Significance
FOXA1 is a critical pioneer factor responsible for facilitating chromatin access of other transcription factors. However, the dynamic nature of FOXA1 binding and the mechanisms governing its reversible binding remains poorly understood. We recently identified that LSD1 can demethylate K270 of FOXA1, thereby stabilizing its chromatin binding. Nevertheless, the methyltransferase that methylates FOXA1 and negatively regulates the FOXA1-LSD1 axis remains unknown. In this study, we have identified SETD7 as the primary methyltransferase of FOXA1, which represses its chromatin binding, and reveals a critical transcription repressor function of SETD7. Our study suggests that maintaining a balance between LSD1 and SETD7 is crucial for FOXA1 chromatin activity, and loss of SETD7 may promote FOXA1 reprogramming and contribute to prostate cancer progression.
Keywords: SETD7, FOXA1, LSD1, lysine methylation, prostate cancer
Abstract
Dysregulation of histone lysine methyltransferases and demethylases is one of the major mechanisms driving the epigenetic reprogramming of transcriptional networks in castration-resistant prostate cancer (CRPC). In addition to their canonical histone targets, some of these factors can modify critical transcription factors, further impacting oncogenic transcription programs. Our recent report demonstrated that LSD1 can demethylate the lysine 270 of FOXA1 in prostate cancer (PCa) cells, leading to the stabilization of FOXA1 chromatin binding. This process enhances the activities of the androgen receptor and other transcription factors that rely on FOXA1 as a pioneer factor. However, the identity of the methyltransferase responsible for FOXA1 methylation and negative regulation of the FOXA1-LSD1 oncogenic axis remains unknown. SETD7 was initially identified as a transcriptional activator through its methylation of histone 3 lysine 4, but its function as a methyltransferase on nonhistone substrates remains poorly understood, particularly in the context of PCa progression. In this study, we reveal that SETD7 primarily acts as a transcriptional repressor in CRPC cells by functioning as the major methyltransferase targeting FOXA1-K270. This methylation disrupts FOXA1-mediated transcription. Consistent with its molecular function, we found that SETD7 confers tumor suppressor activity in PCa cells. Moreover, loss of SETD7 expression is significantly associated with PCa progression and tumor aggressiveness. Overall, our study provides mechanistic insights into the tumor-suppressive and transcriptional repression activities of SETD7 in mediating PCa progression and therapy resistance.
Androgen deprivation therapies (ADTs) are the mainstay of treatment for prostate cancer (PCa). However, patients eventually develop an aggressive metastatic form of cancer, termed castration-resistant PCa (CRPC) (1). The second-generation androgen receptor (AR) signaling inhibition (ARSi) agents, such as enzalutamide and abiraterone, can be used to further treat CRPC (2–4). However, tumor cells can still escape from these treatments through multiple AR-dependent and -independent mechanisms. Many of these mechanisms involve the epigenetic reprogramming of the transcriptional networks caused by alterations of critical epigenetic factors (5).
In particular, the aberrant expression of histone lysine methyltransferases (KMTs) and demethylases (KDMs) has been shown to drive PCa development and progression. Notably, chromatin modifiers like EZH2 and LSD1 gain histone-independent activities, modifying critical transcription factors/cofactors and further altering transcription networks during cancer progression (6, 7). In a recent study, we identified that lysine 270 of FOXA1, a critical linage-specific pioneer factor of AR and ER (estrogen receptor) (8–10), is a nonhistone substrate of LSD1. The LSD1-mediated demethylation of FOXA1 stabilizes its chromatin binding, promoting PCa progression (11, 12). Unlike most Forkhead-domain mutations that commonly affect FOXA1 interaction with DNA (13–15), K270 appears to be a critical site for regulating FOXA1 interaction with core histones (16). Interestingly, another report revealed that EZH2 can methylate FOXA1 at lysine 295, preventing the proteosome-dependent degradation of the FOXA1 protein (17). Given the critical function of FOXA1 in PCa development and its high mutation frequency, particularly in CRPC (10, 11, 13–15, 18, 19), it is plausible that the oncogenic activities of LSD1/EZH2 in CRPC are largely contributed by regulating FOXA1 activity. However, the KMTs that can methylate FOXA1-K270 and repress FOXA1 chromatin activity remain to be determined.
SETD7 (also called KMT7, SET7, or SET9) is a SET domain-containing lysine methyltransferase, and its canonical function is activating gene transcription through methylating H3K4 (20). However, SETD7 can also methylate nonhistone substrates, including AR (K630/632), p53 (K372), E2F1 (K185), and Rb (K873) (21–24). Therefore, the function of SETD7 largely depends on its activity on different substrates, and it can function as either an oncogene or a tumor suppressor in cancer, depending on the cell context and tissue type (25). While SETD7 has been well studied in breast and lung cancers, its function during PCa development is poorly understood. Two early studies suggest that SETD7 can enhance AR activity by directly methylating AR and thus may promote the development of hormone-dependent PCa (24, 26). Additional studies also demonstrate SETD7 regulation on RORα2 and NRF2 in PCa models, and SETD7 knockdown may decrease cell growth (27, 28). However, these studies primarily focus on the function of SETD7 in the early development of PCa, while its role in the progression of CRPC and resistance to ARSi remains unclear.
In this study, we aimed to investigate the role of SETD7 as a methyltransferase of FOXA1-K270, destabilizing FOXA1 chromatin binding and competing with the activity of LSD1. Additionally, we sought to uncover the tumor suppressor function of SETD7 in preventing tumor growth and metastasis in various PCa models under castrated conditions. Through the examination of transcriptional programs in CRPC models, we also identified SETD7’s ability to repress oncogenic transcriptional programs driven by FOXA1, MYC, and E2F. Importantly, our integrated transcriptomic and cistromic analyses indicated that the major activity of SETD7 in PCa involves repressing gene transcription rather than activating it. Furthermore, we found that reduced SETD7 expression in CRPC cells can lead to the redistribution of FOXA1 chromatin binding. Overall, our findings provide molecular insights into the tumor suppressor function of SETD7 during the progression of CRPC, suggesting that the downregulation of SETD7 expression may play a critical role in the reprogramming of FOXA1 activity and adaptation to ARSi in CRPC cells.
Results
SETD7 Methylates Lysine 270 of FOXA1.
FOXA1 plays a pivotal role in promoting PCa progression and adaption to aggressive ARSi (14). Previous studies have indicated the critical role of lysine 270 methylation of FOXA1 in regulating its chromatin binding and pioneer factor activity. Furthermore, the demethylation of K270 by LSD1 has been shown to stabilize FOXA1 binding and confer resistance to ARSi (11, 19). However, the specific methyltransferase responsible for dynamically mediating the methylation of FOXA1 remains unknown.
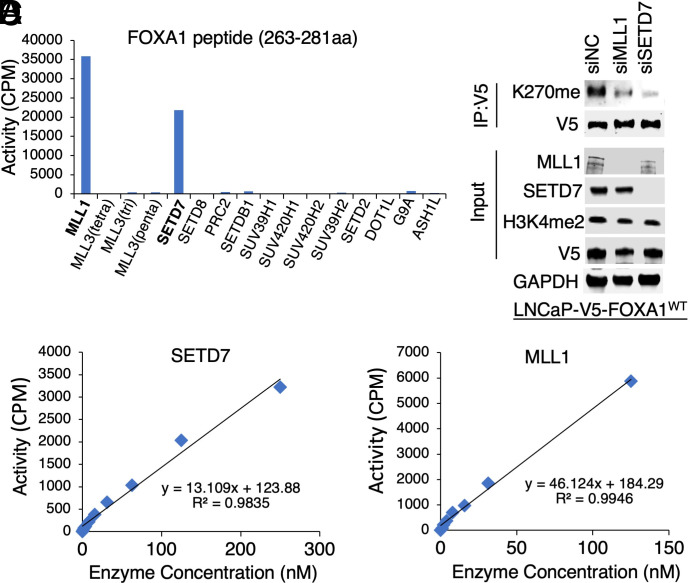
To identify potential candidates, we conducted in vitro methylation assays using a short FOXA1 peptide (aa 263-281) containing K270 and a panel of human KMTs or their functional complexes, which have been rigorously established with the optimal enzymatic conditions for each enzyme using their known histone substrates and has been previously used to profile many substrates and inhibitors (29, 30). As seen in Fig. 1 A–C, both SETD7 and MLL1 demonstrated significant enzymatic activity in methylating the FOXA1 peptide. The enzyme kinetics (Km and kcat) of these enzymes were also comparable to the previously reported kinetics for H3K4 methylation (31, 32) (SI Appendix, Fig. S1 A and B). Interestingly, while both KMTs can methylate H3K4, SETD7 has been reported to possess specific methyltransferase activity toward nonhistone proteins demethylated by LSD1, including p53-K372 and E2F1-K185 (21, 22, 33). Considering that the FOXA1 peptide contains several other lysines, we next determined whether SETD7 and MLL1 are KMTs for K270 using our previously generated FOXA1-K270me-specific antibody (11). In Fig. 1D, we observed reduced FOXA1-K270 methylation in FOXA1-overexpressing LNCaP cells upon silencing either SETD7 or MLLs, with SETD7 knockdown demonstrating a stronger effect. This finding further confirms that both SETD7 and MLL1 can methylate K270 in vivo. However, the levels of methylated H3K4 were not significantly changed, suggesting that these two proteins may not be major H3K4 methyltransferases in PCa cells. Interestingly, while MLL1 appears to interact with FOXA1, no stable interaction between SETD7 and FOXA1 was detected in these cells (SI Appendix, Fig. S1C). Collectively, these data strongly indicate that SETD7 and MLL1 specifically methylate FOXA1-K270. However, we cannot completely exclude the possibility that these KMTs may also weakly methylate other lysine sites on FOXA1.
Fig. 1.
SETD7 methylates lysine 270 of FOXA1. (A) In vitro methylation assay using FOXA1 short peptide (263-281aa) (50 µM) and recombinant lysine methyltransferase proteins or complexes (1 µM). Note: PRC2 is a complex of EZH2, EED, and SUZ12 proteins. (B and C) Validation of SETD7 (B) and MLL1 (C) activity using 5-µM FOXA1 peptide titrated at a series of methyltransferase protein concentrations. (D) Immunoblotting for K270me in LNCaP cells stably expressing V5-FOXA1 (doxycycline pretreated, 0.5 µg/mL, for 2 d) transfected with siMLL, siSETD7, or siNTC (nontarget control).
SETD7 Represses FOXA1 Chromatin Binding.
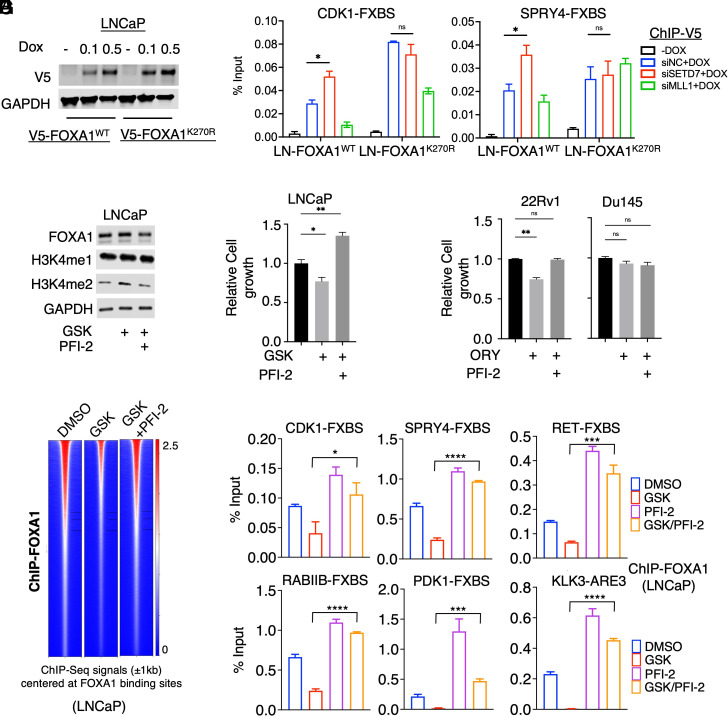
To investigate the impact of SETD7 and MLL1’s K270 methyltransferase activity on FOXA1 chromatin binding, we generated LNCaP stable cells overexpressing either wild-type FOXA1 (FOXA1-WT) or a K270R mutant (FOXA1-K270R) (Fig. 2A) and examined FOXA1 binding upon depletion of SETD7 or MLL1 at two AR-independent FOXA1 binding sites previously identified (11). As shown in Fig. 2B, silencing of SETD7 enhanced the chromatin binding of FOXA1-WT but had no effect on the FOXA1-K270R mutant. On the other hand, MLL1 silencing did not increase the FOXA1 chromatin binding, suggesting that SETD7 is the primary FOXA1 methyltransferase in PCa cells. Consequently, we decided to focus on SETD7 in the subsequent studies.
Fig. 2.
SETD7 represses FOXA1 chromatin binding. (A) Immunoblotting for V5 in LNCaP stable cells overexpressing V5-tagged FOXA1-WT or FOXA1-K270R mutant (doxycycline induced, 0.5 µg/mL, for 2 d). (B) ChIP–qPCR for V5 at two AR-independent FOXA1 binding sites (FXBSs) in these stable cells transfected with siSETD7, siMLL, or siNTC (for 3 d). (C) Immunoblotting for indicated proteins in LNCaP cells treated with LSD1 inhibitor (GSK2879552, 100 µM) alone or in combination with SETD7 inhibitor (PFI-2, 1 µM, for 4 h). (D and E) Proliferation assay for LNCaP (D), 22RV1, or DU145 (E) cells treated with LSD1 inhibitor (100 µM GSK2879552 for LNCaP, 10 µM ORY-1001 for 22Rv1/DU145, for 4 d) alone or in combination with SETD7 inhibitor (PFI-2, 10 µM, for 4 d). (F) Heatmap view for FOXA1 binding intensity (FOXA1 ChIP-seq) in LNCaP cells treated with GSK2879552 (100 µM for 4 h) alone or in combination with PFI-2 (1 µM for 4 h). (G) ChIP–qPCR of FOXA1 at several FOXA1 binding sites.
Since LSD1 inhibition (LSD1-i) broadly disrupts FOXA1 chromatin binding (11), we further examined whether loss of SETD7 activity could counteract the effect of LSD1-i on FOXA1. Using a potent and selective SETD7 inhibitor, PFI-2 (34), which did not affect FOXA1 expression, we found that it fully restored the suppressed cell proliferation caused by LSD1-i (GSK2879552) in LNCaP cells (Fig. 2 C and D). Similar results were observed in 22Rv1 cells using another LSD1-i, ORY1001, but not in FOXA1-negative Du145 cells (Fig. 2E). To gain further insights, we performed ChIP-seq analysis of FOXA1 in LNCaP cells treated with LSD1-i alone or together with PFI-2. As demonstrated in Fig. 2F, consistent with previous findings, LSD1-i led to a significant disruption of FOXA1 chromatin binding, which was rescued by the addition of PFI-2, suggesting that SETD7 may negatively regulate LSD1-mediated FOXA1 chromatin binding. These effects were further confirmed by ChIP-qPCR analysis of FOXA1 at multiple FOXA1 binding sites (Fig. 2G) and by using SETD7 siRNAs (SI Appendix, Fig. S2A). Additionally, there were modest changes in H3K27ac or H3K4me levels (enhancer marks) at some sites in correlation with the alteration of FOXA1 binding (SI Appendix, Fig. S2B).
Considering that FOXA2 shares a nearly identical wing2 region with FOXA1, its K265 residue is also a potential substrate of both LSD1 and SETD7 (11). Indeed, the LSD1-i-mediated repression of FOXA2 binding at multiple identified FOXA2 sites in PC-3 cells (11) could consistently be rescued by SETD7 inhibition but not MLL1 inhibition (SI Appendix, Fig. S3), suggesting that FOXA2 may also be methylated and regulated by SETD7. Collectively, these findings indicate that SETD7 negatively regulates the FOXA1-LSD1 axis in PCa cells.
SETD7 Expression Is Significantly Decreased in CRPC.
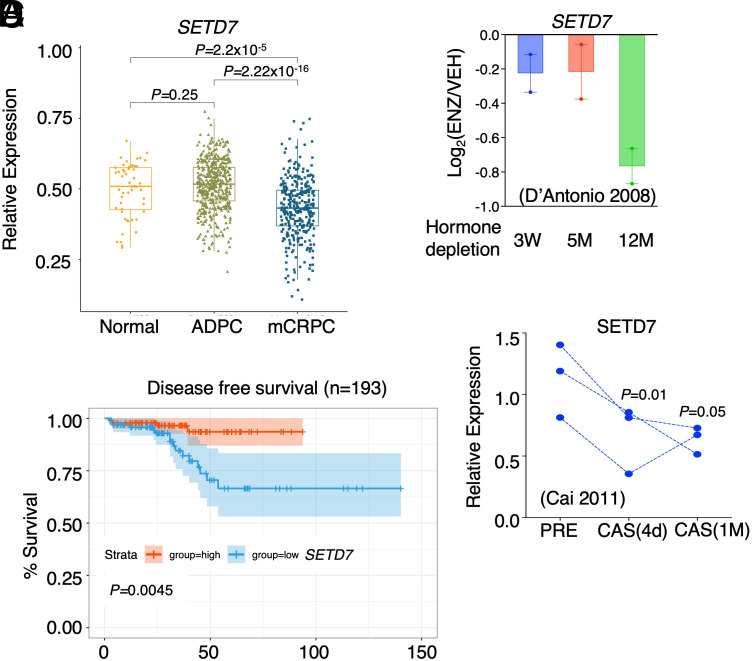
We next investigated the expression of SETD7 in human PCa datasets to understand its relevance in disease. Analysis of the TCGA dataset revealed no significant change in SETD7 expression between primary castration-sensitive PCa samples versus normal prostate samples (Fig. 3A, TCGA dataset) (35). However, in mCRPC samples from the SU2C dataset (36), SETD7 expression was markedly decreased compared to normal or primary PCa samples. Importantly, lower SETD7 expression was significantly associated with poorer clinical outcomes (Fig. 3B and SI Appendix, Fig. S4 A–D).
Fig. 3.
SETD7 expression is significantly increased in CRPC. (A) Box plots for SETD7 expression (ratio to a panel of housekeeping genes) in normal and primary PCa samples (TCGA) and mCRPC samples (SU2C). (B) Kaplan–Meier curve for the overall survival in PCa tumors with higher SETD7 expression (red, the top 25%) versus with lower expression (blue, the bottom 25%). (C) SETD7 mRNA expression in LNCaP cells progressing on ADTs (public dataset, GSE8702). (D) SETD7 mRNA expression in paired VCaP xenografts biopsy at androgen-dependent stage (AD), 4 d after castration (CS), and relapsed tumors (CRPC) (public dataset, GSE31410).
In LNCaP cells progressing on ADTs (37), SETD7 expression showed a modest decrease upon relatively short-term hormone depletion (3 wk to 5 mo) (Fig. 3C). However, a more significant decrease was observed in cells that developed resistance to long-term enzalutamide treatment (12 mo). Furthermore, in LNCaP cells treated with hormone depletion or enzalutamide, SETD7 expression was also modestly decreased (SI Appendix, Fig. S5 A and B). However, this effect is likely an adaption response since SETD7 is not regulated by androgens in LNCaP cells (SI Appendix, Fig. S5C). Conversely, in VCaP PCa cells, SETD7 expression could be rapidly induced by DHT treatment and decreased by castration in the VCaP xenograft model previously generated (38, 39) (Fig. 3D). These findings are consistent with stronger AR binding at the promoter region of SETD7 gene in VCaP cells (SI Appendix, Fig. S5D). Taken together, these data suggest that PCa cells may adapt to ARSi by decreasing SETD7 expression through direct and indirect mechanisms.
Loss of SETD7 Expression Promotes PCa Tumor Growth and Metastasis.
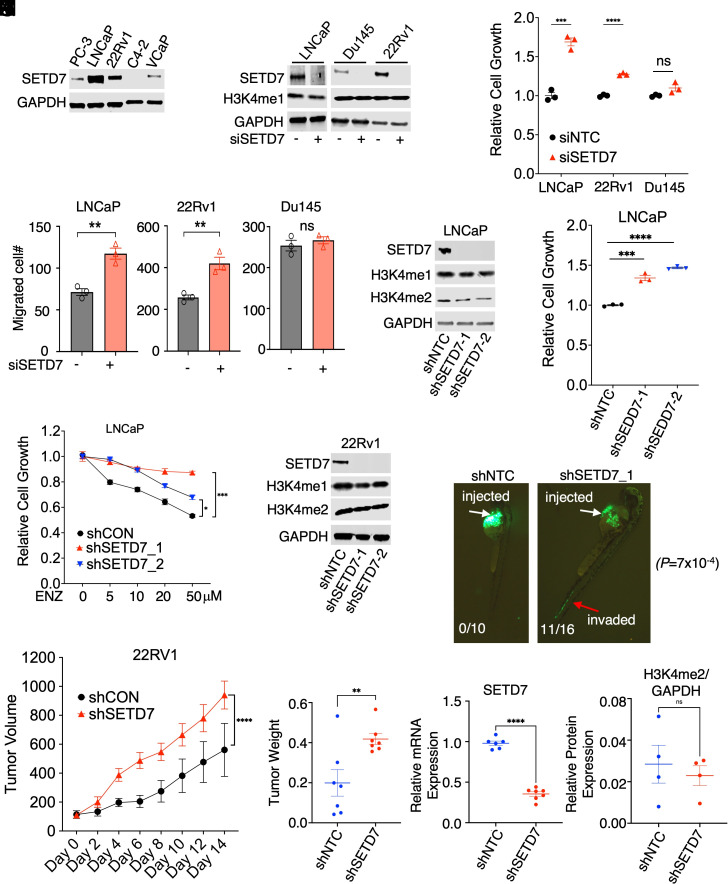
SETD7 protein expression was found to be higher in androgen-dependent LNCaP cells but lower in CRPC cell lines (Fig. 4 A and B). To investigate the function of SETD7, two FOXA1-positive PCa lines (LNCaP and CWR22-Rv1) and a FOXA1-negative line (Du145) were selected. Silencing SETD7 did not affect H3K4 methylation but significantly increased cell growth and migration in LNCaP and 22Rv1 cells cultured under hormone-depleted conditions to mimic ADTs (Fig. 4 C and D). However, this effect was not observed in Du145 cells, suggesting that SETD7 may act as a tumor suppressor in FOXA1-positive PCa cells. The involvement of FOXA1 methylation in this process was supported by weaker cell migration induced by SETD7 silencing in cells expressing FOXA1-K270R mutant (SI Appendix, Fig. S6 A and B). The tumor suppressor activity of SETD7 was further confirmed using two independent shRNAs against SETD7 (Fig. 4 E and F), and the loss of SETD7 expression was shown to desensitize PCa cells to enzalutamide treatment (Fig. 4G).
Fig. 4.
Loss of SETD7 expression promotes PCa tumor growth and metastasis. (A) Immunoblotting for SETD7 in PCa cell lines. (B–D) LNCaP, Du145, and 22Rv1 cells grown in a hormone-depleted medium were transfected with siNTC or siSETD7 (for 3 d) and then subjected to immunoblotting (B), proliferation assay (C), and transwell migration assay (D). (E) Immunoblotting for indicated proteins in LNCaP cells stably expressing lentiviral shRNAs against NTC or SETD7. (F) Proliferation assay in LNCaP-shNTC and -shSETD7 stable cell lines under indicated conditions (for 4 d). (G) Proliferation assay in these stable cells treated with enzalutamide (10 µM) for 4 d. (H) Immunoblotting for SETD7 in 22Rv1 cells stably expressing shNTC and shSETD7. (I) Xenograft tumor growth of 22Rv1 stable cells subcutaneously injected into castrated male mice. (J–L) Tumor weight (J), SETD7 mRNA expression by qRT-PCR (K), and H3K4me2 relative protein expression by immunoblotting (L) in shNTC and shSETD7 xenografts tissue samples. (M) GFP-labeled LNCaP stable cells were injected into zebrafish embryos and immediately examined for tumor cell intravasation.
To investigate the role of SETD7 in CRPC tumor development in vivo, a mouse xenograft model was employed. 22Rv1 lines stably expressing lentiviral shRNAs against SETD7 were generated (Fig. 4H) and subcutaneously injected into castrated male SCID mice to assess tumor development. Loss of SETD7 expression significantly accelerated CRPC tumor development without affecting global H3K4 methylation levels (Fig. 4 I–L). Additionally, a zebrafish embryo model (40) was used to examine the potential of SETD7 silencing to promote metastasis. LNCaP cells expressing lentiviral shNTC or shSETD7 were stably labeled with GFP and injected into zebrafish embryos. While control cells remained within the perivitelline space (0 out of 10 embryos invaded), SETD7-silenced cells rapidly invaded the blood vessel within an hour postinjection (11 out of 16 embryos invaded), indicating a critical role of SETD7 in preventing early steps of the metastasis cascade (Fig. 4M), These in vitro and in vivo findings provide clear evidence for the tumor suppressor function of SETD7 in CRPC progression.
SETD7 Transcriptionally Represses Multiple Oncogenic Programs.
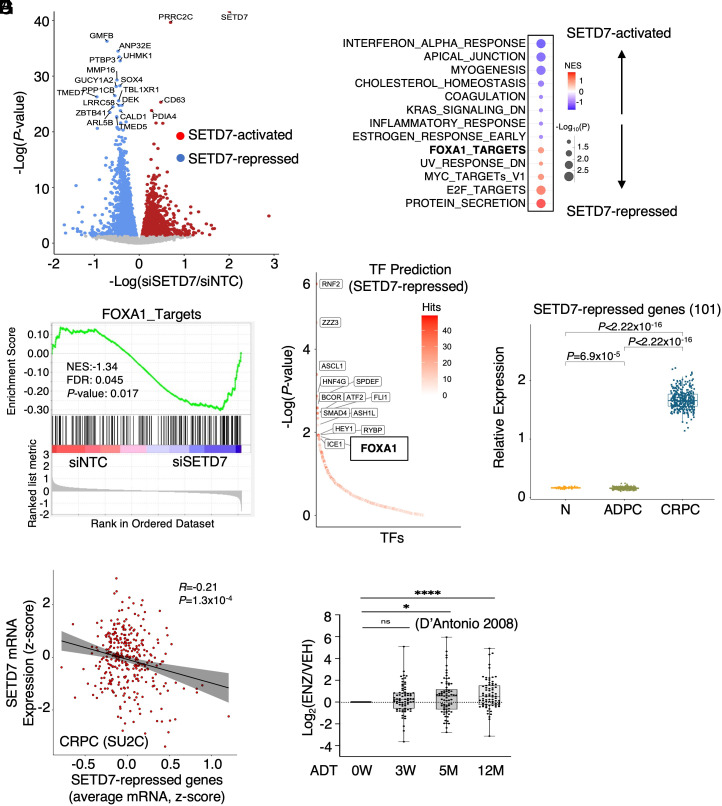
To determine the transcriptional impact of SETD7 silencing in CRPC cells, an RNA-seq analysis was performed in 22Rv1 cells with transient SETD7 silencing under hormone-depleted conditions. SETD7-activated and -repressed genes were identified (Fig. 5A and SI Appendix, Table S1). Gene Set Enrichment Analysis (GSEA) was conducted for hallmark gene sets and PCa-specific signatures, including FOXA1 targets and AR targets (11, 41). The analysis revealed that SETD7-activated genes were significantly enriched in IFN response pathways, while SETD7-repressed genes were enriched in several oncogenic pathways, such as E2F and MYC signaling (Fig. 5B). As expected, SETD7 was found to repress the transcription program of FOXA1 (Fig. 5C), consistent with its role in repressing FOXA1 chromatin binding. It is worth noting that the AR pathway was not enriched in SETD7-regulated genes (SI Appendix, Fig. S7A), despite previous studies suggesting that SETD7 enhances AR activity through methylation (24, 26). Further examination of AR-positive PCa cell lines treated with DHT revealed a modest decrease in some AR-regulated genes upon SETD7 silencing, but there was a significant increase in AR chromatin binding (SI Appendix, Fig. S7 B–E). These findings suggest that SETD7 may have dual effects on AR activity, with methylation enhancing AR nuclear localization but impairing AR chromatin binding through FOXA1 methylation. Notably, the transcriptional impact of SETD7 on many other pathways appears to be opposite to the function of FOXA1 and LSD1 in PCa models (SI Appendix, Fig. S8A), suggesting that SETD7 may function as a global negative regulator of LSD1-mediated oncogenic reprogramming. Using a recently developed CIE platform for the identification and interpretation of regulators of transcriptional response (42), we also predicted RNF2 (a ring finger transcription repressor), ZZZ3 (a zinc finger transcription activator), ASCL1 (a member of the basic helix-loop-helix family of transcription factors), as well as FOXA1 as the top-ranked transcription factors that may be involved in the regulation of SETD7-repressed genes (Fig. 5D).
Fig. 5.
SETD7 transcriptionally represses multiple oncogenic programs. (A) Volcano plot for differentially expressed genes in 22Rv1 cells transfected with siSETD7 versus siNTC. (B) Enrichment of hallmark gene set and our previously defined PCa-specific AR or FOXA1 signatures for SETD7-activated and repressed genes. (C) GSEA plot of previously defined FOXA1 targets (GSE37314) with SETD7-regulated genes. (D) Transcription factors prediction of SETD7-repressed genes by CIE platform. (E) Box plots for SETD7-repressed genes (twofold cutoff) in normal and primary PCa samples (TCGA) and mCRPC samples (SU2C). (F) Correlation of SETD7 and its repressed genes in the SU2C mCRPC dataset. (G) Expression of SETD7-repressed genes in LNCaP cells progressing on ADTs.
Next, we examined the expression of SETD7-repressed genes in the public PCa patient datasets. As shown in Fig. 5E, SETD7-repressed genes were dramatically increased in CRPC (SU2C cohort) compared to normal and primary PCa (TCGA cohort), indicating that this set of genes may play an important role in driving PCa progression. A similar increase in SETD7-repressed genes was also found in another CRPC dataset (43) (SI Appendix, Fig. S8B). Consistently, SETD7 expression was negatively correlated with its repressed genes in the SU2C CRPC dataset (Fig. 5F). Moreover, SETD7 repressed genes were also increased in LNCaP cells adapted to ARSi (Fig. 5G). On the contrary, while MLL1-3 proteins are tumor suppressors in PCa and frequently mutated (loss-of-function mutations) or deleted in CRPC (36), our RNA-seq analysis in 22Rv1 cells with MLL1 silencing did not indicate any enrichment of the FOXA1 pathway in MLL1-regulated genes, although it can similarly decrease E2F and MYC signaling (SI Appendix, Fig. S9 A–C). This suggests that MLL1 regulation on FOXA1 function may be weaker or more complicated compared to SETD7. Overall, these data reveal the transcriptional activity of SETD7 in CRPC cells and suggest that SETD7 may repress multiple oncogenic transcription programs.
SETD7 Chromatin Binding Is Significantly Associated with SETD7-Repressed Genes.
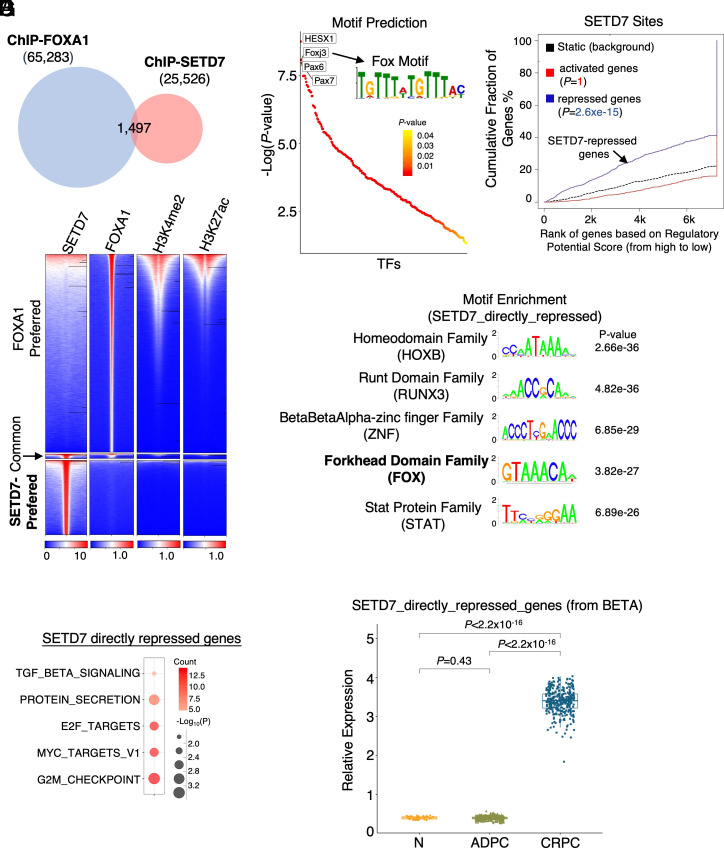
We next determined SETD7 chromatin occupation by performing a ChIP-seq analysis of SETD7 in 22Rv1 cells. Notably, we identified 25,526 high-confidence binding peaks, and these peaks showed very little overlap with FOXA1 chromatin binding peaks in 22Rv1 cells (Fig. 6A). Moreover, these binding peaks were not located at chromatin sites with active enhancer marks such as H3K4me2 and H3K27ac (Fig. 6B), indicating that SETD7-occupied sites are not transcriptionally active. These results are in sharp contrast to previously reported LSD1-occupied chromatin sites, which highly overlap with FOXA1 binding (SI Appendix, Fig. S10 A and B) and serve as transcriptional activation sites (11).
Fig. 6.
SETD7 chromatin binding is significantly associated with SETD7-repressed genes. (A) Venn diagram showing SETD7 and FOXA1 ChIP-seq peaks in 22Rv1 cells. (B) Heatmap view of SETD7 binding with FOXA1 binding and levels of H3K4me2 (public dataset, GSM2135702) and H3K27ac (public dataset, GSM2135704). (C) Motif enrichment analysis for identified SETD7 peaks. (D) BETA to predict the association of SETD7 chromatin binding with SETD7-regulated gene expression. (E) Top-ranked motifs for identified SETD7-directly-repressed genes. (F) GSEA of SETD7-directly-repressed genes (identified from BETA). (G) Box plots for SETD7-directly-repressed genes in normal and primary PCa samples (TCGA) and mCRPC samples (SU2C).
Examining the functional enrichment of SETD7 binding peaks, we found that these sites were located in genes enriched for many cancer-promoting pathways (SI Appendix, Fig. S11). Interestingly, FOXA1 binding motifs were still highly enriched within these SETD7-occupied chromatin sites (Fig. 6C). We then performed Binding and Expression Target Analysis (BETA) (44) to examine the association of SETD7 chromatin binding with its transcriptional output and predict SETD7 direct targets. Surprisingly, SETD7 binding was markedly associated with its repression function but not with its canonical activation function (presumably through H3K4 methylation) (Fig. 6D). These sites were enriched for motifs of HOXB family factors, RUNX3, and FOX family factors (Fig. 6E), suggesting that SETD7 may function globally to prevent FOXA1 binding to unwanted cryptic enhancer sites.
Using BETA, we also predicted a subset of SETD7-directly-repressed genes, which were consistently enriched for E2F and MYC signaling (Fig. 6F). Expression of these SETD7 direct targets was also markedly increased in CRPC patient samples (Fig. 6G) and especially in the metastatic sites (SI Appendix, Fig. S12). Overall, these results clearly demonstrate a noncanonical transcriptional repression function of SETD7, possibly mediated via suppressing FOXA1-induced enhancer activation.
Loss of SETD7 Expression Drives Redistribution of FOXA1 Chromatin Binding.
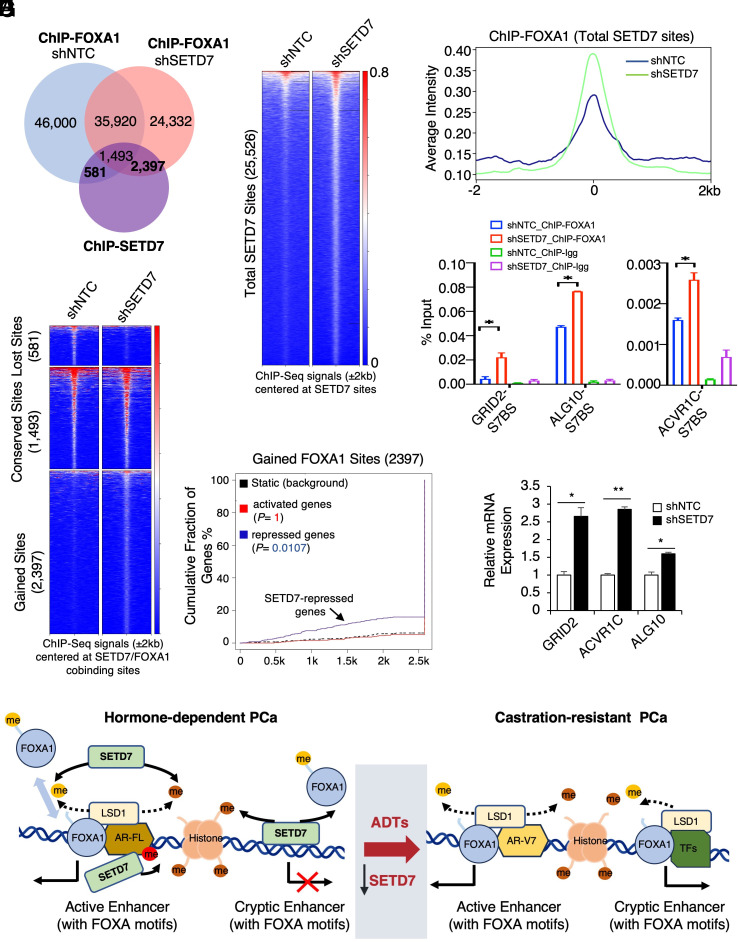
Since SETD7 sites were enriched for FOXA1 motifs, we hypothesize that loss of SETD7 expression in CRPC may globally impact FOXA1 binding. ChIP-seq analyses of FOXA1 were performed in 22Rv1 cells stably expressing shNTC or shSETD7, and the results showed a significant redistribution of FOXA1 binding sites in SETD7-depleted cells (SI Appendix, Fig. S13 A and B). Importantly, at the SETD7-occupied chromatin sites, we observed a notably global increase in FOXA1 binding intensity in response to SETD7 silencing (Fig. 7 A–C). Consistently, only 581 FOXA1/SETD7 cobinding sites were lost after SETD7 was silenced, but 2,397 new FOXA1 sites were established at these SETD7-occupied chromatin regions (Fig. 7D). These redistributed FOXA1 binding sites (2,397) were also significantly associated with SETD7-repressed genes (Fig. 7E), suggesting that the increased FOXA1 binding may be responsible for transcription activation.
Fig. 7.
Loss of SETD7 expression drives redistribution of FOXA1 chromatin binding. (A) Venn diagram for FOXA1 binding sites (FOXA1 ChIP-seq) in 22Rv1 cells stably expressing shNTC or shSETD7 and SETD7 binding sites in parental 22Rv1 cells. (B and C) Heatmap view (B) and average intensity curve (C) for FOXA1 binding intensity at total SETD7 sites. (D) Heatmap view for FOXA1 binding intensity at the altered FOXA1 sites (overlapped with SETD7 sites). (E) BETA to predict the association of gained FOXA1 chromatin binding sites with SETD7-regulated gene expression. (F) ChIP-qPCR of FOXA1 at three identified FOXA1-gained sites. (G) qRT-PCR for mRNA expression of these genes. (H) Graphic summary of the working model.
The increased FOXA1 binding in response to SETD7 silencing or inhibition was further validated by examining FOXA1 binding at a panel of SETD7-occupied sites (Fig. 7F and SI Appendix, Fig. S13C). FOXA1 binding at a classic LSD1/FOXA1-positive site (but SETD7-negative) was also increased (SI Appendix, Fig. S13D). Consistent with the gained FOXA1 binding, the expression of nearby genes was elevated in SETD7-depleted cells (Fig. 7G). These genes include ACVR1C, a member of the activin receptor family, and ALG10, an N-linked glycosylation protein, both of which have been linked to PCa progression (45, 46). To further examine the clinical relevance of this FOXA1 chromatin-binding redistribution, we identified a 22-gene signature as a transcriptional output of SETD7-loss-induced FOXA1 binding at the SETD7-occupied sites (SI Appendix, Fig. S13E). For instance, the increase in GRID2 gene expression caused by SETD7 silencing could be abrogated by silencing FOXA1, indicating a strong dependence on FOXA1 for its expression in SETD7-loss cells (SI Appendix, Fig. S13F). Importantly, these genes exhibited a substantial increase in expression in CRPC samples compared to normal and primary PCa samples (SI Appendix, Fig. S13G). Together, these data indicate that loss of SETD7 expression can induce a transcriptional reprogramming of FOXA1 in CRPC.
Discussion
Our recent finding of LSD1-mediated demethylation of FOXA1 has highlighted the importance of the FOXA1-LSD1 oncogenic axis in driving PCa progression and therapy resistance (11). However, a missing piece of the puzzle is the KMT(s) responsible for methylating FOXA1 to prevent or destabilize FOXA1 chromatin binding and counteract LSD1 activity in PCa cells. The presence of such KMT(s) would allow for dynamic regulation of FOXA1 chromatin binding, preventing overactivation of FOXA1-mediated enhancers in prostate cells. Furthermore, aberrant activity of these KMTs may induce FOXA1-mediated transcriptional reprogramming in cancer cells. In this study, using a biochemistry approach, we identified SETD7 and MLL1 as candidate KMTs fulfilling this role (Fig. 1). Interestingly, many known substrates of SETD7, including H3K4, E2F, p53, and other proteins, are also substrates of LSD1-mediated demethylation (21, 22, 33, 47, 48). While our data clearly demonstrate the negative regulation of FOXA1 chromatin binding and transcriptional output by SETD7, the effects of MLL1 on FOXA1 function were not entirely clear and warrant further investigation.
Notably, we found that SETD7 inhibition globally restored the FOXA1 chromatin binding that was disrupted by LSD1 inhibition (Fig. 2F), suggesting a critical role of SETD7 in establishing the dynamic chromatin binding of FOXA1. Moreover, we also observed that SETD7 can function as a tumor suppressor in PCa, which may seem contradictory to early findings suggesting that SETD7 promotes PCa development as a methyltransferase and coregulator of AR (24, 26). However, our results are not entirely inconsistent with these studies, as we focused on the SETD7 activity under castration conditions where AR activity is suppressed. Interestingly, while SETD7 silencing slightly decreased DHT-stimulated AR activity, it also increased AR chromatin binding, consistent with increased FOXA1 binding. In CRPC, the emergence of AR splice variants, particularly AR-V7, is a major mechanism for restoring AR signaling. However, these truncated AR proteins lack the lysine sites (K630, K632) that are substrates of SETD7, potentially diminishing the positive impact of SETD7 on AR signaling. Consequently, the role of SETD7 in AR signaling may be significantly diminished in CRPC. Indeed, SETD7 expression or its transcriptional repression function does not show significant changes in primary PCa but is dramatically decreased in CRPC, suggesting its specific role in mediating PCa progression and resistance of ARSi, rather than in the early development of PCa.
Another significant finding of this study is that the SETD7 chromatin occupancy is associated with its transcriptional repression function rather than its canonical transcriptional activation function. This indicates that the major chromatin action of SETD7 may be mediated through FOXA1 methylation rather than H3K4 methylation. Interestingly, while LSD1 can directly interact with FOXA1 at chromatin (7), SETD7 does not appear to form a stable protein–protein interaction with FOXA1. Based on our data, we propose that SETD7 may exhibit two modes of activity to alter chromatin accessibility in PCa cells. First, SETD7 can function as a general methyltransferase enzyme in the nucleus to methylate and retain unbound FOXA1 proteins, thus establishing an equilibrium with LSD1’s demethylation activity on FOXA1. Therefore, the loss of SETD7 expression in CRPC may further enhance FOXA1 chromatin binding at some FOXA1-LSD1 co-occupied active enhancer sites. Second, SETD7 can bind to repressive chromatin regions or inactive cryptic enhancers (likely recruited by transcription repressors) containing putative FOXA1 motifs, acting as a gatekeeper to prevent unwanted FOXA1 chromatin binding. This is particularly important for the epigenetic silencing process since unmethylated FOXA1 can access compact chromatin, potentially leading to transcriptional leakage. During PCa progression, decreased SETD7 expression weakens its repressive ability, allowing FOXA1 to access these compact chromatin regions, leading to reactivation of cryptic enhancers and increased transcription of genes that promote CRPC progression. Our working model is presented in Fig. 7H.
In summary, our study provides molecular insights into SETD7 expression and function in CRPC progression and reveals a tumor suppressor activity of SETD7 mediated through FOXA1-K270 methylation and the repression of FOXA1 chromatin binding. Since SETD7 competes with other epigenetic drivers that enhance FOXA1chromatin binding and activity, such as LSD1 and EZH2, the loss of SETD7 expression in CRPC may allow tumor cells to develop oncogenic addiction to these factors, potentially representing a critical mechanism for CRPC progression and tumor adaptation to ARSi treatments.
Materials and Methods
Cell Culture, Transient Transfection, and Establishment of Stable Cell Lines.
LNCaP, 22Rv1, PC-3, Du145, and NCI-H660 cell lines were obtained from ATCC and regularly examined for Mycoplasma contamination using MycoAlert Mycoplasma Detection Kit (Lonza). All cell lines were cultured in RPMI 1640 with 10% FBS, except for NCI-H660, which was cultured with RPMI-1640 base medium with 5% FBS, 10 nM β-estradiol, 10 nM hydrocortisone, and 1% Insulin-Transferrin-Selenium. siRNAs (ON-TARGETplus) and shRNA lentiviral particles (pGIPZ) targeting nontarget control, MLL1, or SETD7 were predesigned and obtained from Dharmacon. Stable cell lines were established by infecting cells with lentivirus and selecting them with puromycin. LNCaP or 22Rv1 cells stably expressing doxycycline-induced V5-tagged FOXA1-WT or K270R were previously generated (11).
Chromatin Immunoprecipitation (ChIP) and ChIP-seq Analysis.
For ChIP preparation, cells were fixed with 1% formaldehyde and chromatin was sheared to ~500 to 800 bp fragments (for ChIP-qPCR) or ~300 bp fragments (for ChIP-seq). Immunoprecipitation was performed using ChIP-grade antibodies. The precipitated protein–DNA complexes were then reverse-cross-linked at 65 °C, followed by DNA purification. The extracted DNA was subjected to ChIP-qPCR using the SYBR green method or ChIP-seq analysis. ChIP-seq libraries were constructed using the SMARTer ThruPLEX DNA-Seq Prep Kit (Takara Bio USA). Next-generation sequencing was performed using Illumina HiSeq2500. ChIP-sequencing reads were mapped to the hg19 human genome, and the significance of enriched peaks was evaluated using MACS2 (version 2.1.4) (49).
RT-qPCR and RNA-seq.
RNA was extracted using the RNeasy kit (Qiagen). Quantitative real-time PCR (qRT-PCR) was performed using Fast 1-step Mix (Thermo Fisher Scientific). All qRT-PCR results were normalized to GAPDH. Taqman primers and probes for SETD7, GRID2, ALG10, ACVR1C, and GAPDH were predesigned and obtained from Thermo Fisher Scientific. For RNA-seq, the library preparation was performed using the TruSeq Strand Total RNA LT (Illumina). Next-generation sequencing was performed using Illumina HiSeq2500. Transcriptome-sequencing reads were aligned to the human reference genome hg19 and all gene counts were processed with the R package Edger (3.36.0) to evaluate differential expression using the Benjamini–Hochberg false discovery rate (FDR)-adjusted P value.
Radioactive Methylation Assay (Scintillation Proximity Assay).
Methylation reactions (10 μL) were carried out in a buffer containing 50 mM Tris–HCl (pH 8.0), 10 mM GSH, and 0.01% Triton X-100, at room temperature using 1.5 μM tritium labeled SAM (PerkinElmer, cat# NET155V250UC), and 50 μM biotinylated FOXA1 peptide (aa 263-281) in 384-well plates in the presence of 1 μM of KMTs. The reactions were incubated for 1 h and then quenched by 10 μL of 7.5M guanidine HCl. Subsequently, 40 μL of buffer (20 mM Tris–HCl, pH 8.0) was added to the quenched samples, and all samples were transferred into a streptavidin and scintillant-coated FlashPlate (PerkinElmer, SMP410), and incubated for 2 h at room temperature. The amount of methylated FOXA1 was quantified by counting the counts per minute (CPM) of radioactivity as measured after 1 h using a TopCount plate reader (PerkinElmer).
Xenograft Study.
All animal experiments were approved by the University of Massachusetts Boston Institutional Animal Care and Use Committee (IACUC) and conducted following institutional and national guidelines. 22Rv1 stable cells were resuspended in serum-free RPMI 1640 medium and mixed in 1:1 ratio with Matrigel (BD Biosciences) prior to subcutaneous implantation (2 × 106 cells per injection) on the flanks of castrated SCID mice (~4 to 6-wk-old, Taconic). Tumor length (L) and width (W) were measured using a caliper at the indicated times, and tumor volumes were calculated as L × W2/2.
Zebrafish Embryo Metastasis Assay.
Zebrafish embryos were generated from AB and TUE wild-type lines by natural spawning. All experiments were performed in 3-d postfertilization embryos following an IACUC-approved protocol. Embryos were dechorionated and anesthetized with 0.04 mg/mL tricaine, and ~100 GFP-expressing cells were microinjected into the perivitelline space of each embryo. After injection, embryos were washed and maintained in 6-well plates, and imaging was performed within 1 h after injection.
Statistical Analysis.
Data in bar graphs represent mean ± SEM of at least three biological repeats. Statistical analysis was performed using Student’s t test by comparing treatment versus vehicle or as otherwise indicated. P value < 0.05 (*, 0.05; **, 0.01; ***, 0.001; ****, 0.0001) was considered statistically significant. The results for immunoblotting are representative of at least three experiments. Boxplots of signature scores and gene expression were compared using the Wilcoxon test for comparison between two groups of samples. The zebrafish metastasis data were analyzed using Fisher’s exact test. The difference in mouse xenograft tumor growth was determined using a two-way ANOVA test. These tests were parametric and based on the assumption of normal distribution and equal variance across all experimental groups.
Note: Detailed information on primers and antibodies, along with additional experimental procedures and methods, can be found in SI Appendix, Materials and Methods.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work is supported by grants from the NIH (R01 CA211350 to C.C. and U54 CA156734 to J.A.M.), DOD (W81XWH-19-1-0361 and W81XWH-21-1-0267 to C.C. and W81XWH-19-1-0777 to S.G.), CIHR (142246 and 15967 to H.H.H., 181755 to S.C., and 154328 to C.H.A.), and Terry Fox New Frontiers Program Project Grant (1090 P3 to H.H.H). C.C. was supported by a Proposal Development Award from the University of Massachusetts Boston. Z.W. and A.B. were supported by CSM (College of Science and Mathematics) Dean’s Doctoral Research Fellowship from the University of Massachusetts Boston. M. Liu was supported by a graduate fellowship from the Integrative Biosciences Programs at the University of Massachusetts Boston. S.C. was supported by a Prostate Cancer Foundation Young Investigator Award (21YOUN06). The Structural Genomics Consortium is a registered charity (no: 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through Ontario Genomics Institute (OGI-196), EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant 875510), Janssen, Merck KgaA (aka EMD in Canada and US), Pfizer and Takeda.
Author contributions
Z.W., J.P., D.H., M.V., C.H.A., H.H.H., and C.C. designed research; Z.W., J.P., M. Liu, S.Z., M. Li, A.B., S.P., K.V., K.R.S., D.H., S.G., M.V., C.H.A., and C.C. performed research; Z.W., J.P., M. Liu, S.C., S.P., K.V., K.R.S., J.A.M., D.H., S.G., M.V., C.H.A., H.H.H., and C.C. analyzed data; and Z.W., J.P., D.H., H.H.H., and C.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Housheng Hansen He, Email: Hansen.He@uhnresearch.ca.
Changmeng Cai, Email: changmeng.cai@umb.edu.
Data, Materials, and Software Availability
The GEO accession for RNA-seq and ChIP-seq data is GSE218094 (50). All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Yuan X., et al. , Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene 33, 2815–2825 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Bono J. S., et al. , Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scher H. I., et al. , Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 367, 1187–1197 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Smith M. R., et al. , Darolutamide and survival in metastatic, hormone-sensitive prostate cancer. N. Engl. J. Med. 386, 1132–1142 (2022), 10.1056/NEJMoa2119115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge R., et al. , Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann. Oncol. 31, 470–479 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Xu K., et al. , EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–1469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai C., et al. , Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep. 9, 1618–1627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao N., et al. , The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol. Endocrinol. 17, 1484–1507 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Carroll J. S., et al. , Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Lupien M., et al. , FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132, 958–970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao S., et al. , Chromatin binding of FOXA1 is promoted by LSD1-mediated demethylation in prostate cancer. Nat. Genet. 52, 1011–1017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M., et al. , LSD1 inhibition disrupts super-enhancer driven oncogenic transcriptional programs in castration-resistant prostate cancer. Cancer Res. 83, 1684–1698 (2023). 10.1158/0008-5472.CAN-22-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S., et al. , Forkhead domain mutations in FOXA1 drive prostate cancer progression. Cell Res. 29, 770–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parolia A., et al. , Distinct structural classes of activating FOXA1 alterations in advanced prostate cancer. Nature 571, 413–418 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu B., et al. , Altered chromatin recruitment by FOXA1 mutations promotes androgen independence and prostate cancer progression. Cell Res. 29, 773–775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwafuchi M., et al. , Gene network transitions in embryos depend upon interactions between a pioneer transcription factor and core histones. Nat. Genet. 52, 418–427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park S. H., et al. , Posttranslational regulation of FOXA1 by Polycomb and BUB3/USP7 deubiquitin complex in prostate cancer. Sci. Adv. 7, eabe2261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams E. J., et al. , FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 571, 408–412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng M., Zhou S., Cai C., Lupien M., He H. H., Pioneer of prostate cancer: Past, present and the future of FOXA1. Protein Cell 12, 29–38 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang C., et al. , The epigenetic regulation of nonhistone proteins by SETD7: New targets in cancer. Front. Genet. 13, 918509 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuikov S., et al. , Regulation of p53 activity through lysine methylation. Nature 432, 353–360 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Kontaki H., Talianidis I., Lysine methylation regulates E2F1-induced cell death. Mol. Cell 39, 152–160 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Munro S., Khaire N., Inche A., Carr S., La Thangue N. B., Lysine methylation regulates the pRb tumour suppressor protein. Oncogene 29, 2357–2367 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Gaughan L., et al. , Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res. 39, 1266–1279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monteiro F. L., Williams C., Helguero L. A., A systematic review to define the multi-faceted role of lysine methyltransferase SETD7 in cancer. Cancers (Basel) 14, 1414 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko S., et al. , Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Mol. Endocrinol. 25, 433–444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C., et al. , Histone methyltransferase Setd7 regulates Nrf2 signaling pathway by phenethyl isothiocyanate and ursolic acid in human prostate cancer cells. Mol. Nutr. Food Res. 62, e1700840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song H., et al. , Isoform-specific lysine methylation of RORalpha2 by SETD7 is required for association of the TIP60 coactivator complex in prostate cancer progression. Int. J. Mol. Sci. 21, 1622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheer S., et al. , A chemical biology toolbox to study protein methyltransferases and epigenetic signaling. Nat Commun 10, 19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor A. P., et al. , Selective, small-molecule co-factor binding site inhibition of a Su(var)3–9, enhancer of zeste, trithorax domain containing lysine methyltransferase. J. Med. Chem. 62, 7669–7683 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Barsyte-Lovejoy D., et al. , (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc. Natl. Acad. Sci. U.S.A. 111, 12853–12858 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vedadi M., et al. , Targeting human SET1/MLL family of proteins. Protein Sci. 26, 662–676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang J., et al. , p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Liu B., et al. , Pharmacological inhibition of SETD7 by PFI-2 attenuates renal fibrosis following folic acid and obstruction injury. Eur. J. Pharmacol. 901, 174097 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research Network, The Molecular Taxonomy of Primary Prostate Cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abida W., et al. , Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 11428–11436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Antonio J. M., Ma C., Monzon F. A., Pflug B. R., Longitudinal analysis of androgen deprivation of prostate cancer cells identifies pathways to androgen independence. Prostate 68, 698–714 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Cai C., Wang H., Xu Y., Chen S., Balk S. P., Reactivation of androgen receptor-regulated TMPRSS2:ERG gene expression in castration-resistant prostate cancer. Cancer Res. 69, 6027–6032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai C., et al. , Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 20, 457–471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teng Y., et al. , Evaluating human cancer cell metastasis in zebrafish. BMC Cancer 13, 453 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin H. J., Zhao J. C., Ogden I., Bergan R. C., Yu J., Androgen receptor-independent function of FoxA1 in prostate cancer metastasis. Cancer Res. 73, 3725–3736 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farahmand S., O’Connor C., Macoska J. A., Zarringhalam K., Causal Inference Engine: A platform for directional gene set enrichment analysis and inference of active transcriptional regulators. Nucleic Acids Res. 47, 11563–11573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nyquist M. D., et al. , Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep. 31, 107669 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang S., et al. , Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protocols 8, 2502–2515 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon D. P., et al. , Activin receptor signaling regulates prostatic epithelial cell adhesion and viability. Neoplasia 11, 365–376 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butler W., Huang J., Glycosylation changes in prostate cancer progression. Front. Oncol. 11, 809170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishioka K., et al. , Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes. Dev. 16, 479–489 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y., et al. , Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y., et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z., Cai C., SETD7 functions as a transcription repressor in prostate cancer via methylating FOXA1. Gene Expression Omnibus. GitHub. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE218094. Deposited 16 November 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The GEO accession for RNA-seq and ChIP-seq data is GSE218094 (50). All study data are included in the article and/or SI Appendix.