Abstract
Background
Candida haemulonii complex-related species are pathogenic yeasts closely related to Candida auris with intrinsic antifungal resistance, but few epidemiological data are available.
Methodology/Principal findings
We analyzed clinical and demographic characteristics of patients with fungemia due to C. haemulonii complex and related species (C. pseudohaemulonii, C. vulturna) reported in France during 2002–2021, and compared them to data of C. parapsilosis fungemia, as they all can be commensal of the skin. We also conducted a study on adult inpatients and outpatients colonized by C. haemulonii complex, managed at the University Hospital of Martinique during 2014–2020. Finally, we performed a literature review of fungemia due to C. haemulonii complex and related species reported in Medline (1962–2022).
In total, we identified 28 fungemia due to C. haemulonii complex in France. These episodes were frequently associated with bacterial infection (38%) and high mortality rate (44%), and differed from C. parapsilosis fungemia by their tropical origin, mainly from Caribbean and Latin America. All isolates showed decreased in vitro susceptibility to amphotericin B and fluconazole. In Martinique, we found that skin colonization was frequent in the community population, while colonization was strongly associated with the presence of foreign devices in ICU patients. The literature review identified 274 fungemia episodes, of which 56 were individually described. As in our national series, published cases originated mainly from tropical regions and exhibited high crude mortality.
Conclusions/Significance
Multidrug-resistant C. haemulonii complex-related species are responsible for fungemia and colonization in community and hospital settings, especially in tropical regions, warranting closer epidemiological surveillance to prevent a potential C. auris-like threat.
Author summary
Yeasts of the C. haemulonii complex (C. haemulonii sensu stricto, C. duobushaemulonii, C. haemulonii var. vulnera) and related species (C. pseudohaemulonii and C. vulturna), are phylogenetically close to Candida auris, but their distribution, pathophysiology and antifungal resistance profiles are poorly known. This work provides the first epidemiological data on these yeasts in France. Fungemia caused by these yeasts were mainly identified in tropical overseas regions (French West Indies, French Guiana), and occurred in patients with risk factors for candidemia, particularly a cutaneous portal of entry. This work highlights the skin carriage of these yeasts in these tropical regions, and their ability to colonize foreign devices. Like for Candida parapsilosis, catheters are the main pathway for fungemia but mortality seems higher with yeasts of the C. haemulonii complex and related. They commonly show in vitro resistance to many antifungal agents, notably fluconazole and amphotericin B, which are still frequently used as first and second-line treatments for candidemia worldwide. The findings from the literature review are consistent with these overall results. These observations justify closer epidemiological surveillance in the concerned regions to prevent a potential C. auris-like threat.
Introduction
Since the 2010s [1], multidrug-resistant Candida auris [2], with its ability to survive on prosthetic materials and spread among patients, has become a spreading healthcare-associated fungus [3–5]. Within the Metschnikowiaceae clade, yeasts of the Candida haemulonii complex (C. haemulonii sensu stricto, C. haemulonii var. vulnera, and C. duobushaemulonii), C. pseudohaemulonii, and C. vulturna are phylogenetically closely related to C. auris and share several pathogenicity-related traits, like adhesion on prosthetic materials [6,7] phenotypic switching [8], and multidrug resistance [7,9–11]. In fact, misidentifications by biochemical methods are frequent, even with updated databases [12]. Few cases of C. haemulonii complex infection have been reported since its description [13,14], but since the 2000s, several fungemia have been reported, especially in tropical areas [10,15–17]. These yeasts present often high MICs for fluconazole [18], the first-line antifungal for treating fungemia in many low-income countries, and exhibit a decreased susceptibility to amphotericin B, often used as salvage therapy. Isolates of this complex are usually susceptible to echinocandins, although echinocandin-resistant strains have been isolated from human samples [19].
In France, rare yeasts are responsible for nearly 7.4% of fungemia [20] but there are no major epidemiological studies focused on this complex. Consequently, risk factors and clinical outcome of fungemia due to Candida haemulonii complex-related species remain largely unknown [21].
In this context, we carried out a study in mainland France and French overseas territories including (i) epidemiology of fungemia caused by C. haemulonii complex, C. pseudohaemulonii and C. vulturna (hereinafter referred to as “fungemia series”), (ii) a case-control study comparing these fungemia to those due to C. parapsilosis, a skin commensal, and (iii) a study of colonization in outpatients and inpatients admitted to intensive care units (ICU) in Martinique. Finally, we conducted a literature review on fungemia caused by these species during the past six decades.
Methods
Ethics statement
The fungemia series was carried out in compliance with French law and the declaration of Helsinki (as adopted in 2000) and was approved by the French Mycoses Study Group scientific council. The surveillance of the NRCMA was approved by the Institut Pasteur Institutional Review Board #1 (#2009–34/IRB) and the "Commission Nationale de l’Informatique et des Libertés" according to the French regulation. The study in Martinique has been approved by the IRB of the Martinique university hospital (#2020/064).
Epidemiology of fungemia in France
We collected clinical data related to fungemia caused by C. haemulonii complex, C. pseudohaemulonii and C. vulturna from two French databases, from their creation until 31st July 2021. Both programs were launched by the French National Reference Centre for Invasive Mycoses & Antifungals (NRCMA): the YEASTS surveillance program, initiated in 2002 with the participation of 27 university or cancer hospitals in the Paris area [22] and the French Surveillance Network of Invasive Fungal Infections (RESSIF) program, initiated in 2012 with the participation of 29 hospitals from mainland France and overseas [20].
Case-control study
The above-described series constitutes the case population. We chose fungemia caused by C. parapsilosis from the RESSIF database as the control population, rather than C. albicans because preliminary information revealed the presence of these study species on human skin.
Colonization in Martinique
We enrolled patients with sample containing C. haemulonii complex yeasts, considered to be non-pathogenic (i.e. not being responsible for infection), at the Martinique University hospital, between April 2014 and August 2020. We retained cases involving adult patients with putative community-acquired colonization (referred to as “outpatients”, including those hospitalized <48h) or hospitalized in ICU. Lack of pathogenicity was retained by the clinician according to clinical context and source of sampling.
Literature review
We performed a literature review on Medline database, from January 1st, 1962 to September 1st, 2022 with the following search terms in all fields: “Candida haemulonii” OR “Torulopsis haemulonii” OR “Candida duobushaemulonii” OR “Candida pseudohaemulonii” OR “Candida vulturna”. We included all reported cases of C. haemulonii complex, C. pseudohaemulonii, and C. vulturna fungemia identified by sequencing, for which individual clinical information and/or susceptibility to antifungals were available.
Isolate characterization
Fungemia series
All bloodstream isolates of C. haemulonii complex and related species were identified at the NRCMA by sequencing of ITS1–5.8S–ITS2 and D1/D2 regions of the ribosomal DNA, using V9D/LS266 and NL1/NL4 primers as previously described [23]. Antifungal susceptibility testing to micafungin, amphotericin B, and fluconazole, was performed using the broth microdilution method published by EUCAST (v 7.3.2 valid from 22 April, 2020), with a modification for micafungin medium as previously described [24].
Colonization in Martinique
From 2014 to 2017, yeasts were identified by culture on chromogenic media chromID Candida agar (BioMerieux, Marcy l’Étoile, France) combined with carbon assimilation testing using the API ID32C system (BioMerieux, Marcy l’Étoile, France). As no API code matches the complex, strains identification was made by matching profiles with those of an internal database of isolates previously identified by sequencing. Furthermore, carbon assimilation identification does not distinguish species within the complex, isolates were identified as C. haemulonii complex-related yeasts.
Since 2017, species identification has been performed routinely by matrix-assisted laser desorption/ionization time of flight mass spectroscopy (MALDI-TOF MS) using the Biotyper system (Bruker Daltonics, Billerica, MA, USA). Nine strains isolated before 2017 were rechecked, showing no discrepancy with biochemical analysis (S1 Table). Nonetheless, since the distinction between C. haemulonii sensu stricto and C. haemulonii var. vulnera is currently not possible without sequencing [25,26], such isolates only identified by MALDI-TOF MS were considered as C. haemulonii sensu lato or C. duobushaemulonii.
Statistical analysis
Analyses were performed on isolates from single episodes of fungemia, each isolate corresponding to a single patient. Associated factors were studied according to the species. We selected the variables significantly associated with any of the two groups at the threshold p<0.20. We developed an explanatory model using logistic regression, with a manual stepwise procedure guided by the Akaike Information Criterion. Explanatory model results were reported as adjusted odds ratios (aOR) with 95% confidence intervals (95%CI). Data were analyzed using R software (version 4.0.2). The map was created from a fla-shop.com background map (https://www.fla-shop.com/svg/, CC BY 4.0 license) modified with inkscape software.
Results
Fungemia series
Between October 1st, 2002, and July 31st, 2021, among the 12 032 candidemia episodes reported in YEASTS and RESSIF, 28 (0.23%) were caused by C. haemulonii complex, C. pseudohaemulonii, or C. vulturna (Table 1). The incidence appears to increase over time (S1 Fig), with 15/28 (53.6%) cases reported between 2017 and 2021. Notifications from the French West Indies and French Guiana accounted for 71.4% (20/28) of all cases.
Table 1. Characteristics of patients with fungemia due to C. haemulonii complex, C. pseudohaemulonii and C. vulturna in France between 2002 and 2021 (YEASTS and RESSIF programs), and C. parapsilosis in France between 2012 and 2021 (RESSIF).
| Characteristics | C. haemulonii complex-related species | C. parapsilosis |
|---|---|---|
| Total | 28 | 942 |
| Hospital (%) | ||
| Martinique university hospital (FWI) | 10 (35.7) | 62 (6.6) |
| Cayenne hospital (French Guiana) | 9 (32.1) | 6 (0.6) |
| Guadeloupe university hospital (FWI) | 1 (3.6) | 1 (0.1) |
| Paris aera hospitals | 5 (17.9) | 200 (21.2) |
| Other hospitals in mainland France | 3 (10.7) | 673 (71.4) |
| Birth continent (%) | ||
| Africa | 1/20 (5.0) | 35/456 (7.6) |
| America | 18/20 (90.0) | 62/456 (13.5) |
| Europe | 1/20 (5.0) | 359/456 (78.2) |
| Travels during the previous year (%) | ||
| Africa | 2/22 (9.1) | 14/216 (6.5) |
| Latin America | 2/22 (9.1) | 1/216 (0.5) |
| West Indies | 4/22 (18.2) | 48/216 (22.2) |
| None | 14/22 (63.6) | 148/216 (68.5) |
| Median age in years [IQR] | 67 [46–72] | 62 [46–72] |
| Male (%) | 16 (57.1) | 625 (66.3) |
| Solid cancer (%) | 8/24 (33.3) | 281 (29.8) |
| Hematological malignancy (%) | 5/26 (19.2) | 118 (12.5) |
| Chronic kidney failure (%) | 3/24 (12.5) | 102 (10.8) |
| Liver cirrhosis (%) | 2/24 (8.3) | 43 (4.6) |
| Diabetes mellitus (%) | 3/24 (12.5) | 129 (13.7) |
| HIV (%) | 2/23 (8.7) | 13/412 (3.2) |
| Corticosteroidsa (%) | 2 (7.1) | 85 (9.0) |
| Other immunosuppressive treatment (%) | 4 (14.3) | 200 (21.2) |
| Surgery during the previous month (%) | 10/26 (38.5) | 358 (38.0) |
| Exposure to antifungal agent in the previous month (%) | ||
| Amphotericin B | 1/24 (4.2) | 11/916 (1.2) |
| Echinocandin | 0 (0.0) | 88/916 (9.6) |
| Fluconazole | 1/24 (4.2) | 9/916 (1.0) |
| Voriconazole | 2/24 (8.3) | 7/916 (0.8) |
| Posaconazole | 0 (0.0) | 1/916 (0.1) |
| Isavuconazole | 0 (0.0) | 5/916 (0.5) |
| None | 20/24 (83.3) | 804/916 (87.7) |
| Central venous catheter (%) | 15/22 (68.2) | 693/753 (92.6) |
| Context of bacterial infection (%) | 11/24 (45.8) | 180 (19.1) |
| Intensive Care Unit (%) | 10/26 (38.5) | 315 (33.4) |
| Shock (%) | 3/24 (12.5) | 52 (5.5) |
| Antifungal treatment (%) | ||
| Amphotericin B | 0/26 (0.0) | 46/922 (5.0) |
| Echinocandin | 15/26 (58.7) | 472/922 (51.2) |
| Fluconazole | 3/26 (11.5) | 313/922 (33.9) |
| Itraconazole | 1/26 (3.8) | 0/922 (0.0) |
| Voriconazole | 0/26 (0.0) | 16/922 (1.7) |
| Posaconazole | 0/26 (0.0) | 3/922 (0.3) |
| None | 7/26 (26.9) | 86/922 (9.3) |
| All-cause mortality at 3 months (%) | 11/25 (44.0) | 192/757 (25.4) |
| Median time to death in days [IQR] | 18 [7–41] | 8 [2–18] |
FWI: French West Indies, IQR: Interquartile Range
a≥ 0.3mg/kg for ≥ 1 month
For each case, demographics, underlying conditions, initial antifungal treatment, and 3-month survival status were collected using a standardized questionnaire through a secure website.
The date of the fungemia corresponded to the date of the first positive blood culture.
Surgery and antifungal pre-exposure were considered in the 30 days prior to fungemia.
Subjects were mostly men (16, 57.1%) and the median age was 67 years (Interquartile Range (IQR) = 46–72); 81.4% of them lived or had stayed in tropical regions. Twenty-six patients (92.9%) had at least one of the following comorbidities: solid cancer, hematological malignancy, recent surgical procedure, central venous catheter, context of bacterial infection, hospitalization in ICU; Nine of them (32.1%) presented at least three of these comorbidities.
Four distinct species were identified. Candida haemulonii sensu stricto was the dominant species (16, 57.1%), followed by C. duobushaemulonii (8, 28.6%) and C. vulturna (3, 10.7%). C. pseudohaemulonii was identified once (1, 3.6%), whereas no C. haemulonii var. vulnera was retrieved. The distribution of species varied according to age, with a median age of 67 and 70 years for C. haemulonii and C. duobushaemulonii, respectively, compared with 46 and 27 years for C. pseudohaemulonii and C. vulturna, respectively.
Among the three patients who presented fungemia due to C. vulturna, two had a history of cancer and were pre-exposed to voriconazole, and 2 had recent surgery. The only C. pseudohaemulonii episode occurred in a 46-year-old woman from Brazil who was hospitalized in French Guiana for gastric linitis. She had no bacterial co-infection or recent surgery.
All-cause mortality at 3 months after diagnosis was 44% (11/25), of which 7 (63%) occurred in the first 30 days and 5 (45%) in the first 15 days. Mortality increased with age, from 3 (27.2%) in the <67 age group to 57.1% in the ≥67 age group. Two patients survived despite the absence of antifungal treatment. They were under 30 years old, had no underlying disease, and benefited from prompt catheter replacement.
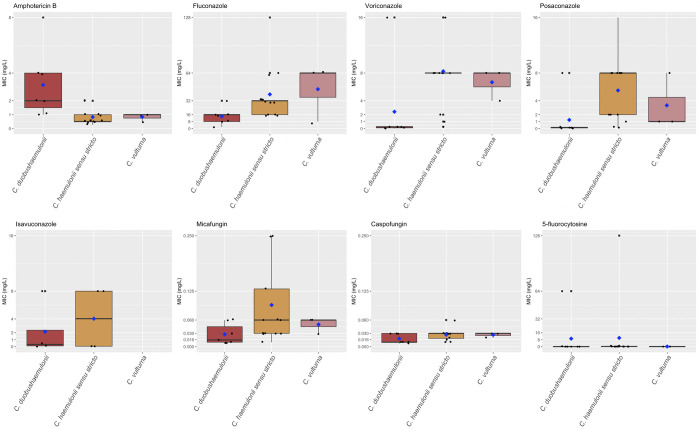
Antifungal susceptibility testing could be performed on 23/28 strains. The median MIC values for amphotericin B are high, especially for C. duobushaemulonii (2 mg/l). Opposingly, those for fluconazole were lower for C. duobushaemulonii, yet still high (16 mg/l). The median MIC values for micafungin were ≤0.250 mg/l for all strains (Fig 1).
Fig 1. Susceptibility profiles to antifungals of the strains of the French fungemia series.
By EUCAST broth micro dilution reference method. C. duobushaemulonii = 7, C. haemulonii sensu stricto = 13, C. vulturna = 3. Bold black bar: median MIC value; Blue diamond: mean MIC value.
Case-control study
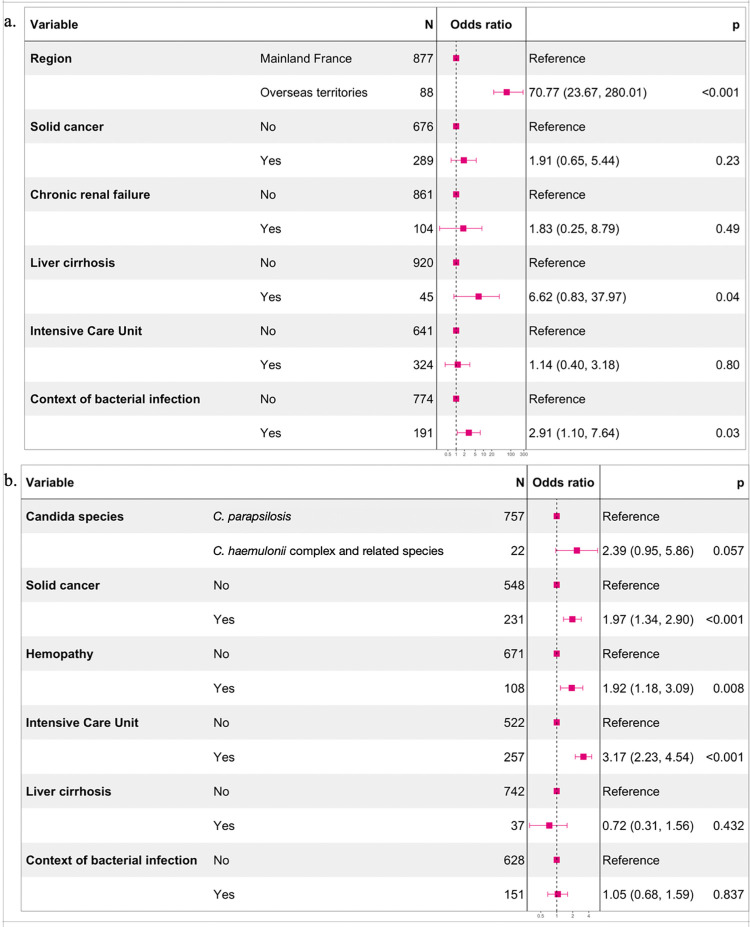
Between 2012 and 2021, 942 episodes of C. parapsilosis fungemia in adults were declared within the RESSIF database (Table 1). The geographical location of the patient in overseas territories was significantly associated with the development of C. haemulonii complex-related species fungemia (aOR = 70.77, 95%CI = 23.67–280.01), followed by the context of bacterial infection (aOR = 2.91, 95%CI = 1.10–7.64) (Fig 2A).
Fig 2. Explanatory models for the occurrence and the mortality of the fungemia.
a. Explanatory model for the occurrence of C. haemulonii complex, C. pseudohaemulonii or C. vulturna fungemia rather than C. parapsilosis (adjusted odds ratio from logistic regression coefficients with 95% confidence interval). b. Explanatory model for the death within 3 months of C. haemulonii complex, C. pseudohaemulonii, C. vulturna or C. parapsilosis fungemia (adjusted odds ratio from logistic regression coefficients with 95% confidence interval).
Three-month all-cause mortality was mainly associated with intensive care management and underlying comorbidities (Fig 2B). After adjusting for patient comorbidities, presence of bacterial infections, and level of care, fatal outcomes tended to be more frequent in C. haemulonii complex-related species fungemia than in C. parapsilosis fungemia (aOR = 2.39, 95%CI = 0.95–5.86).
Colonization in Martinique
We identified 119 C. haemulonii complex colonizations, in 116 adults with consultable medical records (S2 Fig). Of the 87 colonizations in outpatients (Table 2), 61 (70.1%) were cutaneous and 12 (13.8%) were ophthalmologic.
Table 2. Characteristics of patients with C. haemulonii complex colonization in university hospital of Martinique between 2014 and 2020.
| Characteristics | Outpatientsa | Patients in ICUb |
|---|---|---|
| Total of events | 87 | 17 |
| Mean age in years (SD) | 57.8 (16.7) | 62.2 (17.6) |
| Male (%) | 34 (39.1) | 13 (76.5) |
| Birth continent (%) | ||
| America | 81 (93.1) | 15 (88.2) |
| Europe | 6 (6.9) | 2 (11.8) |
| Residence department (%) | ||
| Côtes-d’Armor (Mainland France) | 1 (1.1) | 0 (0.0) |
| Guadeloupe (FWI) | 0 (0.0) | 2 (11.8) |
| Martinique (FWI) | 86 (98.9) | 15 (88.2) |
| Profession (%) | ||
| Catering | 3/77 (3.9) | 0/15 (0.0) |
| Construction | 5/77 (6.5) | 2/15 (13.3) |
| Education | 5/77 (6.5) | 0/15 (0.0) |
| Farmer | 1/77 (1.3) | 1/15 (6.7) |
| Gardener | 0 (0.0) | 1/15 (6.7) |
| Healthcare | 14 (18.2) | 0/15 (0.0) |
| Retired | 24/77 (31.2) | 2/15 (13.3) |
| Unemployed | 12/77 (15.6) | 3/15 (20.0) |
| Other | 13/77 (16.8) | 2/15 (13.3) |
| Travels (%) | 1/57 (1.8) | 0/13 (0.0) |
| Aquatic activity (%) | 2/21 (9.5) | 0/2 (0.0) |
| Gardening (%) | 6/21 (28.6) | 1/2 (50.0) |
| Mean BMI in Kg/m2 (SD) | 26.79 (7.6) | 27.4 (8.2) |
| Solid cancer (%) | 14 (16.1) | 1 (5.9) |
| Hematological malignancy (%) | 4 (4.6) | 1 (5.9) |
| Solid organ transplant (%) | 2 (2.3) | 0 (0.0) |
| Diabetes mellitus (%) | 21 (24.1) | 4 (23.5) |
| Corticosteroids (%) | 3 (3.4) | 2 (11.8) |
| Immunosuppressive treatment (%) | 9 (10.3) | 0 (0.0) |
| HIV (%) | 1 (1.1) | 1 (5.9) |
| Other immunodepression (%) | 9 (10.3) | 1 (5.9) |
| Surgery during the previous month (%) | 1 (1.1) | 4 (23.5) |
| Exposure to antifungal agent in the previous month (%) | 3 (3.4) | 6 (35.3) |
| Median length of stay in hospital when sampling in days [IQR] | 0.2 (0.5) | 38.1 (35.7) |
| Sampling site (%) | ||
| Ear, Nose, or Throat | 4 (4.6) | 1 (5.9) |
| Genital | 1 (1.1) | 0 (0.0) |
| Ophthalmologic | 12 (13.8) | 0 (0.0) |
| Respiratory tract | 9 (10.3) | 12 (70.6) |
| Pusc | 0 (0.0) | 2 (11.8) |
| Skin | 61 (70.1) | 1 (5.9) |
| Urine | 0 (0.0) | 1 (5.9) |
| Species (%)d | ||
| C. duobushaemulonii | 14 (16.1) | 7 (41.2) |
| C. haemulonii sensu lato | 33 (37.9) | 7 (41.2) |
| C. haemulonii sensu lato + C. duobushaemulonii | 7 (8.0) | 1 (5.9) |
| C. haemulonii complex | 33 (37.9) | 2 (11.8) |
SD: Standard deviation; FWI: French West Indies; IQR: Interquartile Range
a Samples collected during ambulatory care (<48 hours of hospitalization)
b Samples collected in ICU at least 48 hours after admission
c Superficial swabbing, considered as skin contaminant
d Identification accuracy depending on the method used (biochemical, MALDI-TOF MS)
Outpatients with skin colonization were predominantly women (41/61, 67.2%); the mean age was 61.3 years; all lived in Martinique and 24.6% (15/61) were unemployed. Their main comorbidities were diabetes mellitus (12, 19.7%), solid cancer (12, 19.7%), and immune deficiency or immunosuppressive treatment (6, 9.8% and 5, 8.2%), respectively. Species identification within the complex was possible in 68.9% (42/61) of cases: 32 (76.2%) of C. haemulonii, 17 (40.4%) of C. duobushaemulonii, and 7 (16.6%) harboring both species. Of note, C. parapsilosis was concomitantly identified in 27 (44.3%) of these samples. Three patients presented 2 positive samples more than 1 month apart, but we could not assert that they were the same species.
Patients with ophthalmologic colonization were mostly women (7/12, 58.3%); the mean age was 35 years; all lived in Martinique and presented no comorbidity, except wearing contact lenses (100%). All had keratitis or corneal abscess due to Pseudomonas aeruginosa or Fusarium solani complex.
The 17 subjects of the ICU group (Table 2) were mostly men (13/17, 76.5%), with a mean age of 62.3 years, living exclusively in the West Indies. The main comorbidities were diabetes mellitus (4, 23.5%), surgery in the previous month (4, 23.5%), and recent exposure to antifungals (6, 35.3%). The median length of hospitalization was 15 days. The most represented specimens were isolated from the respiratory tract (12, 70.6%), including 10 (83.3%) performed on intubated patients. C. haemulonii and C. duobushaemulonii were equally frequent, and one patient had mixed colonization. Notably, C. parapsilosis was also identified in 5 (29.4%) of these samples. Among the 12 patients screened for multi-drug resistant bacteria, 5 (41,6%) were positive. The crude mortality at 3 months was 47.1%.
Literature review
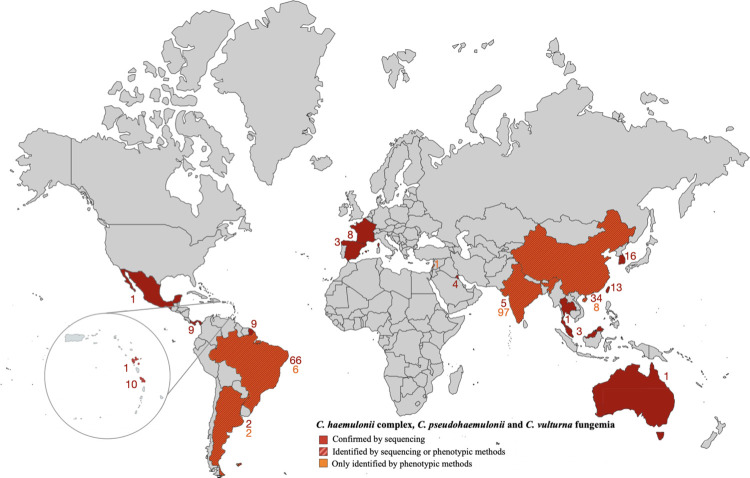
The search yielded 274 fungemia from 14 countries (Fig 3), of which 218 were excluded, mainly due to the lack of individual data (S3 Fig). Finally, 56 cases were selected; all occurred between 2006 and 2022. Cases originated mainly from tropical regions (44/56, 78,6%) (S2 Table). We individualized two age groups: children (n = 19) with a median age of 3.5 years (IQR = 0.1–5), and adults (n = 13), with a median age of 66 years (IQR = 52–82). There was no gender predominance in adults. As in our fungemia series, Candida haemulonii sensu stricto was the dominant species. Among adults, the distribution of species varied according to age, with a median age of 79 years for C. haemulonii sensu stricto compared to 56, 49, and 59 years for C. duobushaemulonii, C. haemulonii var. vulnera, and C. pseudohaemulonii, respectively. Age was known for only one case of C. vulturna (83 years).
Fig 3. World mapping of cases of fungemia due to C. haemulonii complex, C. pseudohaemulonii or C. vulturna.
According to our case series (obtained from the YEASTS program 2002–2021 and the RESSIF Network 2012–2021) and the literature review (Medline, 1962–2022). References are with the S3 Fig. Map created from fla-shop.com (https://www.fla-shop.com/svg/, CC BY 4.0 license) modified with inkscape software.
Although previous antifungal exposure was more frequent in the literature than in our series, the susceptibility profile was similar, with high MIC values for amphotericin B and fluconazole (S4 Fig). The median MIC values of amphotericin B were high, notably for C. pseudohaemulonii (32 mg/l) C. vulturna (8 mg/l), and C. duobushaemulonii (4 mg/l). The median MIC values of fluconazole were high, especially for the C. haemulonii complex yeasts (64 mg/l). Micafungin MIC values were 4 mg/l for 3 strains and ≤0.500 mg/l for all the others.
Among all cases, the crude mortality rate was high (5/12, 41,7%) and comparable to our fungemia series, although the time to death was not specified in most articles.
Discussion
We report a large series of fungemia and colonization caused by emerging multiple resistant yeasts of the C. haemulonii complex, or very closely related species (C. pseudohaemulonii and C. vulturna), and a case-control study with C. parapsilosis as reference. We thereby report the first cases of C. vulturna fungemia in South America and Europe since its description in South-East Asia in 2016 [11]. It is also the most important literature review with individual data of fungemia caused by these yeasts and molecularly proven to avoid misidentification.
Considering our series, these rare fungemia are particularly present in tropical regions. Indeed, among centers of the RESSIF and/or the YEASTS programs, only 9 reported C. haemulonii complex-related species fungemia; of these, 3 declarative centers were located in tropical regions, accounting for 20/28 (71.4%) of the whole series. Living in the tropics is by far the factor most associated with the occurrence of fungemia due to these yeasts rather than with C. parapsilosis.
In our series, 92.9% patients presented risk factors comparable to those of C. parapsilosis fungemia, which also corresponds to that observed elsewhere [27]. Contrastingly, pre-exposure to azoles was uncommon in our series (16.6%), although it is usually considered a significant contributor to non-albicans candidemia [28,29]. The high rate of central venous catheters (68.2%) and the absence of digestive surgical site infections suggest skin as the source for C. haemulonii complex-related species candidemia. This is also supported by our results on skin colonization and the ability of such pathogens to form biofilms on prosthetic materials [6,7].
Fungemia caused by C. haemulonii complex yeasts and related species seems to occur in patients at risk of skin-related fungemia, mainly in tropical regions. Case-control study with C. parapsilosis fungemia and the study of carriage, both in the community and in ICU in Martinique allow us to assume that these fungemia are frequent in tropical regions not because of specific host characteristics, but rather because of the burden causing by these yeasts in the environment, which favors carriage both in the outpatients and inpatients.
The broad antifungal resistance of C. haemulonii complex-related species makes them potentially difficult to treat. Although there are no clinical breakpoints or ECOFFs, the particularly high MIC values for amphotericin B, fluconazole, and to a lesser extent other azoles, suggest intrinsic resistance to these antifungals. Correct identification of C. haemulonii complex-related species is relevant not only for epidemiological surveillance but has also a direct therapeutic impact, with amphotericin B having a particularly low in vitro activity against C. duobushaemulonii and C. pseudohaemulonii isolates for instance [30]. Unlike some previously published studies [10,15,16], no death appeared to be attributable to treatment failure in our series, even for the 3 patients treated with fluconazole or itraconazole (whose strains had exceptionally low MIC values for fluconazole (≤16 mg/L).
All-cause mortality was similar between our series (44%) and the literature review (46%). As already shown for catheter-related candidemia, rapid control of the cutaneous portal of entry may play a critical role in preventing death [31,32].
In Martinique, and probably also in Latin America and other Caribbean territories, cutaneous colonization by C. haemulonii exists, especially outside the hospital setting. This is in contrast with its sibling species C. auris, which is often presented as a pure nosocomial yeast [3]. The three patients with positive skin specimens sampled more than one month apart allow us to suspect chronic colonization. In our study, apart from fungemia, no deep-seated infections were observed. Moreover, the fact that all patients with positive ophthalmologic samples wore contact lenses and that 10 of the 12 patients with respiratory colonization in ICU were intubated, suggests colonization of foreign devices [3].
Our study has several limitations. Missing data, especially on the oldest cases, affected our results. We were notably unable to retrieve the time-to-positivity of blood cultures, the confirmation of systematic catheter removal, and the time required for sterilization of blood cultures. Moreover, no skin fungal mapping was reported, precluding confirmation of the skin colonization hypothesis.
Nevertheless, according to our study, the presence of yeasts in a blood culture of a patient hospitalized or coming from a tropical region, especially if presenting a central veinous catheter, should suggest the diagnosis of C. haemulonii complex-related yeasts fungemia, as for Candida parapsilosis. Such a situation requires management of the infection’s portal of entry and reinforces the utility of probabilistic treatment with echinocandins. Despite the lack of specific clinical trials, there are some in vitro and animal data [33] as well as case reports of treatment failures with azoles or amphotericin B [10,15] that justify the use of echinocandins as first-line therapy. When rapid identification by MALDI-TOF MS is not available, the particular aspect of the cultures on chromogenic media might be sufficient to support the diagnosis [34]. This strategy could limit the delay before initiation of appropriate treatment, which is strongly correlated with mortality in candidemia [31,35]. Horizontal transmission capacity, especially in hospitals, has already been suggested by outbreaks involving the C. haemulonii complex [15,36]. The emergence of these yeasts is also supported at the genomic level: the characteristics shared by C. auris and the C. haemulonii complex-related species (synteny, similar gene family expansions) suggest similarities in the ecology and physiology of these species and the potential for them to also emerge as dangerous pathogens [37].
Historically described as warm sea yeasts [38], the C. haemulonii complex-related yeasts were later identified as saprophytes of tropical aquatic and terrestrial environment [39–41]—for instance, C. vulturna has been isolated from a tropical flower in the Philippines [11]—and as a potential pathogen for some animal species [9,42]. Some countries have recently reported cases in neonatology, suggesting the presence of these yeasts in artificial environments. Although we have no evidence in our series for an epidemic event, other authors have reported a series of clustered cases [15,30], with the same clone of C. pseudohaemulonii, raising the possibility of nosocomial transmission. A scenario of an emerging epidemic clone within these multi-drug resistant yeast populations is therefore quite credible [30], like the C. auris healthcare-associated outbreaks described since the 2010s.
Conclusion
Fungemia caused by C. haemulonii complex and related species yeasts are rarely observed in France and occur mainly in overseas tropical territories. These environmental yeasts colonize the skin of general population, as well as medical devices, favoring skin-related fungemia in patients with risk factors. These multi-drug resistant yeasts share numerous characteristics with C. auris, and deserve to be known and monitored.
Supporting information
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Members of the French Mycoses Study group who contributed to the data are in alphabetical order of the cities.
(PDF)
Data Availability
For data from the RESSIF and YEASTS networks The data used for this publication are considered as pseudonymized personal data. As such the GDPR as well as the French Law pertaining data protection apply to the use of this dataset. For data access request, a requester can contact: Pr Fanny Lanternier, head of NRCMA, cnrma@pasteur.fr The requester shall be informed that access to and reuse of this dataset will require the obtention of (i) an authorization from the French Supervisory Authority regarding data protection (the CNIL) and (ii) an ethics opinion from the competent French ethics committee (the CESREES) For data from the University Hospital of Martinique Access to the data of the University Hospital of Martinique can be requested from the institution's Data Protection Officer (DPO): dpo@chu-martinique.fr.
Funding Statement
This work was supported by Santé Publique France (UF, MDO,YLG, KS, RV, SP, TC, EM, AP, MN,NDG,OL) and Institut Pasteur (UF, MDO,YLG, KS, RV, SP, TC, EM, AP, MN,NDG, OL). The funders had no role in study design, data collection, analysis, or interpretation of data.
References
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009;53: 41–44. [DOI] [PubMed] [Google Scholar]
- 2.Chaabane F, Graf A, Jequier L, Coste AT. Review on antifungal resistance mechanisms in the emerging pathogen Candida auris. Front Microbiol. 2019;10: 2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabino R, Veríssimo C, Pereira ÁA, Antuness F. Candida auris, an agent of hospital-associated outbreaks: which challenging issues do we need to have in mind? Microorganisms. 2020;8. doi: 10.3390/microorganisms8020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Schalkwyk E, Mpembe RS, Thomas J, Shuping L, Ismail H, Lowman W, et al. Epidemiologic shift in candidemia driven by Candida auris, South Africa, 2016–2017. Emerg Infect Dis. 2019;25: 1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO fungal priority pathogens list to guide research, development and public health action. World Health Organization; 25 Oct 2022. Available: https://www.who.int/publications/i/item/9789240060241 [Google Scholar]
- 6.Ramos LS, Oliveira SSC, Souto XM, Branquinha MH, Santos ALS. Planktonic growth and biofilm formation profiles in Candida haemulonii species complex. Med Mycol. 2017;55: 785–789. [DOI] [PubMed] [Google Scholar]
- 7.Ramos LS, Oliveira SSC, Silva LN, Granato MQ, Gonçalves DS, Frases S, et al. Surface, adhesiveness and virulence aspects of Candida haemulonii species complex. Med Mycol. 2020;58: 973–986. [DOI] [PubMed] [Google Scholar]
- 8.Deng Y, Li S, Bing J, Liao W, Tao L. Phenotypic switching and filamentation in Candida haemulonii, an emerging opportunistic pathogen of humans. Microbiol Spectr. 2021;9: e0077921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, et al. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J Clin Microbiol. 2012;50: 3641–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim M, Shin JH, Sung H, Lee K, Kim E, Ryoo N, et al. Candida haemulonii and closely related species at 5 university hospitals in korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis. 2009. pp. e57–e61. doi: 10.1086/597108 [DOI] [PubMed] [Google Scholar]
- 11.Sipiczki M, Tap RM. Candida vulturna pro tempore sp. nov., a dimorphic yeast species related to the Candida haemulonis species complex isolated from flowers and clinical sample. Int J Syst Evol Microbiol. 2016;66. doi: 10.1099/ijsem.0.001302 [DOI] [PubMed] [Google Scholar]
- 12.Ambaraghassi G, Dufresne PJ, Dufresne SF, Vallières É, Muñoz JF, Cuomo CA, et al. Identification of Candida auris by use of the updated Vitek 2 yeast identification system, version 8.01: a multilaboratory evaluation study. J Clin Microbiol. 2019;57. doi: 10.1128/JCM.00884-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavarde V., Daniel F., Saez H., Arnold M., and Faguer B. Peritonite mycosique à Torulopsis haemulonii. Bull Soc Fr Mycol Med. 1984;13: 173–176. [Google Scholar]
- 14.Marjolet M. Torulopsis ernobii, Torulopsis haemulonii: levures opportunistes chez l’immunodéprimé? Bull Soc Fr Mycol Med. 1986;15: 143–146. [Google Scholar]
- 15.Khan ZU, Al-Sweih NA, Ahmad S, Al-Kazemi N, Khan S, Joseph L, et al. Outbreak of fungemia among neonates caused by Candida haemulonii resistant to amphotericin b, itraconazole, and fluconazole. J Clin Microbiol. 2007. pp. 2025–2027. doi: 10.1128/jcm.00222-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruan S-Y, Kuo Y-W, Huang C-T, Hsiue H-C, Hsueh P-R. Infections due to Candida haemulonii: species identification, antifungal susceptibility and outcomes. Int J Antimicrob Agents. 2010. pp. 85–88. doi: 10.1016/j.ijantimicag.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 17.Córdoba S, Vivot W, Bosco-Borgeat ME, Taverna C, Szusz W, Murisengo O, et al. Species distribution and susceptibility profile of yeasts isolated from blood cultures: results of a multicenter active laboratory-based surveillance study in Argentina. Rev Argent Microbiol. 2011;43. doi: 10.1590/S0325-75412011000300003 [DOI] [PubMed] [Google Scholar]
- 18.Desnos-Ollivier M, Lortholary O, Bretagne S, Dromer F. Azole Susceptibility Profiles of More than 9,000 Clinical Yeast Isolates Belonging to 40 Common and Rare Species. Antimicrob Agents Chemother. 2021. doi: 10.1128/AAC.02615-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muro MD, de Araújo Motta F, Burger M, de Azevedo Melo AS, Dalla-Costa LM. Echinocandin resistance in two Candida haemulonii isolates from pediatric patients. J Clin Microbiol. 2012;50: 3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bretagne S, Sitbon K, Desnos-Ollivier M, Garcia-Hermoso D, Letscher-Bru V, Cassaing S, et al. Active surveillance program to increase awareness on invasive fungal diseases: the French RESSIF Network (2012 to 2018). MBio. 2022;13: e0092022. doi: 10.1128/mbio.00920-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bretagne S, Renaudat C, Desnos-Ollivier M, Sitbon K, Lortholary O, Dromer F, et al. Predisposing factors and outcome of uncommon yeast species-related fungaemia based on an exhaustive surveillance programme (2002–14). Journal of Antimicrobial Chemotherapy. 2017. pp. 1784–1793. doi: 10.1093/jac/dkx045 [DOI] [PubMed] [Google Scholar]
- 22.Bretagne S, Desnos-Ollivier M, Sitbon K, Lortholary O, Che D, Dromer F, et al. No impact of fluconazole to echinocandins replacement as first-line therapy on the epidemiology of yeast fungemia (hospital-driven active surveillance, 2004–2017, Paris, France). Front Med. 2021;8: 641965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desnos-Ollivier M, Bretagne S, Boullié A, Gautier C, Dromer F, Lortholary O. Isavuconazole MIC distribution of 29 yeast species responsible for invasive infections (2015–2017). Clin Microbiol Infect. 2019. pp. 634.e1–634.e4. doi: 10.1016/j.cmi.2019.02.007 [DOI] [PubMed] [Google Scholar]
- 24.Desnos-Ollivier M, Bretagne S, Lortholary O, Dromer F, French Mycoses Study Group. Echinocandins susceptibility patterns of 2,787 yeast isolates: importance of the thresholds for the detection of FKS mutations. Antimicrob Agents Chemother. 2022;66: e0172521. doi: 10.1128/aac.01725-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grenfell RC, da Silva Junior AR, Del Negro GMB, Munhoz RB, Gimenes VMF, Assis DM, et al. Identification of Candida haemulonii complex species: use of ClinProTools to overcome limitations of the Bruker Biotyper, Vitek MS IVD, and Vitek MS RUO databases. Front Microbiol. 2016. doi: 10.3389/fmicb.2016.00940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isla G, Taverna CG, Szusz W, Vivot W, García-Effron G, Davel G. Candida haemulonii sensu lato: Update of the determination of susceptibility profile in Argentina and literature review. Curr Fungal Infect Rep. 2017. pp. 203–208. doi: 10.1007/s12281-017-0300-y [DOI] [Google Scholar]
- 27.Lortholary O, Renaudat C, Sitbon K, Desnos-Ollivier M, Bretagne S, Dromer F, et al. The risk and clinical outcome of candidemia depending on underlying malignancy. Intensive Care Med. 2017;43: 652–662. doi: 10.1007/s00134-017-4743-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chow JK, Golan Y, Ruthazer R, Karchmer AW, Carmeli Y, Lichtenberg D, et al. Factors associated with candidemia caused by non-albicans Candida species versus Candida albicans in the intensive care unit. Clin Infect Dis. 2008;46: 1206–1213. [DOI] [PubMed] [Google Scholar]
- 29.Jung DS, Farmakiotis D, Jiang Y, Tarrand JJ, Kontoyiannis DP. Uncommon Candida species fungemia among cancer patients, Houston, Texas, USA. Emerging Infect Dis. 2015. doi: 10.3201/eid2111.150404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gade L, Muñoz JF, Sheth M, Wagner D, Berkow EL, Forsberg K, et al. Understanding the emergence of multidrug-resistant candida: using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front Genet. 2020;0. doi: 10.3389/fgene.2020.00554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andes DR, Safdar N, Baddley JW, Playford G, Reboli AC, Rex JH, et al. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin Infect Dis. 2012. pp. 1110–1122. doi: 10.1093/cid/cis021 [DOI] [PubMed] [Google Scholar]
- 32.Kollef M, Micek S, Hampton N, Doherty JA, Kumar A. Septic shock attributed to Candida infection: importance of empiric therapy and source control. Clin Infect Dis. 2012;54: 1739–1746. doi: 10.1093/cid/cis305 [DOI] [PubMed] [Google Scholar]
- 33.Silva LN, Campos-Silva R, Ramos LS, Trentin DS, Macedo AJ, Branquinha MH, et al. Virulence of Candida haemulonii complex in Galleria mellonella and efficacy of classical antifungal drugs: a comparative study with other clinically relevant non-albicans Candida species. FEMS Yeast Res. 2018;18. doi: 10.1093/femsyr/foy082 [DOI] [PubMed] [Google Scholar]
- 34.de Jong AW, Dieleman C, Carbia M, Mohd Tap R, Hagen F. Performance of two novel chromogenic media for the identification of multidrug-resistant Candida auris compared with other commercially available formulations. J Clin Microbiol. 2021;59. doi: 10.1128/JCM.03220-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garey KW, Rege M, Pai MP, Mingo DE, Suda KJ, Turpin RS, et al. Time to initiation of fluconazole therapy impacts mortality in patients with candidemia: a multi-institutional study. Clin Infect Dis. 2006. pp. 25–31. doi: 10.1086/504810 [DOI] [PubMed] [Google Scholar]
- 36.Ramos R, Caceres DH, Perez M, Garcia N, Castillo W, Santiago E, et al. Emerging multidrug-resistant Candida duobushaemulonii infections in Panama hospitals: importance of laboratory surveillance and accurate identification. J Clin Microbiol. 2018;56. doi: 10.1128/JCM.00371-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muñoz JF, Gade L, Chow NA, Loparev VN, Juieng P, Berkow EL, et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nat Commun. 2018;9. doi: 10.1038/s41467-018-07779-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.None van U, Mc K. Torulopsis haemulonii nov. spec., a yeast from the Atlantic Ocean. Antonie Van Leeuwenhoek. 1962;28. doi: 10.1007/BF02538724 [DOI] [PubMed] [Google Scholar]
- 39.Ferreira N, Belloch C, Querol A, Manzanares P, Vallez S, Santos A. Yeast microflora isolated from brazilian cassava roots: taxonomical classification based on molecular identification. Curr Microbiol. 2010;60. doi: 10.1007/s00284-009-9539-z [DOI] [PubMed] [Google Scholar]
- 40.Pagani DM, Heidrich D, Paulino GVB, de Oliveira Alves K, Dalbem PT, de Oliveira CF, et al. Susceptibility to antifungal agents and enzymatic activity of Candida haemulonii and Cutaneotrichosporon dermatis isolated from soft corals on the Brazilian reefs. Arch Microbiol. 2016;198: 963–971. [DOI] [PubMed] [Google Scholar]
- 41.Tra Bi CY, N’guessan FK, Kouakou CA, Jacques N, Casaregola S, Djè MK. Identification of yeasts isolated from raffia wine (Raphia hookeri) produced in Côte d’Ivoire and genotyping of Saccharomyces cerevisiae strains by PCR inter-delta. World J Microbiol Biotechnol. 2016;32: 1–9. [DOI] [PubMed] [Google Scholar]
- 42.Loosová G, Jindrák L, Kopácek P. Mortality caused by experimental infection with the yeast Candida haemulonii in the adults of Ornithodoros moubata (Acarina: Argasidae). Folia Parasitol. 2013;48: 149–153. [PubMed] [Google Scholar]