Abstract
The expression of the c-jun proto-oncogene is rapidly induced in response to mitogens acting on a large variety of cell surface receptors. The resulting functional activity of c-Jun proteins appears to be critical for cell proliferation. Recently, we have shown that a large family of G protein-coupled receptors (GPCRs), represented by the m1 muscarinic receptor, can initiate intracellular signaling cascades that result in the activation of mitogen-activated protein kinases (MAPK) and c-Jun NH2-terminal kinases (JNK) and that the activation of JNK but not of MAPK correlated with a remarkable increase in the expression of c-jun mRNA. Subsequently, however, we obtained evidence that GPCRs can potently stimulate the activity of the c-jun promoter through MEF2 transcription factors, which do not act downstream from JNK. In view of these observations, we set out to investigate further the nature of the signaling pathway linking GPCRs to the c-jun promoter. Utilizing NIH 3T3 cells, we found that GPCRs can activate the c-jun promoter in a JNK-independent manner. Additionally, we demonstrated that these GPCRs can elevate the activity of novel members of the MAPK family, including ERK5, p38α, p38γ, and p38δ, and that the activation of certain kinases acting downstream from MEK5 (ERK5) and MKK6 (p38α and p38γ) is necessary to fully activate the c-jun promoter. Moreover, in addition to JNK, ERK5, p38α, and p38γ were found to stimulate the c-jun promoter by acting on distinct responsive elements. Taken together, these results suggest that the pathway linking GPCRs to the c-jun promoter involves the integration of numerous signals transduced by a highly complex network of MAPK, rather than resulting from the stimulation of a single linear protein kinase cascade. Furthermore, our findings suggest that each signaling pathway affects one or more regulatory elements on the c-jun promoter and that the transcriptional response most likely results from the temporal integration of each of these biochemical routes.
Activating protein 1 transcription factors (AP-1) are composed of Fos family (c-Fos, FosB, Fra1, and Fra2) (10, 44, 52, 77) and Jun family (c-Jun, JunD, and JunB) (3, 37, 60, 61) proteins. Jun members can form homodimers or heterodimers with any Fos member, as well as with different members of the ATF family of transcription factors (4). The resulting complexes bind to specific DNA sequences known as tetradecanoyl phorbol acetate (TPA)-responsive elements (TRE) or AP-1 sites (2, 50). These sequences are found in the promoter regions of a variety of cellular genes, including the genes for collagenase, stromelysin, metallothionein IIA, interleukin 2, and transforming growth factor β, and some viral genes, including genes from simian virus 40, polyomavirus, and papillomavirus, among others (46).
AP-1 transcription factors are key regulatory molecules that participate in the conversion of extracellular signals into changes in the expression of genetic programs (2). AP-1-dependent promoters are rapidly induced by growth factors, serum, and phorbol esters (2). These transcription factors are likely to play a central role in the control of cell proliferation (34), as suggested by the observation that the microinjection of c-Jun- and c-Fos-specific antibodies can block cell cycle progression of NIH 3T3 fibroblasts (40). Moreover, specific antisense mRNAs inhibit the entry of serum-stimulated cells into the cell cycle (51). In addition, it has been shown that activation of endogenous AP-1 is essential for cellular transformation by a variety of transforming genes, such as v-src, v-H-ras, and activated c-raf (53, 67).
How AP-1 activity is regulated is currently under intense investigation. Available evidence suggests that each AP-1 member is tightly regulated at both the transcriptional and posttranslational levels. Interestingly, as a critical component of AP-1 coding complexes, the expression of c-jun itself is also rapidly and transiently induced by growth factors, serum, and tumor promoters (7, 9, 20, 41, 74). This gene has been described as being among those that display TRE motifs in their promoter regions, thus suggesting that the product of the c-jun gene, c-Jun, regulates its own expression through a positive autoregulatory loop (1). Furthermore, according to this model, another critical step in this process is the activation of preexisting c-Jun by the family of c-Jun NH2-terminal kinases (JNKs), which phosphorylate the transactivating domain of the c-Jun protein on Ser-63 and Ser-73 (21, 35, 47, 48), thereby increasing its transcriptional activity.
Workers in our laboratory have engineered NIH 3T3 murine fibroblasts expressing the m1 class of human muscarinic acetylcholine receptors, and they have used them as a model system to study proliferative signaling through the large family of G protein-coupled receptors (15). In this cellular setting, the m1 receptors can effectively transduce mitogenic signals (66) and can also act as potent agonist-dependent oncogenes if persistently activated (28). By using this biological system, it has been shown that the cholinergic agonist carbachol induces both mitogen-activated protein kinase (MAPK) and JNK activities and that the activation of JNK but not of MAPK correlated with the potent induction of an AP-1-driven reporter gene and the remarkable expression of c-jun mRNA (11). Subsequently, however, evidence has been provided for the existence of a novel signaling pathway initiated by the m1 G protein-coupled receptors at the level of the plasma membrane that converges on distinct response elements on the c-jun promoter (13). In view of these observations, we set out to investigate further the nature of the biochemical routes linking G protein-coupled receptors to the c-jun promoter. We present evidence that G protein-coupled receptors potently activate a number of newly identified MAPK family members and that activation of these kinases is required to stimulate distinct response elements on the c-jun promoter. Taken together, our results suggest that the regulation of c-jun expression by receptors linked to heterotrimeric G proteins involves the integration of numerous signals transduced by a highly complex network of MAPKs, rather than resulting from the stimulation of a linear protein kinase cascade.
MATERIALS AND METHODS
Cell lines.
NIH 3T3 fibroblasts expressing approximately 20,000 human m1 muscarinic receptors per cell, designated NIH 3T3-m1 cells (15), were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Inc.) supplemented with 10% calf serum. NIH 3T3-m1 cells expressing MAPK, JNK, ERK5, p38α, p38γ, and p38δ as influenza virus hemagglutinin HA1 (HA)-tagged protein kinases were maintained under the same culture conditions. The human kidney keratinocytic cell line 293T was maintained in DMEM (Life Technologies, Inc.) supplemented with 10% fetal calf serum.
DNA constructs.
A plasmid containing a luciferase gene driven by a wild-type murine c-jun promoter was kindly provided by R. Prywes (32). The plasmids pJC6, pJC9, pJTX, pJSX, and pJSTX are pBLCAT3-based reporter constructs that carry a chloramphenicol acetyltransferase (CAT) reporter gene controlled by the full-length murine c-jun promoter and its mutants, as previously described (31). ERK5, p38α, p38γ, and p38δ cDNAs were amplified by the PCR technique with human skeletal muscle cDNA (Clontech Laboratories, Inc.) as a template. The sequences of the oligonucleotides utilized will be made available upon request. The amplified DNA fragments were subcloned into pCEFL, a modified pcDNAIII expression vector containing the elongation factor 1 promoter driving the expression of an in-frame N-terminal tag of nine amino acids derived from HA (73). The expression vectors containing HA-tagged MAPK and JNK have been previously described (12, 16). MEK5 cDNA was obtained from Kevin Walton at Cephalon Inc. and was subcloned into pCEFL as a BamHI/NotI fragment. pCEFL-MEK5 DD and -MEK5 AA, dominant-active and dominant-negative forms of MEK5, respectively, were obtained by site-directed mutagenesis (QuickChange kit; Stratagene), replacing serine 311 and threonine 315 by aspartate and alanine, respectively. A kinase-deficient mutant of MKK6, MKK6 KR, was obtained by the same method, replacing a lysine residue in position 82 by arginine (57, 63). Raf CAAX, MEK EE, MEK AA, and MEKK-containing expression vectors have already been described (12–14). Transactivation domains of the ATF2 (amino acids [aa] 1 to 96) (26), Elk-1 (aa 307 to 428) (57), MEF2A (aa 151 to 411), MEF2B (aa 161 to 350), MEF2C (aa 87 to 467), and MEF2D (aa 160 to 515) transcription factors were expressed as Gal4 fusion proteins by subcloning the corresponding sequences in a pcDNAIII vector encoding the DNA binding domain of the yeast transcription factor Gal4. A TATA-Gal4-driven luciferase reporter plasmid, pGal4-Luc, was constructed by inserting six copies of a Gal4 responsive element and a TATA oligonucleotide in place of the simian virus 40 minimal promoter into the pGL3 vector (Promega). GST-MEF2A (aa 151 to 411), -MEF2B (aa 161 to 350), -MEF2C (aa 87 to 467), and -MEF2D (aa 160 to 515) fusion proteins were obtained by PCR, using human MEF2A and murine MEF2B, MEF2C, and MEF2D cDNAs as templates. The sequences of the oligonucleotides utilized will be made available upon request. The amplified DNA fragments were cloned between the BamHI and NotI or EcoRI sites of pGEX4T-3 (Pharmacia Biotech, Piscataway, N.J.), in frame with the glutathione S-transferase (GST) gene. The GST-ATF2 fusion protein has already been described (12).
Transient and stable transfections.
Transient transfections in NIH 3T3 and NIH 3T3-m1 cells were performed by the calcium phosphate precipitation technique or with the Lipofectamine Plus reagent (GIBCO BRL). Stable transfections were performed by the calcium phosphate precipitation technique, and cells were selected in culture medium containing Geneticin (750 μg/ml). 293T cells were transfected by the Lipofectamine Plus reagent (GIBCO BRL) according to the manufacturer’s instructions.
Reporter gene assays.
NIH 3T3-m1 cells were transfected with different expression plasmids, together with 1 μg of pcDNAIII–β-gal (a plasmid expressing the enzyme β-galactosidase) and 1 μg of each of the reporter plasmids, adjusting the total amount of plasmid DNA with empty vector. After overnight incubation, the cells were washed and kept for 24 h in serum-free DMEM. Cells were then stimulated with agonists for an additional 4 h and lysed with reporter lysis buffer (Promega). CAT activity was assayed in the cell extracts by incubation for 16 h in the presence of 0.25 μCi of [14C]chloramphenicol (100 mCi/mmol) and 200 μg of butyryl-coenzyme A per ml in 0.25 M Tris-HCl, pH 7.4. Labeled butyrylated products were extracted with a mixture of xylenes (Aldrich) and were counted as described previously (64). Luciferase activity present in cellular lysates was assayed with d-luciferin and ATP as substrates, and light emission was quantitated with a Monolight 2010 luminometer as specified by the manufacturer (Analytical Luminescence Laboratory). The β-galactosidase activity present in each sample was assayed by colorimetry, and it was used to normalize luciferase activity for transfection efficiency.
Kinase assays.
Phosphorylating activity of epitope-tagged MAPK and JNK was previously described (11). The activity of the epitope-tagged kinases in cells stably transfected with expression vectors for HA-MAPK, HA-JNK, HA-ERK5, HA-p38α, HA-p38γ, and HA-p38δ, as well as in 293T cells transiently transfected with the same expression vectors, was assayed by following a similar protocol. Briefly, cells were seeded at 10% confluence and, 2 days later, were incubated in serum-free medium overnight for MAPK or for 2 h for JNK, p38α, p38γ, and p38δ. After serum starvation, they were stimulated for the time indicated below with 1 mM carbachol, 10 ng of platelet-derived growth factor (PDGF) per ml, or other agonists when they were used as controls. Cells were washed with cold phosphate-buffered saline (PBS) and lysed at 4°C in a buffer containing 25 mM HEPES (pH 7.5), 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 20 mM β-glycerophosphate, 1 mM vanadate, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 20 μg of aprotinin per ml, and 20 μg of leupeptin per ml. Cleared lysates containing HA-tagged kinases were immunoprecipitated at 4°C for 2 h with anti-HA monoclonal antibody (HA.11; Berkeley Antibody Company). Immunocomplexes were recovered with the aid of protein G-Sepharose (Sigma). Beads were washed three times with PBS containing 1% Nonidet P-40 and 2 mM vanadate, once with 100 mM Tris (pH 7.5)–0.5 M LiCl, and once with kinase reaction buffer (12.5 mM morpholinepropanesulfonic acid [MOPS] [pH 7.5], 12.5 mM β-glycerophosphate, 7.5 mM MgCl2, 0.5 mM EGTA, 0.5 mM sodium fluoride, 0.5 mM vanadate). Samples were then resuspended in 30 μl of kinase reaction buffer containing 1 μCi of [γ-32P]ATP per reaction and 20 μM of unlabeled ATP. After 20 min at 30°C, the reactions were terminated by the addition of 10 μl of 5× Laemmli buffer. In vitro kinase assays were performed with 1.5 μg of myelin basic protein (MBP) per μl (Sigma) or 1 μg of purified, bacterially expressed GST-ATF2, -MEF2A, -MEF2B, -MEF2C, or -MEF2D as a substrate. Samples were analyzed by sodium dodecyl sulfate gel electrophoresis on 12% (or 15% for MBP) acrylamide gels, and autoradiography was performed with the aid of an intensifying screen.
Bacterial expression of GST fusion proteins.
The BL 21 Lys strain of Escherichia coli was transformed with the vector pGEX-4T3 encoding the fusion protein GST-ATF2 or GST-MEF2A, -MEF2B, -MEF2C, or -MEF2D. The transformed bacteria were grown in 500 ml of Luria-Bertani medium until the optical density was 0.5, at which time isopropyl-β-thiogalactopyranoside (1 mM final concentration) was added for 3 h. The cells were collected by centrifugation at 3,000 × g for 30 min and were resuspended in buffer containing 10 ml of PBS, 1% Triton X-100, 1 mM EDTA, 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, and 1 mM PMSF. The cell suspension was sonicated and cellular debris was removed by centrifugation at 10,000 × g for 15 min. The supernatant was mixed with 300 μl of glutathione-agarose beads (Pharmacia Biotech) and was centrifuged at 3,000 × g for 5 min. The pellet was washed three times in a buffer containing 1× of PBS, 1% Triton X-100, 1 mM EDTA, 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, and 1 mM PMSF and twice in a solution containing 1× PBS, 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, and 1 mM PMSF. Finally, purified fusion proteins were eluted in a buffer containing 50 mM Tris, 10 mM glutathione, 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, and 1 mM PMSF.
Western blot analysis.
HA immunoprecipitates from stably transfected NIH 3T3-m1 cells carrying HA-MAPK, -JNK, -ERK5, -p38α, -p38γ, and -p38δ cDNAs were analyzed by Western blotting after sodium dodecyl sulfate-polyacrylamide gel electrophoresis with an anti-HA monoclonal antibody (HA.11; Berkeley Antibody Company). Extracts from cells transfected with Gal4-MEF2 proteins were analyzed by the same technique and detected with anti-Gal4 monoclonal antibody RK5C1 (Santa Cruz Biotechnology). Epitope-tagged proteins were visualized by enhanced chemiluminescence detection (kit from Amersham Corp.) with goat anti-mouse immunoglobulin G coupled to horseradish peroxidase as the secondary antibody (Cappel).
Northern blot analysis.
NIH 3T3 cells were grown to 70% confluence in 10-cm-diameter plates and were transfected with pCEFL HA-tagged m1 receptor and expression vectors carrying green fluorescent protein (GFP), MEK5 AA, MKK6 KR (1 μg per plate each), or JNK interacting protein 1 (JIP-1) (0.1 μg per plate). The total transfected DNAs were adjusted to the same amount with empty expression vector. Transiently transfected cells or cells from the NIH 3T3-m1 line were serum starved for 24 h, stimulated with 1 mM carbachol for the times indicated below, and washed with cold PBS. Total RNA was extracted from the cells by homogenization with Trizol (GIBCO BRL) according to the manufacturer’s specifications. For Northern blotting, 10 to 20 μg of total RNA and 10 μg of total RNA from human and mouse brains and hearts (Clontech Laboratories, Inc.) were fractionated in 2% formaldehyde-agarose gels, transferred to nitrocellulose membranes, and hybridized with 32P-labeled DNA probes prepared with the Prime-a-Gene labeling system (Promega). DNA templates were full-length murine c-jun cDNA and fragments from MEF2A (nucleotides [nt] 800 to 1100) (accession no. X63381), MEF2B (nt 487 to 1051) (accession no. D50311), MEF2C (nt 990 to 1400) (accession no. L13171), and MEF2D (nt 710 to 1010) (accession no. S68893) cDNAs. Accuracy in gel loading and transfer was confirmed by fluorescence under UV light after ethidium bromide staining.
RESULTS
The m1 class of G protein-coupled receptors can activate the c-jun promoter in a JNK-independent manner.
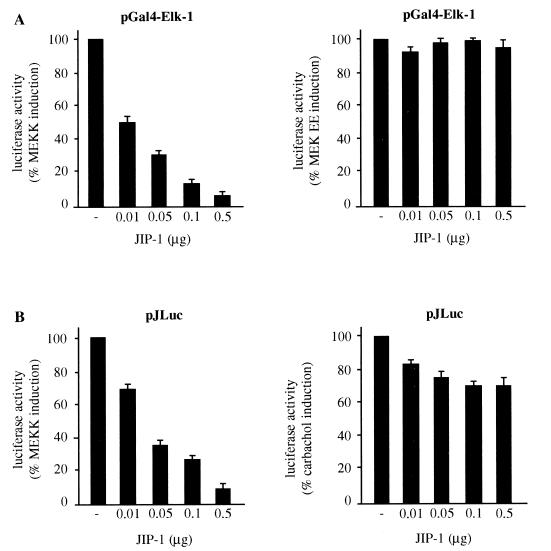
Phosphorylation of the NH2-terminal transactivating domain of c-Jun by JNK has been established as one of the essential mechanisms in the regulation of c-jun expression and AP-1-mediated transcription (35). Consistent with these observations, it has been reported that the stimulation of the m1 G protein-coupled receptor by carbachol induces JNK activity and greatly increases the expression of c-jun mRNA and AP-1 activity (11). However, in recent studies it has been shown that the transcription factor MEF2 can also play a role in c-jun expression (13). As an approach to explore in depth how signaling routes emerging from the m1 receptor control c-jun expression, we first took advantage of the newly discovered scaffolding protein, JIP-1 (70), which, when overexpressed, blocks the nuclear translocation of JNK, thereby impeding JNK-dependent gene expression regulation (22). In order to control the effectiveness and specificity of JIP-1 inhibitory action, we first assessed its effects on the activation of the transcription factor Elk-1 by JNK-dependent and -independent mechanisms. The ternary complex factor protein Elk-1 can be activated by phosphorylation by several members of the MAPK family, including MAPK (23), JNK (71), and p38α (72). For these experiments, we cotransfected NIH 3T3-m1 cells with an expression vector for the transactivation domain of Elk-1 that is fused to the DNA binding domain of Gal4, together with pGal4-Luc, a luciferase reporter gene under the control of six Gal4 responsive elements and a minimum TATA promoter. As shown in Fig. 1, Elk-1-dependent reporter gene expression was potently activated by cotransfection of MAPK and MEK EE, the constitutively active form of MEK1 (14), or MEKK, a truncated JNK kinase kinase, which is a potent activator of JNK (47). Under these experimental conditions, increasing concentrations of JIP-1 blocked in a dose-dependent manner the enhanced transcriptional activity of Gal4–Elk-1 when it was caused by MEKK, but they did not affect significantly the response to the activated form of MEK, MEK EE (Fig. 1A).
FIG. 1.
JIP-1 inhibits only partially the stimulation of the c-jun promoter by m1 G protein-coupled receptors in NIH 3T3 cells. (A) NIH 3T3-m1 cells were cotransfected by the calcium phosphate technique with increasing amounts of JIP-1 expression plasmid together with pcDNAIII–Gal4–Elk-1, pGal4-Luc, pcDNAIII–β-gal (1 μg each), and MEKK (0.1 μg) or MEK EE (1 μg). (B) Cells were transfected as described above with pJLuc and pcDNAIII–β-gal and then were either cotransfected with MEKK or exposed for 4 h to 1 mM carbachol. Lysates were collected 48 h later and were assayed for luciferase and β-galactosidase activities. The data represent luciferase activity normalized by the β-galactosidase activity present in each sample and are expressed as percentages of induction with respect to cells transfected without JIP-1. Results are the averages ± standard errors of triplicate samples from a typical experiment. Similar results were obtained in four independent experiments.
Using the same experimental approach, we compared the effect of JIP-1 on a murine c-jun promoter-driven luciferase reporter gene (pJLuc) (32) stimulated in parallel by carbachol and MEKK. As shown in Fig. 1B, induction of the c-jun promoter by MEKK was strongly inhibited by increasing amounts of JIP-1 in a dose-dependent manner, further supporting the belief that JNK activation enhances the expression from the c-jun promoter. In contrast, the JNK inhibitor reduced only slightly the effect of carbachol, thus strongly suggesting that m1 G protein-coupled receptors can also stimulate JNK-independent pathways controlling the c-jun promoter.
Stimulation of MAPK family members by m1 G protein-coupled receptors.
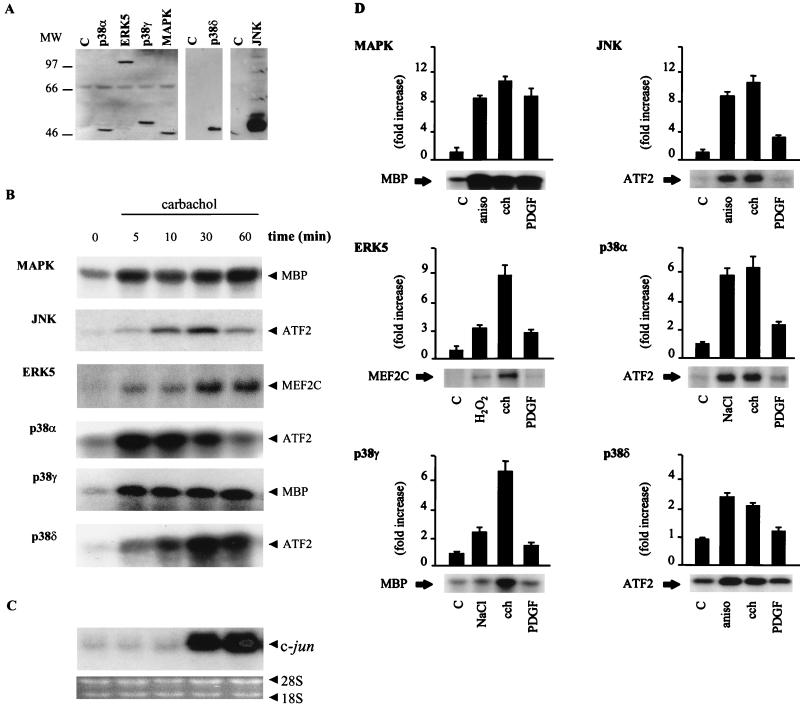
Members of the MAPK family of proline-targeted serine/threonine kinases play an important role in transducing proliferative signaling from G protein-coupled receptors (27). Furthermore, it has recently been shown that in NIH 3T3-m1 cells carbachol activates effectively both MAPK and JNK pathways (11, 13). Thus, in light of our present results, we decided to examine the ability of carbachol-stimulated m1 receptors to regulate the newly identified kinase, ERK5, as well as the new members of the p38 family, p38γ and p38δ. For these experiments, we generated stably transfected NIH 3T3-m1 cells carrying the cDNA for HA-tagged forms of each of these kinases. Lysates from these cell lines were obtained, and expression of epitope-tagged kinases was analyzed by Western blotting with an anti-HA antibody, as shown in Fig. 2A. Stably transfected cells were stimulated with the cholinergic agonist carbachol for 5, 10, 30, and 60 min to establish the temporal pattern of activation for each kinase and to compare it with the pattern of expression of c-jun mRNA. Interestingly, all MAPK family members were strongly activated by carbachol (Fig. 2B), including the recently discovered ERK5, for which the activating pathways are still not well known. Furthermore, kinase stimulation preceded the increased expression of c-jun mRNA (Fig. 2C). Thus, all of these MAPKs could be considered potential candidates to mediate signaling from the m1 receptor to the c-jun promoter.
FIG. 2.
Activation of novel MAPK family members and c-jun expression by m1 G protein-coupled receptors. (A) NIH 3T3-m1 cells were stably transfected with expression vectors containing HA-tagged MAPK, JNK, ERK5, p38α, p38γ, and p38δ. The gels show expression of HA-tagged kinases in lysates from the control (designated C) and each transfectant by Western blot analysis with a specific anti-HA antibody. MW, molecular weight (in thousands). (B) After serum starvation, cell lines were treated with 1 mM carbachol for 5 to 60 min. Nonstimulated cells were used as controls. After stimulation, lysates were immunoprecipitated with anti-HA antibody and used for kinase reactions as described in Materials and Methods. 32P-labeled substrates are indicated. Autoradiograms correspond to representative experiments for each MAPK family member. Similar results were obtained in three to five independent experiments. (C) Total RNA was extracted from NIH 3T3-m1 cells treated with carbachol for the indicated times. Samples containing 10 μg of RNA were fractionated in agarose gels and analyzed by Northern blotting, as described in Materials and Methods, with 32P-labeled murine c-jun cDNA as a probe. The material present in each lane was judged to be equivalent by ethidium bromide staining of rRNAs. (D) NIH 3T3-m1 cells stably transfected with HA-tagged MAPKs were treated with carbachol (cch), 10 ng of PDGF per ml, 10 μg of anisomycin (aniso) per ml, 500 μM H2O2, or 0.3 M NaCl. Treatments were performed for 5 min for MAPK, p38α, p38γ, and p38δ; 15 min for JNK; and 10 min for ERK5. 32P-labeled substrates are indicated. Autoradiograms correspond to representative experiments for each MAPK family member. Data are the means ± standard errors from three to five independent experiments and are expressed as fold induction with respect to nonstimulated cells (controls). C, control.
In order to compare the profiles of activation induced by two different mechanisms, such as G protein-coupled receptors and tyrosine kinase receptors, we also used PDGF, which acts on endogenously expressed tyrosine kinase receptors, as an agonist. The effect of agonists used as positive controls, carbachol, and PDGF on each kinase is shown in Fig. 2D. As previously reported, carbachol, PDGF, and TPA potently induced the MBP-phosphorylating activity of MAPK after 5 min. However, only carbachol induced JNK activity to an extent comparable to that caused by anisomycin when it was used as a positive control. Interestingly, ERK5 was potently activated by carbachol after 5 min, and its activity remained higher than that of the controls for up to 1 h after stimulation (see above); PDGF, however, produced only a limited and transient ERK5 activation that peaked at 10 min and returned to the basal level at 30 min (data not shown). In this case, we used as a specific substrate a GST fusion protein carrying aa 174 to 327 from the transactivation domain of MEF2C. Of interest, another growth factor acting on tyrosine kinase receptors, epidermal growth factor, has been recently shown to activate ERK5; however, this activation was shown in another cell type (39). p38α, p38γ, and p38δ were also potently activated by the cholinergic agonist, but in contrast, PDGF had no significant effect on their kinase activities (Fig. 2D). Taken together, these results establish that whereas MAPK can be stimulated by both tyrosine kinase and G protein-coupled receptors, in these cells, ERK5, p38γ, and p38δ, in addition to JNK and p38α, are specific targets for activating signals downstream from the G protein-linked class of receptors.
m1 G protein-coupled receptor signaling to the c-jun promoter is transduced by JNK and kinases acting downstream of MEK5 and MKK6.
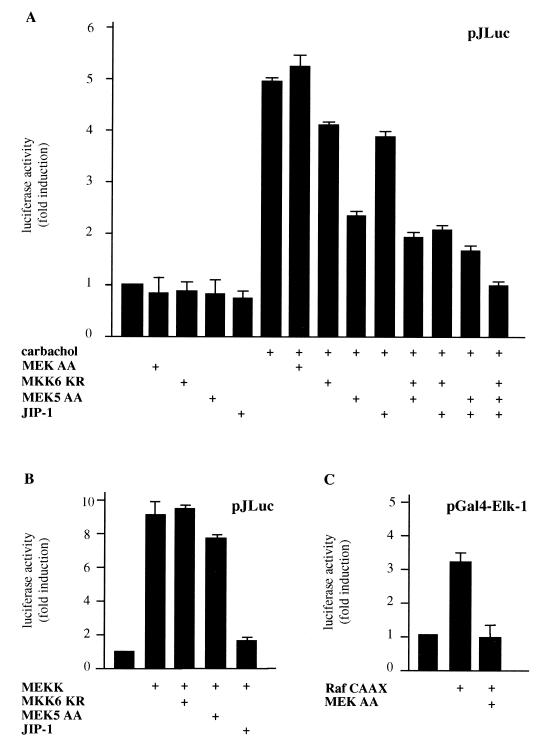
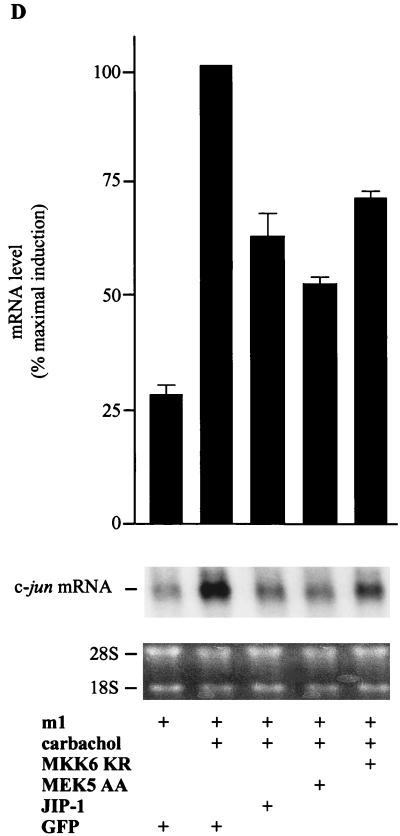
Our findings indicated that a number of MAPK family members could be considered potential candidates to mediate the signal from m1 receptors to the c-jun promoter, with the exception of MAPK, which had previously been demonstrated to fail to activate the c-jun promoter (13). In view of these results, we decided to examine whether the kinases downstream of MEK5 (ERK5) and MKK6 (p38α, p38γ, and p38δ) play a role in signaling from m1 receptors to the c-jun promoter. Thus, we transfected NIH 3T3-m1 cells with pJLuc and JIP-1 or dominant-negative forms of MEK5, MKK6, and MEK, the last of which was a negative control (14, 30, 38). As shown in Fig. 3A, MEK AA had no effect on the c-jun promoter activity, whereas MEK5 AA and MKK6 KR partially inhibited the c-jun promoter-dependent gene expression induced by carbachol. The inhibition was greater when MEK5 AA and MKK6 KR were cotransfected in combination, and it was even greater when they were cotransfected together with JIP-1. Furthermore, when the three inhibitory molecules (MEK5 AA, MKK6 KR, and JIP-1) were transfected together, they completely abolished the stimulatory effect of carbachol on the c-jun promoter. To ensure that MEK5 AA and MKK6 KR did not exert nonspecific inhibitory activities on the JNK pathway, we performed control experiments in parallel which confirmed that neither MKK6 KR nor MEK5 AA prevents the MEKK-induced pJLuc activity. In contrast, JIP-1 almost abolished this response (Fig. 3B). In addition, we confirmed the inhibitory effect of MEK AA by assessing its blocking activity on the activation of Gal4–Elk-1 by Raf CAAX, a membrane-targeted activated form of this MAPK kinase kinase (Fig. 3C). These results indicated that ERK5 and the kinases downstream of MKK6, p38α, p38γ, and p38δ could mediate the JNK-independent stimulation of the c-jun promoter by G protein-coupled receptors. To confirm these data, we studied the effect of these dominant interfering molecules on the expression of the endogenous c-jun mRNA. For these experiments, we transfected NIH 3T3 cells with m1 receptors along with MEK5 AA, MKK6 KR, or JIP-1. Transfection efficiencies were assessed by cotransfection with empty vector and were judged to be nearly identical in each case; mRNA expression levels were adjusted by cotransfection with a GFP-expressing plasmid. As shown in Fig. 3D, the induction of c-jun mRNA expression elicited by carbachol was diminished by JIP-1 and the inhibitory kinases to an extent similar to that observed for the luciferase reporter assays. On the contrary, the dominant-negative MEK, MEK AA, had no effect on the mRNA level (data not shown). Thus, these results provided further evidence that multiple kinase pathways participate in the stimulation of c-jun expression when elicited by G protein-coupled receptors.
FIG. 3.
Kinases downstream from MKK6 and MEK5 mediate the stimulation of the c-jun promoter by carbachol in NIH 3T3 cells expressing m1 G protein-coupled receptors. (A) NIH 3T3-m1 cells were cotransfected by the calcium phosphate technique with pJLuc and pcDNAIII–β-gal reporter plasmid DNAs (1 μg per plate each), along with MEK AA (1 μg), MKK6 KR (1 μg), MEK5 AA (1 μg), or JIP-1 (0.1 μg). Forty-eight hours later, cells were left untreated or exposed for 4 h to 1 mM carbachol. (B) Cells were transfected as described above with MEKK (0.1 μg), pJLuc, pcDNAIII–β-gal, MEK5 AA, MKK6 KR, or JIP-1. (C) Cells were transfected as described above with pGal4–Elk-1, pGal4-Luc, Raf CAAX, and MEK AA (1 μg per plate each). After 48 h, cells were collected and the lysates were assayed for luciferase and β-galactosidase activities. The data represent luciferase activity normalized by the β-galactosidase activity present in each sample, expressed as fold induction relative to the control, and are the averages ± standard errors of triplicate samples from a typical experiment. Similar results were obtained in three independent experiments. (D) NIH 3T3 cells were transfected by the Lipofectamine Plus technique (GIBCO BRL) with pCEFL HA-tagged m1 receptor and GFP, MEK5 AA, MKK6 KR (1 μg per plate each), or JIP-1 (0.1 μg). In all cases, the total transfected DNAs were adjusted to the same amount with empty expression vector. Twenty-four hours after serum starvation, cells were treated with 1 mM carbachol for 30 min and total RNA was extracted as described in Materials and Methods. Samples containing 20 μg of total RNA were fractionated and analyzed by Northern blotting with 32P-labeled murine c-jun cDNA as a probe. The amounts of total RNA present in the lanes were assessed to be equivalent by ethidium bromide staining of rRNAs. Data are the means ± standard errors of replicate samples from two independent experiments and are expressed as percentages of the maximal induction with carbachol. The autoradiogram corresponds to a representative experiment.
Activation of the c-jun promoter by specific members of the MAPK family.
The previous results prompted us to explore whether ERK5, p38α, p38γ, and p38δ were able to activate the c-jun promoter. For these experiments, we cotransfected the reporter plasmid pJLuc, together with the MAPKs, alone or in combination with their upstream activating kinases, using MAPK and JNK as controls. A dominant-active form of MEK5, MEK5 DD, in which the phosphorylation sites at Ser-313 and Thr-317 were replaced by aspartate, was used as an activator for ERK5 (30). MKK6 was cotransfected to activate p38α, p38γ, and p38δ because its specific effect on these kinases had already been established (18, 19, 24, 29). As described above, we used MEK EE and MEKK as activating molecules for MAPK and JNK, respectively. As shown in Fig. 4A, the pathways defined by MEK5 DD-ERK5, MKK6-p38α, and MKK6-p38γ increased the c-jun promoter activity nine-, seven-, and nearly sixfold, respectively, an extent comparable to that caused by MEKK acting on JNK. Instead, MEK EE-MAPK and MKK6-p38δ did not have any effect on the activity of the c-jun promoter. Transfection of the kinases alone served as controls, as none of them affected significantly the promoter activity under our experimental conditions. On the other hand, activating upstream kinases did not activate the promoter at the assayed concentration, although cotransfection with larger amounts of MEKK, MKK6, and MEK5 DD DNA alone were able to increase the promoter activity (data not shown).
FIG. 4.
Stimulation of the c-jun promoter activity by novel members of the MAPK family. (A) NIH 3T3-m1 cells were cotransfected with pJLuc and pcDNAIII–β-gal (1 μg per plate each) plasmid DNAs by the calcium phosphate technique. Expression vectors for MAPK, JNK, ERK5, p38α, p38γ, and p38δ (1 μg each), alone or in combination with their upstream activating kinases MEK EE (1 μg), MEKK (0.1 μg), MEK5 DD (1 μg), and MKK6 (1 μg), were included in the transfection mixtures. (B) Cells were transfected as described above with MKK6 and p38δ, alone or in combination with pcDNAIII-Gal4-ATF2, pGal4-Luc, and pcDNAIII–β-gal (1 μg each). Forty-eight hours after transfection, cells were collected and the lysates were assayed for luciferase and β-galactosidase activities. The data represent luciferase activity normalized by the β-galactosidase activity present in each sample, expressed as fold induction relative to the control. Values are the averages ± standard errors of triplicate samples from a typical experiment. Nearly identical results were obtained in three additional experiments.
To control the functionality of MKK6-p38δ in NIH 3T3-m1 cells, we cotransfected these kinases together with the fusion protein Gal4-ATF2 and the pGal4-Luc reporter gene. As shown in Fig. 4B, ATF2-dependent reporter gene expression was activated by MKK6-p38δ as previously reported (24). Together, these results supported the contention that, in addition to JNK, specific members of the MAPK family, such as ERK5, p38α, and p38γ, but not MAPK or p38δ, participate in the regulation of the c-jun promoter.
Target sequence elements for JNK, ERK5, p38α, and p38γ on the c-jun promoter.
As depicted in Fig. 5A, several responsive elements have been identified on the c-jun promoter, including sites for the transcription factors SP1, CTF, AP-1, and MEF2 (1, 31), as well as two GATAA elements. Among them, the two AP-1-like sites placed at nt −71 to −64 (Jun1TRE) and −190 to −183 (Jun2TRE) seem to be responsible for mediating the UV- and TPA-induced expression of c-jun in HeLa cells (65). The MEF2 family of transcription factors has also been shown to activate the c-jun promoter by binding the MEF2 site at position −59 to −50 (13, 29). In a previous work, it was observed that deletion of sequences upstream of position −80 did not modify the response to carbachol (13). In contrast, both the MEF2 regulatory site and the AP-1-like regulatory site at position −71 to −64 were critical for the regulation of expression from the c-jun promoter in response to signals transmitted by m1 G protein-coupled receptors, since the individual deletion of each element significantly reduced the response to carbachol, whereas the absence of both of them completely abolished it (13). Based on those findings, we next investigated which of these response elements within the c-jun promoter responded to each of the kinases acting downstream from the G protein-coupled receptors. As shown in Fig. 5B, pJC6, which includes the full-length c-jun promoter, was activated by JNK, ERK5, p38α, and p38γ pathways to the same extent as pJLuc (data not shown). Mutations on the AP-1-like site (pJTX) resulted in a complete reduction of JNK and p38α induction, while a MEF2 site-defective promoter (pJSX) lacked any response to ERK5. In contrast, mutations in either the AP-1-like or the MEF2 site only partially diminished the stimulatory effect of p38γ. The activation of the c-jun promoter by each of these kinases was almost abolished when a construct carrying a double mutation at both sites (pJSTX) was used. These results suggest that the AP-1-like site (also termed junATF [9]) is critical for JNK and p38α induction, whereas the MEF2 responsive element is essential for ERK5, and both AP-1 and MEF2 elements can individually mediate activation by p38γ. However, it was noticeable that the stimulation of the pJTX reporter plasmid (lacking the AP-1 site) by ERK5 and p38γ was slightly diminished with respect to that observed for the pJC6 construct (Fig. 5B). Similarly, the induction of the pJSX reporter plasmid (lacking the MEF2 site) by JNK, p38α, and p38γ was also lower than that observed with the pJC6 plasmid, thus suggesting that the presence of both elements (AP-1 and MEF2) is necessary for maximal stimulation. Taken together, these findings indicate that, in addition to JNK, several members of the MAPK family can effectively stimulate the activity of the c-jun promoter and that G protein-coupled receptors signal to the c-jun promoter through a network of MAPKs, each acting, coordinately, on distinct c-jun regulatory elements.
FIG. 5.
Distinct regulatory elements mediate the activation of the c-jun promoter by MAPK family members. (A) Schematic representation of the murine c-jun promoter. (B) NIH 3T3-m1 cells were cotransfected with the reporter plasmid pcDNAIII–β-gal together with pJC6, pJTX, pJSX, or pJSTX (1 μg per plate). Crosses indicate the sites of point mutations in the AP-1-like (pJTX) and the MEF2 (pJSX) binding sites. The plasmid pJSTX contains mutations for both the AP-1-like and MEF2 sites. JNK plus MEKK, MEK5 plus ERK5, MKK6 plus p38α, and MKK6 plus p38γ (1 μg per plate each) were included in the transfection mixtures. Forty-eight hours later, cells were collected and the lysates were assayed for CAT and β-galactosidase activities. The data represent CAT activity normalized by the β-galactosidase activity present in each sample, expressed as the percentages of the pJC6 induction elicited by each kinase, and are the averages ± standard errors of triplicate samples from a typical experiment. Similar results were obtained in three independent experiments.
Expression of MEF2 family members in NIH 3T3 cells.
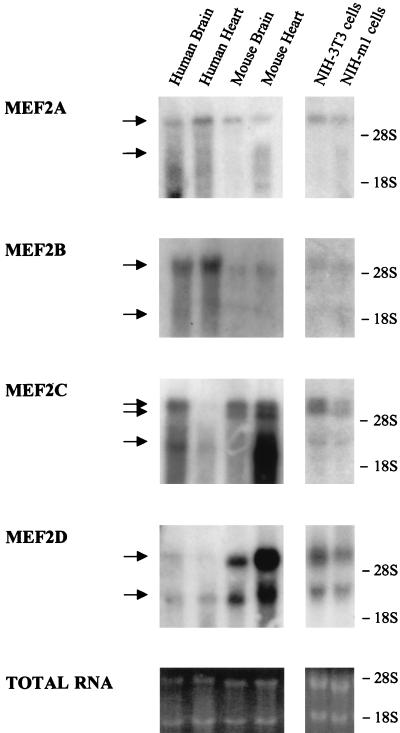
Since the MEF2 response element appears to play a key role in regulating the activity of the c-jun promoter in response to G protein-coupled receptors through ERK5 and p38γ, we set out to investigate which MEF2 family members could be the targets for these MAPKs in NIH 3T3 cells. Currently, the MEF2 family of transcription factors comprises four members, termed MEF2A, -B, -C, and -D (42). Whereas these factors were originally characterized in muscle cells (5, 17, 25), they also appear to be expressed in many other tissues (31, 32, 36, 43, 55, 76). As an approach to examine the expression of MEF2 family members, we analyzed the presence of MEF2 transcripts in NIH 3T3 cells by Northern blot analysis (Fig. 6). As a probe, we used 32P-labeled cDNA fragments corresponding to nonconserved regions of the transactivation domains of MEF2A, -B, -C, and -D, using total RNA from human and mouse skeletal muscles and brains as positive controls. As previously reported (6, 45, 76), transcripts of approximately 6.5 and 3.5 kbp for MEF2A and MEF2B, a doublet around 7.5 and a lower band of 4 kbp for MEF2C, and transcripts of 7 and 4 kbp for MEF2D were observed in positive controls. As shown in Fig. 6, all MEF2 members were expressed in NIH 3T3 as well as in NIH 3T3-m1 cells, albeit to different levels. MEF2D appears to be the most abundant transcript, followed by MEF2A and MEF2C. MEF2B expression was much lower, as it required a prolonged exposure (>48 h) to be detectable. To ensure the specificity of the signal, blots were stripped and rehybridized with probes corresponding to several distinct nonconserved regions, yielding identical results (data not shown).
FIG. 6.
Expression of MEF2A, -B, -C, and -D mRNAs in NIH 3T3 and NIH 3T3-m1 cells. Samples containing 20 μg of total RNA per lane, extracted from NIH 3T3 and NIH 3T3-m1 cells, were fractionated and analyzed by Northern blotting with 32P-labeled DNA fragments that were unique for each MEF2 as probes, as described in Materials and Methods. Ten micrograms of total RNA from human and mouse hearts and brains was used for positive controls. The arrows on the left indicate the detected mRNAs species. rRNA positions are indicated on the right. Ethidium bromide staining of the rRNAs shows the amounts of RNA present in each lane.
The transactivation domains of MEF2 family members are differentially phosphorylated and stimulated by specific MAPKs.
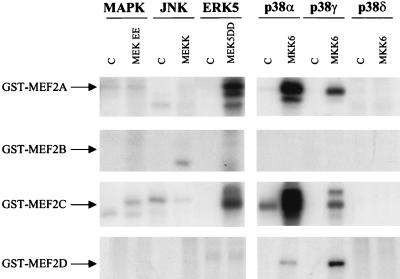
At present, the regulation of the MEF2 proteins is still poorly defined (29, 38, 78). In order to compare the ability of each MAPK family member to phosphorylate the four MEF2 factors, we performed in vitro kinase assays with purified GST-MEF2 fusion proteins containing the transactivation domain of each MEF2 isoform. For these experiments, each MAPK was expressed in 293T cells together with a control plasmid expressing GFP or with the corresponding upstream activating kinase, immunoprecipitated, and tested for kinase activity. The expression of each kinase was confirmed by anti-HA Western blotting (data not shown). As shown in Fig. 7, these MEF2s did not serve as in vitro substrates for MAPK, JNK, and p38δ, whereas MEF2A and -C were phosphorylated by ERK5 and p38α and to a lesser extent by p38γ. In contrast, only p38α and p38γ phosphorylated MEF2D, and none of the assayed kinases phosphorylated MEF2B.
FIG. 7.
Differential in vitro phosphorylation of bacterially expressed GST-MEF2A, -MEF2B, -MEF2C, and -MEF2D fusion proteins by MAPK family members. (A) 293T cells were transfected with expression vectors containing GPF or HA-tagged MAPK, JNK, ERK5, p38α, p38γ, and p38δ and the upstream activating kinases. After serum starvation, cellular lysates were immunoprecipitated with anti-HA antibody and were used for in vitro kinase assays as described in Materials and Methods. The transactivation domains of MEF2A (aa 151 to 411), -B (aa 161 to 350), -C (aa 87 to 467), and -D (aa 160 to 515), expressed as GST fusion proteins, were used as substrates. 32P-labeled products are indicated. Autoradiograms are representative of three independent experiments. C, control.
We next sought to determine whether this differential pattern of phosphorylation by MAPKs was reflected by their ability to stimulate the transcriptional activity of MEF2 proteins in vivo. As an experimental approach, we fused the transactivation domain of each MEF2 protein to the DNA binding domain of Gal4 and assessed their ability to induce the expression from the pGal4-Luc reporter plasmid. As shown in Fig. 8A, all constructs were expressed at comparable levels. We observed that when Gal4-MEF2 fusion proteins were coexpressed with molecules stimulating the different MAPK family members, the ERK5 pathway induced a remarkable increase (∼20-fold) in MEF2A activity and a smaller increase (∼10-fold) in MEF2C activity (Fig. 8B). p38α also stimulated both MEF2A and MEF2C but to a lesser extent than ERK5. In contrast, p38γ activated only MEF2A (∼5-fold); it had no significant effect on MEF2C. p38δ, MAPK, and JNK did not activate any of the MEF2 constructs, which was consistent with the pattern of phosphorylation in the in vitro kinase assays. Remarkably, MEF2B and MEF2D were not activated by any of these kinases (Fig. 8B).
FIG. 8.
Effect of MAPK family members on MEF2 transactivating activity. (A) The expression of Gal4 fusion proteins containing the transactivation domains of MEF2A (aa 151 to 411), -B (aa 161 to 350), -C (aa 87 to 467), and -D (aa 160 to 515) in extracts of transfected cells was determined by Western blotting with anti-Gal4 monoclonal antibody as described in Materials and Methods. MW, molecular weight (in thousands). (B) NIH 3T3-m1 cells were cotransfected with the pDNAIII-Gal4-MEF2 chimeric molecules (0.5 μg), as described above, along with pGal4-Luc and pcDNAIII–β-gal plasmid DNAs (1 μg per plate each) by the calcium phosphate technique. Expression vectors for MAPK, JNK, ERK5, p38α, p38γ, and p38δ (1 μg each), in combination with their upstream activating kinases MEK EE (1 μg), MEKK (0.1 μg), MEK5 DD (1 μg), and MKK6 (1 μg), were included in the transfection mixtures. Forty-eight hours after transfection, cells were collected and the lysates were assayed for luciferase and β-galactosidase activities. The data represent luciferase activity normalized by the β-galactosidase activity present in each sample, expressed as fold induction relative to the control. Values are the averages ± standard errors of triplicate samples from a typical experiment. Nearly identical results were obtained in three additional experiments.
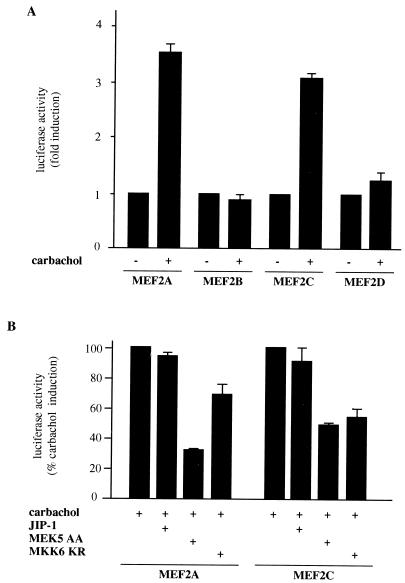
The transactivation domains of MEF2A and MEF2C are activated by m1 G protein-coupled receptors.
Once we established a functional relationship between the different MAPKs and the activation of MEF2 proteins, we investigated whether these transcription factors were stimulated by m1 receptors. As shown in Fig. 9A, expression from the Gal4-driven luciferase reporter was stimulated by carbachol when cells were cotransfected with Gal4-MEF2A and Gal4-MEF2C fusion proteins (Fig. 9A), whereas Gal4-MEF2B and Gal4-MEF2D failed to stimulate transcription. Subsequently, we made use of the dominant interfering molecules to examine whether m1 receptors acted on these transcription factors through MAPK pathways. Indeed, MEK5 AA and MKK6 KR selectively inhibited the luciferase activity induced by carbachol (Fig. 9B). Interestingly, MEK5 AA was a more potent inhibitor of MEF2A transactivation than MKK6 KR, which is in line with previous results supporting a more prominent role for ERK5 in the regulation of MEF2A. In contrast, MEK5 AA and MKK6 KR affected MEF2C to comparable degrees. The absence of inhibition by JIP-1 further supports the idea that JNK does not act on these transcription factors, and it supports the specificity of the inhibitory molecules for the carbachol-induced transactivation. Altogether, these results indicate that both MEF2A and -C could be regulated by m1 receptors through ERK5 and p38s, and they are further evidence that MEF2 proteins participate in the stimulation of the c-jun promoter by G protein-coupled receptors.
FIG. 9.
Relationship between MAPKs and the activation of MEF2 proteins by m1 receptors. The transcriptional activity of MEF2A and MEF2C is increased by carbachol in NIH 3T3 cells expressing m1 G protein-coupled receptors. (A) NIH 3T3-m1 cells were cotransfected by the calcium phosphate technique with pDNAIII-Gal4-MEF2A (aa 151 to 411), -MEF2B (aa 161 to 350), -MEF2C (aa 171 to 328), and -MEF2D (aa 160 to 515) as well as with pGal4-Luc and pcDNAIII–β-gal reporter plasmid DNAs (1 μg per plate). Twenty-four hours later, cells were left untreated (control) (−) or exposed for 20 h to 1 mM carbachol (+) and were assayed for luciferase and β-galactosidase activities. The data represent luciferase activity normalized by the β-galactosidase activity present in each sample, expressed as fold induction relative to the control. Values are the averages ± standard errors of triplicate samples from a typical experiment. Similar results were obtained in three additional experiments. (B) Cells were transfected with pGal4-MEF2A and pGal4-MEF2C, along with JIP-1 (0.1 μg), MEK5 AA (1 μg), or MKK6 KR (1 μg), and were exposed to carbachol as described above. Lysates were collected and assayed for luciferase and β-galactosidase activities. The data represent luciferase activity normalized by the β-galactosidase activity present in each sample, expressed as percentages of induction with respect to control cells exposed to carbachol. Results are the averages ± standard errors of triplicate samples from a representative experiment. Similar results were obtained in two independent experiments.
DISCUSSION
Expression of the c-jun proto-oncogene is one of the earliest nuclear events resulting from exposure of quiescent cells to mitogens, such as serum and growth factors (20, 41, 56, 58, 62). Furthermore, c-jun expression appears to be essential for normal progression of the cell cycle and cell growth in fibroblasts (8, 40). However, the nature of signaling pathways that connect external stimuli to the nuclear regulation of c-jun expression still remains poorly understood. When NIH 3T3 cells expressing m1 G protein-coupled receptors were used, it had previously been observed that the addition of the cholinergic agonist carbachol results in a remarkable increase in c-jun expression, which was correlated with the potent stimulation of the enzymatic activity of JNK (11). These results suggested a linear mechanism connecting m1 receptors to the regulation of c-jun expression through JNK. However, in subsequent studies evidence was obtained that the transcription factor MEF2 plays a critical role in the regulation of the c-jun promoter upon stimulation of G protein-coupled receptors (13). In this study, we confirmed that JNK regulates the activity of the c-jun promoter, using the overexpression of JIP-1, a scaffolding protein for the JNK signaling module (70), as a specific inhibitor of the JNK-nuclear function (22). Indeed, we observed that JIP-1 completely abolished the activation of the c-jun promoter by MEKK, a JNK kinase kinase (47). However, JIP-1 only slightly reduced the elevated c-jun promoter activity caused by carbachol, thus suggesting that biochemical routes, in addition to JNK, may participate in the signaling from G protein-coupled receptors to the c-jun promoter.
Since a number of proline-targeted serine/threonine kinases related to MAPK and JNK have been identified, we next asked whether any of these kinases could play a role in the regulation of c-jun expression by G protein-coupled receptors. Thus, we initially examined the ability of carbachol to stimulate the enzymatic activity of epitope-tagged forms of MAPK, JNK, ERK5, p38α, p38γ, and p38δ stably expressed in NIH 3T3-m1 cells. As previously reported, both MAPK and JNK were potently activated by the cholinergic ligand (11), although only MAPK was activated by endogenously expressed tyrosine kinase receptors for PDGF. Here, we show that only carbachol caused a potent and sustained activation of ERK5, p38α, p38γ, and p38δ. Furthermore, the activation of all of these kinases preceded the remarkable increase in the expression of c-jun mRNA elicited by carbachol. In view of these observations, we considered these MAPK family members potential candidates to mediate the activation of the c-jun promoter by m1 receptors. By using negative interfering mutants of kinases acting upstream of these MAPKs, we obtained evidence indicating that m1 G protein-coupled receptors stimulate the c-jun promoter and c-jun expression through kinases acting downstream of MEK5 and MKK6 in addition to JNK. Moreover, we demonstrated that ERK5, p38α, and p38γ, when activated by their corresponding upstream molecules, were able to stimulate the activity of the c-jun promoter to an extent similar to that of JNK. However, p38δ had no effect on the c-jun promoter activity.
At present, it is difficult to assess the relative contribution of each MAPK in the regulation of c-jun expression, as none of the inhibitory molecules completely abolished the enhanced c-jun promoter activity in response to the G protein-coupled receptor agonist. Nevertheless, activation of ERK5 appears to play a major role in the regulation of c-jun expression, as blockage of activation of the c-jun promoter-containing reporter plasmid by the inhibitory MEK5 mutant was significantly greater than that caused by molecules preventing JNK and p38α or p38γ function. On the other hand, the lack of c-jun promoter activation by PDGF (13) can now be explained by the fact that none of these novel MAPK family members are significantly activated by the tyrosine kinase receptor agonist in this cell type. These observations may have important implications, since the mechanisms of activation of ERK5, p38α, p38γ, and p38δ are still poorly defined. Based on our findings, we can postulate that molecules regulating the activity of these MAPKs are selectively stimulated by G protein-coupled receptors, thus providing a simple experimental model system to investigate the nature of the molecules linking cell surface receptors to each MAPK family member.
The final events in the control of the c-jun expression take place at the level of the responsive elements regulating the activity of the c-jun promoter. In vivo footprinting studies have shown that nuclear factors are bound to the promoter prior to the induction by external stimuli and that no additional interactions occur upon stimulation (33, 58). Fast phosphorylation events of such preformed complexes on the c-jun promoter can explain the rapid and transient transcriptional responses of c-jun to extracellular signals (58, 59). Previously, it was shown that the MEF2 and the AP-1-like sites are involved in the regulation of the c-jun promoter by G protein-coupled receptors (13). In this study, we report that JNK, ERK5, p38α, and p38γ pathways exert a distinct control on these AP-1-like and MEF2 sites: mutations on the former completely abolish the stimulating effect of JNK and p38α, and mutations on the latter impede the activation by ERK5. These data suggest that the transcription factors bound to these DNA sequences are the targets for each of these MAPKs.
For the AP-1-like site, it has been reported that Fos and Jun proteins, along with ATF2 (33, 49), ATF1, and CREB (9), can bind this regulatory element. This could explain the enhanced transcriptional activity from the AP-1-like site, as c-jun and ATF2 can be stimulated by JNK (18, 21) and ATF2 is also a direct target for p38α. Similarly, ATF1 and CREB are substrates of MAPKAP kinase 2, which is a substrate of p38α (68). However, although both JNK and p38α can activate the c-jun promoter through its AP-1 site, available evidence suggests that JNK plays a more prominent role in signaling to this promoter element from the m1 receptor. For example, with a reporter plasmid lacking the MEF2 site, JIP-1 nearly abolishes the response to carbachol, whereas treatment with SB303580, a p38α- and p38β-specific inhibitor (69), diminishes this response less than 30% (data not shown). Furthermore, it has previously been shown that this drug has a very limited effect on the response to carbachol when elicited on the wild-type c-jun promoter (13), although we observed in this study that the dominant-negative MKK6 could diminish this response and c-jun expression. Thus, these results suggest that p38α might participate in the regulation of the c-jun AP1 site, but other SB303580-insensitive p38s, such as p38γ, might also play a role in the regulation of this site, as well as on the activity of the additional c-jun regulatory elements. Ongoing work in our laboratory is aimed at elucidating this issue.
Regarding the MEF2 site, all four members of the myocyte enhancer family (MEF) of transcription factors, MEF2A, -B, -C and -D, can bind this response element. Supershift analysis has revealed that MEF2A and -D are the predominant factors bound to the MEF2 site of the c-jun promoter in C2C12 myocytes and HeLa cells (32, 54), and while this article was under revision, it was demonstrated that the activation of dimers between these factors is carried out by p38α through MEF2A phosphorylation (78). Our results indicate that MEF2A is also phosphorylated in vitro and its transcriptional activity is increased in vivo by ERK5, in agreement with a recently published report (75). Together with the fact that MEF2A and -D are expressed in NIH 3T3 cells, these data strongly suggest that ERK5 might regulate the c-jun promoter activity through MEF2A proteins.
On the other hand, we observed that mutations in either the AP-1-like site or the MEF2 site have a limited effect on p38γ-mediated activation of the c-jun promoter. p38γ can phosphorylate ATF2 and less effectively MAPKAP kinase 2 (24), which could explain the activation of the promoter through the AP-1-like site. Based upon our results, the phosphorylation of MEF2A in vitro and its stimulation by p38γ in vivo might explain the dual effect of this particular kinase on both responsive elements. Interestingly, p38α can phosphorylate MEF2A and MEF2C proteins in vitro and potently activate their transcriptional activity in vivo. Thus, why p38α does not act on the MEF2 site in the c-jun promoter is at present unknown. The composition of the MEF2 dimers and the regulatory phosphorylation sites of each member seem to be cell type dependent (29, 32, 54, 75, 78). Consequently, the accessibility of putative docking sites for MAPKs and phosphorylation sites themselves may vary from cell to cell due to interactions with cell-type-specific factors. Another putative explanation for this variation could be the existence of multiple cell-type-specific spliced isoforms for each MEF2, which might be differentially phosphorylated.
The model emerging from this and other studies is that the pathway linking G protein-coupled receptors to the c-jun promoter is much more complex than initially anticipated, as it involves a number of interrelated signaling pathways rather than a linear series of sequential events (Fig. 10). Furthermore, our findings suggest that each biochemical route might impinge on one or more regulatory elements on the c-jun promoter and that the resulting transcriptional response most likely results from the temporal integration of each of these biochemical routes. Further work will be required to unravel the identity of the molecules connecting G protein-coupled receptors to each MAPK, as well as to understand how the enzymatic activities of these novel MAPK family members affect, alone or in concert, the functional activity of each of the transcription factors regulating the c-jun promoter.
FIG. 10.
Proposed model for m1 G protein-coupled receptor signaling to the c-jun promoter. MAPK family members are activated by heterotrimeric G protein upon m1 receptor stimulation. Activated JNK, ERK5, p38α, and p38γ transduce signals that converge in the nucleus to control the activity of transcription factors bound to the AP-1-like and MEF2 regulatory elements within the c-jun promoter. Known intermediate molecules and pathways are depicted, and they are described in the text. The unknown molecules are indicated by question marks.
ACKNOWLEDGMENTS
We thank Mary May for the critical reading of the manuscript, Kevin Walton at Cephalon Inc. for the gift of MEK5 cDNA, and Omar Coso, Paula Casasco, Hidemi Teramoto, and Muriel Zohar for their help with the MEF2 constructs.
REFERENCES
- 1.Angel P, Hattori K, Smeal T, Karin M. The jun proto-oncogene is positively autoregulated by its product, Jun/AP-1. Cell. 1988;55:875–885. doi: 10.1016/0092-8674(88)90143-2. [DOI] [PubMed] [Google Scholar]
- 2.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 3.Ball A R, Jr, Bos T J, Loliger C, Nagata L P, Nishimura T, Su H, Tsuchie H, Vogt P K. Jun: oncogene and transcriptional regulator. Cold Spring Harbor Symp Quant Biol. 1988;53:687–693. doi: 10.1101/sqb.1988.053.01.078. [DOI] [PubMed] [Google Scholar]
- 4.Benbrook D M, Jones N C. Heterodimer formation between CREB and JUN proteins. Oncogene. 1990;5:295–302. [PubMed] [Google Scholar]
- 5.Braun T, Tannich E, Buschhausen-Denker G, Arnold H-H. Promoter upstream elements of the chicken cardiac myosin light-chain 2-A gene interact with trans-acting regulatory factors for muscle-specific transcription. Mol Cell Biol. 1989;9:2513–2525. doi: 10.1128/mcb.9.6.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitbart R E, Liang C S, Smoot L B, Laheru D A, Mahdavi V, Nadal-Ginard B. A fourth human MEF2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development. 1993;118:1095–1106. doi: 10.1242/dev.118.4.1095. [DOI] [PubMed] [Google Scholar]
- 7.Brenner D A, O’Hara M, Angel P, Chojkier M, Karin M. Prolonged activation of jun and collagenase genes by tumour necrosis factor-alpha. Nature. 1989;337:661–663. doi: 10.1038/337661a0. [DOI] [PubMed] [Google Scholar]
- 8.Carter R, Cosenza S C, Pena A, Lipson K, Soprano D R, Soprano K J. A potential role for c-jun in cell cycle progression through late G1 and S. Oncogene. 1991;6:229–235. [PubMed] [Google Scholar]
- 9.Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Epidermal growth factor induction of the c-jun promoter by a Rac pathway. Mol Cell Biol. 1998;18:1065–1073. doi: 10.1128/mcb.18.2.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen D R, Curran T. fra-1: a serum-inducible, cellular immediate-early gene that encodes a Fos-related antigen. Mol Cell Biol. 1988;8:2063–2069. doi: 10.1128/mcb.8.5.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coso O A, Chiariello M, Kalinec G, Kyriakis J M, Woodgett J, Gutkind J S. Transforming G protein-coupled receptors potently activate JNK (SAPK). Evidence for a divergence from the tyrosine kinase signaling pathway. J Biol Chem. 1995;270:5620–5624. doi: 10.1074/jbc.270.10.5620. [DOI] [PubMed] [Google Scholar]
- 12.Coso O A, Chiariello M, Yu J C, Teramoto H, Crespo P, Xu N, Miki T, Gutkind J S. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 13.Coso O A, Montaner S, Fromm C, Lacal J C, Prywes R, Teramoto H, Gutkind J S. Signaling from G protein-coupled receptors to the c-jun promoter involves the MEF2 transcription factor. Evidence for a novel c-jun amino-terminal kinase-independent pathway. J Biol Chem. 1997;272:20691–20697. doi: 10.1074/jbc.272.33.20691. [DOI] [PubMed] [Google Scholar]
- 14.Cowley S, Paterson H, Kemp P, Marshall C J. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 15.Crespo P, Xu N, Daniotti J L, Troppmair J, Rapp U R, Gutkind J S. Signaling through transforming G protein-coupled receptors in NIH 3T3 cells involves c-Raf activation. Evidence for a protein kinase C-independent pathway. J Biol Chem. 1994;269:21103–21109. [PubMed] [Google Scholar]
- 16.Crespo P, Xu N, Simonds W F, Gutkind J S. Ras-dependent activation of MAP kinase pathway mediated by G-protein beta gamma subunits. Nature. 1994;369:418–420. doi: 10.1038/369418a0. [DOI] [PubMed] [Google Scholar]
- 17.Cserjesi P, Olson E N. Myogenin induces the myocyte-specific enhancer binding factor MEF-2 independently of other muscle-specific gene products. Mol Cell Biol. 1991;11:4854–4862. doi: 10.1128/mcb.11.10.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuenda A, Cohen P, Buee-Scherrer V, Goedert M. Activation of stress-activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPKK3 (MKK6); comparison of the specificities of SAPK3 and SAPK2 (RK/p38) EMBO J. 1997;16:295–305. doi: 10.1093/emboj/16.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cuenda A, Goedert M, Craxton M, Jakes R, Cohen P. Activation of the novel MAP kinase homologue SAPK4 by cytokines and cellular stresses is mediated by SKK3 (MKK6) Biochem Soc Trans. 1997;25:S569. doi: 10.1042/bst025s569. [DOI] [PubMed] [Google Scholar]
- 20.de Groot R P, Pals C, Kruijer W. Transcriptional control of c-jun by retinoic acid. Nucleic Acids Res. 1991;19:1585–1591. doi: 10.1093/nar/19.7.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 22.Dickens M, Rogers J S, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 23.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb M H, Shaw P E. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goedert M, Cuenda A, Craxton M, Jakes R, Cohen P. Activation of the novel stress-activated protein kinase SAPK4 by cytokines and cellular stresses is mediated by SKK3 (MKK6); comparison of its substrate specificity with that of other SAP kinases. EMBO J. 1997;16:3563–3571. doi: 10.1093/emboj/16.12.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gossett L A, Kelvin D J, Sternberg E A, Olson E N. A new myocyte-specific enhancer-binding factor that recognizes a conserved element associated with multiple muscle-specific genes. Mol Cell Biol. 1989;9:5022–5033. doi: 10.1128/mcb.9.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S, Campbell D, Derijard B, Davis R J. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 27.Gutkind J S. The pathways connecting G protein-coupled receptors to the nucleus through divergent mitogen-activated protein kinase cascades. J Biol Chem. 1998;273:1839–1842. doi: 10.1074/jbc.273.4.1839. [DOI] [PubMed] [Google Scholar]
- 28.Gutkind J S, Novotny E A, Brann M R, Robbins K C. Muscarinic acetylcholine receptor subtypes as agonist-dependent oncogenes. Proc Natl Acad Sci USA. 1991;88:4703–4707. doi: 10.1073/pnas.88.11.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han J, Jiang Y, Li Z, Kravchenko V V, Ulevitch R J. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 30.Han J, Lee J D, Jiang Y, Li Z, Feng L, Ulevitch R J. Characterization of the structure and function of a novel MAP kinase kinase (MKK6) J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 31.Han T-H, Lamph W W, Prywes R. Mapping of epidermal growth factor-, serum-, and phorbol ester-responsive sequence elements in the c-jun promoter. Mol Cell Biol. 1992;12:4472–4477. doi: 10.1128/mcb.12.10.4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han T-H, Prywes R. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol Cell Biol. 1995;15:2907–2915. doi: 10.1128/mcb.15.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herr I, van Dam H, Angel P. Binding of promoter-associated AP-1 is not altered during induction and subsequent repression of the c-jun promoter by TPA and UV irradiation. Carcinogenesis. 1994;15:1105–1113. doi: 10.1093/carcin/15.6.1105. [DOI] [PubMed] [Google Scholar]
- 34.Herschman H R. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 35.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 36.Hidaka K, Morisaki T, Byun S H, Hashido K, Toyama K, Mukai T. The MEF2B homologue differentially expressed in mouse embryonal carcinoma cells. Biochem Biophys Res Commun. 1995;213:555–560. doi: 10.1006/bbrc.1995.2167. [DOI] [PubMed] [Google Scholar]
- 37.Hirai S I, Ryseck R P, Mechta F, Bravo R, Yaniv M. Characterization of junD: a new member of the jun proto-oncogene family. EMBO J. 1989;8:1433–1439. doi: 10.1002/j.1460-2075.1989.tb03525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kato Y, Kravchenko V V, Tapping R I, Han J, Ulevitch R J, Lee J D. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato Y, Tapping R I, Huang S, Watson M H, Ulevitch R J, Lee J D. Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature. 1998;395:713–716. doi: 10.1038/27234. [DOI] [PubMed] [Google Scholar]
- 40.Kovary K, Bravo R. The Jun and Fos protein families are both required for cell cycle progression in fibroblasts. Mol Cell Biol. 1991;11:4466–4472. doi: 10.1128/mcb.11.9.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamph W W, Wamsley P, Sassone-Corsi P, Verma I M. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature. 1988;334:629–631. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 42.Martin J F, Miano J M, Hustad C M, Copeland N G, Jenkins N A, Olson E N. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol Cell Biol. 1994;14:1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin J F, Schwarz J J, Olson E N. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc Natl Acad Sci USA. 1993;90:5282–5286. doi: 10.1073/pnas.90.11.5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsui M, Tokuhara M, Konuma Y, Nomura N, Ishizaki R. Isolation of human fos-related genes and their expression during monocyte-macrophage differentiation. Oncogene. 1990;5:249–255. [PubMed] [Google Scholar]
- 45.McDermott J C, Cardoso M C, Yu Y-T, Andres V, Leifer D, Krainc D, Lipton S A, Nadal-Ginard B. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol Cell Biol. 1993;13:2564–2577. doi: 10.1128/mcb.13.4.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mechta F, Lallemand D, Pfarr C M, Yaniv M. Transformation by ras modifies AP1 composition and activity. Oncogene. 1997;14:837–847. doi: 10.1038/sj.onc.1200900. [DOI] [PubMed] [Google Scholar]
- 47.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 48.Minden A, Lin A, Smeal T, Dérijard B, Cobb M, Davis R, Karin M. c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol. 1994;14:6683–6688. doi: 10.1128/mcb.14.10.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morooka H, Bonventre J V, Pombo C M, Kyriakis J M, Force T. Ischemia and reperfusion enhance ATF-2 and c-Jun binding to cAMP response elements and to an AP-1 binding site from the c-jun promoter. J Biol Chem. 1995;270:30084–30092. doi: 10.1074/jbc.270.50.30084. [DOI] [PubMed] [Google Scholar]
- 50.Nakabeppu Y, Ryder K, Nathans D. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell. 1988;55:907–915. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- 51.Nishikura K, Murray J M. Antisense RNA of proto-oncogene c-fos blocks renewed growth of quiescent 3T3 cells. Mol Cell Biol. 1987;7:639–649. doi: 10.1128/mcb.7.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishina H, Sato H, Suzuki T, Sato M, Iba H. Isolation and characterization of fra-2, an additional member of the fos gene family. Proc Natl Acad Sci USA. 1990;87:3619–3623. doi: 10.1073/pnas.87.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okuno H, Suzuki T, Yoshida T, Hashimoto Y, Curran T, Iba H. Inhibition of jun transformation by a mutated fos gene: design of an anti-oncogene. Oncogene. 1991;6:1491–1497. [PubMed] [Google Scholar]
- 54.Ornatsky O I, McDermott J C. MEF2 protein expression, DNA binding specificity and complex composition, and transcriptional activity in muscle and non-muscle cells. J Biol Chem. 1996;271:24927–24933. doi: 10.1074/jbc.271.40.24927. [DOI] [PubMed] [Google Scholar]
- 55.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 56.Quantin B, Breathnach R. Epidermal growth factor stimulates transcription of the c-jun proto-oncogene in rat fibroblasts. Nature. 1988;334:538–539. doi: 10.1038/334538a0. [DOI] [PubMed] [Google Scholar]
- 57.Raingeaud J, Whitmarsh A J, Barrett T, Dérijard B, Davis R J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rozek D, Pfeifer G P. In vivo protein-DNA interactions at the c-jun promoter in quiescent and serum-stimulated fibroblasts. J Cell Biochem. 1995;57:479–487. doi: 10.1002/jcb.240570313. [DOI] [PubMed] [Google Scholar]
- 59.Rozek D, Pfeifer G P. In vivo protein-DNA interactions at the c-jun promoter: preformed complexes mediate the UV response. Mol Cell Biol. 1993;13:5490–5499. doi: 10.1128/mcb.13.9.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryder K, Lanahan A, Perez-Albuerne E, Nathans D. jun-D: a third member of the jun gene family. Proc Natl Acad Sci USA. 1989;86:1500–1503. doi: 10.1073/pnas.86.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryder K, Lau L F, Nathans D. A gene activated by growth factors is related to the oncogene v-jun. Proc Natl Acad Sci USA. 1988;85:1487–1491. doi: 10.1073/pnas.85.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryder K, Nathans D. Induction of protooncogene c-jun by serum growth factors. Proc Natl Acad Sci USA. 1988;85:8464–8467. doi: 10.1073/pnas.85.22.8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 64.Seed B, Sheen J Y. A simple phase-extraction assay for chloramphenicol acyltransferase activity. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 65.Stein B, Angel P, van Dam H, Ponta H, Herrlich P, van der Eb A, Rahmsdorf H J. Ultraviolet-radiation induced c-jun gene transcription: two AP-1 like binding sites mediate the response. Photochem Photobiol. 1992;55:409–415. doi: 10.1111/j.1751-1097.1992.tb04255.x. [DOI] [PubMed] [Google Scholar]
- 66.Stephens E V, Kalinec G, Brann M R, Gutkind J S. Transforming G protein-coupled receptors transduce potent mitogenic signals in NIH 3T3 cells independent on cAMP inhibition or conventional protein kinase C. Oncogene. 1993;8:19–26. [PubMed] [Google Scholar]
- 67.Suzuki T, Murakami M, Onai N, Fukuda E, Hashimoto Y, Sonobe M H, Kameda T, Ichinose M, Miki K, Iba H. Analysis of AP-1 function in cellular transformation pathways. J Virol. 1994;68:3527–3535. doi: 10.1128/jvi.68.6.3527-3535.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tan Y, Rouse J, Zhang A, Cariati S, Cohen P, Comb M J. FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 1996;15:4629–4642. [PMC free article] [PubMed] [Google Scholar]
- 69.Tong L, Pav S, White D M, Rogers S, Crane K M, Cywin C L, Brown M L, Pargellis C A. A highly specific inhibitor of human p38 MAP kinase binds in the ATP pocket. Nat Struct Biol. 1997;4:311–316. doi: 10.1038/nsb0497-311. [DOI] [PubMed] [Google Scholar]
- 70.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 71.Whitmarsh A J, Shore P, Sharrocks A D, Davis R J. Integration of MAP kinase signal transduction pathways at the serum response element. Science. 1995;269:403–407. doi: 10.1126/science.7618106. [DOI] [PubMed] [Google Scholar]
- 72.Whitmarsh A J, Yang S-H, Su M S-S, Sharrocks A D, Davis R J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson I A, Niman H L, Houghten R A, Cherenson A R, Connolly M L, Lerner R A. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 74.Wu B Y, Fodor E J, Edwards R H, Rutter W J. Nerve growth factor induces the proto-oncogene c-jun in PC12 cells. J Biol Chem. 1989;264:9000–9003. [PubMed] [Google Scholar]
- 75.Yang C C, Ornatsky O I, McDermott J C, Cruz T F, Prody C A. Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen-activated protein kinase, ERK5/BMK1. Nucleic Acids Res. 1998;26:4771–4777. doi: 10.1093/nar/26.20.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu Y T, Breitbart R E, Smoot L B, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 77.Zerial M, Toschi L, Ryseck R P, Schuermann M, Muller R, Bravo R. The product of a novel growth factor activated gene, fos B, interacts with JUN proteins enhancing their DNA binding activity. EMBO J. 1989;8:805–813. doi: 10.1002/j.1460-2075.1989.tb03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao M, New L, Kravchenko V V, Kato Y, Gram H, di Padova F, Olson E N, Ulevitch R J, Han J. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]