Abstract
Yeast artificial chromosomes (YACs) are a common tool for cloning eukaryotic DNA. The manner by which large pieces of foreign DNA are assimilated by yeast cells into a functional chromosome is poorly understood, as is the reason why some of them are stably maintained and some are not. We examined the replication of a stable YAC containing a 240-kb insert of DNA from the human T-cell receptor beta locus. The human insert contains multiple sites that serve as origins of replication. The activity of these origins appears to require the yeast ARS consensus sequence and, as with yeast origins, additional flanking sequences. In addition, the origins in the human insert exhibit a spacing, a range of activation efficiencies, and a variation in times of activation during S phase similar to those found for normal yeast chromosomes. We propose that an appropriate combination of replication origin density, activation times, and initiation efficiencies is necessary for the successful maintenance of YAC inserts.
Yeast artificial chromosomes (YACs) are important cloning tools for the analysis of complex genomes such as that of humans. They allow the maintenance, propagation, and analysis of regions of such genomes in an experimentally tractable system, the yeast Saccharomyces cerevisiae. Despite the fact that only one yeast origin of replication is provided by the YAC vector, YACs can maintain cloned inserts of several hundred kilobase pairs (8). However, a major drawback to the use of YACs is the high frequency of regions in the human genome that are unstable in YACs. Such unstable YACs contain inserts that cannot be stably maintained as full-length molecules and are deleted upon propagation in yeast. The instability results in gaps during physical mapping and gene hunting efforts. Examples of regions with unstable YACs or unclonable regions include the Huntington’s disease locus (3), the BRCA1 region (2), and the Bloom’s syndrome region (45).
If replication initiates only from the single origin located close to the end of the YAC molecule, it is unlikely that complete replication of this molecule could occur within the length of a normal S phase (approximately 25 min), given that the replication fork rate in yeast is ∼4 kb/min at 23°C (39). YACs are often as large as native yeast chromosomes, and replication of even the smallest yeast chromosomes, which are less than 300 kb, occurs by initiation at multiple sites throughout the length of the molecule (17, 33, 52). It is therefore possible that sites within the human DNA serve as origins of replication in yeast to complete replication of large YACs and that these origins are essential for stable YAC maintenance.
Early work suggested that mammalian genomes have sequences that are capable of providing autonomous replication of plasmids in yeast (51). Autonomously replicating sequences (ARSs) are identified by their ability to promote high-frequency transformation and stable extrachromosomal maintenance of plasmids (44). These A/T-rich ARSs correspond to plasmid replication origins (6). Restriction fragments from the mouse genome are capable of ARS activity in yeast, and the frequency of these sequences is estimated to be 1 per 24 kb (41). Some human sequences have been shown to have ARS activity in yeast as well (30). Given that the base pair compositions of the human genome and the yeast genome are similar (approximately 60 and 62% A/T, respectively), it may not be surprising to find that human sequences are capable of ARS activity in yeast. However, since many sequences in the yeast genome are capable of ARS activity on plasmids but are weak chromosomal replication origins (17, 52) or are not active chromosomal replication origins at all (13, 33), the ability of human sequences to provide ARS activity on a plasmid is not necessarily an indication that these same sequences act as replication origins in the context of a YAC. Using DNA fiber-fluorescent in situ hybridization on yeast cells with a YAC containing human DNA from the Duchenne muscular dystrophy gene, Rosenberg et al. (40) found structures that suggested the presence of two replication origins within the YAC insert. However, these replication origins were never confirmed by using other techniques, nor were they more precisely localized.
Here we report a systematic study of how yeast replicates a large insert of human DNA in a YAC. We scanned the 240-kb YAC insert to determine whether sequences within it were initiating replication. Replication origins were found. They are discrete units that appear to have the same sequence requirements as yeast chromosomal replicators. In addition, the origin spacing, variation in efficiencies of activation, and variation in times of activation in the S phase are all similar to those of yeast chromosomes. The presence of multiple sites of replication initiation in the human insert of this stable YAC suggests that regions lacking these fortuitous sequences will have a replication defect in yeast that could jeopardize the integrity and maintenance of the molecule and could be a biological basis of YAC instability.
MATERIALS AND METHODS
Strains.
Chromosome 7 YACs were generously provided by Eric Green at the National Human Genome Research Institute, National Institutes of Health. The background yeast strain of these YACs is AB1380 (MATa ade2-1 can1-100 lys 2-1 ura3 trp1 his5 psi+). All of the YACs were examined by pulsed-field gel electrophoresis and Southern analysis, and their sizes were estimated by using a pulsed-field gel marker of lambda concatemers (New England Biolabs). yWSS349 was transformed into yeast strain RM14-3a (MATa cdc7-1 bar1 ura3-52 trp1-289 leu2-3,112 his6) (29). The intact YAC was isolated as previously described (19). Briefly, total chromosomal DNA from yWSS349 in AB1380 was isolated in concentrated agarose plugs and separated in a low-melting-point agarose pulsed-field gel. The gel was run at 14°C for 45 h with a switch time ramped from 20 to 50 s at 200 V. The YAC band was excised, digested with agarase enzyme, and cleared by centrifugation. The YAC was transformed into RM14-3a by the lithium acetate transformation procedure (43), and transformants were selected on plates lacking uracil at 23°C and then restreaked on plates lacking both uracil and tryptophan. Transformants were examined for the presence of intact yWSS349 by pulsed-field gel and Southern analyses.
The URA3 gene on the right vector arm of yWSS349 in RM14-3a was replaced with the LEU2 gene in order to take advantage of the selection and counterselection properties of the URA3 gene (see “ARS analysis” below). To reduce the YAC to single copy, the Leu+ yWSS349 colonies were replica plated several times to nonselective plates and then selected on plates lacking leucine. Colonies were then screened for minimal growth on plates lacking tryptophan. These phenotypically Trp− colonies were examined by pulsed-field gel and Southern analyses and found to contain yWSS349 in single copy.
YAC manipulations.
Vector-insert junctions of yWSS340, yWSS349, yWSS1982, and yWSS4183 were isolated by bubble-vector PCR as previously described (38). The vectorette PCR products were sequenced with the ABI Dye Terminator Cycle Sequencing Core Kit. The DNA sequences of the PCR products were aligned against the sequenced T-cell receptor beta (TCR-β) contig to determine the precise YAC insert endpoints. The left and right endpoints of the yWSS349 insert correspond to bp 623026 and 382799 of TCR-β.
The presence or absence of sequence-tagged sites from the TCR-β contig in the YAC inserts was examined by PCR (22). yWSS349 contains the sequence-tagged sites sWSS602 (129 bp) and sWSS1379 (377 bp).
Fork direction and 2-D gel analyses.
Total yeast DNA was extracted from S phase yWSS349 cells (7), and fork direction gel analysis was performed as previously described (16). Beckman LE agarose was used in the first dimension, and between 40 and 50 μl of enzyme was used for the in-gel digestions. Specific probes were obtained by PCR with unique primers that amplify approximately 1.0 kb from the TCR-β YAC insert sequence. PCRs were performed on a Perkin-Elmer GeneAmp PCR System 9600 with Perkin-Elmer AmpliTaq polymerase. Fork direction gels were quantitated as previously described (48).
Conventional two-dimensional (2-D) gel analysis was performed as previously described (16). The first-dimension gels were 0.4% agarose, and the second-dimension gels were 1.1% agarose. Multiplex 2-D gel analysis was performed as previously described (48). The restriction fragments containing YAC replication origins are as follows: origin A, 6,063-bp NcoI; origin B, 5,099-bp AlwNI; origin C, 3,500-bp NcoI/BglI; origin D, 2,757-bp PstI/PvuII; origin E, 3,332-bp EcoRI/BglII; origin F, 2,600-bp AvaI/MluI; origin G, 3,717-bp NcoI; origin H, 4,870-bp EcoRI.
Density transfer experiments.
Replication times for EcoRI restriction fragments from RM14-3a containing yWSS349 were determined by using synchronous density transfer methods as previously described (29). Quantitation was performed on a Packard InstantImager Electronic Autoradiography system. Replication indices were calculated as previously described (18).
ARS analysis.
Restriction fragments containing sequences from yWSS349 that produced bubble arcs on 2-D gels were subcloned from TCR-β cosmids (generously provided by Lee Rowen) into a yeast ARS test plasmid (pRS306-5 or YIp5-5, both of which contain a yeast centromere [CEN5]).
A 6,037-bp BamHI/HindIII fragment containing origin D was cloned into pUC18, and exonuclease III (ExoIII) deletion series were generated from the BamHI end by using the New England Biolabs Exo-Size Deletion Kit. Fragments from the deletion series were cloned into the yeast test plasmid and assayed for ARS activity.
Site-directed mutations of the sequences with matches to 10 bp of the 11-bp ARS consensus sequence (ACS) (referred to here as 10/11 ACS matches) within the 6,037-bp BamHI/HindIII fragment containing origin D were made by using oligonucleotide-directed mutagenesis as previously described (9). The 10/11 ACS matches were mutated at positions that are most conserved in yeast ARS elements (49). The mutation that abolished origin function changed the 10/11 ACS match from 5′ TTTTGTATTTTT 3′ to 5′ TTTGTATCGAT 3′ and also introduced a ClaI restriction enzyme site. All site-directed mutations were confirmed by DNA sequencing.
The ACS mutation was introduced into the single-copy YAC with LEU2 as the right-arm marker by linearizing an integrating plasmid containing the site-directed mutation and the URA3 gene and selecting for Ura+ transformants. Correct integrants were confirmed by Southern analysis, grown in complete medium for several generations, and plated on plates containing 5-fluoroorotic acid to select for cells that recombined between repeated sequences, resulting in removal of the vector sequences. These colonies were examined by Southern analysis to screen for the presence of the novel restriction enzyme site generated during the mutagenesis.
The helical stability (ΔG) for regions containing yWSS349 origin sequences was determined by using the Thermodyne computer program (32).
RESULTS
YACs from the TCR-β locus.
We have analyzed the replication of a stable YAC containing human DNA from the TCR-β locus on human chromosome 7. This locus was chosen because the sequence of a 685-kb contig containing it has been determined (42) and a number of overlapping YACs were available (20). Having the entire sequence in hand facilitated our search for replication origins by providing a complete restriction enzyme map and obviated the need to sequence any YAC insert sequence that may be acting as a yeast replication origin.
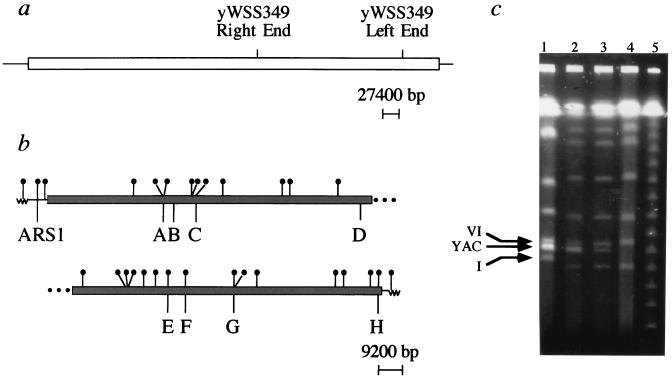
YAC yWSS349 was chosen for study because it is comparable in size (250 kb, including vector sequences) to a small yeast chromosome. The two smallest chromosomes are chromosome I (240 kb) and chromosome VI (280 kb); sizes are for strain S288C (35). To verify that the clone used in our studies had the correct structure, we used bubble-vector PCR (38) to isolate and sequence the YAC vector-insert junctions of yWSS349. This YAC is nonchimeric, and the insert boundaries at the left and right YAC arms correspond to bp 623026 and 382799, respectively, from the 5′ end of the full sequenced TCR-β contig (42). Figure 1a shows the sequenced region of the TCR-β locus with the endpoints of the YAC yWSS349 relative to the contig (compiled from references 20 and 42). The YAC insert is 240,219 bp in length. The insert in yWSS349 contains the expected sequence-tagged sites sWSS602 and sWSS1379 (21) (data not shown).
FIG. 1.
YACs from the sequenced TCR-β contig. (a) The 685-kb sequenced TCR-β contig on human chromosome 7 is represented by the bar. The YAC used in our replication studies is yWSS349, and its insert endpoints are indicated on the TCR-β contig. (b) Locations of 11/11 ACS matches in yWSS349 and locations of replication origins A to H relative to the ACS matches. The dark bar represents the human insert sequences. The perfect ACS matches are indicated by the lollipops. (c) Pulsed-field gel analysis of chromosomes from yeast strains containing the YAC yWSS349. Lane 1, yWSS349 in its original strain, AB1380. Lane 2, yWSS349 after transformation into strain RM14-3a. Lane 3, yWSS349 in strain RM14-3a after reduction of the YAC to single copy. Lane 4, original strain RM14-3a before transformation of yWSS349. Lane 5, lambda ladder pulsed-field gel marker. The pulsed-field gel was run under standard conditions with a switch time ramped from 20 to 50 s for 24 h. The bands corresponding to chromosomes I and VI and the YAC, yWSS349, are indicated by arrows. The karyotype of RM14-3a is slightly different from that of AB1380; in particular, chromosome I is smaller in RM14-3a.
To facilitate the analysis of yWSS349, it was transferred to yeast strain RM14-3a (29), which is amenable to cell cycle synchronization and for which a large amount of information on chromosomal replication is available. The YAC was isolated from a pulsed-field gel and transformed into RM14-3a, selecting initially for the marker on the right vector arm, URA3, and then restreaking transformants on plates selective for both vector markers, URA3 and TRP1. Figure 1c shows a pulsed-field gel with the intact yWSS349 YAC in its original strain, AB1380 (lane 1), and after transformation into RM14-3a (lane 2). As expected, yWSS349 DNA migrates between chromosomes I and VI. However, the ethidium bromide staining is more intense for the YAC, suggesting that it is present in multiple copies per yeast cell. By quantifying Southern blots with a yeast genomic sequence, ARS1, that is present on both the YAC vector arm and in its native location on yeast chromosome IV, we determined that there are approximately five copies of yWSS349 per cell, in RM14-3a as well as in AB1380 (data not shown). We believe that the occurrence of multiple copies of the YAC per cell is an effect of the incomplete promoter for the left vector arm marker, TRP1, which results in inefficient transcription of this gene (36). In synthetic media lacking tryptophan (−Trp), there is selection for cells with multiple copies of the YAC, presumably resulting from rare nondisjunction events, that are capable of more vigorous growth in the −Trp selective media. Yeast cells with multiple YACs do form larger colonies on selective plates than cells with the YAC in single copy (36).
The multicopy YAC strain was reduced to single copy by replica plating cells on nonselective plates several times and screening for colonies that grew more slowly on −Trp plates but grew normally when selection was for other markers on the YAC. To maintain the YAC at single copy, the strain was grown only under selection for the right vector arm marker, URA3 or LEU2. We took advantage of the multicopy YAC strain to facilitate our experiments, and all of the replication assays were performed with this strain unless stated otherwise.
Analysis of fork movement within the YAC insert.
In order to determine whether yeast replication origins exist within the human DNA sequences of yWSS349, we analyzed the direction of replication fork movement within the insert. The direct examination of fork movement provides evidence for whether replication origins exist within the YAC insert without relying on the plasmid ARS assay. In addition, divergent fork movement in two closely spaced regions provides evidence for a bidirectional replication origin located between the two regions (48).
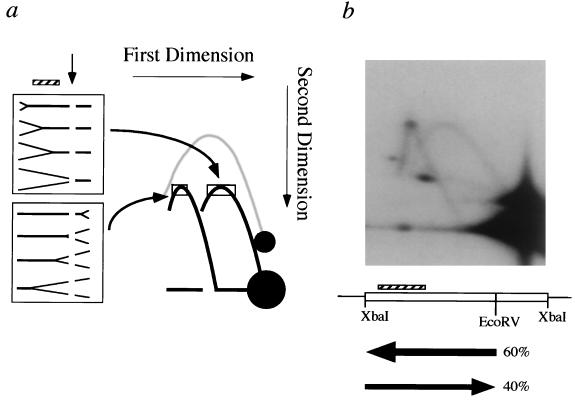
The direction of fork movement can be determined by using a modification of a 2-D agarose gel electrophoresis technique, termed fork direction gels (16). Figure 2a shows a cartoon of a fork direction gel and the arcs that correspond to forks moving in opposite directions through the probed fragment. Chromosomal DNA from cultures in log-phase growth is cleaved with a restriction enzyme and run in a first-dimension gel to separate fragments based on mass. These fragments are then digested within the gel with an enzyme that cuts the fragment of interest asymmetrically. A second-dimension gel that emphasizes differences in shape is run, blotted, and probed for the larger fragment. For a fragment that is passively replicated, this technique distinguishes replication forks traveling through the fragment in one direction from those originating from the opposite direction. The percentage of forks moving in each direction can be quantitated by comparing the hybridization intensities at the tops of the arcs (represented by the open squares in Fig. 2a).
FIG. 2.
Fork direction gel analysis of a region of yWSS349. (a) Determination of the direction of fork movement. In this schematic, the vertical arrow represents the site of in-gel enzyme cleavage and the hatched box represents the sequences used as a probe. The gray arc illustrates the locations of fragments that fail to be cut during the in-gel enzyme digestion and continue to migrate as a simple-Y arc of uncut molecules. The rectangles correspond to the approximate regions along each arc for which 32P counts were obtained with an InstantImager for quantification. (b) Fork direction gel of a fragment from a region of yWSS349 located approximately 180 kb from the left telomere. Chromosomal DNA was digested with XbaI before the first dimension of electrophoresis and digested in the gel with EcoRV. The hatched box represents the probe, which is approximately 2 kb. The XbaI-EcoRV fragment detected by the probe is 3.6 kb. The horizontal arrows illustrate the major direction of fork movement; the percentages were determined from quantitation of the two arcs.
Fork direction gel analysis was performed every 10 to 15 kb along most of the length of the YAC. As an example, Fig. 2b shows the analysis of an XbaI restriction fragment located approximately 180 kb from the left end of the YAC (which contains ARS1) and approximately 70 kb from the right end of the YAC. (This region is located just to the left of origin F in Fig. 1b). In this case, the XbaI fragment was cut in the gel with EcoRV and probed for the larger fragment. If ARS1 were the only functional origin on this YAC, then replication forks would move rightward and generate an arc on the 2-D gel emanating directly from the spot of unbranched fragments. However, there are clearly leftward-moving forks that generated a displaced arc on the 2-D gel. We conclude that there is a replication origin within the human DNA insert located to the right of this XbaI fragment. By quantitating the percentage of forks in each arc, we determined that the 3.6-kb XbaI-EcoRV fragment is replicated by a fork that arose at an origin to the left of the fragment in 40% of cells, but the majority of cells (60%) replicate this fragment by a fork coming from an origin located to the right of this region. There are several explanations for the cell-to-cell variation: an origin may be inherently inefficient in that some cells may use a particular origin and other cells not; alternatively, all origins are efficiently used, but either the rates of fork movement or their times of initiation vary from cell to cell. The kinetics of origin activation (see below and Fig. 4) do not show evidence for an extreme heterogeneity in origin activation, nor do we find any evidence for replication fork barriers or replication fork pause sites within the insert sequences that would alter the rates of replication fork movement.
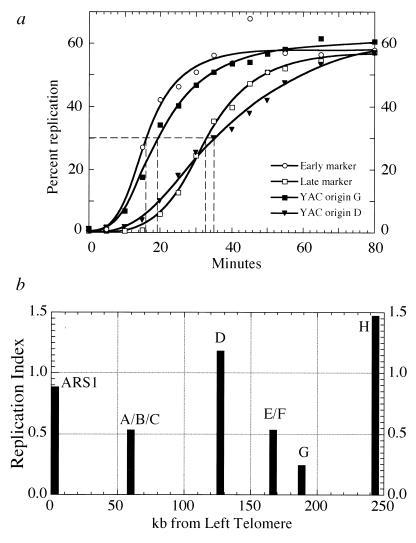
FIG. 4.
Temporal program of replication of YAC origins. (a) Replication kinetics of two of the YAC origins. Minutes in S phase after release from the G1 arrest is shown on the x axis. Percent replication is plotted on the y axis and is determined from the percentages of heavy-heavy DNA and heavy-light DNA for each probed fragment (29). The vertical dashed lines give the Trep values. The early marker fragment contains ARS305 from yeast chromosome III (37), and the late marker, R11, is located near the right end of yeast chromosome V (15). (b) Summary of replication times for the YAC origins. Replication index is plotted on the y axis, and the position of probed fragments along the length of the YAC molecule is plotted on the x axis. Replication index values are obtained by normalizing the Trep for a restriction fragment to the interval between the Treps of the early and late markers in panel a. The early and late markers are defined as having replication indices of 0.0 and 1.0, respectively. The probed EcoRI fragments either contain a YAC origin or are located very close to an origin.
Identification of eight yeast replication origins within yWSS349.
Results of the fork direction gel experiments performed throughout the YAC insert sequences are summarized in Fig. 3a. For almost all of the regions examined, replication forks were moving through the fragment in both directions. The direction of fork movement shown in Fig. 3a is the direction in which the majority of forks are moving. Based on regions that showed divergent fork movement, the fork direction data provided evidence for at least five origins of replication located within the TCR-β human DNA insert of the YAC (Fig. 3a, origins C, D, E, G, and H). Further analysis, discussed below, revealed three additional origins (origins A, B, and F). Origins A and B were identified in the region to the left of where the fork direction gel analysis had been started, while origin F is very close to origin E and was identified upon further analysis of the sequences located between two restriction fragments that displayed divergent fork movement.
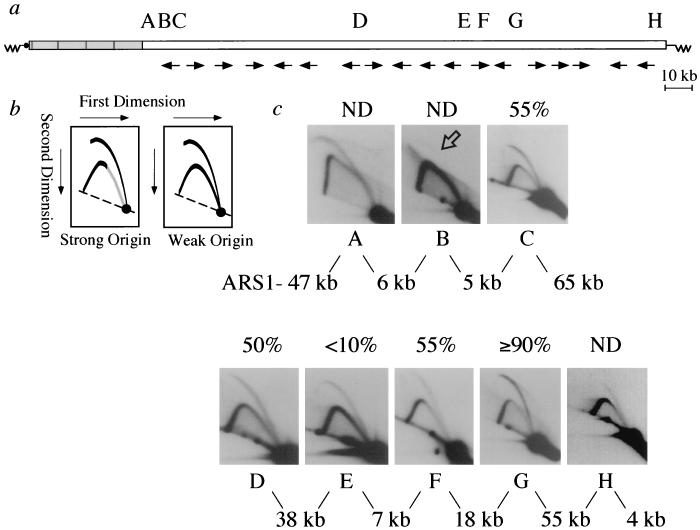
FIG. 3.
Location and characterization of replication origins within the human sequences of yWSS349. (a) Summary of fork direction gel results for sequences within the insert of yWSS349. In the schematic of the YAC, the thin line represents vector sequences, with the dark circle corresponding to the centromere and the zigzag lines corresponding to the telomeres. The wide bar represents the YAC insert. The shaded region corresponds to the tandemly duplicated trypsinogen gene repeats in the first 50 kb of the YAC insert. The arrows indicate the directions in which the majority of forks are moving within each fragment examined with fork direction gels. The approximate locations of origins A through H are shown. (b) Schematic representations of 2-D gel patterns generated by initiation of replication within the restriction fragment. The left panel shows the bubble arc that results when a strong origin is located asymmetrically within the restriction fragment. The right panel shows the bubble and complete simple-Y arcs that result when a weak origin is located within the restriction fragment. The dashed lines in both panels indicate the arc of unreplicating linear molecules. (c) 2-D gels of restriction fragments containing each of the YAC origins, A to H. The approximate efficiency of each origin is indicated above each 2-D gel. The distances between candidate ACS matches within restriction fragments with detected origin activity are indicated below and between the 2-D gels. The distance to the left of origin A refers to the distance from ARS1 on the left vector arm. The distance to the right of origin H refers to the distance to the right telomere. The arrow points to the faint bubble arc visible upon prolonged exposure of the 2-D gel autoradiogram of origin B. ND, not determined.
In order to further define where the replication origin resides between these regions with divergent fork movement, and also to determine whether more than one origin is located in the intervening sequences, we used conventional 2-D gels to identify a restriction fragment that contains an origin of replication (48). Similar to fork direction gels, restriction fragments from replicating cultures are run in a first-dimension gel to separate fragments based on mass. These fragments are directly run in a second-dimension gel that emphasizes differences in shape, and the gel is blotted and probed. If a restriction fragment contains an active origin of replication, the fragment will contain a replication bubble, and the characteristic bubble arc will result on a 2-D gel. Figure 3b shows cartoons of the bubble arcs that result from a fragment containing a replication origin located asymmetrically within the probed fragment. The intensity of the complete simple-Y arc reflects the strength of the replication origin: the stronger the intensity of the complete simple-Y arc, the less efficient the origin. Fragments that produced a bubble arc were found in the regions between all fragments having divergent fork movement. Figure 3c shows 2-D gels of eight YAC insert fragments (origins A to H) that produce a bubble arc and therefore contain a replication origin. Origin B is an extremely weak origin, and the bubble arc (indicated by the arrow) is visible only upon prolonged exposure of the autoradiogram.
Most fragments examined by use of fork direction gels show evidence of leftward- and rightward-moving forks, suggesting that the YAC human insert origins are not 100% efficient. Origins B and E have intense complete simple-Y arcs and relatively faint bubble arcs, suggesting that they are weak origins that only rarely activate replication. While conventional 2-D gels can provide a qualitative estimate of origin efficiency, they are not adequate to obtain a quantitative estimate of origin efficiency for a number of reasons, including the possibility of breakage of molecules containing a bubble (48). In order to obtain a better estimate of origin efficiency, we performed fork direction gel analysis of sequences immediately flanking the origin fragment and quantified the percentage of forks arising from the origin compared to the percentage of forks entering the origin region from the opposite direction and passively replicating it (data not shown). From these quantifications we obtained an estimate, which we believe to be accurate within 10% (48), of origin efficiencies for some of the insert origins (shown above each gel in Fig. 3c). This analysis shows that only one of the YAC insert origins, origin G, is very efficient, being used in approximately 90% of cells in the population (48). The other origins are less efficient and range in use from <10% to approximately 55% of cell cycles.
The spacing between the eight YAC origins is given in Fig. 3c (below each gel in Fig. 3c) and ranges from 5 to 65 kb. If activations from the extremely weak origins B and E are ignored, the average spacing between origins in the human insert sequences of the YAC is approximately 40 kb. This spacing is quite similar to the average spacing between natural yeast chromosomal origins, which is estimated to be between 30 and 50 kb (17, 18, 33, 52).
Fork direction gel analysis was not performed for the first 50 kb from the left end of the YAC, because this region contains tandem repeat units which are roughly 90 to 91% conserved, each containing a trypsinogen gene (42). This region was analyzed with multiplex 2-D gels (48) by using restriction enzyme polymorphisms within the repeats and probes complementary to sequences within each repeat (data not shown). We found no evidence for bubble arcs within the trypsinogen repeat regions themselves, but we did identify origins A and B in the unique sequences 3′ to the trypsinogen repeats (Fig. 3c). Thus, we believe that there are no additional replication origins within the trypsinogen repeat region in the first 50 kb of the YAC.
We examined the replication of the YAC vector arms and confirmed that ARS1 on the left vector arm is used as a replication origin in the YAC (data not shown). Analysis of the right vector arm revealed the existence of an origin (data not shown) that is associated with the Tetrahymena telomeric sequences which have been shown to have ARS activity in yeast (27). Our data show that these Tetrahymena sequences also activate replication within the context of a linear chromosome, although from the intensity of the arc corresponding to forks originating from these sequences in the fork direction gel, it does not appear to be a strong origin of replication. We have not determined whether the left Tetrahymena telomeric sequences also activate replication, but given its proximity to ARS1 on the vector arm and our knowledge of interference between closely spaced origins (5), we hypothesize that initiation from the Tetrahymena telomeric sequence does not play a major role in replication of the left end of the YAC.
YAC origins follow a temporal program of replication.
Having identified eight YAC human insert origins that are spaced similarly to natural yeast chromosomal origins, we wanted to examine whether these origins follow a temporal program of replication. Yeast replication origins initiate replication at different times in S phase, with some sequences initiating replication early in S phase and some doing so later in S phase (14, 17, 18, 37, 52). The time of origin activation is reproducible and is dependent on the chromosomal context of the origin, rather than being intrinsic to the origin itself (15, 18). We currently postulate that early is the default time for origin activation and that surrounding sequences impose late replication on nearby origins by an as-yet-unknown mechanism. We were therefore interested in determining whether the YAC origins all initiate replication early in S phase as the default state or whether they too follow a temporal program of replication initiation.
To determine the time of replication of the YAC insert origins, we performed a density transfer experiment (29). Synchronized cells grown in heavy isotopic medium are transferred to light isotopic medium just before entry into S phase of the cell cycle. Restriction fragments are fractionated on a cesium chloride gradient to separate the heavy-heavy unreplicated DNA from the heavy-light replicated DNA. After fractionation of the gradients, the samples are blotted and probed to detect a restriction fragment that contains or is adjacent to the YAC origin. The percent replication is calculated for each fragment and each time sample. The density transfer experiments were performed for both the multicopy and single-copy YACs with similar results. The data shown in Fig. 4 are from the multicopy strain.
Figure 4a shows the replication kinetics for an early-replicating chromosomal marker from yeast chromosome III and a late chromosomal marker from yeast chromosome V (15, 37). These sequences serve as markers for the onset and duration of S phase. Also shown in Fig. 4a are the replication kinetics for two fragments containing replication origins within the human DNA insert of yWSS349. Origin G initiates replication early in S phase, while origin D initiates replication late in S phase, later than our chromosomal late marker. To obtain a time of replication of a specific restriction fragment, we can calculate a Trep value, the time at which half of the final replication of the fragment has occurred. The Trep for yWSS349 origin G is ∼19 min, while the Trep for origin D is ∼35 min.
Fragments corresponding to all of the replication origins in yWSS349 were probed, and their replication times are summarized in Fig. 4b. Each time is given as a replication index value, where the Trep for a fragment is normalized to the interval between the Treps of the early and late markers in Fig. 4a. The early and late markers are defined as having replication indices of 0.0 and 1.0, respectively. The results show that the YAC has a temporal program of replication where different regions replicate at different times. Two of the YAC origins (origins D and H) have a replication index of >1.0, indicating that these regions are replicated later in S phase than the sequence used as the late yeast chromosomal marker in our experiments. The two YAC origins nearest the two telomeres initiate replication late in S phase. This late time of replication might be expected, since telomeres are known to confer late replication on nearby origins (15); however, we have not demonstrated that origin H’s proximity to the right telomere is the only determinant of its late replication. Given origin H’s apparent inefficiency of activation, passive replication by forks emanating from origin G must also be contributing to origin H’s late time of replication. The most efficient YAC origin, origin G, initiates replication early in S phase, while the rest of the origins replicate in mid- to late S phase.
The moderately efficient origin D, located in the middle of the YAC, is replicated very late in S phase and is flanked by origins that replicate earlier in S phase. The slope of the curve for the origin D data in Fig. 4a is not as steep as the slopes for the data on origin G and on the chromosomal markers. The shallower slope is explained by the fact that origin D is being used as an active origin of replication in only 50% of the cells in the population. In the other 50% of the cells, it is passively replicated by forks initiating elsewhere. The replication kinetics are therefore a composite of those two events: the time of origin activation and the time at which a passive replication fork reaches the origin. Since origin D’s efficiency of activation is similar to those of the earlier replication origins on its flanks, origins C and F, the later time of replication of origin D must reflect its later activation. There is a cluster of late-activated replication origins on yeast chromosome XIV (18); however, it is not known whether the cluster is flanked on both sides by replication origins that initiate earlier in S phase. Thus, origin D is the first example of a solo, internal, late-activated replication origin in a yeast chromosome.
Sequence requirements for YAC insert origins.
Origins of replication in yeast are identified by their ability to act as ARSs on plasmids, although not all ARSs are chromosomal replication origins in their native locations in the yeast genome (14, 25, 33). These origins are well defined and can be narrowed down to a few hundred base pairs. They are modular in nature and consist of a number of different elements, most of which are not conserved at the sequence level. One essential element that is conserved at the sequence level is the ACS, or A element, which consists of an 11-bp A/T-rich sequence (11). Origins of replication in yeast require a match to this ACS, although the match does not need to be perfect: some origins have a 9/11 or 10/11 match (34). An expansion of the ACS to a 17-bp extended ACS has recently been proposed (46). Sequences 3′ to the T-rich strand of the ACS, termed the B domain, are also required for full origin function. Detailed linker-scan mutational analysis of ARS1 from chromosome IV has shown that the B domain can be subdivided into three regions, the B1, B2, and B3 elements (28). These regions bind trans-acting factors (4, 28) and include an easily unwound region (31). The modular nature of ARS1 has been confirmed for other ARS elements in yeast; however, outside the ACS, they are divergent in their flanking sequences (24). Thus, yeast origins are sequence specific although not well enough conserved at the sequence level to be identified by sequence alone.
Figure 1b shows the locations of perfect matches to the 11-bp ACS in yWSS349. Not all 11/11 ACS matches correspond to replication origins. There are no perfect matches in yWSS349 to the 17-bp extended ACS. All of the YAC insert fragments that generated bubble arcs on 2-D gels (Fig. 3c) contain at least one perfect match to the ACS, except for origin D, which lacks any perfect matches. Each fragment also contains multiple 10/11 and 9/11 matches to the ACS. All of the origin fragments for origins C to H were subcloned into an ARS test plasmid, and all fragments are capable of ARS activity on plasmids in yeast. Origins A and B were not directly tested in a plasmid ARS assay.
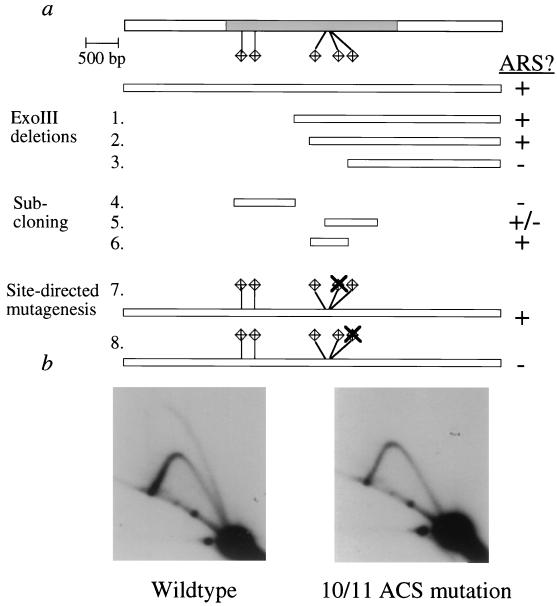
To determine what sequences were essential to origin function, we performed detailed ARS analysis of origin D. The origin D region contains five 10/11 ACS matches that are in two clusters (Fig. 5a). An ExoIII deletion series from the 5′ end of the fragment as well as direct subcloning showed that the first two 10/11 ACS matches on the left are not required for ARS activity (Fig. 5a, constructs 1, 2, and 4). Subcloning of a fragment containing the three clustered 10/11 ACS matches showed that this fragment was sufficient for full ARS activity (construct 6). A fragment containing all three 10/11 ACS matches but only 24 bp 5′ to the first ACS match in the cluster (construct 5) was only a weak ARS, suggesting that this 5′ region is also required for full ARS function. This region contains a set of sequences that are more easily unwound than flanking sequences, with a free energy difference between the duplex and single-stranded states (ΔG) of 105 to 110 kcal/mol (32). Site-directed mutagenesis of the 10/11 ACS matches (constructs 7 and 8) showed that mutation of one of the matches was sufficient to completely abolish ARS activity (construct 8). Thus, a match to the ACS as well as flanking sequences is required for ARS activity of this fragment.
FIG. 5.
ARS analysis of yWSS349 origin D. (a) The top bar represents the 6,037-bp fragment with origin D activity that was subcloned and tested for ARS activity. The diamonds represent the locations of the 10/11 ACS matches. The shaded region corresponds to the fragment that produced a bubble on 2-D gels. The regions retained in the ExoIII deletion series are indicated by bars (constructs 1 to 3). The fragments that were subcloned and tested for ARS activity are represented by the smaller bars (constructs 4 to 6). Site-directed mutagenesis of the 10/11 ACS match is indicated by the X (constructs 7 and 8). (b) 2-D gels of fragments from yWSS349 containing the wild-type origin D sequence (left panel) and the 10/11 ACS mutation (right panel). The restriction fragment analyzed corresponds to the shaded region in panel a.
To determine whether the ACS mutation that abolishes ARS activity also abolishes origin function, we introduced this mutation into yWSS349. Figure 5b shows a 2-D gel of the fragment containing origin D with the wild-type sequence and the fragment that contains the mutation of the 10/11 ACS match. The fragment containing the 10/11 ACS mutation no longer produces a bubble on a 2-D gel. Thus, mutation of this ACS match in human DNA not only abolishes ARS activity but also abolishes origin function.
DISCUSSION
The manner in which yeast replicates human DNA in the context of a large artificial chromosome had not been explored prior to this study. Our knowledge of replication at the level of whole chromosomes in the yeast genome is currently limited to chromosome III (33) and chromosome VI (17, 52), where origins have been identified primarily from ARS assays. The analysis of the 250-kb YAC containing human TCR-β DNA has been the first large-scale analysis in which replication origins were identified by a direct physical assay, fork direction gels, rather than by the plasmid ARS assay. Our analysis of yWSS349 has shown that the replication of this stable YAC is accomplished in a way that mimics natural yeast chromosomal replication.
There are at least six replication origins in yWSS349, with an average spacing among the six moderately or highly active origins of approximately 40 kb. Most of the YAC origins are not used by every cell in the population. Only one of the YAC origins (origin G) is very efficient, being used in at least 90% of cells. The number of inefficient origins in yeast chromosome VI is also quite high, with only one origin, ARS607, being used in >85% of cells in chromosome VI (17, 52). Although the basis of origin inefficiency is unknown, it is possible that the chromatin conformation at the origin sequences results in weaker binding of trans-acting factors required for origin activation. The basis for the inefficiency of the YAC origins and the inefficient chromosome VI origins may be the same. Alternatively, the YAC origins may be more sensitive to chromatin conformation because not all of the sequence elements required for efficient origin function are present in the human DNA or because they are not found at the optimal spacings. In the case of origin D, sequences flanking the 10/11 ACS match are required for full ARS activity. One of the flanking B elements required for full function of yeast ARSs is an easily unwound region, or DNA-unwinding element (47). Sequences immediately flanking origin D’s ACS do have a lower helical stability, but the free energy difference between the duplex and single-stranded states (ΔG) of this region is still higher than the ΔG difference associated with DNA-unwinding elements of yeast ARSs, which range from 71 to 98 kcal/mol (32). Thus, yeast initiation proteins may be interacting with less-than-ideal cis sequences at the YAC origins.
The very weak YAC origins that we have identified (origins B and E) are located close to origins that are more efficient. Origins placed in close proximity (6.5 kb) to each other have been shown to experience origin interference, resulting in decreased efficiency of each origin (5). If origin interference is occurring among the cluster of origins A, B, and C and between origins E and F, then origins B and E appear to be preferentially affected. Also, origins B and E may be inherently weaker origins. The restriction fragment containing origin E that produces a bubble arc on 2-D gels has weak ARS activity on a plasmid unless more flanking sequences are supplied (data not shown). Thus, origin E’s flanking elements may be more widely separated from each other, resulting in weaker binding of trans-acting factors and decreased origin function.
Human chromosomes contain regions of early, middle, and late replication and have distinctive chromosome banding patterns that correlate to these replication timings. Yeast chromosomes, despite their small size, also have regions that replicate at different times during S phase. A temporal program of replication may be one level of control in the cell that ensures proper replication and segregation of linear chromosomes (12). In yeast chromosomes III and VI, both of which are small chromosomes, the telomere-proximal sequences replicate late in S phase, while internal, centromere-proximal regions replicate early (17, 37, 52). More than 150 kb from the left telomere of yeast chromosome XIV is a set of origins that is activated in late S phase (18). Analysis of a small number of late-activated origins in the yeast genome has shown that late replication is a result of chromosomal context (15, 18). Surprisingly, the origins in yWSS349 follow a temporal program to replication similar to that of these natural yeast chromosomes, including a late-activated origin (origin D) located in the middle of the YAC insert. It will be interesting to determine if the late activation of origin D is determined by the flanking human sequences and whether these sequences can also delay the activation of natural yeast chromosomal origins. Alternatively, origin D may be the first instance of an intrinsically late-activated origin. Further analysis of the replication origins in this YAC may provide information regarding the temporal regulation of origin activation in yeast.
The manner in which the TCR-β locus is replicated in human cells has not been analyzed. Indeed, origins of replication in mammalian genomes are far less well understood than yeast origins. Recently, an 8-kb region containing sequences that initiate replication in the human β-globin locus was shown to be capable of initiating replication when moved to an ectopic site (1). This result suggests that mammalian origins of replication are discrete sequence-specific units similar to yeast origins. However, for the dihydrofolate reductase locus of Chinese hamster ovary cells, there still remains some controversy as to whether replication occurs at specific sequences or whether replication initiates relatively nonspecifically within noncoding regions of the genome (10, 23). Mammalian sequences that act as ARSs in yeast have not yet been shown to correlate to replication origins in their native genomes (41), and it has seemed unlikely that these sequences act as replication origins in the mammalian nucleus. However, a number of the recently identified trans-acting factors required for origin initiation in yeast also have functional homologues in humans (26, 50), suggesting that origin function in humans and yeast may be more similar than previously thought. Mammalian replication origins appear to be localized outside coding sequences. The locations of genes, pseudogenes, and gene relics in the human TCR-β locus have been identified by Rowen et al. (42), and it is intriguing that the most efficient origin in yWSS349, origin G, is located within an approximately 15-kb region that does not contain any of these gene-related sequences. However, analysis of replication origins in this region will be complicated by the genomic rearrangements that occur in the generation of the functional TCR genes.
The ability of yeast to accept and replicate large YACs comprised mainly of DNA it has never before encountered has been somewhat taken for granted in the age of physical mapping, gene cloning, and genomic sequencing. A knowledge of how yeast is able to replicate these large stretches of foreign DNA may be important in understanding and addressing the problem of YAC instability. In particular, regions that lack sequences that can act as replication origins in yeast may be highly unstable. The G/C-rich Myxococcus xanthus genome has been shown to be devoid of yeast ARS activity, and there is a limit to the size of inserts of this DNA that can be stably maintained as YACs (27a). With a yeast origin provided on each vector arm, the maximum size of an M. xanthus YAC that could be constructed was approximately 200 kb. If the origin on the right vector arm is removed, this 200-kb YAC is lost at a high rate and is subject to breakage and rearrangement. Thus, it is likely that YACs made with only one yeast origin provided on the vector (such as the pYAC4 vector used to construct YAC libraries [8]) cannot be successfully replicated when the insert has more than 100 kb of sequences that lack fortuitous replication origins. Although our analysis has shown that the frequency of fortuitous yeast origins within human DNA is roughly equivalent to the frequency of natural yeast chromosomal origins, it is possible that human sequences that have an atypical base pair composition, especially gene-rich G/C isochores, may lack these fortuitous origin sequences and be unstable in yeast cells.
ACKNOWLEDGMENTS
We thank Eric Green for providing the chromosome 7 YACs and Lee Rowen for providing the TCR-β cosmids as well as helpful information regarding the TCR-β contig and sequence alignment. We also thank M. K. Raghuraman for critical reading of the manuscript.
This work was supported by National Institute of General Medical Sciences grant 18926 to W.L.F. and B.J.B. and by National Center for Human Genome Research grant 01298 to B.J.B. and W.L.F.
REFERENCES
- 1.Aladjem M I, Rodewald L W, Kolman J L, Wahl G M. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 2.Albertsen H M, Smith S A, Mazoyer S, Fujimoto E, Stevens J, Williams B, Rodriguez P, Cropp C S, Slijepcevic P, Carlson M, Robertson M, Bradley P, Lawrence E, Harrington T, Mei Sheng Z, Hoopes R, Sternberg N, Brothman A, Callahan R, Ponder B A J, White R. A physical map and candidate genes in the BRCA1 region on chromosome 17q12-21. Nat Genet. 1994;7:472–479. doi: 10.1038/ng0894-472. [DOI] [PubMed] [Google Scholar]
- 3.Bates G P, Valdes J, Hummerich H, Baxendale S, Le Paslier D L, Monaco A P, Tagle D, MacDonald M E, Altherr M, Ross M, Brownstein B H, Bentley D, Wasmuth J J, Gusella J F, Cohen D, Collins F, Lehrach H. Characterization of a yeast artificial chromosome contig spanning the Huntington’s disease gene candidate region. Nat Genet. 1992;1:180–187. doi: 10.1038/ng0692-180. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 5.Brewer B J, Fangman W L. Initiation at closely spaced replication origins in a yeast chromosome. Science. 1993;262:1728–1731. doi: 10.1126/science.8259517. [DOI] [PubMed] [Google Scholar]
- 6.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 7.Brewer B J, Lockshon D, Fangman W L. The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell. 1992;71:267–276. doi: 10.1016/0092-8674(92)90355-g. [DOI] [PubMed] [Google Scholar]
- 8.Burke D T, Carle G F, Olson M V. Cloning of large segments of exogenous DNA into yeast by means of artificial chromosome vectors. Science. 1987;236:806–812. doi: 10.1126/science.3033825. [DOI] [PubMed] [Google Scholar]
- 9.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 10.DePamphilis M L. Origins of DNA replication in metazoan chromosomes. J Biol Chem. 1993;268:1–4. [PubMed] [Google Scholar]
- 11.Deshpande A M, Newlon C S. The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4305–4313. doi: 10.1128/mcb.12.10.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diller J D, Raghuraman M K. Eukaryotic replication origins: control in space and time. Trends Biochem Sci. 1994;19:320–325. doi: 10.1016/0968-0004(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 13.Dubey D D, Davis L R, Greenfeder S A, Ong L Y, Zhu J G, Broach J R, Newlon C S, Huberman J A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol Cell Biol. 1991;11:5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson B M, Brewer B J, Reynolds A E, Fangman W L. A yeast origin of replication is activated late in S phase. Cell. 1991;65:507–515. doi: 10.1016/0092-8674(91)90468-e. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson B M, Fangman W L. A position effect on the time of replication origin activation in yeast. Cell. 1992;68:333–339. doi: 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- 16.Friedman K L, Brewer B J. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol. 1995;262:613–627. doi: 10.1016/0076-6879(95)62048-6. [DOI] [PubMed] [Google Scholar]
- 17.Friedman K L, Brewer B J, Fangman W L. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:667–678. doi: 10.1046/j.1365-2443.1997.1520350.x. [DOI] [PubMed] [Google Scholar]
- 18.Friedman K L, Diller J D, Ferguson B M, Nyland S V, Brewer B J, Fangman W L. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 1996;10:1595–1607. doi: 10.1101/gad.10.13.1595. [DOI] [PubMed] [Google Scholar]
- 19.Gnirke A, Huxley C, Peterson K, Olson M V. Microinjection of intact 200 to 500 kb fragments of YAC DNA into mammalian cells. Genomics. 1993;15:659–667. doi: 10.1006/geno.1993.1121. [DOI] [PubMed] [Google Scholar]
- 20.Green E D, Braden V V, Fulton R S, Lim R, Ueltzen M S, Peluso D C, Mohr-Tidwell R M, Idol J R, Smith L M, Chumakov I, Le Paslier D, Cohen D, Featherstone T, Green P. A human chromosome 7 yeast artificial chromosome (YAC) resource: construction, characterization, and screening. Genomics. 1995;25:170–183. doi: 10.1016/0888-7543(95)80123-4. [DOI] [PubMed] [Google Scholar]
- 21.Green E D, Mohr R M, Idol J R, Jones M, Buckingham J M, Deaven L L, Moyzis R K, Olson M V. Systematic generation of sequence-tagged sites for physical mapping of human chromosomes: application to the mapping of human chromosome 7 using yeast artificial chromosomes. Genomics. 1991;11:548–564. doi: 10.1016/0888-7543(91)90062-j. [DOI] [PubMed] [Google Scholar]
- 22.Green E D, Olson M V. Systematic screening of yeast artificial chromosome libraries by use of the polymerase chain reaction. Proc Natl Acad Sci USA. 1990;87:1213–1217. doi: 10.1073/pnas.87.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamlin J L. Mammalian origins of replication. Bioessays. 1992;14:651–659. doi: 10.1002/bies.950141002. [DOI] [PubMed] [Google Scholar]
- 24.Huang R-Y, Kowalski D. Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res. 1996;24:816–823. doi: 10.1093/nar/24.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huberman J A, Zhu J G, Davis L R, Newlon C S. Close association of a DNA replication origin and an ARS element on chromosome III of the yeast, Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:6373–6384. doi: 10.1093/nar/16.14.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang W, Hunter T. Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc Natl Acad Sci USA. 1997;94:14320–14325. doi: 10.1073/pnas.94.26.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiss G B, Amin A A, Pearlman R E. Two separate regions of the extrachromosomal ribosomal deoxyribonucleic acid of Tetrahymena thermophila enable autonomous replication of plasmids in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:535–543. doi: 10.1128/mcb.1.6.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27a.Loehrlein, C. Personal communication.
- 28.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 29.McCarroll R M, Fangman W L. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- 30.Montiel J F, Norbury C J, Tuite M F, Dobson M J, Mills J S, Kingsman A J, Kingsman S M. Characterization of human chromosomal DNA sequences which replicate autonomously in Saccharomyces cerevisiae. Nucleic Acids Res. 1984;12:1049–1068. doi: 10.1093/nar/12.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natale D A, Schubert A E, Kowalski D. DNA helical stability accounts for mutational defects in a yeast replication origin. Proc Natl Acad Sci USA. 1992;89:2654–2658. doi: 10.1073/pnas.89.7.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natale D A, Umek R M, Kowalski D. Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res. 1993;21:555–560. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newlon C S, Collins I, Dershowitz A, Deshpande A M, Greenfeder S A, Ong L Y, Theis J F. Analysis of replication origin function on chromosome III of Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1993;58:415–423. doi: 10.1101/sqb.1993.058.01.048. [DOI] [PubMed] [Google Scholar]
- 34.Newlon C S, Theis J F. The structure and function of yeast ARS elements. Curr Opin Genet Dev. 1993;3:752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- 35.Olson M V. Genome structure and organization in Saccharomyces cervisiae. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. 1991. pp. 1–39. Genome dynamics, protein synthesis, and energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 36.Popov A V, Butzler C, Bruggemann M. Yeast colony size reflects YAC copy number. Nucleic Acids Res. 1997;25:2039–2040. doi: 10.1093/nar/25.10.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reynolds A E, McCarroll R M, Newlon C S, Fangman W L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riley J, Butler R, Ogilvie D, Finniear R, Jenner D, Powell S, Anand R, Smith J C, Markham A F. A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 1990;18:2887–2890. doi: 10.1093/nar/18.10.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rivin C J, Fangman W L. Replication fork rate and origin activation during the S phase of Saccharomyces cerevisiae. J Cell Biol. 1980;85:108–115. doi: 10.1083/jcb.85.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg C, Florijn R J, Van De Rijke F M, Blonden L A, Raap T K, Van Ommen G B, Den Dunnen J T. High resolution DNA fiber-FISH on yeast artificial chromosomes: direct visualization of DNA replication. Nat Genet. 1995;10:477–479. doi: 10.1038/ng0895-477. [DOI] [PubMed] [Google Scholar]
- 41.Roth G E, Blanton H M, Hager L J, Zakian V A. Isolation and characterization of sequences from mouse chromosomal DNA with ARS function in yeasts. Mol Cell Biol. 1983;3:1898–1908. doi: 10.1128/mcb.3.11.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowen L, Koop B F, Hood L. The complete 685-kilobase DNA sequence of the human β T cell receptor locus. Science. 1996;272:1755–1762. doi: 10.1126/science.272.5269.1755. [DOI] [PubMed] [Google Scholar]
- 43.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 44.Stinchcomb D T, Struhl K, Davis R W. Isolation and characterization of a yeast chromosomal replicator. Nature. 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 45.Straughen J, Ciocci S, Ye T-Z, Lennon D N, Proytcheva M, Alhadeff B, Goodfellow P, German J, Ellis N A, Groden J. Physical mapping of the Bloom syndrome region by the identification of YAC and P1 clones from human chromosome 15 band q26.1. Genomics. 1996;35:118–128. doi: 10.1006/geno.1996.0330. [DOI] [PubMed] [Google Scholar]
- 46.Theis J F, Newlon C S. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc Natl Acad Sci USA. 1997;94:10786–10791. doi: 10.1073/pnas.94.20.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umek R M, Kowalski D. Thermal energy suppresses mutational defects in DNA unwinding at a yeast replication origin. Proc Natl Acad Sci USA. 1990;87:2486–2490. doi: 10.1073/pnas.87.7.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Brabant A J, Hunt S Y, Fangman W L, Brewer B J. Identifying sites of replication initiation in yeast chromosomes: looking for origins in all the right places. Electrophoresis. 1998;19:1239–1246. doi: 10.1002/elps.1150190803. [DOI] [PubMed] [Google Scholar]
- 49.Van Houten J V, Newlon C S. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol Cell Biol. 1990;10:3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams R S, Shohet R V, Stillman B. A human protein related to yeast Cdc6p. Proc Natl Acad Sci USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williamson D H. The yeast ARS element, six years on: a progress report. Yeast. 1985;1:1–14. doi: 10.1002/yea.320010102. [DOI] [PubMed] [Google Scholar]
- 52.Yamashita M, Hori Y, Shinomiya T, Obuse C, Tsurimoto T, Yoshikawa H, Shirahige K. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:655–665. doi: 10.1046/j.1365-2443.1997.1530351.x. [DOI] [PubMed] [Google Scholar]