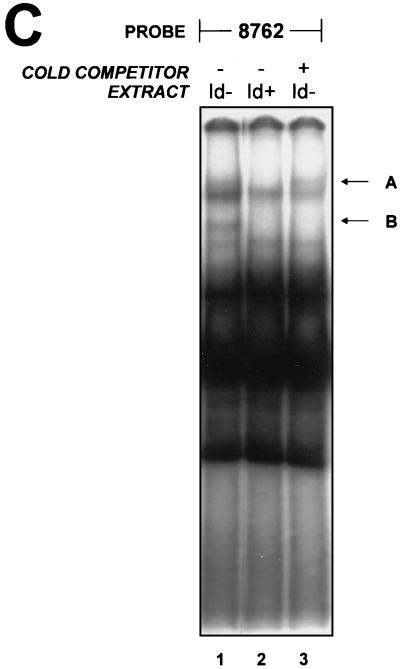
As Id2 functions as a dominant negative regulator of basic HLH (bHLH) proteins by preventing their binding to E-box motifs, we examined our initial screening construct (HS2-γ–luciferase) for likely sites of Id2 activity. Although no E-box elements in the globin promoters have been described, recent studies have documented the importance of conserved E-boxes in HS2 for enhancer function. These elements are located within the HS2 core at positions 8701 and 8762 (15, 24). Our observation that Id2 induced both ɛ- and γ-gene expression, but not α-globin expression, further suggested that HS2 may contain the sequences that mediate the Id2 effect. As a component of the LCR, HS2 is involved in the regulation of all the β-like genes but plays no role in α-globin induction. We therefore ascertained whether the HS2 E-box sequences were critical for Id2-dependent enhancement of γ-promoter inducibility. Id2+ and Id2− K562 cell pools were transfected with a construct containing wild-type HS2 linked to a γ-promoter–luciferase reporter gene hybrid or with constructs in which either the 8701 or 8762 E boxes in HS2 had been mutated and linked to the reporter (Fig. 6A). As shown in Fig. 6B, induction of γ-promoter activity was observed with both the wild-type HS2 and 8701 E-box mutant HS2. In contrast, mutation of the 8762 E-box motif reduced the Id2-dependent induction of γ-promoter activity. This effect was observed without loss of activity of the uninduced construct (reference 24 and data not shown). To examine these findings in the context of protein binding, an EMSA was performed by using the 8762 region as the probe. As seen in Fig. 6C (lane 1), a number of protein-DNA complexes were observed with extract derived from the Id2− K562 cells. These complexes were also observed in extract from Id2+ cells (lane 2) with the exception of complexes A and B, which were absent from the Id2+ K562 cell extract. Previous studies suggested that the presence of complex B may be due to binding of the bHLH protein USF (15). To confirm this, excess unlabelled oligonucleotide containing the USF consensus sequence was added to extract from the Id2− cells. As seen in lane 3, complex B was selectively disrupted by this process. Complex A was not reduced by the USF oligonucleotide and actually appeared to be enhanced in this setting.
FIG. 6.
An E box in HS2 mediates Id2 induction. (A) Diagrammatic representation of the constructs used for transfection into Id2+ and Id2− K562 cells. Wild-type HS2 contains the HindIII-XbaI fragment of HS2 linked to the −260 γ-promoter–luciferase gene hybrid. Conserved and protein binding sites within HS2 are labelled. Mutated sites are denoted by solid black circles. (B) Fold induction of the constructs depicted in panel A. The fold increase in luciferase levels between Id2− and Id2+ K562 cells is shown for each construct. Luciferase values were corrected for the protein concentration of the lysate. Values are the means of at least three separate transfections of more than 50 pools with at least two different plasmid preparations. (C) Binding of proteins to the 8762 E box. EMSA was conducted with nuclear extracts from Id2− (lanes 1 and 3) and Id2+ (lane 2) K562 cells. A USF probe, used as an unlabelled competitor, was added to lane 3 in 50-fold molar excess.
DISCUSSION
We have identified a role for the HLH transcription factor Id2 in the regulation of human globin gene expression. The conduit for this finding was a fungus-derived compound, OSI-2040, which induces γ-gene expression in K562 cells. This compound was defined in a high-throughput drug screen designed to isolate compounds that transcriptionally activate target genes. Based on this, we postulated that the subtraction of differentially expressed genes in OSI-2040-induced and uninduced K562 cells may identify novel factors that mediate the induction of γ-globin gene expression. A similar approach has been employed to identify genes responsive to a variety of stimuli (5, 9, 26). One advantage of our approach is that the starting material for subtraction is derived from identical K562 cells, which differ only in their exposures to OSI-2040. This is critical for minimizing the isolation of false-positive clones. When the powerful RDA subtraction strategy was used, Id2 was isolated as a transcription factor whose expression was coordinately up-regulated with the γ-gene in OSI-2040-induced cells. Although other differentially expressed genes were also identified, they appeared to involve signaling pathways or cell cycle regulation. These genes were presumably involved in drug activation pathways but were unlikely to be involved in the direct activation of the γ-promoter. The absence of γ-globin cDNA amplification in the RDA process was attributable to the addition of γ-globin cDNA to the driver amplicons at each subtraction step.
Our interest in Id2 as a potential globin regulatory protein was heightened by two observations further linking Id2 and γ-gene induction. Firstly, the fetal erythroid cell line, HEL, in which γ-gene induction was not initiated by OSI-2040, also did not express Id2. Secondly, sodium butyrate, a potent fetal-globin-inducing agent, also markedly induced Id2 expression in K562 cells. Sodium butyrate is thought to increase γ-globin expression through a direct transcriptional mechanism in concert with its activity as a histone deacetylase inhibitor (6, 30). Several investigators have localized putative sodium butyrate target sequences in the γ-promoter (30, 36). In addition, a small region of HS2 that includes the core NF-E2 sites has also been implicated in sodium butyrate response (40). Studies of the E-box region of HS2 with sodium butyrate have not been performed, and the demonstration of enhanced Id2 expression is also novel in this context. Interestingly, at the completion of these studies, structural analysis of OSI-2040 revealed that it was identical to a previously described fungal compound, apicidin, which also functions as a potent histone deacetylase inhibitor (13). The mechanism by which these histone-modifying agents induce Id2 expression was not addressed in our study. One possibility is that acetylation of histones in core nucleosomes assembled on the Id2 promoter results in enhanced transcription-factor binding to this region and in consequent gene activation (46). A similar mechanism could also occur on the γ- and ɛ-promoters, resulting in local perturbation of the chromatin structure, which, when combined with the effects of Id2 on protein binding to the LCR, could alter the interaction between enhancer-bound proteins and the promoter (43). Alternatively, the effect of Id2 on the LCR could be reflected in the up-regulation of γ- and ɛ-globin expression, as these genes are permissive in the embryonic and fetal environments of the K562 cell.
In contrast to OSI-2040 and sodium butyrate, hemin induction of γ-gene expression did not involve up-regulation of Id2. This finding suggests that induction of erythroid-cell differentiation alone is insufficient for increasing the levels of Id2. In support of this, observations of cultured human hematopoietic progenitors demonstrate that Id2 expression is down-regulated at the onset of erythroid-cell differentiation (10). Subsequent up-regulation of Id2 occurs only during late erythroid-cell maturation (10, 11).
Conversely, OSI-2040-induced K562 cells and Id2+ K562 cells did not exhibit other features of erythroid-cell differentiation, with no observed increase in glycophorin A, SCL, or GATA-1 levels. These findings are similar to those of previous studies, which demonstrated that enforced overexpression of Id genes inhibits differentiation of a variety of hematopoietic cell lineages (23, 28, 45). Interestingly, addition of Id2 antisense oligonucleotides to human progenitor cells significantly increases erythroid-cell colony formation, suggesting that Id2 has a negative role in erythroid-cell differentiation (10). This finding further strengthens our hypothesis that the increase in embryonic and fetal globin expression is due to a direct transcriptional effect of Id2, rather than to an effect mediated by influences on the erythroid-cell differentiation program.
The transcriptional effects of Id2, a member of the HLH protein family, are thought to be secondary to sequestration of the closely related bHLH family of proteins (4, 44). The bHLH family consists of numerous ubiquitous (E12, E47, and USF) and cell-specific (SCL and MyoD) factors involved in cell fate and differentiation (3, 25, 31, 32). DNA binding of these factors occurs through the formation of obligate homo- or heterodimers on a consensus E-box motif (18, 47). Id2 lacks the basic amino acid domain of the bHLH proteins which is essential for DNA binding, and hence heterodimers of Id2 and bHLH proteins fail to assemble on E-box sites (4). Recent studies have examined the role of E boxes in HS2 in the induction of fetal and embryonic globins (15, 24). They have identified two E-box sequences at positions 8701 and 8762 in the core region of HS2, which are conserved between species. As the original construct used in the screen for OSI-2040 contained HS2 linked to the −260 γ-promoter and as extensive protein binding studies of the γ-promoter have failed to identify bHLH binding sites, we postulated that the effects of Id2 may be mediated, in part, through the E boxes in HS2. The induction of ɛ-gene expression observed with OSI-2040 and the lack of α-gene induction further supported a role for HS2 in this process. Our functional studies suggest that Id2 induction of γ-promoter activity is dependent on the presence of an intact E-box motif at position 8762 in HS2. Although mutation of this site does little to affect the enhancer activity of HS2 with either γ- (our studies and reference 24) or ɛ-promoters (15) in wild-type cells, in Id2+ cells induction of the γ-promoter is lost. One explanation of this finding is that a balance of positive and negative regulators exists at the 8762 site. Mutation of the site ablates binding of both of these classes of factors, and thus, no net effect on enhancer activity is observed (15). In the presence of increased Id2, the balance is altered by the selective sequestration of a negative regulator, thereby mediating increased enhancer activity. Examination of the protein binding data with the 8762 E-box probe supports this hypothesis. Two complexes (A and B) are significantly altered in the Id2+ K562 cells compared to wild-type cells (Fig. 6C). While the identity of complex A remains unknown, complex B contains USF, which has recently been shown to act as a negative regulator in a highly functionally analogous enhancer system. Studies of the immunoglobulin heavy-chain enhancer demonstrate that USF functions as a negative regulator by competing with other bHLH proteins for occupancy of a key E-box element (7). As USF lacks a potent trans-activation domain, its binding to the enhancer E box creates a dominant negative effect. Although homodimeric USF is not one of the bHLH complexes known to be regulated by Id2 (44), the binding of USF to the 8762 site appears to be heterodimeric (15). Thus, the displacement of this protein complex may be mediated by Id2 interacting with the heterodimeric component of the complex. Although no new activator complex is observed with the 8762 probe with the displacement of USF binding, EMSA is performed in the presence of vast probe excess. Therefore, any of the existing complexes defined by Elnitski et al. could play the role of unopposed transcriptional activator (15). The ability of Id2 to stimulate gene transcription has also been examined in studies of the low-affinity nerve growth factor receptor (p75LNGFR) gene promoter. Binding of the bHLH factor ME1a to an E box in the LNGFR promoter specifically represses promoter activity in a variety of cell lines, including PC12, a pheochromocytoma line. Coexpression of Id2 and ME1a in PC12 results in the formation of a non-DNA binding heterodimer with alleviation of ME1a-dependent repression. In this context, promoter activation is achieved through other E-box binding proteins which are unaffected by Id2 (8). These findings are similar to our own and demonstrate that the Id-like transcriptional regulators function as either repressors or activators, depending on the sequence context and on the balance of bHLH proteins which act on a specific site.
Although Id2 is capable of enhancing embryonic and fetal globin expression in cells in which the genes are normally transcribed, it remains to be determined whether Id2 can exert the same effect on transcriptionally inactive ɛ- and γ-genes. Our results, demonstrating a failure of Id2 to induce β-gene expression in K562 cells, suggest that only target promoters that are transcriptionally active will be responsive. Thus, the effects of Id2 are not truly developmentally specific but are dependent on the transcriptional environment of potential target genes. Nevertheless, induction of Id2 expression in the context of active fetal globin expression in primary human cord blood progenitors may generate an HPFH phenotype in which γ-gene expression is unable to be silenced. Ongoing studies will determine the potential of Id2 for genetic therapy approaches to the β-hemoglobinopathies.
ACKNOWLEDGMENTS
We thank Ross Hardison and Robert Hawley for the gift of plasmids and A. W. Nienhuis for continuing support. We thank Adam Chapman for technical assistance. We also thank Ping Cai for natural product purification and Cedric Pieric for fungal fermentation support.
This work was supported by the NHMRC of Australia, The Wellcome Trust (S.M.J.), NIH grant PO1 HL53749-03, Cancer Center Support CORE grant P30 CA 21765, and the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Amrolia P J, Cunningham J M, Jane S M. Maximal activity of an erythroid-specific enhancer requires the presence of specific protein binding sites in linked promoters. J Biol Chem. 1998;273:13593–13598. doi: 10.1074/jbc.273.22.13593. [DOI] [PubMed] [Google Scholar]
- 2.Amrolia P J, Cunningham J M, Ney P, Nienhuis A W, Jane S M. Identification of two novel regulatory elements within the 5′-untranslated region of the human Ag-globin gene. J Biol Chem. 1995;270:12892–12898. doi: 10.1074/jbc.270.21.12892. [DOI] [PubMed] [Google Scholar]
- 3.Begley C G, Aplan P D, Denning S M, Haynes B F, Waldmann T A, Kirsch I R. The gene SCL is expressed during early hematopoiesis and encodes a differentiation-related DNA-binding motif. Proc Natl Acad Sci USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benezra R, Davis R L, Lockshon D, Turner D L, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 5.Braun B S, Frieden R, Lessnick S L, May W A, Denny C T. Identification of target genes for the Ewing’s sarcoma EWS/FLI fusion protein by representational difference analysis. Mol Cell Biol. 1995;15:4623–4630. doi: 10.1128/mcb.15.8.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Candido E P, Reeves R, Davie J R. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- 7.Carter R S, Ordentlich P, Kadesch T. Selective utilization of basic helix-loop-helix–leucine zipper proteins at the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1997;17:18–23. doi: 10.1128/mcb.17.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiaramello A, Neuman K, Palm K, Metsis M, Neuman T. Helix-loop-helix transcription factors mediate activation and repression of the p75LNGFR gene. Mol Cell Biol. 1995;15:6036–6044. doi: 10.1128/mcb.15.11.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu C C, Paul W E. FIG1, an interleukin 4 induced mouse B cell gene isolated by cDNA representational difference analysis. Proc Natl Acad Sci USA. 1997;94:2507–2512. doi: 10.1073/pnas.94.6.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condorelli G, Vitelli L, Valtieri M, Marta I, Montesoro E, Lulli V, Baer R, Peschle C. Coordinate expression and developmental role of Id2 protein and TAL1/E2A heterodimer in erythroid progenitor differentiation. Blood. 1995;86:164–175. [PubMed] [Google Scholar]
- 11.Cooper C L, Brady G, Bilia F, Iscove N N, Quesenberry P J. Expression of the Id family of helix-loop-helix regulators during growth and development in the hematopoietic system. Blood. 1997;89:3155–3165. [PubMed] [Google Scholar]
- 12.Craig J E, Rochette J, Sampietro M, Wilkie A O M, Barnetson R, Hatton C S R, Demenias F, Thein S L. Genetic heterogeneity in heterocellular hereditary persistence of fetal hemoglobin. Blood. 1997;90:428–434. [PubMed] [Google Scholar]
- 13.Darkin-Rattray S J, Gurnett A M, Myers R W, Dulski P M, Crumley T M, Allocco J J, Cannova C, Meinke P T, Colletti S L, Poliskook J D, Schmatz D M. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc Natl Acad Sci USA. 1996;93:13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Economou E P, Antonarakis S E, Kazazian H H, Jr, Serjeant G R, Dover G J. Variation in hemoglobin F production among normal and sickle cell adults is not related to nucleotide substitutions in the gamma promoter regions. Blood. 1991;77:174–177. [PubMed] [Google Scholar]
- 15.Elnitski L, Millers W, Hardison R. Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the b-globin locus control region. J Biol Chem. 1997;272:369–378. doi: 10.1074/jbc.272.1.369. [DOI] [PubMed] [Google Scholar]
- 16.Fibach E, Prasanna P, Rodgers G P, Samid D. Enhanced fetal hemoglobin production by phenylacetate and 4-phenylbutyrate in erythroid precursors derived from normal donors and patients with sickle cell anemia and beta-thalassemia. Blood. 1993;82:2203–2209. [PubMed] [Google Scholar]
- 17.Hawley R G, Lieu F H L, Fong A Z C, Hawley T S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 18.Hsu H-L, Huang L, Tsan J T, Funk W, Wright W E, Hu J-S, Kingston R E, Baer R. Preferred sequences for DNA recognition by the TAL1 helix-loop-helix proteins. Mol Cell Biol. 1994;14:1256–1265. doi: 10.1128/mcb.14.2.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubank M, Schatz D G. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jane S M, Cunningham J M. Molecular mechanisms of hemoglobin switching. Int J Biochem Cell Biol. 1996;28:1197–1209. doi: 10.1016/s1357-2725(96)00058-1. [DOI] [PubMed] [Google Scholar]
- 21.Jane S M, Ney P A, Vanin E F, Gumucio D L, Nienhuis A W. Identification of a stage selector element in the human g-globin gene promoter that fosters preferential interaction with the 5′ HS2 enhancer when in competition with the b-promoter. EMBO J. 1992;11:2961–2969. doi: 10.1002/j.1460-2075.1992.tb05366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jane S M, Nienhuis A W, Cunningham J M. Hemoglobin switching in man and chicken is mediated by a heteromeric complex between the ubiquitous transcription factor CP2 and a developmentally specific protein. EMBO J. 1995;14:97–105. doi: 10.1002/j.1460-2075.1995.tb06979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreider B, Benezra R, Rovera G, Kadesh T. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science. 1992;255:1700–1702. doi: 10.1126/science.1372755. [DOI] [PubMed] [Google Scholar]
- 24.Lam L T, Bresnick E H. A novel DNA-binding protein, HS2NF5, interacts with a functionally important sequence of the human b-globin locus control region. J Biol Chem. 1996;271:32421–32429. doi: 10.1074/jbc.271.50.32421. [DOI] [PubMed] [Google Scholar]
- 25.Lasser A B, Buskin J N, Lockshon D, David R L, Apone S, Hauschka S D, Weintraub H. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatinine kinase enhancer. Cell. 1989;58:823–831. doi: 10.1016/0092-8674(89)90935-5. [DOI] [PubMed] [Google Scholar]
- 26.Lewis B C, Shim H, Li Q, Wu C S, Lee L A, Maity A, Dang C V. Identification of putative c-Myc-responsive genes: characterization of rcl, a novel growth-related gene. Mol Cell Biol. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 28.Lister J, Forrester W C, Baron M H. Inhibition of an erythroid differentiation switch by the helix-loop-helix protein Id1. J Biol Chem. 1995;270:17939–17946. doi: 10.1074/jbc.270.30.17939. [DOI] [PubMed] [Google Scholar]
- 29.Lozzio B B, Lozzio C B. Properties and usefulness of the original K-562 human myelogenous leukemia cell line. Leuk Res. 1979;3:363–370. doi: 10.1016/0145-2126(79)90033-x. [DOI] [PubMed] [Google Scholar]
- 30.McCaffrey P G, Newsome D A, Fibach E, Yoshida M, Su M S-S. Induction of g-globin by histone deacetylase inhibitors. Blood. 1997;90:2075–2083. [PubMed] [Google Scholar]
- 31.Murre C, McCaw P S, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 32.Murre C, McCaw P S, Vaessin H, Caudy M, Jan L Y, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Weintraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 33.Noguchi C T, Rodgers G P, Serjeant G, Schechter A N. Levels of fetal hemoglobin necessary for treatment of sickle cell disease. N Engl J Med. 1988;318:96–99. doi: 10.1056/NEJM198801143180207. [DOI] [PubMed] [Google Scholar]
- 34.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective hematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 35.Orkin S H. Regulation of globin gene expression in erythroid cells. Eur J Biochem. 1995;231:271–281. doi: 10.1111/j.1432-1033.1995.tb20697.x. [DOI] [PubMed] [Google Scholar]
- 36.Pace B S, Li Q, Stamatoyannopoulos G. In vivo search for butyrate responsive sequences using transgenic mice carrying A gamma gene promoter mutants. Blood. 1996;88:1079–1084. [PubMed] [Google Scholar]
- 37.Perkins A C, Sharpe A H, Orkin S H. Lethal b-thalassemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 38.Persons D A, Allay J A, Allay E R, Smeyne R J, Ashmun R A, Sorrentino B P, Nienhuis A W. Retroviral-mediated transfer of the green fluorescent protein gene into murine hematopoietic cells facilitates scoring and selection of transduced progenitors in vitro and identification of genetically modified cells in vivo. Blood. 1997;90:1777–1786. [PubMed] [Google Scholar]
- 39.Poncz M P, Henthorn P, Stoekert C, Surrey S. Globin gene expression in hereditary persistence of fetal hemoglobin and δβo-thalassemia. In: McLean N, editor. Oxford surveys of eukaryotic genes. Oxford, United Kingdom: Oxford University Press; 1989. pp. 163–203. [PubMed] [Google Scholar]
- 40.Safaya S, Ibrahim A, Rieder R F. Augmentation of g-globin gene promoter activity by carboxylic acids and components of the human b-globin locus control region. Blood. 1994;84:3929–3935. [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Stamatoyannopoulos G, Nienhuis A W. Hemoglobin switching. In: Stamatoyannopoulos G, Nienhuis A W, Majerus P W, Varmus H, editors. The molecular basis of blood diseases. W. B. Philadelphia, Pa: Saunders; 1994. pp. 67–105. [Google Scholar]
- 43.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 44.Sun X-H, Copeland N G, Jenkins N A, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991;11:5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun X-H. Constitutive expression of the Id1 gene impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 46.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 47.Wright W E, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol Cell Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]