Abstract
A rapid induction of antiviral genes is critical for eliminating viruses, which requires activated transcription factors and opened chromatins to initiate transcription. However, it remains elusive how the accessibility of specific chromatin is regulated during infection. Here, we found that XAF1 functioned as an epigenetic regulator that liberated repressed chromatin after infection. Upon RNA virus infection, MAVS recruited XAF1 and TBK1. TBK1 phosphorylated XAF1 at serine-252 and promoted its nuclear translocation. XAF1 then interacted with TRIM28 with the guidance of IRF1 to the specific locus of antiviral genes. XAF1 de-SUMOylated TRIM28 through its PHD domain, which led to increased accessibility of the chromatin and robust induction of antiviral genes. XAF1-deficient mice were susceptible to RNA virus due to impaired induction of antiviral genes. Together, XAF1 acts as an epigenetic regulator that promotes the opening of chromatin and activation of antiviral immunity by targeting TRIM28 during infection.
XAF1 directs the specific opening of chromatin during virus infection.
INTRODUCTION
Innate immunity is the natural immune defense formed by hosts during germline development and evolution, and it is the first line of defense against invading pathogens (1, 2). Pathogen-associated molecular patterns (PAMPs) from exogenous pathogens or damage-associated molecular patterns (DAMPs) from hosts can bind to pattern recognition receptors (PRRs) to initiate the downstream innate immune responses (3). There are a variety of PRRs involved in the initiation of innate immune responses, including Toll-like receptors (TLRs) (4), retinoic acid–inducible gene I–like receptors (RLRs) (5), cyclic guanosine 3′,5′-monophosphate–adenosine 5′-monophosphate (GMP-AMP) synthase (cGAS) (6), and NOD-like receptors (7). After sensing the PAMPs or DAMPs, these PRRs recruit adaptor proteins, including stimulator of interferon genes (STING), mitochondrial antiviral signaling protein (MAVS), and TIR domain-containing adaptor-inducing interferon-beta, and then activate TANK binding kinase 1 (TBK1) respectively (8). TBK1 phosphorylates and activates transcription factors interferon regulatory factor 3 (IRF3) and IRF7, and these IRFs initiate transcription of type I interferons (IFNs) (9). Type I IFNs induce antiviral effectors that inhibit viral replication and activate adaptive immune responses (10–13).
IRF3-directed transcription of type I IFNs is vital for antiviral innate immunity. Phosphorylated IRF3 dimerizes and translocates to the nucleus, driving the expression of type I IFNs and IFN-stimulated genes (ISGs) to initiate an IFN-dependent antiviral response (14). ISG expression can be induced by IRF3 directly or secreted type I IFNs via IFN receptor signaling (15).
IRF1 plays a more important role than IRF3 at the very early stage of viral infection. Upon viral infection, peroxisomal MAVS triggers the rapid expression of IFN-independent genes by IRF1 to provide short-term protection, whereas mitochondrial MAVS activates an IFN-dependent signaling pathway by IRF3 with a delayed kinetics, which amplifies and stabilizes the antiviral responses (16). Therefore, IRF1 plays a crucial role in the very early stage, whereas IRF3 plays a critical role after IRF1 activation.
IRF1 induces the transcription of various genes involved in the first response to viral invasion, such as RNA-activated protein kinase (PKR) and 2′,5′-oligoadenylate synthetases (OAS) (17, 18). Previous studies have revealed the roles of IRF1 in the development of various immune cells. IRF1 is expressed broadly and is required for the ontogeny and function of the lymphoid and myeloid compartments of the immune system, including T lymphocytes, natural killer (NK) cells, and macrophages (IRF1 and IRF2 bind to the same regulatory elements of IFN and IFN-inducible genes). Irf1−/− mice are immunodeficient and susceptible to infections. They carry lineage-specific defects in thymocyte development characterized by a marked reduction in CD8+ T cells (19), a decrease in NK cell numbers with associated impaired cytolytic activity (20), and reduced numbers of CD8α+ dendritic cells (DCs) concomitant with a skewed differentiation of CD11b+ cells toward plasmacytoid DCs (21). The diversity of differentiation raises the question of why the expression of the same transcription factor leads to different gene subsets. The combination of gene expression and chromatin accessibility analysis allows the analysis of another level of gene regulation. Pioneering factors that alter chromatin also affect the accessibility of transcription factors to specific genes (22). A recent study showed that IRF1 regulates chromatin accessibility to promote IFNB1 transcription by IRF3, highlighting the importance of IRF1 in the regulation of chromatin accessibility (23).
Regulation of chromatin opening and gene expression underlies a key point during host defense against viral infection, which endows the host with timely and effective expression patterns of antiviral genes. However, how the expression of genes is regulated by chromatin remodeling during viral infection and the mechanisms by which determine the opening of specific chromatin in antiviral immunity remain unclear (24, 25). Tripartite motif-containing (TRIM) 28, as an epigenetic corepressor protein, is ubiquitously expressed. TRIM28 recruits the histone methyltransferase SETDB1 in a SUMO-dependent manner. SETDB1 then creates repressive histone marks at transposable elements (TEs) sites [particularly trimethylation of lysine-9 on histone subunit H3 (H3K9me3)] that recruit heterochromatin protein 1 (HP1) to induce DNA heterochromatinization and TE repression (26). SUMOylation of TRIM28 is lost during viral infection. The loss of SUMOylated TRIM28 leads to derepression of TEs by the removal of DNA heterochromatin (27). In some cases, TE expression may act in cis to promote innate immune responses (26, 28). The mechanism by which TRIM28 SUMOylation is reduced after viral infection is currently unclear.
X-linked inhibitor of apoptosis (XIAP)–associated factor 1 (XAF1) was originally identified as an antagonist of XIAP counteracting its anti-caspase activity by inhibiting the RING domain of XIAP. XAF1 is a nonclassical zinc finger protein and has seven zinc finger domains (29). Frequent epigenetic inactivation of XAF1 in multiple human cancers and its association with the advanced stage and high grade of tumors support the notion that XAF1 alteration is implicated in tumor progression (30–32). Several studies have clarified its functions in tumor suppression and pro-apoptosis, but as an ISG, the function of XAF1 in antiviral immune responses is unknown.
In the present study, we demonstrated that XAF1 was a positive regulator of innate antiviral immunity against RNA viruses. XAF1 deficiency resulted in impaired induction of antiviral genes. Consistently, Xaf1−/− mice were more susceptible to the infection of RNA viruses than wild-type (WT) mice. Mechanistically, upon RNA virus infection, XAF1 was recruited by MAVS and then phosphorylated by TBK1 at serine-252, which initiated its translocation from the cytoplasm to the nucleus. XAF1 then was specifically recruited by IRF1 and bind to TRIM28 to inhibit SUMOylation of TRIM28. This liberated the suppressed chromatin and enhanced the transcription of IRF1-regulated innate immune genes to initiate early antiviral responses. On the other hand, XAF1 and IRF1 facilitated IRF3 to transcribe IFNB1 to initiate IFN-dependent antiviral responses. Therefore, XAF1 plays a key role in the communication between IRF1-mediated early immune responses and IRF3-mediated late IFN-dependent immune responses.
RESULTS
XAF1 facilitates antiviral immune signaling
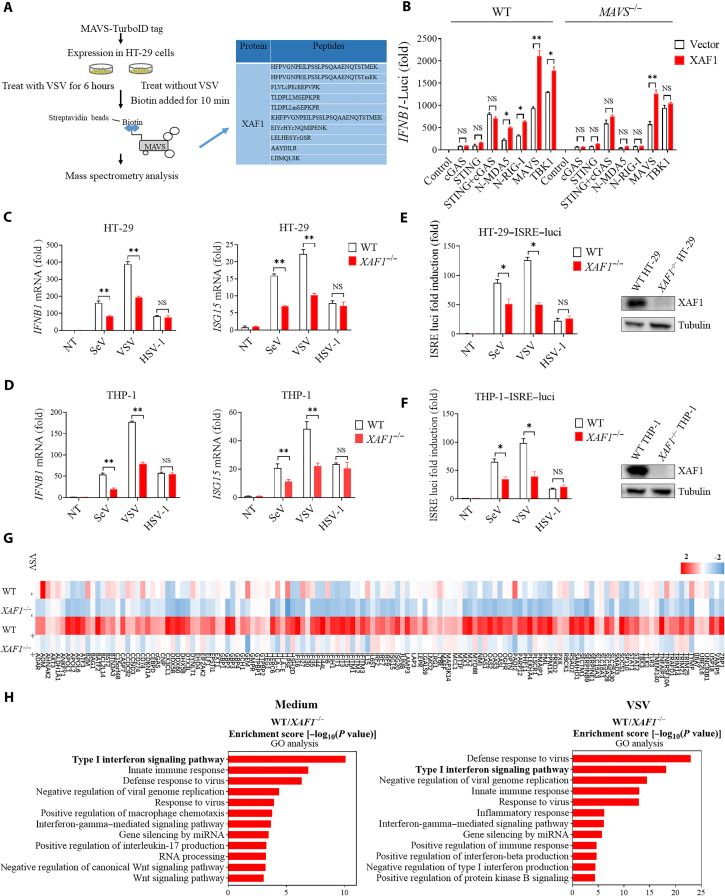
The RLR-MAVS pathway plays a pivotal role in innate immune responses to RNA virus infection. To identify previously unknown cellular regulators of this pathway, we investigated MAVS-interacting proteins by using proximity-dependent biotin identification, which is an effective method for identifying protein-protein interactions in living cells (33). MAVS was cloned into a mammalian expression vector with a TurboID tag for proximity-based labeling and screening of MAVS-interacting proteins (34). HT-29 cells were transfected with the TurboID-tagged MAVS plasmid for 18 hours and then infected with or without vesicular stomatitis virus (VSV) for 6 hours. Following biotin affinity capture, the biotinylated proteins were purified and analyzed by mass spectrometry to identify MAVS-interacting proteins (Fig. 1A). XAF1 was identified as a strong MAVS-binding partner after VSV infection (table S1). The interaction between MAVS and XAF1 was further confirmed by a coimmunoprecipitation (coIP) assay (fig. S1A).
Fig. 1. XAF1 facilitates antiviral immune signaling.
(A) Flow chart depicting the process of MAVS-TurboID proximity-based labeling technology to detect interacting proteins. XAF1 was identified as a MAVS-interacting protein by mass spectrometry. (B) Luciferase (Luci) activity in HEK293T cells transiently transfected with 50 ng of the pRL-TK reporter, 50 ng of luciferase reporter driven by promoters of genes encoding IFN-β, empty vector (Vector), or XAF1 together with expression plasmids for Cgas, STING, cGAS+STING, N-MDA5, N-RIG-I, MAVS, or TBK1 for 24 hours. (C and D) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of IFNB1 and ISG15 was performed after NT (nontreated) or after stimulation with SeV, VSV, or HSV-1 for 12 hours in XAF1−/− HT-29 (C) and THP-1 (D) cells. (E and F) 2fTGH-ISRE (IFN-stimulated response element) cells were treated with supernatant from control and XAF1−/− HT-29 (E) and THP-1 (F) cells, which were NT or stimulated with SeV, VSV, or HSV-1 for 12 hours. After 6 hours of treatment of the supernatant, ISRE luciferase activity was measured. (G) Heatmap for selected ISGs in WT and XAF1−/− HT-29 cells treated with or without VSV for 6 hours. (H) Histogram of the combined score (https://david.ncifcrf.gov/tools.jsp) for biological processes for the up-regulated genes in WT versus XAF1−/− HT-29 cells treated with or without VSV for 6 hours. Data are the means ± SEM (n = 3) from three independent experiments; NS, not significant; *P < 0.05 and **P < 0.01 [Student’s t test, (B) to (F)]. Data are representative of two [(G) and (H)] or three [(B) to (F)] independent experiments with similar results.
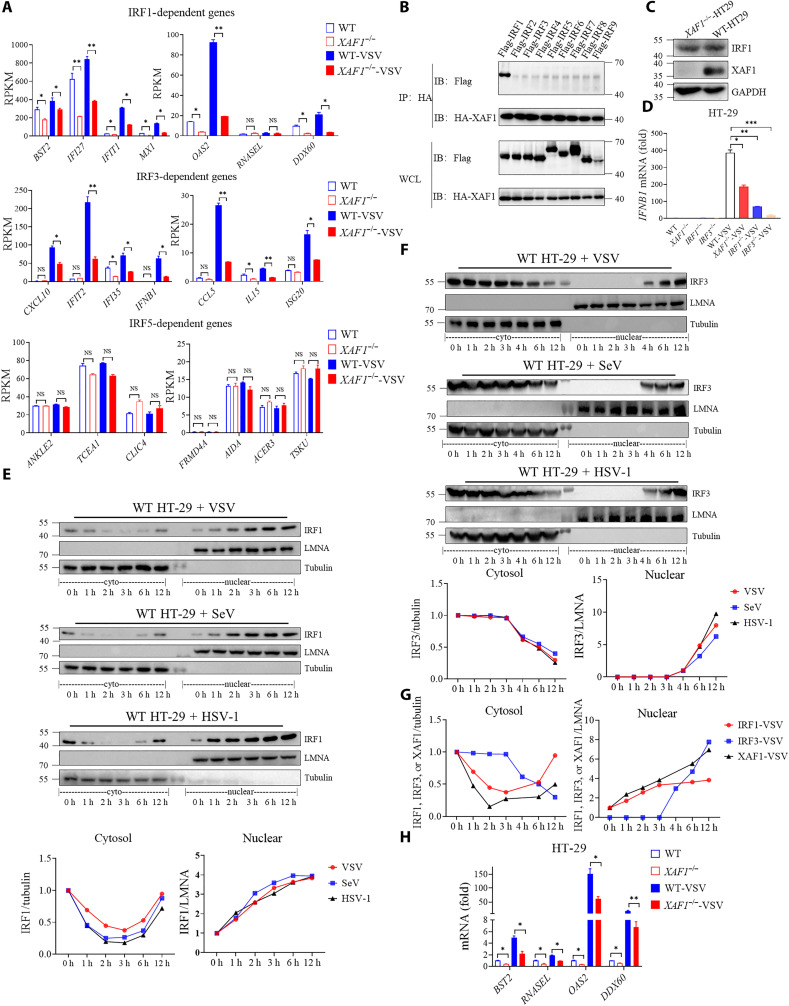
XAF1 is known as a pro-apoptosis protein (29), whose function in innate immunity remains elusive. We first evaluated the effect of XAF1 overexpression on RLR-MAVS and cGAS-STING signaling. Transient overexpression of cGAS, STING, the N-terminal caspase activation and recruitment domains of RIG-I (RIG-N) or MDA5 (MDA5-N), MAVS, or TBK1 alone was sufficient to induce significant IFNB1 expression (Fig. 1B). Notably, simultaneous overexpression of XAF1 further magnified IFNB1 expression stimulated by RIG-N, MDA5-N, MAVS, or TBK1, but not by cGAS or STING in WT human embryonic kidney (HEK) 293 cells. However, this synergistic effect was completely abolished in MAVS−/− cells (Fig. 1B), indicating that XAF1 regulates RLR signaling in a MAVS-dependent manner. Consistently, overexpression of XAF1 enhanced IFNB1 expression during RNA virus infection, including VSV and Sendai virus (SeV) (fig. S1B). Next, we assessed the role of XAF1 in RLR signaling and viral infection using a loss-of-function approach. The XAF1 expression levels are extremely low in many cell lines, but XAF1 is expressed in human colon cancer cell lines such as HT-29 (35, 36). In addition to HT-29 cell line, we also detected XAF1 expression in THP-1 cell line by Western blotting (fig. S1C). We thus knocked out XAF1 in HT-29 and THP-1 cells using a CRISPR-Cas9–mediated genome editing approach. The expression of IFNB1 and ISG15 mRNA during infection with RNA viruses (VSV and SeV) in XAF1−/− cells was significantly lower than that in WT THP-1 and HT-29 cells. However, there was no significant difference in IFNB1/ISG15 expression between WT and knockout cells infected with a DNA virus, herpes simplex virus 1 (HSV-1) (Fig. 1, C and D, and fig. S1D). Moreover, we measured the secretion of type I IFN proteins using a luciferase reporter 2fTGH cell line in which Luc is driven by the IFN-stimulated response element (ISRE) (2fTGH-ISRE-Luc cells). XAF1-deficient HT-29 and THP-1 cells exhibited decreased secretion of type I IFNs after RNA virus infection (Fig. 1, E and F). Consistently, the replication of VSV, but not HSV-1, increased significantly in XAF1-deficient HT-29 and THP-1 cells when compared to WT cells (fig. S1E). These data demonstrated that XAF1 is crucial for the induction of type I IFN responses to RNA, but not DNA viruses.
To assess the impact of XAF1 deficiency on antiviral immune responses in an unbiased manner, we performed genome-wide RNA sequencing (RNA-seq) analysis with RNA from WT and XAF1−/− HT-29 cells after VSV infection. The expression of antiviral genes was apparently reduced in XAF1−/− HT-29 cells (Fig. 1G and fig. S1F). Furthermore, the Gene Ontology (GO) enrichment analysis revealed that most of the down-regulated genes in XAF1 knockout were enriched in the type I IFN signaling pathway, innate immune response, and defense response to virus (Fig. 1H). It is well known that XAF1 promotes apoptosis by inhibiting XIAP. To examine whether XAF1-XIAP axis plays a role in the induction of IFN by viral infection, we knock down XIAP in HT-29 cells. The results showed that XIAP was dispensable for the induction of IFNB1 mRNA during infection of RNA or DNA viruses (fig. S1G). Collectively, these results suggested that XAF1 facilitates host defense against RNA viruses and is critical for the induction of antiviral genes.
XAF1 positively regulates IFN responses in vivo
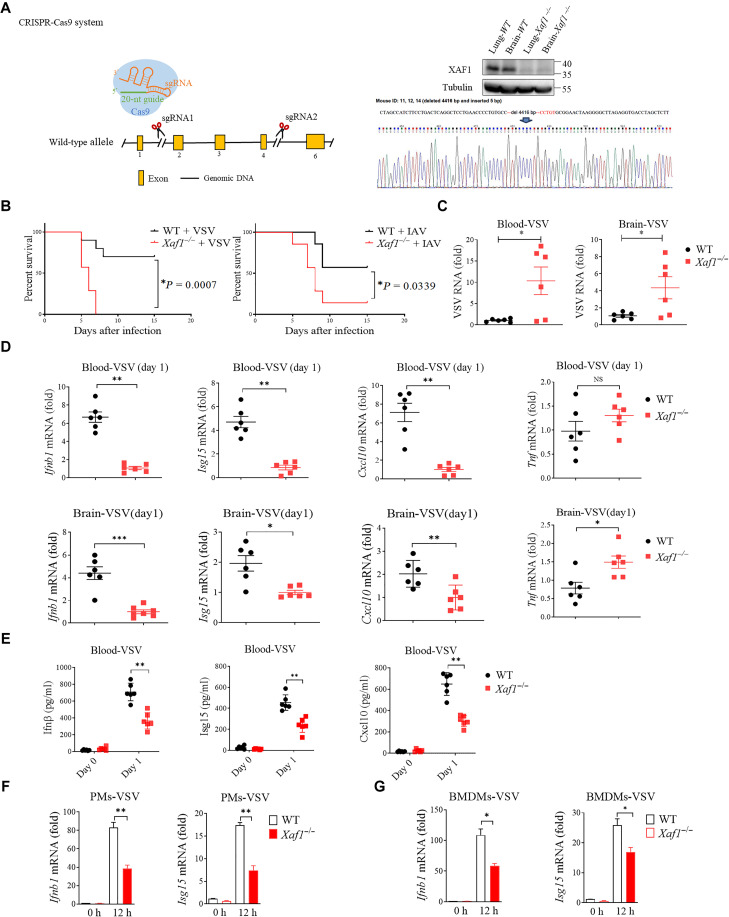
To evaluate the importance of XAF1 in host defense against RNA viruses in vivo, we generated XAF1-deficient mice (Fig. 2A). Then, we infected 8-week-old WT and Xaf1−/− mice with VSV (intravenously), HSV-1 (intravenously), or influenza A virus (IAV) (intranasally) and monitored their survival. The results showed that Xaf1−/− mice were much more susceptible to VSV- and IAV-caused death, but not HSV-1–caused death, than WT mice (Fig. 2B and fig. S2A). Consistently, the VSV loads in the blood and brains of Xaf1−/− mice were significantly higher than those in WT mice (Fig. 2C), whereas the HSV-1 loads were the same between two groups (fig. S2B). In line with this, the mRNA levels of Ifnb1, Isg15, and Cxcl10 were significantly lower in the blood and brains of Xaf1−/− mice than in those of WT mice (Fig. 2D). However, the expression of the inflammatory cytokine Tnf in the blood was not different between WT and Xaf1−/− mice and even higher in in the brains of Xaf1−/− mice (Fig. 2D), suggesting that XAF1 is dispensable for the induction of Tnf during viral infection. Consistent with the reduction of mRNA levels, the secretion of Ifnβ, Isg15, and Cxcl10 proteins, was impaired in Xaf1−/− mice after VSV but not HSV-1 infection (Fig. 2E and fig. S2C). Furthermore, we infected bone marrow–derived macrophages (BMDMs) and peritoneal macrophages (PMs) from WT and Xaf1−/− mice with viruses. The lack of XAF1 significantly dampened the induction of Ifnb1 and Isg15 mRNA by VSV but not by HSV-1 infection (Fig. 2, F and G, and fig. S2, D and E). Together, these data showed that XAF1 is critical for the host defense against RNA virus infection in mouse models.
Fig. 2. XAF1 positively regulates IFN responses in vivo.
(A) Diagram showing CRISPR-Cas9–mediated deletion of the indicated exons to obtain Xaf1−/− mice. (B) WT and Xaf1−/− mice (n = 6 mice per group) were inoculated intravenously with VSV (left) or intranasally IAV (right). The survival rates of the mice were recorded. (C) Four days after VSV infection, the brains and blood of WT and Xaf1−/− mice were retrieved for qRT-PCR analysis of VSV genomic copies. (D) Ifnb1, Isg15, Cxcl10, and Tnf mRNA expression in the brain and blood was assessed by qRT-PCR, 1 day after VSV infection. (E) Effects of XAF1 deficiency on VSV-induced serum levels of Ifnβ, Isg15, and Cxcl10. WT and Xaf1−/− mice were infected with VSV (n = 6) at 107 PFU per mouse for 6 hours before serum was measured by enzyme-linked immunosorbent assay. (F and G) Peritoneal macrophages (PMs) and bone marrow–derived macrophages (BMDMs) isolated from WT and Xaf1−/− mice were infected with VSV for 12 hours. Ifnb1 and Isg15 mRNA was measured by qRT-PCR analysis. Data are means ± SEM (n = 3) from three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 [Student’s t test (C) to (G)]. *P = 0.0007 and *P = 0.0339 [long-rank test (B)]. Data are representative of three independent experiments with similar results. sgRNA, single guide RNA.
XAF1 interacts with MAVS and TBK1
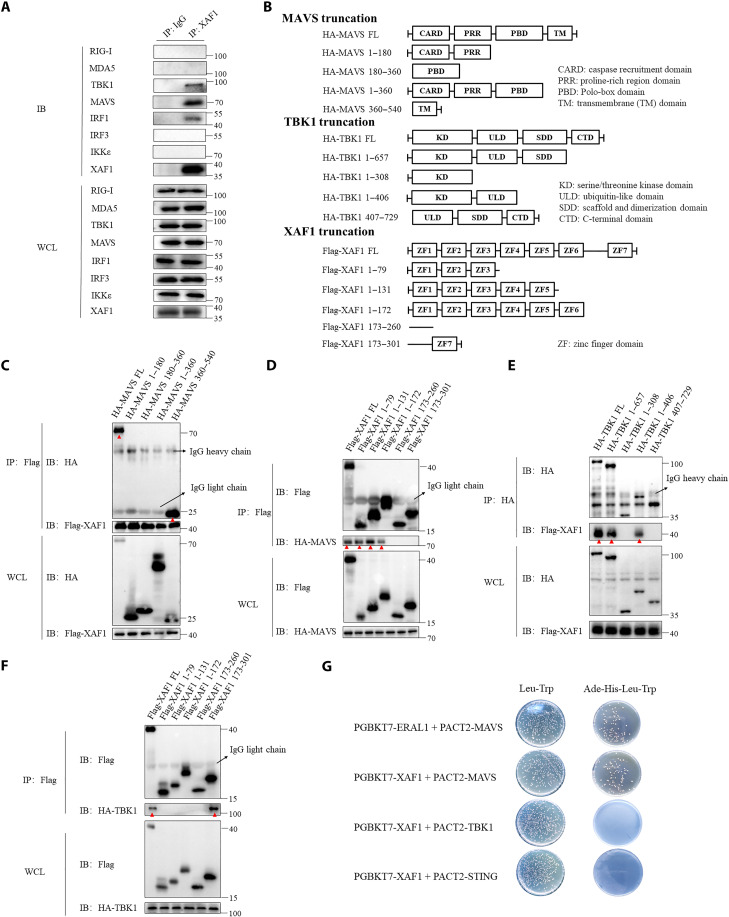
We above showed that XAF1 played an important role in the RLR signaling pathways (Fig. 1B). To clarify the mechanism, we immunoprecipitated and analyzed endogenous XAF1 by Western blotting. Notably, in addition to MAVS, TBK1 and IRF1 were also coprecipitated with XAF1. However, XAF1 did not pull down RIG-I, MDA5, IRF3, or IκB kinase ɛ (IKKɛ) (Fig. 3A). We then generated a series of truncations to identify the respective domains of MAVS, TBK1, and XAF1 mediating their interaction (Fig. 3B). With coIP assays, we observed that full-length XAF1 was co-immunoprecipitated with full-length and the transmembrane domain (corresponding to residues 360 to 540 amino acids) of MAVS (Fig. 3C), and the zinc finger domains 1 to 3 (ZF; 1 to 79 amino acids) of XAF1 were sufficient to pull down MAVS as full-length XAF1 did (Fig. 3D). The ubiquitin-like domain (ULD; 308 to 406 amino acids) of TBK1 interacted with the ZF7 (173 to 301 amino acids) of XAF1 (Fig. 3, E and F) (red arrowheads in C to F point to the desired protein bands). To examine whether these interactions are direct or not, we performed a yeast two-hybrid experiment. We noted that XAF1 directly interacted with MAVS but not TBK1. As a positive control, direct ERAL1-MAVS interaction (37) was recapitulated here. As a negative control, we did not detect the direct interaction between XAF1 and STING (Fig. 3G). Consistently, deletion of individual ZF1, ZF2, ZF3, or ZF7 abolished the ability of XAF1 to enhance MAVS-induced IFNB1 (fig. S3A). Together, our data showed that XAF1 interacted with MAVS and TBK1 to facilitate IFN responses.
Fig. 3. XAF1 interacts with MAVS and TBK1.
(A) coIP assay analysis of the interaction between endogenous XAF1 and RIG-I, MDA-5, TBK1, MAVS, IRF1, IRF3, and IKKe in HT-29 cells. (B) Diagram detailing the series of truncations of MAVS, TBK1, and XAF1. (C and D) coIP assay analysis shows which domains of MAVS and XAF1 interacted. Red arrowheads in (C) and (D) highlight the interaction bands. (E and F) coIP assay analysis shows which domains of TBK1 and XAF1 interacted. Red arrowheads in (E) and (F) highlight the interaction bands. (G) Direct interaction of XAF1 with MAVS, STING, and TBK1 was detected by a yeast two-hybrid assay. Yeast strain AH109 was transfected with PGBKT7-XAF1, PACT2-MAVS, or PACT2-STING and inoculated on the indicated media for 3 days. ERAL1 and MAVS were introduced as positive controls. Data are representative of three independent experiments with similar results. WCL, whole-cell lysate; IP, immunoprecipitation; IB, immunoblot.
TBK1 phosphorylates XAF1 at S252 and promotes early nuclear translocation of XAF1
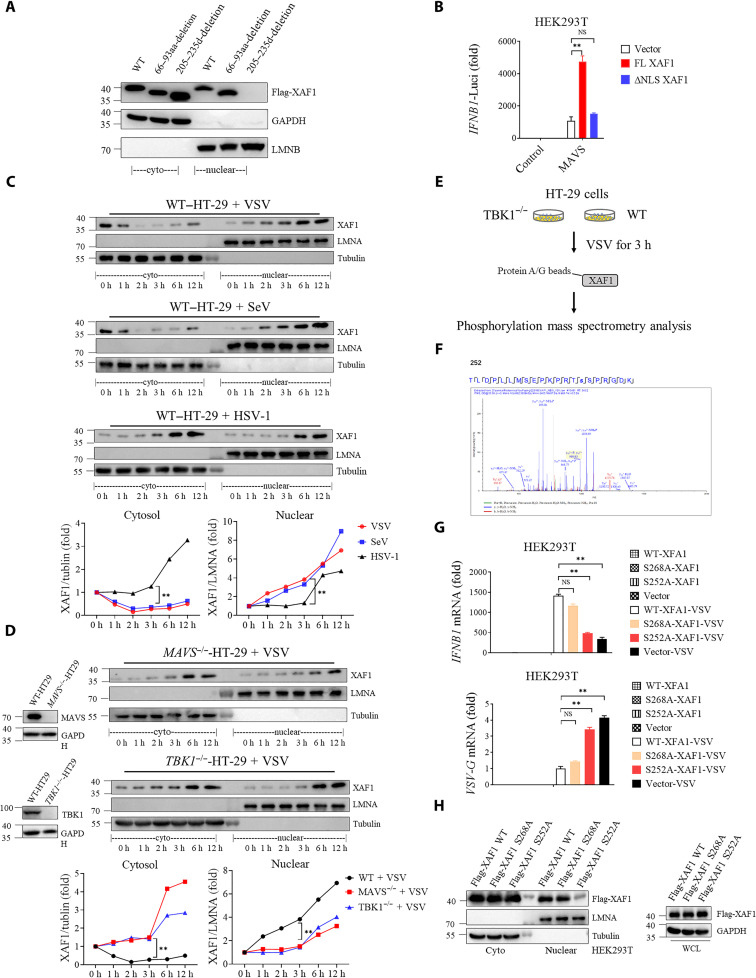
To understand the mechanistic basis by which XAF1 regulates antiviral responses, we explored the subcellular location of XAF1 during virus infection. In unstimulated cells, endogenous XAF1 was found both in the cytosol and nucleus (29). A putative nuclear localization signal (NLS) of XAF1 corresponding to amino acids 205 to 235 was identified by a NLS prediction software (www.compbio.dundee.ac.uk/www-nod/index.jsp). To confirm whether this NLS was functional, we constructed a plasmid in which the putative NLS sequence was deleted. The NLS deletion mutant (ΔNLS) was completely absent from the nucleus (Fig. 4A), and unable to enhance the induction of IFNB1 by MAVS as full-length XAF1 did (Fig. 4B). Moreover, the semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) assay showed that MAVS aggregation was not affected by overexpression of XAF1 (fig. S4A). Consistently, the dimerization, nuclear translocation of IRF3, and phosphorylation of TBK1 were unaffected by deficiency of XAF1 (fig. S4, B and C). These results suggested that XAF1 does not regulate the activation of MAVS-TBK1-IRF3 axis.
Fig. 4. TBK1 phosphorylates XAF1 at S252 and promotes early nuclear translocation of XAF1.
(A) Putative NLS sequences of XAF1 were screened. Western blotting was used to analyze the protein distribution in cytoplasm and nucleus. (B) Luciferase activity in HEK293T cells transfected with 50 ng of pRL-TK reporter and 50 ng of luciferase reporter driven by promoters of genes encoding IFN-β, expression plasmids MAVS together with Vector, full length (FL), or NLS-deletion XAF1 plasmids for 24 hours. (C and D) Cytoplasmic and nuclear fractions were isolated, and the time course of changes in (C) XAF1 levels in each fraction after VSV, SeV, and HSV-1 infection in HT-29 cells and (D) XAF1 levels in each fraction after VSV infection in MAVS−/− (top) or TBK1−/− (bottom) HT-29 cells was tracked by Western blotting. The relative expression of XAF1 [relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or LMNA (Lamin A/C)] curve shows its dynamic changes in cytoplasm and nucleus. (E) A flow chart depicting the process of coIP experiments to detect the phosphorylation site of XAF1 by TBK1 in TBK1−/− and WT HT-29 cells. (F) Phosphorylation site of XAF1 by TBK1 was identified via mass spectrometry. (G and H) HEK293T cells were transfected with indicated Vector, FL-XAF1, S252A-XAF1, and S268A-XAF1 for 24 hours. After 12 hours of VSV infection, qRT-PCR analyses of IFNB1 mRNA (top) and VSVG mRNA (bottom) were performed. Western blotting was used to analyze the protein distribution in cytoplasm and nucleus. Data are means ± SEM (n = 3) from three independent experiments. *P < 0.05 and **P < 0.01 [Student’s t test (B) to (D) and (G)]. Data are representative of two [(E) and (F)] or three [(A) to (D) and (G) and (H)] independent experiments with similar results. aa, amino acid.
Notably, infection with RNA viruses (VSV and SeV) but not DNA virus induced apparent translocation of XAF1 into the nucleus both in HT-29 cells (Fig. 4C) and THP-1 cells (fig. S4D) rapidly as early as 1 hour after infection. In addition, a similar phenomenon was observed when HT-29 cells were stimulated by synthetic RLR agonists, low–molecular weight (LMW) polyinosinic:polycytidylic acid [ploy(I:C)] and high–molecular weight (HMW) ploy(I:C) (fig. S4E). No significant nuclear translocation was noted at the very early stage of HSV-1 infection (1 and 3 hours) (Fig. 4C). However, at later stages of HSV-1 infection (6 and 12 hours), both the cytoplasmic and nuclear XAF1 levels increased equally (Fig. 4C). This is because as an ISG, the expression of XAF1 was significantly induced by IFNs (38). These results demonstrated that XAF1 is rapidly translocated to the nucleus in response to RNA virus infection.
Next, we explored the molecular mechanism driving XAF1 nuclear translocation during RNA virus infection. As above shown, XAF1 interacted with MAVS and TBK1 (Fig. 3A). TBK1 is a key kinase in many PRR pathways. To explore whether MAVS and TBK1 mediate the early nuclear translocation of XAF1, we generated MAVS and TBK1 knockout HT-29 cells. Knockout of MAVS and TBK1 abolished the early nuclear translocation of XAF1 at 1 to 3 hours after infection (Fig. 4D). To determine whether TBK1 activates XAF1 via its kinase activity, we performed mass spectrometry to detect phosphorylation modifications of endogenous XAF1 in TBK1−/− and WT HT-29 cells infected with VSV for 3 hours (Fig. 4E). XAF1 was phosphorylated at S252 and S268 after VSV infection (Fig. 4F, fig. S4F, and table S2). To determine whether phosphorylation of S252 or S268 is crucial for XAF1 function and nuclear translocation, S252A and S268A (mutating serine to alanine prevents potential phosphorylation) mutant XAF1 were constructed. We observed that S252A mutation largely abolished the capacity of XAF1 to promote IFNB1 expression during VSV infection when compared with WT and S268A XAF1. Consistent with this, the S252A mutant failed to suppress VSV replication as WT and S268A XAF1 did (Fig. 4G). Furthermore, the nuclear pool of S252A mutant XAF1 was much less than that of WT and S268A XAF1 (Fig. 4H). Conversely, replacing S252 with D (glutamic acid) to mimic constitutive phosphorylation of XAF1 enhanced IFNB1 expression and XAF1 nuclear translocation compared with WT and S268D XAF1 (fig. S4, G and H). Together, the phosphorylation of XAF1 S252 by TBK1 is essential for the early nuclear translocation and IFN-promoting function of XAF1.
XAF1 is required for induction of IRF1 and IRF3 target genes
Transcription factor enrichment analysis of down-regulated genes from RNA-seq of WT/XAF1−/− HT-29 cells showed that IRF1 and IRF3 were critical transcription factors associated with XAF1 deficiency (fig. S5, A and B). IRF1 induces transcription of early IFN-independent antiviral genes, while IRF3 primarily mediates transcription of type I IFN genes. This is consistent with our above data that XAF1 was essential for induction of Ifnb1 and Isgs, but not Tnf, in mice after infection (Fig. 2). We here further confirmed that the genes regulated by IRF1 and IRF3 were significantly decreased due to deficiency of XAF1, but not the genes regulated by IRF5 (Fig. 5A). Consistent with a previous study (39), we also showed that XAF1 interacted with IRF1 (Fig. 3A), suggesting that XAF1 directly regulates IRF1 activity. However, we observed no physical interaction between IRF3 and XAF1 (Fig. 3A), suggesting that XAF1 indirectly regulates IRF3-mediated transcription. A recent study reported that IRF1 regulated chromatin accessibility to help IRF3-driven transcription of IFNB1 (23). Nonetheless, we further confirmed that XAF1 specifically interacted with IRF1, but not any other IRFs (IRF2 to IRF9), by coIP (Fig. 5B). Earlier studies have shown that overexpression of XAF1 can increase the protein level of IRF1 by reducing the degradation of IRF1 (39); however, we did not observe a difference in the protein level of IRF1 in XAF1−/− HT-29 cells (Fig. 5C), indicating that XAF1 promotes IRF1 activity by a different mechanism.
Fig. 5. XAF1 is required for induction of IRF1- and IRF3- target genes.
(A) Analysis of IRF1-, IRF3-, and IRF5-regulated gene expression levels in WT and XAF1−/− HT-29 cells from RNA-seq data. (B) coIP assay analysis of the interaction between XAF1 and IRF1, IRF2, IRF3, IRF4, IRF5, IRF6, IRF7, IRF8, and IRF9. (C) Western blot showed protein level of endogenous IRF1 in WT and XAF1−/− HT-29 cells. (D) qRT-PCR analysis of IFNB1 after NT or after stimulation with VSV for 12 hours in WT, XAF1−/−, IRF1−/−, and IRF3−/− HT-29 cells. (E and F) Cytoplasmic and nuclear fractions were isolated, and then, the time course of changes in (E) IRF1 levels and (F) IRF3 levels in each fraction after VSV, SeV, and HSV-1 infection in HT-29 cells was tracked by Western blotting analysis. The relative expression of IRF1 and IRF3 (relative to GAPDH or LMNA) curve shows its dynamic changes in the cytoplasm and nucleus. (G) Relative expression of XAF1, IRF1 and IRF3 (relative to GAPDH or LMNA) curve shows its dynamic changes in the cytoplasm and nucleus after VSV infection in HT-29 cells. (H) qRT-PCR analysis of BST2, RNASEL, OAS2, and DDX60 after NT or after stimulation with VSV for 12 hours in WT and XAF1−/− HT-29 cells. Data are means ± SEM (n = 3) from three independent experiments. *P < 0.05 and **P < 0.01, and ***P < 0.001 [Student’s t test (A), (D), and (H)]. Data are representative of two (A) or three [(B) to (H)] independent experiments with similar results. RPKM, reads per kilobase per million mapped reads; HA, hemagglutinin.
Next, we compared the relative contribution of XAF1, IRF1, and IRF3 to IFNB1 expression during RNA virus infection. The IFNB1 levels were progressively decreased in XAF1−/−, IRF1−/−, and IRF3−/− HT-29 cells (Fig. 5D). Moreover, we examined the timing of IRF1 and IRF3 nuclear translocation during viral infection. IRF1 was present in small amounts in the nucleus of resting cells (0 hour) and rapidly entered the nucleus following either DNA or RNA virus treatment as early as 1 hour after infection (Fig. 5E), while IRF3 was only detected in the nucleus at 4 hours and later (Fig. 5F). XAF1 and IRF1 entered the nucleus at the same time and both earlier than IRF3 (Fig. 5G). Moreover, we ruled out the possibility that the entry of XAF1 and IRF1 into the nucleus is reliant on each other. The deficiency of IRF1 or XAF1 did not affect nuclear translocation of each other (fig. S5, C and D). In addition, quantitative polymerase chain reaction (qPCR) confirmed that the expression of IRF1-regulated genes was decreased in XAF1−/− HT-29 cells compared with WT cells with or without VSV infection (Fig. 5H). The expression of IRF3-regulated genes was also significantly down-regulated in XAF1−/− and in IRF1−/− HT-29 cells (fig. S5E), in agreement with a recent report showing that IRF1 regulated chromatin accessibility to aid IRF3-driven transcription (23). All together, these suggested that XAF1 promotes IRF1-mediated transcription directly and IRF3-driven transcription via IRF1.
XAF1 facilitates the immune responses by inhibiting TRIM28
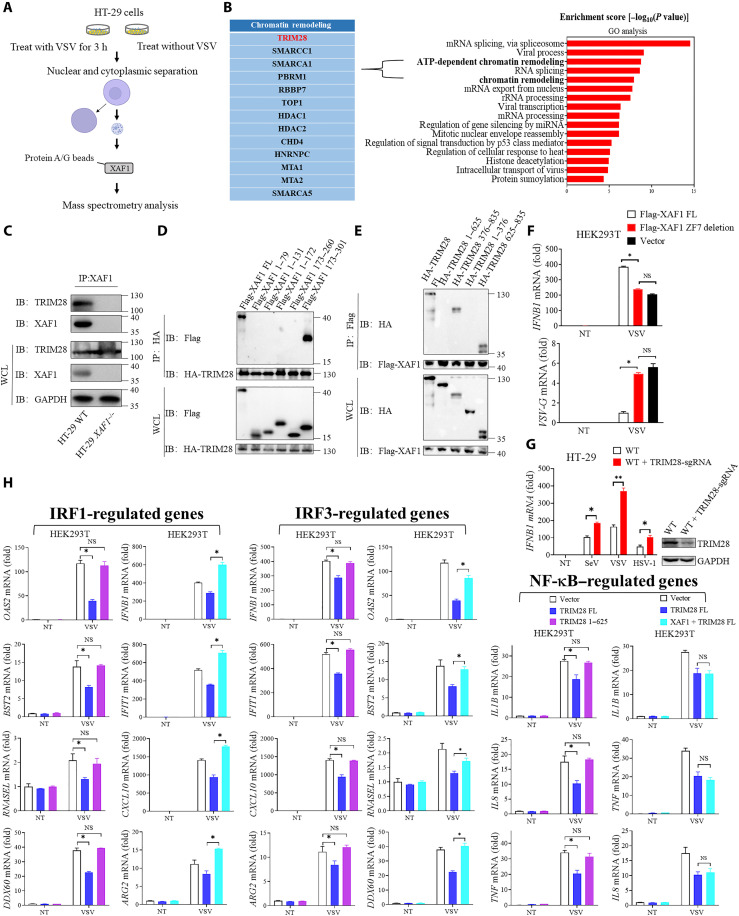
To clarify the molecular mechanism by which XAF1 promotes antiviral gene transcription in the nucleus, we performed co-immunoprecipitating with nuclear extracts and mass spectrometry to identify XAF1-interacting proteins in the nucleus (Fig. 6A and fig. S6A). A total of 190 proteins were identified as potential XAF1 interactors (table S3). GO analysis showed that these proteins were predominantly enriched in the processes of chromatin remodeling (Fig. 6B), suggesting that XAF1 might be involved in chromatin remodeling during viral infection. Among these proteins, TRIM28 was identified as a strong XAF1-binding partner (Fig. 6B). It is known that TRIM28 is a highly SUMOylated protein and recruits the histone methyltransferase SETDB1 in a SUMO-dependent manner. SETDB1 then creates repressive histone marks that recruit HP1 to induce DNA heterochromatinization and TE repression (26). The interaction between endogenous XAF1 and TRIM28 was confirmed by coIP (Fig. 6C). Further immunoprecipitation assays with truncated proteins showed that the plant homeodomain (PHD) and bromodomain (BROMO) domain (corresponding to 625 to 835 amino acids) of TRIM28 interacted with the ZF7 motif (173 to 301 amino acids) of XAF1 (Fig. 6, D and E, and fig. S6B). Consistently, the mutant XAF1 without the ZF7 motif was not able to enhance IFNB1 expression during VSV infection (Fig. 6F).
Fig. 6. XAF1 facilitates the immune responses by inhibiting TRIM28.
(A) A flow chart depicting the process coIP experiments and mass spectrometry to detect proteins interacting with XAF1 in the nucleus. (B) GO analysis of the up-regulated proteins immunoprecipitated by XAF1 after VSV infection. Proteins about chromatin remodeling processes were listed. TRIM28 was identified as an XAF1-interacting protein. (C) coIP assay analysis of the interaction between XAF1 and TRIM28. (D and E) coIP assay analysis shows the protein interaction domains between XAF1 and TRIM28. (F) HEK293T cells were transfected with the indicated Vector FL-XAF1 and ZF7-deletion-XAF1 plasmids for 24 hours. After 12 hours of VSV infection, qRT-PCR analyses of IFNB1 mRNA (top) and VSVG mRNA (bottom) were performed. (G) qRT-PCR analysis of IFNB1 after NT or after stimulation with SeV, VSV, or HSV-1 for 12 hours in WT and WT + TRIM28 sgRNA HT-29 cells. (H) qRT-PCR analysis of IRF1-, IRF3-, and NF-κB–regulated genes after NT or after stimulation with VSV for 12 hours in HEK293T cells transiently transfected with Vector, TRIM28 FL, TRIM28 1 to 625, TRIM28 FL + XAF1. Data are means ± SEM (n = 3) from three independent experiments. *P < 0.05 [Student’s t test (F) to (H)]. Data are representative of three [(C) to (H)] independent experiments with similar results.
Next, we knocked out TRIM28 from HT-29 cells and examined whether it was involved in regulating antiviral immunity. Deficiency of TRIM28 significantly enhanced the induction of IFNB1 by virus infection (Fig. 6G). The PHD domain of TRIM28 has E3 ligase activity and is essential for intramolecular SUMOylation of TRIM28. We thus constructed truncated TRIM28 lacking the PHD domain (TRIM28 1 to 625). We overexpressed full length (TRIM28 FL) and TRIM28 1 to 625 in HEK293 cells. TRIM28 FL but not the truncated form apparently inhibited the expression of IRF1-, IRF3-, or nuclear factor κB (NF-κB)–directed genes during VSV infection (Fig. 6H). Accordingly, chromatin immunoprecipitation (ChIP)–qPCR showed that the chromatin accessibility of those immune genes was suppressed by TRIM28 FL but not TRIM28 1 to 625 (fig. S6C). These data suggested that self-SUMOylation of TRIM28 was required for suppressing antiviral gene transcription. Overexpression of XAF1 was able to offset the inhibitory effect of TRIM28 overexpression on IRF1- or IRF3-directed gene transcription but had no effect on NF-κB–directed gene transcription (Fig. 6H), which was in line with the results in mice (Fig. 2). Consistently, the suppression of the chromatin accessibility of IRF1/IRF3-targeted genes by TRIM28 was relieved by XAF1 overexpression (fig. S6C). On the other hand, the overexpression or knockout of XAF1 did not change IFNB1 expression in TRIM28-deficient cells during RNA/DNA virus infection, suggesting that TRIM28 functions downstream of XAF1 (fig. S6, D to G). These data showed that XAF1 promotes IRF1- and IRF3-regulated genes expression by inhibiting TRIM28.
XAF1 is recruited by IRF1 to inhibit TRIM28
The above results showed that XAF1 interacted with IRF1 and TRIM28 (Figs. 5B and 6C). Notably, both TRIM28 and IRF1 are involved in regulating chromatin compaction and accessibility (23, 26). To explore whether XAF1 specifically regulates the expression of IRF1- and IRF3-regulated genes through chromatin remodeling, we performed a ChIP-seq analysis with an H3 lysine 27 acetylation (H3K27ac) antibody in WT and XAF1 knockout cells. XAF1 deficiency led to an extensive decrease in chromatin accessibility at the promoter (≤1 kb) at 3 hours after VSV infection (fig. S7A). BST2, OAS2, DDX60, and RNASEL encode important antiviral effectors transcriptionally regulated by IRF1. ARG2, IFIT1, CXCL10, and IL15 encode important antiviral effectors directed by IRF3. The virus-induced chromatin accessibility of these genes was significantly decreased in XAF1−/− HT-29 cells compared with WT HT-29 cells (fig. S7B). ATAC-seq (assay for transposase-accessible chromatin using sequencing) analysis confirmed the above results (fig. S7B). In summary, these data demonstrated that XAF1 regulates the expression of IRF1- and IRF3-regulated genes by facilitating chromatin accessibility.
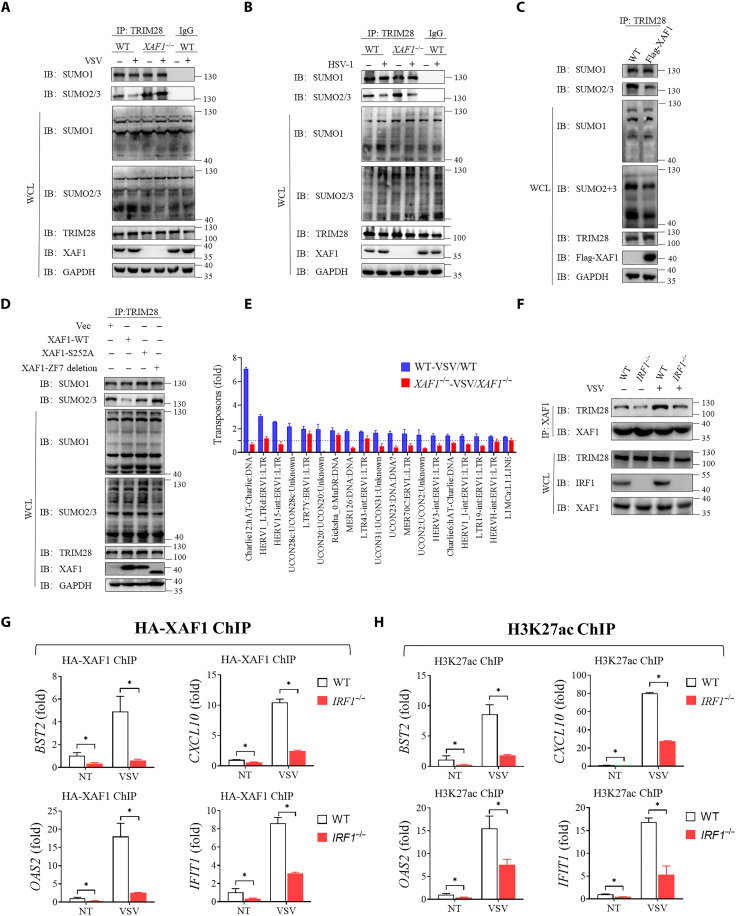
To more clearly determine how XAF1 regulates the chromatin openness through TRIM28, we examined SUMOylation of TRIM28 in WT and XAF1-deficient cells. The endogenous SUMO2/3 but not SUMO1 of TRIM28 decreased after VSV infection in WT cells (Fig. 7A). However, SUMO2/3-TRIM28 was constitutively increased in XAF1−/− compared to WT HT-29 cells before and after VSV infection (Fig. 7A). Intriguingly, knocking out XAF1 did not increase SUMO2/3-TRIM28 levels under HSV-1 infection conditions (Fig. 7B). Conversely, XAF1 overexpression decreased SUMO2/3-TRIM28 levels in HEK293T cells (Fig. 7C). S252A mutation did not affect the interaction between XAF1 and TRIM28 (fig. S7C). Either S252A mutant or ZF7-deleted XAF1 (which lost its interaction with TRIM28) was not able to reduce SUMO2/3 of TRIM28 (Fig. 7D).
Fig. 7. XAF1 is recruited by IRF1 to inhibit TRIM28.
(A) Effects of XAF1 deficiency on endogenous SUMOylation of TRIM28 with or without VSV for 6 hours. (B) Effects of XAF1 deficiency on endogenous SUMOylation of TRIM28 with or without HSV-1 for 6 hours. (C) Western blotting showing the SUMOylation of TRIM28 in HEK293T cells transfected with Vector or XAF1 for 24 hours. (D) Western blotting showing endogenous SUMOylation of TRIM28 in HEK293T cells transfected with Vector, WT-XAF1, S252A-XAF1, or ZF7-deletion-XAF1 for 24 hours. (E) All up-regulated transposons and ERVs after VSV infection in WT HT-29 cells form RNA-seq were showed. Then, these transposons and ERVs change after VSV infection in XAF1−/− HT-29 were also showed. (F) coIP assay analysis of the interaction between XAF1 and TRIM28 in WT HT-29 cells compared with IRF1−/− HT-29 cells with or without VSV infection. (G and H) WT and IRF1−/− HT-29 cells transfected with HA-XAF1 were NT or infected VSV for 3 hours, and ChIP-qPCR signal showing HA-XAF1 (G) or H3K27Ac (H) occupancy of indicated genes. Data are means ± SEM (n = 3) from three independent experiments. *P < 0.05 [Student’s t test (F) to (H)]. Data are representative of three [(A) to (G)] independent experiments with similar results. H3K27ac, H3 lysine 27 acetylation.
In addition to IRF1/3-driven transcription, de-SUMOylation of TRIM28 was shown to unleash transcription of TEs and endogenous retroviruses (ERVs) (26, 27, 40). ERVs could directly activate RLR signaling and subsequent IFN responses (41). Schmidt et al. (27) found that influenza virus–triggered de-SUMOylation of TRIM28 increased the ERVs RNA abundance, leading to enhanced antiviral immunity. Therefore, we examined the expression of TEs and ERVs in WT and XAF1-deficient cells. TEs and ERVs expression was induced by VSV infection in WT cells; however, this was severely impaired by deficiency of XAF1 (Fig. 7E and fig. S7D). These results suggested that XAF1 may also contribute to antiviral immunity via the TRIM28-ERV-RLR axis. Moreover, it is known that newly synthetized RNA was able to bind to and activate scaffold attachment factor A (SAFA; also named HNRNPU) (25, 42). The activated SAFA increased the accessibility of chromatin and promoted induction of antiviral genes (24, 25). SAFA could recognize the stem loop structure of both viral RNA and newly synthetized host RNA and promoted opening of chromatin. Therefore, the newly synthetized ERVs also might be able to bind to and activate SAFA to promote opening of chromatin. To examine this mechanism, we overexpressed XAF1 and then treated cells with α-amanitin to inhibit RNA synthesis (42). Overexpression of XAF1 increased the opening of chromatin, which was completely offset by α-amanitin (fig. S7E). We previously found that TOP1 and SMARCA5 could facilitate the activation of SAFA and opening of chromatin (24). XAF1 failed to enhance antiviral responses after treatment with TOP1 inhibitor or SMARCA5 knockout (fig. S7F). Thus, newly synthesized RNA-SAFA axis was required for promoting opening of chromatin by XAF1.
To explore how XAF1 cooperates with TRIM28 and IRF1, we examined the interaction between XAF1 and TRIM28 in WT and IRF1−/− HT-29 cells with or without VSV infection. In WT cells, the interaction between XAF1 and TRIM28 increased after VSV infection; this phenomenon was not significant in IRF1−/− HT-29 cells (Fig. 7F). Moreover, we performed ChIP-qPCR with hemagglutinin (HA) and H3K27ac antibodies in WT and IRF1−/− HT-29 cells with or without VSV infection. Consistently, IRF1 deficiency decreased the occupancy of XAF1 and H3K27ac at the ISGs regulated by IRF1 or IRF3 (Fig. 7, G and H). Together, these results suggested that XAF1 is recruited by IRF1 to inhibit SUMOylation of TRIM28, thus lifting TRIM28-imposed transcriptional brake on the IRF1/IRF3 target genes.
DISCUSSION
The RLR-MAVS axis is critical for sensing invading RNA viruses and eliciting a rapid innate immune response. MAVS signaling not only rapidly induces antiviral ISGs via IRF1 at the very early stage of viral infection but also sustains ISGs expression via IRF3/7 at later stages (16, 43). However, the mechanisms governing the tightly controlled early and late innate antiviral responses remain to be fully understood. The genome of metazoans is highly organized and packaged with histones to form chromatin. In metazoan cells, the initiation of a gene transcription requires the opening of chromatin and then binding of the transcription factors to its promoter. We previously found that the nuclear matrix protein SAFA remodels chromatin structure through oligomerization with chromatin-associated RNAs for antiviral response (25). In addition, a recent study reported that IRF1 regulates chromatin accessibility to promote IFNB1 transcription by IRF3 (23). Although these studies have proven to a certain extent that the transcription of antiviral genes requires prior chromatin opening, the molecular mechanisms are largely unknown. In this study, we report that XAF1-IRF1 promotes chromatin accessibility of IRF1/3-regulated genes by inhibiting TRIM28 SUMOylation, and this axis coordinates early and late innate antiviral responses.
Most previous studies have shown that XAF1 functions as a tumor suppressor by promoting apoptosis (30–32). Hypermethylation of XAF1 gene in a variety of human malignancies leads to low expression of XAF1, which exacerbates the malignant progression of cancers. Thus, delivery of XAF1 becomes a way to inhibit tumor development (44, 45). Here, we identified XAF1 as a MAVS interacting protein by proximity-based labeling screening (Fig. 1A). coIP experiments and yeast two-hybrid experiments (Fig. 3, A and G) showed that XAF1 directly interacted with MAVS. Then, we revealed a critical role of XAF1 in antiviral signaling and control of RNA virus replication. Mechanistically, upon RNA virus but not DNA virus infection, MAVS recruits XAF1 and TBK1. Then, TBK1 phosphorylates XAF1 at serine-252 to facilitate its translocation from the cytoplasm to the nucleus at the very early stage of viral infection (Fig. 4, C to H). The timing of nuclear translocation of XAF1 after RNA virus infection is similar to that of IRF1 and earlier than that of IRF3 (Fig. 5G). After entering the nucleus, under the guidance of IRF1, phosphorylated XAF1 specifically opens IRF1-targeted chromatin by blocking the SUMOylation of the chromatin repressor TRIM28 in the nucleus (Fig. 7B), and it promotes the expression of IRF1- and IRF3-regulated antiviral genes (Fig. 5A and fig. S5, A and B).
Our study further suggests that XAF1 promotes IRF1-induced, IFN-independent early immune responses and modulates chromatin openness around IRF1 immediately after viral infection. This will also facilitate IRF3 access to its target genes and trigger an IFN-dependent antiviral immune response (Fig. 8). TRIM28 is self-SUMOylated in resting cells, which acts as an epigenetic corepressor protein. It has been reported that viral infection results in de-SUMOylation of TRIM28. However, the mechanism by which TRIM28 is de-SUMOylated is currently unclear. We here found that XAF1 can reduce TRIM28 SUMOylation.
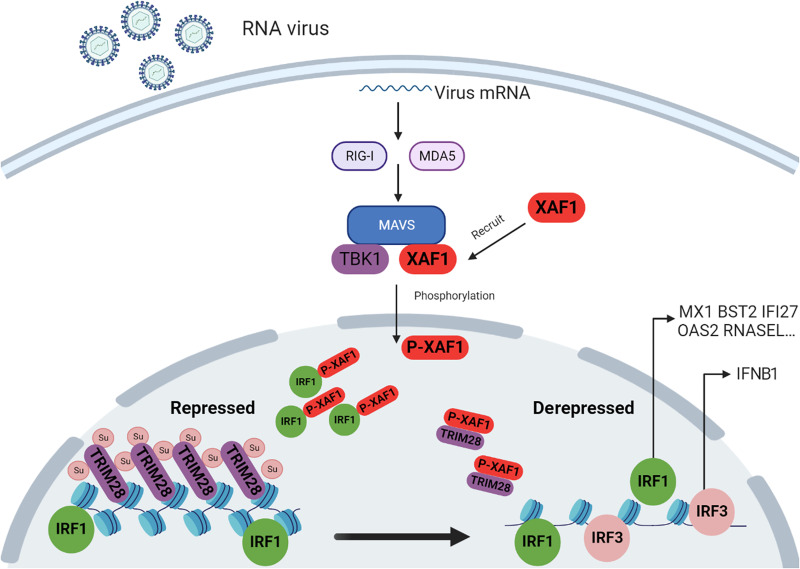
Fig. 8. Model: XAF1 promotes innate immune response against RNA viruses by regulating chromatin accessibility.
Upon RNA virus infection, XAF1 is recruited by MAVS and then phosphorylated by TBK1 at serine-252, which initiates its translocation from the cytoplasm to the nucleus. XAF1 then interacted with IRF1 and specifically liberated the IRF1-targeted chromatin by blocking the SUMOylation of the chromatin repressor TRIM28. This results in the desuppression of chromatin to enhance the transcription of IRF1-regulated innate immune genes to initiate IFN-independent innate immune responses. On the other hand, XAF1 and IRF1 promotes opening of chromatin targeted by IRF3 to initiate an IFN-dependent innate immune response.
Nuclear translocation of XAF1 was observed at very early stage of infection with RNA virus but not DNA virus (Fig. 4C). However, XAF1 entered the nucleus at the late stage during infection with both RNA and DNA viruses (Fig. 4C). Although viral RNA and DNA sensing generally rely on diverse receptors and adaptors, the cross-talk between DNA and RNA sensing is gradually appreciated (46). For example, it has been reported that RIG-I mediated detection of DNA viruses via RNA Pol III (43). Hence, we here speculate that viral RNA transcribed by DNA viruses during replication can activate RNA sensing pathways and promote the translocation of XAF1. As XAF1 is dispensable for antiviral responses against DNA virus, the later nuclear accumulation of XAF1 might not be associated with antiviral immunity directly. Moreover, the differences in early nuclear translocation resulted in differences in the phenotypes against RNA but not DNA viruses in vitro and in vivo (Figs. 1, C and D, and 2B, and fig. S2A). Moreover, knockout of MAVS or TBK1 only abolished the early nuclear translocation of XAF1 from 0 to 3 hours. Nuclear translocation of XAF1 after 3 hours is independent of MAVS or TBK1. Notably, the role of XAF1 translocation to the nucleus independent of MAVS and TBK1 remains unknown and needs to be further explored.
Together, our results provide insights into the communication between IRF1-mediated IFN-independent and IRF3-mediated IFN-dependent antiviral immune responses. XAF1 reprograms specific chromatin regulation and regulates antiviral gene expression during RNA virus infection.
MATERIALS AND METHODS
Cells culture
HT-29, THP-1, HEK293T, HeLa, A549, HepG2, and Vero cells were purchased from American Type Culture Collection (ATCC). 2fTGH-ISRE (human fibrosarcoma cells expressing an ISRE-driven luciferase reporter) was generated by stabilizing the ISRE-luciferase plasmid in 2fTGH. PMs were isolated from the peritoneal cavity of thioglycolate (TG)–elicited mice. BMDMs were isolated from femurs and tibiae of 8- to 12-week-old mice and stimulated with granulocyte-macrophage colony-stimulating factor (GM-CSF) for 7 days. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (HEK293T, HeLa, A549, HepG2, Vero, 2fTGH-ISRE, PMs, and BMDMs) or RPMI 1640 (HT-29, THP-1) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin (100 U/ml).
Reagents and antibodies
Rabbit antibodies against human XAF1 (catalog no. 13805, RRID:AB_2798317), RIG-I (D14G6) (catalog no. 3743, RRID:AB_2269233), MDA5(D74E7) (catalog no. 5321, RRID:AB_10694490), TBK1(D1B4) (catalog no. 3504, RRID:AB_2255663), p-TBK1 Ser172 (catalog no. 5483, RRID:AB_10693472), IKKɛ (catalog no. 3416, RRID:AB_1264180), and IRF3 (D6I4C) (catalog no. 11904, RRID:AB_2722521) were obtained from Cell Signaling Technology. Rabbit antibodies against MAVS (catalog no. CL488-14341, RRID:AB_2883082), IRF1 (catalog no. CL488-11335, RRID:AB_2919026), Lamin A/C (catalog no. 10298-1-AP, RRID:AB_2296961), KAP1 (catalog no. 15202–1-AP, RRID:AB_2209890), SUMO1 (catalog no. 10329-1-AP, RRID:AB_2286872), SUMO2/3 (catalog no. 11251-1-AP, RRID:AB_2198405), LaminB (catalog no. 66095-1-Ig, RRID:AB_11232208), and mouse antibodies against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (catalog no. HRP-60004, RRID:AB_2737588) were purchased from Proteintech. Rabbit antibodies against tubulin (catalog no. AF7011, RRID: AB_2827688) was purchased from Affinity. Mouse antibodies against DDDDK (catalog no. M185–11, RRID: AB_2716804) and Rabbit antibodies against DDDDK (catalog no. PM020, RRID: AB_591224) were purchased from MBL. Rabbit antibodies against Histone H3 (acetyl K27) (catalog no. ab4729, RRID: AB_2118291) was purchased from Abcam. Goat anti-mouse immunoglobulin G (IgG) (H+L) highly cross-adsorbed secondary antibody (catalog no. A32727, RRID: AB_263327) was purchased from Thermo Fisher Scientific. Poly(I:C) HMW (tlrl-pic) and poly(I:C) LMW (tlrl-picw) were purchased from InvivoGen. 3x FLAG peptide (F4799) and agarose type I (A6013-10G) were purchased from Sigma-Aldrich. Pierce Protein A/G Agarose (20422) was purchased from Thermo Fisher Scientific. Recombinant Murine GM-CSF (AF-315-03) was purchased from PeproTech.
Expression plasmid construction
Human cDNAs encoding XAF1, STING, cGAS, TBK1, IRF1, IRF2, IRF3, IRF4, IRF5, IRF6, IRF7, IRF8, IRF9, TBK1 truncation mutants, and XAF1 truncation mutants were cloned into PCMV7.1. Human cDNAs encoding N-RIG-I, MAVS, MAVS-TurboID, RIG-I, and MDA5 were cloned into PCMV14. Human cDNAs encoding XAF1 and TRIM28 truncation mutants were cloned into PLVX. pKH3-TRIM28, porcine cytomegalovirus–VSV glycoprotein (pCMV-VSVG), and psPAX2 were purchased from Addgene.
CRISPR-Cas9 system
MAVS−/−, XAF1−/−, TBK1−/−, IRF1−/−, IRF3−/− XIAP+/−, and TRIM28+/− HT-29 cells as well as XAF1−/− THP-1 cells were constructed by the CRISPR-Cas9 system. Specific guide RNA was ligated into the Bsm BI restriction site of the inducible lentiviral vector (lentiGuide-Puro). Lentivirus particles were produced by cotransfecting HEK293T cells with packaging plasmids psPAX2 (800 ng; Addgene 12260), pCMV-VSVG (800 ng; Addgene 8454), and guide RNA plasmids (2 μg). The medium was changed to fresh DMEM containing 10% FBS at 12 hours after transfection, and viral supernatants were collected at 48 hours. The viral supernatants were used to infect target cells. Infected cells were cultured for 48 hours and then screened by puromycin (2 to 10 μg/ml), and each monoclone was confirmed by sequencing or immunoblot analysis. Cells were negative for mycoplasma. The primers used in this article were listed in the table S4.
Viruses
VSV (Indiana strain) was a gift from J. Rose (Yale University), IAV (PR8 strain) was from F. Qian (Fudan University), HSV-1 (17 strain) was from Z. Jiang (Peking University), and SeV (Cantell strain; VR-907) was purchased from ATCC. The multiplicity of infection (MOI) of the cell culture experiments: 0.1 MOI for VSV, 0.5 MOI for HSV-1, and 0.1 MOI for SeV in all of the figures. The antibodies were diluted 1000 times for immunoblots.
Mice and in vivo virus infection
All animal care and use adhered to the Guide for the Care and Use of Laboratory Animals of the Chinese Association for Laboratory Animal Science. All animal handling procedures were approved by the Animal Care Committee of Peking University Health Science Center (permit number LA 2016240).
Xaf1−/− mice on a C57BL/6J background were purchased from Cyagen Biosciences Inc. WT C57BL/6J mice were purchased from the Department of Laboratory Animal Science of Peking University Health Science Center. All animals were guaranteed adequate clean water and nutritious feed. The primer used is mouse Xaf1 (forward: 5′-GATGGAATGGGTTGGCAGCGTTC-3′; reverse: 5′-CTCCTTGCACACTCATGGGATTG-3′).
Age- and sex-matched C57BL/6 littermates were produced and used in all the in vivo experiments. Eight-week-old mice were infected with HSV-1 at 5 × 106 or VSV at 1 × 108 plaque-forming units (PFU) per mouse by intravenous injection. Eight-week-old mice were intranasally infected with IAV at a dosage of 105 TCID50 (median tissue culture infectious dose). Mice were euthanized 5 to 6 days after infection. Cytokines were detected in the brains and blood collected at days 0 and 1 after infection. Genome copy number was measured in organs harvested 4 to 5 days after infection.
Constructs
Expression constructs generated for this study were prepared by standard molecular biology techniques, and coding sequences were entirely verified. All truncation deletions and mutants were constructed by standard molecular biology techniques. Each truncation, deletion, and mutant were confirmed by sequencing.
Biotin labeling in living cells
Biotin labeling in living cells was conducted as previously described (37). HT-29 cells were plated in 10-cm dishes at 70% confluency and then transfected with MAVS TurboID. After 6 hours of transfection, the culture medium was changed. After 24 hours of transfection, biotin was added at a final concentration of 500 μM for 10 min before harvesting the cells. After gently shaking the cell culture dish for 1 min, the medium was removed, and the cells were washed three times with ice-cold phosphate-buffered saline (PBS). Cells were scraped and transferred to 1.5-ml tubes with ice-cold PBS, spun at 3600 rpm for 5 min, flash-frozen in liquid nitrogen, and stored at −80°C.
Streptavidin magnetic beads enrichment of biotinylated proteins
Cell pellets as described above were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer [0.1% SDS, 0.5% sodium deoxycholate, 150 mM NaCl, 50 mM tris, protease inhibitor cocktail (C0001, TargetMol), 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride (PMSF)] at 4°C for 40 min. The lysates were cleared by centrifugation at 12,000g for 10 min at 4°C. Fifty microliters of each lysate supernatant was reserved for detection of biotinylation activity by Western blotting. Streptavidin magnetic beads were washed twice with RIPA lysis buffer and then mixed with lysate supernatant together with rotation overnight at 4°C. On day 2, the beads were subsequently washed twice with 1 ml of RIPA lysis buffer, once with 1 ml of 1 M KCl, once with 1 ml of 0.1 M Na2CO3, and 1 ml of 2 M urea in 10 mM tris-HCl (pH 8.0). Last, biotinylated proteins were eluted by boiling the beads in 150 μl of elution buffer [55 mM (pH 8.0) tris-HCl, 0.1% SDS, 6.66 mM dithiothreitol (DTT), and 0.66 mM biotin] for 10 min and sent for mass spectrometry.
Luciferase reporter assay
HEK293T cells seeded on 24-well plates were transiently transfected with 50 ng of the pRL-TK reporter (herpes simplex virus thymidine kinase promoter driving renilla luciferase; internal control) and 50 ng of luciferase reporter driven by promoters of genes encoding IFN-β (firefly luciferase; experimental reporter) together with equal amounts of various expression plasmids or empty control plasmids. Then, 24 hours later, without or with virus stimulation, reporter gene activity was analyzed using the Dual-Luciferase Reporter 1000 Assay System (Promega) and measured with a TD-20/20 Luminometer (Turner Designs) according to the manufacturer’s instructions. Data were normalized for transfection efficiency by division of firefly luciferase activity by that of Renilla luciferase.
Type I IFN bioassay
The activity of type I IFN was measured as described (47), with reference to recombinant human IFN-β (R&D Systems) as a standard with 2fTGH cells (for human assays) stably transfected with an IFN-sensitive (ISRE) luciferase construct.
Plaque assay
The viral titer from the cell culture medium was determined by plaque-forming assay as previously described (48). Briefly, virus-containing medium was harvested and serially diluted to infect confluent Vero cells cultured in 24-well plates. After a 1-hour incubation, the supernatants were removed, and the cells were washed twice with PBS. Culture medium containing 2% (w/v) methylcellulose was overlaid for 24 hours with VSV and 48 hours with HSV-1. Then, the overlay was removed, and the cells were fixed with 0.5% (v/v) glutaraldehyde for 30 min and stained with 1% (w/v) crystal violet dissolved in 70% ethanol for 30 min. After washing twice with ddH2O, plaques were counted, and average counts were multiplied by the dilution factor to determine the viral titer as PFU per milliliter (PFU/ml).
Quantitative real-time PCR
Total RNA was isolated from the tissues and cells by TRIzol reagent (TIANGEN Biotech, DP424). Gel electrophoresis and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) were applied to determine the quality and quantity of total RNA. Then, cDNA was conducted from the prepared RNA (600 ng) using the HiScript III First Strand cDNA Synthesis Kit (Vazyme Biotech, R312-02). Quantitative real-time PCR (qRT-PCR) was performed using Applied Biosystems 7500 Real-Time PCR Systems with SYBR qPCR Master mix (Yeasen, catalog no. 11201-11205). The qRT-PCR data were analyzed by the Livak method (2−ΔΔCt). β-Actin was used as a reference gene. The primers for qRT-PCR are listed in table S4.
Whole-genome RNA-seq
Whole-genome RNA-seq was conducted as previously described (49). Total RNA of cells with specific treatment was purified using the RNeasy Mini Kit (QIAGEN, no. 74104). The transcriptome library for sequencing was generated using the VAHTSTM mRNA-seq v2 Library Prep Kit for Illumina (Vazyme Biotech Co. Ltd., Nanjing, China) following the manufacturer’s recommendations. After clustering, the libraries were sequenced on an Illumina HiSeq X Ten platform using the [2 × 150 base pair (bp)] paired-end module. The raw images were transformed into raw reads by base calling using bcl2fastq Conversion Software (CASAVA) (http://pre-support.illumina.com.cn/sequencing/sequencing_software/bcl2fastq-conversion-software.html). Then, raw reads in fastq format were first processed using in-house perl scripts. Clean reads were obtained by removing reads with adapters, reads in which unknown bases were more than 5% and low-quality reads (the percentage of low-quality bases was over 50% in a read, we defined the low-quality base to be the base whose sequencing quality was no more than 10). At the same time, the Q20, Q30, and GC contents of the clean data were calculated (Vazyme Biotech Co., Ltd., Nanjing, China). The original RNA-seq data was uploaded to the Gene Expression Omnibus (GEO) datasets under accession number GES220273 and GSE220274.
coIP assay
HT-29 cells, with or without the indicated virus infection, were lysed in lysis buffer [0.5% Triton X-100, 20 mM Hepes (pH 7.4), 150 mM NaCl, 12.5 mM β-glycerolphosphate, 1.5 mM MgCl2, 2 mM EGTA, 10 mM NaF, 1 mM Na3VO4, and 2 mM DTT] containing protease inhibitors. Lysates were centrifuged and incubated with control IgG or specific antibodies (anti-XAF1, anti-HA, or anti-Flag antibodies) at 4°C overnight. The next day, prewashed protein A/G beads (RIPA) were added and incubated at 4°C for 4 hours. The beads were washed with cold PBS four times and eluted with DTT-containing SDS sample buffer by boiling for 10 min for Western blotting (49).
Yeast two-hybrid experiment
According to protocols recommended by the manufacturer (Clontech), the cDNA fragment of MAVS was cloned into the PACT2 vector. The cDNA fragment of XAF1 was cloned into the pGBKT7 vector. Then, the PACT2 vector with MAVS and pGBKT7-XAF1 were transformed into yeast. Yeasts were grown on SD/–Leu, SD/–Trp, SD/–His/–Trp, and SD/–Ade/–Trp selection media. After mating pGBKT7-XAF1 with PACT2-MAVS, these diploid cells containing both plasmids were cultured on SD/–Leu/–Trp and SD/–Ade/–His/–Leu/–Trp media. Then, the interaction between bait and prey proteins was evaluated.
Nuclear-cytoplasmic fractionation
Cells were washed with ice-cold PBS, lysed in 200 μl of hypotonic buffer [10 mM Hepes (pH 7.4), 10 mM KCl, 1.5 mM MgCl2, and 0.5 mM 2-Mercaptoethanol (ME) supplemented with a protease inhibitor tablet (Roche)], and incubated on ice for 10 min. Then, 10% NP-40 was added until the final concentration was 0.2%. The lysate was incubated on ice for 2 min and immediately centrifuged at 720g for 5 min at 4°C. The pellets were saved as nuclei (Nuc). The supernatants were further centrifuged at 3000g for 10 min to obtain the membrane fraction (Cyt).
Semi-denaturing detergent agarose gel electrophoresis
SDD-AGE was conducted as previously described (37). Cells were lysed with 100 μl of precooled lysis buffer (0.5% Triton X-100, 50 mM tris, 150 mM NaCl, 10% glycerol, and proteinase cocktail and phosphatase cocktail) on ice for 40 min. Supernatants were collected by centrifugation at 13,000 rpm for 10 min at 4°C. Protein-loading buffer (without β-ME) was added to the supernatants and loaded onto a 1.5% agarose gel, vertical agarose electrophoresis was performed for 35 min with a constant voltage of 100 V on ice, and a standard Western blotting procedure was followed.
Native PAGE
Cell pellets as described above were lysed in RIPA lysis buffer [0.1% SDS, 0.5% sodium deoxycholate, 150 mM NaCl, 50 mM tris, protease inhibitor cocktail (C0001, TargetMol), 1% Triton X-100, and 1 mM PMSF] at 4°C for 40 min. The lysates were cleared by centrifugation at 12,000g for 10 min at 4°C. Native sample buffer [62.5 mM tris-Cl (pH 6.8), 15% (v/v) glycerol, and 1% deoxycholate] was added to the supernatants and loaded onto the gel. A 6 to 8% native PAGE gel was used for native protein fractionation. The gel was prerun with native running buffer [25 mM tris and 192 mM glycine (pH 8.4)] with and without 1% (w/v) deoxycholate (Sigma-Aldrich) in the cathode and anode chambers, respectively, for 1 hour at 25 mA.
Mass spectrometry
To identify proteins that may interact with XAF1 and phosphorylation sites of XAF1, samples were sent for mass spectrometry.
ChIP-seq and ChIP-qPCR
ChIP was conducted as previously described (24). Approximately 5 × 107 pretreated cells were crosslinked with 1% formaldehyde at room temperature for 10 min, and the reaction was quenched with 0.125 M glycine for 5 min. The cells were washed twice with PBS, scraped, and pelleted at 2500 rpm for 5 min at 4°C. After lysis and sonication, the majority of the sonicated DNA fragments were sheared to a size of approximately 200 to 600 bp. The sonicated chromatin was spun down at 12,000 rpm for 10 min at 4°C to collect the chromatin. Then, the soluble chromatin was incubated with 3 mg of antibody, and the mixture was rotated at 4°C overnight. After incubation, prewashed Protein G Dynabeads (Invitrogen, 10004d) were added and incubated for 4 hours at 4°C in a rotator. Then, the magnetic Dynabeads were pelleted by placing the tubes in a magnetic rack and washed a total of five times: once with wash buffer A [20 mM tris-HCl (pH 8.0), 500 mM NaCl, 2 mM EDTA, 0.1% SDS, and 1% Triton X-100], once with wash buffer B [10 mM tris-HCl (pH 8.0), 250 mM LiCl, 1 mM EDTA, and 1% NP-40), and three times with wash buffer C [1 mM EDTA and 10 mM tris-HCl (pH 8.0)].
For sequencing, beads were resuspended in 100 ml of elution buffer [50 mM tris-HCl (pH 8.0), 10 mM EDTA, and 1% SDS], followed by incubation at 65°C overnight for reverse crosslinking. DNA was purified from input and Lamin A antibody immunoprecipitation samples from WT and XAF1−/− HT-29 cells treated with or without VSV for 12 hours with the QIAEX II Gel Extraction Kit. The extracted DNA was used for sequencing.
Reads from ChIP-seq experiments were aligned to the human genome GRCh38 using annotated chromosomes and scaffolds. Alignment was performed using bowtie 2. Regions with exceptionally high coverage of ChIP-seq reads were identified using MACS2. Wiggle files representing counts of ChIP-seq reads across the reference genome were created using MACS2. The resulting wiggle files were normalized for sequencing depth by dividing the read counts in each bin by the millions of mapped reads in each sample and were visualized in the UCSC genome browser (The human genome browser at UCSC, 2002) or WashU genome browser (The human epigenome browser at Washington University, 2011) (50). The original ChIP-seq data were uploaded to the GEO datasets under accession numbers GSE220272 and GSE220274.
For ChIP-qPCR, the library construction is the same as the method in ChIP-seq. Then, the libraries were adapted for qRT-PCR with specific primers. The primers used in this article were listed in the table S4.
ATAC-seq
ATAC-seq was conducted as previously described (25). We pellet 50,000 viable sample cells at 500 Relative Centrifugal Force (RCF) at 4°C for 5 min, aspirate all supernatant, and add 50 μl of cold ATAC–resuspension buffer (RSB) containing 0.1% NP40, 0.1% Tween 20, and 0.01% digitonin into the cell pellet and pipette up and down three times and incubate on ice for 3 min. We wash out lysis with 1 ml of cold ATAC-RSB containing 0.1% Tween 20 but no NP-40 or digitonin and invert tube three times to mix. We pellet nuclei at 500 RCF for 10 min at 4°C, aspirate all supernatant, resuspend cell pellet in 50 μl of transposition mixture [25 μl of 2× TD buffer, 2.5 μl of transposase (100 nM final), 16.5 μl of PBS, 0.5 μl of 1% digitonin, 0.5 μl of 10% Tween 20, and 5 μl of H2O] by pipetting up and down six times, and incubate reaction at 37°C for 30 min. Afterward, the DNA was purified with Magen DNA purify kit and amplified with primers containing barcodes by using the TruePrep DNA Library Prep Kit (TD501-01). Subsequent sequencing and data analysis were handed over to GENEWIZ Biotechnology Co., LTD (Suzhou, China). Briefly, all libraries were adapted for high-throughput sequencing (75-bp paired-end) on an Illumina NextSeq 500. Raw sequencing data are collected. After filtering data through sequencing data quality assessment, clean reads are further obtained. After removing adapter sequences and low-quality reads, high-quality reads are processed for further analysis. The peak calling reads are mapped to the human genome and accessible chromatin regions, such as promoters, 3′ untranslated region (3′UTR) and 5′UTR). The data were first merged by the “bedtools merge” tool (https://bedtools.readthedocs.io/en/latest/content/tools/merge.html). The original ATAC-seq data was uploaded to the GEO datasets under accession number GES225587.
Quantification and statistical analysis
The Mantel-Cox (log rank) test was used to determine statistical difference of survival curves. Other results were expressed as the means ± SEM. Data were analyzed by t test with Prism 7 software (GraphPad Software, La Jolla, CA). For the Western blotting analysis, ImageJ software was applied to quantify abundance. All the bar graphs were visualized by Prism 7. Differences in means were considered statistically significant at P < 0.05. Significance levels are *P < 0.05, **P < 0.01, and ***P < 0.001; NS, not significant.
Acknowledgments
We thank Z. Jiang for providing HT-29 cells, J. Rose for VSV (Indiana strain), F. Qiang for IAV (PR8), and A. Iwasaki for HSV-1 (F strain).
Funding: This work was supported by Beijing Natural Science Foundation (Z210014), the National Key Research and Development Program of China (2021YFC2302602, 2020YFA0707800), the National Natural Science Foundation of China (31570891, 31872736, 32022028, 81991505, and 82201928), Peking University Clinical + X (PKU2020LCXQ009), the Peking University Medicine Fund (PKU2020LCXQ009), the Zhuhai Science and Technology Innovation Bureau (ZH22036302200063PWC to Z.Y.), China Postdoctoral Science Foundation (2022 M710265 to H.Y.), a grant from Tianjin Natural Science Foundation of China (no. 21JCQNJC01870 to D.W.), and Incubation fund of Tianjin Third Central Hospital (no. 2019YNR6 to D.W.).
Author contributions: Conceptualization: M.K., F.W., and Y.W. Methodology: M.K., Y.Z., H.Y., S.L., and Y.L. Investigation: M.K., Y.Z., H.Y., T.L., L.C., J.C., Y.L., X.W., Z.Z., and D.W. Visualization: M.K. and X.G. Supervision: F.Y. Writing—original draft: M.K. and F.Y. Writing—review and editing: M.K. and F.Y.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq, ChIP-seq, and ATAC-seq data have been deposited at GEO under accession numbers GSE220272, GES220273, GSE220274, and GES225587. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository with the dataset identifier IPX0005537000/PXD038540.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for tables S1 to S4
Other Supplementary Material for this manuscript includes the following:
Table S1 to S4
REFERENCES AND NOTES
- 1.F. McNab, K. Mayer-Barber, A. Sher, A. Wack, A. O'Garra, Type I interferons in infectious disease. Nat. Rev. Immunol. 15, 87–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.N. Yan, Z. J. Chen, Intrinsic antiviral immunity. Nat. Immunol. 13, 214–222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. J. Desmet, K. J. Ishii, Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 12, 479–491 (2012). [DOI] [PubMed] [Google Scholar]
- 4.T. Kawai, S. Akira, The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010). [DOI] [PubMed] [Google Scholar]
- 5.J. Rehwinkel, M. U. Gack, RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 20, 537–551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.L. Sun, J. Wu, F. Du, X. Chen, Z. J. Chen, Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.X. Cao, Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 16, 35–50 (2016). [DOI] [PubMed] [Google Scholar]
- 8.J. Wu, Z. J. Chen, Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32, 461–488 (2014). [DOI] [PubMed] [Google Scholar]
- 9.O. Takeuchi, S. Akira, Innate immunity to virus infection. Immunol. Rev. 227, 75–86 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. Mukhopadhyay, S. Gordon, The role of scavenger receptors in pathogen recognition and innate immunity. Immunobiology 209, 39–49 (2004). [DOI] [PubMed] [Google Scholar]
- 11.C. Gurtler, A. G. Bowie, Innate immune detection of microbial nucleic acids. Trends Microbiol. 21, 413–420 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J. Hiscott, Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 18, 483–490 (2007). [DOI] [PubMed] [Google Scholar]
- 13.R. Medzhitov, Recognition of microorganisms and activation of the immune response. Nature 449, 819–826 (2007). [DOI] [PubMed] [Google Scholar]
- 14.R. A. Porritt, P. J. Hertzog, Dynamic control of type I IFN signalling by an integrated network of negative regulators. Trends Immunol. 36, 150–160 (2015). [DOI] [PubMed] [Google Scholar]
- 15.S. E. Collins, R. S. Noyce, K. L. Mossman, Innate cellular response to virus particle entry requires IRF3 but not virus replication. J. Virol. 78, 1706–1717 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.E. Dixit, S. Boulant, Y. Zhang, A. S. Y. Lee, C. Odendall, B. Shum, N. Hacohen, Z. J. Chen, S. P. Whelan, M. Fransen, M. L. Nibert, G. Superti-Furga, J. C. Kagan, Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.P. Benech, M. Vigneron, D. Peretz, M. Revel, J. Chebath, Interferon-responsive regulatory elements in the promoter of the human 2′,5′-oligo(A) synthetase gene. Mol. Cell. Biol. 7, 4498–4504 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. Kirchhoff, A. E. Koromilas, F. Schaper, M. Grashoff, N. Sonenberg, H. Hauser, IRF-1 induced cell growth inhibition and interferon induction requires the activity of the protein kinase PKR. Oncogene 11, 439–445 (1995). [PubMed] [Google Scholar]
- 19.J. M. Penninger, C. Sirard, H. W. Mittrücker, A. Chidgey, I. Kozieradzki, M. Nghiem, A. Hakem, T. Kimura, E. Timms, R. Boyd, T. Taniguchi, T. Matsuyama, T. W. Mak, The interferon regulatory transcription factor IRF-1 controls positive and negative selection of CD8+ thymocytes. Immunity 7, 243–254 (1997). [DOI] [PubMed] [Google Scholar]
- 20.S. Taki, T. Sato, K. Ogasawara, T. Fukuda, M. Sato, S. Hida, G. Suzuki, M. Mitsuyama, E. H. Shin, S. Kojima, T. Taniguchi, Y. Asano, Multistage regulation of Th1-type immune responses by the transcription factor IRF-1. Immunity 6, 673–679 (1997). [DOI] [PubMed] [Google Scholar]
- 21.L. Gabriele, A. Fragale, P. Borghi, P. Sestili, E. Stellacci, M. Venditti, G. Schiavoni, M. Sanchez, F. Belardelli, A. Battistini, IRF-1 deficiency skews the differentiation of dendritic cells toward plasmacytoid and tolerogenic features. J. Leukoc. Biol. 80, 1500–1511 (2006). [DOI] [PubMed] [Google Scholar]
- 22.A. Kroger, IRFs as competing pioneers in T-cell differentiation. Cell. Mol. Immunol. 14, 649–651 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R. Song, Y. Gao, I. Dozmorov, V. Malladi, I. Saha, M. M. McDaniel, S. Parameswaran, C. Liang, C. Arana, B. Zhang, B. Wakeland, J. Zhou, M. T. Weirauch, L. C. Kottyan, E. K. Wakeland, C. Pasare, IRF1 governs the differential interferon-stimulated gene responses in human monocytes and macrophages by regulating chromatin accessibility. Cell Rep. 34, 108891 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.L. Cao, S. Liu, Y. Li, G. Yang, Y. Luo, S. Li, H. du, Y. Zhao, D. Wang, J. Chen, Z. Zhang, M. Li, S. Ouyang, X. Gao, Y. Sun, Z. Wang, L. Yang, R. Lin, P. Wang, F. You, The nuclear matrix protein safa surveils viral rna and facilitates immunity by activating antiviral enhancers and super-enhancers. Cell Host Microbe 26, 369–384.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 25.L. Cao, Y. Luo, X. Guo, S. Liu, S. Li, J. Li, Z. Zhang, Y. Zhao, Q. Zhang, F. Gao, X. Ji, X. Gao, Y. Li, F. You, SAFA facilitates chromatin opening of immune genes through interacting with anti-viral host RNAs. PLOS Pathog. 18, e1010599 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.B. G. Hale, Antiviral immunity triggered by infection-induced host transposable elements. Curr. Opin. Virol. 52, 211–216 (2022). [DOI] [PubMed] [Google Scholar]
- 27.N. Schmidt, P. Domingues, F. Golebiowski, C. Patzina, M. H. Tatham, R. T. Hay, B. G. Hale, An influenza virus-triggered SUMO switch orchestrates co-opted endogenous retroviruses to stimulate host antiviral immunity. Proc. Natl. Acad. Sci. U.S.A. 116, 17399–17408 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.M. G. Macchietto, R. A. Langlois, S. S. Shen, Virus-induced transposable element expression up-regulation in human and mouse host cells. Life Sci. Alliance 3, e201900536 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.P. Liston, W. G. Fong, N. L. Kelly, S. Toji, T. Miyazaki, D. Conte, K. Tamai, C. G. Craig, M. W. McBurney, R. G. Korneluk, Identification of XAF1 as an antagonist of XIAP anti-Caspase activity. Nat. Cell Biol. 3, 128–133 (2001). [DOI] [PubMed] [Google Scholar]
- 30.D. S. Byun, K. Cho, B. K. Ryu, M. G. Lee, M. J. Kang, H. R. Kim, S. G. Chi, Hypermethylation of XIAP-associated factor 1, a putative tumor suppressor gene from the 17p13.2 locus, in human gastric adenocarcinomas. Cancer Res. 63, 7068–7075 (2003). [PubMed] [Google Scholar]
- 31.M. G. Lee, J. S. Huh, S. K. Chung, J. H. Lee, D. S. Byun, B. K. Ryu, M. J. Kang, K. S. Chae, S. J. Lee, C. H. Lee, J. I. Kim, S. G. Chang, S. G. Chi, Promoter CpG hypermethylation and downregulation of XAF1 expression in human urogenital malignancies: Implication for attenuated p53 response to apoptotic stresses. Oncogene 25, 5807–5822 (2006). [DOI] [PubMed] [Google Scholar]
- 32.S.-K. Chung, M.-G. Lee, B.-K. Ryu, J.-H. Lee, J. Han, D.-S. Byun, K.-S. Chae, K. Y. Lee, J.-Y. Jang, H.-J. Kim, S.-G. Chi, Frequent alteration of XAF1 in human colorectal cancers: Implication for tumor cell resistance to apoptotic stresses. Gastroenterology 132, 2459–2477 (2007). [DOI] [PubMed] [Google Scholar]
- 33.R. F. Hennigan, J. S. Fletcher, S. Guard, N. Ratner, Proximity biotinylation identifies a set of conformation-specific interactions between Merlin and cell junction proteins. Sci. Signal. 12, eaau8749 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Q. Liu, J. Zheng, W. Sun, Y. Huo, L. Zhang, P. Hao, H. Wang, M. Zhuang, A proximity-tagging system to identify membrane protein-protein interactions. Nat. Methods 15, 715–722 (2018). [DOI] [PubMed] [Google Scholar]
- 35.W. G. Fong, P. Liston, E. Rajcan-Separovic, M. St. Jean, C. Craig, R. G. Korneluk, Expression and genetic analysis of XIAP-associated factor 1 (XAF1) in cancer cell lines. Genomics 70, 113–122 (2000). [DOI] [PubMed] [Google Scholar]
- 36.T. L. Ma, P. H. Ni, J. Zhong, J. H. Tan, M. M. Qiao, S. H. Jiang, Low expression of XIAP-associated factor 1 in human colorectal cancers. Chin. J. Dig. Dis. 6, 10–14 (2005). [DOI] [PubMed] [Google Scholar]
- 37.S. Li, M. Kuang, L. Chen, Y. Li, S. Liu, H. du, L. Cao, F. You, The mitochondrial protein ERAL1 suppresses RNA virus infection by facilitating RIG-I-like receptor signaling. Cell Rep. 34, 108631 (2021). [DOI] [PubMed] [Google Scholar]
- 38.D. W. Leaman, M. Chawla-Sarkar, K. Vyas, M. Reheman, K. Tamai, S. Toji, E. C. Borden, Identification of X-linked inhibitor of apoptosis-associated factor-1 as an interferon-stimulated gene that augments TRAIL Apo2L-induced apoptosis. J. Biol. Chem. 277, 28504–28511 (2002). [DOI] [PubMed] [Google Scholar]
- 39.S. I. Jeong, J. W. Kim, K. P. Ko, B. K. Ryu, M. G. Lee, H. J. Kim, S. G. Chi, XAF1 forms a positive feedback loop with IRF-1 to drive apoptotic stress response and suppress tumorigenesis. Cell Death Dis. 9, 806 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.C. H. Tie, L. Fernandes, L. Conde, L. Robbez-Masson, R. P. Sumner, T. Peacock, M. T. Rodriguez-Plata, G. Mickute, R. Gifford, G. J. Towers, J. Herrero, H. M. Rowe, KAP1 regulates endogenous retroviruses in adult human cells and contributes to innate immune control. EMBO Rep. 19, e45000 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.K. B. Chiappinelli, P. L. Strissel, A. Desrichard, H. Li, C. Henke, B. Akman, A. Hein, N. S. Rote, L. M. Cope, A. Snyder, V. Makarov, S. Buhu, D. J. Slamon, J. D. Wolchok, D. M. Pardoll, M. W. Beckmann, C. A. Zahnow, T. Merghoub, T. A. Chan, S. B. Baylin, R. Strick, Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R. S. Nozawa, L. Boteva, D. C. Soares, C. Naughton, A. R. Dun, A. Buckle, B. Ramsahoye, P. C. Bruton, R. S. Saleeb, M. Arnedo, B. Hill, R. R. Duncan, S. K. Maciver, N. Gilbert, SAF-A regulates interphase chromosome structure through oligomerization with chromatin-associated RNAs. Cell 169, 1214–1227.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.H. Zhou, Y. D. Tang, C. Zheng, Revisiting IRF1-mediated antiviral innate immunity. Cytokine Growth Factor Rev. 64, 1–6 (2022). [DOI] [PubMed] [Google Scholar]
- 44.R. Qi, J. Gu, Z. Zhang, K. Yang, B. Li, J. Fan, C. Wang, Z. He, L. Qiao, Z. Lin, X. Y. Liu, Potent antitumor efficacy of XAF1 delivered by conditionally replicative adenovirus vector via caspase-independent apoptosis. Cancer Gene Ther. 14, 82–90 (2007). [DOI] [PubMed] [Google Scholar]
- 45.L. M. Zhu, D. M. Shi, Q. Dai, X. J. Cheng, W. Y. Yao, P. H. Sun, Y. F. Ding, M. M. Qiao, Y. L. Wu, S. H. Jiang, S. P. Tu, Tumor suppressor XAF1 induces apoptosis, inhibits angiogenesis and inhibits tumor growth in hepatocellular carcinoma. Oncotarget 5, 5403–5415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.C. Cai, Y. D. Tang, G. Xu, C. Zheng, The crosstalk between viral RNA- and DNA-sensing mechanisms. Cell. Mol. Life Sci. 78, 7427–7434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Z. Jiang, P. Georgel, X. du, L. Shamel, S. Sovath, S. Mudd, M. Huber, C. Kalis, S. Keck, C. Galanos, M. Freudenberg, B. Beutler, CD14 is required for MyD88-independent LPS signaling. Nat. Immunol. 6, 565–570 (2005). [DOI] [PubMed] [Google Scholar]
- 48.F. You, P. Wang, L. Yang, G. Yang, Y. O. Zhao, F. Qian, W. Walker, R. Sutton, R. Montgomery, R. Lin, A. Iwasaki, E. Fikrig, ELF4 is critical for induction of type I interferon and the host antiviral response. Nat. Immunol. 14, 1237–1246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. Chen, X. Wei, X. Wang, T. Liu, Y. Zhao, L. Chen, Y. Luo, H. du, Y. Li, T. Liu, L. Cao, Z. Zhou, Z. Zhang, L. Liang, L. Li, X. Yan, X. Zhang, X. Deng, G. Yang, P. Yin, J. Hao, Z. Yin, F. You, TBK1-METTL3 axis facilitates antiviral immunity. Cell Rep. 38, 110373 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Y. Zhao, W. Shu, X. Liu, X. Jiao, J. Li, T. Liu, M. Kuang, R. Li, H. Du, Z. Zhang, Z. Zhou, X. Guo, X. Wei, D. Wang, T. Liu, Y. Li, S. Liu, J. Chen, Y. Luo, H. Xia, L. Cao, W. Tian, X. Sun, W. Dong, G. Zhou, X. Gao, Q. Zhou, F. You, LMNA R527C mutation causes an inflammation driven progeria via triggering DNA-sensing pathways. Res. Square 10.21203/rs.3.rs-1406652/v1, (2022). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for tables S1 to S4
Table S1 to S4