Abstract
We have demonstrated previously that the seven-nucleotide (nt) motif TTTTGTA (the heptamer) that is present within the proximal 3′ untranslated sequences of numerous immediate-early genes is essential for platelet-derived growth factor (PDGF)-stimulated induction of the MCP-1 immediate-early gene. On this basis, the heptamer was suggested to be a conserved regulatory element involved in immediate-early gene expression, although its mechanism of action was unknown. Herein, we demonstrate that the heptamer functions to remove an inhibition of PDGF induction of MCP-1 maintained by two independently acting inhibitory elements present in the MCP-1 5′ flanking sequences (designated I* elements). PDGF treatment relieves the I*-mediated inhibition of MCP-1 expression only if the heptamer is also present. One inhibitory element is contained within a 59-nt portion of MCP-1 5′ flanking sequences and functions in an orientation-independent and heptamer-regulated manner. Significantly, proteins binding to two DNA sequences contribute to the formation of a single multiprotein complex on the 59-nt I* element. The I*-binding complex contains Sp3, an Sp1-like protein, and a novel DNA-binding protein. Moreover, the complex does not form on two 59-nt sequences containing mutations that reverse the inhibition of PDGF induction maintained by the wild-type I* element. We propose to call the multiprotein I*-binding complex a repressosome and suggest that it acts to repress PDGF-stimulated transcription of MCP-1 in the absence of the heptamer TTTTGTA.
Immediate-early genes (IEGs) are a functionally diverse family of genes that have in common induction by growth factors, cytokines, and serum. By definition, they are induced at the transcriptional level in response to a stimulus, and induction is not dependent on new protein synthesis (1, 29, 42, 43, 65). A potentially useful means of subclassifying the growing set of IEGs, and one with apparent mechanistic implications, is to divide the group into genes with fast or slow kinetics of induction. A well-characterized example of a fast-kinetics IEG is c-fos. Serum or platelet-derived growth factor (PDGF) added to quiescent 3T3 cells stimulates transcription of c-fos within 10 min. c-fos expression reaches peak levels within 30 min and returns to baseline levels within 2 h (24, 38). A cluster of three cis-acting regulatory elements contained within the proximal 350 nucleotides (nt) of 5′ flanking sequences of c-fos mediate serum- and growth factor-stimulated induction of c-fos and have proven to be of general interest in problems of growth factor signal transduction. The three functionally distinct c-fos elements include a serum response element, a cyclic AMP response element, and an element responsive to platelet-derived growth factor B-B homodimers known as the sis-inducible element (3, 17, 21–23, 30, 67, 70–73). Furthermore, nuclear trans-acting proteins interacting with each of these cis-acting regulatory elements have been isolated and characterized (14, 31–33, 48, 51, 56, 57, 69). By both sequence analysis and functional analysis, the transcriptional regulatory elements defined initially within c-fos have also been detected within other fast-kinetics IEGs (8, 54, 58).
The c-fos gene, however, does not stand as a prototype for all members of the IEG set. A second subgroup of IEGs exists that is induced with slower kinetics than c-fos and by apparently different mechanisms (27, 28). Included in the slow-kinetics subset of IEGs are the clinically important c-myc oncogene (37) and the CC chemokine gene JE/MCP-1 (for monocyte chemoattractant protein 1; hereafter referred to as MCP-1) (5, 9, 25, 61, 63, 64). In contrast to the rapid but transient response exhibited by c-fos, slow IEGs like c-myc and MCP-1 display a greater than 60-min lag time before initiation of transcription (13, 26, 38). Significantly, no fos-like regulatory elements are found within several kilobases of 5′ or 3′ flanking sequences of the MCP-1 gene or within its coding sequences. The distinct induction kinetics of the MCP-1 gene, and other slow IEGs such as c-myc, therefore reflect the likely action of cis-acting genomic elements distinct from the trio described for c-fos.
We have reported previously that a discontiguous pair of cis-acting elements are both essential for regulated expression of MCP-1. One element, detected initially as a 240-bp fragment found 2.3 kb upstream of the MCP-1 start of transcription, contains four distinct PDGF-regulated elements and acts as a PDGF-regulated enhancer sequence (18–20). The second element required for serum and PDGF induction of MCP-1 was shown to be the seven-base motif TTTTGTA (or heptamer) located in the proximal 3′ MCP-1 untranslated sequences. No single control element has been shown to function in regulated expression of both the fast and slow subclasses of IEGs. Interestingly, identical heptamers are found in the proximal 3′ untranslated sequences of c-myc and at least 25 additional IEGs (20), suggesting that the heptamer could be a novel regulatory sequence playing an essential role in serum-, growth factor-, and cytokine-stimulated expression of both fast- and slow-kinetics IEGs. Until recently, the mechanism of action of the heptamer was unknown.
We have shown more recently that readdition to two non-MCP-1 reporter genes of (i) the PDGF-regulated distal 5′ 240-bp fragment and (ii) a proximal 5′ sequence fragment that does not contain the MCP-1 TATA or CAAT box results in a PDGF-inducible construct in transfection experiments in the absence of the heptamer (68a). These data highlight an apparent paradox, namely, that the TTTTGTA motif is essential for PDGF induction of tagged MCP-1 reporter genes (20) but is apparently dispensable for PDGF induction of two different non-MCP-1 reporter genes. One explanation for this paradox would be if the heptamer functioned to remove an inhibition of PDGF induction of MCP-1. The proposed inhibition of induction by PDGF could be maintained by an inhibitory element(s) present within the MCP-1 sequences (coding or flanking). Heterologous reporter genes lacking the inhibitory element sequence(s) would, in this model, not require the presence of the heptamer for induction by PDGF to occur.
In this report, we demonstrate that (i) a pair of distinct and independently acting inhibitory elements are present within the MCP-1 5′ flanking sequences, (ii) inhibition of PDGF induction of MCP-1 is maintained by an inhibitory element in the absence of the heptamer, and (iii) the more potent of the inhibitory elements is present within a 59-nt portion of MCP-1 5′ sequences and binds a single multiprotein regulatory complex. We demonstrate further that the inhibitory element-binding complex contains the Sp3 transcription factor, an Sp1-like protein, and an apparently novel DNA-binding protein that bind to two distinct DNA-binding sequences within the overall 59-nt inhibitory element sequence. We propose to call the multiprotein, multi-DNA-binding site inhibitory element-binding complex a repressosome.
MATERIALS AND METHODS
Growth factors and reagents.
The recombinant B-B isoform of PDGF was obtained from Intergen. RNase A was from Pharmacia. Proteinase K, RNase T1, calf intestinal phosphatase, and poly(dI-dC) were from Boehringer Mannheim Biochemicals. Bovine calf serum was obtained from Hyclone. Human defibrinogenated platelet-poor plasma was prepared as previously described (55). Recombinant Sp1 was obtained from Promega. A monoclonal antibody to Sp1 (1C6, designated anti-Sp1-2 in Fig. 6) and polyclonal antibodies to Sp1 (PEP2, designated anti-Sp1-1 in Fig. 6), Sp2, Sp3, and Sp4 were obtained from Santa Cruz Biotechnology. Rabbit immunoglobulin G was obtained from Sigma. Synthetic oligonucleotides were obtained from Life Technologies.
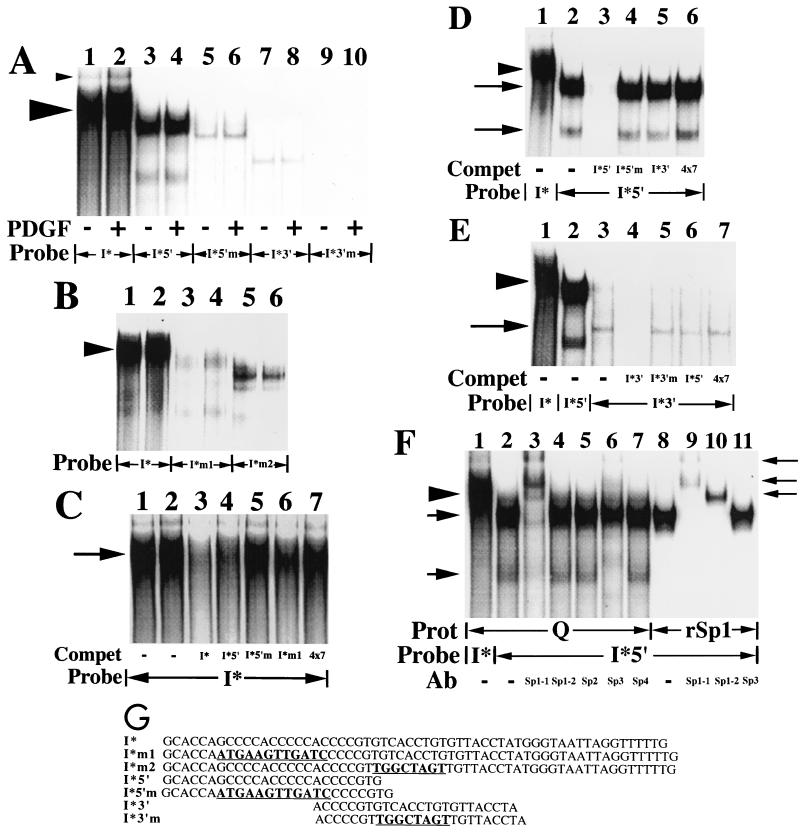
FIG. 6.
A slowly migrating protein complex binds to the 59-nt I* element-containing sequence. (A) Nuclear extracts (20 μg) prepared from quiescent fibroblasts (−) or fibroblasts treated with the B-B isoform of PDGF (30 ng/ml) for 2 h (+) were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. The large arrowhead highlights the predominant complex that binds to the 59-nt fragment (I*) probe. The small arrowhead highlights an additional protein complex, of unclear significance, that also binds to the 59-nt fragment probe. (B) Nuclear extracts (15 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. The arrowhead highlights the predominant complex that binds to the I* probe. (C) Nuclear extracts (15 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probe shown. Unlabeled double-stranded oligonucleotides (300-fold excesses) were used as competitors where indicated. The 4×7 competitor is a double-stranded oligonucleotide containing four copies of the heptamer TTTTGTA. The arrow highlights the predominant complex that binds to the I* probe. Compet, competitor. (D) Nuclear extracts (15 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. Unlabeled double-stranded oligonucleotides (300-fold excesses) were used as competitors where indicated. The arrowhead highlights the predominant complex that binds to the 59-nt fragment (I*) probe. The upper and lower arrows show the positions of two complexes that bind specifically to the I*5′ probe. (E) Nuclear extracts (20 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. Unlabeled double-stranded oligonucleotides (200-fold excesses) were used as competitors where indicated. The arrowhead highlights the predominant complex that binds to the 59-nt fragment (I*) probe. The arrow shows the position of a complex that binds specifically to the I*3′ probe. (F) Nuclear extracts (15 μg) prepared from quiescent fibroblasts (Q) and recombinant Sp1 (rSp1, 1fpu) were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. Antibodies specific for individual members of the Sp1 family of transcription factors (5 μg) were added where indicated. The Sp1-1 and Sp1-2 antibodies bind to distinct, nonoverlapping portions of the Sp1 transcription factor (amino acids 436 to 454 and 520 to 538, respectively). Prot, protein. The arrowhead on the left highlights the predominant complex that binds specifically to the 59-nt fragment (I*) probe. The arrows on the left show the positions of the two complexes that bind specifically to the 5′ subregion (I*5′) probe. The arrows on the right show the positions of complexes supershifted by the anti-Sp1 and anti-Sp3 antibodies (Ab). (G) The sequences of seven oligonucleotides and the relative position of each oligonucleotide within the overall 59-nt footprinted (I*) region are shown. These oligonucleotides were used as double-stranded probes or competitors in the six mobility shift assays whose results are shown here and correspond to the 25-nt (5′ subregion), 24-nt (3′ subregion), and 59-nt (full-length) footprinted regions shown in Fig. 3 (designated I*5′, I*3′, and I*, respectively). Mutant 25- and 24-nt probes, containing the 12- and 8-nt mutations described in the legend to Fig. 4 are designated I*5′m and I*3′m, respectively. Mutant I* probes containing the same 12- and 8-nt mutations are designated I*m1 and I*m2, respectively. The mutated sequences are boldfaced and underlined for each oligonucleotide. Free probe is not shown in these mobility shift assays. No complexes were observed with any of the probes alone in the absence of extract (data not shown).
Cell culture, DNA transfections, stimulation assays, and RNA preparation and analysis.
NIH 3T3 cells were maintained in Dulbecco’s modified Eagle medium (GIBCO Laboratories) supplemented with 10% bovine calf serum. NIH 3T3 cells were used for transient transfections because of their significantly greater transfection efficiency compared with BALB/c-3T3 cells and were transfected as previously described (18, 19). Transfection mixtures included 4 to 5 μg of tagged MCP-1 reporter constructs together with 4 μg of an α-globin reference construct (pSVα-1) per 15-cm-diameter tissue culture plate. After transfection, cells were maintained for 40 to 44 h in 5% platelet-poor plasma and then exposed to the B-B isoform of PDGF (30 ng/ml) for the times indicated in the figure legends. Total RNA was prepared by using Tripure reagent (Boehringer Mannheim) and following the manufacturer’s instructions.
Total RNA samples were analyzed by RNase protection assay as previously described (18). The probe for tagged MCP-1 expression is a 272-bp HincII MCP-1 cDNA fragment spanning portions of the second and third exons. The fragment is elongated by addition of a 33-bp tag into a blunted SauI site and inserted into the SmaI site of pGEM7 (20). The length of the unprotected probe is 377 bases. As a control for equal transfection efficiencies between groups of cells, 15 μg of total RNA was subjected to an RNase protection assay for the presence of a constitutively active cotransfected alpha-globin construct (20). Exposure and quantitation of gels for all experiments were performed on a PhosphorImager (Molecular Dynamics). The statistical significance of differences in PDGF induction obtained with transfected constructs was analyzed, after normalization for differences in induction of the endogenous MCP-1 gene and differences in transfection efficiency, by using the Wilcoxon two-sample test or the Kruskal-Wallis test, as indicated in the figure legends.
Oligonucleotides.
Double-stranded oligonucleotides containing 5′ overhanging GG (top strand) and CC (bottom strand) ends were used. All of the sequences shown are of the top strand. Mutations of the I*, I*5′, and I*3′ sequences are underlined. The I*m1 and I*5′m oligonucleotides include the 12-nt mutation discussed in Results. The I*m2 and I*3′m oligonucleotides include the 8-nt mutation discussed in Results. The sequences are as follows: I*, GCACCAGCCCCACCCCCACCCCGTGTCACCTGTGTTACCTATGGGTAATTAGGTTTTTG; I*m1, GCACCAATGAAGTTGATCCCCCGTGTCACCTGTGTTACCTATGGGTAATTAGGTTTTTG; I*m2, GCACCAGCCCCACCCCCACCCCGTTGGCTAGTTGTTACCTATGGGTAATTAGGTTTTTG; I*5′, GCACCAGCCCCACCCCCACCCCGTG; I*5′m, GCACCAATGAAGTTGATCCCCCGTG; I*3′, ACCCCGTGTCACCTGTGTTACCTA; I*3′m, ACCCCGTTGGCTAGTTGTTACCTA; 4×7, 5′-TTTTGTATTTTGTATTTTGTATTTTGTA.
Preparation of nuclear extracts, electrophoretic mobility shift assays, and DNase I footprinting.
Nuclear extracts from BALB/c-3T3 (clone A31) and NIH 3T3 fibroblasts were prepared as described by Dignam et al. (16). Chemically synthesized oligonucleotides were annealed and labeled with 32P-labeled nucleotides by filling in with the Klenow fragment of DNA polymerase I. Radiolabeled double-stranded oligonucleotides were gel purified prior to use as probes in electrophoretic mobility shift assays. Electrophoretic mobility shift assays were performed as previously described (18, 19). Following 30-min incubations on ice, DNA-protein complexes were electrophoresed on 4% native polyacrylamide gels at 150 V in 0.25× Tris-borate-EDTA buffer for 4.5 h at 4°C. For antibody supershift or competition experiments, antibodies or the indicated excesses of unlabeled double-stranded oligonucleotides were added for a 30-min incubation on ice prior to the addition of a radiolabeled oligonucleotide probe. DNase I footprinting was performed as previously described (19).
Immunoprecipitation and Western blot analysis.
Immunodepletions were performed in the presence of protease inhibitors on ice. After supernatants were cleared, supernatant was transferred to a fresh tube containing 15 μl of a rabbit polyclonal anti-Sp3 antibody. After incubation for 1 h at 4°C, 50 μl of preincubated 10% protein A-Sepharose (Zymed) was added and the mixture was rotated at 4°C for 30 to 60 min. Samples were then centrifuged, and the supernatant was removed for further analysis. Immunoblot analysis, using 30 μg of protein per lane and transfer to Immobilon polyvinylidene difluoride membranes (Millipore), was performed as previously described (68).
Plasmid construction and site-directed mutagenesis.
Site-directed mutagenesis was performed by using a commercially available kit (Quik Change; Stratagene) in accordance with the manufacturer’s instructions. Construct 1 (see Fig. 1, 2, 4, and 5), a 33-bp-tagged MCP-1 reporter gene containing 2.8 kb of MCP-1 5′ flanking sequences and 104 bp of MCP-1 3′ untranslated sequences, has been described previously (20). Construct 2 (see Fig. 1) was created by site-directed mutagenesis of the heptamer into the 3′ untranslated sequences of construct 1 (104 bp 3′ to the translational stop codon). Constructs 3 to 5 (see Fig. 1) were derived from construct 1 following AocI digestion and removal of the 5′ 707-bp AocI-EcoRI, 1,402-bp AocI-SpeI, and 1,936-bp AocI-HincII fragments, respectively, with subsequent religation. Constructs 3 and 4 (see Fig. 2A) are derived from construct 2 by removal of the 1,936-bp AocI-HincII fragment as described above, followed by ligation of the 707-bp AocI-EcoRI and 695-bp SpeI-EcoRI MCP-1 5′ sequence fragments, respectively, into the opened AocI site. Constructs 5 and 6 (see Fig. 2A and B) were created by insertion of the heptamer into construct 4 by site-directed mutagenesis into the 3′ untranslated sequences (104 bp 3′ to the translational stop codon) or 5′ flanking sequences (at −2766 relative to the start of transcription), respectively. Constructs 5m1, m2, and m3 (see Fig. 2B) were created by insertion of mutant heptamer sequences into construct 4 by site-directed mutagenesis into the 3′ untranslated sequences (104 bp 3′ to the translational stop codon). Constructs 5m1 and 5m2 (see Fig. 4) were created by site-directed mutation of nonoverlapping 12- and 8-nt portions of the 695-bp SpeI-EcoRI fragment in construct 3, respectively. The sequences of the 12- and 8-nt mutations are given in the oligonucleotide paragraph. Constructs 4, 5, and 6 (see Fig. 5A) are derived from construct 2 by removal of the 1,936-bp AocI-HincII fragment (see Fig. 1) (construct 5), followed by ligation of the oligonucleotides shown into the opened AocI site. Construct 4(7) (see Fig. 5B) was created by insertion of the wild-type heptamer into construct 4 by site-directed mutagenesis into the 3′ untranslated sequences (104 bp 3′ to the translational stop codon). All constructs were checked for accuracy by DNA sequencing (66).
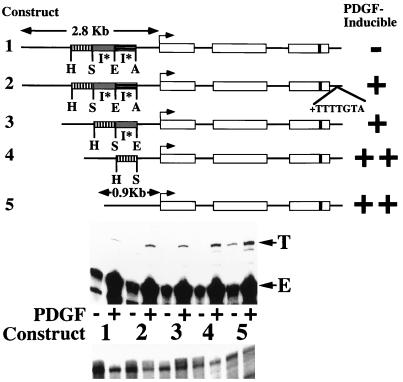
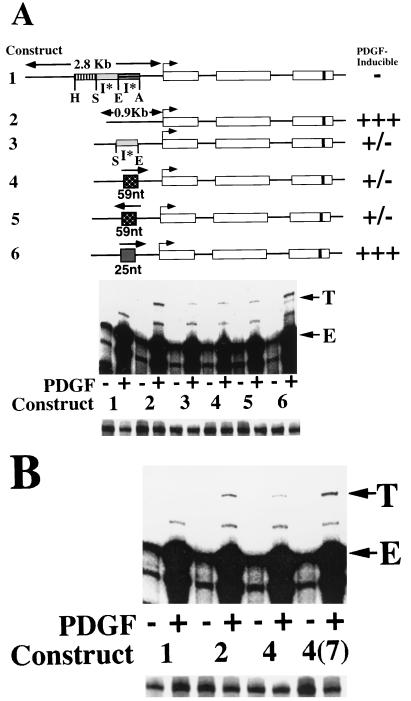
FIG. 1.
Deletion of 5′ flanking sequences results in heptamerless PDGF-inducible MCP-1 reporter genes. At the top is a schematic of the structures of five tagged MCP-1 reporter constructs. Open rectangles represent exons, and introns are represented by the dark lines between the exons. The lengths of 5′ flanking MCP-1 sequences contained within constructs are given in kilobases. The start of transcription is shown by the bent arrow. The 33-bp tag is represented by the dark band in the third exon. Constructs 1, 3, 4, and 5 contain 104 bp of 3′ untranslated sequences (that include the polyadenylation signal but not the heptamer). Only construct 2 contains the heptamer within its 3′ untranslated sequences. The relative positions of AocI (A), EcoRI (E), SpeI (S), and HincII (H) sites, located in the MCP-1 5′ flanking sequences and defining the endpoints of three MCP-1 5′ sequence fragments, are shown. The overall AocI-HincII 5′ fragment includes 1,936 bp. Construct 2 is derived from construct 1 by readdition of the heptamer TTTTGTA to the 3′ untranslated sequences of construct 1. Construct 3 is derived from construct 1 by deletion of the 707-bp EcoRI-AocI fragment. Construct 4 is derived from construct 3 by deletion of the 695-bp SpeI-EcoRI fragment. Construct 5 is derived from construct 4 by deletion of the 534-bp HincII-SpeI fragment. The PDGF-regulated 5′ I* elements are indicated. The PDGF inducibilities of the five constructs in transfection experiments are summarized on the right. In the middle are RNase protection assays of 40 μg of total cellular RNA that was prepared from NIH 3T3 fibroblasts transiently transfected with 5 μg of the constructs shown, allowed to become quiescent, and then not exposed (−) or exposed (+) to the B-B isoform of PDGF (30 ng/ml) for 3 h. The numbers refer to the tagged MCP-1 constructs diagrammed at the top. The positions of the 305- and 241-nt protected fragments corresponding to expression of the transfected and tagged (T) and endogenous (E) MCP-1 genes, respectively, are shown. The experiment was performed four times with similar results. The PDGF inductions obtained with constructs 4 and 5 were 2.6 to 4.0 and 3.0 to 8.1 times greater, respectively, than those obtained with construct 1 in these transfections. The PDGF induction increases observed with constructs 2, 3, 4, and 5, compared to construct 1, are all significant (P < 0.05 by the Wilcoxon two-sample test). At the bottom are RNase protection assays of 15 μg of total cellular RNA taken from the transfections shown above and analyzed with an alpha-globin riboprobe.
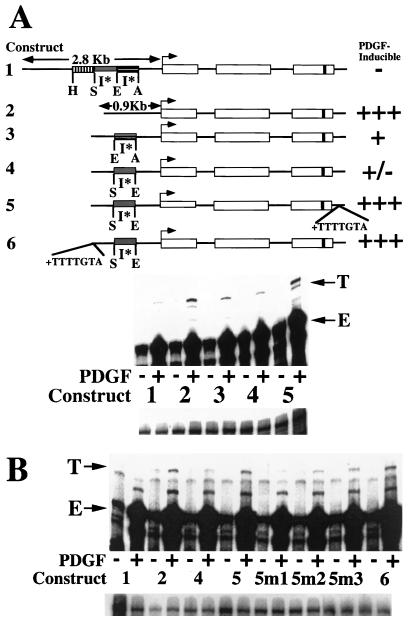
FIG. 2.
PDGF induction of MCP-1 involves interactions between the heptamer TTTTGTA and a 5′ I* element. (A) At the top is a schematic of the structures of six tagged MCP-1 reporter constructs. Constructs 1 through 4 and 6 contain 104 bp of 3′ untranslated sequences (that include the polyadenylation signal but not the heptamer). Only construct 5 contains the heptamer within its 3′ untranslated sequences. The details of the MCP-1 schematics are otherwise as described in the legend to Fig. 1. Constructs 1 and 2 in this figure are identical to constructs 1 and 5, respectively, in Fig. 1. Construct 3 is derived from construct 2 by readdition of the 707-bp EcoRI-AocI fragment. Construct 4 is derived from construct 2 by readdition of the 695-bp SpeI-EcoRI fragment. Construct 5 is derived from construct 4 by readdition of the heptamer to the proximal 3′ untranslated sequences. Construct 6 is derived from construct 4 by readdition of the heptamer to the distal 5′ flanking sequences. The PDGF-regulated 5′ I* elements are indicated. The PDGF inducibilities of the six constructs in transfection experiments are summarized on the right. At the middle are RNase protection assays of 40 μg of total cellular RNA that was prepared from NIH 3T3 fibroblasts transiently transfected with 5 μg of the constructs shown, allowed to become quiescent, and then not exposed (−) or exposed (+) to the B-B isoform of PDGF (30 ng/ml) for 3 h. The numbers refer to the tagged MCP-1 constructs diagrammed at the top. The 305- and 241-nt protected fragments corresponding to expression of the transfected and tagged (T) or endogenous (E) MCP-1 genes, respectively, are indicated. The experiment was performed four times with similar results. The PDGF inductions obtained with constructs 3 and 4 were 10 to 64 and 10 to 15%, respectively, of those obtained with construct 2 in these transfections. The PDGF inductions obtained with construct 5 were 2.1 to 9.7 times greater than those obtained with construct 4 in these transfections. The PDGF induction decreases observed with constructs 3 and 4, compared to construct 2, are both statistically significant (P < 0.05 by the Wilcoxon two-sample test). The PDGF induction increases observed with construct 5, compared to construct 4, are significant (P < 0.05 by the Wilcoxon two-sample test). At the bottom are RNase protection assays of 15 μg of total cellular RNA taken from the transfections shown above and analyzed with an alpha-globin riboprobe. (B) At the top are RNase protection assays of 40 μg of total cellular RNA that was prepared from NIH 3T3 fibroblasts transiently transfected with 4 μg of the constructs shown, allowed to become quiescent, and then not exposed (−) or exposed (+) to the B-B isoform of PDGF (30 ng/ml) for 3 h. The numbers refer to the tagged MCP-1 constructs described at the top, except for constructs 5m1, 5m2, and 5m3. The latter constructs are derived from construct 4 by addition of the mutant heptamers TTTTATG, GGGGGTA, and TTTTGGA, respectively, to the 3′ untranslated sequences. Mutations of the wild-type heptamer sequence are underlined. The 305- and 241-nt protected fragments corresponding to expression of the transfected and tagged (T) and endogenous (E) MCP-1 genes, respectively, are indicated. The experiment was performed four times with similar results. The decreased PDGF inductions obtained with constructs 5m1, 5m2, and 5m3 varied from 6 to 15%, 8 to 25%, and 50 to 60% of those obtained with construct 5, respectively, in these transfections. The PDGF induction differences obtained with constructs 5m1, 5m2, and 5m3, compared to construct 5, are all statistically significant (P < 0.05 by the Wilcoxon two-sample test). The PDGF induction increases observed with constructs 5 and 6, compared to construct 4, are statistically significant (P < 0.05 by the Wilcoxon two-sample test). At the bottom are RNase protection assays of 15 μg of total cellular RNA taken from the transfections shown above and analyzed with an alpha-globin riboprobe.
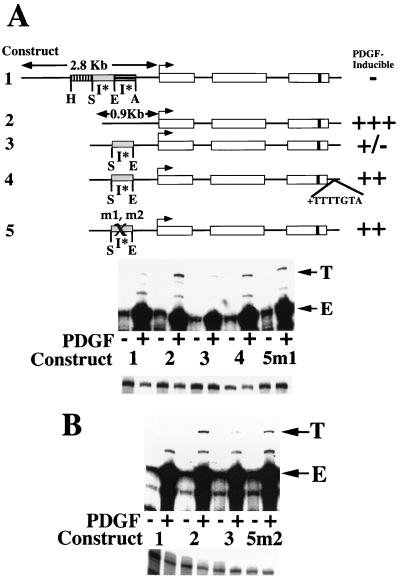
FIG. 4.
Necessity of 12-nt (5′) and 8-nt (3′) footprinted sequences for inhibition of MCP-1 induction. (A) At the top is a schematic of the structures of five tagged MCP-1 reporter constructs. Constructs 1 through 3 and 5 contain 104 bp of 3′ untranslated sequences (that include the polyadenylation signal but not the heptamer). Only construct 4 contains the heptamer within its 3′ untranslated sequences. The details of the MCP-1 schematics are otherwise as described in the legend to Fig. 1. Construct 2 is derived from construct 1 by removal of the 1,936-bp AocI-HincII 5′ fragment. Construct 3 is derived from construct 2 by readdition of the 695-bp SpeI-EcoRI fragment. Construct 4 is derived from construct 3 by addition of the heptamer TTTTGTA to the 3′ untranslated sequences. Constructs 5m1 and m2 are derived from construct 3 by site-directed mutation of nonoverlapping 12-nt (5′ subregion) and 8-nt (3′ subregion) portions, respectively, of the footprinted region shown in Fig. 3 (i.e., mutating the 12-base sequence GCCCCACCCCCA to ATGAAGTTGATC and the 8-base sequence GTCACCTG to TGGCTAGT within the otherwise unaltered 695-bp SpeI-EcoRI fragment). The PDGF-regulated 5′ inhibitory I* elements are indicated. The PDGF inducibilities of the constructs in transfection experiments are summarized on the right. At the middle are RNase protection assays of 40 μg of total cellular RNA that was prepared from NIH 3T3 fibroblasts transiently transfected with 5 μg of the constructs shown, allowed to become quiescent, and then not exposed (−) or exposed (+) to the B-B isoform of PDGF (30 ng/ml) for 3 h. The numbers refer to the tagged MCP-1 constructs described at the top. The 305- and 241-nt protected fragments corresponding to expression of the transfected and tagged (T) and endogenous (E) MCP-1 genes, respectively, are indicated. The experiment was performed four times with similar results. The PDGF inductions obtained with construct 5m1 were 1.6 to 5.2 times greater than those obtained with construct 3 in these transfections. The PDGF induction increases observed with construct 5m1, compared to construct 3, are statistically significant (P < 0.05 by the Wilcoxon two-sample test). At the bottom are RNase protection assays of 15 μg of total cellular RNA taken from the transfections shown above and analyzed with an alpha-globin riboprobe. (B) At the top are RNase protection assays of 40 μg of total cellular RNA prepared from NIH 3T3 fibroblasts transiently transfected with 4 μg of the constructs shown, allowed to become quiescent, and then not exposed (−) or exposed (+) to the B-B isoform of PDGF (30 ng/ml) for 3 h. The numbers refer to the tagged MCP-1 constructs described at the top. The 305- and 241-nt protected fragments corresponding to expression of the transfected and tagged (T) and endogenous (E) MCP-1 genes, respectively, are indicated. The experiment was performed four times with similar results. The PDGF inductions obtained with construct 5m2 were 1.8 to 2.7 times greater than those obtained with construct 3 in these transfections. The PDGF induction increases observed with construct 5m2, compared to construct 3, are statistically significant (P < 0.05 by the Wilcoxon two-sample test). At the bottom are RNase protection assays of 15 μg of total cellular RNA taken from the transfections shown above and analyzed with an alpha-globin riboprobe.
FIG. 5.
A 59-nt sequence is sufficient for inhibition of PDGF induction of MCP-1. (A) At the top is a schematic of the structures of six tagged MCP-1 reporter constructs. All of the constructs contain 104 bp of 3′ untranslated sequences (that include the polyadenylation signal but not the heptamer). The details of the MCP-1 schematics are otherwise as described in the legend to Fig. 1. Construct 2 is derived from construct 1 by removal of the 1,936-bp AocI-HincII 5′ fragment. Construct 3 is derived from construct 2 by readdition of the 695-bp SpeI-EcoRI fragment. Constructs 4 and 5 are derived from construct 2 by readdition of a 59-nt subfragment of the 695-bp SpeI-EcoRI fragment in the in vivo (construct 4) and reverse (construct 5) orientations. The 59-nt subfragment includes the complete footprinted region shown in Fig. 3 and a short stretch of additional MCP-1 sequences immediately 3′ to the footprinted region. Construct 6 is derived from construct 2 by readdition of a 25-nt oligonucleotide (I*5′) containing the 5′ footprinted subregion shown in Fig. 3. The 25-nt sequence is entirely contained within the 59-nt sequences. The PDGF-regulated 5′ I* elements are indicated. The PDGF inducibilities of the six constructs in transfection experiments are summarized on the right. Below the schematic are results from RNase protection assays of 40 μg of total cellular RNA that was prepared from NIH 3T3 fibroblasts transiently transfected with 4 μg of the constructs shown, allowed to become quiescent, and then not exposed (−) or exposed (+) to the B-B isoform of PDGF (30 ng/ml) for 3 h. The numbers refer to the tagged MCP-1 constructs described at the top. The 305- and 241-nt protected fragments corresponding to expression of the transfected and tagged (T) and endogenous (E) MCP-1 genes, respectively, are indicated. The experiment was performed three times with similar results. The PDGF inductions obtained with constructs 3, 4, and 5 varied from 3.5 to 24% of those obtained with construct 2 in these transfections. Differences among the PDGF inductions observed with constructs 3, 4, and 5 are not statistically significant (P = 0.49 by the Kruskal-Wallis test). The PDGF induction decreases observed with constructs 3, 4, and 5, compared to construct 2, are all statistically significant (P < 0.05 by the Wilcoxon two- sample test). At the bottom are RNase protection assays of 15 μg of total cellular RNA taken from the transfections shown above and analyzed with an alpha-globin riboprobe. (B) At the top are RNase protection assays of 40 μg of total cellular RNA that was prepared from NIH 3T3 fibroblasts transiently transfected with 4 μg of the constructs shown, allowed to become quiescent, and then not exposed (−) or exposed (+) to the B-B isoform of PDGF (30 ng/ml) for 3 h. The numbers refer to the tagged MCP-1 constructs described at the top, except for construct 4(7), which is derived from construct 4 by addition of the heptamer TTTTGTA to the 3′ untranslated sequences. The 305- and 241-nt protected fragments corresponding to expression of the transfected and tagged (T) and endogenous (E) MCP-1 genes, respectively, are indicated. The experiment was performed four times with similar results. The PDGF inductions obtained with construct 4(7) were 2.5 to 4.2 times greater than those obtained with construct 4 in these transfections. The PDGF induction increases observed with construct 4(7), compared to construct 4, are statistically significant (P < 0.05 by the Wilcoxon two-sample test). At the bottom are RNase protection assays of 15 μg of total cellular RNA taken from the transfections shown above and analyzed with an alpha-globin riboprobe.
RESULTS
PDGF induction of MCP-1 involves interactions between the heptamer TTTTGTA and two 5′ inhibitory elements.
Progressive deletion of portions of a 1,936-bp AocI-HincII segment of MCP-1 5′ flanking sequences results in heptamerless tagged MCP-1 reporter constructs that are PDGF inducible in transfection experiments (Fig. 1). Deletion of a 707-bp EcoRI-AocI 5′ sequence fragment or readdition of the heptamer to a heptamerless, non-PDGF-inducible MCP-1 reporter gene results in a PDGF-inducible construct in these transfection experiments (compare the PDGF inducibilities of constructs 3 and 2, respectively, with that of construct 1 in Fig. 1). Deletion of an additional 695-bp SpeI-EcoRI 5′ fragment significantly increases the PDGF inducibility of the resulting deletion construct (compare the PDGF inducibilities of constructs 4 and 3 in Fig. 1). In contrast, further deletion of a 534-bp SpeI-HincII 5′ fragment only slightly increases the PDGF inducibility of the resulting deletion construct (compare the PDGF inducibilities of constructs 5 and 4 in Fig. 1). The deletions created did not include portions of any PDGF-regulated cis-acting elements that we have reported previously (18–20).
Readdition of the 707-bp EcoRI-AocI or 695-bp SpeI-EcoRI 5′ fragment, individually, to a heptamerless MCP-1 reporter gene lacking the 1,936-bp AocI-HincII 5′ fragment results in 55 or 82% inhibition of PDGF inducibility, respectively (compare the PDGF inducibilities of constructs 3 and 4 with that of construct 2 in Fig. 2A, arbitrarily setting the PDGF induction obtained with construct 2 at 100%). Readdition of other MCP-1 5′ sequence fragments similar in size did not affect the PDGF inducibility of construct 2 (data not shown), suggesting that the inhibitory effects of the 707- and 695-bp 5′ fragments on PDGF induction are specific. Addition of the heptamer TTTTGTA to the 3′ untranslated sequences of a minimally inducible tagged MCP-1 reporter construct containing the SpeI-EcoRI inhibitory fragment restores PDGF inducibility in transfections (compare the PDGF inducibilities of constructs 5 and 4 with that of construct 2 in Fig. 2A). Similarly, addition of the heptamer to the 3′ untranslated sequences of an MCP-1 reporter construct containing the EcoRI-AocI inhibitory fragment restores PDGF inducibility in transfections (data not shown).
Readdition of the heptamer to the 3′ untranslated or 5′ flanking MCP-1 sequence restores PDGF inducibility to an inhibitory fragment-containing reporter gene with equal effectiveness (compare the PDGF inducibilities of constructs 6 and 5 with that of construct 4 in Fig. 2B). Three distinct mutations of the heptamer, TTTTATG, GGGGGTA, and TTTTGGA, decrease the PDGF inducibilities of the resulting mutant heptamer-containing MCP-1 reporter gene in transfections by 94, 80, and 43%, respectively (compare the PDGF inducibilities of constructs 5m1, 5m2, and 5m3 with that of construct 5 in Fig. 2B).
Taken together, these data are consistent with the existence of independently functioning inhibitory elements present within two nonoverlapping MCP-1 5′ fragments. These data further suggest that PDGF-regulated interactions occur between the wild-type heptamer, TTTTGTA, and the 695-bp SpeI-EcoRI inhibitory element-containing fragment that relieve the inhibition of PDGF induction of MCP-1. Furthermore, these data demonstrate that the inhibitory element-heptamer interactions are heptamer sequence specific. The more potent of the two inhibitory elements is contained within the 695-bp SpeI-EcoRI fragment (construct 4 in Fig. 2A). For these reasons, we will focus on the identity and properties of this inhibitory element in the rest of this report. Throughout this report, the inhibitory element will be designated I*.
A single DNase I-protected sequence is detected within the 695-bp SpeI-EcoRI inhibitory fragment.
DNase I footprinting assays were performed to locate a potential I* element sequence(s) within the 695-bp SpeI-EcoRI inhibitory fragment. A single footprinted region was detected in these assays (Fig. 3). The overall footprint, with endpoints at −1468 and −1434 relative to the start of transcription of the MCP-1 gene, consists of two DNase I-protected regions separated by an apparent DNase I-hypersensitive site (arrow in Fig. 3). The distal (5′) and proximal (3′) protected subregions will be referred to here as the 5′ and 3′ (footprinted) subregions, respectively. Similar footprinted regions are observed with extracts prepared from quiescent and PDGF-treated 3T3 cells (Fig. 3). No additional DNase I-protected sequences are apparent within the rest of the 695-bp fragment (data not shown). A similar footprint was detected by using a bottom-strand labeled probe (data not shown).
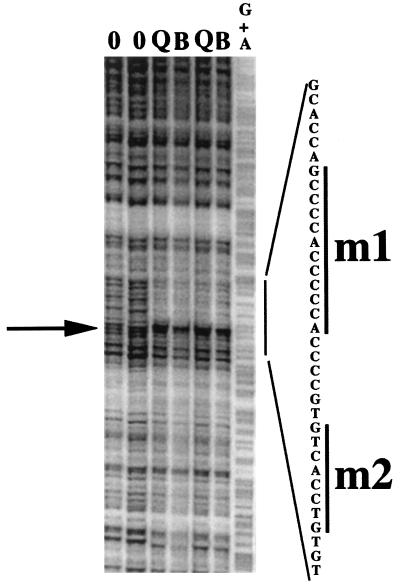
FIG. 3.
A single DNase I-protected sequence is detected within the 695-bp SpeI-EcoRI inhibitory fragment. A top-strand-labeled probe corresponding to the 695-bp inhibitory element-containing SpeI-EcoRI MCP-1 5′ fragment was used in DNase I footprinting assays. Nuclear protein extracts for these assays were prepared from two groups of quiescent fibroblasts (lanes Q) and fibroblasts treated with 30 ng of the B-B isoform of PDGF per ml for 2 h (lanes B). Lanes 0 contained no protein, and lane G+A contained the purine sequence of the top-strand-labeled probe. The nucleotide sequence of the overall protected region is given on the right. The arrow highlights a DNase I-hypersensitive site separating the 5′ and 3′ protected subregions. The 12-nt (m1) and 8-nt (m2) sequences mutated in this study are highlighted by the bars on the right.
A 59-nt sequence is sufficient for inhibition of MCP-1 induction and contains an I* element.
Mutation of 12 nt within the 5′ subregion or 8 nt within the 3′ subregion (bars in Fig. 3) reverses the inhibition maintained by the wild-type SpeI-EcoRI fragment (compare the PDGF inducibilities of constructs 3 and 5m1 or 5m2 in Fig. 4A and B). MCP-1 reporter genes containing either the heptamer within their 3′ untranslated sequences or the 12-nt mutation within the 695-bp inhibitory fragment exhibit similar PDGF inducibilities in these transfection experiments (compare the PDGF inducibilities of constructs 4 and 5m1 in Fig. 4A). Introduction of an identical 12-nt mutation within the 5′ subregion of construct 1 (i.e., a minimally PDGF-inducible reporter gene containing both inhibitory fragments in their proper contexts within the MCP-1 5′ sequences, Fig. 4A) partially reverses the inhibition of PDGF induction observed with wild-type construct 1 (data not shown). In the latter experiments, the reversal of inhibition of PDGF induction is only partial, likely due to the presence and unopposed action of the second (EcoRI-AocI) inhibitory fragment in mutated construct 1.
Readdition of a 25-nt oligonucleotide containing the 5′ footprinted subregion to an I*-lacking PDGF-inducible MCP-1 reporter gene does not recapitulate the inhibitory effect of the 695-bp SpeI-EcoRI fragment in transfections (compare the PDGF inducibilities of constructs 6 and 2 with that of construct 3 in Fig. 5A). In contrast, readdition of a 59-nt oligonucleotide that contains the complete footprinted region highlighted in Fig. 3 to the same reporter gene restores the inhibition of PDGF induction maintained by the 695-bp inhibitory fragment in an orientation-independent manner (compare the PDGF inducibilities of constructs 4 and 5 with that of construct 3 in Fig. 5A). Readdition of the 695-bp inhibitory fragment (construct 3) or the 59-nt oligonucleotide in either orientation (constructs 4 and 5) resulted in 86, 81, or 85% inhibition of the PDGF induction observed with construct 2 in these transfection experiments, respectively (arbitrarily setting the PDGF induction obtained with construct 2 at 100%). Readdition of the heptamer to the 3′ sequences of a 59-nt-containing MCP-1 reporter gene restores PDGF inducibility in transfection experiments [compare the PDGF inducibilities of constructs 4 and 4(7) in Fig. 5B].
In total, these data demonstrate that sequences in both the 5′ and 3′ subregions of the overall footprinted region shown in Fig. 3 are necessary for inhibition of PDGF induction. Furthermore, a 59-nt oligonucleotide containing the complete footprinted region shown in Fig. 3 is sufficient to mediate inhibition of PDGF induction of MCP-1 in an orientation-independent and heptamer-regulated manner; i.e., it appears to contain a functional I* element. By extension, these data suggest that the 5′ and 3′ footprinted subregions act together to effect inhibition of PDGF induction.
A slowly migrating protein complex binds to the I* element.
To detect and characterize potential regulatory proteins binding to the I* element, we performed a series of mobility shift assays using seven oligonucleotide probes. The sequences of the probes and their relative positions within the overall 59-nt I* element are shown in Fig. 6G. A predominant and constitutively binding protein complex was found to bind to a radiolabeled I* probe in mobility shift assays (lanes 1 and 2 in Fig. 6A). Significantly, mutant I* probes containing either of the nonoverlapping 12-nt (5′) or 8-nt (3′) mutations described in Fig. 4 do not bind the slowly migrating complex (compare the greatly diminished binding in lanes 3 and 4 or 5 and 6, respectively, with that in lanes 1 and 2 in Fig. 6B). The all-or-nothing binding exhibited by the wild-type and mutant I* probes suggests that cooperative interactions between the two sites altered in these mutant probes are involved in the formation of the complex on the wild-type I* sequence. Furthermore, since both mutations significantly decreased the inhibition of PDGF induction maintained by the wild-type 695-bp fragment in transfection experiments (constructs 5m1 and 5m2 in Fig. 4), these data suggest that a causal relationship exists between formation of the I* element-binding complex and inhibition of PDGF-stimulated expression of MCP-1 in intact cells.
Binding of the single predominant complex to the I* probe is specific, as it is competed by an excess of an unlabeled wild-type I* oligonucleotide (lane 3 in Fig. 6C) but is not competed by an unrelated oligonucleotide (lane 7 in Fig. 6C). Binding to the I* probe was also quantitatively competed by an excess of a wild-type, but not a mutant, 5′ subregion oligonucleotide (compare binding in lanes 4 and 5, respectively, in Fig. 6C), again demonstrating that protein binding to the I* probe in these mobility shift assays is specific. Finally, binding to the I* probe is partially competed by both a mutant I* oligonucleotide (I*m1) containing the 12-nt mutation of the 5′ subregion sequences (lane 6 in Fig. 6C) or by an oligonucleotide containing the 3′ subregion sequences (the I*3′ oligonucleotide; data not shown). If proteins binding to distinct sequences within the 5′ and 3′ subregions of the overall footprinted (I*) region participate in or are required for the formation of the I* element-binding complex, then using an excess of an unlabeled oligonucleotide corresponding to either the 5′ or 3′ subregion would be expected to bind these proteins, thereby depleting the 3T3 cell nuclear extracts of the free forms of this 5′ or 3′ subregion-binding protein. As was observed in the competition experiments (Fig. 6C), this would result in decreased binding to a labeled I* element probe. Hence, the oligonucleotide competition experiments suggest that sequences within both the 5′ and 3′ subregions of the I* element are required for the formation of the I* element-binding complex. We then sought to detect and identify the proteins binding to the 5′ and 3′ subregions of the I* element.
Distinct and more rapidly migrating constitutively binding complexes bound to the 5′ and 3′ subregion probes (compare the binding in lanes 3 and 4 or 7 and 8, respectively, with that in lanes 1 and 2 in Fig. 6A). Mutant 5′ or 3′ subregion probes containing the 12- and 8-nt mutations described in Fig. 4 did not bind the complexes that bound the two wild-type probes (Fig. 6A). Binding of the two complexes to a 5′ subregion (I*5′) probe is specific, as it was competed by an excess of a wild-type 5′ subregion oligonucleotide but was not competed by a mutant 5′ subregion oligonucleotide or unrelated oligonucleotides (compare the binding in lane 3 or lanes 4 through 6, respectively, with the binding in lane 2 in Fig. 6D). Likewise, binding of a single complex to the 3′ subregion (I*3′) probe is specific, as it was competed by an excess of the wild-type 3′ subregion oligonucleotide but was not competed by a mutant 3′ subregion oligonucleotide or unrelated oligonucleotides (compare the binding in lane 4 or lanes 5 through 7, respectively, with the binding in lane 3 in Fig. 6E). Note that the complexes binding to the 5′ subregion probe are not competed by an excess of an oligonucleotide corresponding to the 3′ subregion (lane 5 in Fig. 6D) and vice versa (lane 6 in Fig. 6E). These data suggest that the single protein complex binding specifically to the 3′ footprinted subregion probe (Fig. 6E) is distinct from the proteins binding to the 5′ subregion probe. Computer database searches did not turn up a binding site match for the 3′ subregion sequences, suggesting that the 3′ subregion-binding protein is a novel DNA-binding factor. In these mobility shifts, binding to the I*3′ probe is significantly weaker than binding to the I* or I*5′ probe (Fig. 6A and E). No significant increase in binding intensity was noted when a labeled I*3′ probe including an additional 5 nt of the 5′ or 3′ flanking sequence was used (data not shown), suggesting that the I*3′ probe used in these experiments does not lack sequences required for binding of the complex.
The 5′ footprinted subregion includes potential binding sites for the Sp1 and Sp3 transcription factors (i.e., the repeated sequence CCCCACCCC) (35), suggesting that one or both of the complexes binding to the wild-type 5′ subregion probe could be these transcription factors. To address this question, antibody supershift mobility shift assays were performed by using a panel of antibodies to individual members of the Sp1 family of transcription factors. Two anti-Sp1 antibodies recognizing different portions of Sp1 were used in these experiments. Only one of the anti-Sp1 antibodies (anti-Sp1-1 recognizing amino acids 436 to 454 of the Sp1 protein) supershifts the more slowly migrating (upper) complex binding to the 5′ subregion probe. In contrast, the anti-Sp1-2 antibody (recognizing amino acids 520 to 538 of the Sp1 protein) does not supershift the upper complex binding to the 5′ subregion probe (lanes 3 and 4 in Fig. 6F). Recombinant Sp1 protein is supershifted quantitatively by both anti-Sp1 antibodies (lanes 9 and 10 in Fig. 6F), demonstrating that both anti-Sp1 antibodies are functional in these experiments. The additional three anti-Sp family antibodies did not supershift the upper complex binding to the 5′ subregion probe (lanes 5 to 7 in Fig. 6F). Additional mobility shift assays were performed by using a series of labeled mutant I*5′ probes, each mutant containing a 4- to 7-base mutation of the sequence immediately 5′ or 3′ of the Sp1 and Sp3 DNA-binding sites present in the wild-type probe. Binding of the upper complex and recombinant Sp1 to this series of mutant I*5′ probes was identical and undiminished compared to their binding to a wild-type I*5′ probe (data not shown). Taken together, these data suggest that the upper complex binding the 5′ subregion probe is an Sp1-like protein that lacks at least the epitope recognized by the Sp1-2 antibody but has a DNA-binding domain in common with recombinant Sp1. Alternatively, the epitope recognized by the Sp1-2 antibody could be masked either by a posttranslational modification of the Sp1 protein or as a result of the binding of another protein to Sp1 in the 3T3 cell nuclear extracts. The anti-Sp1-2 antibody is specific for amino acids 520 to 538 within the C domain of the Sp1 transcription factor, a region that has been shown to be sufficient, but not necessary, to mediate transcriptional activation of a reporter gene (12).
The more rapidly migrating (lower) complex binding to the 5′ subregion probe is supershifted by the anti-Sp3 antibody but not by the anti-Sp1-2, anti-Sp2, or anti-Sp4 antibody, suggesting that it is antigenically related to the Sp3 transcription factor (lanes 4 to 7 in Fig. 6F). All supershifts were abolished by addition of the peptides used to generate the various antibodies (data not shown). The lower complex binding to the 5′ subregion probe was also supershifted by the anti-Sp1-1 antibody (lane 3 in Fig. 6F). The anti-Sp1-1 antibody did not recognize immunoprecipitated Sp3 in a Western blot analysis (data not shown). It is possible that addition of the anti-Sp1-1 antibody distorts the Sp1-like protein-probe complex, thereby allowing concurrent binding of Sp3 and the Sp1-like proteins together on the I*5′ probe.
Taken together, these mobility shift assays suggest that the I*5′ sequences, containing Sp1- and Sp3-binding sites, also bind Sp3 and an Sp1-like protein (Fig. 6F). In transfection experiments, an MCP-1 reporter gene in which just the I*5′ sequences have been added back is strongly PDGF inducible, in contrast to the significantly decreased PDGF inductions observed with reporter constructs containing the complete I* element (compare the PDGF inducibilities of constructs 6 and 2 with those of constructs 4 and 5 in Fig. 5A). Hence, adding back only the I*5′ portion of the full-length I* element to an I*-lacking, PDGF-inducible reporter gene (in these experiments effectively tethering Sp1 and Sp3 to the MCP-1 reporter gene) does not restore inhibition of PDGF induction. Rather, the complete I* element is necessary for inhibition of PDGF induction of MCP-1, suggesting that cooperative interactions occur among the I*-binding proteins and 5′ and 3′ binding sites in vivo in transfection experiments. These findings raise the question of whether the I*5′-binding Sp3 protein, the Sp1-like protein, or both are components of the I* element-binding complex that appears to maintain inhibition of PDGF induction of MCP-1 in the absence of the heptamer.
The I* element-binding complex appears to contain both the Sp3 transcription factor and the Sp1-like protein.
All of the mobility shift assays in this study were performed under conditions of free probe excess (data not shown). Hence, the identification of Sp3 and an Sp1-like protein as both binding to the 5′ subregion (I*5′) probe, but only a single more slowly migrating complex binding to the I* probe (Fig. 6A), suggests that Sp3 and the Sp1-like protein are both contained within the larger complex. Were this not the case, one would have expected to observe either three complexes binding to the I* probe or a single complex binding to the 5′ subregion probe. To address this question further, antibody supershift mobility shift assays were performed to determine the effects of the anti-Sp1 family antibodies on binding to the I* probe. Only the addition of the anti-Sp1-1 antibody greatly diminished the binding of the single complex to the I* element probe (compare binding in lane 2 with that in lane 1 or lanes 3 to 6 in Fig. 7A), consistent with steric hindrance of the formation of the complex or its binding to the I* element in the setting of antibody binding to the Sp1-like protein. The anti-Sp1-1 result was reversed by concurrent addition of the peptide used to generate the antibody (data not shown).
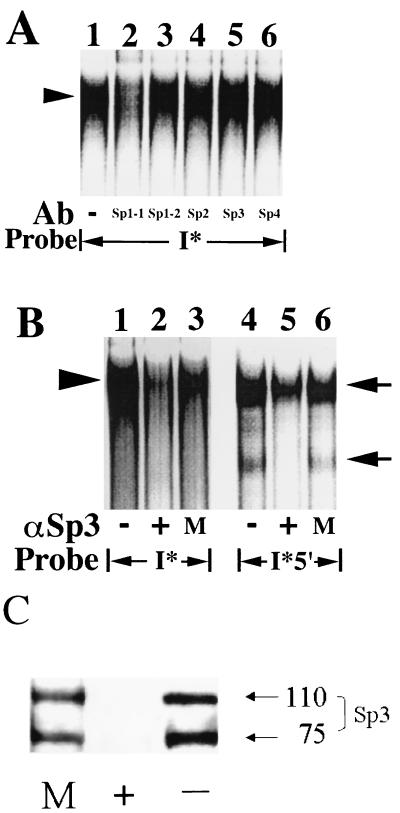
FIG. 7.
The I* element-binding complex contains the Sp3 transcription factor and an Sp1-like protein. (A) Nuclear extracts (12.5 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probe shown. The sequence of the probe is given in Fig. 6G. Antibodies (Ab) specific for individual members of the Sp1 family of transcription factors (2 μg) were added where indicated. The Sp1-1 and Sp1-2 antibodies are as described in the legend to Fig. 6F. The arrowhead on the left highlights the predominant complex that binds specifically to the I* probe. Free probe is not shown. No complexes were observed with the probe alone in the absence of extract (data not shown). (B) Nuclear extracts (12.5 μg) prepared from quiescent fibroblasts were used in mobility shift assays along with the radiolabeled double-stranded oligonucleotide probes shown. The sequences of the probes are given in Fig. 6G. Extracts were immunodepleted initially with anti-Sp3 antibodies (+), mock immunodepleted with rabbit immunoglobulin G (M), or not treated (−). The arrowhead on the left highlights the predominant complex that binds specifically to the I* probe. The arrows on the right highlight the positions of the two complexes that bind specifically to the 5′ subregion (I*5′) probe. Free probe is not shown. No complexes were observed with the probes alone in the absence of extract (data not shown). (C) Immunoblot of an aliquot (30 μg) of the extracts used for panel B with the anti-Sp3 antibody. The designations +, M, and − are as described for panel B. Molecular sizes are given in kilodaltons.
No effect on binding to the I* probe was noted after addition of anti-Sp3 antibody or any of the other anti-Sp1 family antibodies (lanes 3 to 6 in Fig. 7A). The latter data suggest either that Sp3 is not a component of the I*-binding complex or that the Sp3 epitope recognized by the antibody is inaccessible to the antibody by virtue of its being contained within the I*-binding complex. As an alternative means of assessing whether Sp3 is a component of the I* probe-binding complex, anti-Sp3 antibody was used to immunodeplete fibroblast nuclear extracts of Sp3 prior to the use of these extracts in mobility shift assays. A single immunoprecipitation step, using the anti-Sp3 antibody, was sufficient to remove detectable lower complex binding to the 5′ subregion probe in mobility shift assays compared to mock-treated or untreated extracts (compare the lower complex binding in lane 5 to that in lanes 4 and 6 in Fig. 7B), again suggesting that the lower complex binding to the 5′ subregion probe is the Sp3 protein. In contrast, upper complex binding to the 5′ subregion probe was relatively unaffected by the Sp3 immunodepletion procedure (lanes 4 to 6 in Fig. 7B). Western blot analysis was unable to detect residual Sp3 in nuclear extracts after immunodepletion using the anti-Sp3 antibody (Fig. 7C). Immunodepletion of detectable, free Sp3 in fibroblast nuclear extracts greatly reduced the formation or binding of the protein complex to the I* element in mobility shift assays (compare the binding in lane 2 with that in lanes 1 and 3 in Fig. 7B), suggesting that Sp3 is required for the formation of the complex or its binding to the I* probe.
DISCUSSION
A pair of I* elements are present in the MCP-1 5′ flanking sequences and are heptamer regulated.
We have shown previously that PDGF induction of the MCP-1 IEG requires the presence of the 7-nt motif TTTTGTA within the 3′ untranslated sequences of MCP-1. Moreover, MCP-1 reporter genes lacking the heptamer were not PDGF inducible in previous transfection experiments (20). In this study, we have created MCP-1 reporter genes lacking the heptamer that are strongly PDGF inducible through the deletion of a 1,936-bp segment of the MCP-1 5′ flanking sequences (Fig. 1). Hence, while the heptamer is essential for PDGF induction of an intact MCP-1 reporter gene (20), the function of the heptamer is dispensable upon the removal of a defined stretch of 5′ flanking MCP-1 sequences. These data suggest that the apparent positive effect on PDGF inducibility of readdition of the heptamer to the 3′ untranslated sequences results from the heptamer functioning to remove an inhibition of PDGF induction of MCP-1 maintained by what we have termed an inhibitory element(s) present within the 5′ sequences.
Finer 5′ deletional analysis detected the existence of two independently acting inhibitory elements. Although the more potent inhibition of PDGF induction was maintained by a 695-bp SpeI-EcoRI MCP-1 5′ sequence fragment, inhibition maintained by either fragment is reversed by readdition of the heptamer to the 3′ untranslated sequences. These findings demonstrate directly that PDGF-regulated interactions occur between the 3′-located heptamer and the two inhibitory elements present within the MCP-1 5′ flanking sequences in a heptamer sequence-specific manner (Fig. 2B). Since the heptamer is also fully functional when located upstream of the MCP-1 transcription start site, it likely functions as a DNA-regulatory element, presumably as the binding site for a regulatory factor(s). Consistent with this prediction are the results of mobility shift assays detecting a single protein complex binding specifically to a wild-type heptamer probe and not binding to mutant heptamer probes containing the three mutations described in Fig. 2B (data not shown). Computer-aided sequence comparisons suggested that there are no significant regions of sequence homology between the two inhibitory element-containing fragments (data not shown). In total, these data are consistent with the existence of two distinct inhibitory elements present in the MCP-1 5′ flanking sequences that interact with the heptamer TTTTGTA in a PDGF-regulated manner.
Several laboratories have employed nuclear runon assays to demonstrate that PDGF-stimulated expression of the MCP-1 IEG is the result of new transcription (26, 36, 59, 62). Hence, the mechanism whereby the 695-bp SpeI-EcoRI 5′ fragment inhibits PDGF-stimulated expression of MCP-1 in the absence of the heptamer appears to be at the level of repression of transcription. Conversely, the mechanism(s) whereby the heptamer relieves inhibition, in a PDGF-regulated manner, likely occurs through heptamer-mediated reversal of the transcriptional repression of MCP-1 maintained by the 695-bp inhibitory fragment. Other theoretical mechanisms of action for the inhibitory fragment or inhibitory element, e.g., diminished processivity of the RNA polymerase through the MCP-1 coding sequences and/or changes in MCP-1 RNA stability, are rendered unlikely, given the normal position of the SpeI-EcoRI inhibitory fragment within the distal 5′ flanking sequences of the MCP-1 gene, i.e., with endpoints at −1764 and −1069 relative to the MCP-1 start of transcription.
One I* element is contained within a 59-nt MCP-1 sequence and binds a multiprotein complex including Sp3 and an Sp1-like protein.
Evidence presented in this report demonstrates that a 59-nt portion of MCP-1 5′ flanking sequences recapitulates the properties of the full-length 695-bp inhibitory fragment, strongly suggesting that the 59-nt sequence contains one of the I* elements detected in our initial experiments. Significantly, the 59-nt I*-containing sequence was demonstrated to bind a single predominant protein complex with greatly decreased binding or formation of the single complex observed to either of two mutant 59-nt oligonucleotides. Since both mutations significantly decrease the repression of PDGF induction maintained by the wild-type 695-bp fragment in transfection experiments (Fig. 4), these data strongly suggest that a causal relationship exists between the formation or binding of the 59-nt I* element-binding complex and repression of PDGF-stimulated transcription of MCP-1 in intact cells. Note that the 59-nt I* element-binding complex is competed by oligonucleotides corresponding to either the 5′ footprinted subregion alone, a 59-nt oligonucleotide containing a mutation of just the 5′ subregion (I*m1), or the 3′ footprinted subregion alone (Fig. 6C). In total, these data suggest that at least two distinct DNA sequences, contained within the 5′ and 3′ footprinted subregions, comprise the overall 59-nt I* element. By extension, regulatory or architectural proteins binding to the two DNA sites would be predicted to be components of the 59-nt I*-binding complex. In the remainder of this discussion, the designation I* element will refer to the 59-nt oligonucleotide.
Antibody supershift mobility shift assays and immunodepletion experiments suggest that both Sp3 and the Sp1-like protein that we have identified as binding independently to the 5′ subregion probe are contained within the I* element-binding complex (Fig. 7). The apparent coexistence in a multiprotein complex of a transcriptional activator like Sp1 (7, 11, 44, 45, 52) and Sp3, a transcription factor that often antagonizes Sp1 function or binding, thereby effectively repressing transcription (10, 15, 40, 46), is an uncommon occurrence. Transcriptional activation of the neuronal nicotinic acetylcholine receptor β4 subunit gene has been reported to involve interactions between Sp1 and Sp3 on a single Sp1-Sp3 DNA-binding sequence (4). For MCP-1, the apparent juxtaposition of antagonistic transcription factors suggests that the I*-binding complex could, in theory, also perform an activating function in regulating the expression of other genes.
Repressosome model of inhibition of PDGF induction of MCP-1.
The experiments presented in this report are consistent with the existence of a multiprotein complex binding to the I* element that represses PDGF-stimulated transcription of MCP-1 in the absence of the heptamer. Furthermore, the evidence suggests that (i) at least three distinct proteins comprise the I*-binding complex and (ii) regulatory or architectural proteins binding to at least two distinct DNA-binding sites are involved in the formation of the I*-binding complex. We propose that the formation of the I*-binding complex creates a multiprotein repressor surface that maintains inhibition of MCP-1 transcription. Upon PDGF stimulation of cells, and in the presence of the heptamer TTTTGTA, repression of MCP-1 transcription is relieved. A growing body of literature is lending elegant experimental support to the concept of an enhanceosome, i.e., a multiprotein complex formed in a cooperative manner, in which the component proteins bind to independent sites on a short segment of DNA. This results in the formation of a multiprotein activation surface that effectively increases the transcription of a given gene (6, 39, 47, 49). We propose that the I*-binding complex is the first example of the opposite of an enhanceosome, i.e., a multiprotein complex mediating repression of MCP-1 transcription, in which the component proteins bind to distinct sites within the overall I* element. Our results suggest that cooperative interactions occur among the I*-binding proteins and binding sites both in vitro (Fig. 6B) and in vivo (Fig. 5A). We propose that the I*-binding complex be called a repressosome. Our current model of the PDGF-regulated MCP-1 repressosome is shown in Fig. 8.
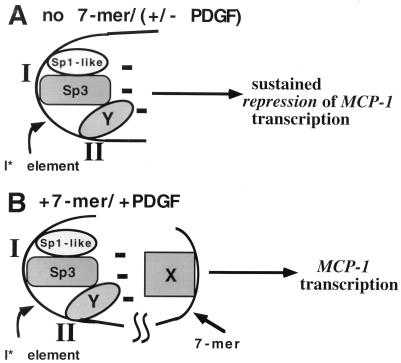
FIG. 8.
Repressosome model of inhibition of PDGF induction of MCP-1. (A) A multiprotein complex (repressosome) binds to the I* element. At least three proteins form the repressosome and bind to two distinct sites on the I* element, designated I and II and corresponding to the I*5′ and I*3′ sequences, respectively. The proteins include the Sp3 transcription factor, an Sp1-like protein, and an apparently novel regulatory protein that binds to site II (represented by oval Y). The net effect is the creation on the I* element of a repressor surface that represses MCP-1 transcription in the absence of the heptamer TTTTGTA (denoted by the multiple minus signs). PDGF stimulation does not result in MCP-1 expression in the absence of the heptamer due to the unopposed action of the repressosome. (B) After PDGF stimulation and in the presence of the heptamer, the effect of the repressor surface is relieved or neutralized and MCP-1 transcription proceeds. An assumption inherent in this model is that the heptamer-binding protein(s) (represented by rectangle X) is altered after PDGF stimulation, thereby allowing it to interact with the repressor surface formed on the I* element. Although Sp3 and the Sp1-like protein are drawn as interacting, this does not have to obtain. This model does not exclude the possibility that additional regulatory or architectural proteins are involved in the formation of the repressosome.
Our results highlight a mechanism of transcriptional repression that differs in several important respects from other reported repression mechanisms, such as the Mad-Max and unliganded nuclear hormone receptor systems. In the former case, increasing levels of the Mad protein appear to titrate the available Myc protein from activator Myc-Max heterodimers and into repressor Mad-Max heterodimers in the setting of cellular differentiation (2). In the latter case, addition of the hormone reverses transcriptional repression by directly binding to its nuclear receptor. In both cases, transcriptional repression appears to be maintained by an additional multiprotein complex that contains a histone deacetylase(s) and that is recruited to proteins (e.g., Mad-Max) binding to a single DNA-binding site (41, 50, 53, 74, 75). In yeast, repression of a-specific genes appears to share some mechanistic details with the model presented in Fig. 8; i.e., repression appears to be maintained by a complex containing the Tup1 and Ssn6 repressors together with the MCM1 and α2 regulatory proteins bound to adjacent sites on the a-specific gene operator (34, 60).
In contrast to the examples cited above, repression of PDGF-stimulated transcription of MCP-1 is not relieved simply by exposure of the cells to the growth factor. Rather, PDGF stimulation of the cells removes the inhibitory effect of the I* sequences only in the presence of the heptamer TTTTGTA. These results suggest that a regulatory protein(s) binding to the heptamer relieves the I*-mediated repression of MCP-1 transcription in response to PDGF treatment. It remains to be determined whether this occurs through the opposed actions of histone acetylases and/or deacetylases, possibly recruited by the heptamer. Other potential mechanisms include (PDGF-regulated proteins binding to) the heptamer masking the proposed multiprotein repressor surface and/or posttranslationally modifying a protein(s) within the repressosome, thereby allowing transcription of MCP-1. The heptamer does not compete binding to the I* element (Fig. 6C), suggesting that it is unlikely that the heptamer functions by ablating repressosome binding to the I* element. Given that the heptamer motif is present within the 3′ untranslated sequences of numerous IEGs in addition to MCP-1 (20), it is tempting to speculate that the repressosome-heptamer interaction may be a shared regulatory mechanism operative for the set of IEGs.
ACKNOWLEDGMENTS
We thank John Alberta, Mike Carey, Rolf G. Freter, Charles Stiles, I. Bernard Weinstein, and Howard J. Worman for critical reading of the manuscript. We also thank Andrea Troxel for help with statistical analysis, Xiaoqin Qu for assistance with the Western analyses, and Christine Chen and Roger Lucas for technical assistance.
This research was supported in part by grants from the NIH (Shannon Award 1R55CA76045-01), the March of Dimes (Basil O’Connor Starter Scholar Award 5-FY97-0031), and the Council for Tobacco Research (to R.R.F.). R.R.F. is a Scholar of the V Foundation for Cancer Research and the James S. McDonnell Foundation. Additional support for this work was provided by grants from the Elsa U. Pardee Foundation for Cancer Research, The Milheim Foundation for Cancer Research, and the Ruth Estrin Goldberg Memorial for Cancer Research (to R.R.F.). H.A.S. is supported by a grant from the NIH (HL03806-01).
REFERENCES
- 1.Almendral J M, Sommer D, Macdonald-Bravo H, Burckhardt J, Perera J, Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988;8:2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayer D E, Eisenman R N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz L A, Riabowol K T, Gilman M Z. Multiple sequence elements of a single functional class are required for cyclic AMP responsiveness of the mouse c-fos promoter. Mol Cell Biol. 1989;9:4272–4281. doi: 10.1128/mcb.9.10.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bigger C B, Melnikova I N, Gardner P D. Sp1 and Sp3 regulate expression of the neuronal nicotinic acetylcholine receptor beta-4 subunit gene. J Biol Chem. 1997;272:25976–25982. doi: 10.1074/jbc.272.41.25976. [DOI] [PubMed] [Google Scholar]
- 5.Boring L, Gosling J, Cleary M, Charo I F. Decreased lesion formation in CCR2 −/− mice reveals a role for chemokines in the initiation of atherosclerosis. Nature. 1998;394:894–897. doi: 10.1038/29788. [DOI] [PubMed] [Google Scholar]
- 6.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Artlett C M, Jimenez S A, Varga J. Modulation of human alpha1 (I) procollagen gene activity by interaction with Sp1 and Sp3 transcription factors in vitro. Gene. 1998;215:101–110. doi: 10.1016/s0378-1119(98)00268-6. [DOI] [PubMed] [Google Scholar]
- 8.Christy B, Nathans D. Functional serum response elements upstream of the growth factor-inducible gene zif268. Mol Cell Biol. 1989;9:4889–4895. doi: 10.1128/mcb.9.11.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cochran B H, Reffel A C, Stiles C D. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell. 1983;33:939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- 10.Conn K J, Rich C B, Jensen D E, Fontanilla M R, Bashir M M, Rosenbloom J, Foster J A. Insulin-like growth factor-I regulated transcription of the elastin gene through a putative retinoblastoma control element. J Biol Chem. 1996;271:28853–28860. doi: 10.1074/jbc.271.46.28853. [DOI] [PubMed] [Google Scholar]
- 11.Costa M W, Atchison M L. Identification of an Sp1-like element within the immunoglobulain kappa 3′ enhancer necessary for maximal enhancer activity. Biochemistry. 1996;35:8662–8669. doi: 10.1021/bi952801y. [DOI] [PubMed] [Google Scholar]
- 12.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 13.Dean M, Levine R A, Ran W, Kindy M S, Sonenshein G E, Campisi J. Regulation of c-myc transcription and mRNA abundance by serum growth factors and cell contact. J Biol Chem. 1986;261:9161–9166. [PubMed] [Google Scholar]
- 14.De Belle I, Walker P R, Smith I C P, Sikorska M. Identification of a multiprotein complex interacting with the c-fos serum response element. Mol Cell Biol. 1991;11:2752–2759. doi: 10.1128/mcb.11.5.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Luca P, Majello B, Lania L. Sp3 represses transcription when tethered to promoter DNA or targeted to promoter proximal RNA. J Biol Chem. 1996;271:8533–8536. doi: 10.1074/jbc.271.15.8533. [DOI] [PubMed] [Google Scholar]
- 16.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fisch T M, Prywes R, Simon M C, Roeder R G. Multiple sequence elements in the c-fos promoter mediate induction by cAMP. Genes Dev. 1989;3:198–211. doi: 10.1101/gad.3.2.198. [DOI] [PubMed] [Google Scholar]
- 18.Freter R R, Alberta J A, Hwang G Y, Wrentmore A L, Stiles C D. Platelet-derived growth factor induction of the immediate-early gene MCP-1 is mediated by NF-κB and a 90 kD phosphoprotein coactivator. J Biol Chem. 1996;271:17417–17424. doi: 10.1074/jbc.271.29.17417. [DOI] [PubMed] [Google Scholar]
- 19.Freter R R, Alberta J A, Lam K K, Stiles C D. A new platelet-derived growth factor-regulated genomic element which binds a serine/threonine phosphoprotein mediates induction of the slow immediate-early gene MCP-1. Mol Cell Biol. 1995;15:315–325. doi: 10.1128/mcb.15.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freter R R, Irminger J C, Porter J A, Jones S D, Stiles C D. A novel 7-nucleotide motif located in 3′ untranslated sequences of the immediate-early gene set mediates platelet-derived growth factor induction of the JE gene. Mol Cell Biol. 1992;12:5288–5300. doi: 10.1128/mcb.12.12.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilman M Z. The c-fos serum response element responds to protein kinase C-dependent and -independent signals but not to cyclic AMP. Genes Dev. 1988;2:394–402. doi: 10.1101/gad.2.4.394. [DOI] [PubMed] [Google Scholar]
- 22.Graham R, Gilman M. Distinct protein targets for signals acting at the c-fos serum response element. Science. 1991;251:189–192. doi: 10.1126/science.1898992. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg M E, Siegfried Z, Ziff E B. Mutation of the c-fos gene dyad symmetry element inhibits serum inducibility of transcription in vivo and the nuclear regulatory factor binding in vitro. Mol Cell Biol. 1987;7:1217–1225. doi: 10.1128/mcb.7.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberg M E, Ziff E B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–437. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 25.Gu L, Okada Y, Clinton S K, Gerard C, Sukhova G K, Libby P, Rollins B J. Absence of monocyte chemoattractant protein-1 reduces atherosclerosis in low density lipoprotein receptor-deficient mice. Mol Cell. 1998;2:275–281. doi: 10.1016/s1097-2765(00)80139-2. [DOI] [PubMed] [Google Scholar]
- 26.Hall D J, Alberta J A, Stiles C D. Labile repressors are involved in the transcriptional control of PDGF-responsive genes. Oncogene Res. 1989;1:127–184. [PubMed] [Google Scholar]
- 27.Hall D J, Jones S D, Kaplan D R, Whitman M, Rollins B J, Stiles C D. Evidence for a novel signal transduction pathway activated by platelet-derived growth factor and by double-stranded RNA. Mol Cell Biol. 1989;9:1705–1713. doi: 10.1128/mcb.9.4.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall D J, Stiles C D. Platelet-derived growth factor-inducible genes respond differentially to at least two distinct intracellular second messengers. J Biol Chem. 1987;262:15302–15308. [PubMed] [Google Scholar]
- 29.Herschman H R. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 30.Hill C S, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 32.Johansen F-E, Prywes R. Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol Cell Biol. 1993;13:4640–4647. doi: 10.1128/mcb.13.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansen F-E, Prywes R. Two pathways for serum regulation of the c-fos serum response element require specific sequence elements and a minimal domain of serum response factor. Mol Cell Biol. 1994;14:5920–5928. doi: 10.1128/mcb.14.9.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson A D. Molecular mechanisms of cell-type determination in budding yeast. Curr Opin Genet Dev. 1995;5:552–558. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 35.Kadonaga J T, Carner K R, Masiarz F R, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 36.Kawahara R S, Deng Z W, Deuel T F. Glucocorticoids inhibit the transcriptional induction of JE, a platelet-derived growth factor-inducible gene. J Biol Chem. 1991;266:13261–13266. [PubMed] [Google Scholar]
- 37.Kelly K, Cochran B H, Stiles C D, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 38.Kelly K, Siebenlist U. Mitogenic activation of normal T cells leads to increased initiation of transcription in the c-myc locus. J Biol Chem. 1988;263:4828–4831. [PubMed] [Google Scholar]
- 39.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-beta enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A, Butler A P. Transcription factor Sp3 antagonizes activation of the ornithine decarboxylase promoter by Sp1. Nucleic Acids Res. 1997;25:2012–2019. doi: 10.1093/nar/25.10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laherty C D, Yang W, Sun J, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 42.Lau L F, Nathans D. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci USA. 1987;84:1182–1186. doi: 10.1073/pnas.84.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau L F, Nathans D. Identification of a set of genes expressed during the G0/G1 transition of cultured mouse cells. EMBO J. 1985;4:3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li N, Seetharam S, Seetharam B. Characterization of the human transcobalamin II promoter. J Biol Chem. 1998;273:16104–16111. doi: 10.1074/jbc.273.26.16104. [DOI] [PubMed] [Google Scholar]
- 45.Majello B, De Luca P, Hagen G, Suske G, Lania L. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. 1994;22:4914–4921. doi: 10.1093/nar/22.23.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majello B, DeLuca P, Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J Biol Chem. 1997;272:4021–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- 47.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN-beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 48.Misra R P, Rivera V M, Wang J M, Fan P, Greenberg M E. The serum response factor is extensively modified by phosphorylation following its synthesis in serum-stimulated fibroblasts. Mol Cell Biol. 1991;11:4545–4554. doi: 10.1128/mcb.11.9.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN-beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 50.Nagy L, Kao H, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 51.Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 52.Pascal E, Tjian R. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 1991;5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- 53.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 54.Perez-Albuerne E D, Schatteman G, Sanders L K, Nathans D. Transcriptional regulatory elements downstream of the JunB gene. Proc Natl Acad Sci USA. 1993;90:11960–11964. doi: 10.1073/pnas.90.24.11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pledger W J, Stiles C D, Antoniades H N, Scher C D. Induction of DNA synthesis in Balb/c-3T3 cells by serum components: reevaluation of the commitment process. Proc Natl Acad Sci USA. 1977;74:4481–4485. doi: 10.1073/pnas.74.10.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prywes R, Roeder R G. Inducible binding of a factor to the c-fos enhancer. Cell. 1986;47:777–784. doi: 10.1016/0092-8674(86)90520-9. [DOI] [PubMed] [Google Scholar]
- 57.Prywes R, Roeder R G. Purification of the c-fos enhancer-binding protein. Mol Cell Biol. 1987;7:3482–3489. doi: 10.1128/mcb.7.10.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qureshi S A, Cao X, Sukatme V P, Foster D A. v-src activates mitogen-responsive transcription factor egr-1 via serum response elements. J Biol Chem. 1991;266:10802–10806. [PubMed] [Google Scholar]
- 59.Rameh L E, Armelin M C S. Downregulation of JE and KC genes by glucocorticoids does not prevent the G0→G1 transition in BALB/3T3 cells. Mol Cell Biol. 1992;12:4612–4621. doi: 10.1128/mcb.12.10.4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Redd M J, Arnaud M B, Johnson A D. A complex composed of Tup1 and Ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 61.Rollins B J. Chemokines. In: Mertelsmann R, Herrmann F, editors. Hematopoietic growth factors in clinical application. 2nd ed. New York, N.Y: Marcel Dekker, Inc.; 1995. pp. 357–380. [Google Scholar]
- 62.Rollins B J, Morrison E D, Stiles C D. A cell-cycle constraint on the regulation of gene expression by platelet-derived growth factor. Science. 1987;238:1269–1271. doi: 10.1126/science.3685976. [DOI] [PubMed] [Google Scholar]
- 63.Rollins B J, Morrison E D, Stiles C D. Cloning and expression of JE, a gene inducible by platelet-derived growth factor and whose product has cytokine-like properties. Proc Natl Acad Sci USA. 1988;85:3738–3742. doi: 10.1073/pnas.85.11.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rollins B J, Stier P, Ernst T, Wong G G. The human homolog of the JE gene encodes a monocyte secretory protein. Mol Cell Biol. 1989;9:4687–4695. doi: 10.1128/mcb.9.11.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rollins B J, Stiles C D. Serum-inducible genes. Adv Cancer Res. 1989;53:1–32. doi: 10.1016/s0065-230x(08)60277-8. . (Review.) [DOI] [PubMed] [Google Scholar]
- 66.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sassone-Corsi P, Visvader J, Ferland L, Mellon P L, Verma I M. Induction of proto-oncogene fos transcription throught the adenylate cyclase pathway: characterization of a cAMP-responsive element. Genes Dev. 1988;2:1529–1538. doi: 10.1101/gad.2.12a.1529. [DOI] [PubMed] [Google Scholar]
- 68.Skopicki H A, Lyons G, Schatteman G, Smith R C, Schirm S, Isner J M I, Walsh K. Embryonic expression of a growth-arrest homeobox gene in cardiac, smooth and skeletal muscle lineages. Circ Res. 1997;80:452–462. doi: 10.1161/01.res.80.4.452. [DOI] [PubMed] [Google Scholar]
- 68a.Sridhar, P., and R. R. Freter. Unpublished data.
- 69.Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 1987;6:2711–2717. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- 71.Treisman R. The SRE: a growth factor responsive transcriptional regulator. Semin Cancer Biol. 1990;1:47–58. [PubMed] [Google Scholar]
- 72.Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 73.Wagner B J, Hayes T E, Hoban C J, Cochran B H. The SIF binding element confers sis/PFGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y, LeRoy G, Seelig H, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]