Abstract
Six checkpoint Rad proteins (Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1) are needed to regulate checkpoint protein kinases Chk1 and Cds1 in fission yeast. Chk1 is required to prevent mitosis when DNA is damaged by ionizing radiation (IR), whereas either kinase is sufficient to prevent mitosis when DNA replication is inhibited by hydroxyurea (HU). Checkpoint Rad proteins are required for IR-induced phosphorylation of Chk1 and HU-induced activation of Cds1. IR activates Cds1 only during the DNA synthesis (S) phase, whereas HU induces Chk1 phosphorylation only in cds1 mutants. Here, we investigate the basis of the checkpoint signal specificity of Chk1 phosphorylation and Cds1 activation. We show that IR fails to induce Chk1 phosphorylation in HU-arrested cells. Release from the HU arrest following IR causes substantial Chk1 phosphorylation. These and other data indicate that Cds1 prevents Chk1 phosphorylation in HU-arrested cells, which suggests that Cds1 actively suppresses a repair process that leads to Chk1 phosphorylation. Cds1 becomes more highly concentrated in the nucleus only during the S phase of the cell cycle. This finding correlates with S-phase specificity of IR-induced activation of Cds1. However, constitutive nuclear localization of Cds1 does not enhance IR-induced activation of Cds1. This result suggests that Cds1 activation requires DNA structures or protein activities that are present only during S phase. These findings help to explain how Chk1 and Cds1 respond to different checkpoint signals.
Genomic integrity is enhanced by cell cycle checkpoints that prevent the onset of mitosis while DNA replication or repair is underway (11, 19–21). Checkpoint defects contribute to genomic instability in human cells; thus, an understanding of checkpoint signaling mechanisms may assist efforts aimed at combating tumor development and other diseases.
Studies of the fission yeast Schizosaccharomyces pombe have played an important role in the unraveling of checkpoint mechanisms (10, 36, 38). These studies have focused on “checkpoint Rad” proteins that are required for both the replication checkpoint elicited by hydroxyurea (HU) and the repair checkpoint activated by DNA-damaging agents, such as ionizing radiation (IR). The list of checkpoint Rad proteins includes Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1. The biochemical functions of checkpoint Rad proteins are poorly understood, but they appear to be involved in sensing stalled replication complexes and damaged DNA. Many of these proteins have human homologs, including Rad3, which has substantial structural and functional similarity to the human ATM and ATR proteins (5). The list of proteins involved in checkpoints continues to expand. Recent studies have identified Cut5/Rad4 and Crb2/Rhp9 as checkpoint proteins in fission yeast (39, 40, 46).
The checkpoint Rad proteins are thought to activate or be components of a signal transduction process that regulates elements of the mitotic control network. One of these signal transduction proteins is Chk1, a protein kinase that is required for the repair checkpoint (2, 44). DNA damage causes phosphorylation of Chk1 by a mechanism that requires checkpoint Rad proteins, but the identity of the Chk1-directed kinase and the effect of phosphorylation remain to be discovered (45). Chk1 phosphorylates Cdc25, the protein phosphatase that dephosphorylates tyrosine-15 of the cyclin-dependent kinase Cdc2 (17, 23, 33, 41). The tyrosine-dephosphorylated form of Cdc2 induces the onset of mitosis. The repair checkpoint decreases Cdc2 tyrosine-15 dephosphorylation in vivo, indicating that Chk1 inhibits Cdc25 (17, 34). This hypothesis was recently confirmed by direct inhibition of Cdc25 by Chk1 in vitro (6, 16). Chk1 also induces association of Cdc25 to 14-3-3 proteins (23, 33, 41, 48). DNA damage induces net nuclear export of Cdc25 by a process that is dependent on Rad24, a 14-3-3 protein (25). Cdc2/cyclin-B, the substrate of Cdc25, is localized in the nucleus; thus, nuclear export of Cdc25 is expected to inhibit mitosis.
The repair and replication checkpoints require the same six checkpoint Rad proteins (2, 12) and regulate tyrosine-15 phosphorylation of Cdc2 (13, 26, 34, 35). However, the two checkpoints differ in their requirements for signaling proteins that function downstream of the checkpoint Rad proteins but upstream of the proteins that regulate Cdc2. Thus, Chk1 is essential for the repair checkpoint activated by DNA damage but not the replication checkpoint elicited by HU. This may be explained if the replication checkpoint has an element of redundancy that is absent in the repair checkpoint. Support for this hypothesis came from recent studies that showed that Chk1 is essential for HU-induced checkpoint arrest in cds1 mutant cells (7, 24, 48). Cds1 is a protein kinase that is activated by a checkpoint Rad protein-dependent process in cells treated with HU (7, 24, 31). Increased production of Cds1 prevented mitosis, which suggested that Cds1 might be an effector of the replication checkpoint (7). Cds1 was proposed to positively regulate Wee1 and Mik1, the two protein tyrosine kinases that phosphorylate Cdc2 on tyrosine-15 (7). Cds1 also regulates Cdc25 by catalyzing phosphorylation on the same sites that are phosphorylated by Chk1 (16, 48). Cds1 also has an HU “recovery” function, because the HU checkpoint is largely intact in cds1 cells and yet cell viability is greatly reduced (31).
A model in which Chk1 and Cds1 act as dual effectors of the replication checkpoint explains why the checkpoint is intact in cds1 and chk1 single mutants but absent in the double mutant. However, the model does not explain why Chk1 remains unphosphorylated in response to HU treatment in wild-type cells (45) or why HU treatment of cds1 mutant cells leads to substantial phosphorylation of Chk1 (24). It was proposed that Cds1 might be required to stabilize replication structures in HU-arrested cells, thus preventing DNA damage that would lead to phosphorylation of Chk1 (24). This model accounts for many findings, but it does not explain why the activity of checkpoint Rad proteins should lead to activation of Cds1 but not phosphorylation of Chk1 in HU-treated cells. Nor does the model explain why IR and other agents that damage DNA induce phosphorylation of Chk1 but not activation of Cds1 in G2, the period of the cell cycle that follows the DNA synthesis (S) phase. Here, we report that IR-induced phosphorylation of Chk1 is actively prevented in HU-arrested cells. This finding suggests that suppression of repair processes that lead to Chk1 phosphorylation might be important in HU-arrested cells. We also report that Cds1 becomes more highly concentrated in the nucleus during S phase and in cells treated with HU. These findings provide a basis for understanding the checkpoint signal specificity of Chk1 phosphorylation and Cds1 activation.
MATERIALS AND METHODS
Strains, plasmids, and general techniques.
The following strains were used in this study: PR109 (wild type), BF1919 (chk1:HAHIS), NR1592 (chk1::ura4+), JMB2274 (chk1:HAHIS cds1::ura4+), JMB2275 (nmt1:GST-cds1+ chk1:HAHIS), NB2276 (cds1-GFP), and NB2342 (cds1-NLSGFP). All strains were leu1-32 ura4-D18. The chk1::ura4+, cds1::ura4+, and nmt1:GST-cds1+ constructs have been described (7, 17, 34). The chromosomal copy of chk1+ was tagged with a sequence encoding two copies of the hemagglutinin (HA) epitope and hexahistidine by using a previously described strategy (42). To make the cds1-GFP construct, the open reading frame of cds1+ (nucleotides 346 to 2016) was amplified by PCR by using the following primers: 5′-CGCCCGCGCCTGCAGCGCATGCTTGATGGTAAG-3′ and 5′-CAGCATGCGGCCGCTACTCGAAGAATTGAGCTG-3′. To make the cds1-NLS-GFP construct, the open reading frame of cds1+ (nucleotides 346 to 2016) was amplified by PCR by using the following primers: 5′-CGCCCGCGCCTGCAGCGCATGCTTGATGGTAAG-3′ and 5′-CAGCATGCGGCCGCTCTTACGCTTCTTCTTAGGACTCGAAGAATTGAGCTG-3′. The PCR products were digested with PstI and NotI and cloned into the vector pXGFP, which placed the cds1+ open reading frame upstream of and in frame with the green fluorescent protein (GFP) open reading frame. Plasmid pXGFP, a plasmid that has the selectable marker ura4+, also contains the nmt1 terminator sequence downstream of GFP (8). The resulting vector was digested with NheI in the cds1 sequence and integrated at the cds1+ locus in PR109. The resultant strain expresses Cds1-GFP from the cds1 locus. The same approach was used to express Cds1-nuclear localization signal (NLS)-GFP from the cds1 locus. Growth media and general methods for S. pombe have been described (30). Unless otherwise indicated, yeast cultures were grown at 30°C in YES medium (glucose, yeast extract, amino acid supplements). HU was used at a concentration of 12 mM. Cells were irradiated at a dose of 100 Gy with a 137Cs source. Growth media and conditions for induction of nmt1-driven constructs have been described (4). Synchronized cultures were made by centrifugal elutriation. Fluorescence-activated cell sorter (FACS) analysis was performed with ethanol-fixed cells at an optical density at 600 nm (OD600) of 1 as described (37).
Immunoblotting and microscopy.
Cells were lysed in buffer A (50 mM Tris [pH 8], 150 mM NaCl, 5 mM EGTA, 10% glycerol, 0.1% Nonidet P-40, 5 mg of leupeptin-aprotinin-pepstatin per ml, and 1 mM phenylmethylsulfonyl fluoride). The protein concentration was normalized by using the OD280 reading, and the proteins were separated by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis (acrylamide:bisacrylamide ratio, 29.2/0.8) and transferred to nitrocellulose membrane. Blots were blocked with 5% milk in TBST (25 mM Tris [pH 7.6], 137 mM NaCl, and 0.2% Tween 20). Chk1HAHIS was precipitated with Ni2+-nitrilotriacetic acid beads and revealed with antibodies to HA, followed by anti-mouse immunoglobulin G antibodies coupled with horseradish peroxidase. Enhanced chemiluminescence detection (Pierce) was used to visualize proteins. The Cds1 kinase assay was performed as described by using glutathione S-transferase (GST)-Wee11–152 and the GST-Wee11–70 truncation product as substrates (7). Cells were photographed by using a Nikon Eclipse E800 microscope equipped with a Photometrics Quantix charge-coupled device camera. Images were acquired with IPLab Spectrum software (Signal Analytics Corporation).
RESULTS
Chk1 is phosphorylated and delays mitosis when cells recover from an HU-induced arrest.
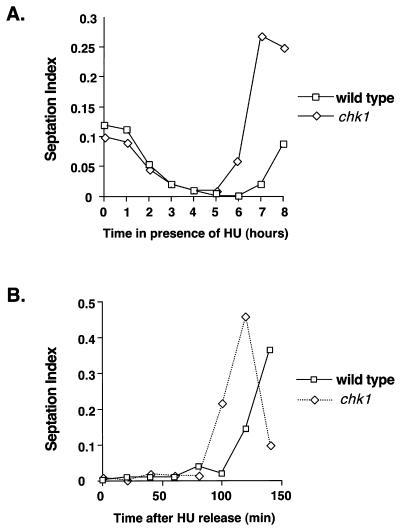
The studies described in this report were prompted by an experiment that carefully monitored the division kinetics of wild-type and chk1 cells that were incubated with 12 mM HU for 8 h at 30°C. In agreement with the results of previous studies (3, 44), the two strains underwent checkpoint arrest with very similar kinetics, as shown by the parallel decreases in septation index (Fig. 1A). Fission yeast cells eventually replicate DNA in medium containing 12 mM HU (31), as indicated by the reappearance of septated cells after approximately 7 h in the wild-type cell culture (Fig. 1A). In this experiment, resumption of division in the chk1 cell culture was advanced approximately 1 h relative to that in the wild type (Fig. 1A). This phenomenon was examined in a different experimental protocol, in which wild-type and chk1 cells were treated with HU for 4 h and then washed in medium lacking HU. In this experiment, division in the chk1 cell culture was advanced approximately 20 min relative to that in the wild type (Fig. 1B).
FIG. 1.
Chk1 delays mitosis during recovery from an HU-induced arrest. (A) Wild-type or chk1 cells were treated with HU, and the septation index was monitored at hourly intervals. The chk1 strain underwent division before the wild-type strain. (B) Wild-type or chk1 strains were treated with HU for 4 h. HU was then removed by washing cells in YES media, and the septation index was monitored every 20 min. Division was advanced in the chk1 cells relative to that in the wild type.
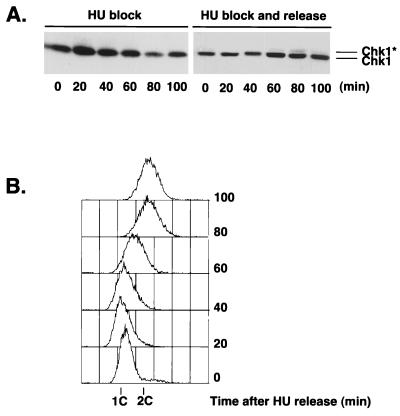
Wild-type and chk1 cells normally divide at the same size when grown in the absence of HU or DNA-damaging agents. Thus, Chk1 apparently has no role in determining the timing of mitosis, except when DNA is damaged. The fact that mitosis is advanced in chk1 cells relative to wild-type cells as these cells recover from an HU treatment suggested that Chk1 might become activated following release from a replication checkpoint arrest. This proposal was explored by performing immunoblot analysis of Chk1 in cells released from a 4-h HU-induced arrest. The form of Chk1 with reduced mobility was not detected in cells held at the HU-induced arrest (Fig. 2), a finding consistent with previous studies (45). However, a small amount of the phosphorylated species of Chk1 was detected at later time points, and the species was most evident at 80 min following the release from the HU-induced arrest (Fig. 2A, right panel). The phosphorylated form of Chk1 was not detected for cells that were maintained in the presence of HU for an additional 100 min (Fig. 2A, left panel). Flow cytometry analysis showed that bulk DNA synthesis was largely completed at between 40 and 80 min in cells that were released from the HU-induced arrest (Fig. 2B), approximately coincident with the appearance of the phosphorylated form of Chk1.
FIG. 2.
Chk1 is phosphorylated as cells recover from an HU-induced arrest. Cells were treated with HU for 4 h at 30°C. HU was removed from half of the culture by washing in YES medium. The other half of the culture was left in the presence of HU. Cells were harvested every 20 min. (A) Samples were processed for immunoblot analysis of Chk1. (B) Samples were processed for FACS analysis to determine DNA content after HU release. ∗, phosphorylated form of Chk1.
Chk1 is not phosphorylated in response to DNA damage in cells held at the HU-induced arrest.
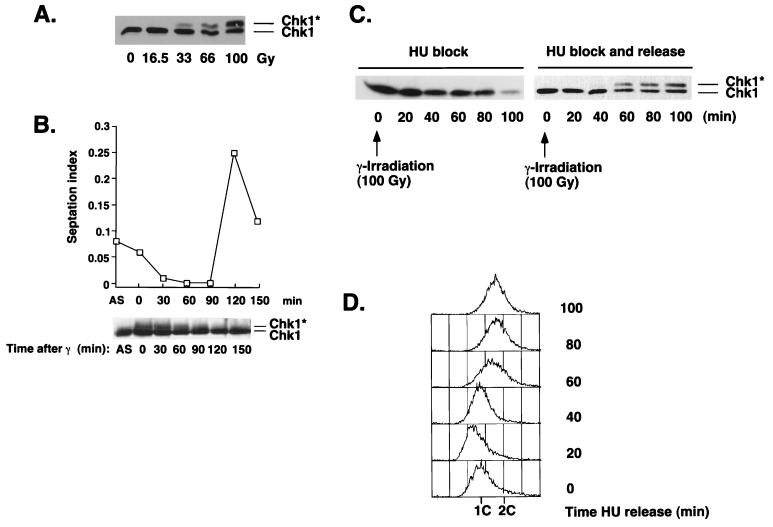
Our studies indicated that the Chk1-dependent repair checkpoint was activated as cells recovered from an HU-induced replication checkpoint but not while the cells were held at the HU-induced arrest. These observations suggested that HU causes DNA damage or at least DNA anomalies that are perceived as damage and that phosphorylation of Chk1 is nevertheless delayed until replication is largely completed. These findings raised the question of whether it was possible to activate the repair checkpoint (as assayed by Chk1 phosphorylation) in cells that were arrested at the replication checkpoint. This question was addressed by using IR to inflict DNA damage on cells that were arrested at the S- to M-phase replication checkpoint with HU. In agreement with previous studies (24, 45), we observed that IR treatment of cells in an asynchronous culture induced substantial phosphorylation of Chk1 (Fig. 3A). In this experiment, cells were harvested immediately after exposure to IR. There was a dose-dependent relationship between the amount of IR and the amount of phosphorylated Chk1. A dose of 100 Gy caused approximately 25 to 50% of the Chk1 to migrate with reduced electrophoretic mobility (Fig. 3A). In cells that were exposed to a 100-Gy dose of IR, the amount of phosphorylated Chk1 decayed with time following completion of irradiation (Fig. 3B). Phosphorylated Chk1 was almost undetectable at 120 min, which coincided with the resumption of cell division (Fig. 3B).
FIG. 3.
IR fails to cause Chk1 phosphorylation in cells arrested with HU. (A) An asynchronous culture of wild-type cells was exposed to γ-irradiation (0 to 100 Gy). Samples were processed immediately for immunoblot analysis of Chk1. (B) Wild-type cells were exposed to a 100-Gy dose of γ-irradiation. Samples were processed for immunoblot analysis of Chk1 and measurement of septation index during a 120-min time course. (C) Cells were treated with HU for 3.5 h at 30°C, followed by exposure to a 100-Gy dose of γ-irradiation. HU was removed from half of the culture by washing in YES medium. The other half of the culture was left in the presence of HU. Cells were harvested every 20 min. (D) Samples from the experiment described in the legend for panel B were processed for FACS analysis to determine the DNA content after HU release. ∗, phosphorylated form of Chk1.
Importantly, we found that a 100-Gy dose of IR was incapable of inducing Chk1 phosphorylation in HU-treated cells (Fig. 3C). However, the slower-mobility form of Chk1 became quite prominent within 60 min after the release from the HU-induced arrest (Fig. 3C, right panel). The phosphorylated form of Chk1 was not detected in cells that were maintained in the presence of HU (Fig. 3C, left panel). In this experiment most of the DNA was replicated at between 40 and 60 min after the release from the HU-induced arrest, apparently coincident with the appearance of the phosphorylated species of Chk1 (Fig. 3D). These data suggest that the repair checkpoint, or at least the signal transduction pathway that leads to Chk1 phosphorylation, cannot be activated while cells are arrested at the replication checkpoint.
Cds1 suppresses phosphorylation of Chk1.
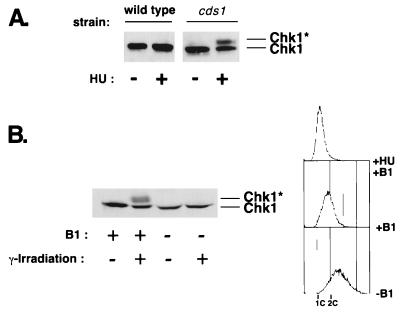
The protein kinase Cds1 is activated in HU-arrested cells and is essential for the replication checkpoint in a chk1 mutant background (7, 24). It was recently reported that Chk1 becomes phosphorylated in cds1 cells that have been treated with HU (24), as we have also observed (Fig. 4A). These findings suggested that Cds1 prevents DNA damage in HU-treated cells, thereby preventing phosphorylation of Chk1 (24). However, our studies revealed that Chk1 is not phosphorylated in HU-arrested cells when IR causes damage (Fig. 3). These findings suggested that Cds1 might actively prevent phosphorylation of Chk1. This idea was explored by determining whether expression of a large amount of Cds1 prevents phosphorylation of Chk1 in response to DNA damage. Expression of a large amount of GST-Cds1 under the control of the thiamine-repressible nmt1 promoter causes a cell cycle arrest (7). We observed that overproduction of GST-Cds1 completely suppressed phosphorylation of Chk1 induced by IR (Fig. 4B). These observations support a model in which activated Cds1 prevents Chk1 phosphorylation.
FIG. 4.
Overproduction of Cds1 prevents phosphorylation of Chk1. (A) HU induces Chk1 phosphorylation in a cds1 background. Wild-type or cds1 cells were HU treated for 3.5 h. Samples were processed for immunoblot analysis of Chk1. (B) GST-Cds1 overexpression in G2 prevents phosphorylation of Chk1 that is induced by DNA damage. Cells that expressed GST-Cds1 under the control of the thiamine-repressible nmt1 promoter were grown in minimal media containing thiamine (+B1; nmt1-repressing conditions) or lacking thiamine (−B1; nmt1-inducing conditions) for 20 h. Cells were γ irradiated or mock irradiated. Samples were processed for FACS analysis to determine DNA content (upper panel) or for immunoblot analysis of Chk1 (lower panel). ∗, phosphorylated form of Chk1.
Chk1 is not required to restrict damage-induced activation of Cds1 to S phase.
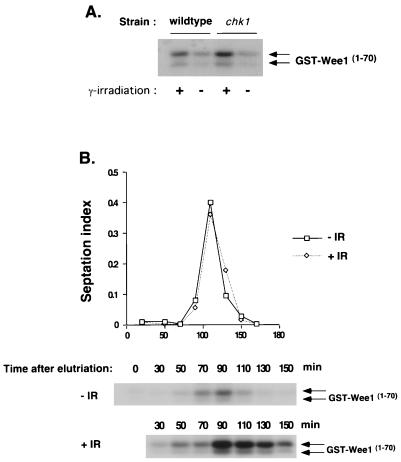
Our findings suggest a model in which activated Cds1 prevents the phosphorylation of Chk1. This model predicts that Cds1 should not be activated in situations in which Chk1 is normally phosphorylated, such as in irradiated cells that are in G2, the period between S phase and mitosis (M). This model receives support from a recent study that showed that Cds1 is activated by DNA damage but that the damage-induced activation of Cds1 is restricted to S phase (24). These observations raised the question of whether Chk1 is required to prevent damage-induced activation of Cds1 during G2. We designed two experiments to answer this question. First, we asked whether elimination of Chk1 leads to increased activation of Cds1 in asynchronous culture, in which ∼70% of cells are in G2. Cds1 activity was measured by using an assay in which a GST fusion protein containing an NH2-terminal fragment of Wee1 (GST-Wee170) is used both as an affinity reagent and as a substrate for Cds1 (7). This assay is highly specific for Cds1 (7). Thus, in this assay, the term activation refers to the ability of Cds1 to both bind and phosphorylate GST-Wee170. Irradiation of asynchronous wild-type and chk1 cells resulted in very similar levels of Cds1 activation (Fig. 5A). There appeared to be a slight increase in Cds1 activity in the chk1 mutant. However, this modest increase in Cds1 activity is probably due to the small number of chk1 cells that should initiate mitosis with damaged DNA and then enter S phase during the 30-min period of irradiation.
FIG. 5.
Chk1 does not prevent Cds1 activation in G2 phase. (A) Asynchronous cultures of wild-type or chk1 cells were irradiated with 100 Gy. Samples were harvested to measure Cds1 kinase activity by using GST-Wee11–152 as a substrate. (B) A chk1 strain was synchronized in G2 by elutriation. Cells were γ irradiated (+IR) or mock irradiated (−IR). Samples were harvested to measure septation index and Cds1 kinase activity by using GST-Wee11–152 as a substrate. The band shown corresponds to the GST-Wee11–70 degradation product as previously described (7). DNA damage incurred during the G2 phase activated Cds1 only after cells had completed mitosis and entered S phase. Arrows indicate two different forms of GST-Wee11–70.
The second experiment used a synchronous culture of chk1 cells in early G2 phase prepared by centrifugal elutriation. These cells were irradiated, or mock irradiated, and samples were harvested at regular intervals. Cell cycle progression was monitored by measurement of the septation index. Cds1 activity was negligible immediately after irradiation (30 min), which corresponds to G2 (Fig. 5). Cds1 activity remained quite low at the following two time points (50 and 70 min). These time points correspond to late G2 and M. A substantial increase in Cds1 activity occurred at 90 min. This time point coincides with the rise in septation index and the onset of S (Fig. 5). Cds1 activity remained high at the following time point and then gradually decreased. In contrast to the irradiated culture, the phosphorylation of GST-Wee170 remained relatively low in extracts made from the mock-irradiated culture. These data show that substantial damage-induced activation of Cds1 occurs only during S, a finding consistent with another recent study (24). Our data also show that Chk1 plays no role in preventing the damage-induced activation of Cds1 during G2.
Cds1 accumulates in the nucleus during S phase.
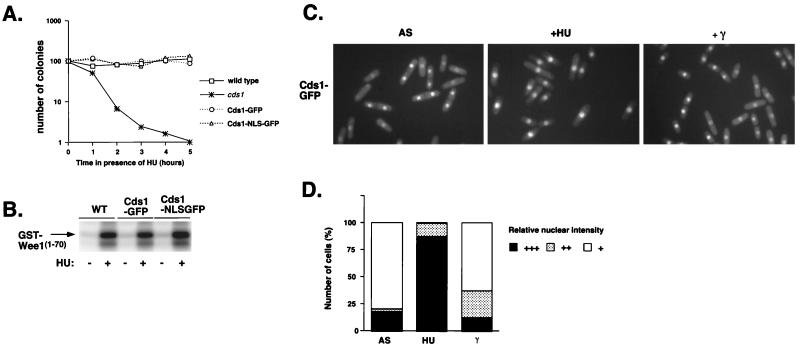
How is the activation of Cds1 by DNA damage restricted to S phase? One hypothesis that might explain this observation is that Cds1 is localized in the nucleus only during S phase. Examination of the localization of Cds1 that was expressed as a GFP fusion protein tested this hypothesis. This experiment used a strain in which the genomic copy of cds1+ was modified to encode Cds1 protein with GFP fused at the C terminus. This strain was identical to the wild type in regard to HU sensitivity (Fig. 6A) and HU-induced cell cycle arrest (8); thus, Cds1-GFP appeared to be fully functional. This conclusion was supported further by the observation that Cds1 and Cds1-GFP were activated equally by HU-treatment in the GST-Wee170 phosphorylation assay (Fig. 6B).
FIG. 6.
Cds1 accumulates in the nucleus during S phase. (A) Addition of GFP or NLS-GFP to the C terminus of Cds1 does not compromise its function. The HU sensitivity of wild-type (WT), Cds1-GFP, Cds1-NLS-GFP, and cds1 cells was determined by monitoring the colony formation during the time course of HU exposure. (B) Modified forms of Cds1 behave the same as the WT in the Cds1 kinase assay following treatment or mock treatment with HU. (C) Cds1-GFP localization was determined by fluorescence microscopy in an asynchronous population (left panel), after 4 h of HU treatment (middle panel), or after a 100-Gy dose of irradiation (right panel). Cds1 is nuclear in septated cells and attached daughters. During HU-induced arrest, Cds1 is strongly nuclear. In contrast, after irradiation, Cds1 is not accumulated in cells arrested at G2. (D) Percentages of cells with relative nuclear staining intensities for Cds1 in an asynchronous population (AS), HU-arrested cells, or 100-Gy-irradiated cells (γ).
The localization of Cds1-GFP was examined in an asynchronous culture containing cells in all phases of the cell cycle (Fig. 6C). Cds1-GFP was detected in the nuclei of all cells, but the nuclear signal appeared to be increased substantially in cells that contained a septum or were attached daughters (Fig. 6D). In asynchronous cultures, these cells are in S phase (29). In uninucleate cells that were in G2 phase, the nuclear Cds1-GFP signal appeared to be slightly higher than the cytoplasmic signal (Fig. 6C and D). In cultures treated with HU for 4 h, 79.5% of the cells presented a strong Cds1-GFP signal in the nucleus (Fig. 6C and D). Immunoblot analysis has shown that the abundance of Cds1 is equal in HU-treated and mock-treated cells (7, 24); thus, the increased nuclear signal in HU-arrested cells is apparently due to increased nuclear localization of Cds1. The cytoplasmic Cds1-GFP signal was not noticeably decreased in HU-arrested cells, but this is probably because the cytoplasmic signal is never substantially above background fluorescence. Thus, it appears that the increased nuclear signal of Cds1-GFP during S phase is driven by changes in the localization of Cds1-GFP. DNA damage appeared to have a slight effect on the localization of Cds1-GFP (Fig. 6C and D).
To explore whether the S-phase-specific activation of Cds1 by DNA damage was explained solely by protein localization, we examined the effect of constitutive nuclear localization of Cds1. This experiment used a strain in which the genomic copy of cds1+ was modified to encode Cds1 protein fused at the C terminus to the simian virus 40 NLS and GFP. As was true for Cds1-GFP, the Cds1-NLS-GFP appeared to be fully functional in the HU sensitivity and GST-Wee170 phosphorylation assays (Fig. 6). Microscopic observation revealed that Cds1-NLS-GFP presented a strong nuclear signal in all cells of an asynchronous culture (Fig. 7A). Importantly, the cells expressing Cds1-NLS-GFP were not abnormally elongated, which indicated that the constitutive nuclear localization of Cds1 did not cause a checkpoint arrest (Fig. 7A). Indeed, the GST-Wee170 phosphorylation assay confirmed that Cds1-NLS-GFP activity was low in asynchronous cells and was greatly stimulated in HU-arrested cells (Fig. 6B). These activities were equivalent to those of Cds1 and Cds1-GFP.
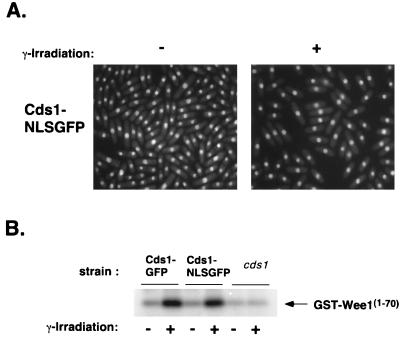
FIG. 7.
Cds1-NLS-GFP that is constitutively present in the nuclei of G2 cells is not activated by irradiation. (A) A strain expressing Cds1-NLS-GFP was grown in minimal media at 30°C. Cds1-NLS-GFP localization was determined by fluorescence microscopy in an asynchronous population (−γ) or after 100 Gy of irradiation (+ γ). Cds1 remains nuclear at all stages of the cell cycle, even during a DNA-damage-induced arrest at G2. (B) Activities of Cds1-GFP and Cds1-NLS-GFP after irradiation were measured with GST-Wee11–70 as a substrate. Cds1-NLS-GFP and Cds1-GFP are both weakly activated by irradiation, while a strain deleted for Cds1 has no detectable activity.
The localization pattern of Cds1-NLS-GFP was unaffected by irradiation (Fig. 7A). Moreover, Cds1-GFP and Cds1-NLS-GFP were activated to similar amounts by irradiation (Fig. 7B). This activity was not detectable in a strain deficient in Cds1 (Fig. 7B). If nuclear localization were limiting for damage-induced activation of Cds1, activation of Cds1-NLS-GFP should have been higher than that of Cds1-GFP. These facts argue that nuclear localization per se is not the key event regulating Cds1. Perhaps Cds1 activation is dependent on interaction with certain DNA structures or protein complexes that are present in the nucleus only during S phase.
DISCUSSION
The goal of this study was to understand how the checkpoint kinases Cds1 and Chk1 can be differentially responsive to distinct checkpoint signals and yet be regulated through a common set of sensor proteins, namely, the checkpoint Rad proteins. Specifically, we wanted to understand why HU fails to induce Chk1 phosphorylation except in a cds1 mutant and why DNA damage fails to induce Cds1 activation except during S phase. Explanations for the former observation are that Chk1 is phosphorylated only in response to DNA damage, HU does not damage DNA, and Cds1 is required to prevent DNA damage in HU-arrested cells (24). This model accounts for most of the data. However, the model does not explain why checkpoint Rad proteins are required for both the repair and replication checkpoints, and for phosphorylation of Chk1, and yet only DNA damage causes Chk1 phosphorylation. Moreover, the model is also apparently inconsistent with studies that showed that HU stimulates recombination (15, 18, 27, 43). Most relevant to our studies is a recent report that showed that a 4-h incubation in HU increases mitotic recombination between two ade6 heteroalleles by ∼80-fold (43). Recombination proceeds via the double-strand breakage of DNA and thus involves repair processes that, in principle, should induce Chk1 phosphorylation. An important test of the model is that IR and other DNA-damaging agents should induce Chk1 phosphorylation in HU-arrested cells. Here, we have shown that this prediction is incorrect. IR failed to induce Chk1 phosphorylation in cells arrested at the replication checkpoint with HU. These findings accord with those presented in another recent report (28).
One explanation for the failure of IR to induce Chk1 phosphorylation in HU-arrested cells could be that these cells are highly efficient at repairing DNA damage. Rad53 and Dun1, Cds1-like proteins present in budding yeast, are apparently involved in the transcriptional induction of repair enzymes (11). If Cds1 has an equivalent role in fission yeast, it is possible that HU-induced activation of Cds1 could result in very rapid repair of DNA damage. This model might also help explain why Chk1 is phosphorylated in HU-arrested cds1 cells, because these cells might be deficient in DNA repair enzymes. However, this model can be excluded on theoretical and factual grounds. First, IR-induced double-strand breakage of DNA is almost exclusively repaired by recombinational mechanisms that require intact homologous chromosomes. HU-treated cells arrest with unreplicated chromosomes; thus, on a theoretical basis, DNA double-strand breaks cannot be repaired in HU-arrested haploid cells. This argument is factually supported by the observation that release of IR-treated cells from an HU-induced arrest leads to a large increase in Chk1 phosphorylation. Thus, IR-induced damage was apparently left unrepaired in HU-arrested cells, but it was unable to induce Chk1 phosphorylation. Once released from the HU-induced cell cycle arrest, IR-exposed cells become greatly elongated and are apparently unable to divide (9). This cell cycle arrest is dependent on Chk1. These observations are consistent with the idea that exposure of HU-arrested cells to IR causes DNA damage that cannot be repaired.
Therefore, our studies show that the question of whether HU induces DNA damage is irrelevant to the observation that HU fails to induce Chk1 phosphorylation, because Chk1 phosphorylation is unresponsive to DNA damage in HU-arrested cells. In fact, the observations that initially led to this study indicate that HU does cause DNA damage. We observed that there is a Chk1-dependent delay of mitosis as cells recover from an HU-induced arrest. This phenomenon is observed as cells complete DNA replication in the presence of HU, as well as when cells complete replication when HU is removed from the growth medium. In the latter experiment, the delay of mitosis correlates with a small but reproducible amount of Chk1 phosphorylation that occurs after HU is removed. Thus, HU appears to cause some DNA damage that leads to increased recombination and Chk1 phosphorylation, but the latter effect is normally delayed until DNA replication is complete.
Why suppress Chk1 phosphorylation in HU-arrested cells?
Why suppress phosphorylation of Chk1 during an HU-induced replication arrest? If Chk1 is considered in isolation, it is difficult to understand why phosphorylation of Chk1, which is presumed to indicate activation of Chk1, would be incompatible with maximizing survival and minimizing damage during an HU-induced arrest. It is crucially important to prevent mitosis when DNA is unreplicated. Preventing mitosis is the sole known purpose of Chk1. This function of Chk1 is underscored by the observation that Chk1 is essential for an HU-induced arrest in cds1 cells. Moreover, cds1 chk1 double mutants exhibit enhanced sensitivity to HU relative to the sensitivity of cds1 cells. These considerations suggest that there is no purpose in specifically preventing Chk1 phosphorylation in HU-arrested cells. Instead, we hypothesize that during S the DNA repair checkpoint signal is suppressed closer to its source, namely, damaged DNA. We propose that phosphorylation of Chk1 requires processing of DNA damage and that this activity is incompatible with DNA replication. One may imagine, for example, that the process of DNA replication yields certain DNA structures that are potential substrates of repair systems. Recruitment of repair systems to these structures might lead to Chk1 phosphorylation but interfere with DNA replication. Thus, it is important that these repair systems be restrained during S phase. Hence, phosphorylation of Chk1 that is induced by HU or irradiation during S is suppressed until DNA replication is complete. We propose that Cds1 keeps these repair systems in check during S phase. Clearly, Cds1 function is not essential when DNA replication proceeds normally, but Cds1 activity is crucial when DNA replication is impaired by treatment of cells with HU.
An alternative model to explain the Cds1-dependent inhibition of Chk1 phosphorylation in HU-arrested cells is that Cds1 and Chk1 compete for the same upstream activators. Activation of Cds1 and phosphorylation of Chk1 are both dependent on checkpoint Rad proteins; therefore, it is possible that Cds1 and Chk1 share the same upstream activators, which might be present in limiting amounts. Perhaps Cds1 is better able to interact with the upstream activators during S phase, thereby preventing Chk1 phosphorylation. We do not favor this model, for the following reason: replacement of the genomic copy of cds1+ with an allele encoding a kinase-inactive form of Cds1 (Cds1-KD) does not have a dominant-negative effect on the HU checkpoint (8). In other words, cds1-KD and Δcds1 strains appear identical. If Cds1-KD interacted with upstream activators, thereby preventing interaction of Chk1 with the upstream activators, the model would predict that cds1-KD cells should behave like Δcds1 Δchk1 cells. These data indicate that the protein kinase activity of Cds1 is required to prevent HU-induced phosphorylation of Chk1. Thus, we think that Cds1 does not prevent Chk1 activation by competing for a common activator. However, a caveat to this conclusion is that we cannot exclude the possibility that Cds1-KD is incapable of interacting with its upstream activators.
Is Cds1 required to reduce DNA damage in HU-arrested cells?
Our studies establish that Chk1 phosphorylation is a poor indicator of DNA damage in HU-arrested cells. Thus, the observation that HU induces substantial Chk1 phosphorylation in cds1 cells cannot be used to conclude that Cds1 prevents DNA damage in HU-arrested cells. So, the question remains, does the absence of Cds1 lead to increased damage of DNA in HU-arrested cells? The answer is almost certainly affirmative, for the following reasons. Release from an HU-induced arrest leads to only a small amount of Chk1 phosphorylation. In contrast, release from an HU-induced arrest that was accompanied by IR leads to a large amount of Chk1 phosphorylation. Thus, HU-arrested cells appear to sustain a small amount of DNA damage (as assayed by Chk1 phosphorylation) that can be greatly increased by IR. This increased quantity of Chk1 phosphorylation is comparable to the amount of Chk1 phosphorylation that is observed in cds1 cells that are arrested with HU. These observations strongly suggest that Cds1 is required to minimize DNA damage in HU-arrested cells. This conclusion is supported by the studies that have shown that following release from an HU-induced arrest, mitosis occurs much later in cds1 cells relative to that in wild-type cells (8). The delay in wild-type cells is presumably due to a damage checkpoint that is enforced by Chk1.
Basis for the S-phase specificity of Cds1 activation.
The other major aim of this study was to understand why DNA damage induces activation of Cds1 only during S phase. We found that Cds1-GFP fusion protein was more highly concentrated in the nuclei of cells that are in S phase (i.e., septated cells or attached daughters) as compared to cells that are in G2. Moreover, the Cds1-GFP nuclear signal increased in cells that were arrested with HU. Thus, enhanced nuclear localization of Cds1 correlates with its activation in S phase by HU treatment or DNA damage. How is Cds1 localization regulated? Cds1 contains one putative NLS and one forkhead-associated (FHA) domain (22). These two motifs are found in a range of nuclear proteins. The NLS-dependent nuclear localization is an active mechanism (32). Perhaps the function of the putative NLS of Cds1 is regulated during the cell cycle. A homologous FHA domain is found in Rad53p, the Saccharomyces cerevisiae homolog of Cds1 (22). This domain might be important for regulating the localization of Cds1. However, nuclear localization of Cds1 is not sufficient for its activation, because the Cds1-NLS-GFP construct, which was constitutively localized in the nucleus, behaved otherwise like Cds1-GFP.
One of the proposed functions of Cds1 is to prevent the collapse of the DNA replication fork during DNA replication block (24). This idea suggests a direct interaction between Cds1 and the replication machinery. Thus, the specific nuclear accumulation of Cds1 during S phase may depend on association with a DNA structure or protein complex that is assembled during DNA replication. Another proposed function of Cds1 is to prevent mitosis by phosphorylating Wee1 (7). Wee1 appears to be localized in the nucleus (1, 47). Thus, colocalization of Cds1 and Wee1 in the nucleus is consistent with the model in which Cds1 regulates Wee1. Cds1 activation is dependent on Rad3, a kinase homologous to the human ATM protein. It is possible that Cds1 is a direct substrate of Rad3. The intracellular localization of Rad3 is unknown, but ATM is associated with chromatin (14). Thus, the nuclear localization of Cds1 might be necessary for interaction with Rad3 or other proteins that might activate Cds1.
ACKNOWLEDGMENTS
We thank N. Rhind for his advice and helpful discussion. We thank also F. Gaits, C. McGowan, and the Scripps Cell Cycle Groups for their support and encouragement.
J.-M.B. was supported by the Association pour la Recherche contre le Cancer. M.N.B. was supported by an NRSA Postdoctoral Fellowship from the National Institutes of Health. This work was funded by a National Institutes of Health grant awarded to P.R.
REFERENCES
- 1.Aligue R, Wu L, Russell P. Regulation of Schizosaccharomyces pombe Wee1 tyrosine kinase. J Biol Chem. 1997;272:13320–13325. doi: 10.1074/jbc.272.20.13320. [DOI] [PubMed] [Google Scholar]
- 2.al-Khodairy F, Carr A M. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 1992;11:1343–1350. doi: 10.1002/j.1460-2075.1992.tb05179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.al-Khodairy F, Fotou E, Sheldrick K S, Griffiths D J, Lehmann A R, Carr A M. Identification and characterization of new elements involved in checkpoint and feedback controls in fission yeast. Mol Biol Cell. 1994;5:147–160. doi: 10.1091/mbc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 5.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, DeMaggio A, Ford J C, Hoekstra M, Carr A M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 6.Blasina A, Van de Weyer I, Laus M C, Luyten W H M L, Parker A E, McGowan C H. A human homolog of the checkpoint kinase Cds1 directly inhibits Cdc25. Curr Biol. 1999;9:1–10. doi: 10.1016/s0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 7.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 8.Boddy, M. N. Unpublished observations.
- 9.Brondello, J.-M. 1999. Unpublished observations.
- 10.Carr A M. Analysis of fission yeast DNA structure checkpoints. Microbiology. 1998;144:5–11. doi: 10.1099/00221287-144-1-5. [DOI] [PubMed] [Google Scholar]
- 11.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 12.Enoch T, Carr A M, Nurse P. Fission yeast genes involved in coupling mitosis to the completion of DNA replication. Genes Dev. 1993;6:2035–2046. doi: 10.1101/gad.6.11.2035. [DOI] [PubMed] [Google Scholar]
- 13.Enoch T, Nurse P. Mutation of fission yeast cell cycle control genes abolishes dependence of mitosis on DNA replication. Cell. 1990;60:665–673. doi: 10.1016/0092-8674(90)90669-6. [DOI] [PubMed] [Google Scholar]
- 14.Flaggs G, Plug A W, Dunks K M, Mundt K E, Ford J C, Quiggle M R E, Taylor E M, Westphal C H, Ashely T, Hoekstra M F, Carr A M. Atm-dependent interactions of a mammalian Chk1 homolog with meiotic chromosomes. Curr Biol. 1997;7:977–986. doi: 10.1016/s0960-9822(06)00417-9. [DOI] [PubMed] [Google Scholar]
- 15.Freudenreich C H, Kantrow S M, Zakian V A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 16.Furnari B, Blasina A, Boddy M N, McGowan C H, Russell P. Cdc25 inhibited in vitro and in vivo by checkpoint kinases Cds1 and Chk1. Mol Biol Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by Chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 18.Galli A, Schiestl R H. Hydroxyurea induces recombination in dividing but not in G1 or G2 cell cycle arrested yeast cells. Mutat Res. 1996;354:69–75. doi: 10.1016/0027-5107(96)00037-1. [DOI] [PubMed] [Google Scholar]
- 19.Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 20.Hartwell L H, Kastan M B. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- 21.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann K, Bucher P. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 23.Kumagai A, Guo Z, Emami K H, Wang S X, Dunphy W G. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsay H, Griffiths D, Edwards R, Christensen P, Murray J, Osman F, Walworth N, Carr A. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 regulated by DNA damage and 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 26.Lundgren K, Walworth N, Booher R, Dembski M, Kirschner M, Beach D. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell. 1991;64:1111–1122. doi: 10.1016/0092-8674(91)90266-2. [DOI] [PubMed] [Google Scholar]
- 27.Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- 28.Martinho R G, Lindsay H D, Flaggs G, DeMaggio A J, Hoekstra M F, Carr A M, Bentley N J. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchison J M. Physiological and cytological methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970;4:131–146. [Google Scholar]
- 30.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 31.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 32.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 33.Peng C Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 34.Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- 35.Rhind N, Russell P. Tyrosine phosphorylation of Cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol Cell Biol. 1998;18:3782–3787. doi: 10.1128/mcb.18.7.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhind N, Russell P. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics. 1998;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell P. Checkpoints on the road to mitosis. Trends Biochem Sci. 1998;24:399–402. doi: 10.1016/s0968-0004(98)01291-2. [DOI] [PubMed] [Google Scholar]
- 39.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of crb2, a protein with BRCT motif, with cut5 and chk1. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez Y, Wong C, Thoma R S, Richman R, Wu Z, Piwnica-Worms H, Elledge S J. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 42.Shiozaki K, Russell P. Stress-activated protein kinase pathway in cell cycle control of fission yeast. Methods Enzymol. 1997;283:506–520. doi: 10.1016/s0076-6879(97)83040-6. [DOI] [PubMed] [Google Scholar]
- 43.Stewart E, Chapman C, Al-Khodairy F, Carr A, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walworth N, Davey S, Beach D. Fission yeast chk1 protein kinase links the rad checkpoint pathway to cdc2. Nature. 1993;363:368–371. doi: 10.1038/363368a0. [DOI] [PubMed] [Google Scholar]
- 45.Walworth N C, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 46.Willson J, Wilson S, Warr N, Watts F Z. Isolation and characterization of the Schizosaccharomyces pombe rhp9 gene: a gene required for the DNA damage checkpoint but not the replication checkpoint. Nucleic Acids Res. 1997;25:2138–2145. doi: 10.1093/nar/25.11.2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu L, Shiozaki K, Aligue R, Russell P. Spatial organization of the Nim1-Wee1-Cdc2 mitotic control network in Schizosaccharomyces pombe. Mol Biol Cell. 1996;7:1749–1758. doi: 10.1091/mbc.7.11.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]