Abstract
Benthic microbial mats dominated by Cyanobacteria are important features of polar lakes. Although culture-independent studies have provided important insights into the diversity of polar Cyanobacteria, only a handful of genomes have been sequenced to date. Here, we applied a genome-resolved metagenomics approach to data obtained from Arctic, sub-Antarctic and Antarctic microbial mats. We recovered 37 metagenome-assembled genomes (MAGs) of Cyanobacteria representing 17 distinct species, most of which are only distantly related to genomes that have been sequenced so far. These include (i) lineages that are common in polar microbial mats such as the filamentous taxa Pseudanabaena , Leptolyngbya , Microcoleus / Tychonema and Phormidium; (ii) the less common taxa Crinalium and Chamaesiphon ; (iii) an enigmatic Chroococcales lineage only distantly related to Microcystis ; and (iv) an early branching lineage in the order Gloeobacterales that is distributed across the cold biosphere, for which we propose the name Candidatus Sivonenia alaskensis. Our results show that genome-resolved metagenomics is a powerful tool for expanding our understanding of the diversity of Cyanobacteria, especially in understudied remote and extreme environments.
Keywords: Cyanobacteria, metagenomics, microbial mats, polar regions
Data Summary
The sequencing data generated in this study have been submitted to the European Nucleotide Archive (ENA) under the BioProject PRJEB59431. Individual accession numbers for raw reads and genomic bins are listed in Tables S1 and S3, available in the online version of this article, respectively. Genomic bins can also be downloaded from doi.org/10.6084 /m9 .figshare.22003967[1]. The bioinformatics workflow used throughout this study is available in github.com/igorspp/polar-cyanobacteria-MAGs.
Impact Statement.
Cyanobacteria are photosynthetic microorganisms that play important roles in polar lakes. Many Cyanobacteria are difficult to grow in the laboratory, particularly in isolation from other organisms, which makes it challenging to sequence their genomes. As such, considerably fewer genomes of Cyanobacteria have been sequenced so far compared to other bacteria. In this study, we used a metagenomics approach to recover novel genomes of Cyanobacteria from Arctic and Antarctic microbial mats without the need to isolate the organisms. The community DNA was extracted and sequenced, and the genomes of individual populations were separated using bioinformatics tools. We recovered the genomes of 17 different species of Cyanobacteria, many of which have not been sequenced before. We describe in more detail an interesting lineage of ancestral Cyanobacteria in the order Gloeobacterales , for which we propose the name Candidatus Sivonenia alaskensis. Our study shows that genome-resolved metagenomics is a valuable approach for obtaining novel genomes of Cyanobacteria, which are needed to improve our understanding of life in the polar regions and the planet at large.
Introduction
Microbial mats are highly successful and productive systems found in a wide range of environments since the dawn of life on Earth [2, 3]. Microbial mats commonly comprise a vast diversity of microorganisms such as auto- and heterotrophic bacteria, fungi, microalgae, and heterotrophic protists embedded in an exopolysaccharide matrix [4]. Benthic microbial mats represent an important survival strategy against the harsh environmental conditions in polar and alpine lakes, and often have Cyanobacteria as their primary source of organic carbon and nitrogen [5, 6]. In addition to aquatic microbial mats, Cyanobacteria are also important members of terrestrial and epi- and supraglacial communities in polar environments [7, 8].
Despite their importance, knowledge on the diversity and ecology of Cyanobacteria in polar environments is fragmentary [9]. Studies on the diversity of polar Cyanobacteria have mostly focused on microscopic identification and strain isolation [10–16], analysis of environmental 16S rRNA gene sequences [17–22] or a combination of these methods [23–25]. On the one hand, the microscopic identification of Cyanobacteria is hindered by the high plasticity of taxonomic markers such as cell dimensions and division patterns and the relative paucity of morphological characters [26]. In addition, morphology-based assessments underestimate the diversity of Cyanobacteria in the environment compared to molecular approaches based on environmental DNA [20]. Molecular approaches, in turn, are hampered by the scarcity of cyanobacterial genomes stored in public databases, which are largely underrepresented compared to other microbial phyla and heavily biased towards the marine Prochlorococcus / Synechococcus clade [27, 28].
The genomic catalogue of polar Cyanobacteria is currently limited to a handful of strains, including Pseudanabaena sp. BC1403 and Phormidesmis priestleyi BC1401 from Greenland [29], Leptolyngbya sp. Cla-17 from the Canadian Arctic [30], and the Antarctic strains Phormidesmis priestleyi ULC007 [31], Leptolyngbya sp. BC1307 [32], Synechococcus sp. SynAce01 [33] and Nostoc sp. SO-36 [34]. Twelve other low-quality genomes obtained by a metagenome-like assembly approach of non-axenic strains are also available [35]. Genome-resolved metagenomics has been established in recent years as a powerful approach to obtain microbial genomes, as it circumvents the difficulties associated with culturing microorganisms by reconstructing microbial genomes directly from environmental DNA [36–38]. Many genomes of uncultured polar Cyanobacteria have been obtained recently using this approach, including several novel lineages of early branching Cyanobacteria in the order Gloeobacterales [39–41].
In this study, we aimed to expand the genomic catalogue of polar Cyanobacteria. To achieve this, we applied a genome-resolved metagenomics approach to data obtained from microbial mats from Arctic, sub-Antarctic and Antarctic lakes spanning a wide geographical and limnological range. Our results include the recovery of novel genomes of polar Cyanobacteria and the description of an early branching lineage that is distributed across polar and alpine environments.
Methods
Description of samples and sampling procedure
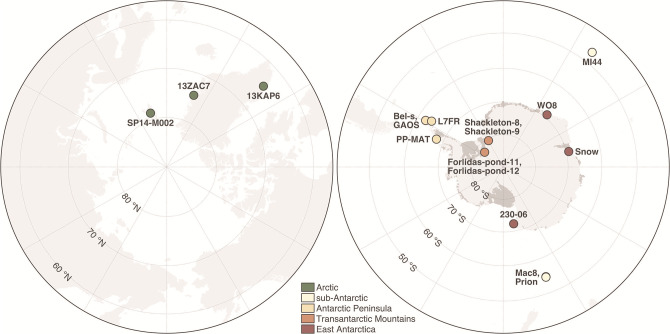
We analysed 17 microbial mat samples obtained from 15 Arctic, sub-Antarctic and Antarctic lakes (Fig. 1). The studied lakes were selected from a larger set of 216 polar lakes (B. Tytgat et al., submitted) and encompass the main polar biogeographical regions, namely the Arctic (Greenland and Svalbard), sub-Antarctic (Macquarie Island in the South Pacific Ocean and Marion Island in the South Indian Ocean), Antarctic Peninsula, Transantarctic Mountains and East Antarctica. The Antarctic lakes are distributed across five Antarctic Conservation Biological Regions (ACBRs) [42]. We aimed to include in our sampling design at least three samples from each main biogeographical region. Furthermore, with the exception of lakes in the Transantarctic Mountains, the samples were selected based on comparable values of pH (7.4±0.8) and conductivity (0.2±0.1 mS cm–1) (Table S1), since these parameters are known to be major drivers of polar microbial community composition [43]. The pH and conductivity of the Transantarctic Mountains lakes are 8.1±0.6 and 36.9±60.9 mS cm–1, respectively. A macroscopic description of each sample can be found in Table S1.
Fig. 1.
Location of the Arctic (left) and Antarctic (right) lakes where microbial mats were sampled for this study. Maps were created with public open access data from the Norwegian Polar Institute (Tromsø, Norway). More information about the samples can be found in Table S1.
The samples were taken following a standardized protocol [44, 45] in the course of several coordinated national and international limnological research programmes. In general, visible mats or biofilms were sampled in the littoral ice-free zone of the lakes (20–50 cm from the lake shore) using a sterilized spatula. Benthic microbial mats at depths >1 m in the Antarctic Peninsula were sampled using a Glew surface sediment corer, and the upper 1 cm of the core was aseptically collected using a sterilized spatula. In the Transantarctic Mountains, benthic samples were collected in the littoral (moat) zone below 10–15 cm of lake ice, as well as from the deepest part of the lakes after drilling through the ice using a Jiffy Drill. All samples were stored immediately in the dark and kept frozen at –20 °C until processing.
DNA extraction and metagenome sequencing
We used the DNeasy PowerBiofilm DNA Isolation kit (Qiagen) to extract DNA from ca. 0.5 g of each microbial mat sample and checked the concentration and quality of the DNA extracts using the Qubit dsDNA BR Assay kit (Thermo Fisher Scientific). We used the Nextera XT kit (Illumina) to prepare the metagenomic libraries, which were then sent to Eurofins Genomics for sequencing using the Illumina HiSeq 2500 platform (2×100 bp). We checked the quality of the raw sequencing data with fastQC v0.11.9 (bioinformatics.babraham.ac.uk/projects/fastqc) and multiQC v1.8 [46], and used Cutadapt v1.16 [47] to trim adapters and low-quality base calls (Phred score <20), and to discard short reads (<50 bp). Finally, we used METAXA v2.2 [48] to extract reads matching the 16S rRNA gene, which were then classified with mothur v1.44.3 [49] using the silva database release 138.1 [50] and the Naïve Bayesian Classifier with a confidence cut-off of 80 % [51].
Metagenome assembling and binning
We assembled and binned each metagenome individually and as two co-assemblies with MEGAHIT v1.1.1.2 [52]. One co-assembly was done by grouping the Arctic (n=3) and sub-Antarctic (n=3) samples. The second co-assembly comprised the remaining samples from the Antarctic Peninsula, Transantarctic Mountains and East Antarctica (n=11). For each individual and co-assembly, we used anvi’o v7.0 [53] to bin contigs ≥2500 bp into metagenome-assembled genomes (MAGs) as previously described [37, 38]. In brief, we used Prodigal v2.6.3 [54] to find gene calls, HMMER v.3.3 [55] to identify a set of 71 bacterial and 76 archaeal single-copy genes [56], and DIAMOND v0.9.14 [57] to assign taxonomy to the single-copy genes according to the Genome Taxonomy Database (GTDB) release 04-RS89 [58]. We used bowtie v2.4.2 [59] to map the quality-filtered reads from all samples to the contigs and SAMtools v1.1 [60] to sort and index the mapping output. We then used the anvi-interactive interface of anvi’o to manually sort the contigs into genomic bins based on differential coverage and tetranucleotide frequency. Bins that were ≥50 % complete according to the presence of single-copy genes [56] were manually curated using the anvi-refine interface of anvi’o. We refined the bins by removing outlying contigs according to coverage, tetranucleotide frequency and taxonomic signal of single-copy genes. We assigned taxonomy to the refined bins based on 122 archaeal and 120 bacterial single-copy genes with GTDB-Tk v1.3.0 [61] and the GTDB release 05-RS95 [58]. Bins assigned to the phylum Cyanobacteria that were ≥50 % complete and ≤10 % redundant – hereafter referred as MAGs – were kept for downstream analyses. We used fastANI v1.32 [62] to compute the genome-wide average nucleotide identity (ANI) between the Cyanobacteria MAGs. We considered that MAGs sharing ≥95 % ANI belong to the same species according to Konstantinidis et al. [63].
Phylogenetic analysis
We used a concatenated alignment of 38 ribosomal proteins to place the MAGs in a phylogenetic tree alongside all genomes assigned to the Cyanobacteria/Melainabacteria group in GenBank (NCBI:txid1798711, accessed on 17 November 2022). We used ncbi-genome-download v0.3.1 (github.com/kblin/ncbi-genome-download) to recover the genomes from GenBank. In anvi’o v7.0 [53], we retrieved the translated amino acid sequence of each ribosomal protein with HMMER v3.3 [55] and aligned them with muscle v3.8.1551 [64]. We concatenated the alignments of the 38 ribosomal proteins and built a maximum-likelihood tree with IQ-TREE v2.1.4 [65] using the automatic model selection and 1000 ultrafast bootstrap approximation replicates. We also used fastANI v1.32 [62] to calculate the genome-wide ANI between MAGs and GenBank genomes. For better visualization, we computed a more compact maximum-likelihood tree including only the MAGs, their closest neighbours in GenBank, strains from the Pasteur Culture Collection of Cyanobacteria (PCC) and other selected genomes. We classified the MAGs based on their phylogenetic placement following the taxonomic system of Komárek et al. [66].
Gene annotation
In anvi’o v7.0 [53], we annotated the gene calls identified by Prodigal v2.6.3 [54] against the KOfam [67] and the Pfam [68] databases with HMMER v3.3 [55] and the COG [69] database with DIAMOND v0.9.14 [57]. We also used tblastn (web interface, available at blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM = tblastn and PAGE_TYPE = BlastSearch) to search for additional genes involved in mechanisms of resistance to stress. Only hits with an e-value <10–5 and bitscore >50 were considered, according to Pearson [70].
Distribution analyses
We used metagenomic read recruitment to compute the relative abundance of each MAG across the 17 microbial mat samples. Prior to this, we used dRep v3.2.2 [71] to dereplicate the MAGs based on a ≥99 % ANI threshold. We then used CoverM v0.6.1 (github.com/wwood/CoverM) to map the quality-filtered reads to the MAGs with minimap v2.17 [72] and compute relative abundances based on the proportion of reads recruited by the MAGs. For this, we considered only matches with ≥95 % identity and ≥75 % coverage. We also used sourmash branchwater [73, 74] and IMNGS [75] to search the two Gloeobacterales MAGs against metagenomic and amplicon sequencing datasets in the Sequence Read Archive (SRA), respectively. For the first, we used the mastiff implementation of sourmash branchwater (github.com/sourmash-bio/2022-search-sra-with-mastiff). The datasets where significant matches were found (containment ≥20 %) were downloaded from SRA with fasterq-dump v3.0.1 (github.com/ncbi/sra-tools) and mapped back to the two Gloeobacterales MAGs with CoverM v0.6.1 as described above. For the analysis of amplicon sequencing datasets, we used the web interface of IMNGS (imngs.org) and only considered datasets where significant matches (≥99 % similarity) accounted for ≥0.1 % of the sequences.
Results and discussion
Taxonomic profiling of the microbial mat communities
We obtained around 500 million paired-end metagenomic reads (99.3 Gb) from 17 Arctic, sub-Antarctic and Antarctic microbial mat samples (Fig. 1, Table S1). Taxonomic profiling based on reads matching the 16S rRNA gene revealed Cyanobacteria as the second most abundant microbial phylum after Proteobacteria (mean relative abundance of 20.8 and 24.0 %, respectively) (Table S2). The dominance of these two phyla is commonly observed in polar microbial mats [76–78]. However, we observed differences in taxonomic composition across the samples (Table S2). For example, Cyanobacteria made up 63.5 % of the microbial community in the sample ‘Forlidas-pond-12’, a microbial mat taken from the hypersaline brine layer at the bottom of Forlidas Pond (Transantarctic Mountains) which has been known to harbour relatively simple communities [20, 21, 25]. On the other hand, Cyanobacteria were virtually absent in samples ‘Prion’ (sub-Antarctic), ‘GAOS’ (Antarctic Peninsula), ‘Shackleton-9’ (Transantarctic Mountains) and ‘WO8’ (East Antarctica). Rather than being related to geographical and climatic aspects of the sampling sites, the observed differences in taxonomic composition probably reflect the heterogenous characteristics of the microbial mats analysed in this study (see Table S1 for a macroscopic description of the samples). Although our sampling design includes a diverse set of Arctic, sub-Antarctic and Antarctic lakes, it is evident that we have covered only a minute fraction of the entire limnological and biological diversity of polar and sub-polar regions.
Recovery of cyanobacterial genomes from metagenomic data
We assembled the metagenomes with MEGAHIT [52] and obtained 176 097 and 72 514 contigs ≥1000 bp for the Antarctic and Arctic/sub-Antarctic co-assemblies, respectively. The total assembled length was 447.6 and 182.2 Mb, respectively. The output of the individual assemblies ranged from 218 contigs/0.3 Mb (sample ‘13ZAC7’) to 96 722 contigs/262.4 Mb (sample ‘PP-MAT’) (Table S1). The assembly of sample ‘Forlidas-pond-11’ did not yield any contigs due to the very low sequencing depth achieved for this sample. After assembling the metagenomes, we used anvi’o [53] to manually bin and curate MAGs. Taxonomic classification based on the GTDB release 05-RS95 [58] assigned 37 MAGs to the phylum Cyanobacteria (Fig. 2, Table 1). Most MAGs originated from the individual assemblies of Antarctic samples (n=20) and the Antarctic co-assembly (n=15) (Table S3). We did not recover any MAG from the individual assemblies of Arctic samples despite the high abundance of Cyanobacteria in these samples (5.9–28.6 % of the reads matching the 16S rRNA gene) (Table S2) and the high sequencing depth (3.6–7.5 Gb) (Table S1). The disparity in the number of MAGs recovered from each sample is probably related to differences in sequencing depth which, together with the level of complexity of the underlying communities, affect the number of genomes that are sufficiently represented in a metagenomic dataset [79].
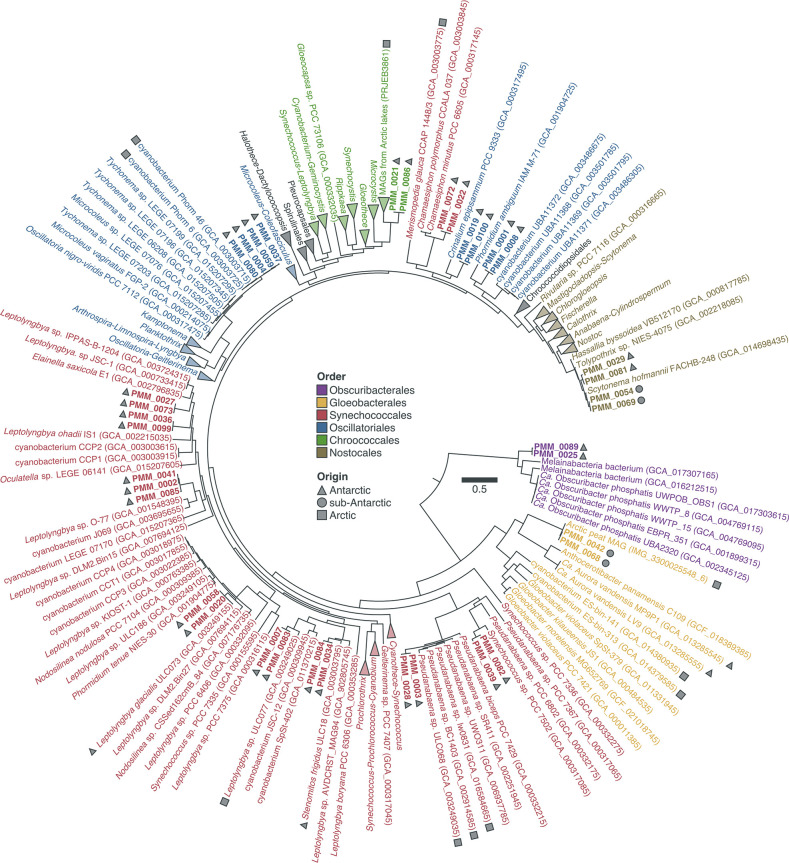
Fig. 2.
Phylogenetic analysis of 37 metagenome-assembled genomes (MAGs) assigned to the phylum Cyanobacteria , including both Cyanobacteria stricto sensu (clade Oxyphotobacteria) and the Melainabacteria. Maximum-likelihood tree (LG+R8 model) based on a concatenated alignment of 38 ribosomal proteins from the MAGs (in bold), their closest neighbours in GenBank, PCC strains and other selected genomes. The geographical origins of polar MAGs and strains are indicated. Order-level classification is shown according to the taxonomic system of Komárek et al. [66]. The scale bar indicates the number of amino acid changes per site.
Table 1.
Information on 37 metagenome-assembled genomes (MAGs) of Cyanobacteria stricto sensu (clade Oxyphotobacteria) and Melainabacteria recovered from polar microbial mats.
|
Group |
MAG |
Species cluster* |
Size (Mb) |
Completion (%)† |
Redundancy (%)† |
GC (%) |
|---|---|---|---|---|---|---|
|
PMM_0025 |
sp.1 |
6.1 |
90.1 |
8.5 |
47.7 |
|
|
PMM_0089 |
sp.1 |
4.7 |
77.5 |
5.6 |
47.9 |
|
|
PMM_0042 |
sp.2 |
2.9 |
97.2 |
0 |
49.3 |
|
|
PMM_0068 |
sp.2 |
2.8 |
95.8 |
0 |
48.8 |
|
|
PMM_0039 |
sp.3 |
1.5 |
57.7 |
0 |
41.5 |
|
|
PMM_0082 |
sp.3 |
2.7 |
85.9 |
0 |
40.3 |
|
|
PMM_0003 |
sp.4 |
3.1 |
78.9 |
5.6 |
42.9 |
|
|
PMM_0028 |
sp.4 |
3.7 |
84.5 |
8.5 |
42.6 |
|
|
PMM_0034 |
sp.5 |
3.3 |
66.2 |
0 |
51.3 |
|
|
PMM_0084 |
sp.5 |
4.8 |
81.7 |
4.2 |
51.8 |
|
|
PMM_0007 |
sp.6 |
5.2 |
94.4 |
1.4 |
49 |
|
|
PMM_0083 |
sp.6 |
4.6 |
94.4 |
1.4 |
48.4 |
|
|
PMM_0020 |
sp.7 |
3.2 |
90.1 |
2.8 |
57.2 |
|
|
PMM_0058 |
sp.7 |
2.8 |
84.5 |
1.4 |
56.9 |
|
|
PMM_0002 |
sp.8 |
2.9 |
90.1 |
1.4 |
52.5 |
|
|
PMM_0041 |
sp.8 |
2.5 |
50.7 |
1.4 |
52.4 |
|
|
PMM_0085 |
sp.8 |
3.7 |
81.7 |
4.2 |
52.5 |
|
|
PMM_0036 |
sp.9 |
4.6 |
66.2 |
4.2 |
49.4 |
|
|
PMM_0099 |
sp.9 |
4.7 |
67.6 |
5.6 |
49.3 |
|
|
PMM_0027 |
sp.10 |
2.2 |
73.2 |
1.4 |
55 |
|
|
PMM_0073 |
sp.10 |
3.6 |
85.9 |
5.6 |
55.3 |
|
|
PMM_0022 |
sp.11 |
2.5 |
80.3 |
2.8 |
44.9 |
|
|
PMM_0072 |
sp.11 |
2.9 |
83.1 |
1.4 |
44.4 |
|
|
PMM_0004 |
sp.12 |
4.4 |
60.6 |
7 |
45.6 |
|
|
PMM_0037 |
sp.12 |
5.2 |
67.6 |
8.5 |
45.3 |
|
|
PMM_0059 |
sp.12 |
4.9 |
90.1 |
5.6 |
45.6 |
|
|
PMM_0080 |
sp.12 |
6.4 |
94.4 |
2.8 |
45.1 |
|
|
PMM_0019 |
sp.13 |
5.7 |
95.8 |
2.8 |
45.2 |
|
|
PMM_0100 |
sp.13 |
3.8 |
80.3 |
2.8 |
45.4 |
|
|
PMM_0001 |
sp.14 |
5.8 |
94.4 |
2.8 |
45.4 |
|
|
PMM_0008 |
sp.14 |
5.8 |
94.4 |
2.8 |
45.4 |
|
|
PMM_0021 |
sp.15 |
4 |
77.5 |
2.8 |
39 |
|
|
PMM_0086 |
sp.15 |
3.8 |
80.3 |
2.8 |
38.9 |
|
|
PMM_0029 |
sp.16 |
2.7 |
73.2 |
7 |
42 |
|
|
PMM_0081 |
sp.16 |
4.4 |
91.5 |
2.8 |
42.1 |
|
|
PMM_0054 |
sp.17 |
5.1 |
90.1 |
2.8 |
42.2 |
|
|
PMM_0069 |
sp.17 |
3 |
76.1 |
0 |
41.6 |
MAG dereplication based on a ≥95 % ANI threshold [63] grouped the 37 Cyanobacteria MAGs into 17 species-level clusters (Table 1). Phylogenetic analysis based on a concatenated alignment of 38 ribosomal proteins assigned 35 of the 37 Cyanobacteria MAGs to the clade Oxyphotobacteria (Cyanobacteria stricto sensu) (Fig. 2, Table 1). These belong to five orders according to the taxonomic system of Komárek et al. [66]: Gloeobacterales (n=2), Synechococcales (n=19), Oscillatoriales (n=8), Chroococcales (n=2) and Nostocales (n=4). The other two MAGs belong to the order Obscuribacterales of the Melainabacteria, a sister lineage to the Oxyphotobacteria that lacks the photosynthetic machinery [80]. Indeed, annotation of protein-coding genes revealed that the two Obscuribacterales MAGs do not appear to encode proteins of the Calvin cycle (Rbc), photosystems I and II (Psa and Psb), cytochrome b6f complex (Pet), or phycobilisomes (Apc, Cpc, Cpe, and Pec) (Fig. 3). Interestingly, the presence of genes for the small and large subunits of nitric oxide reductase (NorC and NorB, respectively) indicates a potential role of this lineage in the production of the greenhouse gas nitrous oxide [37].
Fig. 3.
Presence of genes involved in carbon fixation, photosynthesis, nitrogen cycle and circadian clock in 37 metagenome-assembled genomes (MAGs) of Cyanobacteria stricto sensu (clade Oxyphotobacteria) and Melainabacteria.
Genome-resolved metagenomics is a reliable tool for the investigation of cyanobacterial diversity
In general, we observed a good correspondence between individual and co-assembly MAGs, that is, closely related genomic bins with ≥99 % ANI were recovered from the two assembly types (Table S3). The robustness of our metagenomic approach is further illustrated by the high similarity that some of the MAGs share with genomes available in GenBank. In particular, two MAGs obtained from different assemblies (‘PMM_0058’ and ‘PMM_0020’) are almost identical (99.7–99.8% ANI) to the genome of the strain Leptolyngbya glacialis ULC073 (Fig. 2, Table S3). This is not surprising given that the three genomes originate from the same hypersaline brine layer in the benthos of Forlidas Pond, Transantarctic Mountains [25]. Leptolyngbya glacialis ULC073 and Leptolyngbya antarctica ULC047 (Ace Lake, Princess Elizabeth Land) [13], which share identical 16S rRNA gene sequences, are representative strains of an ubiquitous morpho-/genotype in Antarctic lakes belonging to the Leptolyngbya–Nodosilinea clade [13, 21, 24, 25]. Despite the importance of this lineage, the genome of Leptolyngbia glacialis ULC073 currently available in GenBank (accession GCA_003249155.1), which was obtained from a non-axenic unialgal culture using a metagenome-like approach [35], is very fragmented (650 contigs, N 50=10.7 kb) and somewhat redundant (7.0 % according to our analysis of 71 single-copy genes). Based on these parameters, the MAGs ‘PMM_0058’ and ‘PMM_0020’, which are 84.5–90.1% complete, 1.4–2.8% redundant, comprise 290–339 contigs and have an N 50 of 12.1–13.0 kb (Tables 1 and S3), can be considered better representatives of this important lineage of Antarctic Cyanobacteria.
Other MAGs that are closely related to strains are the Nostocales MAGs ‘PMM_0054’ and ‘PMM_0069’. Genome-wide analysis revealed that they share 93.8–94.5% ANI with their closest genome on GenBank, Scytonema hofmannii FACHB-248 (Fig. 2, Table S3). However, their 16S rRNA gene is 99.4 % similar to the sequence of Dactylothamnos antarcticus CENA433 isolated from a freshwater biofilm in the Antarctic Peninsula [81], for which genomic information is currently lacking. Finally, the Gloeobacterales MAGs ‘PMM_0042’ and ‘PMM_0068’ share 97.2 % ANI with the MAG ‘IMG_3300025548_6’ recovered from peat soil in Alaska [39] (Fig. 2, Table S3).
Metagenomics reveals novel genomic diversity of polar Cyanobacteria
Phylogenetic placement and genome-wide comparison with sequences from GenBank revealed that most MAGs differ from genomes that have been sequenced so far (Fig. 2, Table S3). In particular, 19 of the 37 MAGs have <80 % ANI with genomes currently available in GenBank and 12 are only distantly related to existing genomes (80.1–93.2% ANI) (Table S3). Interestingly, phylogenetic placement clustered 16 and eight MAGs alongside polar and alpine strains, respectively (Fig. 2). This is in agreement with previous studies showing that many lineages of Cyanobacteria are distributed across the cold biosphere [21, 82, 83]. Most MAGs are affiliated with filamentous taxa in the orders Synechococcales (n=17), Oscillatoriales (n=8) and Nostocales (n=4) (Fig. 2, Table 1), highlighting the importance of filamentous Cyanobacteria as the builders of polar microbial mat ecosystems [5, 6, 84, 85]. Moreover, Cyanobacteria belonging to the order Nostocales often dominate the microbial communities in oligotrophic polar environments due to their ability to fix atmospheric nitrogen [5–8]. As observed previously (e.g. Olson et al. [86]), genes encoding the different subunits of the nitrogenase enzyme (NifHDK) involved in nitrogen fixation were exclusive to the four Nostocales MAGs (Fig. 3).
Most Synechococcales MAGs (n=13) are phylogenetically related to strains that have been traditionally classified as Leptolyngbya , which is a morphological group comprising Cyanobacteria with a thin, simple filamentous morphotype that includes many different genera according to molecular data [28, 66]. Our Leptolyngbya MAGs can be broadly categorized into four major lineages (Fig. 2): (i) Leptolyngbya stricto sensu (‘PMM_0007’ and ‘PMM_0083’), (ii) Leptolyngbya–Stenomitos (‘PMM_0034’ and ‘PMM_0084’), (iii) Leptolyngbya–Nodosilinea (‘PMM_0020’ and ‘PMM_0058’), and (iv) Leptolyngbya–Oculatella–Elainella (‘PMM_0085’, ‘PMM_0002’, ‘PMM_0041’, ‘PMM_0099’, ‘PMM_0036’, ‘PMM_0073’ and ‘PMM_0027’). The other four MAGs of filamentous Synechococcales are affiliated with the early branching Pseudanabaena (Fig. 2). Two of these (‘PMM_0003’ and ‘PMM_0028’) are most closely related (80.9–81.0% ANI) to the strain Pseudanabaena sp. ULC068 isolated from a lake in the Canadian sub-Arctic (W. Vincent, unpublished) (Table S3), and also clustered alongside the strains BC1403 from Greenland [29] and lw0831 from Svalbard [87] (Fig. 2). The other two Pseudanabaena MAGs (‘PMM_0039’ and ‘PMM_0082’) are distantly related (<80 % ANI) to Synechococcus sp. PCC 7502, a unicellular strain isolated from an alpine Sphagnum bog that clusters with the early branching Pseudanabaena [27].
Other MAGs of filamentous Cyanobacteria are affiliated with the order Oscillatoriales (n=8) (Fig. 2). Four of these (‘PMM_0004’, ‘PMM_0037’, ‘PMM_0059’ and ‘PMM_0080’) are most closely related (90.1–93.2% ANI) to the strain Phorm 46 isolated from a lake in the Canadian Arctic [88] (Table S3), and also clustered alongside strains of Tychonema and Microcoleus vaginatus (Fig. 2). The other two MAGs of filamentous Oscillatoriales (‘PMM_0001’ and ‘PMM_0008’) are distantly related (<80 % ANI) to the strain Phormidium ambiguum IAM M-71, which has an uncertain phylogenetic placement. Phylogenetic analysis of the amplified 16S rRNA gene sequence (accession AB003167) originally placed Phormidium ambiguum IAM M-71 alongside other Oscillatoriales such as Oscillatoria and Lyngbya [89, 90]. However, a later 16S rRNA phylogeny [91] and a phylogenomic tree based on 834 single-copy genes [92] both placed the strain IAM M-71 in a similar phylogenetic position as the one inferred here, that is, basal to the Nostocales (Fig. 2). A blast analysis suggests that the AB003167 sequence is chimeric with Phormidium muscicola IAM M-221, but a phylogenetic artefact based on long branch attraction is also possible given the lack of related genomes. Interestingly, the P. ambiguum MAGs were the most widespread MAGs in our dataset, being detected in five samples in the Antarctic Peninsula, Transantarctic Mountains and East Antarctica (Table S4). Finally, the two remaining Oscillatoriales MAGs (‘PMM_0019’ and ‘PMM_0100’) clustered alongside Crinalium epipsammum PCC 9333 (Fig. 2). Crinalium is a filamentous genus of Cyanobacteria with unusual elliptical trichomes [93]. Sequences related to Crinalium have been recovered from different alpine habitats [94] and a new species, Crinalium glaciale, has been described from cryoconite pools in Antarctica on the basis of morphological identification [95].
In addition to filamentous taxa, we also recovered MAGs related to unicellular Cyanobacteria in the orders Gloeobacterales (n=2), Synechococcales (n=2) and Chroococcales (n=2) (Fig. 2). All except the two Gloeobacterales MAGs were distantly related (<80 % ANI) to genomes currently available in GenBank (Table S3). The two Synechococcales MAGs (‘PMM_0022’ and ‘PMM_0072’) clustered alongside Chamaesiphon minutus PCC 6605 and Chamaesiphon polymorphus CCALA 037 (Fig. 2). Chamaesiphon is a cosmopolitan genus that is often reported in polar and alpine terrestrial and aquatic environments, and includes two species potentially endemic to Antarctica (Chamaesiphon arctowskii and Chamaesiphon austro-polonicus) [16, 20, 21, 84, 85, 96]. Finally, the two Chroococcales MAGs (‘PMM_0021’ and ‘PMM_0086’) formed a distinct lineage related to Microcystis and several MAGs recovered from Arctic lakes (BioProject PRJEB38681) (Fig. 2).
Description of Candidatus Sivonenia alaskensis
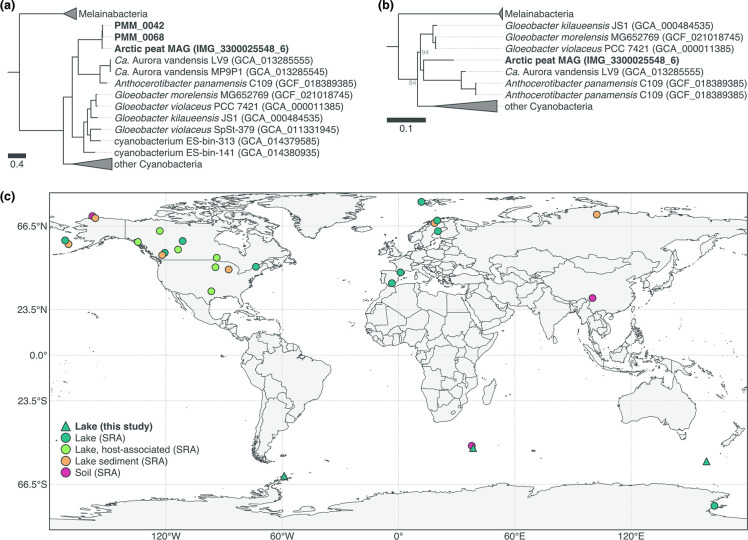
We investigated in more detail the two Gloeobacterales MAGs ‘PMM_0042’ and ‘PMM_0068’ given the importance of this group as the most basal lineage of extant Cyanobacteria [97–100]. Phylogenetic analysis based on a concatenated alignment of 38 ribosomal proteins placed both MAGs alongside the MAG ‘IMG_3300025548_6’ [39] (Fig. 4a), with which they share 97.2 % ANI (Table S3). This is above the ≥95 % ANI threshold commonly used for delineating microbial species [63], which thus suggests that the three MAGs (‘PMM_0042’, ‘PMM_0068’ and ‘IMG_3300025548_6’) belong to the same species. Furthermore, their phylogenetic placement and low ANI (<80 %) with other Gloeobacterales indicate that they constitute a distinct genus in this order. Separation from the other Gloeobacterale s is also supported by analysis of the 16S rRNA gene of the MAG ‘IMG_3300025548_6’, which is 91.8–92.0 %, 90.0% and 89.3 % similar to the sequences of Gloeobacter spp., Candidatus Aurora vandensis and Anthocerotibacter panamensis, respectively (Fig. 4b). We consider that the MAGs ‘PMM_0042’, ‘PMM_0068’ and ‘IMG_3300025548_6’ represent a novel lineage in the order Gloeobacterales and propose the name Candidatus Sivonenia alaskensis (Sivonenia: in honour of our colleague and Cyanobacteria expert Dr Kaarina Sivonen, professor emerita of the University of Helsinki; alaskensis: relative to the geographical origin of the MAG ‘IMG_3300025548_6’, which is proposed here as the nomenclatural type for this species according to the SeqCode initiative [101]).
Fig. 4.
Candidatus Sivonenia alaskensis, a lineage of early branching Cyanobacteria in the order Gloeobacterales . (a) Maximum-likelihood tree (LG+R8 model) based on a concatenated alignment of 38 ribosomal proteins from the three Ca. Sivonenia alaskensis MAGs (in bold) and other Gloeobacterales and selected genomes from GenBank. All nodes have bootstrap support ≥95 %. The scale bar indicates the number of amino acid changes per site. (b) Maximum-likelihood tree (GTR+F+R8 model) of the 16S rRNA gene of Ca. Sivonenia alaskensis. Nodes have bootstrap support ≥95 % unless shown otherwise. The scale bar indicates the number of nucleotide changes per site. (c) Geographical distribution of Ca. Sivonenia alaskensis based on significant matches with metagenomic and 16S rRNA gene amplicon sequencing datasets in SRA (≥20 % containment and ≥0.1 % relative abundance, respectively).
In silico analysis indicates that Ca. Sivonenia alaskensis is a thylakoid-less cyanobacterium
Analysis of the protein-coding genes of the Ca. Sivonenia alaskensis MAGs revealed many similarities with other Gloeobacterales , thus supporting their phylogenetic placement within this order of early branching Cyanobacteria (Fig. 4ab). For instance, strains of Gloeobacter spp. and Anthocerotibacter panamensis differ significantly from other Cyanobacteria by the lack of thylakoid membranes and the presence of a reduced photosynthetic apparatus [102–105]. These traits are considered ancestral features of oxygenic photosynthesis given the basal position of Gloeobacterales in the evolution of Cyanobacteria and plastids [97–99]. Like the genomes of other Gloeobacterales [39, 105–108], the Ca. Sivonenia alaskensis MAGs seem to lack the genes for several subunits of the photosystems I (PsaI, PsaJ, PsaK and PsaX) and II (PsbY, PsbZ and Psb27), the circadian clock (KaiA, KaiB and KaiC) and the thylakoid membrane (SqdB and SqdX) (Fig. 3, Table S5). Moreover, similarly to Anthocerotibacter panamensis and Ca. Aurora vandensis but unlike Gloeobacter spp [105]., Ca. Sivonenia alaskensis seems to lack two subunits of the photosystem II (PsbM and PsbU) and the cytochrome b6f complex (PetM and PetN), and does not appear to contain any gene involved in the synthesis of phycoerythrin (Pec). Overall, in silico analysis of the proteome of Ca. Sivonenia alaskensis suggests that this lineage comprises organisms without thylakoid membranes and with a reduced photosynthetic machinery, both of which are the defining characteristics of the order Gloeobacterales [66]. Moreover, the predicted structure of the photosystem II, cytochrome b6f and phycobilisome machineries of Ca. Sivonenia alaskensis holds more similarities with Anthocerotibacter panamensis and Ca. Aurora vandensis than with Gloeobacter spp., supporting the evolutionary relationship inferred from the analysis of ribosomal proteins and the 16S rRNA gene (Fig. 4ab). Whether the unique characteristics of Gloeobacter spp. reflect the ancestral state of the phylum Cyanobacteria has been an open question in the study of the early evolution of this group for many decades [100]. The discovery of Ca. Sivonenia alaskensis, Anthocerotibacter panamensis [105] and Ca. Aurora vandensis [40] suggests that characteristics such as the lack of thylakoid membranes and a reduced photosynthetic apparatus are indeed a pervasive trait of the early branching Gloeobacterales .
Ca. Sivonenia alaskensis is distributed across the cold biosphere
Read recruitment analysis revealed that the two Ca. Sivonenia alaskensis MAGs are found in four microbial mat samples from the sub-Antarctic (Macquarie Island and Marion Island) and Antarctic Peninsula (King George Island), where they constitute up to 1.0 % of the metagenomes (Table S4). To gain further insights into the ecology of Ca. Sivonenia alaskensis, we used sourmash branchwater [73, 74] to search metagenomic datasets in SRA for sequences matching the Ca. Sivonenia alaskensis MAGs. We also searched its 16S rRNA gene in amplicon sequencing datasets in SRA using IMNGS [75]. This extensive search, which included collectively ca. 1.3 million public datasets from around the globe, revealed sequences related to Ca. Sivonenia alaskensis in lakes, sediments and soils across many polar, sub-polar and alpine environments, and to a minor extent at lower latitude and non-alpine regions (Fig. 4c, Table S6). Sequences matching the Ca. Sivonenia alaskensis MAGs were particularly abundant (0.4–8.0 %) in amplicon sequencing datasets of active communities (that is, derived from RNA molecules) in the sediment of thermokarst lakes near Barrow (Alaska) [109], as well as in metagenomic datasets from the sediment of Lake Hill (St. Paul Island, Alaska) [110] (0.8–3.1 % of the reads). Interestingly, sequences matching the 16S rRNA gene of Ca. Sivonenia alaskensis were found in several datasets obtained from the gut microbiome of stickleback fishes (Actinopterygii: Gasterosteidae) and mayflies (Insecta: Ephemeroptera) [111–113].
Despite their importance for the study of the evolution of oxygenic photosynthesis, little is known about the ecology of the early branching Gloeobacterales compared to the other Cyanobacteria [88, 97, 105, 114]. Gloeobacter , which was for many decades the only described genus in this order, is typically found in low-light, wet rock habitats [102, 115, 116]. Amplicon sequencing studies have also reported 16S rRNA gene sequences loosely related to Gloeobacter spp. in Arctic [18, 22] and temperate [117] soil crusts, and in Arctic [83, 118] and Antarctic [119] microbial mats. The phylogenetic and ecological range of the order Gloeobacterales has expanded recently with the discovery of Ca. Aurora vandensis from Antarctic lakes [40], A. panamensis associated with a tropical bryophyte [105], and five MAGs from different ecosystems including the Ca. Sivonenia alaskensis MAG ‘IMG_3300025548_6’ recovered from Arctic peat soil [39]. Apart from A. panamensis, Gloeobacterale s appear to show a preference for low-light environments. This has been linked to their slow growth which, in turn, appears to be a consequence of their reduced photosynthetic apparatus [105]. Furthermore, our results suggest that Ca. Sivonenia alaskensis is distributed across cold regions, especially polar and alpine lakes and sediments (Fig. 4c, Table S6). Its high abundance in an RNA-derived amplicon sequencing dataset of lake sediments in Alaska [109] suggests that Ca. Sivonenia alaskensis forms active populations in this habitat. By contrast, the detection of sequences matching the 16S rRNA gene of Ca. Sivonenia alaskensis in the microbiome of stickleback fishes does not mean that they are active members of gut communities. These sequences probably represent cells that were ingested either incidentally or collaterally via zooplankton that is consumed by the fish (D. Bolnick, personal communication).
Analysis of resistance mechanisms to environmental stress in Ca. Sivonenia alaskensis
To obtain insights regarding the distribution of Ca. Sivonenia alaskensis across the cold biosphere, we searched the MAGs for genes involved in resistance mechanisms to environmental stress. We found 75 genes related to mechanisms to cope with desiccation, cold and ultraviolet radiation (UVR) stresses in at least one of the Ca. Sivonenia alaskensis MAGs (Table S7). Among these are genes involved in the Wzy- and ABC transporter-dependent pathways for the assembly and export of extracellular polymeric substances (EPS). The production of an EPS matrix is a mechanism that is commonly employed by Cyanobacteria to cope with desiccation and freezing [120]. Genes involved in the synthase-dependent pathway of EPS production were not found. Scytonemin and mycosporine-like amino acids (MAAs) are often produced by Cyanobacteria as UVR-screening compounds [121]. Despite having several of the genes involved in the production of scytonemin and MAAs, the genes encoding the key proteins ScyC, ScyD, EboA, EboB, EboC and MysC were not found. As such, the production of these compounds by Ca. Sivonenia alaskensis is unlikely. We identified other mechanisms of resistance to cold in the Ca. Sivonenia alaskensis MAGs, including proteins involved in regulation of cell membrane fluidity, regulation of replication and translation, and RNA metabolism (Table S7). Although these proteins are part of the general cell functioning, the up-regulation of their genes has been reported as a cold-shock response in Cyanobacteria [122, 123]. Finally, mechanisms of DNA repair include the base excision repair pathway for several glycosylases, the homologous recombination pathway for single-stranded breaks and one of the subtypes of the nuclear excision repair pathway.
Conclusion
We investigated 17 polar microbial mat metagenomes and recovered 37 MAGs of Cyanobacteria representing 17 species at different levels of phylogenetic novelty: around half of the MAGs are very distant (<80 % ANI) to genomes currently available in GenBank; the other half are related to polar and alpine strains with varying levels of genome similarity (80.1–99.8% ANI). Among the latter, we describe the phylogenetic, metabolic potential and ecological characteristics of a lineage in the early branching Gloeobacterales . In silico analyses indicate that this lineage – which we name Ca. Sivonenia alaskensis – is a thylakoid-less cyanobacterium that is found across cold environments and harbours common mechanisms of resistance to environmental stress. Our study shows that genome-resolved metagenomics is a reliable and straightforward way of recovering novel genomes of Cyanobacteria without the need for strain isolation. However, strain isolation is still useful for many purposes and may in fact benefit from genomic information obtained from MAGs to design protocols for targeted isolation. Based on the ≥95 % ANI threshold commonly used for delineating microbial species [63], most of the MAGs obtained represent different species or even genera from those currently represented by genomes in GenBank. Comparison with strains without genome data was not possible as only one of the 37 MAGs included the 16S rRNA gene, which is the most widely used molecular marker for the taxonomy of Cyanobacteria [28]. Assembling and binning 16S rRNA genes from short-read metagenomic data is difficult due to the highly conserved nature of this gene and its skewed coverage and sequence composition signals compared to the rest of the genome. The use of long-read technologies (e.g. Oxford Nanopore and PacBio SMRT sequencing) could help alleviate this issue [124, 125].
Supplementary Data
Funding information
This work was supported by the Belgian Federal Science Policy Office (BELSPO) (projects AMBIO – SD/BA/01A, CCAMBIO – SD/BA/03A and PORTAL – B2/212 /P1/PORTAL), the Belgian National Fund for Scientific Research (FRS-FNRS) (grants 2.4570.09 and CR.CH.10-11-1.5139.11), and the EU-Interact project MiBiPol. I.S.P. and J.H. were supported by the Academy of Finland grant 1314114, R.V.P. by the Doctoral Programme in Microbiology and Biotechnology (University of Helsinki), B.D. and V.S. by the FRS-FNRS, B.R.R. by the Special Funds for Research (University of Liège), the IPD-STEMA Programme, and the Junta de Andalucía (PAIDI-DOCTOR 21_00571), and A.W. is Senior Research Associate of the FRS-FNRS. Open access was funded by the Helsinki University Library.
Acknowledgements
The authors would like to acknowledge Sofie D’Hondt (UGent) for help with DNA extraction and library preparation, the IT Centre for Science – CSC (Finland) for providing the computational resources used in the study, and Kaarina Sivonen, Daniel Bolnick, Danillo Alvarenga and Tânia Shishido for comments. We also thank Dominic A. Hodgson, Steve J. Roberts, Wim Van Nieuwenhuyze, Koen Sabbe, Dagmar Obbels, Otakar Strunecký, Kate Kopalová, Jan Kavan, Josef Elster, Pieter Vanormelingen and Eveline Pinseel for help during sampling campaigns or/and sharing samples.
Author contributions
I.S.P., Y.L., B.T., E.V. and A.W. conceived the experiments. I.S.P. performed most of the analysis, and R.V.P., V.S. and B.R.R. contributed with minor parts. I.S.P. and R.V.P. wrote the manuscript. All authors provided important feedback, helped shape the study and contributed to the writing of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANI, average nucleotide identity; EPS, extracellular polymeric substance; MAA, mycosporine-like amino acid; MAG, metagenome-assembled genome; UVR, ultraviolet radiation.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Seven supplementary tables are available with the online version of this article.
References
- 1.Pessi I. Polar cyanobacteria MAGs. figshare. Figshare. 2023. [DOI]
- 2.Riding R. In: Modern Stromatolites: A Review. Riding R, editor. Berlin, Heidelberg: Springer; 1991. Calcareous algae and Stromatolites ; pp. 541–561. [DOI] [Google Scholar]
- 3.Stal LJ. In: Ecology of Cyanobacteria II. Whitton BA, editor. Dordrecht: Springer Netherlands; Cyanobacterial mats and stromatolites; pp. 65–125. [DOI] [Google Scholar]
- 4.Bolhuis H, Cretoiu MS, Stal LJ. Molecular ecology of microbial mats. FEMS Microbiol Ecol. 2014;90:335–350. doi: 10.1111/1574-6941.12408. [DOI] [PubMed] [Google Scholar]
- 5.Singh SM, Elster J. In: Algae and Cyanobacteria in Extreme Environments. Seckbach J, editor. Dordrecht: Springer Netherlands; Cyanobacteria in Antarctic lake environments: a mini-review; pp. 303–320. [DOI] [Google Scholar]
- 6.Vincent WF, Quesada A. In: Ecology of Cyanobacteria II. Whitton BA, editor. Dordrecht: Springer Netherlands; Cyanobacteria in high latitude lakes, rivers and seas; pp. 371–385. [DOI] [Google Scholar]
- 7.Quesada A, Vincent WF. In: Ecology of Cyanobacteria II. Whitton BA, editor. Dordrecht: Springer Netherlands; Cyanobacteria in the cryosphere: snow, ice and extreme cold; pp. 387–399. [DOI] [Google Scholar]
- 8.Van Goethem MW, Cowan DA. In: The Ecological Role of Micro-Organisms in the Antarctic Environment. Castro-Sowinski S, editor. Cham: Springer International Publishing; Role of Cyanobacteria in the ecology of polar environments; pp. 3–23. [DOI] [Google Scholar]
- 9.Chrismas NAM, Anesio AM, Sánchez-Baracaldo P. The future of genomics in polar and alpine cyanobacteria. FEMS Microbiol Ecol. 2018;94:fiy032. doi: 10.1093/femsec/fiy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vincent WF, Downes MT, Castenholz RW, Howard-Williams C. Community structure and pigment organisation of cyanobacteria-dominated microbial mats in Antarctica. Eur J Phycol. 1993;28:213–221. doi: 10.1080/09670269300650321. [DOI] [Google Scholar]
- 11.Elster J, Komarek O. Ecology of periphyton in a meltwater stream ecosystem in the maritime Antarctic. Antartic science. 2003;15:189–201. doi: 10.1017/S0954102003001226. [DOI] [Google Scholar]
- 12.Elster J, Lukesová A, Svoboda J, Kopecky J, Kanda H. Diversity and abundance of soil algae in the polar desert, Sverdrup Pass, central Ellesmere Island. Polar Record. 1999;35:231–254. doi: 10.1017/S0032247400015515. [DOI] [Google Scholar]
- 13.Taton A, Grubisic S, Ertz D, Hodgson DA, Piccardi R, et al. Polyphasic study of Antarctic cyanobacterial strains. J Phycol. 2006;42:1257–1270. doi: 10.1111/j.1529-8817.2006.00278.x. [DOI] [Google Scholar]
- 14.Palinska KA, Schneider T, Surosz W. Phenotypic and phylogenetic studies of benthic mat-forming cyanobacteria on the NW Svalbard. Polar Biol. 2017;40:1607–1616. doi: 10.1007/s00300-017-2083-6. [DOI] [Google Scholar]
- 15.Strunecky O, Raabova L, Bernardova A, Ivanova AP, Semanova A, et al. Diversity of cyanobacteria at the Alaska North Slope with description of two new genera: Gibliniella and Shackletoniella . FEMS Microbiol Ecol. 2020;96:fiz189. doi: 10.1093/femsec/fiz189. [DOI] [PubMed] [Google Scholar]
- 16.Taton A, Hoffmann L, Wilmotte A. Cyanobacteria in microbial mats of Antarctic lakes (East Antarctica) a microscopical approach. Algol Stud. 2008;126:173–208. doi: 10.1127/1864-1318/2008/0126-0173. [DOI] [Google Scholar]
- 17.Namsaraev Z, Mano M-J, Fernandez R, Wilmotte A. Biogeography of terrestrial cyanobacteria from Antarctic ice-free areas. Ann Glaciol. 2010;51:171–177. doi: 10.3189/172756411795931930. [DOI] [Google Scholar]
- 18.Pushkareva E, Pessi IS, Wilmotte A, Elster J. Cyanobacterial community composition in Arctic soil crusts at different stages of development. FEMS Microbiol Ecol. 2015;91:fiv143. doi: 10.1093/femsec/fiv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pushkareva E, Pessi IS, Namsaraev Z, Mano M-J, Elster J, et al. Cyanobacteria inhabiting biological soil crusts of a polar desert: Sør Rondane Mountains, Antarctica. Syst Appl Microbiol. 2018;41:363–373. doi: 10.1016/j.syapm.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Pessi IS, Maalouf PDC, Laughinghouse HD, Baurain D, Wilmotte A. On the use of high-throughput sequencing for the study of cyanobacterial diversity in Antarctic aquatic mats. J Phycol. 2016;52:356–368. doi: 10.1111/jpy.12399. [DOI] [PubMed] [Google Scholar]
- 21.Pessi IS, Lara Y, Durieu B, Maalouf P de C, Verleyen E, et al. Community structure and distribution of benthic cyanobacteria in Antarctic lacustrine microbial mats. FEMS Microbiol Ecol. 2018;94 doi: 10.1093/femsec/fiy042. [DOI] [PubMed] [Google Scholar]
- 22.Pessi IS, Pushkareva E, Lara Y, Borderie F, Wilmotte A, et al. Marked succession of cyanobacterial communities following glacier retreat in the high Arctic. Microb Ecol. 2019;77:136–147. doi: 10.1007/s00248-018-1203-3. [DOI] [PubMed] [Google Scholar]
- 23.Taton A, Grubisic S, Brambilla E, De Wit R, Wilmotte A. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Appl Environ Microbiol. 2003;69:5157–5169. doi: 10.1128/AEM.69.9.5157-5169.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taton A, Grubisic S, Balthasart P, Hodgson DA, Laybourn-Parry J, et al. Biogeographical distribution and ecological ranges of benthic cyanobacteria in East Antarctic lakes. FEMS Microbiol Ecol. 2006;57:272–289. doi: 10.1111/j.1574-6941.2006.00110.x. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Carazo R, Hodgson DA, Convey P, Wilmotte A. Low cyanobacterial diversity in biotopes of the Transantarctic mountains and Shackleton range (80-82°S), Antarctica. FEMS Microbiol Ecol. 2011;77:503–517. doi: 10.1111/j.1574-6941.2011.01132.x. [DOI] [PubMed] [Google Scholar]
- 26.Wilmotte A, Golubić S. Morphological and genetic criteria in the taxonomy of Cyanophyta/Cyanobacteria. Algol Stud. 1991;64:1–24. [Google Scholar]
- 27.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci. 2013;110:1053–1058. doi: 10.1073/pnas.1217107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mareš J. Multilocus and SSU rRNA gene phylogenetic analyses of available cyanobacterial genomes, and their relation to the current taxonomic system. Hydrobiologia. 2018;811:19–34. doi: 10.1007/s10750-017-3373-2. [DOI] [Google Scholar]
- 29.Chrismas NAM, Barker G, Anesio AM, Sánchez-Baracaldo P. Genomic mechanisms for cold tolerance and production of exopolysaccharides in the Arctic cyanobacterium Phormidesmis priestleyi BC1401. BMC Genomics. 2016;17:533. doi: 10.1186/s12864-016-2846-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Péquin B, Tremblay J, Maynard C, Wasserscheid J, Greer CW. Draft whole-genome sequences of the polar Cyanobacterium Leptolyngbya sp. strain Cla-17 and its associated flavobacterium. Microbiol Resour Announc. 2022;11:e0005922. doi: 10.1128/mra.00059-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lara Y, Durieu B, Cornet L, Verlaine O, Rippka R, et al. Draft genome sequence of the Axenic strain Phormidesmispriestleyi ULC007, a Cyanobacterium isolated from Lake Bruehwiler (Larsemann Hills, Antarctica) Genome Announc. 2017;5:e01546-16. doi: 10.1128/genomeA.01546-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chrismas NAM, Williamson CJ, Yallop ML, Anesio AM, Sánchez-Baracaldo P. Photoecology of the Antarctic cyanobacterium Leptolyngbya sp. BC1307 brought to light through community analysis, comparative genomics and in vitro photophysiology. Mol Ecol. 2018;27:5279–5293. doi: 10.1111/mec.14953. [DOI] [PubMed] [Google Scholar]
- 33.Tang J, Du L-M, Liang Y-M, Daroch M. Complete genome sequence and comparative Analysis of Synechococcus sp. CS-601 (SynAce01), a cold-adapted Cyanobacterium from an oligotrophic Antarctic habitat. Int J Mol Sci. 2019;20:152. doi: 10.3390/ijms20010152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Effendi DB, Sakamoto T, Ohtani S, Awai K, Kanesaki Y. Possible involvement of extracellular polymeric substrates of Antarctic cyanobacterium Nostoc sp. strain SO-36 in adaptation to harsh environments. J Plant Res. 2022;135:771–784. doi: 10.1007/s10265-022-01411-x. [DOI] [PubMed] [Google Scholar]
- 35.Cornet L, Bertrand AR, Hanikenne M, Javaux EJ, Wilmotte A, et al. Metagenomic assembly of new (sub)polar Cyanobacteria and their associated microbiome from non-axenic cultures. Microbial Genomics. 2018;4:212. doi: 10.1099/mgen.0.000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delmont TO. Discovery of nondiazotrophic Trichodesmium species abundant and widespread in the open ocean. Proc Natl Acad Sci. 2021;118:e2112355118. doi: 10.1073/pnas.2112355118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pessi IS, Viitamäki S, Virkkala A-M, Eronen-Rasimus E, Delmont TO, et al. In-depth characterization of denitrifier communities across different soil ecosystems in the tundra. Environ Microbiome. 2022;17:30. doi: 10.1186/s40793-022-00424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pessi IS, Rutanen A, Hultman J. Candidatus Nitrosopolaris, a genus of putative ammonia-oxidizing archaea with a polar/alpine distribution. FEMS Microbes. 2022;3 doi: 10.1093/femsmc/xtac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grettenberger CL. Novel Gloeobacterales spp. from diverse environments across the globe. mSphere. 2021;6:e0006121. doi: 10.1128/mSphere.00061-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grettenberger CL, Sumner DY, Wall K, Brown CT, Eisen JA, et al. A phylogenetically novel cyanobacterium most closely related to Gloeobacter . ISME J. 2020;14:2142–2152. doi: 10.1038/s41396-020-0668-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lumian JE, Jungblut AD, Dillion ML, Hawes I, Doran PT, et al. Metabolic capacity of the Antarctic Cyanobacterium Phormidium pseudopriestleyi that sustains oxygenic photosynthesis in the presence of hydrogen sulfide. Genes. 2021;12:426. doi: 10.3390/genes12030426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terauds A, Lee JR, Heikkinen R. Antarctic biogeography revisited: updating the Antarctic Conservation Biogeographic Regions. Diversity Distrib. 2016;22:836–840. doi: 10.1111/ddi.12453. [DOI] [Google Scholar]
- 43.Verleyen E, Sabbe K, Hodgson D, Grubisic S, Taton A, et al. Structuring effects of climate-related environmental factors on Antarctic microbial mat communities. Aquat Microb Ecol. 2010;59:11–24. doi: 10.3354/ame01378. [DOI] [Google Scholar]
- 44.Sabbe K, Hodgson DA, Verleyen E, Taton A, Wilmotte A, et al. Salinity, depth and the structure and composition of microbial mats in continental Antarctic lakes. Freshwater Biol. 2004;49:296–319. doi: 10.1111/j.1365-2427.2004.01186.x. [DOI] [Google Scholar]
- 45.Hodgson D, Vyverman W, Verleyen E, Sabbe K, Leavitt P, et al. Environmental factors influencing the pigment composition of in situ benthic microbial communities in east Antarctic lakes. Aquat Microb Ecol. 2004;37:247–263. doi: 10.3354/ame037247. [DOI] [Google Scholar]
- 46.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 48.Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, et al. METAXA2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour. 2015;15:1403–1414. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 49.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, et al. Introducing mothur: open-source, platform-independent, community-supported Software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 53.Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat Microbiol. 2021;6:3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee MD. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics. 2019;35:4162–4164. doi: 10.1093/bioinformatics/btz188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 58.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, et al. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH, Hancock J. GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics. 2020;36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Konstantinidis KT, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Komárek J, Kaštovský J, Mareš J, Johansen JR. Taxonomic classification of Cyanoprokaryotes (Cyanobacterial genera) 2014, using a polyphasic approach. Preslia. 2014;86:295–335. [Google Scholar]
- 67.Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, et al. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2252. doi: 10.1093/bioinformatics/btz859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galperin MY, Wolf YI, Makarova KS, Vera Alvarez R, Landsman D, et al. COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 2021;49:D274–D281. doi: 10.1093/nar/gkaa1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pearson WR. An introduction to sequence similarity (“homology”) searching. Curr Protoc Bioinforma. 2013;42:3. doi: 10.1002/0471250953.bi0301s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olm MR, Brown CT, Brooks B, Banfield JF. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–2868. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics. 2016;32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Irber L, Pierce-Ward NT, Brown CT. Sourmash branchwater enables lightweight petabyte-scale sequence search. Bioinformatics. doi: 10.1101/2022.11.02.514947. [DOI]
- 74.Lumian J, Sumner D, Grettenberger C, Jungblut AD, Irber L, et al. Biogeographic distribution of five Antarctic Cyanobacteria using large-scale k-mer searching with sourmash branchwater. BioRxiv. doi: 10.1101/2022.10.27.514113. [DOI] [PMC free article] [PubMed]
- 75.Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, et al. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep. 2016;6:33721. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaikova E, Goerlitz DS, Tighe SW, Wagner NY, Bai Y, et al. Antarctic relic microbial mat community revealed by metagenomics and metatranscriptomics. Front Ecol Evol. 2019;7:1. doi: 10.3389/fevo.2019.00001. [DOI] [Google Scholar]
- 77.Slattery M, Lesser MP. Allelopathy-mediated competition in microbial mats from Antarctic lakes. FEMS Microbiol Ecol. 2017;93 doi: 10.1093/femsec/fix019. [DOI] [PubMed] [Google Scholar]
- 78.Varin T, Lovejoy C, Jungblut AD, Vincent WF, Corbeil J. Metagenomic analysis of stress genes in microbial mat communities from Antarctica and the High Arctic. Appl Environ Microbiol. 2012;78:549–559. doi: 10.1128/AEM.06354-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen L-X, Anantharaman K, Shaiber A, Eren AM, Banfield JF. Accurate and complete genomes from metagenomes. Genome Res. 2020;30:315–333. doi: 10.1101/gr.258640.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soo RM, Skennerton CT, Sekiguchi Y, Imelfort M, Paech SJ, et al. An expanded genomic representation of the phylum cyanobacteria. Genome Biol Evol. 2014;6:1031–1045. doi: 10.1093/gbe/evu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Komárek J, Genuário DB, Fiore MF, Elster J. Heterocytous cyanobacteria of the Ulu Peninsula, James Ross Island, Antarctica. Polar Biol. 2015;38:475–492. doi: 10.1007/s00300-014-1609-4. [DOI] [Google Scholar]
- 82.Chrismas NAM, Anesio AM, Sánchez-Baracaldo P. Multiple adaptations to polar and alpine environments within cyanobacteria: a phylogenomic and Bayesian approach. Front Microbiol. 2015;6:1070. doi: 10.3389/fmicb.2015.01070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jungblut AD, Lovejoy C, Vincent WF. Global distribution of cyanobacterial ecotypes in the cold biosphere. ISME J. 2010;4:191–202. doi: 10.1038/ismej.2009.113. [DOI] [PubMed] [Google Scholar]
- 84.Velichko N, Smirnova S, Averina S, Pinevich A. A survey of Antarctic cyanobacteria. Hydrobiologia. 2021;848:2627–2652. doi: 10.1007/s10750-021-04588-9. [DOI] [Google Scholar]
- 85.Davydov D. Cyanobacterial diversity of Svalbard Archipelago. Polar Biol. 2021;44:1967–1978. doi: 10.1007/s00300-021-02931-3. [DOI] [Google Scholar]
- 86.Olson JB, Steppe TF, Litaker RW, Paerl HW. N2-fixing microbial consortia associated with the ice cover of Lake Bonney, Antarctica. Microb Ecol. 1998;36:231–238. doi: 10.1007/s002489900110. [DOI] [PubMed] [Google Scholar]
- 87.Su H-N, Wang Q-M, Li C-Y, Li K, Luo W, et al. Structural insights into the cold adaptation of the photosynthetic pigment-protein C-phycocyanin from an Arctic cyanobacterium. Biochim Biophys Acta BBA Bioenerg. 2017;1858:325–335. doi: 10.1016/j.bbabio.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 88.Moore KR, Magnabosco C, Momper L, Gold DA, Bosak T, et al. An expanded ribosomal phylogeny of Cyanobacteria supports a deep placement of plastids. Front Microbiol. 2019;10:1612. doi: 10.3389/fmicb.2019.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ishida T, Yokota A, Sugiyama J. Phylogenetic relationships of filamentous cyanobacterial taxa inferred from 16S rRNA sequence divergence. J Gen Appl Microbiol. 1997;43:237–241. doi: 10.2323/jgam.43.237. [DOI] [PubMed] [Google Scholar]
- 90.Honda D, Yokota A, Sugiyama J. Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus strains. J Mol Evol. 1999;48:723–739. doi: 10.1007/pl00006517. [DOI] [PubMed] [Google Scholar]
- 91.Ishida T, Watanabe MM, Sugiyama J, Yokota A. Evidence for polyphyletic origin of the members of the orders of Oscillatoriales and Pleurocapsales as determined by 16S rDNA analysis. FEMS Microbiol Lett. 2001;201:79–82. doi: 10.1111/j.1574-6968.2001.tb10736.x. [DOI] [PubMed] [Google Scholar]
- 92.Chen M-Y, Teng W-K, Zhao L, Hu C-X, Zhou Y-K, et al. Comparative genomics reveals insights into cyanobacterial evolution and habitat adaptation. ISME J. 2021;15:211–227. doi: 10.1038/s41396-020-00775-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Winder B, Stal LJ, Mur LR. Crinalium epipsammum sp. nov.: a filamentous cyanobacterium with trichomes composed of elliptical cells and containing poly- -(1,4) glucar (cellulose) J Gen Microbiol. 1990;136:1645–1653. doi: 10.1099/00221287-136-8-1645. [DOI] [Google Scholar]
- 94.Bohunická M, Mareš J, Hrouzek P, Urajová P, Lukeš M, et al. A combined morphological, ultrastructural, molecular, and biochemical study of the peculiar family Gomontiellaceae (Oscillatoriales) reveals a new cylindrospermopsin-producing clade of cyanobacteria. J Phycol. 2015;51:1040–1054. doi: 10.1111/jpy.12354. [DOI] [PubMed] [Google Scholar]
- 95.Broady PA, Kibblewhite AL. Morphological characterization of Oscillatoriales (Cyanobacteria) from Ross Island and Southern Victoria Land, Antarctica. Antartic science. 1991;3:35–45. doi: 10.1017/S095410209100007X. [DOI] [Google Scholar]
- 96.Komárek J. Phenotypic and ecological diversity of freshwater coccoid cyanobacteria from maritime Antarctica and Islands of NW Weddell Sea. II. Czech Polar Rep. 2014;4:17–39. doi: 10.5817/CPR2014-1-3. [DOI] [Google Scholar]
- 97.Demoulin CF, Lara YJ, Cornet L, François C, Baurain D, et al. Cyanobacteria evolution: Insight from the fossil record. Free Radic Biol Med. 2019;140:206–223. doi: 10.1016/j.freeradbiomed.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seo P-S, Yokota A. The phylogenetic relationships of cyanobacteria inferred from 16S rRNA, gyrB, rpoC1 and rpoD1 gene sequences. J Gen Appl Microbiol. 2003;49:191–203. doi: 10.2323/jgam.49.191. [DOI] [PubMed] [Google Scholar]
- 99.Gupta RS, Mathews DW. Signature proteins for the major clades of Cyanobacteria. BMC Evol Biol. 2010;10:24. doi: 10.1186/1471-2148-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Raven JA, Sánchez-Baracaldo P. Gloeobacter and the implications of a freshwater origin of Cyanobacteria. Phycologia. 2021;60:402–418. doi: 10.1080/00318884.2021.1881729. [DOI] [Google Scholar]
- 101.Hedlund BP, Chuvochina M, Hugenholtz P, Konstantinidis KT, Murray AE, et al. SeqCode: a nomenclatural code for prokaryotes described from sequence data. Nat Microbiol. 2022;7:1702–1708. doi: 10.1038/s41564-022-01214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rippka R, Waterbury J, Cohen-Bazire G. A cyanobacterium which lacks thylakoids. Arch Microbiol. 1974;100:419–436. doi: 10.1007/BF00446333. [DOI] [Google Scholar]
- 103.Guglielmi G, Cohen-Bazire G, Bryant DA. The structure of Gloeobacter violaceus and its phycobilisomes. Arch Microbiol. 1981;129:181–189. doi: 10.1007/BF00425248. [DOI] [Google Scholar]
- 104.Bryant DA, Cohen-Bazire G, Glazer AN. Characterization of the biliproteins of Gloeobacter violaceus . Arch Microbiol. 1981;129:190–198. doi: 10.1007/BF00425249. [DOI] [Google Scholar]
- 105.Rahmatpour N, Hauser DA, Nelson JM, Chen PY, Villarreal A JC, et al. A novel thylakoid-less isolate fills a billion-year gap in the evolution of Cyanobacteria. Curr Biol. 2021;31:2857–2867. doi: 10.1016/j.cub.2021.04.042. [DOI] [PubMed] [Google Scholar]
- 106.Nakamura Y. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Research. 2003;10:137–145. doi: 10.1093/dnares/10.4.137. [DOI] [PubMed] [Google Scholar]
- 107.Saw JH, Cardona T, Montejano G. Complete genome sequencing of a novel Gloeobacter species from a waterfall cave in Mexico. Genome Biol Evol. 2021;13:evab264. doi: 10.1093/gbe/evab264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saw JHW, Schatz M, Brown MV, Kunkel DD, Foster JS, et al. Cultivation and complete genome sequencing of Gloeobacter kilaueensis sp. nov., from a lava cave in Kīlauea Caldera, Hawai’i. PLoS One. 2013;8:e76376. doi: 10.1371/journal.pone.0076376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Matheus Carnevali PB, Herbold CW, Hand KP, Priscu JC, Murray AE. Distinct microbial assemblage structure and archaeal diversity in sediments of Arctic Thermokarst Lakes differing in methane sources. Front Microbiol. 2018;9:1192. doi: 10.3389/fmicb.2018.01192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Graham RW, Belmecheri S, Choy K, Culleton BJ, Davies LJ, et al. Timing and causes of mid-Holocene mammoth extinction on St. Paul Island, Alaska. Proc Natl Acad Sci. 2016;113:9310–9314. doi: 10.1073/pnas.1604903113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bolnick DI, Snowberg LK, Caporaso JG, Lauber C, Knight R, et al. Major histocompatibility complex class IIb polymorphism influences gut microbiota composition and diversity. Mol Ecol. 2014;23:4831–4845. doi: 10.1111/mec.12846. [DOI] [PubMed] [Google Scholar]
- 112.Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Knight R, et al. Individuals’ diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch) Ecol Lett. 2014;17:979–987. doi: 10.1111/ele.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Org E, et al. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun. 2014;5:4500. doi: 10.1038/ncomms5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Delwiche CF. Microbial biodiversity: a newly isolated cyanobacterium sheds light on the evolution of photosynthesis. Curr Biol. 2021;31:R843–R845. doi: 10.1016/j.cub.2021.05.059. [DOI] [PubMed] [Google Scholar]
- 115.Mareš J, Hrouzek P, Kaňa R, Ventura S, Strunecký O, et al. The primitive thylakoid-less cyanobacterium Gloeobacter is a common rock-dwelling organism. PLoS One. 2013;8:e66323. doi: 10.1371/journal.pone.0066323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Golubic S, Campbell SE. Analogous microbial forms in recent subaerial habitats and in Precambrian cherts: Gloethece coerulea eitler and Eosynechococcus moorei Hofmann. Precambrian Res. 1979;8:201–217. doi: 10.1016/0301-9268(79)90029-9. [DOI] [Google Scholar]
- 117.Williams L, Loewen-Schneider K, Maier S, Büdel B, Baldrian P. Cyanobacterial diversity of western European biological soil crusts along a latitudinal gradient. FEMS Microbiol Ecol. 2016;92:fiw157. doi: 10.1093/femsec/fiw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lionard M, Péquin B, Lovejoy C, Vincent WF. Benthic cyanobacterial mats in the igh Arctic: multi-layer structure and fluorescence responses to osmotic stress. Front Microbio. 2012;3:140. doi: 10.3389/fmicb.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakai R, Abe T, Baba T, Imura S, Kagoshima H, et al. Microflorae of aquatic moss pillars in a freshwater lake, East Antarctica, based on fatty acid and 16S rRNA gene analyses. Polar Biol. 2012;35:425–433. doi: 10.1007/s00300-011-1090-2. [DOI] [Google Scholar]
- 120.Pereira SB, Mota R, Vieira CP, Vieira J, Tamagnini P. Phylum-wide analysis of genes/proteins related to the last steps of assembly and export of extracellular polymeric substances (EPS) in cyanobacteria. Sci Rep. 2015;5:14835. doi: 10.1038/srep14835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gao Q, Garcia-Pichel F. Microbial ultraviolet sunscreens. Nat Rev Microbiol. 2011;9:791–802. doi: 10.1038/nrmicro2649. [DOI] [PubMed] [Google Scholar]
- 122.Sinetova MA, Los DA. Systemic analysis of stress transcriptomics of Synechocystis reveals common stress genes and their universal triggers. Mol Biosyst. 2016;12:3254–3258. doi: 10.1039/c6mb00551a. [DOI] [PubMed] [Google Scholar]
- 123.Gao X, Zhu Z, Xu H, Liu L, An J, et al. Cold adaptation in drylands: transcriptomic insights into cold-stressed Nostoc flagelliforme and characterization of a hypothetical gene with cold and nitrogen stress tolerance. Environ Microbiol. 2021;23:713–727. doi: 10.1111/1462-2920.15153. [DOI] [PubMed] [Google Scholar]
- 124.Kim CY, Ma J, Lee I. HiFi metagenomic sequencing enables assembly of accurate and complete genomes from human gut microbiota. Nat Commun. 2022;13:6367. doi: 10.1038/s41467-022-34149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sereika M, Kirkegaard RH, Karst SM, Michaelsen TY, Sørensen EA, et al. Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat Methods. 2022;19:823–826. doi: 10.1038/s41592-022-01539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.