Abstract
Ubiquitin-fold modifier 1 (UFM1) is a ubiquitin-like protein covalently conjugated with intracellular proteins through ufmylation, similar to ubiquitylation. Ufmylation is involved in processes such as endoplasmic reticulum (ER)–associated protein degradation, ribosome-associated protein quality control (RQC) at the ER (ER-RQC), and ER-phagy. However, it remains unclear how ufmylation regulates such distinct ER-related functions. Here, we provide insights into the mechanism of the UFM1 E3 complex in not only ufmylation but also ER-RQC. The E3 complex consisting of UFL1 and UFBP1 interacted with UFC1, UFM1 E2, and, subsequently, CDK5RAP3, an adaptor for ufmylation of ribosomal subunit RPL26. Upon disome formation, the E3 complex associated with ufmylated RPL26 on the 60S subunit through the UFM1-interacting region of UFBP1. Loss of E3 components or disruption of the interaction between UFBP1 and ufmylated RPL26 attenuated ER-RQC. These results provide insights into not only the molecular basis of the ufmylation but also its role in proteostasis.
Structure prediction using AlphaFold2 reveals operating principles of ufmylation and a role of UFM1 E3 complex in ER-RQC.
INTRODUCTION
Posttranslational modifications (PTMs) amplify limited genomic information and extend the functions of single proteins to enable their participation in diverse cellular processes. The best characterized PTM is protein modification by ubiquitin or ubiquitin-like modifiers (UBLs) (1–4). There are numerous modification processes in which single or multiple ubiquitins or UBLs bind to proteins, and these act as signal converters that regulate various cellular functions. Protein modifications by ubiquitin and UBLs are catalyzed by a highly regulated and elaborate cellular system and are carried out by sequential reactions of multiple enzymes comprising an activating enzyme (E1), a conjugating enzyme (E2), and a ligase enzyme (E3). Initially, the specific E1 for each ubiquitin or UBL forms a high-energy thioester bond with the ubiquitin or UBL via adenylation in an adenosine triphosphate–dependent manner. The ubiquitin or UBL activated by an E1 is then transferred to a specific E2 via thioester bonds. In some cases, E2s can directly transfer ubiquitins or UBLs to substrate proteins in an isopeptide linkage. However, an E2 generally requires the participation of an E3 to achieve substrate-specific ubiquitylation or UBL conjugation in cells. An E3, which contains a core domain such as RING, HECT, or U-box domain, is defined as the enzyme required to recognize the target proteins, and it plays a central role in ubiquitination or UBL conjugation (5). While there are only 2 E1s and approximately 30 E2s in humans, the number of E3s is estimated to exceed 600 for ubiquitination. Ubiquitination and most UBL conjugation reactions are reversible, with deubiquitinating enzymes removing ubiquitin and UBLs from target proteins and regulating ubiquitination and UBLs binding. Covalent modification of intracellular proteins by ubiquitin mediates selective protein degradation, mainly by the 26S proteasome and autophagy, while modification of intracellular proteins by UBLs regulates protein localization, protein-protein and protein-DNA interactions, and biochemical activity.
Ubiquitin-fold modifier 1 (UFM1) is a UBL covalently bound to intracellular proteins by ufmylation, a reaction similar to ubiquitination (6, 7). UFM1 is synthesized as a pro-form, and the two amino acids at the C terminus of pro-UFM1 are cleaved by specific proteases, UFSP1 and UFSP2 (8, 9). The result is a mature form in which the glycine residue essential for conjugation is exposed. The mature form of UFM1 is activated by forming a high-energy thioester bond with UBA5, a UFM1-specific E1 enzyme. Activated UFM1 is transferred to the UFM1-specific E2 enzyme, UFC1, which is covalently bound to the target protein via a UFM1-specific E3 enzyme complex comprising UFL1, UFBP1 (also called DDRGK1 or C20orf116), and CDK5RAP3 (10–13). Of these E3 components, UFL1 and UFBP1 are essential for ufmylation (14, 15), while CDK5RAP3 may be involved in the regulation of E3 activities such as polyufmylation and direction of ufmylation on the ribosomal protein RPL26 (13). This reaction is reversible, as covalently bound UFM1 is cleaved by the cysteine protease UFSP2 (8, 16). In mice, disruption of the UFM1 system results in abnormalities in erythroid differentiation (11, 12, 17, 18), liver development (12), and neurogenesis (19, 20). Genetic mutations in human UFM1, UBA5, and UFC1 result in reduced ufmylation and hereditary paediatric encephalopathy (19, 21–23). Mutations in UFSP2 also cause the autosomal dominant disorders Beukes hip dysplasia (24) and vertebral epiphyseal dysplasia (25).
UFBP1 has a signal peptide at its N terminus that anchors it to the endoplasmic reticulum (ER) and a transmembrane helix, localized on the ER (10, 26, 27), and UFL1 forms a stable complex with UFBP1 (13, 28), suggesting that the UFM1 system may play roles involving the ER. The UFM1 system has long been associated with ER stress and ER-associated degradation (ERAD) (7). Further, growing lines of evidence indicate that ufmylation plays important roles in ribosome-associated protein quality control at the ER (ER-RQC) and/or nascent peptide quality control on the ER (29–31), as well as in selective autophagy of the ER (ER-phagy) (27, 32, 33). The protein quality control at ER seems to be mediated by the ufmylation of ribosomal proteins, such as RPL26 (29, 30), and the ER-phagy by the ufmylation of the UFM1 E3 component, UFBP1, and an ER-anchoring protein, CYB5R3 (10, 27, 34). However, besides the x-ray crystal structures of UFM1 (35), UBA5 (36), UFC1 (37), and their complex (38, 39), the structure of the UFM1 E3 complex is unknown, and the UFM1 E3 components have no similarity to any previously identified E3 of ubiquitin or UBLs, making it difficult to infer the structure. Therefore, the molecular mechanisms through which the UFM1 E3 complex regulates diverse functions on the ER, such as ER-RQC and ER-phagy, are largely unknown. While it has been suggested that ER-RQC products might undergo non-ERAD non-proteasomal degradation (31), a recent study proposed that ufmylation is required for the LTN1-mediated proteasomal degradation of ER-RQC substrates and not cytosolic substrates (40). Hence, the precise orchestration of ER-RQC substrates by the UFM1 system remains largely elusive.
Here, we provide mechanistic insights into the role of the UFM1 E3 ligase complex in ufmylation and its association with the 60S ribosomal subunit through biochemical analyses and structure prediction using AlphaFold2 (AF2) (41, 42) with the AlphaFold-Multimer mode (43). We found that while UFL1 and UFBP1 are the main constituents of the UFM1 E3-ligase, CDK5RAP3, an accessory component of the E3 complex, alters the specificity of the E3 complex from UFBP1 to RPL26, thereby switching from ER-phagy to ER-RQC. Further, we showed that upon treatment with anisomycin, an inducer of disome formation, the UFM1 E3 complex binds to ufmylated RPL26 on the 60S ribosomal subunit through the UFM1-interacting region of UFBP1, which is indispensable for ER-RQC. Our findings indicate that the UFM1 E3 complex is a ligase and also acts as an essential factor for ER-RQC.
RESULTS
Prediction of the UFL1-UFBP1-CDK5RAP3 E3 complex structure
Highly accurate protein structure prediction by AF2 is radically changing life science research. Although the structure of the three components constructing the E3 ligase for ufmylation (UFL1, UFBP1, and CDK5RAP3) has never been determined experimentally, we can download their predicted structure from the AlphaFold Protein Structure Database (fig. S1) (44). UFL1 is composed of two N-terminal α helices, five tandem winged-helix (WH) domains, and a C-terminal helical domain that contains an intrinsically disordered region (IDR), whereas UFBP1 is composed of one N-terminal transmembrane helix, an IDR containing a coiled-coil, one WH domain, and two C-terminal α helices. CDK5RAP3 is mainly composed of α helices. Two α helices (residues 109 to 168 and 427 to 493) form an antiparallel coiled-coil with each other, which is as long as ~10 nm, and two globular domains are located at both ends of the coiled-coil, creating a dumbbell-like shape. One globular domain is composed of ~120 N-terminal residues and ~30 C-terminal residues and, thus, is named the terminal dumbbell (T-dumbbell), whereas another globular domain is named the central dumbbell (C-dumbbell). The C-dumbbell has an IDR composed of 80 residues.
Although AF2 was initially not good at predicting the structures of heterocomplexes, the recent implementation of the AlphaFold-Multimer mode in AF2 (43) now enables us to predict their structure with much greater accuracy. Using AF2 with the AlphaFold-Multimer mode (all AF2 predictions mentioned henceforth were performed using this mode), we predicted the structure of the UFL1-UFBP1-CDK5RAP3 E3 complex. It has been reported that UFL1 interacts with the proteasome-COP9-initiation factor domain–containing region of UFBP1 (229 to 273 amino acids), while UFC1 and UFBP1 interact with the N-terminal region of UFL1 (1 to 212 amino acids) (10, 33, 45). Therefore, we initially used truncated forms of UFL1 (residues 1 to 302) and UFBP1 (residues 209 to 314) and obtained a well-converged predicted model of the UFL1(1-302)-UFBP1(209-314)-CDK5RAP3 complex (fig. S2A). After that, we superimposed the full-length AF2 structures of UFL1 and UFBP1 on each truncated form and obtained the full-length UFL1-UFBP1-CDK5RAP3 E3 complex model (Fig. 1A). Upon complex formation, the second N-terminal α helix of UFL1 and the two C-terminal α helices of UFBP1 fold into one WH domain, thus forming a complex with seven consecutive WH domains. The UFBP1 WH domain is named WH1, the five WH domains of UFL1 are named WH3 to WH7 (counted from the N terminus), and the WH domain formed upon complex formation is named WH2. These predictions are consistent with the structure of the UFL1-UFBP1 complex recently predicted using AF2 (13, 28). In the predicted ternary complex, CDK5RAP3 interacts with the six tandem WH domains (WH1 to WH6) of the UFL1-UFBP1 complex and the N-terminal helix of UFL1 so that the long axis of CDK5RAP3 and the WH1-WH6 portion of the UFL1-UFBP1 complex become approximately parallel (Fig. 1A).
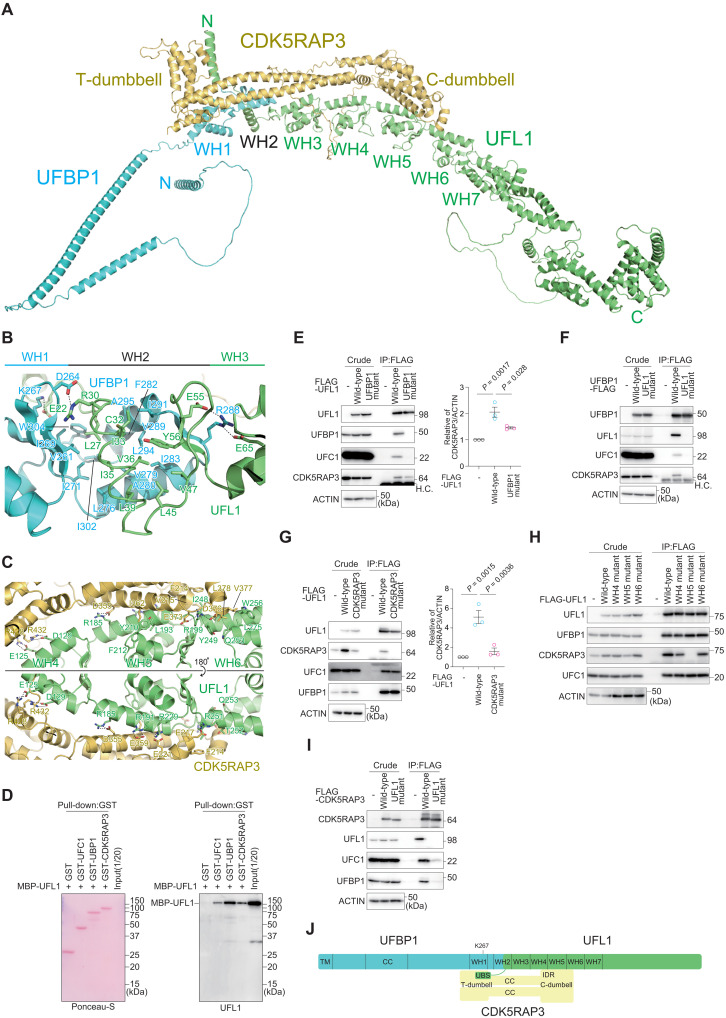
Fig. 1. Structure and interaction modes of the UFM1 E3 complex.
(A) Predicted three-dimensional structure of the full-length UFL1-UFBP1-CDK5RAP3 E3 complex. (B and C) Predicted recognition mode of UFL1 by UFBP1 (B) and by CDK5RAP3 (C). The side chains involved in the interactions are shown with a stick model, where oxygen and nitrogen atoms are colored red and blue, respectively. Broken lines indicate possible salt bridges. (D) Direct interaction of UFL1 with UFC1, UFBP1, and CDK5RAP3. Glutathione S-transferase (GST), GST-UFC1, GST-UFBP1, or GST-CDK5RAP3 was immobilized on glutathione Sepharose column. The Sepharose and recombinant MBP-UFL1 were incubated, and the pulled-down products were then subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting with anti-UFL1 antibody. Data shown are representative of three separate experiments. (E to I) Biochemical characterization of the binding among the E3 components. (E) FLAG-tagged wild-type UFL1 and UFBP1 interaction–defective UFL1 mutant (UFL1UFBP1 mutant) were transfected into UFL1-deficient HEK293T cells. (F) FLAG-tagged wild-type UFBP1 and UFL1 interaction–defective UFBP1 (UFBP1UFL1 mutant) were transfected into UFBP1-deficient HEK293T cells. H.C. indicates IgG heavy chain. (G) FLAG-tagged wild-type UFL1 and CDK5RAP3 interaction–defective UFL1 mutant (UFL1CDK5RAP3 mutant) were transfected into UFL1-deficient HEK293T cells. (H) FLAG-tagged wild-type UFL1 and UFL1 mutants harboring different WH domains (UFL1WH4, UFL1WH5, and UFL1WH6 mutant) were transfected into UFL1-deficient HEK293T cells. (I) FLAG-tagged wild-type CDK5RAP3 and UFL1-interaction defective CDK5RAP3 (CDK5RAP3UFL1 mutant) were transfected into CDK5RAP3-deficient HEK293T cells. Forty-eight hours after transfection, cells were lysed and immunoprecipitated with anti–FLAG-M2 gel; then, the immunoprecipitants were subjected to immunoblot analysis with the indicated antibodies. Bar graph shows the results of quantitative densitometric analysis of CDK5RAP3 relative to actin (n = 3). Data are means ± SE. Statistical analysis was performed by Welch’s t test. Data shown are representative of three separate experiments. (J) Schematic binding model of the UFM1 E3 complex. TM, transmembrane helix; CC, coiled-coil; WH, winged helix; UBS, UFC1-binding sequence.
Interactions leading to construction of the UFM1 E3 complex
As described above, UFL1-UFBP1 interaction results in creation of the WH2 domain, which is composed of three α helices (one from UFL1 and two from UFBP1) and a three-stranded β sheet (two strands from UFL1 and one from UFBP1). In detail, Cys32, Ile33, Val36, Leu39, Leu45, Val47, and Tyr56 of UFL1 form hydrophobic interactions with Leu276, Val279, Ala280, Ile283, Ile291, Leu294, and Ala295 of UFBP1, and these interactions form the hydrophobic core of the WH2 domain (Fig. 1B). The WH2 domain closely interacts with the WH1 domain via hydrophobic interactions formed between Leu27 and Ile35 of UFL1 and Val261, Ile269, Ile271, Ile302, and Trp304 of UFBP1. In addition to hydrophobic interactions, four possible salt-bridge pairs are observed between UFL1 (Glu22, Arg30, Glu55, and Glu65) and UFBP1 (Asp264, Lys267, and Arg288). Besides these, dozens of hydrogen bonds are formed between UFL1 and UFBP1. These hydrophobic and hydrophilic interactions bury a 1350-Å2 surface area of each protein.
Both the C- and T-dumbbells of CDK5RAP3 interact with the UFL1-UFBP1 complex. The C-dumbbell binds to the WH4, WH5, and WH6 domains of UFL1 (Fig. 1C). Eight possible salt-bridge pairs are observed between UFL1 (Glu125 and Asp129 from WH4; Arg185, Arg191, Arg199, and Arg229 from WH5; and Arg251 from WH6) and CDK5RAP3 (Glu217, Glu221, Asp355, Glu359, Glu373, Asp376, Arg428, and Arg432). Besides these, dozens of hydrogen bonds are formed between UFL1 and the C-dumbbell. In addition, several hydrophobic interactions are observed, including those between Ile248, Tyr249, Thr252, Gln253, Trp256, and Leu275 of the WH6 domain and Phe214, Val215, Val377, and Leu378 of CDK5RAP3 and between Leu193, Tyr210, and Phe212 of WH5 and Ile362 of CDK5RAP3. These interactions bury a 1780-Å2 surface area of each protein. In addition to the C-dumbbell–mediated interactions, CDK5RAP3 uses the T-dumbbell to bind to the N-terminal helix of UFL1 and the WH1 domain of UFBP1, mainly through hydrophilic interactions, which buries 630- and 490-Å2 surface areas, respectively (fig. S2B). These observations suggest that CDK5RAP3 binds to the UFL1-UFBP1 complex mainly via recognition of UFL1 WH4 to WH6 by the C-dumbbell.
A central role of UFL1 in E3 complex formation
To first establish whether UFL1 binding to UFBP1, CDK5RAP3, or UFC1 is direct, we purified recombinant UFL1, UFBP1, CDK5RAP3, and UFC1 from Escherichia coli and confirmed the direct interaction between UFL1 and each protein in in vitro pull-down assays (Fig. 1D). Next, to confirm the recognition mode biochemically, we constructed UFL1 and UFBP1 mutants in which amino acids involved in the hydrophobic interactions were replaced with Ala (UFL1L27A C32A I35A L39A, UFL1UFBP1 mutant and UFBP1I271A L276A I302A W304A, UFBP1UFL1 mutant). To exclude the effect of endogenous UFL1, we generated UFL1-deficient human embryonic kidney (HEK) 293T cells (fig. S3A). FLAG-tagged wild-type UFL1 and UFL1UFBP1 mutant were expressed in the UFL1-deficient HEK293T cells, and we verified that the expression levels were similar (Fig. 1E). An immunoprecipitation assay with anti-FLAG antibody revealed that wild-type UFL1, but not UFL1UFBP1 mutant, interacted with endogenous UFBP1 (Fig. 1E). In addition to UFBP1 and CDK5RAP3 (10–13), UFL1 directly interacts with UFC1 (10). The immunoprecipitant from cells expressing the UFL1UFBP1 mutant contained endogenous UFC1 and CDK5RAP3, but the levels of both were less than those from wild-type UFL1–expressing cells (Fig. 1E). FLAG-tagged wild-type UFBP1 and UFBP1UFL1 mutant were expressed into UFBP1 knockout (KO) HEK293T cells (27), and the cell lysates were immunoprecipitated with anti-FLAG antibody. The immunoprecipitant prepared from wild-type UFBP1–expressing cells contained endogenous UFL1, UFC1, and CDK5RAP3 (Fig. 1F). UFL1, UFC1, and CDK5RAP3 were not detected in the immunoprecipitants of cells expressing UFBP1UFL1 mutant (Fig. 1F).
In the next series of experiments, to biochemically investigate the interaction of UFL1 with CDK5RAP3, we constructed UFL1 and CDK5RAP3 mutants in which the amino acids involved in the electrostatic interaction between the two proteins were substituted with Ala (UFL1D129A R185A R191A R199A R251A, UFL1CDK5RAP3 mutant and CDK5RAP3E217A D355A E359A E373A R432A, CDK5RAP3UFL1 mutant). FLAG-tagged wild-type UFL1 and UFL1CDK5RAP3 mutant were expressed in the UFL1−/− HEK293T cells, and we verified that both wild-type and mutant UFL1 were expressed at similar levels (Fig. 1G). The amount of endogenous CDK5RAP3 increased when wild-type UFL1, but not UFL1CDK5RAP3 mutant, was introduced into UFL1-defeicient cells (Fig. 1G). Immunoprecipitation with anti-FLAG antibody showed that wild-type UFL1, but not UFL1CDK5RAP3 mutant, bound to endogenous CDK5RAP3 (Fig. 1G). Like wild-type UFL1, the UFL1 mutant showed the ability to bind to UFC1 and UFBP1 (Fig. 1G). The UFL1CDK5RAP3 mutant has mutations in the WH4, WH5, and WH6 domains. To clarify which domain contributes most to the interaction between UFL1 and CDK5RAP3, we constructed FLAG-tagged WH4 (UFL1D129A), WH5 (UFL1R185A R191A R199A), and WH6 (UFL1R251A) mutants then expressed each in UFL1-deficient cells and carried out immunoprecipitation assays with anti-FLAG antibody. As shown in Fig. 1H, only immunoprecipitants prepared from UFL1-deficient cells expressing the WH5 mutant did not contain endogenous CDK5RAP3. To investigate the effect of CDK5RAP3 mutant on the interaction with UFL1, we used CDK5RAP3 KO HEK293T cells (27) and expressed FLAG-tagged wild-type CDK5RAP3 and CDK5RAP3UFL1 mutant. The cell lysates were immunoprecipitated with anti-FLAG antibody, and the immunoprecipitants were subjected to immunoblot analysis with anti-UFL1 and anti-CDK5RAP3 antibodies. The expression levels of wild-type CDK5RAP3 and the mutant were comparable (Fig. 1I). The immunoprecipitation assay showed that wild-type but not mutant CDK5RAP3 interacted with endogenous UFL1 (Fig. 1I). The immunoprecipitants from cells expressing the CDK5RAP3 mutant contained hardly any UFC1 or UFBP1 (Fig. 1I). These results indicate that UFL1 plays a central role in the interactions between UFC1, UFBP1, and CDK5RAP3 (Fig. 1J).
Structural basis of UFC1-UFL1 interaction
E3 promotes ubiquitylation-like conjugation reactions through direct interaction with E2. We next used AF2 to predict how E3 components recognize UFC1. Structural prediction of the full-length UFL1-UFC1 binary complex suggests that UFC1 is mainly recognized by the N-terminal α helix (residues 1 to 21) of UFL1, which we hereafter refer to as the UFC1-binding sequence (UBS) (fig. S4A). All of the top five prediction models showed similar interaction between UFL1UBS and UFC1, although the relative orientation between UBS and the other region of UFL1 was variable (fig. S4B). Structural prediction of the UFL1UBS-UFC1 binary complex gives us almost the same interaction model (Fig. 2A and fig. S4C). These predictions are consistent with our previous report that UFL1 directly recognizes UFC1(Fig. 1D) (10). In addition, AF2 predicted the direct interaction of UFBP1 with UFC1 (the best model is shown in fig. S4D); however, the top five prediction models each show distinct UFBP1-UFC1 interactions (fig. S4E), suggesting that these models are unreliable. Structural prediction of the UFL1-UFBP1-UFC1 ternary complex provides us a combined model of the UFL1-UFC1 and UFBP1-UFC1 complexes (Fig. 2B and fig. S4F), in which UFL1-UFC1 and UFBP1-UFC1 interactions bury 961- and 555-Å2 surface areas of each protein, respectively. UFC1 binding did not affect the UFL1-UFBP1 complex structure in these two proteins, including the formation of the WH2 domain, but changed the location of the UBS so that the UBS-bound UFC1 could interact with UFBP1. UFBP1 interacts with UFC1 via WH1, mainly through hydrophilic interactions that include salt bridges between UFBP1 Arg265, Lys267 and UFC1 Asp50 and between UFBP1 Glu232 and UFC1 Lys164 (fig. S4D). These structural predictions suggest that UFL1UBS plays a central role in recruiting E2 to the E3 complex, and this recruitment is enhanced by UFBP1 WH1.
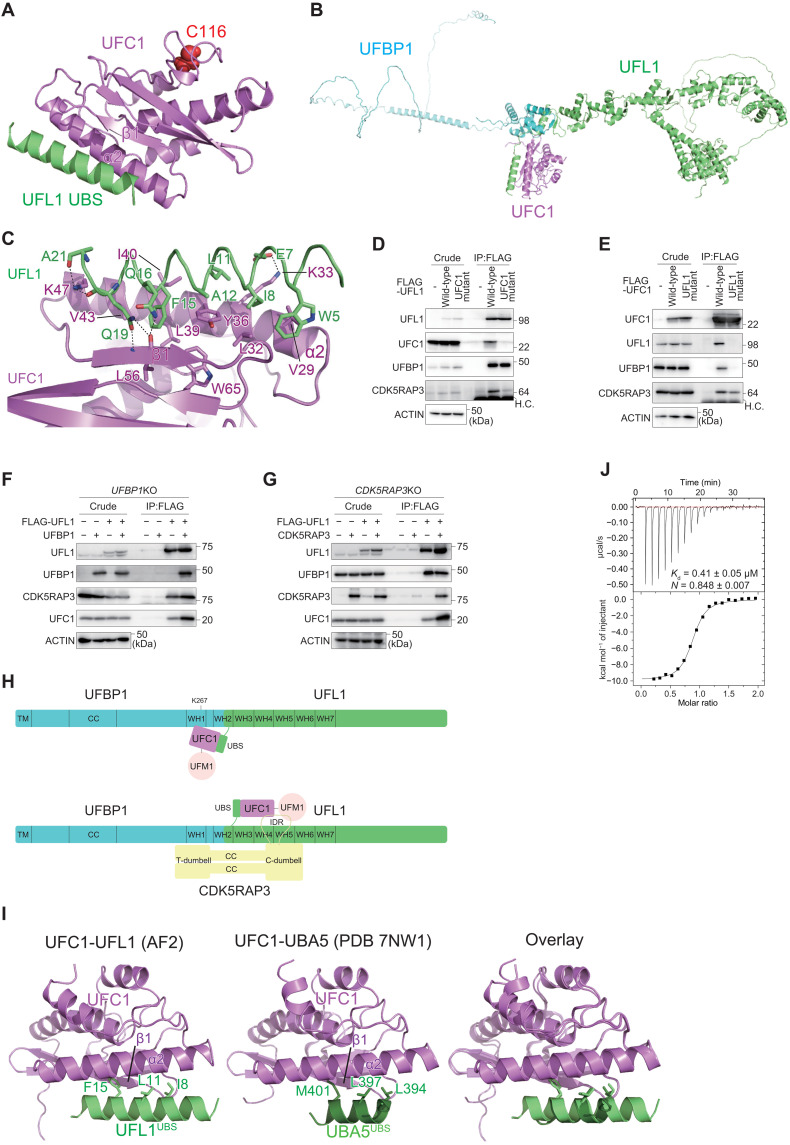
Fig. 2. Structure of the UFM1 E3 component UFL1 and the E2 UFC1 complex.
(A) Structural prediction of the UFL1UBS-UFC1 binary complex. Catalytic Cys116 of UFC1 is shown with a space-filling model. (B) Structural prediction of the UFL1-UFBP1-UFC1 ternary complex. (C) Recognition mode of UFL1UBS by UFC1. The side chains involved in interactions are shown with a stick model, where oxygen and nitrogen atoms are colored red and blue, respectively. Broken lines indicate possible electrostatic interactions. (D and E) Biochemical characterization of the binding between UFL1 and UFC1. (D) FLAG-UFL1 and UFC1 interaction–defective UFL1UFC1 mutant were transfected into UFL1-KO cells. (E) FLAG-UFC1 and UFL1 interaction–defective UFC1UFL1 mutant were transfected into UFC1-KO cells. Forty-eight hours after transfection, cells were lysed and immunoprecipitated with anti–FLAG-M2 gel; then, the immunoprecipitants were subjected to immunoblot with the indicated antibodies. Data shown are representative of three separate experiments. (F and G) Integrity of UFBP1 and CDK5RAP3 within the E3 complex influences the UFL1-UFC interaction. (F) FLAG-UFL1 was cotransfected with empty vector or UFBP1 into UFBP1-KO cells. (G) FLAG-UFL1 was cotransfected with empty vector or CDK5RAP3 into CDK5RAP3-KO cells. Forty-eight hours after transfection, cells were lysed and immunoprecipitated with anti–FLAG-M2 gel; then, the immunoprecipitants were subjected to immunoblot with the indicated antibodies. Data shown are representative of three separate experiments. (H) Schematic model of the binding of the UFM1 E3 complex (UFBP1-UFL1) to UFM1-charged UFC1, in the absence (top) and presence (bottom) of the E3 accessory protein CDK5RAP3. (I) Structural comparison between UFL1UBS-UFC1 (left) and UBA5UBS-UFC1 (middle) complexes. The side chains of the three hydrophobic residues of UFL1UBS and UBA5UBS that are important for UFM1 binding are shown with a stick model. The right shows the superimposition of the two complexes. (J) Binding affinity of UFL1UBS to UFC1 was measured by ITC.
Detailed interaction between UFL1UBS and UFC1 is shown in Fig. 2C. Previous crystallographic studies revealed that UFC1 is composed of three β strands (β1 to β3), four α helices (α1 to α4), and one 310 helix (37), which together fold into a canonical E2 fold, although this fold has an additional α helix at the N terminus and lacks the two C-terminal α helices observed in other E2 proteins. The predicted structure of UFC1 alone is essentially similar to the crystal structure, except for the six C-terminal residues, and is also similar to the predicted structure of UFC1 complexed with UFL1 (fig. S4G). This suggests that the prediction of the UFC1 structure is accurate in principle and that UFL1 binding has little effect on UFC1 folding. UFL1UBS is bound to α2 and β1 of UFC1, which are located at opposite sides of the catalytic cysteine (Cys116); hence, UFL1UBS has no apparent effect on the catalytic site structure or its accessibility (Fig. 2A). The UFL1UBS-UFC1 interaction arises mainly from hydrophobic interactions (Fig. 2C). The side chains of Trp5, Ile8, Leu11, Ala12, and Phe15 of UFL1 interact with those of Val29, Leu32, Tyr36, Leu39, Ile40, and Val43 of UFC1. In addition, UFL1 Glu7 forms a salt bridge with UFC1 Lys33 and the side chains of UFL1 Gln16, and Gln19 forms three hydrogen bonds with the main chain of UFC1 Leu56. These interactions bury the 770-Å2 surface area of each protein.
Structural mechanism of CDK5RAP3 recruitment to the E3 complex
To verify the interaction mode of UFL1 with UFC1 biochemically, we constructed UFL1 and UFC1 mutants, in which amino acids involved in the hydrophobic interactions of each other were substituted with Ala (UFL1W5A E7A E16A E19A F15A, UFL1UFC1 mutant and UFC1K33A Y36A I40A, UFC1UFL1 mutant). FLAG-tagged wild-type UFL1 or UFL1UFC1 mutant was expressed in the UFL1 KO cells, and cell lysates were subjected to immunoprecipitation assay with anti-FLAG antibody followed by immunoblots with anti-UFL1, anti-UFC1, anti-UFBP1, and anti-CDK5RAP3 antibodies. Both wild-type and mutant UFL1 were expressed at similar levels in the UFL1-deficient cells (Fig. 2D). As expected, wild-type UFL1 but not mutant UFL1 interacted with endogenous UFC1 (Fig. 2D). While UFL1UFC1 mutant has the ability to bind to UFBP1, it showed lower binding affinity for CDK5RAP3 (Fig. 2D). Next, we expressed FLAG-tagged wild-type UFC1 or UFC1UFL1 mutant in UFC1 KO HEK293T cells (23). As shown in Fig. 2E, the expression levels of wild-type and mutant UFC1 were comparable. The immunoprecipitant prepared from the UFC1-deficient cells expressing wild-type UFC1 contained endogenous UFL1 (Fig. 2E) but not that prepared from the mutant-expressing cells (Fig. 2E). Endogenous CDK5RAP3 was present in immunoprecipitants of cells expressing wild-type UFC1 but not the UFL1-interaction defective UFC1 mutants (Fig. 2E). To investigate whether the integrity of UFBP1 and/or CDK5RAP3 affected the interaction of UFL1 with UFC1, we investigated this interaction in UFBP1- and CDK5RAP3 KO cells. When FLAG-tagged UFL1 was coexpressed with UFBP1 in UFBP1 KO cells, endogenous UFC1 was found in the immunoprecipitants of FLAG-UFL1 cells (Fig. 2F). Even when only FLAG-UFL1 was expressed in UFBP1 KO cells, endogenous UFC1 was detected in the immunoprecipitants of FLAG-UFL1 cells but in smaller amounts than when coexpressed with UFBP1 (Fig. 2F). Similar results were also observed in CDK5RAP3 KO cells. When FLAG-tagged UFL1 was coexpressed with CDK5RAP3 in CDK5RAP3 KO cells, endogenous UFC1 was found in the immunoprecipitants of FLAG-UFL1 cells (Fig. 2G). Even when FLAG-UFL1 was solely expressed in CDK5RAP3 KO cells, endogenous UFC1 was detected in the immunoprecipitants of FLAG-UFL1 cells but in smaller amounts than when coexpressed with UFBP1 (Fig. 2G). These results suggest that while UFL1 is mainly responsible for UFC1 binding, both UFBP1 and CDK5RAP3 in the E3 complex also contribute to the interaction with UFC1. This is supported by AF2 models showing that UFBP1 interacts with UFC1 (fig. S4D), and CDK5RAP3 interacts with both UFC1 and UFM1 (most probably a UFM1-UFC1 intermediate) (refer to fig. S6) (Fig. 2H).
Structural mechanism of switching from the E1-E2 to the E2-E3 complex
Analysis of the crystal structure of UFC1 complexed with the UBA5UBS suggests that UFL1 and UBA5 interact with UFC1 in very similar ways (Fig. 2I) (39). In both UBSs, one α helix binds to α2 and β1 of UFC1, mainly through hydrophobic interactions. Binding to UFC1 is quite similar between the side chains of L394, L397, and M401 of UBA5UBS and those of I8, L11, and F15 of UFL1UBS. The helix of UBA5UBS is shorter than that of UFL1UBS and thus has fewer interactions with UFC1, which is partly compensated for by the interactions mediated by the N-terminal loop of the UBS helix (39). Since there is complete overlap between UFL1 and UBA5 in terms of their binding sites on UFC1 (Fig. 2I), it is obvious that the E1 (UBA5) and E3 (UFL1-UFBP1) enzymes compete with each other for E2 (UFC1) binding. This competitive binding is also conserved in other Ub/Ubl conjugation systems and enables the switch from the E1-E2 complex to the E2-E3 complex to allow conjugation reactions to proceed (46, 47). To confirm the binding of UFL1UBS to UFC1 and compare its affinity with that of UBA5UBS, we performed isothermal titration calorimetry (ITC) between UFC1 and UFL1UBS. As indicated in Fig. 2J, the dissociation constant (Kd) value between UFL1UBS and UFC1 was 0.41 μM, confirming that UFL1UBS binds to UFC1, and its affinity was 2- to 2.5-fold higher than that of full-length UBA5 (36) and 6-fold higher than that of UBA5UBS (39). These observations suggest that during ufmylation, UFC1 changes its binding partner from UBA5 to UFL1 to complete the conjugation reaction.
Structural mechanism of substrate specificity switching by CDK5RAP3
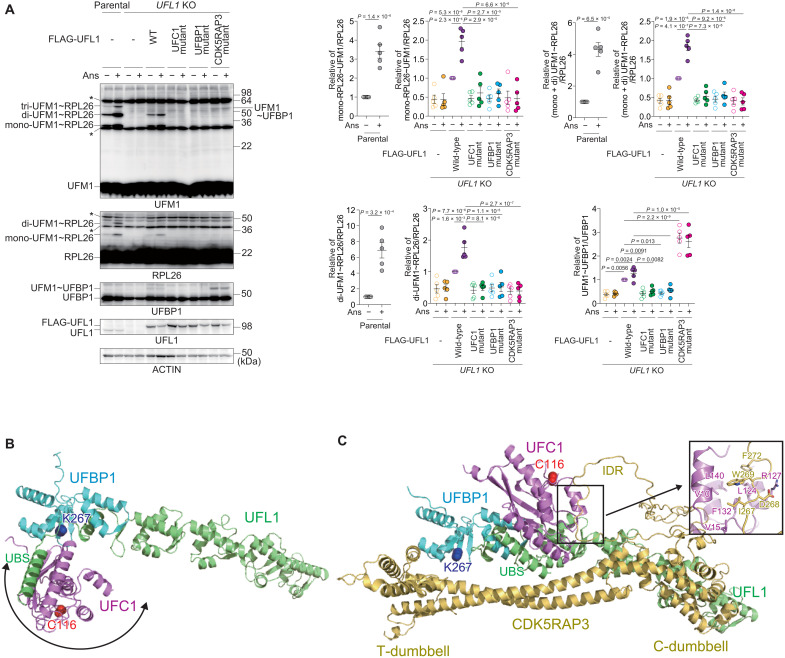
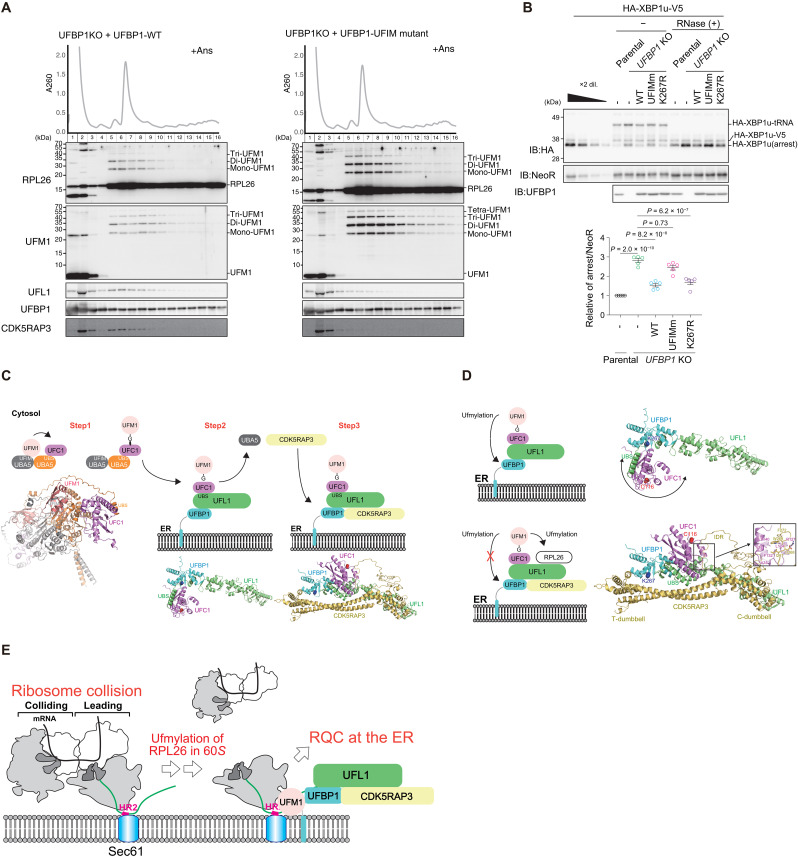
RPL26 is a major target of ufmylation, and the UFL1-UFBP1-CDK5RAP3 complex plays a critical role in this process (13, 29, 30). RPL26 ufmylation is enhanced by treatment with anisomycin, which prevents protein synthesis by inhibiting the peptidyl transferase center A site (30, 48). We confirmed that anisomycin treatment increased the levels of both di- and mono-ufmylated RPL26 (Fig. 3A). To investigate the effect of UFL1 mutants characterized by defective interaction with UFC1, UFBP1, or CDK5RAP3 during anisomycin-induced RPL26 ufmylation, we expressed each mutant in UFL1 KO HEK293T cells. While ufmylation was completely suppressed by UFL1 ablation, it was restored by expression of wild-type UFL1 (Fig. 3A). This restoration was not observed following the expression of any of the mutants (Fig. 3A), indicating that UFL1 interactions with UFC1, UFBP1, and CDK5RAP3 are required for the ufmylation of RPL26. UFBP1 is also ufmylated at Lys267, which enhances its ligase activity toward two other UFM1 substrates, ASC1 (45) and CYB5R3 (27). Unlike RPL26, ufmylation of UFBP1 was not enhanced by the treatment of anisomycin (Fig. 3A). We noticed that UFL1CDK5RAP3 mutant, which showed defective interaction with CDK5RAP3, promoted the ufmylation of UFBP1 (Fig. 3A), which occurred even following anisomycin exposure (Fig. 3A). These results suggest that CDK5RAP3 may change the substrate specificity of the E3 enzyme. To study the role of CDK5RAP3 in substrate specificity, we predicted the structure of the UFL1-UFBP1-CDK5RAP3-UFC1 complex using AF2. In the absence of CDK5RAP3, UFBP1 Lys267 was exposed, and UFC1 bound to UFL1UBS was identified near UFBP1 Lys267 (Fig. 3B). On the other hand, CDK5RAP3 binding markedly changed the location of UFC1 so that it was distal to UFBP1 Lys267 by (i) narrowing the space around UFBP1 Lys267 via the T-dumbbell and (ii) binding to UFC1 using the IDR (Fig. 3C).
Fig. 3. Substrate specificity switching by the UFM1 E3 component CDK5RAP3.
(A) The role of CDK5RAP3 in the ufmylation of RPL26 and UFBP1. Wild-type UFL1 and UFC1 interaction–defective (UFL1UFC1 mutant), UFBP1 interaction–defective (UFL1UFBP1 mutant), and CDK5RAP3 interaction–defective (UFL1CDK5RAP3 mutant) UFL1 mutants were transfected in UFL1-deficient HEK293T cells. Forty-eight hours after transfection, the cells were treated with 200 nM anisomycin (Ans) for 1 hour and then lysed. The cell lysates were subjected to SDS-PAGE followed by immunoblot analysis with the indicated antibodies. Bar graphs show the results of quantitative densitometric analysis of ufmylated RPL26 (mono- and di-ufmylated RPL26) relative to free RPL26 (n = 5) and of ufmylated UFBP1 relative to free UFBP1 (n = 5). Data are means ± SE. Statistical analysis was performed by Welch’s t test. Data shown are representative of five separate experiments. (B and C) Predicted structures of the UFL1(1-302)-UFBP1(209-314)-UFC1 complex (B) and the UFL1(1-302)-UFBP1(209-314)-CDK5RAP3-UFC1 complex using AF2 (C). The positioning of UFC1 in (B) is variable due to the flexible nature of UFL1UBS. The side chains of UFBP1 Lys267 and UFC1 Cys116 are shown with a space-filling model. The inlet in (C) indicates the detailed interactions between the CDK5RAP3 IDR and UFC1.
We constructed CDK5RAP3 mutants lacking the C-dumbbell, T-dumbbell, or IDR (ΔC-dumbbell, ΔT-dumbbell, and ΔIDR, respectively) and then performed an immunoprecipitation assay. As expected, the ΔIDR mutant hardly bound to UFC1 (fig. S5A). While the ΔT-dumbbell mutant still had the ability to bind to UFC1, the ΔC-dumbbell mutant showed decreased binding (fig. S5A). This is because the C-dumbbell is essential for interaction with UFL1 (Fig. 1, A, C, and I), and UFL1 is primarily responsible for interaction with UFC1. An in vitro pull-down assay showed that recombinant CDK5RAP3, but not the UFC1-interacting defective mutants (CDK5RAP3I267A W269A, CDK5RAP3UFC1 mutant), bound to recombinant UFC1 (fig. S5B). In addition, while the immunoprecipitant prepared from CDK5RAP3-deficient cells expressing FLAG-tagged CDK5RAP3 contained endogenous UFC1, the level was much lower in the case of CDK5RAP3UFC1 mutant–expressing cells (fig. S5C). These structural models, together with biochemical analyses, suggest that CDK5RAP3 impairs the ufmylation of UFBP1 Lys267 by positioning UFC1 away from the residue and driving the ufmylation toward RPL26.
Interaction of CDK5RAP3 with UFM1
We recently showed that UFBP1 has a UFM1-interacting motif (UFIM) at the IDR N-terminal of the WH1 domain and binds to UFM1 in a manner similar to UBA5UFIM (fig. S6, A and B) (27, 49). Unexpectedly, the E3 component CDK5RAP3, but not UFL1, was also predicted by AF2 to bind to UFM1 using the IDR inserted in the C-dumbbell (fig. S6C). This binding process is similar to that of UFBP1UFIM and UBA5UFIM, forming an intermolecular β sheet with UFM1 β2 and inserting two hydrophobic residues into the hydrophobic pockets of UFM1 (fig. S6D). Thus, this binding sequence in CDK5RAP3 was also named UFIM. An in vitro pull-down assay revealed that recombinant CDK5RAP3, but not the UFIM mutants (CDK5RAP3I321A, CDK5RAP3UFIM mutant), bound to recombinant UFM1 (fig. S6E). To investigate the interaction of CDK5RAP3 with UFM1 in cells, we expressed wild-type CDK5RAP3 or CDK5RAP3UFIM mutant in CDK5RAP3-deficient cells and then performed an immunoprecipitation assay. As shown in fig. S6F, the immunoprecipitants from the cells expressing wild-type CDK5RAP3, but not CDK5RAP3UFIM mutant, contained both free UFM1 and UFC1. We hypothesized that in addition to the interaction of CDK5RAP3 with UFC1 (Fig. 3C and fig. S5), an intermediate consisting of a UFC1-UFM1 thioester binds to CDK5RAP3. To prove this, we used UFC1C116S, in which the active site cysteine is substituted with serine. Instead of the thioester bond, the UFC1C116S formed an O-ester bond with UFM1, which was stable even under reducing conditions (6). When MYC-tagged UFC1C116S and green fluorescent protein (GFP)–tagged UFM1 were coexpressed with wild-type or UFIM mutant CDK5RAP3 in CDK5RAP3 KO cells, CDK5RAP3 interacted with GFP-UFM1-MYC-UFC1C116S (fig. S6G). The binding affinity was lower in the case of the CDK5RAP3UFIM mutant (fig. S6G).
CDK5RAP3 has the ability to interact with both UFC1 and UFM1-charged UFC1 (figs. S5 and S6). These interactions may have a positive effect on the RPL26 ufmylation. However, both UFC1- and UFM1-binding mutants of CDK5RAP3 promoted di- and tri-ufmylation of RPL26 upon anisomycin treatment (fig. S6H). This suggests that excessive stabilization of the UFL1-UFBP1-CDK5RAP3-UFM1-UFC1 pentameric complex via CDK5RAP3 UFIM-UFM1 interaction may make the complex too stable and thereby lower the turnover frequency. This hypothesis is consistent with the results of an in vitro ufmylation assay of RPL26 (13).
Association of the E3 UFM1 complex with the 60S ribosomal subunit
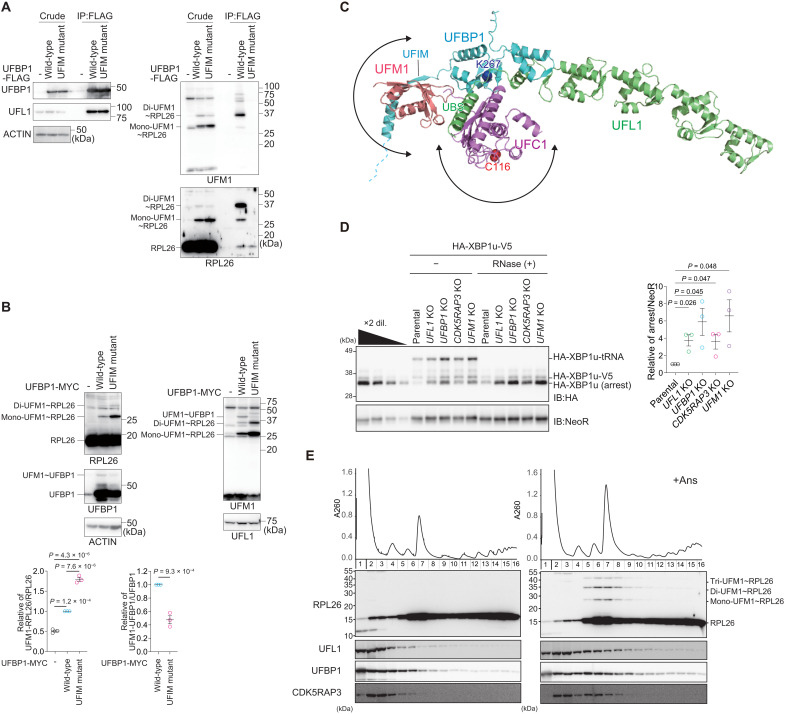
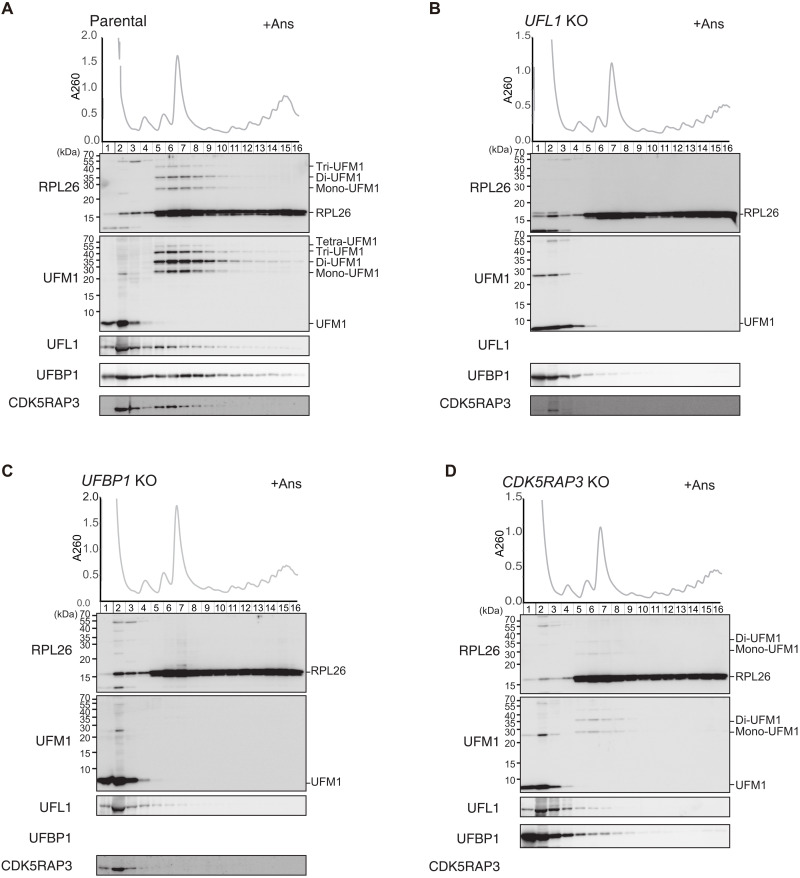
Next, to test the interaction of UFBP1 with UFM1 in cells, we generated UFBP1 UFSP2 double knockout (DKO) HEK293T cells (fig. S3B) and used them to express wild-type UFBP1 and the UFIM mutant (UFBP1F196A V198A, UFBP1UFIM mutant). Both proteins were expressed at a similar level (Fig. 4A). The ufmylated proteins accumulated in cells lacking UFSP2 (16). When wild-type UFBP1 was expressed in the UFBP1 UFSP2 DKO HEK293T cells, we observed ufmylated RPL26 even without anisomycin treatment (Fig. 4A). An immunoprecipitation assay showed that wild-type UFBP1, but not the UFBP1UFIM mutant, interacted with di- and mono-ufmylated RPL26, particularly di-ufmylated RPL26 (Fig. 4A). Both wild-type UFBP1 and the UFBP1UFIM mutant bound to free RPL26 (Fig. 4A), suggesting that UFBP1 originally had an affinity for the ribosome that was strengthened when RPL26 was ufmylated. We therefore sought to determine the effect of the UFBP1 UFIM mutant on ufmylation of RPL26 and UFBP1. When this mutant was overexpressed in UFBP1-deficient cells, UFBP1 ufmylation was impaired, and this enhanced the ufmylation of RPL26 (Fig. 4B). This suggests that UFBP1 UFIM has two opposing functions: promoting the ufmylation of UFBP1 and inhibiting the ufmylation of RPL26. The former is predicted by the AF2 structure of the UFL1-UFBP1-UFC1-UFM1 complex, in which UFM1 binding to UFBP1 UFIM restricts the localization of UFM1 (and the UFM1-UFC1 intermediate) to the vicinity of UFBP1 Lys267, thereby promoting its ufmylation (Fig. 4C). The latter function cannot be predicted by AF2 because it fails to predict the E3-RPL26 complex structure. The stable association of UFBP1 with ufmylated RPL26 via UFIM may trap the UFM1 E3 on the ribosome and then suppress further RPL26 ufmylation.
Fig. 4. The association of the E3 complex with the 60S ribosome.
(A) The interaction of UFBP1 with both non-ufmylated and ufmylated RPL26. FLAG-UFBP1 or UFM1 interaction–defective UFBP1UFIM mutant was transfected into UFBP1- and UFSP2-DKO cells. Forty-eight hours after transfection, cells were lysed and immunoprecipitated with anti–FLAG-M2 gel; then, the immunoprecipitants were subjected to immunoblot with the indicated antibodies. Data shown are representative of three separate experiments. (B) Increased ufmylation of RPL26 by UFBP1UFIM mutant. MYC-UFBP1 or UFBP1UFIM mutant was transfected into UFBP1-KO cells. Forty-eight hours after transfection, cells were lysed, and the lysates were subjected to immunoblot with indicated antibodies. Bar graphs show the results of quantitative densitometric analysis of mono- and di-ufmylated RPL26 relative to free RPL26 (n = 3) and of ufmylated UFBP1 relative to free UFBP1 (n = 3). Data are means ± SE. Statistical analysis was performed by Welch’s t test. (C) Structural prediction of the UFL1-UFBP1-UFC1-UFM1 quaternary complex, which is generated by combining the AF2 structure of the UFL1-UFBP1-UFC1 and UFBP1-UFM1 complexes. (D) The indicated KO cells were transfected with the HA-XBP1u-V5. Twenty-four hours after transfection, the cells were lysed. The lysates were subjected to neutral PAGE followed by immunoblot with indicated antibodies (left). The free peptide and the peptidyl-tRNA (pep-tRNA) arrest products were detected by immunoblot with indicated antibodies. Samples treated with ribonuclease (RNase) to digest the tRNA moiety of pep-tRNA are indicated by (+) (right). The bar graph shows the results of quantitative densitometric analysis of the arrest products relative to NeoR (n = 3). Data are means ± SE. Statistical analysis was performed by Welch’s t test. (E) Cells were treated with anisomycin (Ans, 0.1 μg/ml) for 30 min and then lysed. Ribosomes were separated by ultracentrifugation through sucrose density gradients. Protein samples prepared from gradient fractions were analyzed by immunoblot with indicated antibodies.
What is the significance of the interaction between UFBP1 and ufmylated RPL26? We examined the role of the UFL1-UFBP1-CDK5RAP3 E3 complex in RQC-mediated degradation of nascent polypeptides derived from stalled ribosomes. Ribosome stalling during the translation of XBP1u mRNA contributes to the efficient targeting of XBP1u mRNA to the ER membrane (50, 51). Ribosome stalling at the XBP1u stalling sequence induces ER-RQC, and nascent polypeptides derived from stalled ribosomes are degraded in an LTN1-dependent manner (52). To evaluate the role of the UFL1-UFBP1-CDK5RAP3 E3 complex in ER-RQC, we investigated LTN1-mediated degradation of nascent XBP1u polypeptides in UFM1 E3 KO cells. The levels of these polypeptides were increased in UFL1, UFBP1, CDK5RAP3, and UFM1 KO cells (Fig. 4D), and the polypeptides were stabilized in those KO cells (fig. S7A). In contrast, the levels of cytosolic RQC substrate were not increased in any of the cells lacking E3 components (fig. S7B). These results indicate that ufmylation is required for ER-RQC but not for cytosolic RQC, as previously reported (31, 40).
We next asked whether the association of the UFL1-UFBP1-CDK5RAP3 E3 complex with the 60S ribosomal subunit is facilitated by anisomycin treatment, which increases both ribosome collisions (53) and the levels of di- and mono-ufmylated RPL26 (Fig. 3A). To verify the interaction of the subunits of the UFM1 E3 complex with ribosomes, we performed polysome analysis. The association of E3 components with the 60S subunit was estimated by Western blotting using the factions obtained after centrifugation in sucrose gradients. Without anisomycin treatment, UFL1 and CDK5RAP3 were distributed mainly in the ribosome-free fraction, whereas UFBP1 was found in both the ribosome-free and 60S fractions, probably due to its affinity for non-ufmylated RPL26 (Fig. 4A). Upon anisomycin treatment, the distribution of these factors in the 60S fraction was substantially increased (Fig. 4E). Given that the interaction of UFL1 with CDK5RAP3 was observed even without anisomycin treatment (Figs. 1 and 2), and UFL1 formed a stable complex with the ER-localizing protein UFBP1 (13, 28), we propose that the E3 UFM1 complex associates with the 60S subunit and that this association is facilitated by anisomycin treatment.
A crucial role of the UFBP1-UFM1 interaction in ER-RQC
We next investigated which component of the UFM1 E3 complex is indispensable for the anisomycin-induced association of E3 complex with the 60S ribosome. Polysome analysis revealed that deletion of one of the three E3 components disrupted the association of the other components with the 60S subunit (Fig. 5, A to D). In UFL1 KO cells, the expression level of CDK5RAP3 was substantially reduced, as was the association of UFBP1 with the 60S subunit (Fig. 5B). In UFBP1 KO cells, neither UFL1 nor CDK5RAP3 was associated with the 60S subunit (Fig. 5C). In CDK5RAP3 KO cells, the association of UFBP1 and UFL1 with the 60S subunit was reduced (Fig. 5D), suggesting that ufmylation by the E3 complex facilitates the interaction of the E3 complex with the 60S subunit. Since UFBP1 associates with ufmylated RPL26 via UFBP1UFIM-UFM1 interaction, we next examined whether the defect of the interaction affects the association of the UFM1 E3 complex with the 60S subunit. Compared with wild-type UFBP1 expression, the association of UFL1 and CDK5RAP3 with the 60S subunit was substantially reduced in UFBP1-deficient cells expressing UFBP1UFIM mutant (Fig. 6A), indicating that the interaction of UFBP1 with ufmylated RPL26 via the UFIM contributes to the association of the E3 complex with the 60S subunit. We assessed the role of binding of UFBP1 to ufmylated RPL26 in RQC and found that UFBP1-deficient cells expressing UFBP1UFIM mutant exhibited elevated levels of nascent XBP1u polypeptides (Fig. 6B). Meanwhile, UFBP1 KO cells expressing UFBP1K267R mutant, which does not undergo ufmylation, did not show such increase (Fig. 6B). These results indicate that interaction of UFBP1 with ufmylated RPL26 is essential for ER-RQC, while ufmylation of UFBP1 is not.
Fig. 5. UFBP1 is indispensable for the association of E3 components with the 60S ribosomal subunit upon anisomycin treatment.
Parental HEK293T cells (A) or UFL1- (B), UFBP1- (C), and CDK5RAP3- (D) KO cells were treated with anisomycin (Ans, 0.1 μg/ml) for 30 min and then lysed. Ribosomes were separated by ultracentrifugation through sucrose density gradients. Protein samples were prepared from gradient fractions and analyzed by Western blotting using antibodies against RPL26, UFM1, UFL1, UFBP1, and CDK5RAP3.
Fig. 6. The interaction of UFBP1 with ufmylated RPL26 is indispensable for ER-RQC.
(A) UFBP1 KO cells were transfected with wild-type UFBP1 or UFM1 interaction–defective UFBP1. Twenty-four hours after transfection, the cells were treated with anisomycin (Ans, 0.1 μg/ml) for 30 min and then lysed. Ribosomes were separated by ultracentrifugation through sucrose density gradients. Protein samples prepared from gradient fractions were analyzed by immunoblot with indicated antibodies. Data shown are representative of three separate experiments. (B) UFBP1-KO cells expressing wild-type UFBP1, UFBP1UFIM mutant, or UFBP1K267R mutant were transfected with the HA-XBP1u-V5. Twenty-four hours after transfection, the cells were lysed. Top: The cell lysates were subjected to neutral PAGE followed by immunoblot with indicated antibodies. The free peptide and the peptidyl-tRNA (pep-tRNA) arrest products were detected by immunoblot with an anti–hemagglutinin (HA) antibody. Samples treated with RNase to digest the tRNA moiety of pep-tRNA are indicated by (+). Bottom: The relative levels of the HA-XBP1u arrest products with P values were determined by five independent experiments. (C) Schematic model of the enzymatic cascades during RPL26 ufmylation. Each protomer within the UBA5 homodimer binds UFM1 and UFC1 via UFIM and UBS, respectively, and catalyzes the formation of the UFM1-UFC1 thioester intermediate. UFL1-UFBP1 anchored to the ER recruits the UFM1-UFC1 intermediate via UFL1UBS binding to UFC1. Last, CDK5RAP3 is recruited to the E3 complex to complete the organization of the pentameric E3 complex on the ER. (D) Schematic model of substrate switching of the UFM1 E3 ligase. In the absence of CDK5RAP3, the UFM1-UFC1 intermediate can freely change its position and thereby access and ufmylate UFBP1 K267. Upon CDK5RAP3 binding, the position of the UFM1-UFC1 intermediate is locked away from UFBP1 K267, preventing its ufmylation and promoting RPL26 ufmylation instead. (E) Schematic model of ER-RQC through the UFM1 E3 ligase.
DISCUSSION
On the basis of multiple structural analyses of the ternary UFM1 E3 complex, E2, and UFM1 using AF2, as well as biochemical analyses, we propose a sequential organization model of the E3 complex on the ER as follows. A complex consisting of UBA5 and UFC1, which are the E1 and E2 enzymes associated with UFM1, respectively, initially generates a UFM1-UFC1 thioester intermediate. Next, a complex composed of the E3 components UFL1 and UFBP1 receives the UFM1-UFC1 intermediate from UBA5 at the ER by competitive binding of UFL1UBS to UFC1. Last, the UFL1-UFBP1 complex with the UFM1-UFC1 intermediate recruits CDK5RAP3 to create the UFL1-UFBP1-CDK5RAP3-UFM1-UFC1 pentameric complex on the ER, which relocates the UFM1-UFC1 intermediate away from UFBP1 Lys267 to promote ufmylation of RPL26 (Fig. 6C). On the other hand, if CDK5RAP3 is unavailable to bind to the UFL1-UFBP1 complex, then the UFM1-UFC1 intermediate can freely change its position relative to the E3 complex to ufmylate UFBP1 Lys267. This model suggests that CDK5RAP3 functions as an accessory component of the E3 complex and switches the E3 complex specificity from UFBP1 to RPL26 (Fig. 6D). While the ufmylation of RPL26 is involved in ER-RQC and/or nascent peptide quality control on the ER (29–31), that of UFBP1 participates in ER-phagy (27). It is possible that when a certain ER-RQC threshold is exceeded, ER-phagy removes portions of the ER that contain a number of stalled ribosomes.
In plants and mammals, the ufmylation of collided ribosomes is followed by autophagic trafficking of substrates to the lysosome (29, 30, 32). Recent studies demonstrated that ufmylation of the ribosomal protein RPL26 mediates ER-RQC but not cytosolic RQC (40). Depending on whether ribosome stalling is present or not, RQC components are recruited to the ER membranes, and substrates with various topologies are targeted for degradation by LTN1. The LTN1 RING domain provides access for ubiquitination of the target lysine residue of the nascent chain on the 60S ribosomal subunit. It has been proposed that C-terminal alanine and threonine tail exposes buried lysine residues, thereby facilitating LTN1-mediated ubiquitination (54). The gap between the 60S subunit and the translocon should be sufficiently large to enable access (55), and RPL26 ufmylation is thought to expand the gap for LTN1-mediated ubiquitination by recruiting a UFM1-interacting protein (40). In this study, we found that the UFL1-UFBP1-CDK5RAP3 E3 complex stably bound to ufmylated RPL26 via UFBP1UFIM, a function of this complex that occurred in addition to its ligase activity.
What is the significance of the stable association of UFBP1 with the 60S ribosomal ribosome? In UFBP1-deficient cells expressing the UFBP1UFIM mutant, the impairment of LTN1-mediated degradation of products by ribosome stalling on XBP1u mRNA was evident (Fig. 6B). However, an interesting observation was made in Fig. 6A, where the ufmylation of RPL26 in the 60S subunit was actually promoted. These results indicate that the stable association of the E3 complex with the 60S subunit via UFBP1 binding to UFM1-RPL26, but not the ligase activity of this complex, is crucial for ER-RQC (Fig. 6E). The mechanism by which the LTN1-mediated degradation of the polypeptide on the 60S subunit depends on the interaction of UFBP1 with the 60S subunit remains unknown. Given that RPL26 is located close to the translocon (56), the recruitment of NEMF and/or LTN1 to the 60S subunit may require the interaction of the UFBP1 with the 60S subunit through ufmylated RPL26. The ufmylation of the 60S subunit containing peptidyl-tRNA, an LTN1 substrate, leads to the interaction of UFBP1 with ufmylated RPL26 through the UFIM, resulting in a stable association between the UFM1 E3 complex with the 60S ribosome as well as the binding of LTN1 to the 60S subunit for the ubiquitination of the peptidyl-tRNA on this subunit. Structural analysis of the 60S-UFM1 E3 complex will provide fundamental information to clarify the crucial role of UFM E3 in quality control that eliminates aberrant products derived from ribosome collision.
MATERIALS AND METHODS
Structural prediction using AF2 with the AlphaFold-Multimer mode
Complex structures were predicted using AF2 v2.2.0 (downloaded on 16 May 2022 from https://github.com/deepmind/alphafold) installed on a local computer (Sunway Technology Co. Ltd., Tokyo, Japan) (57). The predictions were run using the AlphaFold-Multimer mode (43), with five models and a single seed per model, and default multiple sequence alignment generation using the MMSeqs2 server (58). The unrelaxed predicted models were subjected to an Amber relaxation procedure, and the relaxed model with the highest confidence based on predicted LDDT scores was selected as the best model and used for figure preparation (57). Structural figures were prepared using PyMOL (http://pymol.org/pymol).
Cell culture
HEK293T cells (American Type Culture Collection CRL- 3216) were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum, 2 mM l-glutamine, penicillin (5 U/ml), and streptomycin (50 μg/ml). To introduce expression vectors, HEK293T cells were transfected with PEI MAX (Polysciences Inc., Warrington, PA, USA). UFL1 (5′-CCAGCGGGCGCAGTTCGCCG-3′) or UFBP1 (5′-GTAGCGGCGGCTCTGCTAGT′) guide RNA was designed using the CRISPR Design tool (https://crispr.dbcls.jp/) and subcloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene, #42230), a human codon–optimized SpCas9 and chimeric guide RNA expression plasmid. To generate UFL1, UFC1, CDK5RAP3, and UFBP1 UFSP2 KO HEK293T cells, HEK293T or UFSP2 KO HEK293T cells (16) were transfected the aforementioned pX330 vectors together with pEGFP-C1 (#6084-1, Clontech Laboratories, Mountain View, CA, USA) and cultured for 2 days. GFP-positive cells were sorted and expanded. Ablation of UFL1, UFC1, or CDK5RAP3 was confirmed by a heteroduplex mobility assay followed by immunoblot analysis with anti-UFL1, anti-UFC1, or anti-CDK5RAP3 antibody. UFM1- (23), UFC1- (23), UFBP1- (27), CDK5RAP3- (27), and UFSP2-deficient HEK293T cells (16) were used in this study. HEK293T and HeLa cells were authenticated by short-tandem repeat profile. All cell lines were tested for mycoplasma contamination.
Immunoblot and immunoprecipitation analysis
Following procedures previously described in (27), we performed immunoblot and immunoprecipitation analyses. Briefly, cells were lysed with ice-cold TNE (50 mM tris-HCl [pH7.5], 150 mM NaCl, 1mM EDTA) buffer containing 1% NP-40, 1% TX-100, and protease inhibitors. The lysates were centrifuged at 20,000g for 10 min at 4°C, and the resulting supernatants were used as samples for immunoblot analysis. Samples were subjected to SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a polyvinylidene difluoride (PVDF) membrane (IPVH00010, Merck Millipore, Burlington, MA, USA). Antibodies against UFM1 (ab109305, Abcam, Cambridge, UK; 1:1000), UFL1 (A303-456A; Bethyl Laboratories, Montgomery, TX, USA; 1:1000), UFBP1 (21445-1-AP, Proteintech, Rosemont, IL, USA; 1:1000), CDK5RAP3 (H00080279-M01; Novus Biologicals, Englewood, CO, USA; 1:500), UFSP2 (ab185965, Abcam; 1:1000), PRL26 (ab59567, Abcam; 1:1000), ACTIN (A1978; Sigma-Aldrich, Burlington, MO, USA; 1:2000), and FLAG (M185-3 L, Medical & Biological Laboratories, Tokyo, Japan; 1:1000) were purchased from the indicated suppliers. Blots were incubated with horseradish peroxidase–conjugated goat anti-mouse immunoglobulin G (IgG) (H + L) (115-035-166, Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA; 1:10,000) or goat anti-rabbit IgG (H + L) (111-035-144, Jackson ImmunoResearch Laboratories Inc.; 1:10,000) and visualized by chemiluminescence. Band density was measured using the software Multi Gauge V3.2 (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan). For immunoprecipitation analysis, cells were lysed in 300 μl of immunoprecipitation (IP) buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 1% TX-100] containing protease inhibitor cocktail (Roche, Basel, Switzerland), and the lysates were then centrifuged at 20,000g for 10 min at 4°C to remove debris. In the next step, 200 μl of IP buffer and 10 μl of anti-FLAG M2 Affinity Agarose Gel (A2220, Merck Millipore) were added to the 200 μl of lysate, and the mixture was mixed under constant rotation for 3 hours at 4°C. The immunoprecipitates were washed five times with ice-cold IP buffer. The complex was boiled for 5 min in SDS sample buffer in the presence of β-mercaptoethanol to elute proteins.
Isothermal titration calorimetry
For ITC experiments, UFC1 prepared for pull-down assay and synthesized UFL1 (1MADAWEEIRRLAADFQRAQFA21, from Toray Research Center, Tokyo, Japan) were subjected to size exclusion chromatography with 20 mM Hepes (pH 6.8) and 150 mM sodium chloride using a Superdex 200 column (Cytiva, Tokyo, Japan). ITC experiments were performed using a Microcal iTC200 calorimeter (Malvern Panalytical, Malvern, Worcestershire, UK), with stirring at 750 rpm at 25°C. The cell and syringe were filled with 30 μM UFC1 and 300 μM UFL1 (1–21), respectively. The titration involved 18 injections of 2 μl of the syringe sample at intervals of 120 s into a cell after one injection of 0.4 μl of syringe sample. MicroCal Origin 7.0 software was used for data analysis. Thermal measurement data of the first syringe sample injections were removed from the analysis. Thermal titration data were fit to a single-site binding model, which determines thermodynamic parameter the enthalpy (ΔH), Kd, and stoichiometry of binding (N). The error of each parameter shows the fitting error.
Pull-down assay
Recombinant maltose-binding protein (MBP)–fused UFL1 and glutathione S-transferase (GST)–fused UFM1, UFC1, UFBP1, UFBP1F196A V198A, CDK5RAP3, CDK5RAP3I321A, and CDK5RAP3I267A W269A were produced in E. coli and purified by chromatography on glutathione-Sepharose 4B resin (Amersham Biosciences, Arlington Heights, IL, USA) and amylose resin (New England Biolabs, Ipswich, MA, USA), respectively. GST-UFM1 and GST-UFC1 were cleaved the GST tag by precision protease (Amersham Biosciences). The purified proteins were mixed in TNE buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, and 1 mM EDTA] containing 1% NP-40 for 1 hour at 4°C and then precipitated with glutathione Sepharose. The mixtures were washed five times with ice-cold TNE. The bound proteins were analyzed by SDS-PAGE followed by Ponceau S staining or immunoblot analysis.
RQC assay using XBP1u-staller sequence
RQC activity was monitored by the detection of the arrest product derived from a reporter HA-XBP1u-V5 plasmid (52). The UFL1, UFBP1, CDK5RAP3, and UFM1 KO cells were transfected with HA-XBP1u-V5 plasmid, and cell lysates were prepared after 24 hours. A retrovirus expression system was used to express wild-type UFBP1 and the mutants in the UFBP1 KO cells. HEK293T cells were plated on six-well plates at 8 × 105 cells per well. Gag-Pol, VSV-G, and retroviral vector (pMRX-IRES-puro-UFBP1, pMRX-IRES-puro-UFBP1UFIM mutant, or pMRX-IRES-puro-UFBP1K267R) were transfected into HEK293T cells using Lipofectamine LTX (Thermo Fisher Scientific, Waltham, MA, USA, 15338100). Twenty-four hours after transfection, the medium was replaced with 1 ml of DMEM containing 10 mM Hepes. Twenty-four hours after medium change, virus-containing medium was harvested. The filtered viral supernatant was used to infect HEK293T UFBP1-deficient cells with polybrene (8 μg/ml), and infected cells were selected using puromycin (1 μg/ml). Protein samples of cell lysates with or without ribonuclease (RNase) treatment were separated by neutral PAGE followed by Western blotting to detect the HA-XBP1u arrest products using the anti–hemagglutinin (HA) antibody to detect peptidyl-tRNAs (52, 59). The levels of the HA-XBP1u arrest products and the control NeoR proteins were quantified using dilution series. The relative levels of the HA-XBP1u arrest products with P values were calculated by three independent experiments.
For a cycloheximide chase assay, parental HEK293T, UFL1-, UFBP1-, CDK5RAP3-, and UFM1-deficient HEK293T cells were transfected with HA-XBP1u-V5 plasmid. After 24 hours, the cells were treated with cycloheximide (100 μg/ml) for 0, 30, 60, 90, or 120 min and then lysed. The cell lysates were treated with RNase, and the samples were then subjected to neutral PAGE followed by immunoblot analysis with antibodies against HA and NeoR. The relative levels of the HA-XBP1u arrest products with P values were calculated by five independent experiments.
Lentivirus production and infection in a cytosolic RQC assay
Following procedures previously described in (59), we performed lentivirus production and infection. Briefly, 293FT cells were grown to around 60% confluency on 6-cm plates. A transfection mixture [14.7 μl of Lipofectamine 2000 (Thermo Fisher Scientific, 11668-019), 1.2 μg of pCAG-HIVgp (RDB04394), 1.2 μg of pCMV-VSVG-RSV-Rev (RDB04393), and 2.5 μg of CSII-CMV-MCS-IRES2-Bsd (RDB04385)] containing V5-EGFP-K(AAA)0 or 20-RFP-HA reporter was prepared in 500 μl of Opti-MEM–reduced serum medium (Thermo Fisher Scientific, 31985-062). The mixture was incubated at room temperature for 15 min and gently added to the 293FT cells for 48-hour incubation (37°C, 5% CO2). Forty-eight hours after the start of the transfection, lentivirus-containing cell culture supernatants were collected and filtered (Merck Millipore, MILLEX GV 0.45 μm). On the day before infection, the resultant virus solution (4.5 ml) was supplemented with polybrene (Sigma-Aldrich, H9268, 5 μg/ml) and added to HEK293T cells that were seeded at 4 × 105 cells per 6-cm dish. Forty-eight hours after infection, one-fifth of the infected cells was seeded on new 10-cm plates following trypsinization and further incubated in the presence of blasticidin-S (FUJIFILM Wako Pure Chemical Corporation, 029-18701, 10 μg/ml) for 48 hours to select infected cell populations. Cells were washed with phosphate-buffered saline (PBS) and lysed with radioimmunoprecipitation assay buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 10 mM sodium fluoride, 10 mM β-glycerophosphate, 1% Triton X-100, 0.1% SDS, and 0.5% sodium deoxycholate]. Protein samples were separated by SDS-PAGE and were transferred to a PVDF membrane (#IPVH00010, Merck Millipore).
Sucrose density gradient ultracentrifugation and Western blotting
Following procedures previously described in (59), we performed sucrose density gradient ultracentrifugation and Western blotting. Briefly, cells were treated with or without anisomycin at the indicated concentrations for 30 min (37°C, 5% CO2). Cells were washed with PBS twice and lysed with lysis buffer [50 mM tris-HCl (pH 6.8), 100 mM NaCl, 10 mM MgCl₂, 1% NP-40, 2 mM 2-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride] supplemented with protease inhibitors (Thermo Fisher Scientific) and centrifuged at 1500g for 5 min at 4°C. The HEK293 cell extracts were layered on top of the sucrose gradients and centrifuged at 201,000g in a SW40 rotor for 1.5 hours at 4°C. The gradients were then fractionated with a BioComp Piston Gradient Fractionator. The polysome profiles were generated by continuous absorbance measurement at 260 nm using a single-path UV-1 optical unit (ATTO Biomini UV monitor, ATTO, Tokyo, Japan) connected to a chart recorder (ATTO digital mini recorder). For the Western blots, 500 μl of each fraction was mixed with 55.6 μl of 100% trichloroacetic acid and incubated for 20 min at 4°C. After centrifugation (15,000g, 15 min, 4°C), the supernatant was removed, and the pellet was washed with acetone and dissolved in 30 μl of 2× SDS sample buffer [100 mM tris (pH 6.8), 4% (w/v) SDS, 20% glycerol, 0.02% bromophenol blue, and 50 mM dithiothreitol (DTT)]. Protein samples were separated by SDS-PAGE and transferred to a PVDF membrane (#IPVH00010, Merck Millipore).
For neutral PAGE, cells were lysed with lysis buffer and centrifuged at 20,000g for 10 min. Supernatants were collected, and equal amounts of total proteins were used as protein samples. For RNase(+) samples, RNase A (QIAGEN, Hilden, Germany) was added at a final concentration of 0.05 mg/ml and incubated on ice for 30 min; for RNase(−) samples, Milli-Q water was added instead. After incubation, 1× sample buffer [50 mM tris (pH 6.8), 2% (w/v) SDS, 10% glycerol, 0.01% Bromophenol Blue (BPB), and 50 mM DTT] was added and heated for 5 min at 70°C. Proteins were separated by 15% PAGE under neutral pH conditions (pH 6.8) for 5 hours with a 150-V constant voltage in MES-SDS buffer (50 mM MES, 50 mM tris base, 3.465 mM SDS, and 1 mM EDTA) and were transferred to a PVDF membrane (#IPVH00010, Merck Millipore).
Statistical analysis
Values, including those displayed in the graphs, are presented as means ± SE. Statistical analysis was performed using the unpaired t test (Welch’s test) or Šidák’s multiple comparisons test with GraphPad Prism version 9.2.0 software (GraphPad software, Boston, MA, USA). A P value less than 0.05 was considered to indicate statistical significance.
Acknowledgments
We would like to thank H. Morishita, R. Kurusu, and Y. Ichimura for critically reading this manuscript.
Funding: This work was supported by JSPS KAKENHI grant number JP22K06931 (to R.I.); JP21H04163 (to S.K-H.); JP19H05281, JP21H05277, and JP22H00401 (to T.I.); JP19H05707 (to N.N.N.); JP19H05706 and JP21H004771 (to M.K.); AMED grant numbers JP20gm1110010 and JP223fa627001 (to T.I.), JP22gm1410004h0003 (to M.K.); JST CREST grant number JPMJCR20E3 (to N.N.N.); and by the Takeda Science Foundation (to T.I., N.N.N., and M.K.).
Author contributions: T.I., N.N.N., and M.K. designed and directed the study. R.I., S.I., and G.M. carried out the biochemical experiments. S.K.-H. generated KO cell lines. N.N.N. conducted structural analyses with AF2. T.I., N.N.N., and M.K. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
REFERENCES AND NOTES
- 1.A. Hershko, A. Ciechanover, The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 2.O. Kerscher, R. Felberbaum, M. Hochstrasser, Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006). [DOI] [PubMed] [Google Scholar]
- 3.L. Cappadocia, C. D. Lima, Ubiquitin-like protein conjugation: Structures, chemistry, and mechanism. Chem. Rev. 118, 889–918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.K. N. Swatek, D. Komander, Ubiquitin modifications. Cell Res. 26, 399–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.N. Zheng, N. Shabek, Ubiquitin Ligases: Structure, function, and regulation. Annu. Rev. Biochem. 86, 129–157 (2017). [DOI] [PubMed] [Google Scholar]
- 6.M. Komatsu, T. Chiba, K. Tatsumi, S. Iemura, I. Tanida, N. Okazaki, T. Ueno, E. Kominami, T. Natsume, K. Tanaka, A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 23, 1977–1986 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Y. Gerakis, M. Quintero, H. Li, C. Hetz, The UFMylation system in proteostasis and beyond. Trends Cell Biol. 29, 974–986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.S. H. Kang, G. R. Kim, M. Seong, S. H. Baek, J. H. Seol, O. S. Bang, H. Ovaa, K. Tatsumi, M. Komatsu, K. Tanaka, C. H. Chung, Two novel ubiquitin-fold modifier 1 (Ufm1)-specific proteases, UfSP1 and UfSP2. J. Biol. Chem. 282, 5256–5262 (2007). [DOI] [PubMed] [Google Scholar]
- 9.D. Millrine, T. Cummings, S. P. Matthews, J. J. Peter, H. M. Magnussen, S. M. Lange, T. Macartney, F. Lamoliatte, A. Knebel, Y. Kulathu, Human UFSP1 is an active protease that regulates UFM1 maturation and UFMylation. Cell Rep. 40, 111168 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.K. Tatsumi, Y.-S. Sou, N. Tada, E. Nakamura, S.-I. Iemura, T. Natsume, S. H. Kang, C. H. Chung, M. Kasahara, E. Kominami, M. Yamamoto, K. Tanaka, M. Komatsu, A novel type of E3 ligase for the Ufm1 conjugation system. J. Biol. Chem. 285, 5417–5427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Y. Cai, W. Pi, S. Sivaprakasam, X. Zhu, M. Zhang, J. Chen, L. Makala, C. Lu, J. Wu, Y. Teng, B. Pace, D. Tuan, N. Singh, H. Li, UFBP1, a key component of the Ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLOS Genet. 11, e1005643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R. Yang, H. Wang, B. Kang, B. Chen, Y. Shi, S. Yang, L. Sun, Y. Liu, W. Xiao, T. Zhang, J. Yang, Y. Zhang, M. Zhu, P. Xu, Y. Chang, Y. Jia, Y. Huang, CDK5RAP3, a UFL1 substrate adaptor, is crucial for liver development. Development 146, dev169235 (2019). [DOI] [PubMed] [Google Scholar]
- 13.J. J. Peter, H. M. Magnussen, P. A. DaRosa, D. Millrine, S. P. Matthews, F. Lamoliatte, R. Sundaramoorthy, R. R. Kopito, Y. Kulathu, A non-canonical scaffold-type E3 ligase complex mediates protein UFMylation. EMBO J. 41, e111015 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.B. Qin, J. Yu, S. Nowsheen, M. Wang, X. Tu, T. Liu, H. Li, L. Wang, Z. Lou, UFL1 promotes histone H4 ufmylation and ATM activation. Nat. Commun. 10, 1242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.H. Zhu, B. Bhatt, S. Sivaprakasam, Y. Cai, S. Liu, S. K. Kodeboyina, N. Patel, N. M. Savage, A. Sharma, R. J. Kaufman, H. Li, N. Singh, Ufbp1 promotes plasma cell development and ER expansion by modulating distinct branches of UPR. Nat. Commun. 10, 1084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R. Ishimura, M. Obata, S. Kageyama, J. Daniel, K. Tanaka, M. Komatsu, A novel approach to assess the ubiquitin-fold modifier 1-system in cells. FEBS Lett. 591, 196–204 (2017). [DOI] [PubMed] [Google Scholar]
- 17.K. Tatsumi, H. Yamamoto-Mukai, R. Shimizu, S. Waguri, Y. S. Sou, A. Sakamoto, C. Taya, H. Shitara, T. Hara, C. H. Chung, K. Tanaka, M. Yamamoto, M. Komatsu, The Ufm1-activating enzyme Uba5 is indispensable for erythroid differentiation in mice. Nat. Commun. 2, 181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M. Zhang, X. Zhu, Y. Zhang, Y. Cai, J. Chen, S. Sivaprakasam, A. Gurav, W. Pi, L. Makala, J. Wu, B. Pace, D. Tuan-Lo, V. Ganapathy, N. Singh, H. Li, RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 22, 1922–1934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.M. Muona, R. Ishimura, A. Laari, Y. Ichimura, T. Linnankivi, R. Keski-Filppula, R. Herva, H. Rantala, A. Paetau, M. Pöyhönen, M. Obata, T. Uemura, T. Karhu, N. Bizen, H. Takebayashi, S. M. Kee, M. J. Parker, N. Akawi, J. M. Rae, M. E. Hurles, D. D. D. Study, O. Kuismin, M. I. Kurki, A.-K. Anttonen, K. Tanaka, A. Palotie, S. Waguri, A.-E. Lehesjoki, M. Komatsu, Biallelic variants in UBA5 link dysfunctional UFM1 ubiquitin-like modifier pathway to severe infantile-onset encephalopathy. Am. J. Hum. Genet. 99, 683–694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.J. Zhang, H. Zhu, S. Liu, M. Quintero, T. Zhu, R. Xu, Y. Cai, Y. Han, H. Li, Deficiency of murine UFM1-specific E3 ligase causes microcephaly and inflammation. Mol. Neurobiol. 59, 6363–6372 (2022). [DOI] [PubMed] [Google Scholar]
- 21.E. Colin, J. Daniel, A. Ziegler, J. Wakim, A. Scrivo, T. B. Haack, S. Khiati, A.-S. Denommé, P. Amati-Bonneau, M. Charif, V. Procaccio, P. Reynier, K. A. Aleck, L. D. Botto, C. L. Herper, C. S. Kaiser, R. Nabbout, S. N'Guyen, J. A. Mora-Lorca, B. Assmann, S. Christ, T. Meitinger, T. M. Strom, H. Prokisch; FREX Consortium, A. Miranda-Vizuete, G. F. Hoffmann, G. Lenaers, P. Bomont, E. Liebau, D. Bonneau, Biallelic variants in UBA5 reveal that disruption of the UFM1 cascade can result in early-onset encephalopathy. Am. J. Hum. Genet. 99, 695–703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.E. M. C. Hamilton, E. Bertini, L. Kalaydjieva, B. Morar, D. Doǰáková, J. Liu, A. Vanderver, J. Curiel, C. M. Persoon, D. Diodato, L. Pinelli, N. L. van der Meij, B. Plecko, S. Blaser, N. I. Wolf, Q. Waisfisz, T. E. M. Abbink, M. S. van der Knaap; Recessive H-ABC Research Group , UFM1 founder mutation in the Roma population causes recessive variant of H-ABC. Neurology 89, 1821–1828 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M. S. Nahorski, S. Maddirevula, R. Ishimura, S. Alsahli, A. F. Brady, A. Begemann, T. Mizushima, F. J. Guzmán-Vega, M. Obata, Y. Ichimura, H. S. Alsaif, S. Anazi, N. Ibrahim, F. Abdulwahab, M. Hashem, D. Monies, M. Abouelhoda, B. F. Meyer, M. Alfadhel, W. Eyaid, M. Zweier, K. Steindl, A. Rauch, S. T. Arold, C. G. Woods, M. Komatsu, F. S. Alkuraya, Biallelic UFM1 and UFC1 mutations expand the essential role of ufmylation in brain development. Brain 141, 1934–1945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.C. M. Watson, L. A. Crinnion, L. Gleghorn, W. G. Newman, R. Ramesar, P. Beighton, G. A. Wallis, Identification of a mutation in the ubiquitin-fold modifier 1-specific peptidase 2 gene, UFSP2, in an extended South African family with Beukes hip dysplasia. S. Afr. Med. J. 105, 558–563 (2015). [DOI] [PubMed] [Google Scholar]
- 25.M. Di Rocco, M. Rusmini, F. Caroli, A. Madeo, M. Bertamino, G. Marre-Brunenghi, I. Ceccherini, Novel spondyloepimetaphyseal dysplasia due to UFSP2 gene mutation. Clin. Genet. 93, 671–674 (2018). [DOI] [PubMed] [Google Scholar]
- 26.K. Lemaire, R. F. Moura, M. Granvik, M. Igoillo-Esteve, H. E. Hohmeier, N. Hendrickx, C. B. Newgard, E. Waelkens, M. Cnop, F. Schuit, Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic beta cells from ER stress-induced apoptosis. PLOS ONE 6, e18517 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R. Ishimura, A. H. El-Gowily, D. Noshiro, S. Komatsu-Hirota, Y. Ono, M. Shindo, T. Hatta, M. Abe, T. Uemura, H.-C. Lee-Okada, T. M. Mohamed, T. Yokomizo, T. Ueno, K. Sakimura, T. Natsume, H. Sorimachi, T. Inada, S. Waguri, N. N. Noda, M. Komatsu, The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3. Nat. Commun. 13, 7857 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Banerjee, J. K. Varga, M. Kumar, O. Schueler-Furman, R. Wiener, AlphaFold2-based fusion design deciphers crucial role of the E3 UFL1 N-terminal helix in E2 UFC1 binding and ufmylation. bioRxiv 2022.09.15.508077 (2022).
- 29.C. P. Walczak, D. E. Leto, L. Zhang, C. Riepe, R. Y. Muller, P. A. DaRosa, N. T. Ingolia, J. E. Elias, R. R. Kopito, Ribosomal protein RPL26 is the principal target of UFMylation. Proc. Natl. Acad. Sci. U.S.A. 116, 1299–1308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.L. Wang, Y. Xu, H. Rogers, L. Saidi, C. T. Noguchi, H. Li, J. W. Yewdell, N. R. Guydosh, Y. Ye, UFMylation of RPL26 links translocation-associated quality control to endoplasmic reticulum protein homeostasis. Cell Res. 30, 5–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.L. Wang, Y. Xu, S. Yun, Q. Yuan, P. Satpute-Krishnan, Y. Ye, SAYSD1 senses UFMylated ribosome to safeguard co-translational protein translocation at the endoplasmic reticulum. Cell Rep. 42, 112028 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.M. Stephani, L. Picchianti, A. Gajic, R. Beveridge, E. Skarwan, V. S. de Medina Hernandez, A. Mohseni, M. Clavel, Y. Zeng, C. Naumann, M. Matuszkiewicz, E. Turco, C. Loefke, B. Li, G. Dürnberger, M. Schutzbier, H. T. Chen, A. Abdrakhmanov, A. Savova, K.-S. Chia, A. Djamei, I. Schaffner, S. Abel, L. Jiang, K. Mechtler, F. Ikeda, S. Martens, T. Clausen, Y. Dagdas, A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. eLife 9, e58396 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.J. R. Liang, E. Lingeman, T. Luong, S. Ahmed, M. Muhar, T. Nguyen, J. A. Olzmann, J. E. Corn, A genome-wide ER-phagy screen highlights key roles of mitochondrial metabolism and ER-resident UFMylation. Cell 180, 1160–1177.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.L. Pirone, W. Xolalpa, J. O. Sigurethsson, J. Ramirez, C. Pérez, M. González, A. R. de Sabando, F. Elortza, M. S. Rodriguez, U. Mayor, J. V. Olsen, R. Barrio, J. D. Sutherland, A comprehensive platform for the analysis of ubiquitin-like protein modifications using in vivo biotinylation. Sci. Rep. 7, 40756 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.P. Padala, W. Oweis, B. Mashahreh, N. Soudah, E. Cohen-Kfir, E. A. Todd, C. E. Berndsen, R. Wiener, Novel insights into the interaction of UBA5 with UFM1 via a UFM1-interacting sequence. Sci. Rep. 7, 508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.N. Soudah, P. Padala, F. Hassouna, M. Kumar, B. Mashahreh, A. A. Lebedev, M. N. Isupov, E. Cohen-Kfir, R. Wiener, An N-terminal extension to UBA5 adenylation domain boosts UFM1 activation: Isoform-specific differences in ubiquitin-like protein activation. J. Mol. Biol. 431, 463–478 (2019). [DOI] [PubMed] [Google Scholar]
- 37.T. Mizushima, K. Tatsumi, Y. Ozaki, T. Kawakami, A. Suzuki, K. Ogasahara, M. Komatsu, E. Kominami, K. Tanaka, T. Yamane, Crystal structure of Ufc1, the Ufm1-conjugating enzyme. Biochem. Biophys. Res. Commun. 362, 1079–1084 (2007). [DOI] [PubMed] [Google Scholar]
- 38.W. Oweis, P. Padala, F. Hassouna, E. Cohen-Kfir, D. R. Gibbs, E. A. Todd, C. E. Berndsen, R. Wiener, Trans-binding mechanism of ubiquitin-like protein activation revealed by a UBA5-UFM1 complex. Cell Rep. 16, 3113–3120 (2016). [DOI] [PubMed] [Google Scholar]
- 39.M. Kumar, P. Padala, J. Fahoum, F. Hassouna, T. Tsaban, G. Zoltsman, S. Banerjee, E. Cohen-Kfir, M. Dessau, R. Rosenzweig, M. N. Isupov, O. Schueler-Furman, R. Wiener, Structural basis for UFM1 transfer from UBA5 to UFC1. Nat. Commun. 12, 5708 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.F. Scavone, S. C. Gumbin, P. A. Da Rosa, R. R. Kopito, RPL26/uL24 UFMylation is essential for ribosome-associated quality control at the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 120, e2220340120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. W. Senior, R. Evans, J. Jumper, J. Kirkpatrick, L. Sifre, T. Green, C. Qin, A. Zidek, A. W. R. Nelson, A. Bridgland, H. Penedones, S. Petersen, K. Simonyan, S. Crossan, P. Kohli, D. T. Jones, D. Silver, K. Kavukcuoglu, D. Hassabis, Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020). [DOI] [PubMed] [Google Scholar]
- 42.K. Tunyasuvunakool, J. Adler, Z. Wu, T. Green, M. Zielinski, A. Žídek, A. Bridgland, A. Cowie, C. Meyer, A. Laydon, S. Velankar, G. J. Kleywegt, A. Bateman, R. Evans, A. Pritzel, M. Figurnov, O. Ronneberger, R. Bates, S. A. A. Kohl, A. Potapenko, A. J. Ballard, B. Romera-Paredes, S. Nikolov, R. Jain, E. Clancy, D. Reiman, S. Petersen, A. W. Senior, K. Kavukcuoglu, E. Birney, P. Kohli, J. Jumper, D. Hassabis, Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R. Evans, M. O’Neill, A. Pritzel, N. Antropova, A. Senior, T. Green, A. Žídek, R. Bates, S. Blackwell, J. Yim, O. Ronneberger, S. Bodenstein, M. Zielinski, A. Bridgland, A. Potapenko, A. Cowie, K. Tunyasuvunakool, R. Jain, E. Clancy, P. Kohli, J. Jumper, D. Hassabis, Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021.10.04.463034 (2021).
- 44.M. Varadi, S. Anyango, M. Deshpande, S. Nair, C. Natassia, G. Yordanova, D. Yuan, O. Stroe, G. Wood, A. Laydon, A. Žídek, T. Green, K. Tunyasuvunakool, S. Petersen, J. Jumper, E. Clancy, R. Green, A. Vora, M. Lutfi, M. Figurnov, A. Cowie, N. Hobbs, P. Kohli, G. Kleywegt, E. Birney, D. Hassabis, S. Velankar, AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.H. M. Yoo, S. H. Kang, J. Y. Kim, J. E. Lee, M. W. Seong, S. W. Lee, S. H. Ka, Y. S. Sou, M. Komatsu, K. Tanaka, S. T. Lee, D. Y. Noh, S. H. Baek, Y. J. Jeon, C. H. Chung, Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol. Cell 56, 261–274 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Z. M. Eletr, D. T. Huang, D. M. Duda, B. A. Schulman, B. Kuhlman, E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat. Struct. Mol. Biol. 12, 933–934 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Y. Qiu, K. Hofmann, J. E. Coats, B. A. Schulman, S. E. Kaiser, Binding to E1 and E3 is mutually exclusive for the human autophagy E2 Atg3. Protein Sci. 22, 1691–1697 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.N. Garreau de Loubresse, I. Prokhorova, W. Holtkamp, M. V. Rodnina, G. Yusupova, M. Yusupov, Structural basis for the inhibition of the eukaryotic ribosome. Nature 513, 517–522 (2014). [DOI] [PubMed] [Google Scholar]
- 49.S. Habisov, J. Huber, Y. Ichimura, M. Akutsu, N. Rogova, F. Loehr, D. G. McEwan, T. Johansen, I. Dikic, V. Doetsch, M. Komatsu, V. V. Rogov, V. Kirkin, Structural and functional analysis of a novel interaction motif within UFM1-activating Enzyme 5 (UBA5) required for binding to ubiquitin-like proteins and Ufmylation. J. Biol. Chem. 291, 9025–9041 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.K. Yanagitani, Y. Kimata, H. Kadokura, K. Kohno, Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science 331, 586–589 (2011). [DOI] [PubMed] [Google Scholar]
- 51.S. Kanda, K. Yanagitani, Y. Yokota, Y. Esaki, K. Kohno, Autonomous translational pausing is required for XBP1u mRNA recruitment to the ER via the SRP pathway. Proc. Natl. Acad. Sci. U.S.A. 113, E5886–E5895 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.P. Han, Y. Shichino, T. Schneider-Poetsch, M. Mito, S. Hashimoto, T. Udagawa, K. Kohno, M. Yoshida, Y. Mishima, T. Inada, S. Iwasaki, Genome-wide survey of ribosome collision. Cell Rep. 31, 107610 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.C. C.-C. Wu, A. Peterson, B. Zinshteyn, S. Regot, R. Green, Ribosome collisions trigger general stress responses to regulate cell fate. Cell 182, 404–416.e14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.K. K. Kostova, K. L. Hickey, B. A. Osuna, J. A. Hussmann, A. Frost, D. E. Weinberg, J. S. Weissman, CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science 357, 414–417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.K. von der Malsburg, S. Shao, R. S. Hegde, The ribosome quality control pathway can access nascent polypeptides stalled at the Sec61 translocon. Mol. Biol. Cell 26, 2168–2180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.K. Braunger, S. Pfeffer, S. Shrimal, R. Gilmore, O. Berninghausen, E. C. Mandon, T. Becker, F. Förster, R. Beckmann, Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 360, 215–219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli, D. Hassabis, Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.M. Mirdita, M. Steinegger, J. Söding, MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics 35, 2856–2858 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. Narita, T. Denk, Y. Matsuo, T. Sugiyama, C. Kikuguchi, S. Ito, N. Sato, T. Suzuki, S. Hashimoto, I. Machova, P. Tesina, R. Beckmann, T. Inada, A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation. Nat. Commun. 13, 6411 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7