Abstract
In contrast to mice, zebrafish have an exceptional yet elusive ability to replenish lost β cells in adulthood. Understanding this framework would provide mechanistic insights for β cell regeneration, which may be extrapolated to humans. Here, we characterize a krt4-expressing ductal cell type, which is distinct from the putative Notch-responsive cells, showing neogenic competence and giving rise to the majority of endocrine cells during postembryonic development. Furthermore, we demonstrate a marked ductal remodeling process featuring a Notch-responsive to krt4+ luminal duct transformation during late development, indicating several origins of krt4+ ductal cells displaying similar transcriptional patterns. Single-cell transcriptomics upon a series of time points during β cell regeneration unveil a previously unrecognized dlb+ transitional endocrine precursor cell, distinct regulons, and a differentiation trajectory involving cellular shuffling through differentiation and dedifferentiation dynamics. These results establish a model of zebrafish pancreatic endocrinogenesis and highlight key values of zebrafish for translational studies of β cell regeneration.
krt4+ ductal progenitors and dlb+ endocrine precursors serve as a direct origin for β cells in the zebrafish pancreas.
INTRODUCTION
During murine and human embryonic development of the pancreas, the ductal cells reside in the trunk of the branched epithelium and can up-regulate the transcription factors Neurog3 and Fev to induce early and late endocrine progenitor cells, respectively (1–5). The late progenitor cells are dedicated to specific committed endocrine cell types based on a sophisticated balance among a wide range of lineage-specific transcription factors for final cell fate acquisition (3, 6–8). This differentiation process is sustained until the neonatal stage and markedly declines soon after birth. In mouse, upon β cell injury, which can be induced by numerous genetic and pharmacological strategies, the proliferation of remnant β cells serves as the primary source of regenerative β cells, indicated by pulse-chase experiment and lineage fate mapping studies (9, 10). Notably, the induction of massive β cell loss, overexpression of a key transcription factor cocktail using genetic approaches, or inhibition of cell differentiation blockades can stimulate a variety of cell types to up-regulate insulin expression, highlighting the astonishing cell plasticity in a broad range of cells for reprogramming toward pancreatic β cell fate (11–19). Some studies suggest that the pancreatic duct might harbor a latent progenitor population to give rise to insulin-producing cells under diseased conditions in adults (20–24). However, this is still under debate due to a lack of reliable ductal cell–specific lineage tracing tools in mouse models and speculated endocrine cell contaminations in human samples (20, 25, 26).
Zebrafish, a widely used model organism for regeneration studies, can fully recover from extreme β cell loss in both larval and adult stages within 1 month (27, 28). Lineage tracing experiments with temporal labeling of β cells indicated that although proliferation of preexisting β cells can be accelerated by drug treatment, there are other sources contributing to the restoration of the majority of insulin-producing cells upon β cell ablation (29, 30). The main sources of developing β cells and regenerative insulin–producing cells have yet to be determined by genetic fate mapping. However, prevailing evidence from lineage tracing or similar studies using transgenic zebrafish strains indicates that a small fraction of endocrine cells in the secondary islets can be formed from Notch-responsive cells (also referred to as intrapancreatic ductal cells or centroacinar cells) (31–33); however, they do not explain the total contribution for the endocrine pancreas. Thus, broader or different ductal/progenitor cell–specific Cre lines are needed to expand the genetic fate mapping to include the majority of endocrine cell formation (30, 34–38). Moreover, recent studies suggested that sst1.1+ endocrine cells might convert to insulin-producing cells upon β cell ablation (37, 39). However, evidence from lineage tracing is still lacking.
During zebrafish development, the early pancreatic endocrine cells arise from the dorsal pancreatic bud and culminate at around 36 hours after fertilization (40). The ventral pancreatic bud subsequently engulfs the dorsal bud and gives rise to exocrine tissues and additional endocrine cells that coalesce with the principal islet in the head of the pancreas (40). At around 5 days postfertilization (dpf), in parallel with the start of food intake, secondary islets in the pancreatic tail region occasionally appear, and more β cells will progressively form in both the principal and secondary islets through adulthood as a response to increased nutrient consumption (41). Using pharmacological and genetic approaches, researchers have shown that the downregulation of Notch signaling can lead to a substantial increase in β cells in the secondary islets (31, 42, 43). Lineage fate mapping experiments using the tp1 (a Notch-responsive viral element) promoter to drive CreERT2 commonly label around 75% of Notch-responsive cells but fail to label the majority of endocrine cells in the anterior region of the principal and the secondary islets, indicating that other surrogate cell sources might exist with a unique reprogramming mechanism underlying the differentiation process (32).
To resolve this apparent disparity, we combined a variety of state-of-the-art tools including single-cell RNA sequencing (RNA-seq), newly generated knock-in and transgenic lines for lineage fate mapping, and targeted cell ablation to characterize a previously undefined ductal cell type, i.e., krt4+ ductal cells that function as the ductal progenitors of endocrine cells. This type of ductal cell has a transcriptional signature, morphological profile, and organization distinct from the putative Notch-responsive ductal cells. Briefly, these krt4+ ductal cells display an alternative Notch-signaling state and assemble together to gradually form luminal structures throughout the pancreas. These ductal cells, coupled with a subset of neurod1+ endocrine cells, can convert to bihormonal (also known as ins+&sst1.1+ hybrid) cells after β cell ablation. Moreover, the krt4+. luminal duct has several independent origins and eventually forms large connecting tubular structures, with a common transcriptional pattern in the process of ductal tree morphogenesis. Last, by sequencing the krt4 lineage at the single-cell level in the basal state and after β cell ablation, we uncovered dlb+ endocrine progenitor cells and several transitional cell states along the newly defined differentiation trajectory toward bihormonal and mature cells. By harnessing the up-regulated molecular events in the transitional cell clusters, e.g., by inhibiting the phosphatidylinositol 3-kinase (PI3K)–AKT–mammalian target of rapamycin (mTOR) pathway, we can induce krt4+ duct–derived insulin-producing cells. In all, we generate a blueprint of zebrafish endocrinogenesis and put forward a redefined model of endocrine cellular origin that encompasses an emerging concept linking it with ductal remodeling. Our single-cell analyses show that endocrinogenesis is largely conserved when comparing adult zebrafish with embryonic mouse and human and uncover previously unknown key molecular events governing β cell formation and regeneration.
RESULTS
Single-cell transcriptomics identifies ductal heterogeneity and krt4+ duct in adult zebrafish
For a preliminary view of the cellular composition of the islet and the surrounding niche, we first analyzed one public single-cell RNA-seq dataset of the adult zebrafish principal islet (44). After filtering and quality control, we identified 17 clusters, two of which showed expression of various well-known ductal markers including sox9b, nkx6.1, cftr, and anxa4 (Fig. 1, A and B). We noted that these two clusters differ in terms of Notch-signaling responsive genes, with the larger cluster expressing her6 and her9 to a low extent, while the smaller cluster preferentially expressed her15.1. Both clusters are id2a+, indicating that they are both bone morphogenetic protein (BMP) responsive (Fig. 1C). Next, we demonstrated that epcam, cldn7b, cd9b, cx30.3, and onecut1 were expressed in both types of ducts, while clcn1b and igf2a presented highest expression in her15.1+ duct (fig. S1, A to H). We also identified vasnb, cdh17, and cldnc, which code for membrane-bound proteins, as previously undefined pan-ductal marker genes (Fig. 1, B and C). Immunostaining for Vasnb, Cdh17, and Cldnc, as well as hybridization chain reaction (HCR) 3.0 in situ immunofluorescence of anxa4 in the Tg(Tp1bglob:eGFP)um14Tg [abbreviated as Tg(tp1:EGFP)] larvae at 6 dpf, displayed two types of morphologically distinct ductal cells with or without green fluorescent protein (GFP) labeling, consolidating the prior in silico analyses (Fig. 1, D to G) (45, 46).
Fig. 1. Ductal heterogeneity in the adult zebrafish pancreas.
(A) Uniform Manifold Approximation and Projection (UMAP) plot showing the different cell clusters. The classifications were based on known marker genes highly expressed in each cell type in the zebrafish pancreas. The two clusters of pancreatic ductal cell types are denoted by dashed lines and arrows. NK, natural killer. (B and C) Feature plot shown in UMAP embedding of selected ductal markers (B), Notch and BMP downstream genes enriched in pancreatic ducts, and novel ductal markers (C). (D) Immunostaining using a Vasnb antibody in Tg(tp1:EGFP) transgenic larva showing the ducts in the hepatopancreatic biliary system. The dashed lines outline the organs in different colors, i.e., pancreas (yellow), gallbladder (magenta), liver (cyan), and intestine (white). Scale bars, 200 μm. (E to G) Representative confocal images of immunostaining of Cdh17 and Cldnc in Tg(tp1:EGFP) larvae (E) and (F) and in situ hybridization for anxa4 in Tg(tp1:EGFP) larvae (G) with Z-projection (E′) and (E″), (F′) and (F″), and (G′). The pancreas (yellow), islet (cyan), and non–Notch responsive ducts (white) were outlined by dashed lines in different colors. The Notch-responsive intrapancreatic ducts are indicated by arrows (orange). Scale bars, 200 and 40 μm (magnification). (H) Double transgenic for tp1:EGFP and tp1:H2BmCherry with anti-Vasnb staining showing the labeling pattern in the duct and islets in the larvae pancreas. The pancreata, extrapancreatic duct, intermediate duct, and islet are highlighted with yellow, cyan, white, and orange dashed lines, respectively. Scale bars, 200 μm (left) and 40 μm (right). A, anterior; P, posterior; D, dorsal; V, ventral. (I) In situ hybridization for krt4 showing the extrapancreatic-to-intermediate duct system with bifurcation in 6 dpf larva. The insets are magnified Z-projections with white dashed lines outlining the tree-like structure of the intermediate duct. Scale bars, 200 and 40 μm (insets).
Immunostaining of Tg(tp1:EGFP);Tg(tp1:H2BmCherry) larvae at 22 dpf showed that the ducts in the anterior region of the pancreas present as a direct continuum of the extrapancreatic duct and have a luminal structure, as well as being devoid of enhanced GFP (EGFP) signal [although contained some H2BmCherry, which accumulates since histone 2B (H2B) stabilizes mCherry] (Fig. 1H). As these cells are positioned in between the extrapancreatic duct and intrapancreatic duct, we named them intermediate duct for ease of description. Conversely, the predefined Notch-responsive intrapancreatic ducts present long protrusions surrounding the principal islet and extending toward the pancreatic tail region that express higher mCherry signal. Notably, we also saw that a substantial number of the endocrine cells residing in the principal islet are weakly mCherry+, which is in line with the single-cell RNA-seq data showing the expression of her15.1 in a subset of endocrine cells. Furthermore, we noted that a cytokeratin gene, krt4, is a marker gene of the large ductal cluster (Fig. 1C). Using HCR3.0 in situ hybridization, we found that extrapancreatic duct and intermediate duct are both krt4+. The intermediate ducts branch out to form a tree-like structure with a bifurcation toward the principal islet (Fig. 1I).
To better explore the pancreatic ductal system, we performed immunostaining in existing reporter lines or in situ hybridization in 6 dpf larvae for markers identified in the single-cell RNA-seq data from the adult zebrafish principal islet (i.e., not a direct validation due to different stages). The immunostaining of Vasnb in Tg(-2.5cadherin17:mCherry) [abbreviated as Tg(cdh17:H2BmCherry)] and Tg(nkx2.2a:mEGFP) suggested mosaic cdh17 and nkx2.2 expression patterns in the intermediate duct (fig. S2, A and B). We also observed yap activity and pdx1 expression in both ducts and in a subset of the endocrine cells (fig. S2, C to E). In situ hybridization and immunofluorescent staining also demonstrated the expression of id2a, sox9b, hnf1ba, and onecut1 in the intermediate duct (fig. S1, I to L). Notably, the ciliated cell master regulator, foxj1a, is restricted in the extrapancreatic duct as shown by both the transgenic reporter and the in situ hybridization (fig. S2F). Hence, the krt4+ duct can be also molecularly classified into foxj1a+ or foxj1a− categories on top of the anatomical architecture. Together, we are able to visualize and molecularly define the whole ductal system in the zebrafish pancreas and distinguish novel ductal cell types, highlighting a ductal cell type showing specific krt4 expression and differential Notch target gene activation pattern.
Lineage tracing using canonical duct cell tracers only tracked a limited number of endocrine cells
To address from which origin new postembryonic endocrine cells arise, we first used the Tg(EPV.Tp1-Mmu.Hbb:Cre-ERT2,cryaa:mCherry)s959 [abbreviated as Tg(tp1:CreERT2)] line, which has been commonly used as a tracer of Notch-responsive cells and crossed it with two different responder lines: Tg(ubi:loxp-EGFP-stop-loxp-mCherry) [abbreviated as Tg(ubi:Switch)] and Tg(ubi:loxp-CFP-stop-loxp-H2BmCherry) [abbreviated as Tg(ubi:CSHm)]. We treated larvae with 4-hydroxytamoxifen (4-OHT) using maximum dosage (20 μM) at 2 or 3 dpf for 24 hours and analyzed the fate mapping results at 6 dpf. Apart from labeling the Notch-responsive intrapancreatic duct, neither extrapancreatic nor intermediate duct could be lineage traced or genetically labeled during development (Fig. 2, A and B). By using a nitroreductase/metronidazole (NTR/MTZ)–mediated cell ablation model (ins:flagNTR), we saw a similar result after β cell ablation with no lineage-traced insulin or somatostatin1.1 (Sst1.1)–positive cells, and the extrapancreatic and intermediate duct also appeared devoid of lineage tracing (Fig. 2, C and D). However, long-term lineage tracing (23 dpf) showed that a small proportion of the ductal cells located in the duct with a luminal structure could be genetically labeled (Fig. 2, E and F), in accordance with previous work and that we can see a small proportion of endocrine cells being labeled in a similar set up (fig. S10, B and D) (31). These results suggested that the tp1 driven ductal tracer is useful in tracking the Notch-responsive cell lineage, whereas it is not qualified to trace the other ductal cells. It also prompts us to examine whether the previously uncharacterized ductal cell type might be a major source of endocrine cells in the pancreas.
Fig. 2. Lineage tracing showed limited Notch-responsive duct–to–endocrine cells conversion at the principal islet.
(A and B) Tg(tp1:CreERT2) was crossed to two different color-switch lines under the control of ubiquitin B promoter. H2BmCherry/mCherry expression was specifically induced in tp1+ Notch-responsive ducts and their progenies with 20 μM 4-OHT administered from 3 to 4 dpf, as visualized with Z-projections 6 dpf. Yellow, white, and cyan dashed lines indicate the pancreata, Notch-responsive intrapancreatic ducts, and non–Notch-responsive duct anterior to the principal islet, respectively. Scale bars, 100 μm. (C and D) Schematics and confocal Z-projections of tp1 lineage–traced cells under β cell ablation condition. Tentative overlap in the Z-projection (the mCherry+ cell indicated by orange arrow) was confirmed to not coexpress Sst1.1 and insulin (Ins) in single planes. Scale bars, 40 μm. The yellow and blue dashed lines indicate the principal islet and extra- and intermediate pancreatic duct, respectively. (E and F) Long-term lineage tracing in tp1:CreERT2;ubi:Switch with 4-OHT treatment 3 to 4 dpf. Confocal Z-projections of pancreatic head (E) and tail (F) 23 dpf. Yellow dashed lines indicate the principal islet. Scale bars, 40 μm. (G and H) cftr:Gal4;UAS:EGFP;tp1:H2BmCherry indicate cftr+ cells 6 dpf (G) and 22 dpf (H). The ductal cells are highlighted using cyan dashed lines. Scale bars, 40 μm. (I and J) cftr:Gal4;UAS:EGFP;UAS:Cre;ubi:Switch indicate cftr+ lineage traced cells 14 dpf (I) and 20 dpf (J). Yellow dashed lines in (I) indicate the pancreatic head (magnifications in right panels). The yellow arrow points to lineage-traced endocrine cells. Yellow arrows in (J) indicate lineage-traced Gcg+ cells in head and tail regions. Scale bars, 40 μm.
To test our hypothesis, we used another duct tracer driven by the cftr:Gal4 transgene. Staining for Vasnb in TgBAC(cftr:Gal4);Tg(UAS:EGFP);Tg(tp1:H2BmCherry) indicated a mosaic labeling pattern in the intermediate duct by the cftr:Gal4 transgene (Fig. 2, G and H). Long-term lineage tracing experiments using TgBAC (cftr:Gal4);Tg(UAS:EGFP);Tg(UAS:Cre);Tg(ubi:Switch) juveniles demonstrated that a large number of intermediate duct can be genetically labeled, together with scattered fate-mapped endocrine cells sitting in proximity to the duct (Fig. 2, I and J). However, the mosaicism due to the nature of the cftr transgene and/or Gal4/UAS system may underestimate the number of new endocrine cells from the non–Notch-responsive duct, i.e., the intermediate duct. In sum, our lineage tracing results suggested that a surrogate source other than Notch-responsive duct serve as ductal progenitors with neogenic competence.
Genetic fate mapping and targeted cell ablation using krt4+ knock-in iCre/CreERT2 lines
Since the non–Notch-responsive ductal cells are krt4+ and to lineage trace these ducts and their descendants, we leveraged two recently published CRISPR-Cas9 3′ knock-in tracers generated in our laboratory, i.e., TgKI(krt4-p2a-mNeonGreen-t2a-iCre) [abbreviated as TgKI(krt4:iCre)] and TgKI(krt4-p2a-EGFP-t2a-CreERT2) [abbreviated as TgKI(krt4:CreERT2)] (47). The different Cre lines complement each other in that the iCre line gives high recombination efficiency but may have minor leakage, whereas the CreERT2 line enables tight control but with lower recombination efficiency. These knock-in lines have been shown to recapitulate krt4 expression in the skin and intestine, and we also confirmed the fluorescent signals in the pancreatic ducts with luminal structure, specifically in the extrapancreatic duct in 6 dpf larvae and later also in all large luminal ducts throughout the pancreatic tail (Fig. 3, A and B). For the lineage tracing experiment, we first crossed the TgKI(krt4:iCre) with the Tg(ins:loxP-CFP-stop-loxP-H2BmCherry)KI141 [abbreviated as Tg(ins:CSHm)] responder (Fig. 3C). The rationale for this is that, in the krt4-expressing ductal cells, the genetic cassettes in between two loxP sites will be excised and only the lineage-traced β cells would have H2BmCherry expressed in the nucleus. Therefore, the first phase of β cell differentiation derived from the dorsal bud and other krt4− sources would retain the cyan fluorescent protein (CFP) expression, whereas the β cells derived from the krt4+ duct would be H2BmCherry+. We observed that the CFP-labeled old β cells are mostly located at the posterior region of the principal islet (Fig. 3, D and E), in line with previous findings (30). In contrast, the krt4-derived β cells (which in 3-to-4-week-old larva made up >70% of all β cells) are mostly located anterior to the β cells derived from a krt4− origin (table S1). With staining for Vasnb, we could readily visualize that the krt4-derived β cells (magenta) are budding out from the intermediate duct (Fig. 3, D and E). Moreover, we saw that β cells within the secondary islets are mostly H2BmCherry+, although intermingled with a few scattered CFP+ β cells (indicative of Notch-responsive duct–derived β cells) (Fig. 3F).
Fig. 3. Lineage tracing and targeted cell ablation using krt4 knock-in iCre line.
(A and B) Confocal images of krt4+ ductal cells in 6 dpf larvae (A) and 30 dpf juvenile fish (B). The brown, white, and cyan dashed lines indicate the pancreata, intestine, and the principal islet, respectively. The yellow arrows indicate large luminal krt4+ ducts throughout the pancreas, i.e., even in the pancreatic tail as the fish grow older. Scale bars, 100 μm. (C) The Cre/loxP strategy used to demonstrate krt4+ ductal cell–derived β cells. krt4-p2a-mNeonGreen-t2a-iCre was crossed to a color-switch line under the control of the insulin promoter. H2BmCherry is specifically induced in krt4-derived β cells, while β cells from other origins express CFP. (D and E) Z-projection showing differential distribution of β cells from krt4− (green) and krt4+ (magenta) origins, 21 dpf (D) and 25 dpf (E). The ductal trees are visualized with anti-Vasnb staining. Scattered β cells budding out from the intermediate duct are highlighted with red arrows. White dashed lines indicate the principal islet. Scale bars, 40 μm. (F) Confocal images of krt4-derived β cells (magenta) in the secondary islets. Scale bars, 40 μm. (G to I) Design of targeted cell ablation of krt4-derived β cells by induction of DTA (G) with representative confocal images [(H) and (I)]. Cyan and yellow dashed lines in the Z-projection (H) indicate the area with surviving β cells at the posterior part of the principal islet and the β cell debris at the anterior part, respectively. The cell with orange arrow in (I) and magnified image (I′) is activated Caspase-3–positive, shown in single plane, indicating apoptosis. Scale bars, 80 μm.
To validate the above findings, we designed a complementary targeted cell ablation approach using a transgenic line expressing an inducible diphteria toxin chain alpha (DTA) suicide transgene under the control of ins promoter (Fig. 3G). The rationale is that if krt4+ ducts serve as the progenitor cells, then their β cell descendants would be specifically ablated, while β cells of other origins would stay intact. Given that the ablation efficiency is highly dependent on the activity of ins promoter, we applied this system only in the basal state, as the ins promoter activity will diminish during regeneration and thereby not be sufficient to induce effective ablation. We crossed the TgKI(krt4:iCre) with Tg(ins:loxp-dsRed-stop-loxp-DTA) and costained the juvenile pancreata with glucagon and Sst1.1 antibodies to depict the principal islet. The β cells in the anterior regions of the principal islet were apoptotic, as displayed by the scattered dsRed debris, while a small number of β cells (indicative by intact dsRed signal) at the posterior part of principal islet were retained (Fig. 3H). Moreover, immunostaining of activated Caspase-3 indicated scattered apoptotic cells lining the border of the principal islet (Fig. 3I).
For further confirmation and to avoid possible false conclusions based on potential leakage in the iCre line, we temporally labeled the krt4+ cells in TgKI(krt4:CreERT2);Tg(ubi:CSHm) with 4-OHT from 1 to 2 dpf (Fig. 4A and fig. S3, A and B). Immunostaining of 35 dpf juveniles showed that the krt4-traced H2BmCherry+ β cells are predominantly located at the anterior region of the principal islet (Fig. 4B). Moreover, we saw extrapancreatic and intermediate duct H2BmCherry labeling, while the fluorescent signals disappeared distal to the ductal bifurcation region. This suggests that the luminal ducts along the tail of the pancreas, despite forming a direct continuum with the intermediate duct around the principal islet, originate from a source other than the intermediate duct (Fig. 4, C to F).
Fig. 4. Spatiotemporal-controlled lineage tracing of krt4+ ducts.
(A) Workflow of the Cre/loxP system used for spatiotemporal lineage tracing of krt4+ ductal cells. Fish carrying krt4-p2a-EGFP-t2a-CreERT2 (identified by green skin) are crossed to fish carrying ubi:CSHm transgenic lines (identified by blue skin). The 4-OHT is administered from 1 to 2 dpf, and we analyzed 35 dpf juvenile fish. (B) The maximum Z-projection of confocal images showing the islets from TgKI(krt4:CreERT2);Tg(ins:H2BGFP);Tg(ins:CSHm) line stained with Vasnb antibody (white) and 4′,6-diamidino-2-phenylindole (DAPI). All β cells are displayed with green fluorescence (from the ins:H2BGFP transgene) in the nucleus, and krt4-derived endocrine and ductal cells that can be traced back to krt4+ duct origin expressed H2BmCherry. Notably, the luminal duct in the pancreatic tail and Notch-responsive ductal cells are devoid of H2BmCherry. The fluorescence in krt4+ ducts is not visible because of its low intensity and the used confocal microscopy setting. (C to F) Single-plane confocal images showing the labeling pattern in three different layers/regions, with the cyan and yellow outlined regions (C) magnified in (D) to (F). Scale bars, 80 μm [(B) and (C)] and 40 μm [(D) to (F)].
Unlike mammals, neurog3 does not function as a pioneering factor for β cell neogenesis in zebrafish. Instead, previous findings showed that two other basic helix-loop-helix transcription factors, ascl1b and neurod1, play combinatorial roles as endocrine progenitors in zebrafish (48). Thereby, we assumed that the real ductal progenitor cells will up-regulate the expression of the above two transcription factors in the context requiring accelerated neogenesis, i.e., in the chemogenetic β cell ablation model [using Tg(ins:FLAG-NTR,cryaa:mCherry)s950, abbreviated as Tg(ins:flagNTR)]. We observed that upon β cell ablation, there are a substantial number of ascl1b:EGFP+ cells either positioned in the intermediate duct region or budding out from the ductal tree, a process sustained at least until 1 day postablation (dpa) in larvae (fig. S4, A and B, and tables S2 and S3). The immunostaining results using the neurod1:EGFP reporter showed similar results, consistent with the prior time-lapse imaging study indicating the presence of endocrine cells in/along the duct anterior to the principal islet during early development (fig. S4, C and D) (49).
In older larvae, to our surprise, we noted that even without β cell ablation, a large number of cells in the duct with luminal structure are ascl1b:EGFP+ and tp1:H2BmCherry−, indicating that those luminal ductal cells with lower Notch activity might be primed for differentiation (fig. S4E). We also found that the expression of ascl1b is restricted to the ductal cells (mainly intermediate duct) upon β cell ablation (fig. S4F). Immunostaining using neurod1:EGFP transgenics in the basal state showed that large numbers of EGFP+ cells are located at both the proximal and distal luminal ducts (fig. S4, G and H). Z-stack movies showed the neogenic endocrine cells (indicated by neurod1:EGFP) residing in, along, and closely attaching to the duct (movies S1 to S3). Nevertheless, when treated with low dosage of Notch inhibitor (LY411575, 1 μM), ascl1b:EGFP was markedly expressed only in the tp1-driven Notch-responsive duct, indicating that the naturally occurring neogenic program differs from the pharmacological Notch inhibition and that different ductal cell types engage in the processes (fig. S4, I and J). Together, our results provide concrete evidence demonstrating that the krt4+ ducts are progenitors giving rise to the majority of postembryonic endocrine cells.
Lineage tracing β cell regeneration
To further interrogate the regenerative framework of β cell neogenesis in zebrafish, we specifically ablate β cells from 3 to 4 dpf and allow β cell regeneration for additional 2 days. To better quantify the contribution of krt4+ duct to β cell regeneration, we switched to Tg(ubi:CSHm) and counted the number of traced insulin-producing cells in the principal islet (fig. S5A). We observed that around 40% insulin-producing cells in the principal islet can be lineage-traced and are located in the region close to the krt4+ ductal bifurcation (fig. S5, B to E, and tables S4 and S5). Notably, these cells also express sst1.1, demonstrating a bihormonal state. However, the other half of regenerated insulin-producing cells are distributed mainly at the distal region of principal islet and are also sst1.1+. The long-term lineage tracing experiment showed that most insulin– and Sst1.1–double positive cells turn into monohormonal cells (>95%), at least when the β cells were ablated at the larval stage, implicating an adaptation to the basal physiological states in developing/growing zebrafish (fig. S5F and table S6). Last, constitutive and temporal lineage tracing experiments also indicated that two other major endocrine cell types in the islet, Gcg+ and Sst1.1+ cells, can also be derived from the krt4+ duct origin (fig. S5, G and H).
To have a comprehensive understanding of different cellular behaviors during insulin-producing cell recovery, we leveraged multiple endocrine cell tracers for lineage tracing experiments during β cell regeneration. We first applied a pan-endocrine tracer: Tg(neurod1:CreERT2, cryaa:Venus)KI137 [abbreviated as Tg(neurod1:CreERT2)] with temporal labeling before β cell ablation. Although the neurod1 tracer resulted in mosaic labeling of the endocrine cells, there is no ectopic labeling (leakage) in the ductal cells (fig. S3C). We found that neurod1-derived insulin–producing cells are dispersedly distributed in the principal islet after 2 days of regeneration in both 6 dpf larvae and 31 dpf juveniles (fig. S6, A to G). These cells are all insulin– and Sst1.1–double positive. Further, to rule out the cell fate conversion from δ (sst2+), α, and gip+ cells, we performed fate mapping using Tg(sst2:Cre; cryaa:YFP)s963 [abbreviated as Tg(sst2:Cre)], TgBAC(arxa:Cre), and Tg(gip:iCre), respectively, and only observed a very limited number of insulin-producing cells that could be traced back to their origins (fig. S7, A to I, and tables S7 to S9). Furthermore, we did not see significant changes of lineage-traced insulin-expressing cells, indicating that these three cell types were not highly responsive to injury and do not initiate cellular conversion upon β cell ablation (fig. S7, J to L, and tables S10 to S12). In sum, these data support the previous findings regarding sst1.1+ endocrine cells converting to insulin-producing cells and highlight a cellular plasticity in multiple cell types, which might account for the exceptional regenerative capacity of β cells in zebrafish.
Concurrent ductal remodeling and transcriptional changes
As we showed that the krt4+ duct in the pancreatic tail does not originate from the krt4+ extrapancreatic and intermediate duct (Fig. 4, B to F), our next question was where and how the krt4+ ducts in different regions are formed and assembled to build a continuous organized luminal ductal system. We used our recently generated nkx6.1 knock-in CreERT2 line [TgKI(nkx6.1-p2A-EGFP-t2A-CreERT2)KI132, abbreviated as TgKI(nkx6.1:CreERT2)] and tp1:CreERT2 transgenics to lineage trace their descendants from 1 to 2 dpf (for nkx6.1) and 2 to 3 dpf (for tp1), respectively. For TgKI(nkx6.1:CreERT2), the administration of 4-OHT achieves maximum labeling efficiency in the Notch-responsive intrapancreatic duct, while the labeling in the extrapancreatic and intermediate duct has high variability from almost no labeling to >50% labeling (fig. S8, A to C, and table S13). We took advantage of this varied labeling pattern in the larval stage to better delineate the allocation of ducts in different regions of the adults. The long-term lineage tracing results showed that the luminal duct located in the pancreatic head and tail regions is mostly mCherry+ with a large number of lineage-traced β cells attached to it, while there is no lineage-traced β cells sitting close to the Notch-responsive duct (Fig. 5A and tables S14 to S16). The luminal ductal cells in the junction region, i.e., the intermediate duct anterior to the principal islet, are mostly mCherry− in half of the samples, while the other half of the samples can obtain mCherry labeling in extrapancreatic and intermediate ducts (fig. S8). These results indicated that the Notch-responsive intrapancreatic duct might undergo a marked transformation and gradually assemble to form a luminal tubular structure in the pancreatic tail (Fig. 5B and tables S17 and S18).
Fig. 5. Spatiotemporal-controlled lineage tracing of nkx6.1+ ducts.
(A) Single-plane confocal images showing the labeling pattern in TgKI(nkx6.1:CreERT2);Tg(ubi:CSHm) treated with 4-OHT from 1 to 2 dpf, displaying five distinct regions with high magnifications shown on the right and bottom, respectively. The extrapancreatic and intermediate ducts are devoid of labeling, while the luminal ducts in the pancreatic tail and Notch-responsive intrapancreatic ducts can be traced back to the nkx6.1+ cell origin. The ductal cells residing in between the intermediate duct and the luminal duct in the tail regions are devoid of labeling. The quantification results of the proportion of lineage traced ductal cells are displayed in (A′) (extra-intermediate duct region), (A″) (proximal luminal duct), and (A‴) (distal luminal duct). (B) Single-plane confocal images showing luminal duct and lineage-traced β cells in a secondary islet. The luminal ducts in the pancreatic tails are H2BmCherry+. The coexpression of H2BGFP and H2BmCherry indicate β cells within the secondary islet that can be traced back to the nkx6.1+ cell origin. The quantification results of ins:H2BGFP–single positive cells and ins:H2BGFP/H2BmCherry–double positive cells are shown in (B′), with the proportion of lineage traced ins:H2BGFP+ cell shown in (B″). Five size-matched secondary islets were used for the quantification. Anti-Vasnb (white) and ins:H2BGFP transgene (green) were used to visualize the ductal tree and the islets. Scale bars, 80 μm.
For further validation, we performed a long-term lineage tracing experiment using tp1 tracer restricted to the Notch-responsive duct. We confirmed that the tp1-driven ductal line can barely label extrapancreatic and intermediate ducts in the larval stage (fig. S8, D and E). We noted that the luminal duct in the tail region shows mosaic labeling, while there is an absence of mCherry signal in the intermediate ductal cells and in the extrapancreatic duct at the early juvenile stage (fig. S10). Moreover, we also confirmed that the tp1-traced descendant cells in the luminal ducts overlap with krt4, further confirming the remodeling process (fig. S11). Last, immunostaining of the foxj1a:EGFP reporter and Tg(foxj1a:iCre);Tg(ubi:CSHm)-traced juveniles showed that the foxj1a+ cells and their descendants only appeared in the extrapancreatic duct, demonstrating that there might be at least three different duct origins (krt4+/foxj1a+, krt4+/foxj1a−, and krt4−/tp1+) that develop independently and coordinate with each other to build an intricate krt4+ luminal ductal system with cells of similar morphology, transcriptional signatures, and neogenic competence (fig. S12).
Single-cell transcriptomics specifies a differentiation trajectory and endocrine progenitor cells
To further explore the cellular and molecular events underlying duct-to-endocrine cell neogenesis, we performed single-cell RNA-seq of adult zebrafish pancreata (6 to 9 months old) isolated from TgKI(krt4:iCre);Tg(ubi:CSHm);Tg(ins:flagNTR) adult fish in the basal state and after β cell ablation. We performed fluorescence-activated cell sorting (FACS) to sort out mCherry+/DAPI− cells, followed by droplet-based sequencing using the 10× platform (fig. S13A). To elucidate the complexity of cellular behavior in both the acute and sustained phase of regeneration, we selected 2, 5, or10 days after the first dose of MTZ (presented as 2, 5, or 10 dpa, single ablation) and 2 or 5 days after the second dose of MTZ (presented as 2 or 5 dpa, double ablation). We left 1 month interval between the first and second MTZ dose because within this time, the adult fish could accomplish full recovery of hyperglycemia and the replenished insulin-producing cells had up-regulated ins promoter transcriptional activity (i.e., to enable an efficient second ablation) (fig. S13, B and C).
We sequenced >40,000 cells, and after regular quality control, cell filtering, dimension reduction, and graph-based clustering, we identified 21 clusters in total (Fig. 6A). We applied well-defined cell markers based on prior knowledge and identified endocrine cells (neurod1), pancreatic duct (cftr, krt4, cdh17, and vasnb), stromal cells (tagln and acta2), smooth muscle cells and pericytes (pdgfra and pdgfrb), endothelial cells and lymphatic cells (pecam1 and lyve1a), leukocytes (ptprc and coro1a), erythrocytes/red blood cells (hbaa1), B cells (ighv1 to ighv4), peripheral nervous system neurons (elavl3 and elavl4), peripheral nervous system glia (sox10), and innate lymphoid cells type 2 (ILC2)/T helper cells (TH2) (runx3 and il4) (Fig. 6B; https://olovanderssonlab.shinyapps.io/neogenesis/). We also defined one small cluster as intestinal epithelial cell contamination due to very specific expression of the tight junction gene cldn15la (50). Moreover, we identified a small cluster with specific expression of krt5 and many other epithelial cells markers. We also included this cluster in the downstream analyses as we ruled out the possibility of skin cell (tp63 and krtt1c19e) and gallbladder cell (sstr5) contamination (51). To extrapolate the detailed cell embeddings of pancreatic duct and endocrine cells, we manually subselected cells belonging to the endocrine, pancreatic duct, and krt5+ cell clusters (~10,000 cells) and re-ran the analytical pipeline (Fig. 6, C and D).
Fig. 6. Single-cell transcriptomics highlight distinct molecular signatures in various cell types.
(A) t-distributed stochastic neighbor embedding (t-SNE) plot showing the cell type assignment of all single cells (~40,000). The classifications were based on previously known marker genes that were significantly enriched in each cluster (highlighted in parentheses). TH2, T helper 2 cells; ILC2, innate lymphoid cells type 2. (B) t-SNE plots of well annotated cell markers, colored by the normalized gene expression levels. (C and D) UMAP plot of subclustering including pancreatic duct, krt5+ cells, and endocrine cells (~10,000). (E) UMAP plots of various marker genes, colored by the normalized gene expression levels. (F and G) UMAP plot and 3D PCA plot showing the embedding of krt5+ cell and all endocrine cells. (H) Heatmap highlighting the key lineage-committed transcription factors and hormones in each cluster, with colors displayed on column-scaled mean expression. The rows and clusters were ordered by hierarchical clustering of scaled expression values.
The pancreatic ductal cells can be further categorized into two major clusters, with the majority showing high expression of krt4, her6, and her9 (her6/9 are zebrafish homologs of HES1/4), indicating krt4+ luminal ductal cells. The smaller ductal cluster is defined as Notch-responsive duct because it displayed preferential her15.1 and her15.2 (zebrafish homologs of HES5) expression (Fig. 6E). Such a molecular signature is generally in line with the previous single-cell RNA-seq dataset. In addition, we noted that igf2a and igf2b are marker genes for these two types of ducts, respectively. The krt5+ cell cluster presents a distinct expression pattern indicated by several transcription factors, including ascl1b, myt1b, and pou2f3 (Fig. 6E). Further, these krt5+ cells harbor many molecular signature genes that have been described as markers of goblet cells and paneth cells in the intestine (including agr2, spry2, galnt7, tnfrsf11a, and sox8b) (52), indicating functional similarities. The endocrine clusters can be subdivided into four major categories using low resolution (0.2), i.e., ins/sst1.1 hybrid cells, gcga/ghrl cells, δ cells (sst2+), and gip cells. We managed to obtain a more delicate clustering using higher resolution (0.7) and identified a new cluster that was previously part of the ins/sst1.1 hybrid cells (Fig. 6D). These cells were featured by specific expression of the Notch ligand, dlb (the orthologs of Dll1/DLL1), which could mediate lateral inhibition in a differentiating endocrine progenitor (53, 54). Moreover, these cells up-regulated the Notch negative regulator hes6, suggesting diminished Notch activity. To better display the lineage path of dlb+ cells to ins/sst1.1 hybrid cells without becoming skewed from a large number of pancreatic ductal cells, we performed one more subclustering to exclude krt4+ duct and Notch-responsive duct clusters (Fig. 6F). We identified eight clusters, with some remaining krt4+ ductal cells positioned in the center and dlb+ cells positioned in between krt4+ duct and ins/sst1.1 hybrid cells of the Uniform Manifold Approximation and Projection (UMAP) embedding. Likewise, the three-dimensional (3D) principal components analysis (PCA) plot also showed a trajectory from krt4+ duct to dlb+ cells to ins/sst1.1 hybrid cells in the 3D space (Fig. 6G). Notably, the heatmap of canonical transcription factors in pancreatic cell fate commitment and pancreatic hormones showed the up-regulation of many endocrine-related genes in dlb+ cells, including pax4, pax6b, isl1, myt1a, sox11a, sox11b, and fev (Fig. 6H). Considering that these cells are multihormonal, as well as dlb+ and hes6+, we propose that they stand for a novel and rare transitional cell population with endocrine precursor (EP) properties.
Pseudotime and regulon reveal gene regulatory networks along the differentiation trajectory
To unravel the molecular events underpinning the cell state transition between dlb+ cells and ins/sst1.1 hybrid cells, we first subset these cells and performed pseudotime analyses using different algorithms (slingshot and Monocle 3). We identified a unified lineage path starting from the dlb+ cells toward ins/sst1.1 hybrid cells. To better uncover the distinct molecular pathways enriched in different cells, we set a new resolution as 0.3 and redefined the cells within the trajectory into early transition, late transition, and ins/sst1.1 hybrid cells (Fig. 7, A and B). Differential gene expression analyses among these three clusters highlighted a large number lineage-committed transcription factors (insm1a, atf3, sox4b, insm1b, gata6, klf6b, pax4, arxa, and ets2), injury responsive transcription factors (jun, jund, junba, fosb, foxl1a, cebpd, and cebpa) (55, 56), epigenetic regulators (tet3), and other DNA binding/RNA splicing proteins (hmga1a, hmgb2b, hmgn6, and hmgn2) that are highly expressed in the early transition cells (Fig. 7C). Furthermore, the early transition cells also retain the expression of various cell membrane/junctional proteins that are indicative of epithelial cell properties (cldn7b, anxa4, epcam, tm4sf4, and cldn15a). These genes gradually decline when cells transit toward ins/sst1.1 hybrid cells. Gene enrichment analyses revealed that genes involved in protein translation and ribosomal biogenesis are highly expressed in the early transition cells, while the up-regulated genes in the late transition cells are engaged in protein folding, mRNA splicing, and pancreas development (Fig. 7D). The ins/sst1.1 hybrid cells are enriched in genes related to carbohydrate and nucleotide metabolism. When we ordered the DNA binding proteins and lineage-committed transcription factors based on the expression levels (regardless of whether they are differentially expressed), we found that most of them are relatively highly expressed in the early and late transition and are down-regulated when cells become ins/sst1.1 hybrid cells (Fig. 7E).
Fig. 7. In silico analyses of transition-to-ins+/sst1.1+ hybrid cells.
(A) UMAP plot displaying three clusters of transition-to-ins+/sst1.1+ hybrid cells, color-coded by subpopulation. The numbers and proportions of each cell type were displayed in a pie chart. (B) The Slingshot and Monocle 3 analyses reveal a linear trajectory, with early transition cells predominantly early in pseudotime (left), late transition cells in the middle, and ins+/sst+ hybrid cells predominantly later in pseudotime (right). (C) Heatmap of the top 150 differentially expressed genes per cluster, with cell markers gene, DNA binding genes, tight junction genes, membrane protein gene, and mitochondrial genes highlighted on the right. (D) Gene Ontology (GO) analysis of differential expressed genes in each cell cluster along the transition-to-ins+/sst1.1+ hybrid cells. Dot plots showing the differentially enriched GO terms colored by adjusted P value and sized by gene ratio. (E) Ternary plot shows the expression of DNA binding genes in early transition, late transition, and ins+/sst1.1+ hybrid cells. Genes with highest percentage of expression in early transition cells are in blue, Late transition in green, and ins+/sst1.1+ hybrid cells in red. (F) Heatmaps showing the enriched regulons among the three cell states in the trajectory. The regulons were ordered by hierarchical clustering. (G) Heatmap showing the hierarchical clustering of identified regulon and four regulon modules. (H) The UMAP for transition-to-ins+/sst+ hybrid cells based on the regulon activity scores (RASs), each cell is color-coded on the basis of the cell states assignment. (I) UMAP projection of transition-to-ins+/sst1.1+ hybrid cells overlayed by the expression level of transcription factors (left) and RAS (right).
These results further prompted us to perform regulon analyses to find the key transcriptional regulatory networks leading to the cell state transition. We found 12 regulons, which could be clustered into six modules, with crema, atf3, creb3l1, fev, bach1b, and jun regulons enriched in the early transition cells, cebpa, cebpd, and tcf21 regulons featured in the late transition cells, and pdx1, klf11b and tp53 regulons, are mostly functional in the ins/sst1.1 hybrid cells (Fig. 7, F and G, and fig. S14). In addition, we used the regulon score to recluster the transition-to-ins/sst1.1 hybrid cell lineage and noted that early transition cells were very well separated from the other cells, whereas the other two clusters intermingled with each other, suggesting that the transcriptional state of the late transition cells is more similar to that of the ins/sst1.1 hybrid cells (Fig. 7H). We also mapped the gene expression level and the regulon score in parallel on UMAP and obtained consolidated evidence of the leading regulons, i.e., atf3, fev, jun, and crema for early transition cells, cebpd for late transition cells, as well as pdx1 and klf11b in ins/sst1.1 hybrid cells (Fig. 7I).
Velocity-based methods identify dedifferentiation and differentiation events
Next, to further dissect the lineage path of the transition-to-ins/sst1.1 hybrid cells, we performed multiple analyses based on the mRNA splicing information. Velocity analyses showed that most cells have velocity vectors pointing toward the early transition cell state. We identified two distinct vectors in the late transition cluster, with a subset of cells in the upper cluster showing fate direction toward the ins/sst1.1 hybrid cells, while the lower cluster was going backward (Fig. 8A). To distinguish the varied cellular behavior, we proceeded with further subclustering using high resolution (0.7) and identified eight clusters, named early transitions 1, 2, and 3; late transitions 1 and 2; ins/sst1.1 hybrid cells 1, 2, and 3 (Fig. 8B). In addition, the PAGA algorithm identified that ins/sst1.1 hybrid cell 2 and early transition 1 cells serve as the major root of dedifferentiation and differentiation, respectively (Fig. 8C). Consistently, the CellRank algorithm also determined two roots located at early transition cells and ins/sst1.1 hybrid cells, respectively (Fig. 8, D and E).
Fig. 8. Velocity-based analyses and pathway validation.
(A) UMAP colored by latent time progression. Cells later in the trajectory (determined by RNA velocity) are yellow. The UMAP is overlaid by arrows indicating the extrapolated future cell states identified through RNA velocity analysis, suggesting that both differentiation and dedifferentiation processes occur simultaneously during insulin-producing cell recovery. (B) UMAP projection of transition-to-ins+/sst1.1+ hybrid cells with eight subclusters (three early transition cell states, two late transition cell states, and three ins+/sst1.1+ hybrid cell states). (C) The PAGA analysis indicates dedifferentiation process. (D and E) The root identification and velocity-based pseudotime results by CellRank. (F) Graphical summary using Sankey plots demonstrating the relationship between cell states (left) and condition (right). (G) Representative single-plane confocal image of hes6+ cells from the immunostaining of the knock-in hes6-p2a-EGFP-t2a-CreERT zebrafish line. The yellow arrows point to two EGFP+ cells adjacent to the duct. (H) Scatterplots showing the level of unspliced versus spliced transcripts of cell state specific genes. (I) GO analysis of up-regulated genes expressed in late transition 1 and late transition 2 cells, with enriched gene shown below. Dot plots showing the differentially enriched GO terms colored by adjusted P value and sized by gene ratio. MAPK, mitogen-activated protein kinase. (J) Violin plots and dot plots highlighting the differential gene set scores between late transition 1 and late transition 2 cells using single-cell gene set variation analysis. (K) UMAP plot showing the statistically significant gene activity scores in each cell. IL-6, interleukin-6; Jak, Janus kinase; Stat3, signal transducers and activators of transcription 3; AUC, area under the curve. (L) Representative single-plane confocal images and quantifications of β cells following treatment with either dimethyl sulfoxide (DMSO) or the PI3K inhibitor wortmannin. Treatment with wortmannin increased the number of newly generated krt4-derived β cells (mCherry+/insulin+; Wilcoxon test) in the basal state (without β cell ablation). Scale bars, 40 μm.
We also found that cells isolated from the 2 dpa double ablation condition contributed to the majority of cells in early and late transition states (Fig. 8F). Cells in the transitional states up-regulate dlb and hes6. For biological validation, we generated a knock-in at the hes6 locus TgKI(hes6-p2a-EGFP-t2a-CreERT2), visualizing its expression. Immunostaining of juvenile fish revealed hes6+ cells residing just adjacent to the luminal duct (Fig. 8G), a position consistent with the expected nature of the transitional cells. The scatterplot revealed that the late transition 1 cells have a higher number of cells with more unspliced insulin mRNA versus the spliced form, suggesting that these cells are undergoing differentiation toward β cells, while the late transition 2 cells show the opposite pattern (Fig. 8H). Cells in early transition showed more abundant unspliced dlb and hes6 gene expression, indicating a progenitor status, while cells in the ins/sst1.1 hybrid cells demonstrated higher amounts of unspliced fev, indicating a precursor status (Fig. 8H). Notably, we found that the late transition 1 cells mostly originate from the 2 dpa double ablation group, indicating that the differentiation process is mainly initiated upon the acute phase in the double ablation condition (Fig. 8H).
Differential gene expression between the two late transition clusters revealed that late transition 1 cells are characterized by the up-regulation of a variety of immune-related genes (Fig. 8I). Contrarily, the late transition 2 cells showed elevated expression of epigenetic regulators and chromatin remodelers (Fig. 8I). Single-cell gene set variation analyses and pathway activity analyses further demonstrated that the late transition 1 cells contain molecular signatures of inflammation, epithelial-to-mesenchymal transition (EMT), and an apical surface, while late transition 2 cells have a higher molecular signature of genes for pancreas development (Fig. 8J). This indicated that the late transition 1 cells are more similar to the progenitor state and the late transition 2 cells preserve more molecular properties of endocrine cells (Fig. 8J). Further, we interrogated the activity scores of the hallmark gene sets and showed that the PI3K-Akt-mTOR and gene sets involved in inflammatory response activities are highest in early transition and gradually declined along the trajectory (Fig. 8K). On the basis of a bimodal model, we set 0.15 as the threshold for activated PI3K-Akt-mTOR signaling and find that >40% (42.3 and 41.4%) cells in early or late transition cells are PI3K-Akt-mTORhigh, while the percentage was 27% cells in insulin+/Sst1.1+ hybrid cells. Considering that insulin signaling is involved in the activation of the PI3K-Akt-mTOR pathway, we assumed that the general inhibition of PI3K-Akt-mTOR might promote the transition-to-ins/sst1.1. We tested our hypothesis by treating krt4 lineage–traced larvae with the broad PI3K inhibitor wortmannin and observed an increased number of krt4 lineage–traced insulin+ cells (Fig. 8L and table S35). These data correlate with previous findings (57) and serve as proof of concept for how induction of β cell neogenesis targets can be derived from a comprehensive single cell transcriptomics analysis.
Cellular conversion shares similarities in transcriptomic changes with neogenesis
Since the insulin-producing cells we captured were mostly multihormonal, we leveraged a recently published single-cell dataset that was specifically enriched for β cells with or without ablation (fig. S15) (37). Aiming to reveal the different transcriptional profiles between mature β cells and regenerative insulin–producing cells, we filtered out other cell types (including multihormonal cells) and only kept the β cells and ins/sst1.1 hybrid cells. We identified a UMAP embedding with cells demonstrating a sequential course from early regeneration stage to late regeneration stage to mature β cells (fig. S15, A to D). We manually defined them into five clusters, named Hybrid 1/2/3 and Beta 1/2 cells. The Hybrid 1 cells are mostly composed of hybrid cells isolated from samples at 2 dpa. Differential gene expression showed the up-regulation of injury responsive genes (cebpb, cebpd, and socs3a) and immune-related genes (c9, cxcl14, and il4r.1) in Hybrid 1 cells, while pdx1 expression peaks in Hybrid 2 cells (fig. S15E). The expression levels of ins and mnx1, however, are most highly expressed in Beta 1 cell (fig. S15E). Regulon analyses identified 16 regulons; the Beta 1/2 cells can be easily distinguished from Hybrid 1/2/3 cells based on the regulons (fig. S15F). We also demonstrated that the estrogen receptor esr1 regulon dominates in the two clusters of mature β cells derived from samples in the basal state (fig. S15G). On the basis of the regulon, the β cells can be very well separated from the hybrid cells, indicating that the transcriptional state of the hybrid cells are distinct from the β cells (fig. S15, H and I). We also observed that both fev expression and fev regulon are up-regulated in the Hybrid cells, highlighting the function of fev in driving cells toward a precursor-like multihormonal state (fig. S15J), which indicate similar transcriptional regulation in cellular conversion and neogenesis.
DISCUSSION
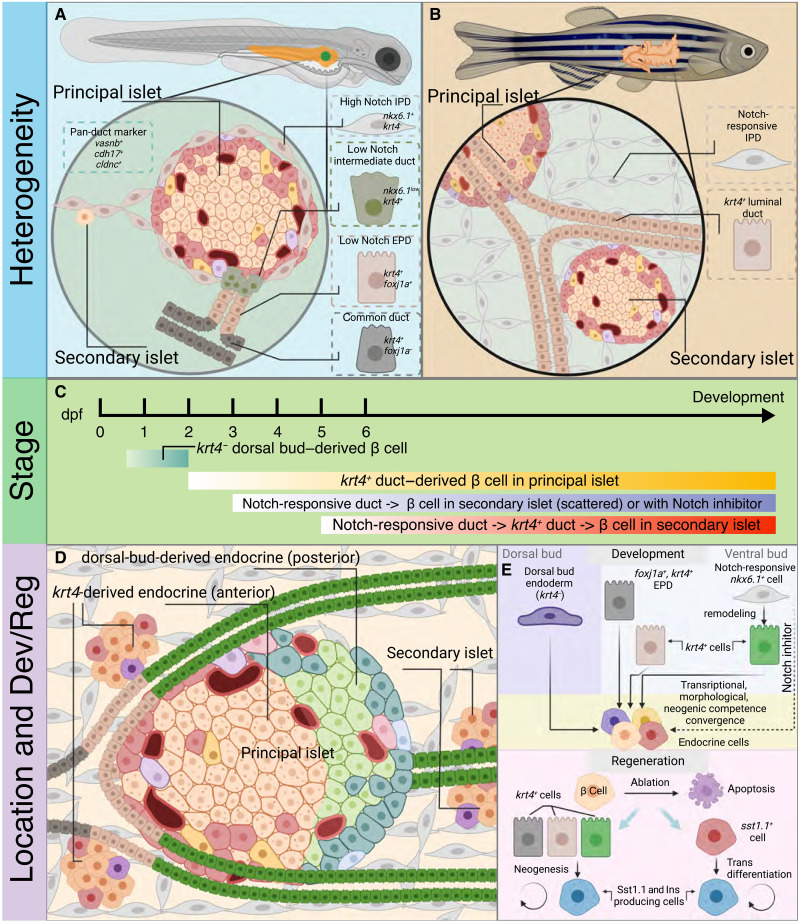
By leveraging single-cell transcriptomics and a multitude of novel knock-in and transgenic lines for lineage tracing and targeted cell ablation, our study reveals several major discoveries and proposes a redefined model of the development and regeneration of endocrine cells in zebrafish (Fig. 9). First, we unraveled ductal heterogeneity in the zebrafish pancreas and found that the krt4+ extrapancreatic and intermediate ducts (in larvae) and all luminal ducts (in a continuum throughout pancreatic tail in juveniles and adults) serve as the direct source of endocrine cells. This finding is in agreement with the previous lineage tracing study shown by spatiotemporal labeling using Tg(ins:CreERT2, cryaa:Venus)KI138 [abbreviated as Tg(ins:CreERT2)] and confirmed the hypothesis of the existence of unknown progenitor cells residing anterior to the principal islet (30). In this regard, anxa4 has been proposed to mark plastic duct anterior to the principal islet in larvae, but since anxa4 is also expressed in endocrine cells, it is unusable for lineage tracing experiments (58, 59) despite having a similar ductal expression to krt4 (at least in embryonic stages). Further studies are needed to clarify differences between anxa4+ and krt4+ ductal compartments in various stages of development and the adult. Nevertheless, our current study provides extensive and novel molecular and cellular insights that add to the mechanistic understanding of the krt4+ duct and its plasticity. It also expands the prior discoveries showing the dynamics of extrapancreatic duct–to–β cell differentiation based on time-lapse imaging and BMP/fibroblast growth factor signaling inhibition described in the embryonic stage (49, 58, 60). Moreover, we revealed a ductal cell remodeling process occurring in developed juvenile fish, indicated by complementary temporally controlled fate mapping experiments.
Fig. 9. Graphical summary describes the landscape of endocrinogenesis in zebrafish pancreas.
(A) Illustration of zebrafish pancreas in larval and juvenile/adult stage. The schematic showing that in larval stage, the zebrafish ductal tree is composed of extrapancreatic duct (EPD) (foxj1a+/krt4+), intermediate duct (foxj1−/krt4+), and Notch-responsive intrapancreatic duct (IPD) (tp1+/krt4−); while in juvenile and adult, the krt4+ duct forms luminal structure extending to the pancreatic tail, which also contain Notch-responsive ductal cells located in the peripheral regions. (B) The sketch of zebrafish endocrinogenesis suggesting multiple phases of neogenesis that take place at various stages. (C) Schematic summary of the divergent origins and contributions of the krt4+duct in juvenile and adult pancreata. (D and E) The working model of β cell differentiation and insulin-producing cell regeneration. During development, the primitive islet is formed from the dorsal pancreatic bud, followed by krt4-derived duct-to-endocrine differentiation in the principal islet. The Notch-responsive duct does not naturally serve as the direct progenitor of endocrine cells but gradually undergoes transformation and assembles to become krt4+ luminal duct, which has neogenic competence and can differentiate into endocrine cells in the secondary islets throughout the development until the adult stage. After massive loss of β cells, the neurod1+/sst1.1+ endocrine cells would initiate reprogramming and up-regulate insulin expression for functional compensation.
The divergent sources of ductal cells found at different stages can be gradually reprogrammed to transcriptionally and morphologically similar states and retain the capability to differentiate. Such a model has previously been implicated in oligodendrocyte precursor cells in mouse development, indicating a universal principle across different organs and species (61). In addition, these experiments resolved fundamental questions regarding duct-to-endocrine neogenesis in zebrafish. The prevailing perspective suggests that the tp1-driven Notch-responsive ductal cells, also known as centroacinar cells, are the progenitors of endocrine cells (32, 33, 42, 43). However, the tp1-driven lineage tracing results either labeled a minority of the regenerated β cells or was without temporal control and not specific to the Notch-responsive ductal cells (expression of which could be induced either in the intrapancreatic duct or the differentiated cells) (30, 32, 43). In our revised model, we reconcile the old model with the new knowledge presented in the current study. We propose that Notch-responsive intrapancreatic ductal cells (with a fibroblast-appearing morphology) can be naturally reprogrammed to form krt4+ luminal ducts throughout the pancreas, and, thus, a subset of endocrine cells in the secondary islets can be traced back to the Notch-responsive duct and a nkx6.1+ cell origin (32, 62). These observations are in concordance with ductal remodeling in the liver upon lumen formation and Notch-signaling deficiency (63–65). Further, there are other krt4+ ductal cells from surrogate sources, i.e. extrapancreatic and intermediate duct, that account for the endocrine cell formation in the anterior part of principal islet. We observed that the Notch-responsive ducts and krt4+ luminal ducts have marked differences in transcriptional states, in terms of the Notch-target genes, with her15.1/her15.2 being specifically expressed in the Notch-responsive duct and her6/her9 mainly in the krt4+ duct. These differential Notch signatures reflect a complex Notch activation pattern and might explain the limited effects of Notch inhibition on krt4+ duct cells using canonical γ-secretase inhibitors. Further investigation is needed to unveil the distinct roles of these Notch-target genes in ductal cell identity maintenance and progenitor cell property settlement.
Using several different Cre/CreERT2 lines, we provide concrete evidence showing multiple coordinated cellular behaviors during β cell regeneration. Our comprehensive analyses address multiple potential cell sources of regenerative insulin–producing cells and provide detailed lineage paths of endocrine cell transdifferentiation and duct neogenesis of β cells during regeneration. We applied a temporally controlled lineage tracing strategy (based on neurod1-driven expression of CreERT2) to consolidate the concept of endocrine cell transdifferentiation serving as the major source of regenerative insulin–producing cells (37, 39). Combining various Cre lines to track specific endocrine cell types, we ruled out α-to-β, δ-to-β, and gip-to-β conversion as the predominant contributors. We also showed that both neurod1- and krt4-derived regenerative insulin–producing cells coexpress Sst1.1 (the ortholog of mouse and human Sst), indicating that a distinct gene regulatory network was reconstructed in the unique regenerative microenvironment. The comparative analysis of gene regulatory networks between bona fide β cells and regenerative β cells indicated that the esr1 (estrogen receptor 1) regulon might play a pivotal role in functional maturation of β cells. This is markedly interesting as recent integrative single-cell RNA-seq data also showed a specific expression pattern of ESR1 in only adult human β cells, indicating a potential cross-species conservation in β cell maturation (https://singlecell.broadinstitute.org/single_cell/study/SCP1526/functional-metabolic-and-transcriptional-maturation-of-human-pancreatic-islets-derived-from-stem-cells) (66). Moreover, Esr1 has been intensively investigated in rodent models and is shown to play essential roles in β cell iron metabolism, promoting β cell survival, preventing β cell apoptosis, and maintaining mitochondrial and endoplasmic reticulum functions (67–73). Functional assays, novel in vivo models or 3D cocultured systems are needed to reveal the environmental cues that drive the differential transcriptional states in between the mono- and bihormonal cells and figure out the permissive niche signals to promote β cell maturation.
To dissect the lineage path at the molecular level, we performed single-cell RNA-seq using pancreata isolated from adult zebrafish in the physiological state and in a series of time points during regeneration. We comprehensively characterized the molecular signatures of the krt4 lineage cells using several algorithms and managed to delineate a conserved lineage path, i.e., the transition-to-ins/sst1.1 hybrid cells. Notably, we found that the dlb+ cells demonstrated a unique transcriptional state featured by up-regulation of multiple lineage-committed transcription factors (myt1a, fev, and pax4), as well as a multihormonal expression pattern (ins, sst1.1, gcga, gcgb, sst2, and ghrl). In addition, these cells specifically express the Notch ligand dlb and up-regulate the Notch negative regulator hes6, indicating that Notch inhibition and lateral inhibition might be important events involved in krt4+ duct-to-endocrine conversion. We note that dlb is the zebrafish ortholog of the human DLL1 gene. These results are consistent with previous findings in mouse and human embryonic pancreas development, with genes featured in trunk EPs, including Hes6 and Dll1, getting up-regulated in the EP stage, followed by downregulation during further endocrine differentiation (6, 7). Although it is still under debate whether adult mice harbor pancreatic ductal progenitors, our results nevertheless suggest an evolutionarily conserved duct-to-endocrine differentiation between adult zebrafish and embryonic mouse and human (22, 25, 74, 75).
The dlb+ cells also maintain a certain level of epithelial cell markers, such as cdh17, cldnh, cldnc, and epcam, suggesting that they are transitional between krt4+ duct and endocrine cells. These transcriptional features implicate these cells as being in a primed state for further differentiation to endocrine cells. Our regulon analyses showed that these dlb+ cells are enriched with atf3, fev, crema, and creb3l1 regulons. Neurog3 and Fev have been recognized to play sequential roles in mouse endocrinogenesis (3, 7), and our study suggests that the endocrine progenitor cell state in zebrafish is also governed by fev. The sst1.1+ endocrine cell conversion to insulin-producing cells also involves the up-regulation of fev (regulon) to form hybrid cells. Thus, these results highlight the conserved functional role of fev in endocrinogenesis and its distinct function in direct reprogramming (76, 77). Whether the hybrid cells resolve into monohormonal cells could be dependent on their origin and timing; when we ablated the β cells in larvae, we observed mostly monohormonal krt4 lineage–traced cells in the juvenile stage, whereas when Carril Pardo et al. (39) ablated β cells in the adult, the sst1.1-derived hybrid cells remained bihormonal. Thus, it appears that the origin of insulin-producing cells, i.e., from krt4+ duct in development/growth versus sst1.1+ cells after β cell ablation, has different abilities to become monohormonal.
Our velocity-based analyses further revealed that both dedifferentiation and differentiation processes occur during recovery from β cell loss, although dedifferentiation seems to dominate this dynamic process. We identified two late transitional cell states that, in the trajectory, sit in between the dlb+ cells and the regenerative insulin–producing cells. The cells showing up-regulation of various immune responsive genes are only enriched in the acute phase after two consecutive rounds of β cell ablation. A subset of these cells had more unspliced than spliced insulin mRNA, with velocity vectors pointing toward the insulin-producing cell fate. These cells also up-regulated EMT-related genes, indicating an ongoing differentiation process (78). The cells showing dedifferentiation vectors up-regulate a broad collection of epigenetic remodeling genes (79). These results suggest that the inflammatory response following β cell loss might play an essential role in the course of differentiation. We speculate that the dysregulation of epigenetic factors might serve as either a protective factor in response to cellular aging/stress or a detrimental factor in preventing insulin-producing cell maturation. Considering the latter case, in combination with our regulon results, reversing the behavior of these cells by targeting the key regulons might be a promising strategy to promote redifferentiation and increase functional β cell mass (79). We noted that a wide variety of activating protein 1 (AP-1) genes (jun, jund, junba, and fosb) are highly expressed in the early transition cells. Since the AP-1 complex has been shown to function in the signal transduction of proregenerative responses in numerous tissues (e.g., zebrafish and salamander ependymo-radial glia, mouse subventricular zone, and zebrafish lateral line hair cells), we propose that the inflammatory response could provide a favorable microenvironment to trigger progenitor cells to differentiate (56, 80–86). This is supported by the regulon analysis showing that the injury responsive genes (cebpa and cebpd) and regulons are dominating in late transition 1 cells. However, further functional studies are needed to unveil whether and how much inflammatory signaling is required to promote induction of endocrine cells upon injury (87).
We also applied pharmaceutical approaches to investigating the potential pathways that are crucial for differentiation to insulin-producing cells. By repressing the canonical PI3K-Akt-mTOR pathway using wortmannin, we observed an increase in krt4+ lineage–traced β cells in the basal state, suggesting that PI3K signaling regulates cell differentiation or maturation (57).
Last, we would have been unable to make these discoveries without developing novel knock-in zebrafish tools. The previously published findings in zebrafish were based on classical transgenic lines that could be compromised by their natural disadvantages, such as the ectopic labeling (transgene leakage issue) or a lack of labeling. In particular, we need to be cautious in drawing definitive conclusions when only using reporters of signaling pathways. This is due to the complexity of signaling pathways (a wide range of downstream responsive genes responds differently to various environmental stimuli) making it questionable whether the cloned sequence elements can recapitulate the complex activation pattern of a signaling pathway. Our study serves as proof of concept for using a new generation of knock-in tracers (targeting key transcription factors or structural proteins) to resolve lineage-related questions in zebrafish. This could lead to a new standard of genetic fate mapping for both confirming old and generating new discoveries in the zebrafish field.
In conclusion, our study provides solid evidence for a new model of β cell differentiation and recovery in zebrafish. We identify a ductal remodeling process that fills the gaps in the previous model. The comprehensive molecular profiling at the single-cell level suggested both distinct and conserved aspects of β cell neogenesis in zebrafish, highlighting the advantage of using zebrafish and opening new avenues for understanding and treating human diseases.
MATERIALS AND METHODS
Zebrafish lines and husbandry
The following published transgenic and knock-in zebrafish lines were used in this study: Tg(Tp1bglob:H2BmCherry)S939 (43) abbreviated as Tg(tp1:H2BmCherry), Tg(tp1:EGFP) (43), Tg(tp1:CreERT2) (41), TgKI(krt4-p2A-mNeonGreen-t2A-iCre)KI128 (47) abbreviated as TgKI(krt4:iCre), TgKI(krt4-p2A-EGFP-t2A-CreERT2)KI129 (47) abbreviated as TgKI(krt4:CreERT2), TgKI(krt4-p2a-mNeonGreen)KI127 (47), TgKI(nkx6.1:CreERT2) (47), TgBAC(cftr:Gal4)pd1101 (88), Tg(UAS:GFP)zf82 (89), Tg(5xUAS:EGFP)nkuasgfp1a (89) abbreviated as Tg(UAS:Cre), Tg(-3.5ubb:loxP-EGFP-loxP-mCherry)cz1701 (90) abbreviated as Tg(ubi:Switch), Tg(UBB:loxP-CFP-STOP-Terminator-loxP-hmgb1-mCherry)jh63 (64) abbreviated as Tg(ubi:CSHm), Tg(ins:flagNTR) (29), TgBAC(neurod1:EGFP)nl1 (91), TgBAC(pdx1:EGFP)bns13 (92), TgBAC(ascl1b:EGFP-2A-Cre-ERT2)ulg006 (62) abbreviated as Tg(ascl1b:EGFP), Tg(nkx2.2a:mEGFP)vu17 (93), Tg(ins:Hsa.HIST1H2BJ-GFP)s960 (94) abbreviated as Tg(ins:H2BGFP), TgBAC(arxa:Cre)bns250 (92), Tg(sst2:Cre) (34), and Tg(Hsa.CTGF:NLS-mCherry)ia49 (95), as referred as yap reporter.
Several lines were reestablished by reinjecting the following constructs: Tg(cdh17:H2Bmcherry) (plasmid from B. He) (96), and Tg(−5.2foxj1a:EGFP) abbreviated as Tg(foxj1a:EGFP) (plasmid from B. Ciruna) (97).
The strategy of generating TgKI(onecut1-p2a-EGFP-t2a-CreERT2)KI145 and TgKI(hes6-p2a-EGFP-t2a-CreERT2)KI146 lines has been described elsewhere, using polymerase chain reaction (PCR)–amplified 5′-AmC6–modified double-stranded DNA as the donor (47). The single-guide RNA (sgRNA) and primers used for generating the knock-in were the following: sgRNA, GCACTTGTACCAAAGCATGA(TGG) (onecut1) and AGGTGAGCATCTGTCTACCA(GGG) (hes6); onecut1, GCCAACTCCTCCTCCTCCAGCACTTGTACCAAAGCAGGAAGCGGAGCTACTAACTTCAGC (forward) and TTTTTTCTTTTTTTCCCCACAATGCAGCCGTTACTCCATCAAGCTGTGGCAGGGAAACCC (reverse); hes6, ACATTATACACCAACAAGTCCATTTGGAGACCCTGGGGAAGCGGAGCTACTAACTTCAGC (forward) and ACTTGAATGTTCATGAATCGGCGGAGGTGAGCATCTGTCTAAGCTGTGGCAGGGAAACCC (reverse).
The following lines were newly generated by the Tol2 transposon system Tg(neurod1:CreERT2), Tg(ins:CreERT2), and Tg(foxj1a:iCre; cryaa:Venus)KI139 abbreviated as Tg(foxj1a:iCre). The injected constructs were built by Multisite Gateway cloning using LR Clonase II Plus (Thermo Fisher Scientific). We followed the protocol as described by C. Mosimann (dx.doi.org/10.17504/protocols.io.b4xdqxi6) of recombining entry clones p5E, pME, and p3E into a destination vector, pDEST. The following entry clones were used in this study: p5E: p5E-ins; p5E-neurod1 (gift from A. Nechiporuk), p5E-foxj1a (gift from B. Ciruna), pME: pME-iCre, pME-CreERT2, p3E: p3E-poly A, and pDest: pDEST-Tol2-gy. The mosaic F0s and founders were selected on the basis of yellow eye marker at 3 dpf.
The following lines were newly generated by the I-SceI system: Tg(ins:loxP-dsRed-terminator-stop-loxP-DTA)KI140 abbreviated as Tg(ins:CSDTA), Tg(gip:iCre)KI136, and Tg(ins:CSHm). The double-stranded DNA sequence of DTA was synthesized by Integrated DNA Technologies. The final constructs were generated by In-Fusion cloning (Takara Bio). The mosaic F0s and founders were selected on the basis of the fluorescent signals in the islets at 3 dpf. Tg(gip:iCre)KI136 was generated by replacing Venus with iCre in Tg(gip:Venus) using In-Fusion cloning (98).
Males and females aged between 3 months and 2 years were used for breeding. Zebrafish larvae were incubated at 28.5°C in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, and 0.33 mM MgSO4 at pH 7.4 with NaHCO₃) until 6 dpf. Juvenile and adult fish were maintained on a 14:10-hour light/dark cycle at 28°C. All studies involving zebrafish were approved by the Karolinska Institutet Animal Care and Use Committee, performed in accordance with local guidelines and regulations, and approved by regional authorities.
Lineage tracing with or without tamoxifen-inducible Cre recombinase
We used both Cre and CreERT2 lines for genetic fate mapping. The knock-in Cre lines were crossed with Tg(ubi:CSHm) for the quantitative analyses. For a more strict approach with temporal labeling, we treated the zebrafish larvae carrying the CreERT2 cassette as well as the ubi:CSHm or ubi:Switch transgene with 20 μM 4-OHT unless otherwise stated (Sigma-Aldrich) in E3 medium in 24-well plates, with four to eight embryos/larvae per well, for 24 hours without refreshment to accomplish maximum labeling without compromising the survival rate. We treated TgKI(nkx6.1:CreERT2) and TgKI(krt4:CreERT2) with 4-OHT from 1 to 2 dpf, while we performed the treatment for Tg(tp1:CreERT2), Tg(ins:CreERT2), and Tg(neurod1:CreERT2) from 2 to 3 dpf.
Sample fixation for immunostaining
Before fixation, we euthanized the zebrafish with tricaine (Sigma-Aldrich) in E3 medium, followed by washing three times with distilled water. We then fixed the samples in 4% formaldehyde (Sigma-Aldrich) in phosphate-buffered saline (PBS) (Thermo Fisher Scientific) at 4°C for at least 24 hours. We stopped the fixation by washing three times with PBS and then removed the skin, crystallized yolk (of larvae), and liver (if needed) to expose the pancreas for immunostaining.
Immunostaining
We followed the immunostaining protocol that was previously described by Liu et al. (99). Briefly, we incubated the samples in blocking solution [0.3% Triton X-100 and 4% bovine serum albumin (BSA) in PBS] at room temperature for at least 1 hour, followed by overnight incubation with primary antibodies at 4°C. After removing the primary antibodies, we washed the samples using washing buffer (0.3% Triton X-100 in PBS) at least 10 times at room temperature. Next, we incubated the samples in blocking solution with fluorescent dye–conjugated secondary antibodies and the nuclear counterstain 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific) at 4°C overnight. On the following day, we removed the solution and washed the samples with washing buffer 10 times at room temperature. The following primary antibodies were used: chicken anti-GFP (1:500; Aves Labs, GFP-1020), goat anti-tdTomato (1:500; MyBioSource, MBS448092), rabbit anti-insulin (1:100; Cambridge Research Biochemicals, customized), mouse anti-glucagon (1:50; Sigma-Aldrich, G2654), rat anti-Sst1.1 (1:50; GeneTex, GTX39061), rabbit anti-Cdh17 (1:1000; customized sera, gift from Y. Cao, Tongji University), rabbit anti-Cldnc (1:200; customized sera, gift from Y. Cao, Tongji University), rabbit anti-Vasnb (1:1000; customized sera, gift from P. Panza), and rabbit anti-activated Caspase-3 (1:100; Chemicon).
Hybridization chain reaction
HCR3.0 was performed following the protocol described by Choi et al. (100). Briefly, the larvae were fixed in 4% paraformaldehyde (PFA) at 4°C overnight, with subsequent perforation step in 100% methanol at −20°C for at least 24 hours. After that, we performed gradient rehydration steps using 75, 50, and 25% methanol, followed by washing five times using 100% PBS with 0.1% Tween 20 (PBST). The larvae were then treated with a series of preparation steps including further permeabilization [using proteinase K (30 μg/ml) for 45 min at room temperature], postfixation (4% PFA for 20 min), five washes with PBST, and prehybridization (using hybridization buffer at 37 °C for 30 min). The larvae were then incubated overnight at 37 °C in the hybridization buffer containing 2 pmol of the probe set. On the next day, the solution was washed off four times with washing buffer at 37°C, followed by 4× saline‑sodium citrate with 0.1% Tween 20 (SSCT) washes at room temperature for 15 min each. Next, the larvae were incubated in the amplification buffer with 15 pmol of fluorescently labeled hairpin amplifier overnight at room temperature. On the next day, the larvae were washed twice with 5× SSCT, followed by one incubation step with DAPI (1:1000; in 5× SSCT) for 30 min at room temperature. Last, we performed additional washes three times with 5× SSCT. Probe sequences were designed by the manufacturer (Molecular Instruments). The probe sets used were as follows: dr_krt4-B1, dr_cftr-B1, dr_sox9b-B1, dr_hnf1ba-B1, dr_cx30.3-B1, dr_clcn1b-B1, dr_cldn7b-B1, dr_cd9d-B1, dr_igf2a-B1, dr_anxa4-B1, dr_id2a-B1, and dr_foxj1a-B1. The conjugated hairpin amplifier was B1, with the fluorophore 647.
Confocal imaging
Before confocal imaging, we mounted the stained samples in VECTASHIELD Antifade Mounting Medium (Vector Laboratories) on microscope slides with the pancreas facing the cover slips. We imaged the pancreas with the Leica TCS SP8 platform using 40× objectives for larvae until 7 dpf and 20× objectives for juveniles later than 30 dpf. The images were analyzed by Fiji (101).
Chemical ablation of β cells and drug treatment
Ablation of β cells in the zebrafish larvae was performed by incubating the larvae carrying the ins:flagNTR transgene for 24 hours with 10 mM MTZ (Sigma-Aldrich) diluted in 1% dimethyl sulfoxide (VWR) in E3 solution supplemented with 0.2 mM 1-phenyl-2-thiourea (Acros Organics) from 3 to 4 dpf. For the juvenile and adult stages, β cell ablation was achieved by incubating the zebrafish with 1 mM MTZ for 24 hours. Chemical treatment of zebrafish larvae was performed by adding the chemicals to E3 buffer for 48 hours unless otherwise stated. The chemicals used were 50 nM wortmannin (Selleckchem, S2758) and 1 μM LY-411575 (Selleckchem, S2714).
Adult zebrafish pancreas dissection and FACS
We euthanized the adult fish by putting each one into ice-cold Hanks’ balanced salt solution (HBSS; no calcium and no magnesium) for 10 min. Next, we used blunt forceps to carefully remove the skin, kidney, eggs (in females), liver, and gallbladder to expose the pancreata. This step is critical for avoiding krt4+ cell contamination from other organs. We also removed adipose tissue as much as possible. Then, we cut at the anterior region of the intestinal bulb and hindgut and transferred the intestine and pancreas together to a new dish. We used blunt dissection tools to carefully separate the pancreata from the intestine. Meanwhile, we also removed the spleen (appearing in dark red), which is also attached to the intestine. We moved and immersed the whole pancreata into 5 ml of HBSS kept on ice. For each condition, we pooled four to eight samples together for an enzymatic digestion with 600 μl of 1× TrypLE (Thermo Fisher Scientific) supplemented with 60 μl of 100× Pluronic F-68 (Thermo Fisher Scientific) at 37°C on a shaker at 125 rpm for 45 to 60 min to prevent tissue adhesion. We also pipetted the tissue up and down every 5 min for better digestion. We added 6 ml of precooled 2% BSA to end the digestion and centrifuged the sample at 500g for 5 min. After removing the supernatant, we washed the pellets with 300 μl of precooled solution containing 1% BSA, 0.1% Pluronic F-68, and 0.1% DAPI and used a Corning Falcon cell strainer (Corning, 352235) to filter out not fully digested tissues.
During FACS, we selected single-cell populations based on forward scatter and side scatter signals. Next, we performed negative selection with the DAPI channel to remove dead cells and cell debris. Last, we used the Cherry channel for the subsequent gating to collect all cells of the krt4-derived lineage in a new tube as the single-cell suspension.
Cell encapsulation, library preparation, and sequencing
Droplet-based single-cell RNA-seq was performed using the Chromium Single Cell 3′ Library and Gel Bead Kit v3 (10× Genomics) and Chromium Single Cell 3′ Chip G (10× Genomics). Approximately 8000 to 10,000 cells from each condition were loaded and encapsulated in a single v3 reaction. GEM generation and library preparation were performed according to manufacturer’s instructions. Cells were partitioned into gel beads in emulsion in the controller for cell lysis and reverse transcription. The 10× single-cell RNA-seq libraries were PCR-amplified (13 cycles), pooled, denatured, and diluted in before paired-end sequencing on a NextSeq 500 according to manufacturer’s recommendations. Sequencing data were aligned to the zebrafish reference genome (GRCz11) with addition of the mCherry sequence using Cell Ranger v5.0.1 (10× Genomics) to generate a gene-by-cell count matrix with default parameters.
Data preprocessing
The single-cell RNA-seq data of adult zebrafish principal islet datasets were downloaded from the Gene Expression Omnibus under accession numbers GSE106121 and GSE166052. The unique molecular identifier (UMI) counts matrix was imported into R and processed using the Seurat R package version 4.0.4 (102). Cells with a detected gene number of <400 or above 7000 genes were considered as low-quality cells or doublets and removed before downstream analysis. The count matrix was then normalized and scaled with default parameters, followed by highly variable gene selection. Further doublet filtering was performed using the Doubletfinder R package (103). After regular dimension reduction using PCA, we selected 8 to 30 PCs (depending on the anticipated cell types) for graph-based clustering (Louvain). We used the UMAP algorithm to display the relationships within and between different clusters and the Harmony algorithm for batch correction (104). To unravel the molecular details and cellular trajectory in krt4-derived duct-to-endocrine and endocrine lineages, we manually annotated each cell cluster based on well-known verified marker genes and made subsets of clusters of interest, followed by a series of normalization, scaling, dimensional reduction, batch correction, and graph-based clustering. The lists of genes encoding DNA binding proteins, membrane proteins, cell junctions, and mitochondrial proteins are retrieved from human protein atlas (www.proteinatlas.org/), and we used “biomaRt” R package to perform human-to-zebrafish ortholog conversion. The marker genes and genes with differential expression were listed in tables S6 to S21.
SCENIC analysis
The SCENIC pipeline was performed using the python package pySCENIC (105, 106). We first selected the 3000 most high variable genes and used their original count as input. We took advantage of the recent study in zebrafish that provided us a list of known transcription factors and the motif annotation for the construction of adjacency matrix (indicating the gene expression correlation between the transcription factors and the putative target genes) refined gene regulatory network after pruning targets (107). Last, cells were scored for each regulon and obtained a regulon activity score (RAS) using AUCell (www.bioconductor.org/packages/release/bioc/html/AUCell.html) (105). We applied the RAS matrix to generate t-distributed stochastic neighbor embedding (t-SNE) and UMAP embedding. In downstream analyses, we followed the analytical pipeline that was previously described by Suo et al. (108). Briefly, we first calculated the regulon specific score matrix to identify the cell type/state–specific regulon. We also used hierarchical clustering built on the regulon to generate a connection specificity index matrix and define individual regulon modules, which exhibit the relationship between the regulon and regulon module. Last, we measured the regulon module activity scores (average RAS on regulons in each module) for all cells and averaged them on the basis of cell type to identify the correlation between the regulon modules and cell types. The list of transcription factors and the motif annotations can be accessed from https://db.cngb.org/stomics/zesta/.
Pseudotime trajectory and in silico fate mapping using scVelo and CellRank
Psuedotime analysis was conducted with slingshot (version 1.8.0) (109) and Monocle 3 (version 1.2.9) (110) after batch correction. We used the default settings and defined the early transition as the root node for pseudotime calculations.
scVelo was used to calculate RNA velocity in transition-to-ins/sst1.1 hybrid cells (5). We first selected cells based on barcoding and filtered gene expression matrix to only include the 3000 most highly variable genes. Then, we computed the moments for velocity estimation using n = 15 nearest neighbors and the top 8 PCs. We calculated the latent time using the cells from the uninjured samples as root. Last, RNA velocity was estimated on the basis of latent time scores, using the dynamical model, and we mapped the velocity vectors on the prior UMAP embedding. We leveraged the directional information from RNA velocity and computed the initial and terminal states of the lineage path using the function “cr.tl.initial_states” and “cr.tl.initial_states” in CellRank with default settings (8).
Enrichment analysis
We selected differentially expressed genes among different clusters found by “FindMarkers” function with parameter (min.pct = 0.25, logfc.threshold = 0.25) and filtered by pvalue.adj < 0.05. Gene enrichment analyses were accomplished using the function “enrichGO” with pvalueCutoff = 0.01, qvalueCutoff = 0.05 (OrgDb = org.Dr.eg.db, pAdjustMethod = “BH”) from the R package clusterProfiler (V3.18.1, https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (111). To compare pathway activity between different cell states along transition-to-ins/sst1.1 hybrid cell lineage, we fetched the normalized data of the groups and calculated the pathway activity score based on the 50 hallmark gene sets listed in Molecular Signatures Database (MSigDB) v7.5.1 (www.gsea-msigdb.org/gsea/msigdb/genesets.jsp?collection=H) using the function “gsva” in the GSVA R package (www.bioconductor.org/packages/release/bioc/html/GSVA.html) (112). The limma package was used to identify the differentially enriched pathways between the two selected groups. To decipher and visualize the pathway activity of individual cells along the transition-to-ins/sst1.1 hybrid cell path, we also used the hallmark gene sets in MSigDB and calculated the pathway activity score of each module using function “AddModuleScore” from Seurat.
Statistical analysis
Similar experiments were performed at least two times independently. For immunostaining and HCR3.0 in situ hybridization experiments, the replicates were performed and the expression patterns were confirmed in at least five individual samples, i.e., where only representative images were presented in the figure. For juvenile and adult lineage tracing experiments, results were confirmed in at least two independent experiments performed on different days. No statistical method was used to predetermine sample size. No data were excluded from the analyses. The number of cells in the confocal microscopy images was all quantified manually with the aid of Fiji. The scheme of knock-in strategy was illustrated by using “IBS” software (113). Statistical analyses were carried out by two-tailed Mann Whitney U tests (comparing two groups) or by Kruskal-Wallis tests (comparing three groups) unless otherwise stated. The results were presented as the mean values ± SEM, and P ≤ 0.05 is considered statistically significant. The n number represents the number of zebrafish in each group for each experiment. All statistical analyses and data visualization were performed on R platform (version 4.0.2) using “ggplot2” and “ggpubr” packages.
Acknowledgments
We thank K. Mamounis (Karolinska Institutet) for critical reading of the manuscript and C. Karampelias (Karolinska Institutet) and L. Ren (Karolinska Institutet) for helpful discussion. We also thank Y. Cao (Tongji University) and P. Panza (Max Planck Institute for Heart and Lung Research) for antibodies as gifts; D. Stainier (MPI-HLR), B. Appel (U Colorado), B. He (Karolinska Institutet), M. Parsons (UC Irvine), N. Ninov (CRTD Dresden), B. Peers (GIGA, ULG), A. Nechiporuk (OHSU), B. Ciruna (SickKids, Toronto), M. Bagnat (DUKE), and I. Adameyko (MedUni Wien and Karolinska Institutet) for sharing the fish lines and constructs. We also thank M. N. Zarif from the Biomedicum flow cytometry core facility for the assistance in FACS sorting. We acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure, especially K. Wallenborg, S. Masoumi, and A. Glaros for technical support in single-cell RNA-seq. We thank U. Wargh for the assistance with fish care. The illustration of the schematics was created by Biorender.com.
Funding: Research in the laboratory of O.A. was supported by funding from: The European Research Council under the Horizon 2020 research and innovation programme (grant no. 772365), The Swedish Research Council, the Novo Nordisk Foundation, and Strategic Research Programmes at the Karolinska Institutet (SRP Diabetes and StratRegen). J.M. was supported by China Scholarship Council (CSC).
Author contributions: Conceptualization: J.M. and O.A. Methodology: J.M., K.-C.L., and O.A. Investigation: J.M. Visualization: J.M. Supervision: O.A. Writing—original draft: J.M. Writing—review and editing: J.M., K.-C.L., and O.A.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: Raw and processed single-cell RNA-seq data (raw data and h5) are accessible at the Gene Expression Omnibus with the accession number (GSE226841). The UMAP plots and violin plots can be queried in our shiny web-browser (https://olovanderssonlab.shinyapps.io/neogenesis/). Source data are provided with this paper. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
Legends for movies S1 to S3
Legends for tables S1 to S35
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S35
Movies S1 to S3
REFERENCES AND NOTES
- 1.E. D. Bankaitis, M. E. Bechard, C. V. Wright, Feedback control of growth, differentiation, and morphogenesis of pancreatic endocrine progenitors in an epithelial plexus niche. Genes Dev. 29, 2203–2216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.A. Mamidi, C. Prawiro, P. A. Seymour, K. H. de Lichtenberg, A. Jackson, P. Serup, H. Semb, Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature 564, 114–118 (2018). [DOI] [PubMed] [Google Scholar]
- 3.L. E. Byrnes, D. M. Wong, M. Subramaniam, N. P. Meyer, C. L. Gilchrist, S. M. Knox, A. D. Tward, C. J. Ye, J. B. Sneddon, Lineage dynamics of murine pancreatic development at single-cell resolution. Nat. Commun. 9, 3922 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S. Horn, S. Kobberup, M. C. Jorgensen, M. Kalisz, T. Klein, R. Kageyama, M. Gegg, H. Lickert, J. Lindner, M. A. Magnuson, Y. Y. Kong, P. Serup, J. Ahnfelt-Ronne, J. N. Jensen, Mind bomb 1 is required for pancreatic β-cell formation. Proc. Natl. Acad. Sci. U.S.A. 109, 7356–7361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.V. Bergen, M. Lange, S. Peidli, F. A. Wolf, F. J. Theis, Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020). [DOI] [PubMed] [Google Scholar]
- 6.X. X. Yu, W. L. Qiu, L. Yang, Y. Zhang, M. Y. He, L. C. Li, C. R. Xu, Defining multistep cell fate decision pathways during pancreatic development at single-cell resolution. EMBO J. 38, e100164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.X. X. Yu, W. L. Qiu, L. Yang, Y. C. Wang, M. Y. He, D. Wang, Y. Zhang, L. C. Li, J. Zhang, Y. Wang, C. R. Xu, Sequential progenitor states mark the generation of pancreatic endocrine lineages in mice and humans. Cell Res. 31, 886–903 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. Lange, V. Bergen, M. Klein, M. Setty, B. Reuter, M. Bakhti, H. Lickert, M. Ansari, J. Schniering, H. B. Schiller, D. Pe'er, F. J. Theis, CellRank for directed single-cell fate mapping. Nat. Methods 19, 159–170 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Y. Dor, J. Brown, O. I. Martinez, D. A. Melton, Adult pancreatic β-cells are formed by self-duplication rather than stem-cell differentiation. Nature 429, 41–46 (2004). [DOI] [PubMed] [Google Scholar]
- 10.H. Zhao, X. Huang, Z. Liu, W. Pu, Z. Lv, L. He, Y. Li, Q. Zhou, K. O. Lui, B. Zhou, Pre-existing beta cells but not progenitors contribute to new beta cells in the adult pancreas. Nat. Metab. 3, 352–365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.V. Cigliola, L. Ghila, F. Thorel, L. van Gurp, D. Baronnier, D. Oropeza, S. Gupta, T. Miyatsuka, H. Kaneto, M. A. Magnuson, A. B. Osipovich, M. Sander, C. E. V. Wright, M. K. Thomas, K. Furuyama, S. Chera, P. L. Herrera, Pancreatic islet-autonomous insulin and smoothened-mediated signalling modulate identity changes of glucagon+ α-cells. Nat. Cell Biol. 20, 1267–1277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.K. Furuyama, S. Chera, L. van Gurp, D. Oropeza, L. Ghila, N. Damond, H. Vethe, J. A. Paulo, A. M. Joosten, T. Berney, D. Bosco, C. Dorrell, M. Grompe, H. Raeder, B. O. Roep, F. Thorel, P. L. Herrera, Diabetes relief in mice by glucose-sensing insulin-secreting human α-cells. Nature 567, 43–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.S. Chera, D. Baronnier, L. Ghila, V. Cigliola, J. N. Jensen, G. Gu, K. Furuyama, F. Thorel, F. M. Gribble, F. Reimann, P. L. Herrera, Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature 514, 503–507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.F. Thorel, V. Nepote, I. Avril, K. Kohno, R. Desgraz, S. Chera, P. L. Herrera, Conversion of adult pancreatic α-cells into β-cells after extreme β-cell loss. Nature 464, 1149–1154 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Perez-Frances, L. van Gurp, M. V. Abate, V. Cigliola, K. Furuyama, E. Bru-Tari, D. Oropeza, T. Carreaux, Y. Fujitani, F. Thorel, P. L. Herrera, Pancreatic Ppy-expressing γ-cells display mixed phenotypic traits and the adaptive plasticity to engage insulin production. Nat. Commun. 12, 4458 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.W. Li, C. Cavelti-Weder, Y. Zhang, K. Clement, S. Donovan, G. Gonzalez, J. Zhu, M. Stemann, K. Xu, T. Hashimoto, T. Yamada, M. Nakanishi, Y. Zhang, S. Zeng, D. Gifford, A. Meissner, G. Weir, Q. Zhou, Long-term persistence and development of induced pancreatic beta cells generated by lineage conversion of acinar cells. Nat. Biotechnol. 32, 1223–1230 (2014). [DOI] [PubMed] [Google Scholar]
- 17.W. Li, M. Nakanishi, A. Zumsteg, M. Shear, C. Wright, D. A. Melton, Q. Zhou, In vivo reprogramming of pancreatic acinar cells to three islet endocrine subtypes. eLife 3, e01846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.C. R. Xu, L. C. Li, G. Donahue, L. Ying, Y. W. Zhang, P. Gadue, K. S. Zaret, Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. EMBO J. 33, 2157–2170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D. E. Metzger, C. Liu, A. S. Ziaie, A. Naji, K. S. Zaret, Grg3/TLE3 and Grg1/TLE1 induce monohormonal pancreatic β-cells while repressing α-cell functions. Diabetes 63, 1804–1816 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. Inada, C. Nienaber, H. Katsuta, Y. Fujitani, J. Levine, R. Morita, A. Sharma, S. Bonner-Weir, Carbonic anhydrase II-positive pancreatic cells are progenitors for both endocrine and exocrine pancreas after birth. Proc. Natl. Acad. Sci. U.S.A. 105, 19915–19919 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R. Sancho, R. Gruber, G. Gu, A. Behrens, Loss of Fbw7 reprograms adult pancreatic ductal cells into α, δ, and β cells. Cell Stem Cell 15, 139–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.C. Gribben, C. Lambert, H. A. Messal, E. L. Hubber, C. Rackham, I. Evans, H. Heimberg, P. Jones, R. Sancho, A. Behrens, Ductal Ngn3-expressing progenitors contribute to adult β cell neogenesis in the pancreas. Cell Stem Cell 28, 2000–2008.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.E. Dirice, G. Basile, S. Kahraman, D. Diegisser, J. Hu, R. N. Kulkarni, Single-nucleus RNA-seq reveals singular gene signatures of human ductal cells during adaptation to insulin resistance. JCI Insight 7, e153877 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.E. Dirice, D. F. De Jesus, S. Kahraman, G. Basile, R. W. Ng, A. El Ouaamari, A. K. K. Teo, S. Bhatt, J. Hu, R. N. Kulkarni, Human duct cells contribute to β cell compensation in insulin resistance. JCI Insight 4, e99576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.J. Magenheim, M. A. Maestro, N. Sharon, P. L. Herrera, L. C. Murtaugh, J. Kopp, M. Sander, G. Gu, D. A. Melton, J. Ferrer, Y. Dor, Matters arising: Insufficient evidence that pancreatic β cells are derived from adult ductal Neurog3-expressing progenitors. Cell Stem Cell 30, 488–497.e3 (2023). [DOI] [PubMed] [Google Scholar]
- 26.C. Gribben, R. Sancho, A. Behrens, Response to Magenheim et al.: Ductal Ngn3-expressing progenitors contribute to adult beta cell neogenesis in the pancreas. Cell Stem Cell 30, 345–347 (2023). [DOI] [PubMed] [Google Scholar]
- 27.J. B. Moss, P. Koustubhan, M. Greenman, M. J. Parsons, I. Walter, L. G. Moss, Regeneration of the pancreas in adult zebrafish. Diabetes 58, 1844–1851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Curado, D. Y. Stainier, R. M. Anderson, Nitroreductase-mediated cell/tissue ablation in zebrafish: A spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat. Protoc. 3, 948–954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O. Andersson, B. A. Adams, D. Yoo, G. C. Ellis, P. Gut, R. M. Anderson, M. S. German, D. Y. Stainier, Adenosine signaling promotes regeneration of pancreatic β cells in vivo. Cell Metab. 15, 885–894 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.S. P. Singh, S. Janjuha, T. Hartmann, O. Kayisoglu, J. Konantz, S. Birke, P. Murawala, E. A. Alfar, K. Murata, A. Eugster, N. Tsuji, E. R. Morrissey, M. Brand, N. Ninov, Different developmental histories of beta-cells generate functional and proliferative heterogeneity during islet growth. Nat. Commun. 8, 664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Y. Wang, M. Rovira, S. Yusuff, M. J. Parsons, Genetic inducible fate mapping in larval zebrafish reveals origins of adult insulin-producing β-cells. Development 138, 609–617 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.F. Delaspre, R. L. Beer, M. Rovira, W. Huang, G. Wang, S. Gee, C. Vitery Mdel, S. J. Wheelan, M. J. Parsons, Centroacinar cells are progenitors that contribute to endocrine pancreas regeneration. Diabetes 64, 3499–3509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. L. Beer, M. J. Parsons, M. Rovira, Centroacinar cells: At the center of pancreas regeneration. Dev. Biol. 413, 8–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.L. Ye, M. A. Robertson, D. Hesselson, D. Y. Stainier, R. M. Anderson, glucagon is essential for alpha cell transdifferentiation and beta cell neogenesis. Development 142, 1407–1417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.J. Lu, K. C. Liu, N. Schulz, C. Karampelias, J. Charbord, A. Hilding, L. Rautio, P. Bertolino, C. G. Ostenson, K. Brismar, O. Andersson, IGFBP1 increases β-cell regeneration by promoting α- to β-cell transdifferentiation. EMBO J. 35, 2026–2044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.C. Karampelias, H. Rezanejad, M. Rosko, L. Duan, J. Lu, L. Pazzagli, P. Bertolino, C. E. Cesta, X. Liu, G. S. Korbutt, O. Andersson, Reinforcing one-carbon metabolism via folic acid/Folr1 promotes β-cell differentiation. Nat. Commun. 12, 3362 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.S. P. Singh, P. Chawla, A. Hnatiuk, M. Kamel, L. D. Silva, B. Spanjaard, S. E. Eski, S. Janjuha, P. Olivares-Chauvet, O. Kayisoglu, F. Rost, J. Blasche, A. Krankel, A. Petzold, T. Kurth, S. Reinhardt, J. P. Junker, N. Ninov, A single-cell atlas of de novo β-cell regeneration reveals the contribution of hybrid β/δ-cells to diabetes recovery in zebrafish. Development 149, dev199853 (2022). [DOI] [PubMed] [Google Scholar]
- 38.I. Manfroid, X. Caubit, S. Kerridge, L. Fasano, Three putative murine Teashirt orthologues specify trunk structures in Drosophila in the same way as the Drosophila teashirt gene. Development 131, 1065–1073 (2004). [DOI] [PubMed] [Google Scholar]
- 39.C. A. Carril Pardo, L. Massoz, M. A. Dupont, D. Bergemann, J. Bourdouxhe, A. Lavergne, E. Tarifeno-Saldivia, C. S. Helker, D. Y. Stainier, B. Peers, M. M. Voz, I. Manfroid, A δ-cell subpopulation with a pro-β-cell identity contributes to efficient age-independent recovery in a zebrafish model of diabetes. eLife 11, e67576 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.H. A. Field, P. D. Dong, D. Beis, D. Y. Stainier, Formation of the digestive system in zebrafish. II. Pancreas morphogenesis. Dev. Biol. 261, 197–208 (2003). [DOI] [PubMed] [Google Scholar]
- 41.N. Ninov, D. Hesselson, P. Gut, A. Zhou, K. Fidelin, D. Y. Stainier, Metabolic regulation of cellular plasticity in the pancreas. Curr. Biol. 23, 1242–1250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.M. J. Parsons, H. Pisharath, S. Yusuff, J. C. Moore, A. F. Siekmann, N. Lawson, S. D. Leach, Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898–912 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.N. Ninov, M. Borius, D. Y. Stainier, Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development 139, 1557–1567 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.B. Spanjaard, B. Hu, N. Mitic, P. Olivares-Chauvet, S. Janjuha, N. Ninov, J. P. Junker, Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat. Biotechnol. 36, 469–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.P. Panza, A. A. Sitko, H. M. Maischein, I. Koch, M. Flotenmeyer, G. J. Wright, K. Mandai, C. A. Mason, C. Sollner, The LRR receptor Islr2 is required for retinal axon routing at the vertebrate optic chiasm. Neural. Dev. 10, 23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.C. Karampelias, K. Watt, C. L. Mattsson, A. F. Ruiz, H. Rezanejad, J. Mi, X. Liu, L. Chu, J. W. Locasale, G. S. Korbutt, M. Rovira, O. Larsson, O. Andersson, MNK2 deficiency potentiates β-cell regeneration via translational regulation. Nat. Chem. Biol. 18, 942–953 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.J. Mi, O. Andersson, Efficient knock-in method enabling lineage tracing in zebrafish. Life Sci. Alliance 6, e202301944 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.L. C. Flasse, J. L. Pirson, D. G. Stern, V. Von Berg, I. Manfroid, B. Peers, M. L. Voz, Ascl1b and Neurod1, instead of Neurog3, control pancreatic endocrine cell fate in zebrafish. BMC Biol. 11, 78 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R. A. Kimmel, L. Onder, A. Wilfinger, E. Ellertsdottir, D. Meyer, Requirement for Pdx1 in specification of latent endocrine progenitors in zebrafish. BMC Biol. 9, 75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.C. C. Murdoch, S. T. Espenschied, M. A. Matty, O. Mueller, D. M. Tobin, J. F. Rawls, Intestinal serum amyloid A suppresses systemic neutrophil activation and bactericidal activity in response to microbiota colonization. PLOS Pathog. 15, e1007381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.J. Peloggia, D. Munch, P. Meneses-Giles, A. Romero-Carvajal, M. E. Lush, N. D. Lawson, M. McClain, Y. A. Pan, T. Piotrowski, Adaptive cell invasion maintains lateral line organ homeostasis in response to environmental changes. Dev. Cell 56, 1296–1312.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.J. Wen, G. P. Mercado, A. Volland, H. L. Doden, C. R. Lickwar, T. Crooks, G. Kakiyama, C. Kelly, J. L. Cocchiaro, J. M. Ridlon, J. F. Rawls, Fxr signaling and microbial metabolism of bile salts in the zebrafish intestine. Sci. Adv. 7, eabg1371 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.A. Apelqvist, H. Li, L. Sommer, P. Beatus, D. J. Anderson, T. Honjo, M. Hrabe de Angelis, U. Lendahl, H. Edlund, Notch signalling controls pancreatic cell differentiation. Nature 400, 877–881 (1999). [DOI] [PubMed] [Google Scholar]
- 54.J. Jensen, E. E. Pedersen, P. Galante, J. Hald, R. S. Heller, M. Ishibashi, R. Kageyama, F. Guillemot, P. Serup, O. D. Madsen, Control of endodermal endocrine development by Hes-1. Nat. Genet. 24, 36–44 (2000). [DOI] [PubMed] [Google Scholar]
- 55.G. N. Huang, J. E. Thatcher, J. McAnally, Y. Kong, X. Qi, W. Tan, J. M. DiMaio, J. F. Amatruda, R. D. Gerard, J. A. Hill, R. Bassel-Duby, E. N. Olson, C/EBP transcription factors mediate epicardial activation during heart development and injury. Science 338, 1599–1603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Baek, N. T. T. Tran, D. C. Diaz, Y. Y. Tsai, J. N. Acedo, M. E. Lush, T. Piotrowski, Single-cell transcriptome analysis reveals three sequential phases of gene expression during zebrafish sensory hair cell regeneration. Dev. Cell 57, 799–819.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.L. Ye, M. A. Robertson, T. L. Mastracci, R. M. Anderson, An insulin signaling feedback loop regulates pancreas progenitor cell differentiation during islet development and regeneration. Dev. Biol. 409, 354–369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.P. D. Dong, C. A. Munson, W. Norton, C. Crosnier, X. Pan, Z. Gong, C. J. Neumann, D. Y. Stainier, Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nat. Genet. 39, 397–402 (2007). [DOI] [PubMed] [Google Scholar]
- 59.D. Zhang, V. S. Golubkov, W. Han, R. G. Correa, Y. Zhou, S. Lee, A. Y. Strongin, P. D. Dong, Identification of Annexin A4 as a hepatopancreas factor involved in liver cell survival. Dev. Biol. 395, 96–110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.W. S. Chung, O. Andersson, R. Row, D. Kimelman, D. Y. Stainier, Suppression of Alk8-mediated Bmp signaling cell-autonomously induces pancreatic β-cells in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 107, 1142–1147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.S. Marques, D. van Bruggen, D. P. Vanichkina, E. M. Floriddia, H. Munguba, L. Varemo, S. Giacomello, A. M. Falcao, M. Meijer, A. K. Bjorklund, J. Hjerling-Leffler, R. J. Taft, G. Castelo-Branco, Transcriptional convergence of oligodendrocyte lineage progenitors during development. Dev. Cell 46, 504–517.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.A. P. Ghaye, D. Bergemann, E. Tarifeno-Saldivia, L. C. Flasse, V. Von Berg, B. Peers, M. L. Voz, I. Manfroid, Progenitor potential of nkx6.1-expressing cells throughout zebrafish life and during beta cell regeneration. BMC Biol. 13, 70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.C. Zhao, J. Matalonga, J. J. Lancman, L. Liu, C. Xiao, S. Kumar, K. P. Gates, J. He, A. Graves, J. Huisken, M. Azuma, Z. Lu, C. Chen, B. S. Ding, P. D. S. Dong, Regenerative failure of intrahepatic biliary cells in Alagille syndrome rescued by elevated Jagged/Notch/Sox9 signaling. Proc. Natl. Acad. Sci. U.S.A. 119, e2201097119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D. Zhang, K. P. Gates, L. Barske, G. Wang, J. J. Lancman, X. I. Zeng, M. Groff, K. Wang, M. J. Parsons, J. G. Crump, P. D. S. Dong, Endoderm Jagged induces liver and pancreas duct lineage in zebrafish. Nat. Commun. 8, 769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.M. I. Thestrup, S. Caviglia, J. Cayuso, R. L. S. Heyne, R. Ahmad, W. Hofmeister, L. Satriano, D. G. Wilkinson, J. B. Andersen, E. A. Ober, A morphogenetic EphB/EphrinB code controls hepatopancreatic duct formation. Nat. Commun. 10, 5220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.D. Balboa, T. Barsby, V. Lithovius, J. Saarimaki-Vire, M. Omar-Hmeadi, O. Dyachok, H. Montaser, P. E. Lund, M. Yang, H. Ibrahim, A. Naatanen, V. Chandra, H. Vihinen, E. Jokitalo, J. Kvist, J. Ustinov, A. I. Nieminen, E. Kuuluvainen, V. Hietakangas, P. Katajisto, J. Lau, P. O. Carlsson, S. Barg, A. Tengholm, T. Otonkoski, Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cells. Nat. Biotechnol. 40, 1042–1055 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.J. P. Tiano, V. Delghingaro-Augusto, C. Le May, S. Liu, M. K. Kaw, S. S. Khuder, M. G. Latour, S. A. Bhatt, K. S. Korach, S. M. Najjar, M. Prentki, F. Mauvais-Jarvis, Estrogen receptor activation reduces lipid synthesis in pancreatic islets and prevents β cell failure in rodent models of type 2 diabetes. J. Clin. Invest. 121, 3331–3342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.C. Le May, K. Chu, M. Hu, C. S. Ortega, E. R. Simpson, K. S. Korach, M. J. Tsai, F. Mauvais-Jarvis, Estrogens protect pancreatic β-cells from apoptosis and prevent insulin-deficient diabetes mellitus in mice. Proc. Natl. Acad. Sci. U.S.A. 103, 9232–9237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.B. Xu, C. Allard, A. I. Alvarez-Mercado, T. Fuselier, J. H. Kim, L. A. Coons, S. C. Hewitt, F. Urano, K. S. Korach, E. R. Levin, P. Arvan, Z. E. Floyd, F. Mauvais-Jarvis, Estrogens promote misfolded proinsulin degradation to protect insulin production and delay diabetes. Cell Rep. 24, 181–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.S. Liu, C. Le May, W. P. Wong, R. D. Ward, D. J. Clegg, M. Marcelli, K. S. Korach, F. Mauvais-Jarvis, Importance of extranuclear estrogen receptor-α and membrane G protein-coupled estrogen receptor in pancreatic islet survival. Diabetes 58, 2292–2302 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.W. P. Wong, J. P. Tiano, S. Liu, S. C. Hewitt, C. Le May, S. Dalle, J. A. Katzenellenbogen, B. S. Katzenellenbogen, K. S. Korach, F. Mauvais-Jarvis, Extranuclear estrogen receptor-α stimulates NeuroD1 binding to the insulin promoter and favors insulin synthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 13057–13062 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Z. Zhou, V. Ribas, P. Rajbhandari, B. G. Drew, T. M. Moore, A. H. Fluitt, B. R. Reddish, K. A. Whitney, S. Georgia, L. Vergnes, K. Reue, M. Liesa, O. Shirihai, A. M. van der Bliek, N. W. Chi, S. K. Mahata, J. P. Tiano, S. C. Hewitt, P. Tontonoz, K. S. Korach, F. Mauvais-Jarvis, A. L. Hevener, Estrogen receptor α protects pancreatic β-cells from apoptosis by preserving mitochondrial function and suppressing endoplasmic reticulum stress. J. Biol. Chem. 293, 4735–4751 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.T. Fuselier, P. Mota de Sa, M. M. F. Qadir, B. Xu, C. Allard, M. M. Meyers, J. P. Tiano, B. S. Yang, V. Gelfanov, S. H. Lindsey, R. D. Dimarchi, F. Mauvais-Jarvis, Efficacy of glucagon-like peptide-1 and estrogen dual agonist in pancreatic islets protection and pre-clinical models of insulin-deficient diabetes. Cell Rep. Med. 3, 100598 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.M. Sjoqvist, E. R. Andersson, Do as I say, Not(ch) as I do: Lateral control of cell fate. Dev. Biol. 447, 58–70 (2019). [DOI] [PubMed] [Google Scholar]
- 75.P. A. Seymour, C. A. Collin, A. R. Egeskov-Madsen, M. C. Jorgensen, H. Shimojo, I. Imayoshi, K. H. de Lichtenberg, R. Kopan, R. Kageyama, P. Serup, Jag1 modulates an oscillatory Dll1-Notch-Hes1 signaling module to coordinate growth and fate of pancreatic progenitors. Dev. Cell 52, 731–747.e8 (2020). [DOI] [PubMed] [Google Scholar]
- 76.E. Duvall, C. M. Benitez, K. Tellez, M. Enge, P. T. Pauerstein, L. Li, S. Baek, S. R. Quake, J. P. Smith, N. C. Sheffield, S. K. Kim, H. E. Arda, Single-cell transcriptome and accessible chromatin dynamics during endocrine pancreas development. Proc. Natl. Acad. Sci. U.S.A. 119, e2201267119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.d. L. O. Sean, Z. Liu, H. Sun, S. K. Yu, D. M. Wong, E. Chu, S. A. Rao, N. Eng, G. Peixoto, J. Bouza, Y. Shen, S. M. Knox, A. D. Tward, A. L. Gloyn, J. B. Sneddon, Single-cell multi-omic roadmap of human fetal pancreatic development. bioRxiv 2022.02.17.480942 [Preprint]. 18 February 2022. 10.1101/2022.02.17.480942 [DOI]
- 78.N. Sharon, R. Chawla, J. Mueller, J. Vanderhooft, L. J. Whitehorn, B. Rosenthal, M. Gurtler, R. R. Estanboulieh, D. Shvartsman, D. K. Gifford, C. Trapnell, D. Melton, A peninsular structure coordinates asynchronous differentiation with morphogenesis to generate pancreatic islets. Cell 176, 790–804.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.T. T.-H. Lu, S. Heyne, E. Dror, E. Casas, L. Leonhardt, T. Boenke, C.-H. Yang, Sagar, L. Arrigoni, K. Dalgaard, R. Teperino, L. Enders, M. Selvaraj, M. Ruf, S. J. Raja, H. Xie, U. Boenisch, S. H. Orkin, F. C. Lynn, B. G. Hoffman, D. Grün, T. Vavouri, A. M. Lempradl, J. A. Pospisilik, The polycomb-dependent epigenome controls β cell dysfunction, dedifferentiation, and diabetes. Cell Metab. 27, 1294–1308.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.L. Bernardino, F. Agasse, B. Silva, R. Ferreira, S. Grade, J. O. Malva, Tumor necrosis factor-alpha modulates survival, proliferation, and neuronal differentiation in neonatal subventricular zone cell cultures. Stem Cells 26, 2361–2371 (2008). [DOI] [PubMed] [Google Scholar]
- 81.L. Cavone, T. McCann, L. K. Drake, E. A. Aguzzi, A. M. Oprisoreanu, E. Pedersen, S. Sandi, J. Selvarajah, T. M. Tsarouchas, D. Wehner, M. Keatinge, K. S. Mysiak, B. E. P. Henderson, R. Dobie, N. C. Henderson, T. Becker, C. G. Becker, A unique macrophage subpopulation signals directly to progenitor cells to promote regenerative neurogenesis in the zebrafish spinal cord. Dev. Cell 56, 1617–1630.e6 (2021). [DOI] [PubMed] [Google Scholar]
- 82.N. Kyritsis, C. Kizil, S. Zocher, V. Kroehne, J. Kaslin, D. Freudenreich, A. Iltzsche, M. Brand, Acute inflammation initiates the regenerative response in the adult zebrafish brain. Science 338, 1353–1356 (2012). [DOI] [PubMed] [Google Scholar]
- 83.J. Ohnmacht, Y. Yang, G. W. Maurer, A. Barreiro-Iglesias, T. M. Tsarouchas, D. Wehner, D. Sieger, C. G. Becker, T. Becker, Spinal motor neurons are regenerated after mechanical lesion and genetic ablation in larval zebrafish. Development 143, 1464–1474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.L. L. Leach, N. J. Hanovice, S. M. George, A. E. Gabriel, J. M. Gross, The immune response is a critical regulator of zebrafish retinal pigment epithelium regeneration. Proc. Natl. Acad. Sci. U.S.A. 118, e2017198118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.S. P. Hui, D. Z. Sheng, K. Sugimoto, A. Gonzalez-Rajal, S. Nakagawa, D. Hesselson, K. Kikuchi, Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 43, 659–672.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Y. Fang, V. Gupta, R. Karra, J. E. Holdway, K. Kikuchi, K. D. Poss, Translational profiling of cardiomyocytes identifies an early Jak1/Stat3 injury response required for zebrafish heart regeneration. Proc. Natl. Acad. Sci. U.S.A. 110, 13416–13421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.S. J. Burke, H. M. Batdorf, D. H. Burk, T. M. Martin, T. Mendoza, K. Stadler, W. Alami, M. D. Karlstad, M. J. Robson, R. D. Blakely, R. L. Mynatt, J. J. Collier, Pancreatic deletion of the interleukin-1 receptor disrupts whole body glucose homeostasis and promotes islet β-cell de-differentiation. Mol. Metab. 14, 95–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.A. Navis, M. Bagnat, Loss of cftr function leads to pancreatic destruction in larval zebrafish. Dev. Biol. 399, 237–248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.K. Asakawa, M. L. Suster, K. Mizusawa, S. Nagayoshi, T. Kotani, A. Urasaki, Y. Kishimoto, M. Hibi, K. Kawakami, Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. U.S.A. 105, 1255–1260 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.C. Mosimann, C. K. Kaufman, P. Li, E. K. Pugach, O. J. Tamplin, L. I. Zon, Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169–177 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.N. Obholzer, S. Wolfson, J. G. Trapani, W. Mo, A. Nechiporuk, E. Busch-Nentwich, C. Seiler, S. Sidi, C. Sollner, R. N. Duncan, A. Boehland, T. Nicolson, Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J. Neurosci. 28, 2110–2118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.C. S. M. Helker, S. T. Mullapudi, L. M. Mueller, J. Preussner, S. Tunaru, O. Skog, H. B. Kwon, F. Kreuder, J. J. Lancman, R. Bonnavion, P. D. S. Dong, M. Looso, S. Offermanns, O. Korsgren, F. M. Spagnoli, D. Y. R. Stainier, A. Whole organism small molecule screen identifies novel regulators of pancreatic endocrine development. Development 146, dev172569 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.B. B. Kirby, N. Takada, A. J. Latimer, J. Shin, T. J. Carney, R. N. Kelsh, B. Appel, In vivo time-lapse imaging shows dynamic oligodendrocyte progenitor behavior during zebrafish development. Nat. Neurosci. 9, 1506–1511 (2006). [DOI] [PubMed] [Google Scholar]
- 94.N. Tsuji, N. Ninov, M. Delawary, S. Osman, A. S. Roh, P. Gut, D. Y. Stainier, Whole organism high content screening identifies stimulators of pancreatic beta-cell proliferation. PLOS ONE 9, e104112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.M. Astone, J. K. H. Lai, S. Dupont, D. Y. R. Stainier, F. Argenton, A. Vettori, Zebrafish mutants and TEAD reporters reveal essential functions for Yap and Taz in posterior cardinal vein development. Sci. Rep. 8, 10189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.N. Abu Seman, B. He, J. R. Ojala, W. N. Wan Mohamud, C. G. Ostenson, K. Brismar, H. F. Gu, Genetic and biological effects of sodium-chloride cotransporter (SLC12A3) in diabetic nephropathy. Am. J. Nephrol. 40, 408–416 (2014). [DOI] [PubMed] [Google Scholar]
- 97.D. T. Grimes, C. W. Boswell, N. F. Morante, R. M. Henkelman, R. D. Burdine, B. Ciruna, Zebrafish models of idiopathic scoliosis link cerebrospinal fluid flow defects to spine curvature. Science 352, 1341–1344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.L. Chu, M. Terasaki, C. L. Mattsson, R. Teinturier, J. Charbord, E. Dirice, K. C. Liu, M. G. Miskelly, Q. Zhou, N. Wierup, R. N. Kulkarni, O. Andersson, In vivo drug discovery for increasing incretin-expressing cells identifies DYRK inhibitors that reinforce the enteroendocrine system. Chem. Biol. 29, 1368–1380.e5 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.K. C. Liu, A. Villasenor, M. Bertuzzi, N. Schmitner, N. Radros, L. Rautio, K. Mattonet, R. L. Matsuoka, S. Reischauer, D. Y. Stainier, O. Andersson, Insulin-producing β-cells regenerate ectopically from a mesodermal origin under the perturbation of hemato-endothelial specification. eLife 10, e65758 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.H. M. T. Choi, M. Schwarzkopf, M. E. Fornace, A. Acharya, G. Artavanis, J. Stegmaier, A. Cunha, N. A. Pierce, Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Y. Hao, S. Hao, E. Andersen-Nissen, W. M. Mauck III, S. Zheng, A. Butler, M. J. Lee, A. J. Wilk, C. Darby, M. Zager, P. Hoffman, M. Stoeckius, E. Papalexi, E. P. Mimitou, J. Jain, A. Srivastava, T. Stuart, L. M. Fleming, B. Yeung, A. J. Rogers, J. M. McElrath, C. A. Blish, R. Gottardo, P. Smibert, R. Satija, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.C. S. McGinnis, L. M. Murrow, Z. J. Gartner, DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.I. Korsunsky, N. Millard, J. Fan, K. Slowikowski, F. Zhang, K. Wei, Y. Baglaenko, M. Brenner, P. R. Loh, S. Raychaudhuri, Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.S. Aibar, C. B. Gonzalez-Blas, T. Moerman, V. A. Huynh-Thu, H. Imrichova, G. Hulselmans, F. Rambow, J. C. Marine, P. Geurts, J. Aerts, J. van den Oord, Z. K. Atak, J. Wouters, S. Aerts, SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.B. Van de Sande, C. Flerin, K. Davie, M. De Waegeneer, G. Hulselmans, S. Aibar, R. Seurinck, W. Saelens, R. Cannoodt, Q. Rouchon, T. Verbeiren, D. De Maeyer, J. Reumers, Y. Saeys, S. Aerts, A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 107.C. Liu, R. Li, Y. Li, X. Lin, K. Zhao, Q. Liu, S. Wang, X. Yang, X. Shi, Y. Ma, C. Pei, H. Wang, W. Bao, J. Hui, T. Yang, Z. Xu, T. Lai, M. A. Berberoglu, S. K. Sahu, M. A. Esteban, K. Ma, G. Fan, Y. Li, S. Liu, A. Chen, X. Xu, Z. Dong, L. Liu, Spatiotemporal mapping of gene expression landscapes and developmental trajectories during zebrafish embryogenesis. Dev. Cell 57, 1284–1298.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 108.S. Suo, Q. Zhu, A. Saadatpour, L. Fei, G. Guo, G. C. Yuan, Revealing the critical regulators of cell identity in the mouse cell atlas. Cell Rep. 25, 1436–1445.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.K. Street, D. Risso, R. B. Fletcher, D. Das, J. Ngai, N. Yosef, E. Purdom, S. Dudoit, Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.X. Qiu, Q. Mao, Y. Tang, L. Wang, R. Chawla, H. A. Pliner, C. Trapnell, Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.G. Yu, L. G. Wang, Y. Han, Q. Y. He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.S. Hanzelmann, R. Castelo, J. Guinney, GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.W. Liu, Y. Xie, J. Ma, X. Luo, P. Nie, Z. Zuo, U. Lahrmann, Q. Zhao, Y. Zheng, Y. Zhao, Y. Xue, J. Ren, IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15
Legends for movies S1 to S3
Legends for tables S1 to S35
Tables S1 to S35
Movies S1 to S3