Significance
cDC1 are crucial for immune responses against tumors as well as viruses. Here, we found that human cDC1 consist of XCR1− and XCR1+ cells. On transcriptomic level, XCR1+ cDC1 show a more mature phenotype in the steady state than XCR1− cDC1. After TLR stimulation, only XCR1+ cDC1 can secret high amounts of inflammatory cytokines and elicit strong antiviral immune responses, while XCR1− cDC1 must first acquire XCR1 expression in a terminal differentiation step. Thus, XCR1 is a marker of terminally differentiated cDC1 that licenses the antiviral effector functions of human cDC1, while XCR1− cDC1 seem to represent a late immediate precursor of cDC1.
Keywords: cDC1, XCR1, DC differentiation, IL-12, antiviral immunity
Abstract
Dendritic cells (DCs) are major regulators of innate and adaptive immune responses. DCs can be classified into plasmacytoid DCs and conventional DCs (cDCs) type 1 and 2. Murine and human cDC1 share the mRNA expression of XCR1. Murine studies indicated a specific role of the XCR1–XCL1 axis in the induction of immune responses. Here, we describe that human cDC1 can be distinguished into XCR1− and XCR1+ cDC1 in lymphoid as well as nonlymphoid tissues. Steady-state XCR1+ cDC1 display a preactivated phenotype compared to XCR1− cDC1. Upon stimulation, XCR1+ cDC1, but not XCR1− cDC1, secreted high levels of inflammatory cytokines as well as chemokines. This was associated with enhanced activation of NK cells mediated by XCR1+ cDC1. Moreover, XCR1+ cDC1 excelled in inhibiting replication of Influenza A virus. Further, under DC differentiation conditions, XCR1− cDC1 developed into XCR1+ cDC1. After acquisition of XCR1 expression, XCR1− cDC1 secreted comparable level of inflammatory cytokines. Thus, XCR1 is a marker of terminally differentiated cDC1 that licenses the antiviral effector functions of human cDC1, while XCR1− cDC1 seem to represent a late immediate precursor of cDC1.
Dendritic cells (DCs) are key players of the immune system capable to induce long-lasting immune responses to foreign pathogens, but also to maintain peripheral tolerance to self (1, 2). DCs in mice and humans arise from a common DC progenitor and can be classified into conventional DCs (cDCs) and plasmacytoid DCs (pDCs) (3–6). In both species, cDCs share a common progenitor called pre-cDCs, which is precommitted to develop into cDC type 1 or 2 (7–9). While several markers for the identification of cDCs differ between the human and mouse system, both cDC1 and cDC2 share transcriptional activators (10–15), in which cDC1 are dependent on the expression of the transcription factors IRF8 and Batf3, and cDC2 partially depend on the transcription factor IRF4 (14, 16–21). In humans, cDC1, also known as CD141+ DCs, are identified by the high expression of CD141 as well as CLEC9A, CADM1, and CD272 (BTLA), while cDC2, also known as CD1c+ DCs, express high level of CD1c, CLEC10A, FcεR1A, and SIRPα (12, 22–25).
Notably, murine DC subpopulations show strong functional differences in vivo (12, 26, 27). While cDC1 excel in their capacity to cross-present antigens and thereby induce cytotoxic T cell responses, cDC2 induce stronger CD4+ T cell responses and are able to polarize T cells into a variety of T helper cells (14, 26–28). The capacity of murine cDC1 to induce cytotoxic T cell responses was shown to depend on the expression of XCR1 as CD8+ T cells secrete XCL1 after activation leading to sustained interactions with XCR1+ cDC1 (29–31). Further, abrogation of the interaction of cDC1 and CD8+ T cells via the XCR1–XCL1 axis leads to diminished T cell responses and impaired protection in several viral and bacterial infection models (30, 32). In addition to CD8+ T cells, also NK cells were shown to produce XCL1 and to use the XCR1–XCL1 axis to interact with cDC1 in antiviral immunity (33–36).
While transcriptomic data show a high and specific expression of XCR1 in human cDC1, a systematic evaluation of protein expression and function of XCR1 for human cDC1 is missing. In the here presented study, we show that XCR1+ and XCR1− cDC1 can be identified in different human lymphoid tissues in similar proportions. We further demonstrate that XCR1+ cDC1 are superior in the production of inflammatory cytokines. Moreover, while both XCR1+ and XCR1− cDC1 are able to induce T cell responses, XCR1+ cDC1 excel in the activation of NK cells and the induction of an antiviral state in epithelial cells. When sorted XCR1+ and XCR1− cDC1 were cultured with stroma cells, we found that XCR1− cDC1 acquired XCR1 expression. Only after acquisition of XCR1 expression, cDC1 showed the capacity to secrete inflammatory cytokines leading to strong antiviral immune responses. Thus, XCR1 expression is a marker for terminal differentiation of human cDC1 and is linked to their full repertoire of effector functions, especially antiviral properties.
Results
Human cDC1 Contain XCR1+ and XCR1− Cells.
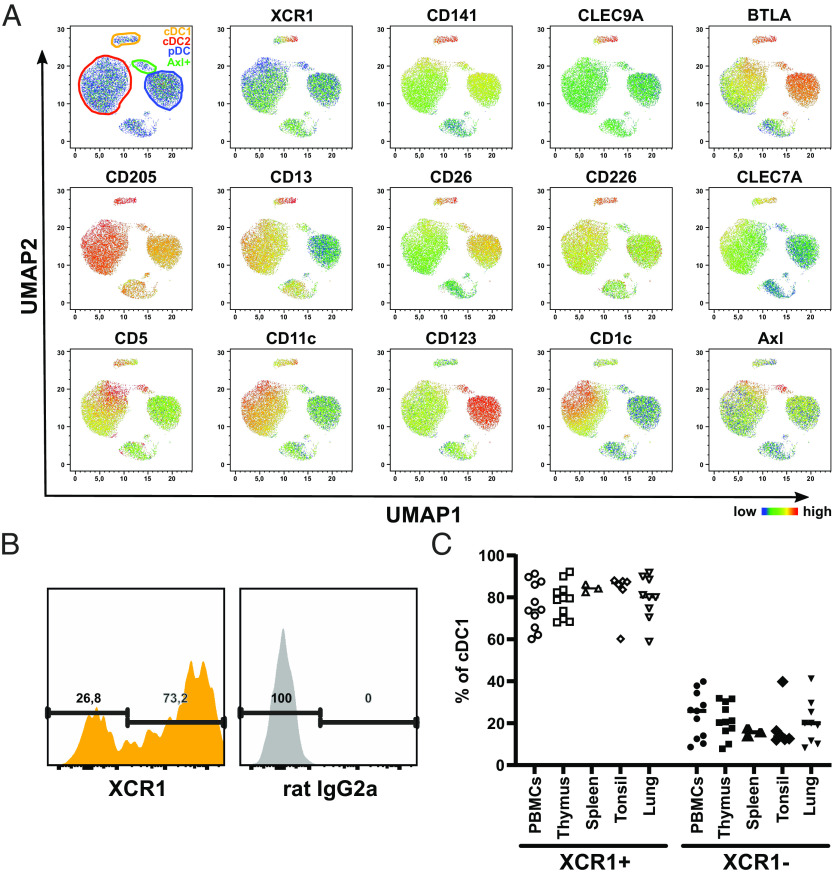
In the murine system, XCR1 has been suggested to be exclusively expressed on cross-presenting cDC1 (29, 37). Transcriptionally, XCR1 is also described as marker for human cDC1 (12, 38–40). Analyzing our previously published microarray data of various immunologic cell populations, including sorted human cDC1, cDC2, and pDCs from blood, spleen, and thymus, confirmed the specific mRNA expression of XCR1 in human cDC1 (SI Appendix, Fig. S1A) (12). As mRNA expression does not necessarily reflect protein expression, we validated the exclusive expression of XCR1 on human cDC1 by flow cytometry (SI Appendix, Fig. S1 B and C). Therefore, we stained PBMCs and single-cell suspensions of splenic, thymic, and tonsillar tissue with a panel of antibodies to identify immune cell populations including cDC1, cDC2, and pDCs (SI Appendix, Fig. S2). Indeed, the expression of XCR1 was restricted to cDC1 both in blood as well as in the lymphoid tissues thymus, spleen, and tonsils (SI Appendix, Fig. S1C). We then incorporated XCR1 into our flow cytometry panel and performed a high-dimensional analysis of the human DC compartment using Uniform Manifold Approximation and Projection (41) for dimension reduction (UMAP, Fig. 1A). While the DC subpopulations as well as Axl+ DCs were separated from each other, cDC1 showed a split up into two populations, which correlated with the expression of XCR1 (Fig. 1A). This was irrespective of the analyzed tissue, as we found that the cDC1 population was not uniformly positive for XCR1 but always contained a negative subpopulation of around 20% in all tested lymphoid as well as nonlymphoid tissues (Fig. 1 B and C). In order to confirm that XCR1− cDC1 truly represented cDC1, we analyzed other known markers for cDC1 (Fig. 1A and SI Appendix, Fig. S3). Indeed, XCR1− cDC1 showed comparable expression of the canonical cDC1 markers CLEC9A, BTLA, DEC205, and CD226 (Fig. 1A and SI Appendix, Fig. S3). However, XCR1− cDC1 displayed slightly lower expression of CD13, CD26, and CLEC7A (Dectin-1), while they expressed higher level of CD5 (SI Appendix, Fig. S3). Thus, we conclude that XCR1 is an exclusive marker for the cDC1 subpopulation and that human cDC1 can be distinguished into XCR1− and XCR1+ cDC1.
Fig. 1.
The cDC1 compartment comprises XCR1− and XCR1+ cells. (A) Single-cell suspensions of human blood were stained with a panel of fluorochrome-coupled antibodies (SI Appendix, Table S5). Samples were acquired using a BD LSRFortessa and analyzed using FlowJo software. After gating for living cells as shown in SI Appendix, Fig. S2, T cells, B cells, NK cells, and monocytes were excluded. The remaining HLA-DR+ cells were clustered using the algorithm UMAP. Scatter plots show UMAP with gated cDC1 (yellow-orange), cDC2 (red), pDC (blue) and Axl+ DC (green). Color-coded heat maps depict the expression of select markers for the identification of the different DC subpopulations. (B and C) Gated (B and C) blood, (C) lymphoid tissue and lung cDC1 as shown in SI Appendix, Fig. S2 were stained with PE-coupled anti-XCR1 (clone: S15046E, BioLegend) or appropriate isotype control (rat IgG2a). Cells were segregated into XCR1+ and XCR1− cells based on the signal in the isotype control. (B) Histograms show one representative blood donor. (C) Scatter plot (horizontal line: mean + SD) shows the ratio of XCR1+ and XCR1− cDC1 in blood, spleen, thymus, tonsil, and lung donors (n = 3 to 11).
XCR1+ cDC1 Show a More Mature Phenotype on the Transcriptional Level.
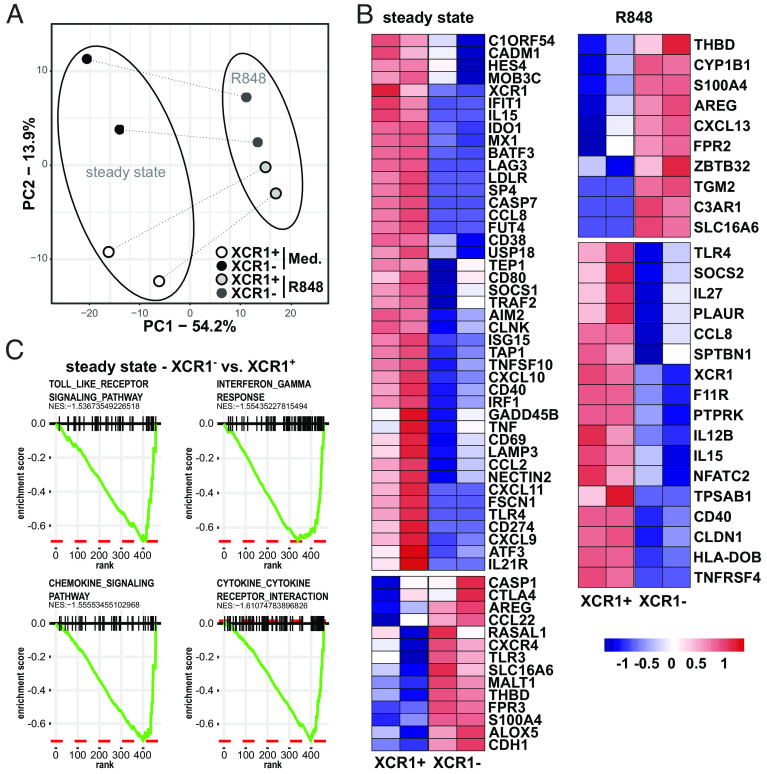
As recent studies on human DC subpopulations using single-cell RNAseq techniques showed that human cDC2 (also known as CD1c+ DCs) consist of at least two subpopulations with differing functions, termed DC2 and DC3 (24, 25, 42) or cDC2a and cDC2b (43), we were interested whether also XCR1− and XCR1+ cDC1 might resemble subpopulations with different functional properties. Therefore, we enriched DCs from PBMCs by depletion of unwanted cells, such as T cells, B cells, and NK cells, followed by cell sorting of blood XCR1− and XCR1+ cDC1 fractions (SI Appendix, Fig. S4). Then, purified XCR1− and XCR1+ cDC1 were stimulated for 3 h with R848 or kept in medium as a control. Subsequently, we determined the mRNA expression of 800 genes by Nanostring assay using the Myeloid Innate Immunity Panel with 30 additional spiked-in genes. Via principal component analysis (PCA), we identified the principal component (PC) 1, which accounted for 50% of transcriptional differences in the dataset and mainly separated TLR-stimulated from steady-state XCR1− and XCR1+ cDC1, and PC2, which comprised the main difference between XCR1− and XCR1+ cDC1 (Fig. 2A). We found that XCR1− and XCR1+ cDC1 were clearly separated in the steady state. However, after stimulation with the TLR-ligand R848 the transcriptional differences between XCR1− and XCR1+ cDC1 decreased (Fig. 2A). Thus, TLR stimulation seems to induce a comparable transcriptional program in XCR1− and XCR1+ cDC1 and, thereby, reduces the transcriptional differences between XCR1− and XCR1+ cDC1. Indeed, when performing hierarchical clustering of the dataset, TLR-treated XCR1+ and XCR1− cDC1 clustered together and showed comparable changes after TLR stimulation compared to the steady-state condition (SI Appendix, Fig. S5A). In both, XCR1+ and XCR1− cDC1, stimulation with R848 induced upregulation of molecules involved in intracellular signaling (NFKBIA, NFKBIZ, IKBKE, NFKB1, STAT4, STAT5A, IRF7, MAPK11, and MAP2K1), costimulation (CD40, CD70, CD80, CD86, and CD274), as well as cytokine and chemokine secretion (CCL2-4, CXCL2, CXCL8-11, IFNB1, IFNA2, IL1B, IL6, IL12B, IL15, IL23A, and IL27) (SI Appendix, Fig. S5A).
Fig. 2.
Transcriptional differences of XCR1− and XCR1+ cDC1. XCR1− and XCR1+ cDC1 were sorted as shown in SI Appendix, Fig. S4. Subsequently, cells were stimulated or not with 5 µg/mL R848 for 3 h. mRNA expression of 800 genes was determined using NanoString technology (770 genes from the nCounter Human Myeloid Innate Immunity V2 Panel and 30 DC- and inflammation-specific genes) as described before (44). (A) Eight samples (two donors, two subpopulations, two conditions) of 800 gene features were clustered using principal components analysis (PCA). Samples of XCR1− cDC1 are shown as black (steady state) and dark gray (R848) circles, while samples of XCR1+ cDC1 are depicted as white (steady state) and light-gray (R848) circles. (B) Heatmap shows DEGs between XCR1− and XCR1+ cDC1 in the steady state (cultured in medium for 3 h) or after stimulation with R848. (C) Gene expression values in (B) were analyzed by Gene Set Enrichment Analysis. Shown Pathways were significantly enriched in XCR1+ cDC1 compared to XCR1− cDC1 in the steady state (cutoff: a normalized enrichment score lower than −1.5 and P-value lower than 0.05).
Next, to determine differentially expressed genes (DEGs), we compared mRNA expression of XCR1− and XCR1+ cDC1 in the steady state as well as after TLR stimulation using a threshold of false-discovery rate (FDR)–adjusted P value of 0.01 and a log2fold change (FC) of 1.5 (Fig. 2B and SI Appendix, Fig. S5A). In the steady state, we found that the mRNA expression of previously identified markers for human cDC1, such as C1ORF54, CADM1, BATF3, CLNK, and XCR1 (9, 12, 42), were enhanced in XCR1+ cDC1, while XCR1− cDC1 showed enhanced mRNA expression of the cDC1-associated molecules THBD (CD141) and TLR3 (Fig. 2B). Further, steady-state XCR1+ cDC1 seemed to be much more mature on transcriptional level, as they expressed a higher level of several costimulatory molecules, cytokines, and chemokines, such as CD40, CD80, CD274, IL15, CCL2, CCL8, CXCL9, CXCL10, and TNF (Fig. 2B). In addition, they showed an enrichment of genes involved in antiviral immunity (ISG15, IFIT1, USP18, AIM2, IRF1, MX1, and LAMP3). This was confirmed by pathway analysis of DEGs of steady-state XCR1− and XCR1+ cDC1 showing an enrichment of genes of Toll-like receptor signaling, Interferon-γ response, chemokine signaling, and cytokine–cytokine receptor interaction pathways in XCR1+ cDC1 (Fig. 2C). In contrast, XCR1− cDC1 rather expressed higher levels of genes involved in proliferation (AREG, RASAL1, CXCR4) and immunoregulation (CTLA4 and CCL22). As XCR1− cDC1 showed markedly lower expression of cDC1 markers, we wanted to verify that they transcriptionally belong to the cDC1 and not cDC2 lineage as in mice XCR1−/low cDC1 were described with phenotypic or transcriptional similarities to cDC2 (45, 46). Thus, we compared the transcriptional profile to published mRNA data of human cDC2 (44), which were generated with the same Myeloid Innate Immunity Panel by NanoString technology. After identification of DEGs between steady-state XCR1− cDC1 and cDC2, the genes were hierarchically clustered and plotted as heatmap (SI Appendix, Fig. S5B). Indeed, XCR1− cDC1 showed an enrichment of cDC1 signature genes, such as BATF3, THBD, CADM1, TLR3, IRF8, CLNK, FLT3, DPP4, CD226, and C1ORF54, compared to cDC2, while DEGs enriched in cDC2 belonged to the cDC2 lineage, such as FCER1A, CLEC10A, FCGR2A, ITGAX, S100A4, S100A10, S100A11, and inflammasome-related molecules [NLRP3, NLRC4, CASP1, CASP4, and MEFV (SI Appendix, Fig. S5b)]. Thus, these data confirm that XCR1− cDC1 transcriptionally belong to the cDC1 lineage.
Under inflammatory conditions, R848-treated XCR1− and XCR1+ cDC1 differed only in few genes. We identified an enrichment of several key genes for immune responses induced by DCs in XCR1+ cDC1, such as IL12B, IL15, and CD40 (Fig. 2B). In summary, our data clearly show that XCR1+ cDC1 displayed a more mature phenotype with stronger expression of signature cDC1 genes as well as costimulatory molecules, cytokines and chemokines. However, XCR1− cDC1 were also able to react to TLR stimulation on the transcriptional level with the upregulation of molecules involved in immune responses.
In order to verify our transcriptional data on XCR1− and XCR1+ cDC1, we analyzed published scRNAseq data of Saichi et al. (47). In the dataset, we could identify a distinct cluster of CLEC9a+ cDC1 (SI Appendix, Fig. S6A). XCR1 expression could dissect the cluster into XCR1+ and XCR1− cells (SI Appendix, Fig. S6B). We then extracted the CLEC9a+ cDC1 and analyzed them in more detail by displaying a XCR1+ module consisting of DEGs identified in steady-state XCR1+ cDC1 and a XCR1− module consisting of the DEGs identified in steady-state XCR1− cDC1 (SI Appendix, Fig. S6C). Based on these expression values, CLEC9a+ DCs were clustered into XCR1− and XCR1+ cDC1 (SI Appendix, Fig. S6D). Indeed, XCR1+ cDC1 showed enhanced expression of CADM1, XCR1, IDO1, AIM2, CLNK, CD40, and CXCL9, while XCR1− cDC1 demonstrated higher expression of CASP1, MALT1, and S100A4 (SI Appendix, Fig. S6D). Thus, XCR1− and XCR1+ cDC1 are also present in previously published scRNAseq data.
XCR1+ cDC1 Are Superior in the Secretion of Inflammatory Cytokines and Chemokines.
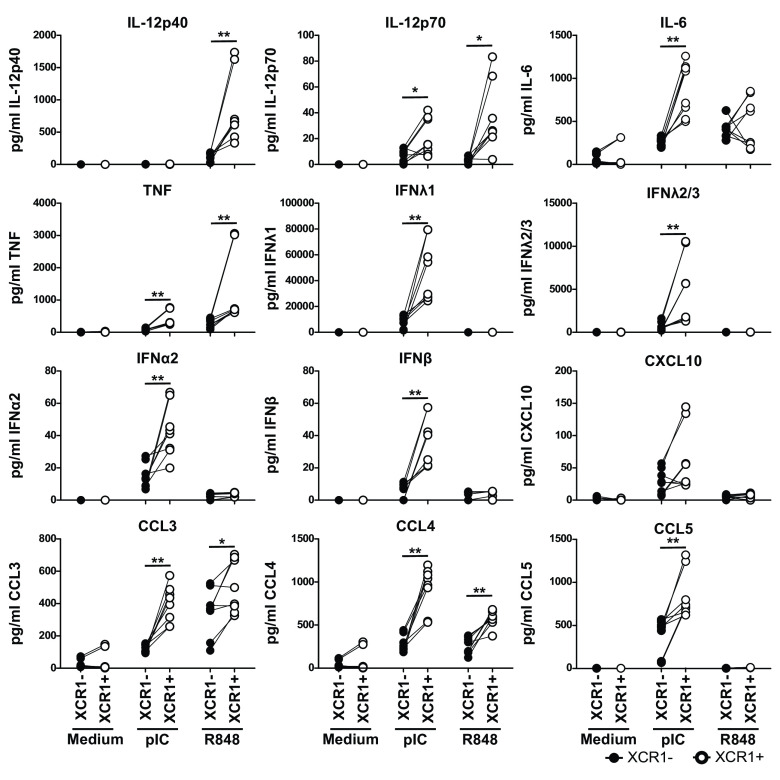
As XCR1+ cDC1 demonstrated enhanced expression of genes important in regulating immune responses, we were interested in whether they differed also on protein level. Therefore, we sorted blood XCR1− and XCR1+ cDC1 and stimulated the cells with the TLR-ligands pIC (TLR3) and R848 (TLR7/8), previously shown to strongly activate human cDC1 (48, 49). As XCR1+ cDC1 showed a mature phenotype on transcriptional level, we analyzed the expression of costimulatory molecules after 3-h stimulation by flow cytometry. Indeed, XCR1+ cDC1 excelled in the expression of CD40, CD86 as well as HLA-DR compared to XCR1− cDC1 (SI Appendix, Fig. S7A). Further, we harvested the supernatants of the cells for analysis of secreted cytokines after 12 h of stimulation and analyzed the cells for expression of costimulatory molecules/activation markers (Fig. 3 and SI Appendix, Fig. S7B). TLR stimulation of XCR1− and XCR1+ cDC1 induced higher expression of the costimulatory molecules CD40 and OX40L as well as of the coinhibitory molecule PD-L1 on XCR1+ cDC1, while CD86 was comparably expressed on XCR1− and XCR1+ cDC1 (SI Appendix, Fig. S7B). Further, stimulation led to an almost complete downregulation of XCR1 on XCR1+ cDC1 (SI Appendix, Fig. S7B). However, analyzing secreted cytokines by XCR1− and XCR1+ cDC1 showed that mainly XCR1+ cDC1 secreted inflammatory cytokines and chemokines in response to TLR stimulation, while XCR1− cDC1 were rather inert (Fig. 3). Notably, stimulation of TLR3 and TLR8 with pIC and R848, respectively, induced completely different cytokine responses by XCR1+ cDC1 (Fig. 3). While stimulation of TLR3 induced high secretion of type III IFN and low amounts of type I IFN, it was unable to induce IL-12/IL-23p40 (Fig. 3). Contrary, stimulation of TLR8 induced high amounts of IL-12/IL-23p40 but was unable to elicit type I or III interferons by cDC1 (Fig. 3). Furthermore, while both TLR3 and TLR8 treatment induced secretion of the monocyte-recruiting chemokines CCL3 and CCL4, only TLR3 stimulation led to measurable secretion of CXCL10 (CXCR3) and CCL5 (CCR5) (Fig. 3). As signaling of XCR1 might be involved in the superior secretion of cytokines and chemokines by XCR1+ cDC1, we wondered whether additional stimulation of XCR1 with XCL1 would influence the activation of XCR1− or XCR1+ cDC1. Thus, we cultured XCR1− and XCR1+ cDC1 with or without XCL1 and stimulated the cells either with pIC or R848 or kept them in medium as a control. Our data revealed that additional treatment with XCL1 did not influence costimulatory molecule expression or cytokine secretion by XCR1− or XCR1+ cDC1 (SI Appendix, Fig. S8). Overall, XCR1+ cDC1 showed a significantly enhanced secretion of several cytokines and chemokines such as IL-12, IL-6, TNF, type I and III interferons, as well as CCL-3, -4, and -5. Thus, we conclude from these data that XCR1+ cDC1 excel in cytokine and chemokine secretion, compared to XCR1− cDC1.
Fig. 3.
XCR1+ cDC1 are superior in the secretion of inflammatory cytokines. XCR1− and XCR1+ cDC1 were sorted as shown in SI Appendix, Fig. S4. Subsequently, cells were stimulated or not either with 5 µg/mL R848 or 5 µg/mL pIC for 12 h. Supernatants of the cells were harvested and analyzed for the secretion of IL-12/IL-23p40, IL-12p70, IL-6, TNF, IFNλ1, IFNλ2/3, IFNα2, IFNβ, CXCL10, CCL3, CCL4, and CCL5 using the LEGENDplex Human Anti-Virus Response Panel (BioLegend). Samples were acquired using a BD LSRFortessa and data were analyzed using the LEGENDplex software suit (BioLegend). Scatter plots (black, filled circles: XCR1−; open circles: XCR1+) depict individual donors. Data for XCR1− and XCR1+ cDC1 from the same donor are connected by line. *P < 0.05; **P < 0.01 (N = 6, paired Student’s t test).
XCR1− and XCR1+ cDC1 Show Comparable Capacity to Induce T Cell Responses.
In the murine system, XCR1 expression is linked to the cross-presentation of antigens to CD8+ T cells as only XCR1+ DCs can efficiently cross-present antigens in vivo (29). In addition to the identified distinct pattern in cytokine and chemokine secretion of XCR1+ to XCR1− cDC1 (Fig. 3), we hypothesized that the capacity of the cDC1 subsets to cross-present antigens differs. In order to measure cross-presentation, we made use of an HLA-A2-dependent TCR recognizing a specific peptide of the gp100 protein, which was transfected into CD8+ T cells of HLA-A2+ blood donors as published before (50–52). Simultaneously to the transfection of the T cells, we isolated XCR1− and XCR1+ cDC1 from the very same donor, stimulated them with the TLR-ligands pIC and R848 for 3 h and then either fed them with a long peptide containing the peptide for the gp100-specific TCR, pulsed them with the already processed gp100-peptide as positive control or kept them in medium without any peptide as a negative control. Then, they were cocultured with the TCR-transfected T cells for 18 h, followed by harvesting of the supernatant for analysis of secreted cytokines and flow cytometric analysis of activation markers on the T cells. The analysis revealed that—against our hypothesis—both XCR1− and XCR1+ cDC1 were able to cross-present epitopes derived from long peptides to CD8+ T cells with equal induction of IFNγ and IL-2 secretion (SI Appendix, Fig. S9A). From these data, we conclude that XCR1+ and XCR1− cDC1 can cross-present antigens equally well to antigen-specific CD8+ T cells.
Next, to test the capacity of XCR1− and XCR1+ cDC1 to induce naïve T cell responses, we performed mixed leukocyte reactions (MLR). Therefore, we FACS-sorted XCR1− and XCR1+ cDC1 from one donor. Then, we stimulated the DCs with either pIC or R848, and coincubated the DCs with isolated naïve CD8+ or CD4+ T cells of an HLA-mismatched donor for 7 d. Our data revealed that both, XCR1− and XCR1+ cDC1 could induce the proliferation of naïve CD8+ and CD4+ T cells (SI Appendix, Fig. S9 B and C). It seemed that the stimulation of XCR1− cDC1 with pIC was more effective in inducing IFNγ as well as IL-2 production in CD8+ T cells compared to R848 stimulation (SI Appendix, Fig. S9B). The MLR with naïve CD4+ T cells gave comparable results to the MLR with naïve CD8+ T cells as both cDC1 subsets induced similar amounts of T cell proliferation (SI Appendix, Fig. S9C). Further, they showed a mixed Th1 (IFNγ)/Th2 (IL-4) CD4+ T cell phenotype (SI Appendix, Fig. S9C). In summary, although XCR1+ cDC1 excelled in the production of T cell–shaping cytokines such as IL-12, both XCR1− and XCR1+ cDC1 showed a comparable capacity to induce CD4+ and CD8+ T cell responses.
XCR1+ cDC1 Excel in the Induction of Antiviral Immunity.
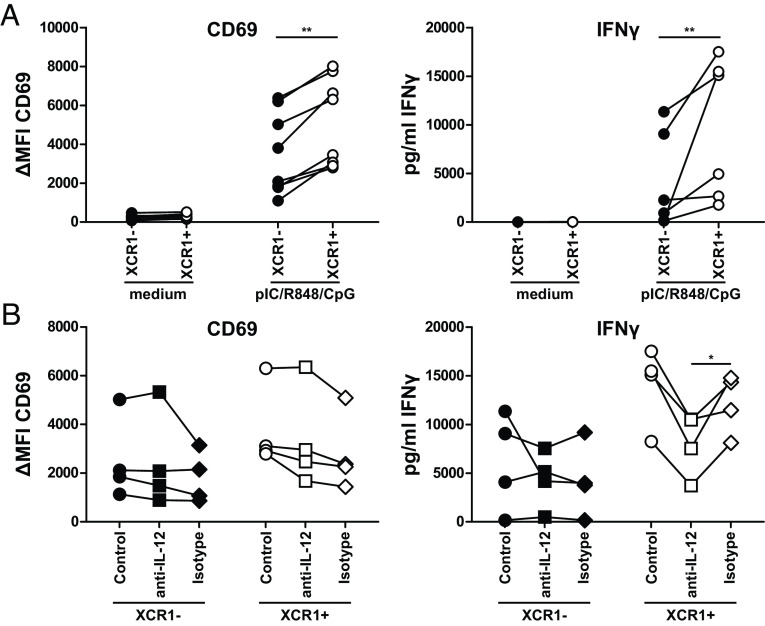
As XCR1+ cDC1 excelled in the secretion of cytokines involved in antiviral immunity (Fig. 3) as well as showed an enrichment of genes involved in the IFNγ response on the transcriptional level (Fig. 2B), we wondered whether this would influence their capacity to induce antiviral immunity via other cells than T cells. As NK cells have been shown to interact with cDC1 and are important in the immediate response to viral infections, we cocultured TLR-stimulated and control-treated XCR1− and XCR1+ cDC1 with autologous NK cells for 18 h (SI Appendix, Fig. S10A). Subsequently, we analyzed the activation of NK cells by expression of the activation marker CD69 and by measuring the concentration of secreted IFNγ (Fig. 4A). Our data revealed that TLR-treated XCR1+ cDC1 induced significantly stronger activation of NK cells than XCR1− cDC1 evidenced by enhanced CD69 expression as well as stronger secretion of IFNγ (Fig. 4A). This activation could not be observed for NK-DC cultures, in which control-treated XCR1− or XCR1+ cDC1 were used.
Fig. 4.
XCR1+ cDC1 excel in the activation of NK cells due to IL-12 secretion. Sorted XCR1− or XCR1+ cDC1 were cocultured with NK cells in a 1:5 ratio in presence or not of 2.5 µg/mL R848, 2.5 µg/mL pIC and 2.5 µg/mL CpG (Type A) as described in SI Appendix, Fig. S10A. After 18 h, supernatants were harvested and analyzed for the secretion of IFNγ by CBA Flex-Set (BD Biosciences) as well as NK cells for the expression of CD69 by flow cytometry. Samples were acquired using a BD LSRFortessa and analyzed using FlowJo (CD69 expression) or FCAP Array 3.1 (IFNγ secretion). (A) Scatter plots show ΔMFI for CD69 (N = 8, each donor connected by line) and secretion of IFNγ (N = 6, each donor connected by line). (B) Assay was performed as described in (A) with the addition of 10 µg/mL of a neutralizing anti-IL-12/IL-23p40 antibody (clone: C11.5) or an appropriate isotype control during the 18 h coculture. Scatter plots show ΔMFI for CD69 (N = 4, each donor connected by line) and secretion of IFNγ (N = 4, each donor connected by line). *P < 0.05; **P < 0.01 (paired Student’s t test).
As DC-derived IL-12 is crucial for activating NK cells in response to pathogens, we further investigated whether IL-12 plays a role in the enhanced NK cell activation by XCR1+ cDC1 (53, 54). Therefore, we blocked IL-12 during the coculture of DCs and NK cells using a neutralizing antibody against IL-12p40 (Fig. 4B). Indeed, our data showed that neutralizing IL-12 led to significantly impaired secretion of IFNγ by NK cells cocultured with TLR-treated XCR1+ but not XCR1− cDC1. This was not the case when the control antibody was applied (Fig. 4B). In accordance to data of Messlinger et al., we did not observe an effect of IL-12 neutralization on the up-regulated expression of CD69 on NK cells (53). In conclusion, our data show that XCR1+ cDC1 excelled in activating NK cells in an IL-12-dependent manner. However, we still observed residual activation of NK cells after neutralization of IL-12 (Fig. 4B). Thus, we wanted to understand whether secreted cytokines by cDC1 alone are needed for the activation of the NK cells or whether cell–cell contact is necessary. Thus, we separated NK cells and XCR1− or XCR1+ cDC1 using a Transwell insert with a pore size of 0.4 µm (SI Appendix, Fig. S11A) or cultured NK cells with supernatants of pIC and R848-stimulated XCR1− and XCR1+ cDC1 (SI Appendix, Fig. S11B) and compared it to XCR1+ and XCR1− cDC1 and NK cells from the same donor in the direct coculture in F-bottom wells (SI Appendix, Fig. S10). While we observed upregulation of CD69 by NK cells in the Transwell assay (SI Appendix, Fig. S11A) as well as after culture with supernatants of XCR1− and XCR1+ cDC1 (SI Appendix, Fig. S11B), it was significantly lower compared to the direct coculture (SI Appendix, Fig. S10). We conclude that in addition to cytokines also direct cell–cell contact is necessary for full activation of NK cells.
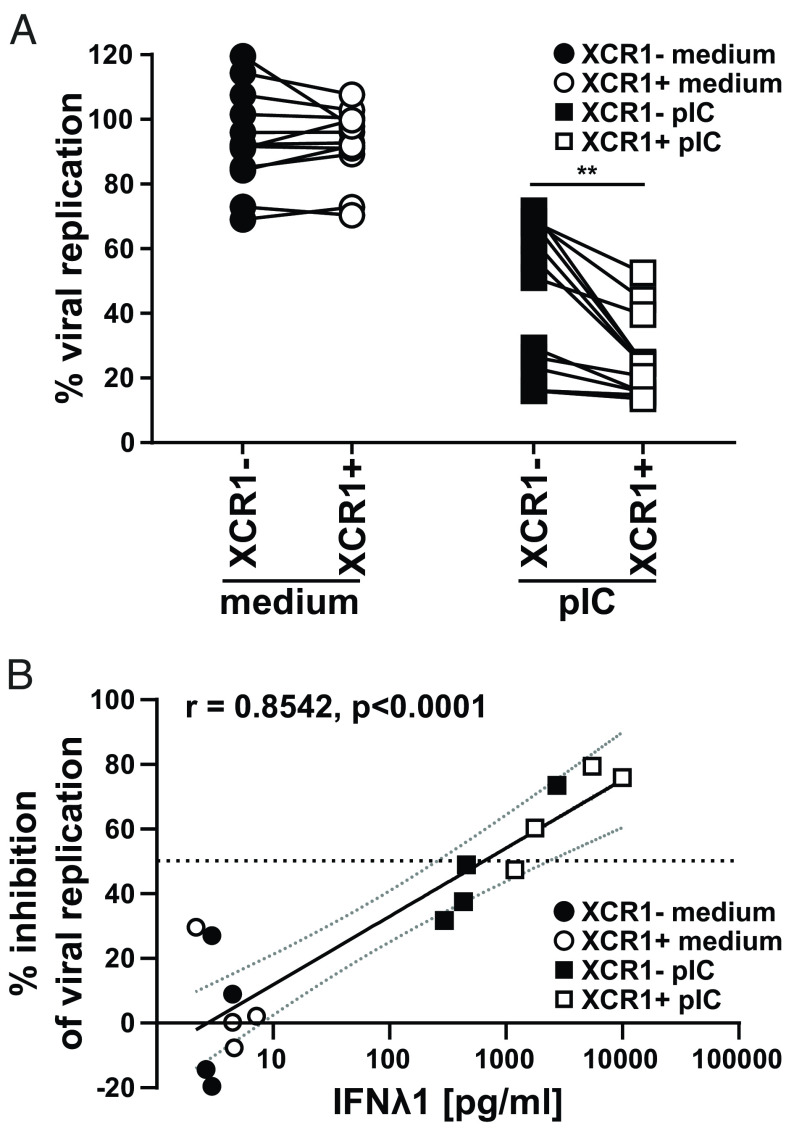
In addition to IL-12, we also found a strongly enhanced secretion of type I and type III interferons, especially IFNλ1, in XCR1+ cDC1 compared to XCR1− cDC1 (Fig. 3). While IFNλ has no direct effect on the activation of NK cells (55, 56), we hypothesized that cDC1 induce an antiviral state in epithelial cells by IFNλ. To prove our hypothesis that IFN production has a critical impact on the XCR1+ cDC1 mediated immune response, we performed virus infection experiments of an influenza virus-susceptible cell line (U2OS). We tested whether XCR1− and XCR1+ cDC1 differ in the capacity to inhibit viral replication in these cells. Therefore, we sorted XCR1+ and XCR1− cDC1 from blood of healthy donors and stimulated them with the IFN-inducing TLR3 ligand pIC (Fig. 3 and SI Appendix, Fig. S10B). Next, U2OS cells were pretreated with the supernatant of either control- or pIC-treated XCR1− or XCR1+ cDC1 for 24 h. Then, U2OS cells were challenged with an influenza virus type A strain expressing luciferase (PR8-luc) for 48 h and infection of the cells was detected by luciferase assay. Our data revealed that both, XCR1− and XCR1+ cDC1, were able to inhibit viral replication of influenza virus type A in U2OS cells, when they were stimulated with pIC as they showed strongly reduced bioluminescence (Fig. 5A). Of note, we found that the supernatant of XCR1+ cDC1 was superior in inhibiting the replication of influenza virus type A compared to the supernatant of XCR1− cDC1 (Fig. 5A). Control-treated (medium) XCR1− and XCR1+ cDC1 could not inhibit viral replication in U2OS cells (Fig. 5A). When correlating the secreted IFNλ1 amount per condition to the virus blocking effect, we found that the strongest block of replication was induced by pIC-treated XCR1+ cDC1, as high IFNλ1 secretors, and a partial viral block was induced by pIC-treated XCR1− cDC1, as low IFNλ1 secretors (Figs. 3 and 5 A and B). Only when XCR1− cDC1 were able to secrete high amounts IFNλ1 (i.e., more than 1,000 pg/mL IFNλ1 induced >50% inhibition of viral replication), they showed a comparable capacity to induce an antiviral state in U2OS cells (Fig. 5B). Thus, we conclude that due to higher secretion of IL-12 and IFNλ1, XCR1+ cDC1 are more effective in inducing antiviral immunity than XCR1− cDC1.
Fig. 5.
XCR1+ cDC1 are superior in the induction of an antiviral state in epithelial cells. Sorted XCR1− and XCR1+ cDC1 were stimulated or not with 5 µg/mL pIC as described in SI Appendix, Fig. S10B Supernatants (SN) were harvested and used for the pretreatment of U2OS cells. After overnight culture, U2OS cells were challenged with luciferase-expressing Influenza virus type A (strain PR8-Luc). (A) Scatter plots show viral replication (% compared to nontreated infected cells) in U2OS cells after treatment with SN of XCR1− and XCR1+ cDCs ± stimulation with 5 µg/mL pIC. XCR1− (filled black circles: medium-treated; filled squares: pIC-treated) and XCR1+ cDC1 (open circles: medium-treated; open squares: pIC-treated) from the same donor are connected by lines (N = 12; **P < 0.01; paired Student’s t test). (B) SN from four donors were used for pretreatment of U2OS cells as in (A) and analyzed for the secretion of IFNλ1 by CBA assay (LEGENDplex Human Anti-Virus Response Panel). Scatter plots show inhibition of viral replication in U2OS cells correlated with the amount of IFNλ1 in the used SN for pretreatment (N = 4; XCR1− cDC1: black symbols, XCR1+ cDC1: white symbols; control-treated: circles, pIC-treated: squares, horizontal line depicts 50% inhibition of viral replication). Semilog line was fitted and Pearson Correlation Coefficient r and P value (two-tailed, exact) determined using GraphPad Prism9.
Capacity of cDC1 to Secrete Antiviral Cytokines Is Linked to the Expression of XCR1.
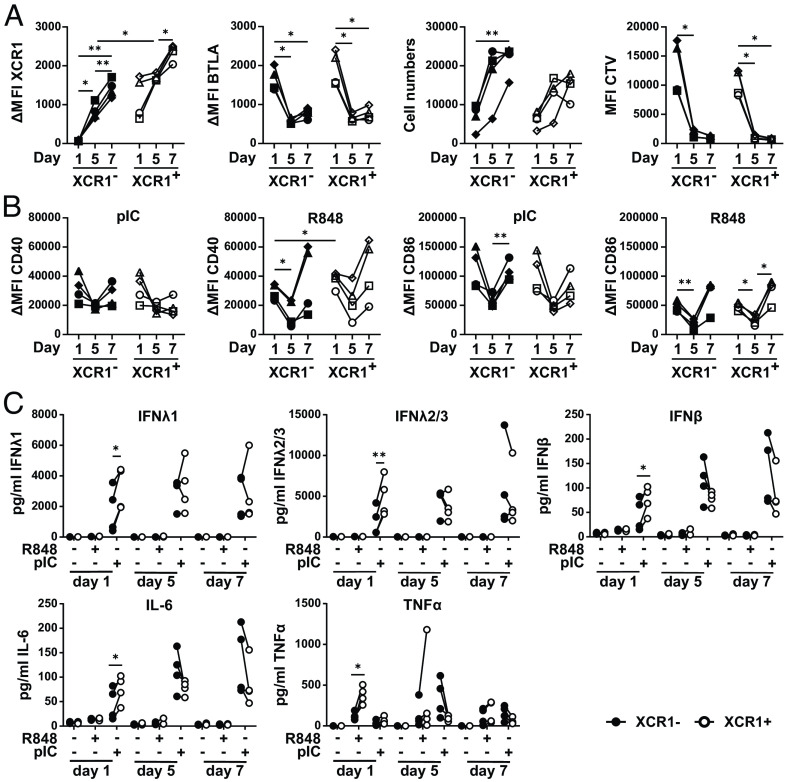
Balan et al. recently reported that Notch signaling and GM-CSF are crucial for cDC1 development in vitro and that an immediate XCR1− precursor arises during the differentiation of pre-DCs into cDC1 (23). Thus, we were interested in whether XCR1− cDC1 would acquire XCR1 expression during culture and whether this would enable the cells to secrete high amounts of inflammatory cytokines, such as IFNλ. Therefore, we isolated XCR1− and XCR1+ cDC1 from peripheral blood and cultured them on Mitomycin C-treated MS5 stroma cells in the presence of GM-CSF, FLT3L, and SCF for 7 d to enable precursor cell differentiation into cDC1 (SI Appendix, Fig. S10C). In order to monitor the differentiation, we harvested XCR1− and XCR1+ cDC1 after 1, 5, and 7 d of the culture. Our flow cytometry analyses revealed that XCR1− and XCR1+ cDC1 showed similar expression of BTLA during the culture, while the expression of XCR1 on XCR1− cDC1 increased to the level of XCR1+ cDC1 in the beginning of the culture, although still slightly less compared to XCR1+ cDC1 at the end of the culture (Fig. 6A). While Balan et al. reported that mainly XCR1− cDC1 proliferated in their culture system (23), we found a substantial proliferation of XCR1− and XCR1+ cDC1 evidenced by dilution of the proliferation dye CellTrace Violet as well as an increase in cell numbers (Fig. 6A).
Fig. 6.
Differentiation of XCR1− cDC1 into XCR1+ cDC1 enables secretion of inflammatory cytokines. Sorted XCR1− and XCR1+ cDC1 were cultured in a DC differentiation assay as described in SI Appendix, Fig. S10C. On days 0, 4, and 6, cells were stimulated for 12 h or not with either 5 µg/mL pIC or R848. SN were harvested and cells were analyzed by flow cytometry for XCR1, BTLA, CD40, and CD86 expression. SN were analyzed for cytokine and chemokine secretion by CBA assay (LEGENDplex Human Anti-Virus Response Panel). (A) Scatter plots show XCR1 and BTLA expression (ΔMFI values), cell numbers as well as MFI value of CTV signal and (B) CD40 and CD86 expression (ΔMFI values) (filled black symbols: XCR1− cDC1; open symbols: XCR1+ cDC1; each individual donor has a distinct symbol and is connected by lines; N = 4; *P < 0.05, **P < 0.01; Two-way ANOVA). (C) Harvested SN of the samples in (B and C) were analyzed for the secretion of IFNλ1, IFNλ2/3, IL-6, TNFα, and IFNβ secretion by CBA assay (LEGENDplex Human Anti-Virus Response Panel). Concentration of the cytokines is shown as scatter plots (filled black circles: XCR1− cDC1; open circles: XCR1+ cDC1; individual donors connected by lines; N = 4; *P < 0.05, **P < 0.01; paired Student’s t test).
As XCR1− and XCR1+ cDC1s showed a strong difference in the ability to secrete inflammatory cytokines, we were interested in whether XCR1− cDC1 would acquire cytokine secretion capacity together with the expression of XCR1. Therefore, we stimulated FACS-isolated XCR1− and XCR1+ cDC1 in the beginning (day 0), during (day 4), and at the end of the culture (day 6) with the TLR-ligands pIC or R848 for 12 h (SI Appendix, Fig. S10C). The cDC1 subsets were analyzed by flow cytometry to measure the expression of costimulatory molecules (Fig. 6B) as well as by CBA assay for cytokine production (Fig. 6C). While at the beginning of the culture XCR1+ cDC1 expressed higher amounts of the costimulatory molecule CD40 compared to XCR1− cDC1 after stimulation with R848, this was not the case at day 7 of the culture (Fig. 6B). Further, when stimulated with pIC or R848 on day 0 of the culture, XCR1+ cDC1 secreted significantly more IFNλ1, IFNλ2/3, IFNβ, and IL-6 (pIC) or TNFα (R848) than XCR1− cDC1 (Fig. 6C). This effect was abolished when XCR1− cDC1 expressed comparable amounts of XCR1 starting at day 5 of culture (Fig. 6C). Thus, we conclude that cDC1 acquire the capacity to secrete inflammatory cytokines in a terminal differentiation step, while other functions such as the ability to induce T cell responses are already present during earlier states.
Discussion
Here, we showed that human cDC1 can be distinguished into XCR1− and XCR1+ cDC1 throughout lymphohematopoietic as well as nonlymphoid tissues. Our data further revealed that XCR1+ cDC1 displayed a preactivated phenotype on mRNA level compared to XCR1− cDC1 and excelled after TLR stimulation in cytokine secretion. This led to enhanced NK cell activation and better protection of epithelial cells against influenza infection. As shown before, XCR1− cDC1 could acquire XCR1 expression during culture under DC differentiation conditions (23). We could further show that the acquisition of XCR1 expression by XCR1− cDC1 was accompanied with the ability to secrete comparable levels of inflammatory cytokines. Thus, only XCR1+ cDC1 seem to possess full effector functions (e.g. antiviral immunity) of cDC1, while XCR1− cDC1 have to acquire these functions in a terminal differentiation step.
In mice, XCR1 expression on cDC1 is linked to their superior capacity to cross-present antigens and induce cytotoxic CD8+ T cell immune responses. Here, it was described that shortly after recognizing their antigen, CD8+ T cells secrete high amounts of XCL1 potentially leading to stronger interactions with cDC1. Further data from Dorner at al. suggested that increased proliferation of CD8+ T cells as well as enhanced secretion of IFNγ is induced by XCR1 expressing cDC1 (29). However, in the interaction of CD8+ T cells and cDC1, the expression of XCR1 is especially important during the later stages of the immune response. While initial priming of CD8+ T cells can occur on XCR1− cDC1, activated XCL1-secreting T cells were described to recruit XCR1+ cDC1. These were then able to cross-present antigens derived from migratory DCs dying in the lymph node after acquiring antigens in the periphery (30, 31). During these processes, XCR1+ cDC1 appear to serve also as a platform for CD4+ T cell help for CD8+ cytotoxic T cells. The recruitment of XCR1+ immature cDC1 to other dying DCs interacting with T cells might be an important way to initiate effective antiviral responses. This is of special importance as human cDC1, in contrast to human cDC2, are quite resistant to infection with certain viruses (57). Thereby, cDC1 rely on the cross-presentation of antigens (57). As shown before, human cDC1 are superior in the cross-presentation of dead cell–derived antigens due to the expression of the F-actin recognizing receptor CLEC9A (11, 13, 39, 58, 59). As XCR1 expression in this context is only important during later stages of the interaction between T cells and DCs, this might explain why we did not observe differences in the capacity of XCR1− and XCR1+ cDC1 to induce T cell responses or cross-prime CD8+ T cells in vitro (SI Appendix, Fig. S9).
Besides CD8+ T cells, also NK cells are described to secrete XCL1 in the steady state and after activation (33–36). Here, we could show that XCR1+ cDC1 were superior in the activation of NK cells compared to XCR1− cDC1, which could be attributed to the enhanced secretion of IL-12 by XCR1+ cDC1 following TLR stimulation (Figs. 3 and 4). It was shown before that interaction of cDC1 and NK cells is crucial for immunity against infections as well as tumors (32, 33, 60–67). While NK cells recruit cDC1 to tumors via the chemokines XCL1 and CCL5 (61, 65), cDC1 secrete the chemokines CXCL9 and CXCL10 (32, 68), the ligands for CXCR3 expressed on NK cells (69–71). Except for reciprocal recruitment, cDC1-derived IL-12 is crucial for IFNγ secretion by NK cells and augments their killing efficacy (53, 54, 67, 72). Furthermore, NK cell–derived IFNγ boosts IL-12 and CXCL9/10 secretion by cDC1, thereby inducing a positive feedback loop (32). This cDC1–NK cell axis is required for memory responses against different viruses as well as intracellular bacteria and is also a marker for good prognosis in several types of cancer (32, 61, 65). In addition, the interaction of XCR1+ cDC1 with XCL1-secreting NK cells is crucial for the upregulation of the homing receptor CCR7 on cDC1 in murine MCMV infection models (33). Here, NK cell–derived GM-CSF induced expression of CCR7 leading to the subsequent migration of XCR1+ cDC1 to the T cell zone and the induction of naïve T cell responses (33). Notably, our data suggest that mainly human XCR1+ cDC1 can secrete IL-12 as well as CXCL10 (Fig. 3), making the XCR1+ cDC1 superior for the induction of NK cell responses (Fig. 4). Thus, our findings propose that the expression of XCR1 is not only important for the recruitment of cDC1 to the tumor microenvironment or infected cells but also for the activation of NK cells. Further, NK cells were shown to be the major producer of Flt3l and the level of FLT3L in tumors was associated with the amount of cDC1 in the tumor and with the overall survival of the patients (65). As NK cells also provide GM-CSF to cDC1 during viral infection shown in murine studies (33), NK cells could even drive the differentiation of XCR1− cDC1 into XCR1+ cDC1, which is relying on GM-CSF (23) as well as FLT3L (Fig. 6). Thereby, XCR1− cDC1 might replenish the pool of XCR1+ cDC1 leaving the tissue to induce T cell responses in secondary lymphoid organs. Whether this system might be manipulated by the tumor, has to be tested in future studies.
In this study, we could identify an immediate precursor cell of terminally differentiated cDC1 characterized by the expression of the cDC1 marker CD141, CLEC9A, BTLA, DEC205, CD13, CD26, and CD226 to a similar level as terminally differentiated cDC1 but without the expression of XCR1 (Figs. 1 and 6 and SI Appendix, Fig. S3). Although the cDC1 compartment appeared homogeneous in previous scRNAseq analysis, we could further identify these cells in published scRNAseq data from Saichi et al. (47) based on the DEGs identified in our Nanostring dataset (Fig. 2 and SI Appendix, Fig. S5). This might be due to smaller transcriptional differences between XCR1− and XCR1+ cDC1 than between DC2 and DC3, which were identified before by scRNAseq (42). XCR1−/low cDC1 were also identified in mice (45, 46). However, DC1b as defined by Hongo et al. showed reduced expression of all cDC1-defining markers, such as CD8, Clec9a, CD24, DEC205, and BTLA, while they coexpressed low level of cDC2-defining markers, such as SIRPα, CD11b, and CD4 (45). Thus, these cells seem to have an intermediate phenotype between cDC1 and cDC2. In addition, Wylie et al. described CD8+XCR1− DCs that are transcriptionally and functionally related to pDC as well as cDC2 and lack the expression of cDC1 signature genes (46). However, human XCR1− cDC1 can be transcriptionally clearly distinguished from cDC2 and belong to the cDC1 lineage (SI Appendix, Fig. S5B). Thus, how and whether human and murine XCR1− cDC1 belong to each other, is not clear yet. See et al. described human pre-DCs expressing the pDC-specific markers CD123 and CD303 as well as Axl and Siglec-6 (9). The pre-DCs give rise to cDC1-committed precursors, which still lack the expression of cDC1-specific surface markers such as CD141 and BTLA (9). In line with our data, the early human pre-DCs and pre-cDC1 did not produce notable amounts of IL-12 and TNF in response to TLR3 and TLR7/8 stimulation compared to cDC1 (9). However, also here, pre-DCs and pre-cDC1 were equally able to induce naïve CD4+ T cell proliferation (9). Thus, XCR1 expression seems to be a marker for fully functional and differentiated cDC1 able to secrete high amounts of cytokines and to induce antiviral immunity. Therefore, XCR1− cDC1 seem to represent a direct intermediate between pre-cDC1 and functional terminally differentiated cDC1. As XCR1− cDC1 are present in blood, lymphoid as well as nonlymphoid tissues, they might serve as a reservoir to rapidly replenish the cDC1 pool under certain conditions, such as infections.
Materials and Methods
Human Tissue Preparation.
Leukocyte reduction system cones were retrieved from anonymous healthy adult donors. Thymus samples were retrieved from cardiac surgeries of otherwise healthy children. The sources of spleen and tonsil samples were patients requiring therapeutic splenectomy and tonsillectomy, respectively. Lung samples were retrieved from patients undergoing partial resection of the lung. This study was approved by the local ethics committee (Ethikkommission der Friedrich-Alexander-Universität Erlangen-Nürnberg; 185_12 B, 22-252-Bp, 192-21 B, 3761, 136_16 B, 4163), and informed written consents were obtained for all samples in accordance with the Declaration of Helsinki.
Blood and all tissues were freshly processed as described before (12, 22, 44, 73). In brief, blood product from leukocyte reduction system cones was transferred into a 50-ml tube and diluted with RPMI1640 to 40 mL. Lung tissue was injected with 5 mL RPMI1640 containing 0.1% human sera type AB, 400 U/mL collagenase D (Serva) and 100-µg deoxyribonuclease I (Sigma) using a syringe to facilitate the digestion of the tissue. Thymic, splenic, tonsillar, and lung tissue was chopped into small pieces using forceps and scalpel. Then, the tissue was transferred into C-tubes (Miltenyi Biotech), filled with 5 mL RPMI1640, further mechanically disrupted using a Gentle MACS tissue dissociator (Miltenyi Biotech), and enzymatically digested with 400 U/mL collagenase D (Serva) and 100 µg (spleen, tonsil) or 300 µg (thymus) deoxyribonuclease I (Sigma). After filtering the cell suspension twice, the cell suspension of splenic, tonsillar, thymic, and lung tissue was diluted with RPMI1640. Then, 20 mL of diluted cell suspension of blood, spleen, thymus, tonsil, or lung was overlaid onto 14 mL Human Pancoll (ρ = 1.077 g/mL; Pan Biotech), and a density gradient centrifugation (520 × g, 25 min, RT, no deceleration) was performed as described before (12, 22, 44). After the centrifugation, the interphase containing the mononuclear cells was collected, washed twice with RPMI1640, and used for experiments.
Cell Lines.
Murine MS-5 stromal cells (ACC 441, DSMZ) were cultured in Alpha medium with 10% FCS, 2 mM L-Glutamine, 2 mM sodium pyruvate, and 1% Penicillin/Streptomycin (MS-5 medium) in 75 cm2 cell culture flasks at 37 °C with 95% humidity and 5% CO2. Confluent culture was split every third day 1:5 using trypsin/EDTA.
Human Bone Osteosarcoma Epithelial (U2OS) Cells were derived from a 15-y-old human female suffering from osteosarcoma and were cultured in Dulbecco’s Modified Eagle’s with 10% FCS and 1% Penicillin/Streptomycin at 37 °C in 95% humidity and 5% CO2. Confluent culture was split every 2 to 3 d 1:10 using trypsin/EDTA.
Madin-Darby Canine Kidney (MDCK) type II cells were cultured in Dulbecco’s Modified Eagle’s with 10% FCS and 1% Penicillin/Streptomycin at 37 °C in 95% humidity and 5% CO2. Confluent culture was split every 2 to 3 d 1:10 using trypsin/EDTA.
Virus Strains.
PR8-luc virus is based on lvPR8, with segments 4 (HA) and 6 (NA) from hvPR8 (74). Segment 8 contains reporter gene Gluc fused to NS1 and linked by a 2A peptide to NEP (75). The original virus stock was kindly provided by Michael Winkler (German Primate Center, Göttingen, Germany) and further amplified on MDCK II cells (RRID:CVCL_0424).
Microarray and scRNAseq Data Analysis.
Published microarray data [Gene Expression Omnibus (GEO) database: accession number GSE77671] were analyzed for relative expression of XCR1 (12). Transcriptome data of whole human genome oligo microarray (Agilent) of human CD1c+ DCs (cDC2), CD141+ DCs (cDC1), and pDCs from three blood, spleen, and thymus donors as well as blood monocytes, B cells, and CD4+ and CD8+ T cells were used. Raw values generated by automated feature extraction have been RMA background corrected and quantile normalized using R (Windows, x64, 3.3.1). Relative expression values were plotted taking advantage of the gplots package of R.
Published scRNAseq data (GEO database: accession number GSE169346, discovery dataset) were analyzed for the composition of the cDC1 compartment (47). cDC1 were extracted from the dataset, normalized by a LogTransform method, and integrated using a canonical correlation analysis (CCA) method. XCR1− and XCR1+ clusters in the cDC1 compartment were identified by “FindCluster” with a resolution parameter of 0.8. Gene expression module analysis was performed using Seurat. DEGs of steady-state XCR1+ cDC1 (XCR1+ module) and XCR1− cDC1 (XCR1- module) were included for the analysis (Fig. 2B), where only DEGs having a logFC higher than 0.3 or lower than −0.3 were analyzed (pseudobulk average feature expression values inferred by Seurat “AverageExpression”).
Flow Cytometric Analysis of XCR1 Expression.
Flow cytometric analysis of single-cell suspensions of blood, spleen, tonsil, thymus, and lung were performed on a BD LSRFortessa and analyzed using FlowJo software (BD Biosciences). 5*106 (blood) to 8*106 (thymus, spleen, tonsil, lung) cells were stained either with a PE-labeled antibody directed against XCR1 (clone S15046E, BioLegend) or with a PE-labeled rat IgG2a isotype control (RTK2758, BioLegend) for 15 min at 37 °C. After washing, the cells were stained with an antibody cocktail to discriminate different immune cell populations for 15 min at 4 °C (blood, spleen, thymus, tonsil: SI Appendix, Table S1; lung: SI Appendix, Table S2). After washing, the cells were resuspended in PBS+2% human sera+0.1 mg/mL DAPI and acquired using a BD LSRFortessa.
Enrichment of DCs.
To perform cell sorts, blood DCs were enriched with the EasySep Pan-DC Pre-Enrichment Kit (Stemcell Technologies) as described in the manufacturer’s manual. Briefly, PBMCs were diluted in PBS+2% human sera+1 mM EDTA and stained with anti-human CD32 (Fc gamma RII) Blocker (Stemcell Technologies) and the EasySep Pan-DC Pre-Enrichment Cocktail for 45 min at room temperature (RT). Then, magnetic Dextran Rapidspheres were added, and cells were incubated for 10 min at RT followed by incubation for 5 min inside “The Big Easy” EasySep Magnet (Stemcell Technologies). The unlabeled DCs were poured off into a new tube and stained for cell sorts.
Cell Sorting of Human DC Subpopulations.
DC-enriched cell suspensions were stained with an antibody cocktail in PBS+2% human sera for 30 min on ice (SI Appendix, Tables S3 and S4). After washing of the cells, cells were resuspended in PBS+2% human sera+0.1 mg/mL DAPI and cell sorted using a BD FACSAria II cell sorter into CD1c+ cDC2 (CD3−CD14−CD19−CD20−CD56−CD123−NKp46−HLA-DR+CD11c+CD1c+), XCR1+ cDC1 (CD1c−CD3−CD14−CD19−CD20−CD56−CD123−NKp46−HLA-DR+CD11cintCD141+XCR1+), XCR1− cDC1 (CD1c−CD3−CD14−CD19−CD20−CD56−CD123−NKp46−HLA DR+CD11cintCD141+XCR1−), and pDCs (CD1c−CD3−CD11b−CD14−CD19−CD20−CD56−NKp46−HLA DR+CD123+CD303+. The purity of sorted cell populations was reanalyzed and routinely above 95%.
Differentiation of cDC1 on Stroma Cells.
MS-5 stroma cells were treated with 10 µg/mL Mitomycin C for 3 h. After washing with PBS twice, 2,5*104 cells were seeded into the wells of a tissue culture–treated 96-well plate (F-bottom). On the next day, freshly sorted XCR1+ and XCR1− cDC1 were labeled with 5 µM Cell Trace Violet for 20 min at 37 °C. After washing, 10,000 DCs were added to each well and cultured in the presence of 160 ng/mL GM-CSF, 160 ng/ml SCF, and 800 ng/mL FLT3L for up to 7 d. On days 0, 1, 5, and 7, cells were analyzed for the expression of XCR1 and for the dilution of CellTrace Violet. Further, before harvesting the cells, Precision Count Beads™ (BioLegend) were added to each well to determine the exact cell number after flow cytometric analysis. In some experiments, cells were stimulated on days 0, 4, and 6 with 5 µg/mL pIC or 5 µg/mL R848. After 12 h, supernatants were harvested and analyzed with the LEGENDplex Antihuman Viral Response Panel (BioLegend) for secretion of cytokines. Cells were analyzed by flow cytometry for the expression of costimulatory molecules (CD40, CD86) as well as cDC1 markers (XCR1, BTLA). Acquisition of the samples was performed using a BD LSRFortessa and data were analyzed using FlowJo (XCR1, BTLA, CD40, CD86) or LEGENDplex software (VigeneTech; cytokine secretion).
Stimulation of Sorted DCs.
Cell-sorted DC subpopulations were resuspended in DC medium (RPMI1640+5% human sera+5% Pannexin NTA+5% Pannexin NTS+1% Glutamine+1% sodium pyruvate+1% Penicillin/Streptomycin) to a concentration of 1 to 2*105 cells/ml dependent on the experiment. Cells were cultured in sterile 96-well plates (v-bottom) and stimulated either with one of the TLR-ligands pIC (5 µg/mL) or R848 (5 µg/mL). TLR-ligands were purchased from Invivogen. For some experiments, recombinant XCL1 (Peprotech) was added in different concentrations (100 ng/mL, 1 µg/mL) in the beginning of the culture. After 3 h or 12 h of stimulation, cells were analyzed by flow cytometry for the expression of XCR1 (XCR1-PE, clone: S15046E, BioLegend) and/or activation markers (CD40-A647, clone: 5C3, BioLegend; CD86-PE-CF594, 2331(FUN-1), BD Biosciences; OX40L-PE, 11C3.1, BioLegend; PD-L1-BV421, MIH3, BioLegend). Supernatants of stimulated cells were harvested and stored at −80 °C until analysis with the LEGENDplex Human Anti-Virus Response Panel (BioLegend). Acquisition of the samples was performed using a BD LSRFortessa, and data were analyzed using FlowJo (XCR1, CD40, CD86, OX40L, and PD-L1) or LEGENDplex software (VigeneTech; cytokine secretion).
Cross-Priming Assays.
CD8+ T cells and DC subpopulations were isolated from PBMCs of HLA-A2+ healthy donors. DCs were enriched and isolated as described above. CD8+ T cells were isolated using the EasySep Human CD8+ T Cell Isolation Kit (Stemcell Technologies) as described in the manufacturer’s instructions. Isolated CD8+ T cells were then electroporated with mRNA encoding a TCR that is specific for the HLA-A*02:01-resticted gp100-peptide YLEPGPVTA. The electroporation was performed as described before (50–52), and more details are available in SI Appendix. After incubation for 4 h at 37 °C, T cells were harvested and cocultured with sorted DC subpopulations in a 1:1 ratio. Before the coculture, DCs were fed either with a long or short gp100-peptide and stimulated with 5 µg/mL pIC and R848 for 3 h. After 18 and 36 h of cocultivation, T cells were analyzed for the expression of CD69 and CD25 by flow cytometry and the supernatant was analyzed for the secretion of IFNγ, IL-2, and TNF by CBA assay (BD Biosciences). Acquisition of the samples was performed using a BD LSRFortessa, and data were analyzed using FlowJo (CD69, CD25) or FCAP Array v3.0 software (BD Bioscience; cytokine secretion).
Mixed Leukocyte Reactions.
Naïve CD4+ or CD8+ T cells were isolated with EasySep Human Naïve CD4+ T Cell Isolation Kit II (Stemcell Technologies) or EasySep Human Naïve CD8+ T Cell Isolation Kit II (Stemcell Technologies), respectively, as described in the manufacturer’s instruction. Purified T cells were labeled with CFSE (5 µM; Life Technologies) for 15 min at 37 °C. Afterward T cells and DCs of HLA-mismatched donors were cocultured in different ratios. DCs were stimulated either with R848 (1 µg/mL) or pIC (1 µg/mL). After 6 d, T cells were restimulated with 50 ng/mL PMA and 500 ng/mL Ionomycin for 6 h in presence of 5 µg/mL Brefeldin A. T cells were harvested and analyzed by flow cytometry for activation (CD25) markers and intracellular cytokines [IL-2, IL-4 (CD4+ T cells only), IL-10 (CD4+ T cells only), IL-17A (CD4+ T cells only), IFNγ, and TNFα]; more details are available in SI Appendix. Acquisition of the samples was performed using a BD LSRFortessa, and data were analyzed using FlowJo.
DC-NK Cell Coculture.
NK cells were enriched from PBMCs by the depletion of unwanted cells using biotinylated antibodies and MoJo Streptavidin Nanobeads (BioLegend); more details are available in SI Appendix. The enriched NK cells were stained with CD3-BV605 (clone: UCHT-1, BioLegend), CD19-BV605 (clone: SJ25C1, BioLegend), CD20-BV605 (clone: 2H7, BioLegend), HLA-DR-BV510 (clone: L243, BioLegend), CD56-BV421 (clone: 5.1H11, BioLegend) and NKp46-BV421 (clone: 9E2, BioLegend) and sorted as CD56+NKp46+CD3−CD19−CD20−HLA-DR− cells. Sorted XCR1− and XCR1+ cDC1 and NK cells were cocultured in a 1:5 ratio and either stimulated with a cocktail of pIC (2.5 µg/mL), R848 (2.5 µg/mL), and CpG (2.5 µg/mL) or cultured in DC-medium alone. For some experiments, DCs and NK cells were separated in Transwell plates (96 well, 0.4 µm insert). NK cells were cultured in the well, while DCs were added to insert. In order to analyze the influence of secreted cytokines on NK cells, NK cells were cultured alone and supernatants of stimulated DCs were added corresponding to a 1:5 DC:NK cell ratio. After 18 h, cells were harvested and analyzed by flow cytometry for activation of NK cells (CD69). The supernatants were stored at −80 °C until analysis with LEGENDplex Human Interferon Panel (BioLegend) for secretion of IFNγ.
Inhibition of Viral Replication.
XCR1+ and XCR1− cDC1 were sorted from PBMCs of healthy donors as described above. Then, cells were stimulated for 3 h with 5 µg/mL pIC. After washing to remove excess pIC, cells were cultured for further 12 h. Subsequently, supernatants were harvested and stored at −80 °C until use in viral replication assays. U2OS cells were cultured in 100 µL in 96-well plates (flat bottom) and pretreated with 20-µL supernatant of pIC- or control-treated XCR1+ or XCR1− cDC1 for 24 h. Then, cells were infected with a multiplicity of infection of 12 with an influenza A virus expressing luciferase (PR8-luc). After 48 h, supernatants of U2OS cells were collected and luciferase activity measured to determine rate of infection. As a control, infected, untreated as well as not infected, untreated cells were used.
Nanostring Assay.
XCR1+ and XCR1− cDC1 were sorted from PBMCs of healthy donors as described above. Afterward, cells were stimulated for 3 h at 37 °C with 5 µg/mL R848 or control treated. Then, cells were lysed in 1/3 RLT buffer (Qiagen) and sample mRNAs were stored at −80 °C until further analysis. Nanostring assay with the nCounter® Human Myeloid Innate Immunity V2 Panel (NanoString Technologies) with 30 additional genes (CASP4, CASP8, NLRP1, NLRP12, NLRC4, NAIP, AIM2, MEFV, GSDMD, ANPEP, IFNL1, IFNL2, IFNL3, CADM1, NLRC5, RAB15, ZNF366, CD226, TEAD4, TCF7L2, EPAS1, LHX6, TCF7, CLNK, C1ORF54, ABCA1, ZBTB32, GZMK, AXL, and SIGLEC6) was performed using the manufacturer’s instructions (more details are available in SI Appendix). Data were subsequently analyzed and exported using the nSolver software (version 4.0, NanoString Technologies). Data were further processed with RUVg (76) to remove unwanted variation. Eventually, they were analyzed in R using NanoStringNorm (77) and edgeR (78), and DEGs were determined with a threshold of false-discovery rate (FDR)–Benjamini–Hochberg adjusted P value of 0.01 and a log2 fold change (FC) of 1.5.
Quantification and Statistical Analysis.
Students t test or one-way or two-way ANOVA with Šídák’s multiple comparisons test was performed using Prism 9.0 (GraphPad). Results were displayed as mean ± SD or scatter plots with one dot representing a single donor. Differences were deemed significant when the P value (Student’s t test) or the adjusted P value (ANOVA with post hoc test for multiple testing) was lower than 0.05. Statistical details, such as the used test and the exact number of donors (N), are found in the Figure legend of each Figure.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank the Core Unit Cell Sorting and Immunomonitoring for cell-sorting support and the Department of Transfusion Medicine of the University Hospital Erlangen. We further also thank all members of the Dudziak laboratory for their critical comments and advice. We would like to thank Michael Winkler from the German Primate Center (Göttingen) for providing the original influenza virus stocks. This work was partly supported by grants from the German Research Foundation [Deutsche Forschungsgemeinschaft (DFG)] to D.D. (CRC1181-TPA7 261193037, DU548/5-1 420943261, TRR305-TPB05 429280966) and a proposal funded by the Agence Nationale de la Recherche (ANR) and the DFG (DU548/6-1 431402787). D.D. received support from Interdisziplinäres Zentrum für Klinische Forschung (IZKF) (IZKF-A80). L. Heger was supported by Erlanger Leistungsbezogene Anschubfinanzierung und Nachwuchsförderung (ELAN) (DE-17-09-15-1-Heger). D.D. was funded by the Bavarian State of ministry of Science and Art, Bayresq.Net. M.K. received Era-Net grant 01KT1801, CompLS program grant 031L0262C from the German Federal ministry of Education and Research (BMBF) and funding by the German Research Foundation (GRK 2740 B02). M.K. and C.L. were funded by the German Research Foundation (SFB TR221/INF 324392634).
Author contributions
L. Heger and D.D. designed research; L. Heger, L. Hatscher, C.H.K.L., L.A., J.J.L., T.K., R.N., N.S., J.D., P.I., M.T., and E.S. performed research; N.S., J.D., S.E.A., A.P., H.S., A.H., C.A., and R.C. contributed new reagents/analytic tools; L. Heger, C.L., C.H.K.L., M.T., M.K., and D.D. analyzed data; and L. Heger and D.D. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Lukas Heger, Email: lukas.heger@uk-erlangen.de.
Diana Dudziak, Email: diana.dudziak@uk-erlangen.de.
Data, Materials, and Software Availability
The nanostring data are available in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/gds) under the accession number GSE237715 (79). Flow cytometry data are available in the FlowRepository (http://flowrepository.org/) under the accession numbers FR-FCM-Z6KP (80), -Z6KQ (81), -Z6L7 (82), -Z6KV (83), and -Z6MY (84). Previously published data were used for this work: Published microarray data were analyzed for relative expression of XCR1 and are available in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/gds) under the accession number GSE77671 (12). Published scRNAseq data were analyzed for the composition of the cDC1 compartment and are available in the Gene Expression Omnibus database under the accession number GSE169346 (47). Published mRNA expression data (NanoString technology) were analyzed and are available in the Gene Expression Omnibus database under accession number GSE162407 (44).
Supporting Information
References
- 1.Banchereau J., Steinman R. M., Dendritic cells and the control of immunity. Nature 392, 245–252 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Steinman R. M., Banchereau J., Taking dendritic cells into medicine. Nature 449, 419–426 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Geissmann F., et al. , Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merad M., Sathe P., Helft J., Miller J., Mortha A., The dendritic cell lineage: Ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31, 563–604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amon L., et al. , Transcriptional control of dendritic cell development and functions. Int. Rev. Cell Mol. Biol. 55–151 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Amon L., Lehmann C. H. K., Heger L., Heidkamp G. F., Dudziak D., The ontogenetic path of human dendritic cells. Mol. Immunol. 120, 122–129 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Schlitzer A., et al. , Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16, 718–728 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Grajales-Reyes G. E., et al. , Batf3 maintains autoactivation of Irf8 for commitment of a CD8α+ conventional DC clonogenic progenitor. Nat. Immunol. 16, 708–717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.See P., et al. , Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356, eaag3009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robbins S. H., et al. , Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9, R17 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulin L. F., et al. , Characterization of human DNGR-1+ BDCA3+ leukocytes as putative equivalents of mouse CD8alpha+ dendritic cells. J. Exp. Med. 207, 1261–1271 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidkamp G. F., et al. , Human lymphoid organ dendritic cell identity is predominantly dictated by ontogeny, not tissue microenvironment. Sci. Immunol. 1, eaai7677 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Jongbloed S. L., et al. , Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 207, 1247–1260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlitzer A., et al. , IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control Mucosal IL-17 cytokine responses. Immunity 38, 970–983 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watchmaker P. B., et al. , Comparative transcriptional and functional profiling defines conserved programs of intestinal DC differentiation in humans and mice. Nat. Immunol. 15, 98–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meredith M. M., et al. , Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 209, 1153–1165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildner K., et al. , Batf3 deficiency reveals a critical role for CD8+ dendritic cells in cytotoxic T cell immunity. Science (80-) 322, 1097–1100 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiavoni G., et al. , ICSBP is essential for the development of mouse type I interferon-producing cells and for the generation and activation of CD8alpha(+) dendritic cells. J. Exp. Med. 196, 1415–1425 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aliberti J., et al. , Essential role for ICSBP in the in vivo development of murine CD8alpha+ dendritic cells. Blood 101, 305–310 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Persson E., et al. , IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity 38, 958–969 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Suzuki S., et al. , Critical roles of interferon regulatory factor 4 in CD11bhighCD8 - dendritic cell development. Proc. Natl. Acad. Sci. U.S.A. 101, 8981–8986 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heger L., et al. , CLEC10A is a specific marker for human CD1c+ dendritic cells and enhances their toll-like receptor 7/8-Induced cytokine secretion. Front. Immunol. 9, 744 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balan S., et al. , Large-scale human dendritic cell differentiation revealing notch-dependent lineage bifurcation and heterogeneity. Cell Rep. 24, 1902–1915.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutertre C. A., et al. , Single-cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory dendritic cells. Immunity 51, 573–589.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Bourdely P., et al. , Transcriptional and functional analysis of CD1c+ human dendritic cells identifies a CD163+ subset priming CD8+CD103+ T cells. Immunity, 1–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.den Haan J. M. M., Lehar S. M., Bevan M. J., Cd8+ but not Cd8− dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192, 1685–1696 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dudziak D., et al. , Differential antigen processing by dendritic cell subsets in vivo. Science 315, 107–111 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Tussiwand R., et al. , Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity 42, 916–928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorner B. G., et al. , Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 31, 823–833 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Eickhoff S., et al. , Robust anti-viral immunity requires multiple distinct T cell-dendritic cell interactions. Cell 162, 1322–1337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brewitz A., et al. , CD8+ T cells orchestrate pDC-XCR1+ dendritic cell spatial and functional cooperativity to optimize priming. Immunity 46, 205–219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexandre Y. O., et al. , XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J. Exp. Med. 213, 75–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghilas S., et al. , Natural killer cells and dendritic epidermal γδ T cells orchestrate type 1 conventional DC spatiotemporal repositioning toward CD8+ T cells. iScience 24, 103059 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorner B. G., et al. , Coordinate expression of cytokines and chemokines by NK cells during murine cytomegalovirus infection. J. Immunol. 172, 3119–3131 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Hedrick J. A., et al. , Lymphotactin is produced by NK cells and attracts both NK cells and T cells in vivo. J. Immunol. 158, 1533–1540 (1997). [PubMed] [Google Scholar]
- 36.Yamazaki C., et al. , Conservation of a chemokine system, XCR1 and its ligand, XCL1, between human and mice. Biochem. Biophys. Res. Commun. 397, 756–761 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Bachem A., et al. , Expression of XCR1 characterizes the Batf3-dependent lineage of dendritic cells capable of antigen cross-presentation. Front. Immunol. 3, 1–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crozat K., et al. , The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 207, 1283–1292 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachem A., et al. , Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 207, 1273–1281 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tel J., et al. , Human plasmacytoid dendritic cells efficiently cross-present exogenous Ags to CD8+ T cells despite lower Ag uptake than myeloid dendritic cell subsets. Blood 121, 459–467 (2013). [DOI] [PubMed] [Google Scholar]
- 41.McInnes L., Healy J., Melville J., UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv [Preprint] (2018). 10.48550/arXiv.1802.03426 (Accessed 6 December 2018). [DOI]
- 42.Villani A.-C., et al. , Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown C. C., et al. , Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell 179, 846–863.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hatscher L., et al. , Select hyperactivating NLRP3 ligands enhance the T H 1- and T H 17-inducing potential of human type 2 conventional dendritic cells. Sci. Signal. 14, eabe1757 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Hongo D., et al. , Identification of two subsets of murine DC1 dendritic cells that differ by surface phenotype, gene expression, and function. Front. Immunol. 12, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wylie B., et al. , CD8+XCR1neg dendritic cells express high levels of toll-like receptor 5 and a unique complement of endocytic receptors. Front. Immunol. 10, 1–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saichi M., et al. , Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity. Nat. Cell Biol. 23, 538–551 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Nizzoli G., et al. , Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood 122, 932–942 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Hémont C., Neel A., Heslan M., Braudeau C., Josien R., Human blood mDC subsets exhibit distinct TLR repertoire and responsiveness. J. Leukoc. Biol. 93, 1–11 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Schaft N., et al. , A new way to generate cytolytic tumor-specific T cells: Electroporation of RNA coding for a T cell receptor into T lymphocytes. Cancer Immunol. Immunother. 55, 1132–1141 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoyer S., et al. , Braf and mek inhibitors affect dendritic-cell maturation and t-cell stimulation. Int. J. Mol. Sci. 22, 11951 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerer K. F., Hoyer S., Dörrie J., Schaft N., Electroporation of mrna as universal technology platform to transfect a variety of primary cells with antigens and functional proteins. Methods Mol. Biol. 1499, 165–178 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Messlinger H., et al. , Monocyte-derived signals activate human natural killer cells in response to Leishmania parasites. Front. Immunol. 9, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferlazzo G., et al. , Distinct roles of IL-12 and IL-15 in human natural killer cell activation by dendritic cells from secondary lymphoid organs. Proc. Natl. Acad. Sci. U.S.A. 101, 16606–16611 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morrison M. H., et al. , IFNL cytokines do not modulate human or murine NK cell functions. Hum. Immunol. 75, 996–1000 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Tian Z., et al. , Involvement of NK cells in IL-28B–mediated immunity against influenza virus infection. J. Immunol. 199, 1012–1020 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Silvin A., et al. , Constitutive resistance to viral infection in human CD141+ dendritic cells. Sci. Immunol. 2, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sancho D., et al. , Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature 458, 899–903 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ahrens S., et al. , F-actin is an evolutionarily conserved damage-associated molecular pattern recognized by DNGR-1, a receptor for dead cells. Immunity 36, 635–645 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Mittal D., et al. , Interleukin-12 from CD103+ Batf3-dependent dendritic cells required for NK-cell suppression of metastasis. Cancer Immunol. Res. 5, 1098–1108 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Böttcher J. P., et al. , NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 172, 1022–1028.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Eck J., et al. , Clinically applicable CD34+-derived blood dendritic cell subsets exhibit key subset-specific features and potently boost anti-tumor T and NK cell responses. Cancer Immunol. Immunother. 70, 3167–3181 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.El Weizman O., et al. , ILC1 Confer Early Host Protection at Initial Sites of Viral Infection. Cell 171, 795–808.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.López-Yglesias A. H., et al. , T-bet-dependent ILC1- And NK cell-derived IFN-γ mediates cDC1-dependent host resistance against Toxoplasma gondii. PLoS Pathog. 17, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barry K. C., et al. , A natural killer–dendritic cell axis defines checkpoint therapy–responsive tumor microenvironments. Nat. Med. 24, 1178–1191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bödder J., et al. , Harnessing the cDC1-NK cross-talk in the tumor microenvironment to battle cancer. Front. Immunol. 11, 1–13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacobs B., et al. , Characterization and manipulation of the crosstalk between dendritic and natural killer cells within the tumor microenvironment. Front. Immunol. 12, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Groom J. R., et al. , CXCR3 chemokine receptor-ligand interactions in the Lymph Node optimize CD4+ T helper 1 cell differentiation. Immunity 37, 1091–1103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Campbell J. J., et al. , Unique subpopulations of CD56+ NK and NK-T peripheral blood lymphocytes identified by chemokine receptor expression repertoire. J. Immunol. 166, 6477–6482 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Berahovich R. D., Lai N. L., Wei Z., Lanier L. L., Schall T. J., Evidence for NK cell subsets based on chemokine receptor expression. J. Immunol. 177, 7833–7840 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Wennerberg E., Kremer V., Childs R., Lundqvist A., CXCL10-induced migration of adoptively transferred human natural killer cells toward solid tumors causes regression of tumor growth in vivo. Cancer Immunol. Immunother. 64, 225–235 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Groen R. A., et al. , IFN-λ-mediated IL-12 production in macrophages induces IFN-γ production in human NK cells. Eur. J. Immunol. 45, 250–259 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Heger L., et al. , Guidelines for DC preparation and flow cytometric analysis of human lymphohematopoietic tissues. Eur. J. Immunol. e2249917 (2022). [DOI] [PubMed]
- 74.Grimm D., et al. , Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl. Acad. Sci. U.S.A. 104, 6806–6811 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eckert N., et al. , Influenza A virus encoding secreted Gaussia luciferase as useful tool to analyze viral replication and its inhibition by antiviral compounds and cellular proteins. PLoS One 9, e97695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Risso D., Ngai J., Speed T. P., Dudoit S., Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Waggott D., et al. , NanoStringNorm: An extensible R package for the pre-processing of nanostring mRNA and miRNA data. Bioinformatics 28, 1546–1548 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCarthy D. J., Chen Y., Smyth G. K., Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heger L., et al. , XCR1 expression distinguishes human conventional dendritic cells type 1 with full effector functions from their immediate precursors. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE237715. Deposited 18 July 2023. [DOI] [PMC free article] [PubMed]
- 80.Heger L., Dudziak D., Expression of cDC1-associated surface markers on XCR1- and XCR1+ cDC1. FlowRepository. http://flowrepository.org/id/FR-FCM-Z6KP. Deposited 14 July 2023.
- 81.Heger L., Dudziak D., Differentiation of XCR1- cDC1 into XCR1+ cDC1. FlowRepository. http://flowrepository.org/id/FR-FCM-Z6KQ. Deposited 13 July 2023.
- 82.Heger L., Dudziak D., Interaction of XCR1- and XCR1+ cDC1 with NK cells. FlowRepository. http://flowrepository.org/id/FR-FCM-Z6L7. Deposited 20 July 2023.
- 83.Heger L., Dudziak D., Cytokine secretion by sorted XCR1- and XCR1+ cDC1. FlowRepository. http://flowrepository.org/id/FR-FCM-Z6KV. Deposited 17 July 2023.
- 84.Heger L., Dudziak D., Inhibition of viral replication by XCR1- and XCR1+ cDC1. FlowRepository. http://flowrepository.org/id/FR-FCM-Z6MY. Deposited 28 July 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The nanostring data are available in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/gds) under the accession number GSE237715 (79). Flow cytometry data are available in the FlowRepository (http://flowrepository.org/) under the accession numbers FR-FCM-Z6KP (80), -Z6KQ (81), -Z6L7 (82), -Z6KV (83), and -Z6MY (84). Previously published data were used for this work: Published microarray data were analyzed for relative expression of XCR1 and are available in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/gds) under the accession number GSE77671 (12). Published scRNAseq data were analyzed for the composition of the cDC1 compartment and are available in the Gene Expression Omnibus database under the accession number GSE169346 (47). Published mRNA expression data (NanoString technology) were analyzed and are available in the Gene Expression Omnibus database under accession number GSE162407 (44).