Significance
The nucleobase/ascorbic acid transporter (NAT) family comprises proteins in all kingdoms of life. Their substrates, nucleobases and their analogs, have special attention in therapeutic applications, such as the treatment of solid tumors, lymphoproliferative diseases, viral infections, and inflammatory diseases. Here, we present the crystal structure of a bacterial NAT member, PurT of Colwellia psychrerythraea (PurTCp) at 2.80 Å resolution that, together with functional data, provide insight into the transport mechanism of NAT family members.
Keywords: PurTCp, NAT gene family, H+-coupled transport, nucleobases, X-ray crystallography
Abstract
Members of the nucleobase/ascorbic acid transporter (NAT) gene family are found in all kingdoms of life. In mammals, the concentrative uptake of ascorbic acid (vitamin C) by members of the NAT family is driven by the Na+ gradient, while the uptake of nucleobases in bacteria is powered by the H+ gradient. Here, we report the structure and function of PurTCp, a NAT family member from Colwellia psychrerythraea. The structure of PurTCp was determined to 2.80 Å resolution by X-ray crystallography. PurTCp forms a homodimer, and each protomer has 14 transmembrane segments folded into a transport domain (core domain) and a scaffold domain (gate domain). A purine base is present in the structure and defines the location of the substrate binding site. Functional studies reveal that PurTCp transports purines but not pyrimidines and that purine binding and transport is dependent on the pH. Mutation of a conserved aspartate residue close to the substrate binding site reveals the critical role of this residue in H+-dependent transport of purines. Comparison of the PurTCp structure with transporters of the same structural fold suggests that rigid-body motions of the substrate-binding domain are central for substrate translocation across the membrane.
The nucleobase/ascorbate transporter (NAT) family encompasses proteins that are responsible for the uptake of nucleobases in all kingdoms of life. The NAT family of transporters also mediates the uptake of vitamin C (L-ascorbic acid) in vertebrates. Nucleobases and their analogs (e.g., allopurinol, 5-fluorouracil, 6-mercaptopurine, and acyclovir) have gained special interest in therapeutic applications as they are used in the treatment of solid tumors, lymphoproliferative diseases, viral infections such as hepatitis and AIDS, and inflammatory diseases such as Crohn’s disease and gout, and as antiparasitic drugs such as trypanocides (1–6). In vertebrates, ascorbic acid, the other substrate of NAT proteins, is central in several vital enzymatic reactions and protects tissues from oxidative damage by scavenging free radicals (7–9).
Intestinal and renal (re)absorption of vitamin C is mediated by the epithelial Na+-dependent L-ascorbic acid cotransporter SVCT1 (SLC23A1), whereas the homologous SVCT2 (SLC23A2) mediates vitamin C transport in metabolically active cells (e.g., in the adrenal glands, pituitary gland, thymus, corpus luteum, retina, and cornea) (10, 11). Furthermore, rat SNBT1 was identified as the first Na+-dependent nucleobase transporter in mammals (12).
In contrast to these three mammalian NAT members that mediate Na+-coupled transport of L-ascorbic acid and nucleobases, the transport of nucleobases by evolutionarily distant eubacteria, archaea, filamentous fungi, plants, insects, and nematodes has been described as being driven by a proton gradient (i.e., H+-dependent symport) (1, 13, 14). The crystal structures of the NAT members UraA, the uracil/5-fluorouracil transporter of Escherichia coli (15, 16), and UapA, the uric acid/xanthine H+ symporter of Aspergillus nidulans (17) were obtained in substrate (uracil and xanthine, respectively) bound, inward-open state. These structures have been used as a template for computational studies that propose mechanistic models for the entire NAT family (18, 19). However, the mechanistic understanding of H+-coupled transport by UraA (15, 20) or UapA (17) remains limited, thereby hampering the interpretation of functional data of NAT proteins in a structural context. Here, we present the structure of a bacterial NAT member, PurT of Colwellia psychrerythraea (PurTCp) at 2.80 Å resolution that, in conjunction with flux and binding studies, provides insight into the mechanism of transport by this family of transport proteins.
Results
Purification and Functional Characterization of PurTCp.
A homolog of human SLC23A1 from the bacterium C. psychrerythraea 34H was cloned, expressed, and purified (Materials and Methods). The protein elutes as a single peak on size-exclusion chromatography, and the elution volume is consistent with it being a homodimer (SI Appendix, Fig. S1A). This conclusion is also consistent with a crosslinking study which shows dimer formation (SI Appendix, Fig. S1B). Dimeric assembly was also reported for UraA (16) and UapA (17).
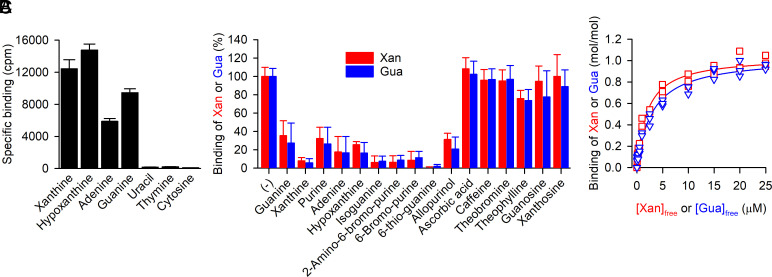
To assess the activity of the purified, detergent-solubilized SLC23A1 homolog, we measured direct binding of radiolabeled purines and pyrimidines with the scintillation proximity assay (SPA) (21). Fig. 1A shows that the purified protein binds the purines guanine, adenine, xanthine, and hypoxanthine, whereas the pyrimidines uracil, thymine, or cytosine did not interact with the candidate protein. To further investigate the substrate specificity of the protein, we performed competition binding assays in which the binding of 3H-xanthine or 3H-guanine was measured in the presence of a 500-fold concentration excess of nonradiolabeled compounds (Fig. 1B). Consistent with the results shown in Fig. 1A, the nonlabeled purines guanine, xanthine, adenine, and hypoxanthine inhibited the binding of 3H-xanthine or 3H-guanine by ≥80%, suggesting competitive binding to a shared binding site. Similar inhibitory effects were observed with 2-amino-6-bromopurine, 6-bromopurine, 6-thioguanine, allopurinol, isoguanine, and purine. Other purines (caffeine, theobromine, and theophylline) reduced binding of 3H-guanine or 3H-xanthine by ≤40%, whereas the purine nucleosides guanosine or xanthosine, or ascorbic acid, the substrate for the mammalian NAT members, failed to elicit an inhibitory effect. Further elucidation of the binding kinetics for 3H-xanthine and 3H-guanine with the SPA revealed dissociation constants (Kd) of about 2 µM and 3 µM, respectively, and a nucleobase (xanthine or guanine)-to-protein molar binding ratio of ~1 (Fig. 1C, see also SI Appendix, Fig. S2), revealing the presence of a single nucleobase binding site in the purine transporter of C. psychrerythraea 34H that we refer to as PurTCp.
Fig. 1.
Substrate specificity of PurTCp. (A) Binding of 0.5 µM 3H-xanthine, 3H-hypoxanthine, 3H-adenine, 3H-guanine, 3H-uracil, 3H-thymine, or 3H-cytosine (all at a specific radioactivity of 5 Ci/mmol) was measured with the SPA using 100 ng of purified PurTCp in 200 mM Tris/MES, pH 7.0, 10% glycerol, 0.1 mM TCEP, and 0.1% decyl-β-D-maltopyranoside for 16 h at 4 °C. Specific binding was determined by subtracting the nonspecific counts per minute (cpm) determined in the presence of 800 mM imidazole, which competes with the PurTCp-His tag for binding to the Cu2+-His tag SPA beads, from the cpm measured in the absence of imidazole. (B) Binding of 0.5 µM 3H-xanthine (red) or 3H-guanine (blue) to 100 ng of purified PurTCp in the presence or absence of 250 µM of the indicated compound in assay buffer composed of 200 mM Tris/MES, pH 8.0, 20% (v/v) glycerol, 1 mM TCEP, and 0.1% DDM. Data were normalized to the cpm measured in the absence of compounds for 3H-xanthine or 3H-guanine binding. Data in panels A and B are the mean ± SEM of n ≥ 3 (each performed as a technical replicate). (C) Saturation binding of 3H-xanthine (red) or 3H-guanine (blue) to 100 ng of purified PurTCp was performed in the same buffer as used in the experiments shown in panel B and yielded a dissociation constant (Kd) for xanthine binding of 2.02 ± 0.2 µM and a Kd for guanine binding of 3.02 ± 0.33 µM with a molar substrate-to-PurTCp binding ratio of 1.04 ± 0.02 and 1.04 ± 0.03 for xanthine and guanine binding, respectively. Data of three independent experiments each performed in triplicate were subjected to global nonlinear regression fitting in Prism 8.
Structure of PurTCp.
The PurTCp structure was determined to a resolution of 2.80 Å and refined (Rwork/Rfree of 21/25%). The asymmetric unit contains two protomers of PurTCp, and residues 1 to 165 and 172 to 457 are resolved in each protomer (Materials and Methods and SI Appendix, Table S1). Both the amino (N) and carboxy (C) termini are located to the cytosolic side based on the positive-inside rule (22).
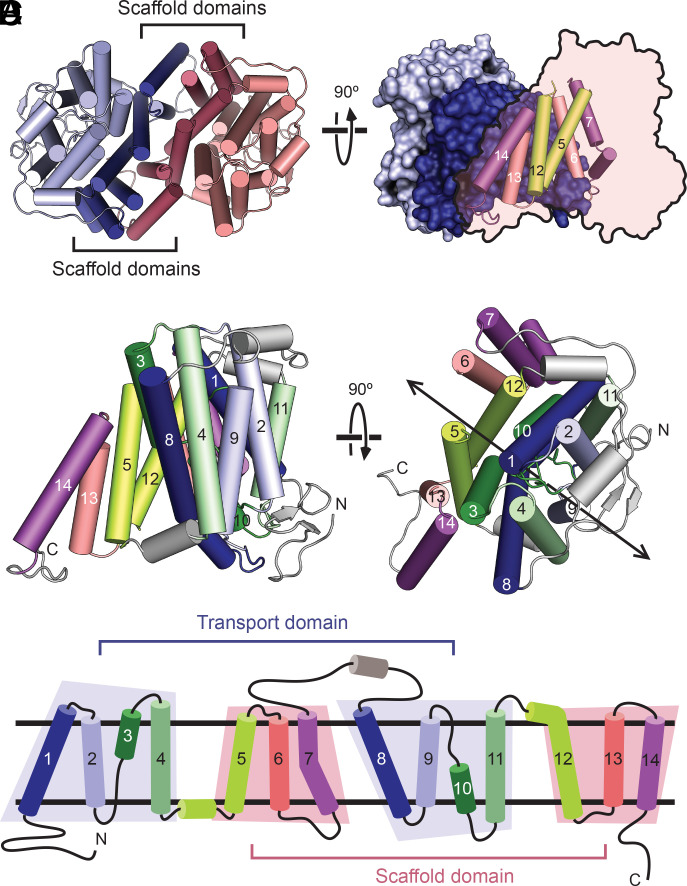
PurTCp forms a dimer with an extensive interface (Fig. 2 A and B). Each protomer has 14 transmembrane segments (TMs), and like many other secondary solute transporters, the first half of the molecule is related to the second half by a pseudo-two-fold symmetry (Fig. 2 C and D). The two halves are connected by a long periplasmic loop. The 14 TMs are largely alpha-helical with the exception of TMs 3 and 10, and they fold into two structurally distinct domains that we refer to as transport and scaffold domains to indicate their perceived functional roles and consistent with the definitions used in a bacterial homolog of glutamate transporters (23, 24). Transport and scaffold domains have been named as core and gate domains, respectively, in the structures of UraA (15, 16). The scaffold domain is composed of TMs 5 to 7 from the first half and 12 to 14 from the second half of the protomer, and the six helices have few interactions among them and are arranged almost side by side similar to a picket fence. The scaffold domains from the two protomers form the dimer interface with a buried surface area of 3,709 Å2.
Fig. 2.
Fold and oligomeric structure of PurTCp. (A) The PurTCp dimer viewed from the extracellular side, with the scaffold and transport domains colored in lighter or darker shades, respectively. (B) A view of the dimer from within the plane of the membrane. One protomer is shown with a surface view, and the other is shown as a transparent outline with helices from the scaffold domain shown as cylinders. (C) Two perpendicular views of a protomer from the PurTCp structure colored in pairs of symmetry-equivalent transmembrane helices. The twofold pseudosymmetry axis is marked on the right. (D) A topology diagram colored according to the same scheme as in panel C.
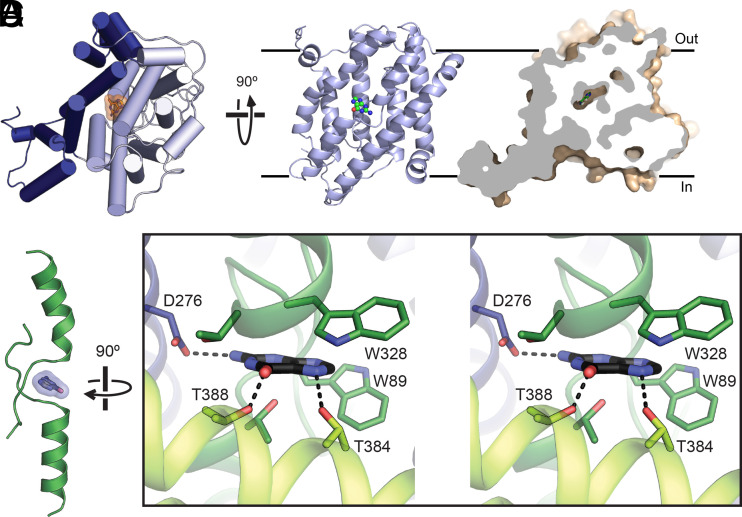
The transport domain is formed by TMs 1 to 4 from the first half and TMs 8 to 11 from the second half of the protomer, and the eight helices are arranged into a compact unit (Fig. 2 C and D). TM 3 and its pseudosymmetry mate TM 10 are both composed of an alpha helix preceded by an extended beta-strand, and the two TMs cross each other at roughly the middle of the membrane (Fig. 3 A and B). The cross-over region is known to bind substrates based on previous structures of the NAT family of transporters (15–17). A nonprotein electron density was found near the cross-over region, and a guanine nucleobase was built into the density based on functional studies, although the current resolution is not sufficient to unambiguously define its identity (SI Appendix, Fig. S3 C and D). The substrate is contained within the transport domain between the ends of the half membrane-spanning helices in TM 3 and TM 10 and is occluded from the solvent on both sides of the membrane (Fig. 3 A–C), similar to the bound uracil in the UraA structure (SI Appendix, Fig. S4). Two Trp residues flank the bound guanine, and potential hydrogen bonds are provided by a number of hydrophilic residues from TMs 3, 8, 10, and 12, including a highly conserved Asp residue (Asp276) on TM 8 (Fig. 3D and SI Appendix, Fig. S5).
Fig. 3.
Substrate binding site. (A) A protomer of PurTCp viewed from the extracellular side (Left) and the transport domain viewed from the membrane (Right), with the bound substrate molecule. (B) Cutaway surface representation of a PurTCp monomer. (C) The bound substrate (blue) in PurTCp is shown in relation to the TM3 and TM10 transmembrane passes. (D) Stereo view of the substrate-binding site in PurTCp, with nearby residues shown as sticks and potential hydrogen bonds marked with dashed lines.
Effect of the pH on PurTCp Activity.
Nucleobase transport by nonmammalian NAT members was proposed to be thermodynamically coupled to the flux of H+, i.e., electrogenic H+/nucleobase symport (25). Since residues with acidic side chains, i.e., Asp or Glu, oftentimes play central roles in the translocation of H+ during the symport process, we hypothesized that a conserved Asp or Glu in close proximity of the identified purine substrate is involved in the proposed H+-dependent transport process. We identified Asp276 as the only conserved acidic amino acid in our sequence alignment of NAT members (SI Appendix, Fig. S5). Notably, Asp276 participates in the coordination of the bound purine substrate identified in our structure (Fig. 3D). We replaced Asp276 with Ala or Asn to test whether the side chain at position 276 is the potential H+ binding site in PurTCp that undergoes protonation/deprotonation during the transport process.
pH dependence on the binding activity of PurTCp.
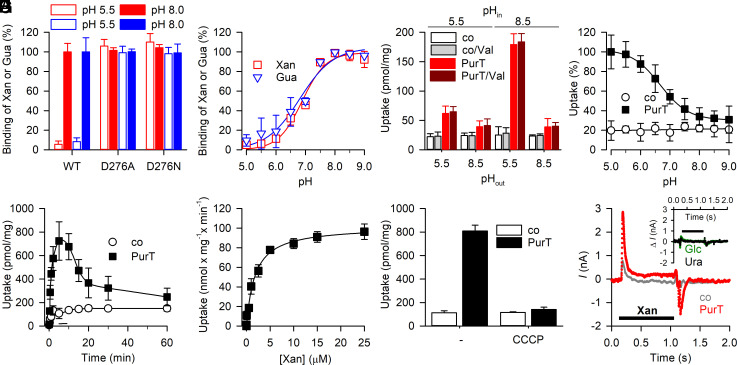
Equilibrium binding of 0.1 µM 3H-xanthine or 3H-guanine by PurTCp-D276A and -D276N was measured at pH 5.5 and pH 8.0 and compared to the binding activity by PurTCp-wild-type (WT) under the same experimental conditions. Whereas purine binding by the two Asp276 mutants was similar to that observed for PurTCp-WT at pH 8.0, lowering the pH to 5.5 almost abolished 3H-xanthine and 3H-guanine binding by the WT but did not affect the binding activity for the two Asp276 mutants (Fig. 4A).
Fig. 4.
Functional characterization of PurTCp. (A) SPA-based binding of 0.5 µM 3H-xanthine (red) or 3H-guanine (blue) binding by PurTCp-WT or -D276A or D276N measured at pH 5.5 and pH 8.0. (B) pH dependence of 0.5 µM 3H-xanthine or 3H-guanine binding by PurTCp-WT. (C) The uptake of 1 µM 3H-xanthine by PurTCp-containing proteoliposomes requires an inwardly directed H+ gradient. Proteoliposomes containing PurTCp or control liposomes devoid of protein were prepared in either 20 mM HEPES-KOH, pH 8.5 or 20 mM MES-KOH, pH 5.5 and 100 mM KCl, 2 mM β-mercaptoethanol (pHin) and transport was measured for 30 s in 20 mM HEPES-KOH, pH 8.5 or 20 mM MES-KOH, pH 5.5 and 100 mM NaCl, 2 mM β-mercaptoethanol (pHout) in the presence or absence of 1 µM of the K+ ionophore valinomycin (Val) as indicated. (D) pH dependence of 1 µM 3H-xanthine transport by proteoliposomes containing PurTCp or control liposomes (pHin = 8.5) measured for 1 min in assay buffer with pHout between 5.0 and 9.0. Data were normalized to the maximum transport activity observed for PurTCp-containing proteoliposomes at pHout = 5.0. (E) Time course of 1 µM 3H-xanthine in proteoliposomes containing PurTCp or control liposomes (pHin = 8.5; pHout = 5.5). (F) Transport kinetics of PurTCp incorporated into proteoliposomes (pHin = 8.5; pHout = 5.5) revealed a Michaelis–Menten constant (Km) of 1.78 ± 0.23 µM and a maximum velocity of transport (Vmax) of 102.0 ± 3.3 nmol × mg−1 × min−1. (G) Substrate uptake is dependent on the proton motive force (pmf). The uptake of 1 µM 3H-xanthine in proteoliposomes containing PurTCp or control liposomes was measured for 5 min in the presence or absence of 5 µM of the protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP). Data in panels A–G are means ± SEM of three independent experiments performed as technical triplicates. (H) Transport of xanthine is accompanied by H+ flux. Representative recording (n = 8) of a SSM electrophysiological measurement performed with proteoliposomes containing PurTCp (red) or with control liposomes (gray) using the SURFE2R N1 (Nanion Technologies, Inc.). Ten micromolar xanthine was added to the assay buffer as indicated by the bar. The Inset shows the net difference between currents (ΔI) elicited by 10 µM of the nontransported pyrimidine uracil (Ura, black) or glucose (Glc, green) in PurTCp-containing proteoliposomes and control liposomes. Data of a representative experiment (n = 3 to 6) are shown. All proteoliposome preparations were tested for H+ leakiness using an assay by Tsai and Miller (26) (SI Appendix, Fig. S6c).
Testing the effect of the pH on 3H-xanthine and 3H-guanine binding by PurTCp-WT revealed a steep pH dependence (with halfway point ~pH 6.8) with the highest activities observed at pH ≥ 7.5 (Fig. 4B). Since PurTCp-D276A and -D276N lost the pH dependence of purine binding and exhibited similar binding activities at pH 5.5 and pH 8.0 (Fig. 4A), this result implies that the highest substrate binding activity occurs when the side chain at position 276 is unprotonated.
pH dependence on the transport activity of PurTCp.
Based on the strict pH dependence observed for binding, we tested the transport of 3H-xanthine with PurTCp-WT reconstituted into proteoliposomes of defined H+ gradients across the liposomal membrane. Fig. 4C shows that an inwardly directed H+ gradient (i.e., [H+]out > [H+]in or pHout < pHin; ) yielded the highest uptake activity. In contrast, generation of an electrical membrane potential (∆Ψ) through a valinomycin-induced K+ diffusion potential (inside negative) did not serve as the sole driving force for xanthine uptake, nor did it stimulate H+ gradient-driven xanthine accumulation. Likewise, a K+ diffusion potential-generated inverse membrane potential (outside negative) did not significantly affect the transport activity (SI Appendix, Fig. S6).
Notably, xanthine uptake was impaired for PurTCp-D276A and -D276N, indicating that conserved Asp276 plays a critical role in H+ binding and/or the regulation of substrate binding and transport by H+ (SI Appendix, Fig. S6). Testing the effect of the external pH on xanthine transport (Fig. 4D) revealed an inverse pH dependence pattern to that observed for binding (Fig. 4B). Here, the highest activity was observed at pH ≤ 5.5, and the halfway point of the curve was about pH 6.7 (Fig. 4D).
Measuring the time course of 3H-xanthine transport (Fig. 4E) under optimized conditions (pHin = 8.5, pHout = pH 5.5) showed that xanthine accumulation peaked within about 5 min before reaching the steady state of transport (~1 h), a phenomenon often observed in secondary active transporters when the driving forces of the transport reaction (here and the xanthine concentration gradient) become exhausted (27, 28). Determining the concentration dependence of the initial rates of uptake (measured at 3H-xanthine concentrations between 10 nM and 25 µM for 10-s periods to avoid reverse transport) yielded a Michaelis–Menten constant (Km) of ~1.8 µM and a maximum velocity of transport (Vmax) of ~100 nmol × mg of PurTCp−1 × min−1 (Fig. 4F). This Vmax translates to a catalytic turnover number (kcat) of ~0.1 s−1, which is similar to the kcat observed for other secondary transporters (28, 29).
To elucidate whether the observed pH dependence of xanthine transport is reflective of H+/purine symport, we tested the effect of the protonophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP) on the pH-dependent transport of 3H-xanthine. Fig. 4G shows that dissipation of the transmembrane H+ gradient impairs transport by PurTCp. In further support of the notion that PurTCp mediated H+/purine symport, solid-supported membrane (SSM)-based electrophysiology using PurTCp-containing proteoliposomes (pHin = 8.5) showed that the addition of 10 µM xanthine to an assay medium with a pH of 5.5 elicited a change in the membrane potential which was reflected in the upward transient current as the result of an inward flux of positive charges. The transient current component for PurTCp-containing proteoliposomes was ~6 times larger in magnitude than that observed in control liposomes lacking PurTCp (Fig. 4H). The removal of xanthine from the assay medium elicited an inverse current transition in PurTCp-containing proteoliposomes. Performing the assay in the presence of 10 µM of the nontransported pyrimidine uracil or glucose yielded SSM traces for PurTCp-containing proteoliposomes and control liposomes that were indistinguishable from each other (Fig. 4 H, Inset).
Discussion
The structure of PurTCp, together with functional data presented here, provides further insight into the underlying elements of ion-coupled substrate transport by the H+-dependent members of the NAT family. The purine binding site is located to the cross-over region of TM3 and 10, and this feature is conserved in transporters of the same structural fold (SI Appendix, Fig. S7). The PurTCp structure was captured in an occluded conformation in which the substrate does not have unimpeded access to either side of the membrane.
Our functional data support H+/substrate symport as the underlying mechanism of nucleobase transport by PurTCp. While the transport of nucleobases is strictly dependent on the presence of a transmembrane H+ gradient ( ), SSM-based electrophysiological measurements employing PurTCp-containing proteoliposomes reveal a change in the membrane potential as a result of H+ influx into the proteoliposomes concomitant with the translocation of nucleobases. This notion is supported by the finding that the protonophore CCCP dissipates PurTCp-mediated nucleobase transport.
Our functional data further highlight the essential character of conserved Asp276, a residue that was identified to interact with the bound guanine (the carboxy oxygen of Asp276 binds the guanine amino group) in the PurTCp structure. H+-coupled nucleobase transport was impaired with PurTCp variants in which Asp276 was replaced with Ala or Asn. However, in contrast to PurTCp-WT, binding of nucleobases by these variants was pH independent, thus hinting to a transport mechanism that involves Asp276 in the coordinated association and dissociation of the symported H+. Under the equilibrium binding conditions in the SPA (i.e., the lack of across the membrane, i.e., the extra- and intracellular faces of PurTCp are exposed to the same pH), nucleobase binding by purified PurTCp-WT decreased with increasing H+ concentrations, suggesting that protonation of Asp276 interferes with nucleobase binding under those conditions. This notion is supported by the fact that PurTCp variants with amino acid side chains at position 276 that cannot be protonated feature virtually indistinguishable nucleobase binding activities at all pH values tested. Since these Asp276 mutants fail to mediate H+-coupled nucleobase symport, it appears possible that protonation of Asp276 in PurTCp-WT occurs after binding of the purine substrate and is the trigger for conformational changes that coordinate the dissociation of bound substrate and H+ from their respective binding sites to the cytoplasmic side of PurTCp.
It is thus feasible to speculate that H+ binding to Asp276 may induce conformational changes to nucleobase-bound PurTCp that lead to the release of H+ and nucleobase substrate in the cytoplasm. This model supports the common paradigm of the alternating access mechanism according to which the binding of the coupling cation induces a conformation that is required for the coordinated translocation of the substrate molecule across the membrane.
Materials and Methods
Expression, Purification, and Crystallization.
A total of 100 bacterial homologs of nucleobase/cation symporter 2 (NCS2) proteins were cloned into a modified pET28 plasmid (Novagen) with either an N- or a C-terminal deca-histidine tag and a tobacco etch virus (TEV) protease recognition site at the central facility of the New York Consortium on Membrane Protein Structure (NYCOMPS) as described in Love et al. (30). Expression vectors containing gene of interest were transformed into BL21-Gold (DE3) competent cells (Agilent), and the cells were cultured to a density of ~1 OD600/mL at 37 °C. Overexpression of protein was then induced by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM at 20 °C for overnight. For expression screening, 10 mL of cell cultures was harvested, and cell pellets were resuspended in 1 mL of lysis buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 2 mM β-mercaptoethanol, and 1 mM phenylmethanesulfonyl fluoride (PMSF). Cells were lysed by sonication, and the protein was extracted with 30 mM n-dodecyl-β-D-maltopyranoside (DDM) in the lysis buffer for 2 h at 20 °C. The cell lysate was centrifuged at 40,000 × g for 45 min at 4 °C, and the supernatant was loaded onto a cobalt affinity column (Clontech). After washing the column with 20-bed volume of 20 mM HEPES, pH 7.5, 150 mM NaCl, 10% (v/v) glycerol, 2 mM β-mercaptoethanol, 5 mM DDM, and 20 mM imidazole, pH 8.0, bound protein was eluted in the same washing buffer except with 300 mM imidazole pH 8.0. Protein samples were then subjected to SDS-PAGE and Coomassie staining. Expressed clones were scaled-up to 2- to 12-L cultures, and purified proteins were analyzed by size-exclusion chromatography. One clone from C. psychrerythraea 34H (PurTCp) with an N-terminal deca-histidine tag yielded ~1 mg protein/liter culture and displayed a monodispersed peak when eluted from a size-exclusion column, representing a suitable candidate for crystallization trials.
Purification of PurTCp was modified slightly from the above described as the last elution step was omitted. Instead, PurTCp bound on a cobalt column was released by TEV protease cleavage for 1 h at 20 °C to separate cleaved PurTCp from uncleaved protein. Purified PurTCp was then loaded onto a Superdex 200 10/300 GL column (Cytiva Life Sciences) equilibrated in 20 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM β-mercaptoethanol, 12 mM n-nonyl-β-D-maltopyranoside (NM), and 4 mM 3-([3-cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO).
Cross-Linking.
Purified PurTCp or UraA (~1 mg/mL) was incubated with indicated concentrations of disuccinimidyl glutarate (DSG) at room temperature for 10 min and loaded onto SDS-PAGE. Protein bands were visualized by Coomassie staining.
Crystallization.
Purified PurTCp protein was concentrated to ~10 mg/mL as approximated by ultraviolet absorbance, and crystallization was set up using the sitting-drop vapor diffusion method. The best PurTCp crystals were grown at 4 °C in 30% (v/v) PEG-400, 0.1 M MES pH 6.0, 3 mM Na-glycochenodeoxycholate, and 0.5 mM 6-bromopurine and reached full size within 7 d. The crystals were cryoprotected by raising PEG-400 (v/v) concentrations gradually to 38% with an increment of 2% over a course of 16 h before being flash frozen in liquid propane.
X-ray Data Collection and Processing.
X-ray data were collected at the NE-CAT beamlines 24-ID-C and 24-ID-E at the Advanced Photon Source at Argonne National Laboratory. A native dataset was collected at a wavelength of 0.9150 Å with a resolution of 2.85 Å. The diffraction data were processed with HKL2000 (31). The crystal belongs to the space group P21212 (No. 18) with unit cell dimensions of 131.713 Å, 135.993 Å, and 79.169 Å. Molecular replacement using the core domain of either the AlphaFold2 prediction or UraA (PDB ID 5XLS) as a search model led to a solution for the initial phase, which was improved by iterations of modeling building in COOT (32) and refinement in PHENIX (33). Protein geometry was validated with MolProbity (34).
Scintillation Proximity Assay.
Binding of nucleobases was performed by means of the scintillation proximity assay (SPA) using His-tagged PurTCp in conjunction with copper His-Tag YSi beads (Perkin Elmer, RPNQ0096). One hundred nanogram of purified PurTCp was immobilized on 125 µg SPA beads per 100-µL assay in 200 mM Tris/MES at the indicated pH, 20% (v/v) glycerol, 1 mM tris(2-carboxyethyl)phosphine (TCEP), and 0.1% DDM unless otherwise stated. 3H-labeled xanthine (5.2 Ci/mmol), hypoxanthine (9 Ci/mmol), adenine (29.7 Ci/mmol), guanine (21.2 Ci/mmol), or uracil (38.7 Ci/mmol) were used for the experiments (all radiochemicals were purchased from American Radiolabeled Chemicals, Inc.).
Transport Measurements.
Purified PurTCp-WT, -D276A, or -D279N was reconstituted into preformed liposomes made of total E. coli lipids (Avanti) at a 1:100 (w/w) ratio as described (30, 35). Liposomes were prepared in i) 20 mM HEPES-KOH, pH 8.5, 100 mM KCl, and 2 mM β-mercaptoethanol, ii) 20 mM MES-KOH, pH 5.5, 100 mM KCl, and 2 mM β-mercaptoethanol, iii) 20 mM HEPES-KOH, pH 8.5, 100 mM NaCl, and 2 mM β-mercaptoethanol, or iv) 20 mM MES-KOH, pH 5.5, 100 mM NaCl, and 2 mM β-mercaptoethanol. The same buffers (in all possible cis/trans permutations) were also used as assay buffer to test the influence of the membrane potential on the transport reaction by generating a valinomycin-mediated K+ diffusion potential. For assessing the influence of the external pH on the uptake activity, the external buffer was composed of 20 mM MES-KOH, pH 5.5 to 6.5 or 20 mM HEPES-KOH, pH 7.0 to 9.0 and 100 mM KCl, 2 mM β-mercaptoethanol. The uptake of 3H-xanthine was measured with a rapid filtration assay using 0.45 µM nitrocellulose filters (Millipore). Reactions were incubated at 23 °C for the indicated periods of time and quenched by the addition of ice-cold 100 mM potassium phosphate, pH. 6.0, and 100 mM LiCl before filtration. The radioactivity retained on the filters was determined with scintillation counting using the dried filters. Known amounts of radioactivity were used to convert decays per minute (dpm) to mol.
Microscale Thermophoresis.
Binding of either guanine or xanthine by purified PurTCp was measured by microscale thermophoresis (MST) (36) using the Monolith NT.LabelFree and Monolith NT.115 (NanoTemper Technologies, Inc.) to detect the thermophoretic signals of native tryptophan and tagged fluorescence in PurTCp, respectively. For label-free binding measurements, 500 nM PurTCp was mixed with guanine or xanthine at a concentration range of 0.47 to 94 µM in MST assay buffer containing 20 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 10% v/v glycerol, 2 mM β-mercaptoethanol, and 0.1% w/v DM (n-decyl-β-D-maltopyranoside). The mixture was incubated at room temperature for 20 min and then loaded into Monolith NT.LabelFree capillaries. Measurements were conducted at high (60%) MST power and 15% excitation power using the MO.Control v1.4.4 software. For binding assays using the Monolith NT.115, prior to the MST measurement, purified PurTCp was labeled with RED fluorescent dye NT-647 (RED-tris-NTA; NanoTemper Technologies, Inc.) following the manufacturer’s protocol. As for label-free measurements, assays were performed using 50 nM RED-labeled PurTCp in MST assay buffer in Monolith NT.115 Premium capillaries, and the thermophoresis reaction was set at high MST power. MST raw data, after evaluation in MO. Control, were transferred, normalized, and analyzed using nonlinear regression fitting in GraphPad Prism 8.
SSM Electrophysiology.
SSM electrophysiological measurements were performed using the SURFE2R N1 (Nanion Technologies, Inc.) according to published protocols (37). Briefly, the sensors were filled with 1.5 μL of the lipid solution (1,2-diphytanoyl-sn-glycero-3-phosphocholine in n-decane), 50 μL of nonactivating buffer (20 mM MES, pH 5.5, and 100 mM NaCl) and 10 μL of PurTCp-containing proteoliposomes (after being extruded and sonicated). The activating solution (20 mM MES, pH 5.5, 100 mM NaCl, and 10 μM xanthine) was applied with the single-solution exchange protocol (activating buffer incubation for 1 s). Four different datasets from individual sensors were recorded. Peak currents were corrected by subtracting the peak currents recorded with control liposomes (devoid of PurTCp). To obtain the PurTCp-elicited charge (Coulomb) movement associated with xanthine transport, the area under the curve (current as function of time) was analyzed with GraphPad Prism 8.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the US NIH grant R01 GM119396 (M.Q. and M.Z.), DK122784 (M.Z.), GM145316 (M.Z.) and HL086392 (M.Z.). We are indebted to NYCOMPS (and its successor organization COMPPÅ) for their invaluable contribution.
Author contributions
J.W., X.Z., P.W., M.Z., and M.Q. designed research; J.W., X.Z., P.W., Z.R., K.C., E.G.-I., M.Z., and M.Q. performed research; M.Z. and M.Q. contributed new reagents/analytic tools; J.W., X.Z., P.W., Z.R., K.C., E.G.-I., M.Z., and M.Q. analyzed data; and M.Z. and M.Q. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Ming Zhou, Email: mzhou@bcm.edu.
Matthias Quick, Email: mq2102@cumc.columbia.edu.
Data, Materials, and Software Availability
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, https://www.rcsb.org/ under accession ID 7TAK (38). All other data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.de Koning H., Diallinas G., Nucleobase transporters (review). Mol. Membr. Biol. 17, 75–94 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Nagai K., Nagasawa K., Kihara Y., Okuda H., Fujimoto S., Anticancer nucleobase analogues 6-mercaptopurine and 6-thioguanine are novel substrates for equilibrative nucleoside transporter 2. Int. J. Pharm. 333, 56–61 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Haines M. L., et al. , Clinical usefulness of therapeutic drug monitoring of thiopurines in patients with inadequately controlled inflammatory bowel disease. Inflamm. Bowel. Dis. 17, 1301–1307 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Luscher A., Lamprea-Burgunder E., Graf F. E., de Koning H. P., Maser P., Trypanosoma brucei adenine-phosphoribosyltransferases mediate adenine salvage and aminopurinol susceptibility but not adenine toxicity. Int. J. Parasitol. Drugs. Drug. Resist 4, 55–63 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nigam S. K., What do drug transporters really do? Nat. Rev. Drug Discov. 14, 29–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang Y., Li S., Chen L., The physiological role of drug transporters. Protein Cell 6, 334–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michels A. J., Frei B., Myths, artifacts, and fatal flaws: Identifying limitations and opportunities in vitamin C research. Nutrients 5, 5161–5192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lykkesfeldt J., Michels A. J., Frei B., Vitamin C. Adv. Nutr. 5, 16–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padayatty S. J., et al. , Vitamin C as an antioxidant: Evaluation of its role in disease prevention. J. Am. College Nutr. 22, 18–35 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Tsukaguchi H., et al. , A family of mammalian Na+-dependent L-ascorbic acid transporters. Nature 399, 70–75 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Burzle M., et al. , The sodium-dependent ascorbic acid transporter family SLC23. Mol. Aspects Med. 34, 436–454 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto S., et al. , Identification and functional characterization of the first nucleobase transporter in mammals: Implication in the species difference in the intestinal absorption mechanism of nucleobases and their analogs between higher primates and other mammals. J. Biol. Chem. 285, 6522–6531 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niopek-Witz S., Deppe J., Lemieux M. J., Mohlmann T., Biochemical characterization and structure-function relationship of two plant NCS2 proteins, the nucleobase transporters NAT3 and NAT12 from Arabidopsis thaliana. Biochim. Biophys. Acta 1838, 3025–3035 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Karatza P., Frillingos S., Cloning and functional characterization of two bacterial members of the NAT/NCS2 family in Escherichia coli. Mol. Membr. Biol. 22, 251–261 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Lu F., et al. , Structure and mechanism of the uracil transporter UraA. Nature 472, 243–246 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Yu X., et al. , Dimeric structure of the uracil:proton symporter UraA provides mechanistic insights into the SLC4/23/26 transporters. Cell Res. 27, 1020–1033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alguel Y., et al. , Structure of eukaryotic purine/H+ symporter UapA suggests a role for homodimerization in transport activity. Nat. Commun. 7, 11336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalli A. C., Sansom M. S., Reithmeier R. A., Molecular dynamics simulations of the bacterial UraA H+-uracil symporter in lipid bilayers reveal a closed state and a selective interaction with cardiolipin PLoS Comput. Biol. 11, e1004123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosti V., Lambrinidis G., Myrianthopoulos V., Diallinas G., Mikros E., Identification of the substrate recognition and transport pathway in a eukaryotic member of the nucleobase-ascorbate transporter (NAT) family. PLoS One 7, e41939 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen P. S., Frees D., Fast R., Mygind B., Uracil uptake in Escherichia coli K-12: Isolation of uraA mutants and cloning of the gene. J. Bacteriol. 177, 2008–2013 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quick M., Javitch J. A., Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc. Natl. Acad. Sci. U.S.A. 104, 3603–3608 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson H., von Heijne G., Membrane protein topology: Effects of ΔµH+ on the translocation of charged residues explain the "positive inside" rule Embo J. 13, 2267–2272 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reyes N., Ginter C., Boudker O., Transport mechanism of a bacterial homologue of glutamate transporters. Nature 462, 880–885 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akyuz N., Altman R. B., Blanchard S. C., Boudker O., Transport dynamics in a glutamate transporter homologue. Nature 502, 114–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopkins P., Shaw R., Acik L., Oliver S., Eddy A. A., Fluorocytosine causes uncoupled dissipation of the proton gradient and behaves as an imperfect substrate of the yeast cytosine permease. Yeast 8, 1053–1064 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Tsai M. F., Miller C., Substrate selectivity in arginine-dependent acid resistance in enteric bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, 5893–5897 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quick M., Wright E. M., Employing Escherichia coli to functionally express, purify, and characterize a human transporter. Proc. Natl. Acad. Sci. U.S.A. 99, 8597–8601 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malinauskaite L., et al. , A mechanism for intracellular release of Na+ by neurotransmitter/sodium symporters. Nat. Struct. Mol. Biol. 21, 1006–1012 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Z., Itokawa M., Uhl G. R., Dopamine transporter proline mutations influence dopamine uptake, cocaine analog recognition, and expression. Faseb J. 14, 715–728 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Love J., et al. , The New York Consortium on Membrane Protein Structure (NYCOMPS): A high-throughput platform for structural genomics of integral membrane proteins. J. Struct. Funct. Genomics 11, 191–199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zehnpfennig B., Wiriyasermkul P., Carlson D. A., Quick M., Interaction of α-lipoic acid with the human Na+/multivitamin transporter (hSMVT). J. Biol. Chem. 290, 16372–16382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wienken C. J., Baaske P., Rothbauer U., Braun D., Duhr S., Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 100 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Bazzone A., Barthmes M., Fendler K., SSM-based electrophysiology for transporter research. Methods Enzymol. 594, 31–83 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Weng J., Zhou X., Ren Z., Chen K., Zhou M., Structure of a NAT transporter. Protein Data Bank https://www.rcsb.org/structure/7TAK. Deposited 21 December 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, https://www.rcsb.org/ under accession ID 7TAK (38). All other data are included in the article and/or SI Appendix.