Abstract
Tetraspanins regulate a variety of cellular functions. However, the general cellular mechanisms by which tetraspanins regulate these functions remain poorly understood. In this article we collected the observations that tetraspanins regulate the formation and/or development of various tubular structures of cell membrane. Because tetraspanins and their associated proteins (1) are localized at the tubular structures, such as the microvilli, adhesion zipper, foot processes, and penetration peg, and/or (2) regulate the morphogenesis of these membrane tubular structures, tetraspanins probably modulate various cellular functions through these membrane tubular structures. Some tetraspanins inhibit membrane tubule formation and/or extension, while others promote them. We predict that tetraspanins regulate the formation and/or development of various membrane tubular structures: (1) microvilli or nanovilli at the plasma membranes free of cell and matrix contacts, (2) membrane tubules at the plasma membrane of cell-matrix and cell-cell interfaces, and (3) membrane tubules at the intracellular membrane compartments. These different membrane tubular structures likely share a common morphogenetic mechanism that involves tetraspanins. Tetraspanins probably regulate the morphogenesis of membrane tubular structures by altering (1) the biophysical properties of the cell membrane such as curvature and/or (2) the membrane connections of cytoskeleton. Since membrane tubular structures are associated with cell functions such as adhesion, migration, and intercellular communication, in all of which tetraspanins are involved, the differential effects of tetraspanins on membrane tubular structures likely lead to the functional difference of tetraspanins.
Keywords: Adhesion zipper, Membrane tubular structures, Membrane curvature, Membrane microdomain, Microvillus, Tetraspanin
Tetraspanins
Tetraspanins exist in eukaryotes ranging from yeast to humans. In animals, approximately 2–3 dozen tetraspanin genes are found in species such as worm, fly, mouse, and human, implying ancient, diverse, and important roles of tetraspanins in evolution and development.
Tetraspanin proteins are characterized by four transmembrane segments that contain approximately half of the total amino acid residues [1]. Because of the homology in amino acid sequences and the sharing of a well-conserved CCG sequence, they form a superfamily [2, 3]. Tetraspanins associate with each other and with other transmembrane proteins such as integrins, immunoglobulin superfamily proteins, and proteases to form tetraspanin-enriched microdomains (TEMs) [4, 5]. Importantly, growth factor receptors such as EGFR and c-Met and membrane-bound growth factors such as heparin-binding EGF (HB-EGF) are linked to TEM. Signaling molecules such as Pl 4-kinase, activated PKC, syntenin, and ezrin/radixin/moesin (ERM) family proteins are found in TEMs [6, 7]. At the molecular level, tetraspanins are known as molecular facilitators, by modulating the function and activity of their associated proteins [3–5].
Tetraspanins engage in a wide variety of biological functions such as cell migration, cell-cell fusion, cell-cell adhesion, cell-matrix adhesion, cell spreading, cell proliferation, cell signaling, intracellular vesicle trafficking, peri-cellular proteolysis, viral entry and release, immune response, vascular morphogenesis and remodeling, tumor progression and metastasis, neurite navigation, and thrombosis [3–7]. With the expanding repertoire of tetraspanin-involved, -regulated, or -required functions, the biochemical and/or biophysical nature that governs tetraspanins to engage in all of these physiological and pathological events remains basically unknown. Based on the same ancestry shared by tetraspanin genes and the sequence homology shared by tetraspanin proteins, a universal biological mechanism that governs most of tetraspanin-related functions likely exists.
Membrane tubular structures
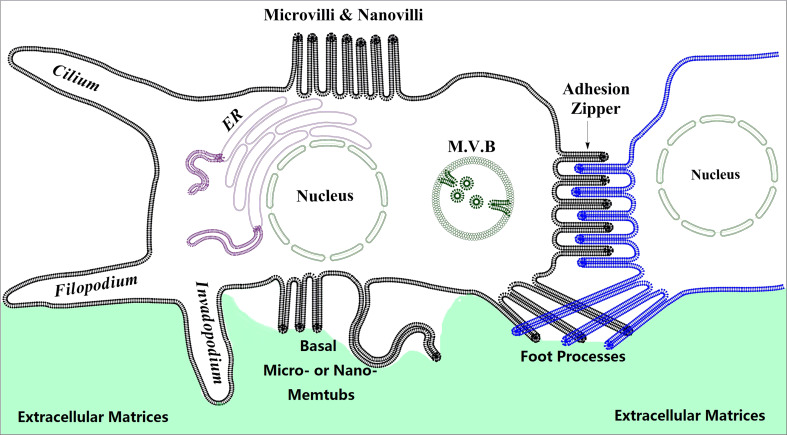
The membrane tubular structures are the tube- or tubule-like structures of the cell membrane (Fig. 1). In these structures, the longitudinal axis is significantly longer than the latitudinal axis. The membrane tubular structures, including axons, dendrites, and spikes, are well recognized and elucidated in neuronal cells. The functional importance of these structures is also fully recognized in the nervous system. We focus on various membrane tubules from non-neuronal cells and their relationship with tetraspanins in this article. On the one hand, these structures in non-neuronal cells have not been systematically elucidated, and therefore their functional importance has not drawn sufficient attention. On the other hand, tetraspanins are not well studied in the nervous system, without saying that they are not important in nerve functions.
Fig. 1.
The membrane tubular structures (memtubs) of cells. We depict various membrane tubules from non-neuronal cells. In neurons, the membrane tubular structures, including axons, dendrites, and spines, are well elucidated. For non-neuronal cells, tubular or rod-like extrusive membrane structures include “macro”extrusions such as filopodia, invadopodia, cilia, and penetrative pegs (not shown), and “micro”extrusions such as microvilli, foot processes, basal microextrusions, and lateral microextrusions. Lateral microextrusions could form the adhesion zipper when cells contact each other. Micro-memtubs may also be found in intracellular organelles such as the multivesicular body (MVB), a form of late endosomes, and ER
Functionally, the membrane tubular structures are involved in sense, effect, and communication. First, the membrane tubular structures can sense mechanical or chemical signals. For example, the basal cells in epithelia can send the membrane tubular structures to reach the lumen and sense the luminal environmental changes [8], and so do the intestinal dendritic cells [9]. Second, the membrane tubular structures could be the effector device for cells. For example, invadopodia and filopodia directly lead cell movement, while others such as renal podocyte foot processes and intestinal epithelial microvilli exert mechanical force onto the glomerular basement membrane and release microvesicles to the extracellular environment, respectively. The membrane tubular structures are likely involved in cell retraction, especially when they are positioned at the trailing or concave edge of cells. Sometimes the membrane tubular structures probably carry dual functions. For example, microvilli may sense the extracellular environment first and then enclose to uptake solutes through macropinocytosis [10]. Third, the membrane tubular structures are the communication channels between cells. For example, tunneling nanotubules and cytonemes exchange the contents of the cytoplasm and plasma membrane and transmit viruses between cells [11, 12]. Thus, the membrane tubular structures of the cells from non-nerve systems can actually engage the functions that have been considered for the axons, dendrites, and spines of the cells of the nerve system. Mechanistically, the participation in sense, effect, and communication strongly underscores the notion that the membrane tubular structures are the platform of signaling machinery.
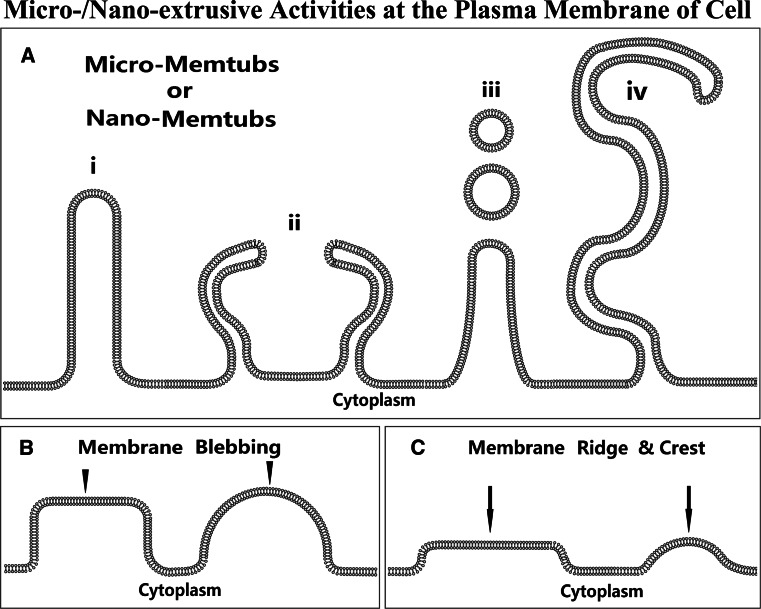
Microvilli, microprotrusions, and foot processes are the protrusive membrane tubular structures at the “micro” scale (Fig. 2a), relative to the “macro” protrusive membrane tubular structures such as filopodia, invadopodia, and cilia [13–16]. However, the micro-membrane tubular structure could also result from cell retraction processes (Fig. 3). Morphologically, microvilli or microprotrusions are miniature filopodia [13, 14]. However, the number of microvilli or microprotrusions in a given cell is typically higher than that of filopodia. Unlike another macroprotrusive membrane structure, lamellipodia, the lengths of microprotrusion and filopodia, which are typically rod- or tubule-like (Fig. 1), are significantly larger than their widths. Microprotrusions and filopodia likely share a similar morphogenetic mechanism because the membrane extrusion activity occurs at the defined spots of the plasma membrane compared with lamellipodia. Of note, microprotrusion symbolizes only one form of the membrane protrusive activities, which also result in fewer projecting membrane structures including membrane blebbing, ridges, and crests (Fig. 2b, c), although these could occur at the “macro” scale and involve relatively large areas of membrane.
Fig. 2.
The micro- and nano-extrusive activities at the plasma membrane can result in at least three kinds of microextrusive structures such as micro-memtubs (a), membrane blebbing (b) or podosomes (not shown), and membrane ridges or crests (c), based on the shape of extrusion structure and the magnitude of extrusion activity. In the category of micro-memtubs (a), microvilli or nanovilli are probably the most commonly seen micro- or nano-extrusions (i). The adjacent microvilli may become enclosing to engulf extracellular solutes (ii) [10]. Microvilli can release microvesicles to extracellular environments (iii) [13]. The thin and long micro- or nano-extrusions represent the memtubs called nanotubules and nanopodia (iv) [24]
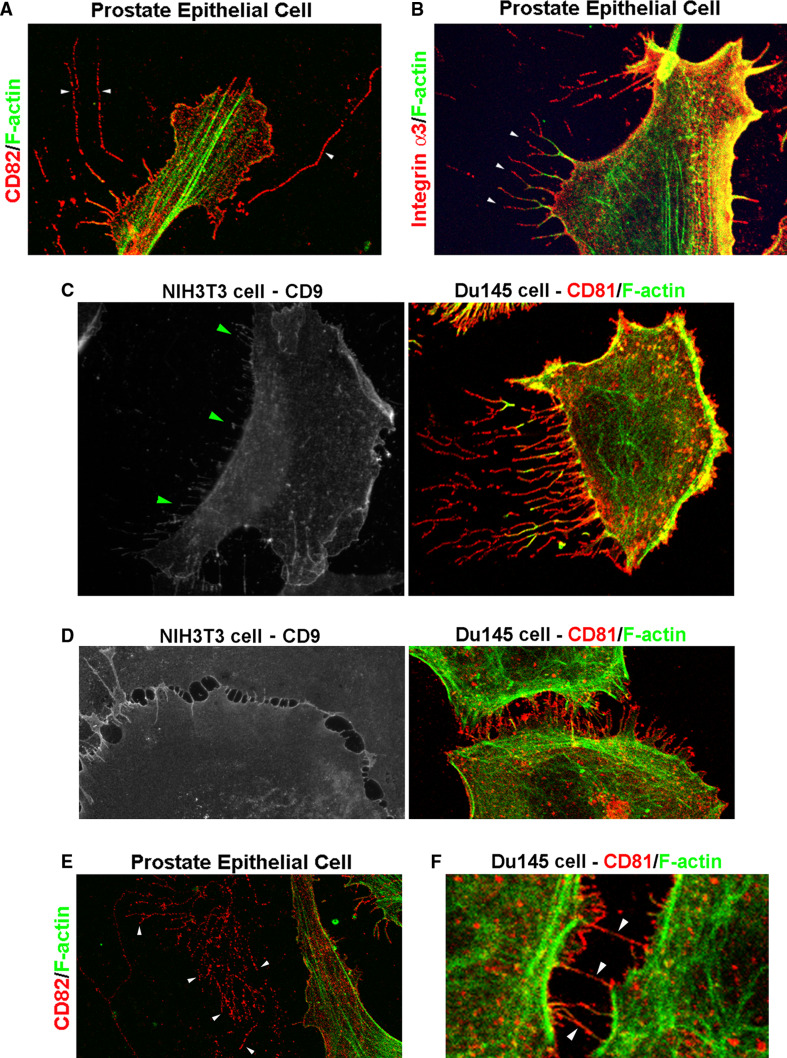
Fig. 3.
Tetraspanin-containing membrane tubular structures (memtubs) in various cellular behaviors. a Tetraspanin CD82 was detected in thin and long micro-memtubs of human prostate epithelial cells. b Tetraspanin-associated proteins such as integrin α3 are found in the micro-memtubs, as indicated by the arrowheads, of human prostate epithelial cells. Although actin fibers were detectable in these micro-memtubs, F-actin was present in the cell-proximal portion of these structures. Notably, the integrin α3-positive micro-memtubs are branched. c Tetraspanins are found in the micro-memtubs associated with cell retraction during cell movement or cell front-rear polarization. For example, CD9-positive micro-memtubs are present at the trailing edge of migrating NIH3T3 murine fibroblast cells, as indicated by the arrowheads; while CD81-positive micro-memtubs are present at the trailing edge of migrating Du145 human prostate cancer cells. The memtubs formed at the leading edges are typically filopodia. d Tetraspanins CD9 and CD81 are found in the micro-memtubs during the formation of the cell-cell adhesion zipper in NIH3T3 and Du145 cells, respectively. e The micro-memtubs can be remained on extracellular matrices after cells have either moved away or become detached. The image shows the wreckage of CD82-positive micro-memtubs, as indicated by the arrowheads. Note that F-actin is deprived of these wreckages. f Tetraspanin CD81-positive micro-memtubs form nanotubules between Du145 cells
Tetraspanins are present at the membrane tubular structures
Tetraspanins and their associated proteins such as integrins and IgSF proteins are enriched or detectable in various membrane tubular structures (Fig. 3). When localized at the cell surface beyond cell-cell or cell-matrix contact, the micro-membrane tubular structures are microvilli or nanovilli (Fig. 1). For example, the microvilli at the apical surface of intestinal epithelium project into the intestinal lumen. Tetraspanins such as CD82, CO-029, and Tspan-1 are found at the lumenal vesicles secreted from the intestinal microvilli [17], strongly suggesting that these tetraspanins are originally present in the microvilli. Tetraspanin CD9 is enriched at the microvilli on egg cells [18–20], and CD9 and tetraspanin CD63 are found on the microprotrusions of activated platelets [21, 22]. Tetraspanin CD81 is localized at the microvilli at the inner surface of retinal pigment epithelial cells [23]. Tetraspanins TM4SF1/Ly6, CD81 and CD82 are found at extremely long microvilli projected from endothelial cells [24] (Fig. 3).
The micro-membrane tubular structures localized at the cell surface that interfaces with another cell are transient, dynamic structures. These membrane tubules may form the temporary contact between cells or the “adhesion zipper,” i.e., interdigitating microprotrusions at the cell-cell contacts (Figs. 1, 3). They occur when cells initially interact with or contact each other. For example, the interdigitating microprotrusions are a transition contact mechanism for epithelial cells before the adherens junctions appear and stabilize the epithelial cell-cell contacts [25–27]. However, we recently also found that endothelial cells form an adhesion zipper when they contact each other [28]. Tetraspanins CD9 and CD81 can be localized at the adhesion zipper [28, 29] (Fig. 3c). The association of tetraspanins with this cell-cell contact mechanism is consistent with the co-emergence of tetraspanin and multicellular organisms in evolution [30, 31].
The membrane tubular structures are also found at the cell-matrix contact (Fig. 1). Their morphologies vary from tissue to tissue. For example, kidney podocytes generate the intertwined microprotrusions called foot processes toward the glomerular basement membrane. Notably, tetraspanin CD151 and its associated integrin α3β1 regulate the foot process morphogenesis, evidenced by the foot process effacement in CD151- or α3-null podocytes [32–34]. Another membrane protrusion that interfaces the cell and matrix is the invadopodia, which penetrate into pericellular matrices and are often found in invasive tumor cells [15, 16]. Although the role of tetraspanins in the morphogenesis of invadopodia has not been investigated, fungal tetraspanin PLS1 is required for the formation of penetration pegs [35], a cell wall-penetrative structure similar to invadopodia. Because tetraspanin emerges from fungi in evolution, the cellular function of PLS1 likely denotes the primordial function of tetraspanins at the cellular level.
When initiated at the membrane of intracellular compartments such as endosomes and endoplasmic reticulum (ER), the membrane tubular structures are either intrusions or extrusions, which can further develop into vesicles and cisternae (Fig. 1). For example, the invagination or inward projection at the membrane of late endosomes leads to the formation of inner vesicles in the multivesicular bodies, while the outward projection from the ER membrane leads to the formation of ER tubules. Several tetraspanins such as CD63, CD81, CD82, and CD151 are enriched in the inner vesicles of late endosomes [36, 37]. Although the roles of these tetraspanins in the morphogenesis of inner vesicles remain to be determined, these inner vesicles are apparently derived from the inward projection of the limiting membrane of late endosomes.
Tetraspanins regulate the morphogenesis of membrane tubular structures
Lines of evidence indicate that the appearance or disappearance of the membrane tubular structure correlates with the gain, loss, or fluctuation of tetraspanin expressions. Tetraspanins and their associated proteins not only are found in various membrane tubular structures, but also regulate the formation and development of these structures. Likely, tetraspanins and their associated proteins regulate the morphogenesis of all kinds of membrane tubular structures described above. We predict that the regulation of membrane tubular structure formation and/or development is a general property of tetraspanins, by which they affect various cellular behaviors.
Different tetraspanins apparently play different roles in the formation and/or development of membrane tubular structures. Tetraspanin CD81 appears to promote the formation and extension of microvilli [10, 38]. In addition, CD81 may increase the membrane curvature of microvilli, evidenced by the presence of CD81 proteins in microvilli and thinner microvilli found in CD81-overexpressing cells [10]. Hence, CD81 appears to function as a positive regulator for the microprotrusion formation and/or development by facilitating membrane bending. Since microprotrusions are still found in CD81-null cells, CD81 is not required for the generation of microprotrusions [10]. Other tetraspanins such as CD9 and CD151 may behave like CD81 in microprotrusion morphogenesis. For example, the development of microvilli at the surface of egg cells is impaired when CD9 is ablated [19, 20], while CD9 overexpression promotes the formation of microvilli [39]. CD151 in epithelial and endothelial cells facilitates the formation of the adhesion zipper when the cells are in contact [28, 40], and the engagement of CD151 with its Ab induces microprotrusions in isolated epithelial cells [40]. A recent study revealed that tetraspanin TM4SF1/Ly6 promotes the formation of nanopodia, extremely long micro-membrane tubular structures, in endothelial cells [24].
Tetraspanin CD82 probably exerts an opposite effect on membrane microprotrusive activities. In our earlier study, we found that, in both Du145 and PC3 prostate cancer cells, CD82 overexpression inhibits the formation and development of “dorsal” microprotrusion or microvilli, and reduces the number of pericellular “microvesicles” [10, 41], which could be either the cross section of microprotrusions or microprotrusion-derived microvesicles. PC3 cells overexpressing CD82 also exhibited the decreased membrane curvature or increased radii of the microvillar tips [10]. Thus, CD82 functions as a negative regulator for the formation and/or development of “dorsal” microprotrusions or microvilli. However, the effect of CD82 on ventral or basal microprotrusions, which likely initiate the adhesion zipper formation, remains to be determined.
Tetraspanins engage various cellular functions by regulating membrane tubular structures.
Tetraspanins play important roles in cell adhesions [1–7]. Membrane tubular structure could serve as the structural base for both cell-cell and cell-matrix adhesions. For example, microvilli on egg cells, where the cell surface CD9 proteins are enriched, are the anchorage or attachment sites for the sperm that has penetrated through the zona pellucida [18–20]. After fertilization, fewer CD9 proteins are present in microvilli, and the surface distribution of microvilli becomes less uniform [42]. The infertility of CD9-null female mice likely results from the aberrant microprotrusive activities of the CD9-null egg plasma membrane, i.e., the formations of microvilli and pericellular microvesicles. Such aberrancy in turn leads to the abnormal adhesive interactions between sperm and CD9-null eggs, although the sperm can still bind CD9-null eggs [18, 43]. In addition, CD9-positive mircoprotrusions form an adhesion zipper when cells contact each other in the presence of CD9 mAb [29], as mentioned before. For the membrane tubular structures that are enriched with cell-matrix adhesion molecules, they may facilitate cell-matrix adhesion through mechanisms such as cell adhesion strengthening. The foot processes of glomerular podocytes intertwine with each other, anchor at the glomerular basement membrane, and are critical for both podocyte-podocyte and podocyte-matrix adhesions. CD151 and its associated integrin α3β1 are localized at the end or tips of foot processes, where podocytes anchor to the glomerular basement membrane [34].
It is well documented that tetraspanins regulate cell movement. Tetraspanins likely regulate cell movement by modulating the formation and/or development of microprotrusions. For example, the prototypic fungal tetraspanin PSL1 directs the formation of a cellular protrusive structure called penetration peg, which is important for fungus invasiveness [35]. And the Drosophila tetraspanin Late Bloomer facilitates the transformation of invading axon growth cones into pre-synaptic arbors at the neuromuscular junction [44] by functioning as a negative regulator of membrane protrusive activity. As mentioned before, CD82 inhibits the formation and development of microvilli and pericellular microvesicles, which correlate with its motility-inhibitory activity [41]. CD82 TM polar residues crucial for motility-inhibitory activity are also needed for the proper formations of these structures [41]. Interestingly, immobilized CD82 mAb induces the formation of elongated cellular extensions [6], morphologically similar to microvilli and filopodia. However, it remains unknown whether such extension results from the increased protrusion or decreased retraction activity. Since both cellular protrusion and retraction become attenuated upon CD82 overexpression (our unpublished data), the cellular extension induced by the immobilized CD82 mAb could result from the decreased cellular retraction. Although the role of microprotrusions in cell movement has not been systematically studied, it is likely that the formation of microprotrusions in isolated individual cells correlates with cell motility. Conversely, if microprotrusions form adhesion zipper between cells, more microprotrusions may reflect less cell motility. Hence, microprotrusive activities of the plasma membrane per se do not reflect cell motility. For example, there are conflicting reports regarding the role of CD9 in cell movement: quite a few studies found that CD9 inhibits cell migration, but some showed that CD9 promotes cell migration [7]. We predict that CD9 inhibits cell movement when cells are in contact but promotes movement when cells are isolated.
Tetraspanins are also involved in endocytosis and exocytosis [37, 45, 46]. Microprotrusions might reflect the endocytic and/or exocytic activities of cells. For example, microprotrusions could reflect the early stage of macropinocytosis, a form of endocytosis. In macropinocytosis, a pair of enfolding microprotrusions encompasses the extracellular solutes, suggesting the formation of endocytic structure [10]. This phenomenon is possibly combined with or followed by the local invagination of the cell membrane between two enfolding microprotrusions. However, the two enfolding microprotrusions in the transmission EM image could reflect the side view of enclosing membrane folding or ruffles. Notably, the number of pericellular microvesicles was substantially reduced when microprotrusions were largely lost upon CD82 overexpression [10, 41] or CD9 ablation [20], highlighting the roles of tetraspanins and microprotrusions in excytosis or vesicle release. Indeed, recent progress indicates that intestinal microvilli, in which tetraspanins CO-029, CD82, and Tspan1 are present, generate microvesicles [17]. These observations are also consistent with the involvement of tetraspanin in viral entry and release [5]. Coincidently, one of the most frequent changes in virally infected cells is the formation of microvilli [47, 48], suggesting the contribution of microvilli to the viral budding or release process. For viral release, membrane tubules could be the location or device for membrane budding or represent a transition structure of dynamic budding. Similarly, membrane tubules could facilitate the attachment of virus to the cell surface prior to viral entry. Hence, based on our hypothesis, facilitating microvillus formation by CD81 is likely important for its function as a key entry factor for hepatitis C virus and influenza virus.
It is well recognized that tetraspanins function at the molecular level as “molecular facilitators” [3] by regulating the functions of their associated proteins. Tetraspanins associate with each other and with other transmembrane proteins, which are typically cell adhesion proteins such as integrins and immunoglobulin superfamily proteins, to form the structurally tangible membrane microdomain [49, 50] called the tetraspanin web [4] or tetraspanin-enriched microdomain (TEM) [5]. The tetraspanin web describes the organization of tetraspanins at the molecular level, while the membrane tubular structures denote the effect of tetraspanins on the cell membrane at the cellular level. We emphasize that membrane tubules are the functional manifestation of TEMs at the cell membrane.
How tetraspanins regulate the morphogenesis of membrane tubular structures
Tetraspanins may alter the membrane curvature
For example, the tips of microprotrusions become less bending when CD9 is not present [19], suggesting that CD9 promotes the outward curvature of the plasma membrane. CD81 plays a similar role [10]. We predict that the female infertility of CD81-deficient mice is also due to less outward curvature of the egg cell membrane. In contrast, CD82 overexpression diminishes the outward curvature of the plasma membrane and often induces concave cell peripheries [10]. Tetraspanin RDS is exclusively localized at the disc rim, the curved membrane region, of the rod photoreceptor outer segments [51, 52], and is necessary for forming and maintaining the proper morphology of the rim [53]. The presence of RDS in membrane vesicles causes the flattening of these vesicles [54], a membrane-bending process.
If no other cellular factor is involved, more or thinner microvilli reflect higher outward intrinsic curving activity of the plasma membrane, while fewer or thicker microvilli reflect relatively less activity. Likewise, the membrane inward curving activity leads to the invagination of the plasma membrane. Biophysically, tetraspanin webs could be the membrane domains or areas with altered curvature, relative to the regular areas of the cell membrane. Dependent upon the tetraspanin composition, tetraspanin webs may (1) form more positive, (2) form more negative, or (3) deform membrane curvature. Consequently the different membrane curvature leads to either promoting or inhibiting the formation and/or extension of membrane tubular structures, membrane blebbing, or membrane ridge or crest (Fig. 2). Although tetraspanins per se appear not to fall into the category of membrane-curving proteins based on the current understanding [55, 56], they may still bend the membrane when they become dimerized or polymerized, and then further form tetraspanin web through a scaffolding mechanism. The posttranslational modifications of tetraspanins such as glycosylation, lipidation, and disulfide bond formation could contribute to or even determine this membrane-bending mechanism. Another possibility is that tetraspanins may physically associate with membrane-curving proteins or lipids, different tetraspanins have differential abilities to bind to these molecules, and such a difference results in different membrane curving activities. Functionally, we predict that the local curvature of tetraspanin web causes altered connection between the plasma membrane and the cytoskeleton, and changes the spatial presentation of segregated adhesion, signaling, and enzymatic machineries.
Tetraspanin may alter membrane-dependent cytoskeletal reorganization
As the master regulators of the actin cytoskeleton, Rho small GTPases Cdc42, Rac, and Rho determine the formation of membrane macroprotrusive structures such as lamellipodia and filopodia. Although the roles of Rho small GTPases in membrane microprotrusive activities remain to be determined, membrane protrusive structures are likely driven by the outward actin polymerization. Tetraspanins may regulate the membrane microprotrusive activities through modifying actin reorganization in a Rho small GTPase-dependent manner since the signaling connection between tetraspanins and Rho small GTPases is well recognized [8]. Ezrin/radixin/moesin (ERM) proteins are needed for the membrane microprotrusive activity that induces the microvillus formation [57, 58]. Tetraspanins are found to either directly or functionally associate with ERM proteins [59, 60]. Hence, ERM proteins could serve as the adaptors, directly linking tetraspanins to the actin cytoskeleton, and thereby regulate the formation and/or development of membrane tubular structures.
In reality, both membrane bending and actin cytoskeleton reorganization are probably involved in the formation and development of membrane tubular structures. In some cases, one may play a more important role than another. For example, the membrane tubular structures sometimes contain an electron-dense core, which is likely to be a cytoskeleton, under transmission electron microscopy (our unpublished data). While the membrane tubular structures sometimes exhibit even density in cross section without an electron-dense core, suggesting that, at least, no continuous cytoskeleton is involved (our unpublished data). Also, tetraspanin TM4SF1/Ly6-induced extremely long microvilli projected from endothelial cells are poor in F-actin [24] (Fig. 3a). We found that the micro-membrane tubular structures at the retraction edges of cells also contain less F-actin (Fig. 3b, c).
Perspective
Compared with membrane blebbing and membrane ridges or crests, the membrane tubular structures of cells represent the maximal activity of membrane deformation (Fig. 2). It is not surprising if tetraspanins are involved in the formation and development of membrane blebbing and membrane ridges or crests. In this article, we focused on the membrane tubular structures that extrude from the plasma membrane of cells. The roles of tetraspanins in (1) other membrane tubular structures such as membrane tubular invagination and (2) intrusive deformation of the plasma membrane such as membrane pits remain to be determined. Given the facts that tetraspanins typically regulate various cell functions oppositely, we would predict that some tetraspanins facilitate the formation and development of those membrane tubular structures, while some tetraspanins inhibit these processes.
Tetraspanins have been considered as molecular facilitators [3–5]. How tetraspanins function as molecular facilitators still remains unclear. Promoting or inhibiting the formation and development of membrane tubular structures could be the subcellular mechanism by which tetraspanins modulate the activity of their associated molecules. For example, the localization of tetraspanin-associated cell adhesion proteins at microvilli makes these adhesion molecules more accessible to their ligands and counter-receptors. Thus, regulating membrane tubular structure at the subcellular level coincides well with the facilitator function of tetraspanins at the molecular level.
Acknowledgments
This work was supported by National Institutes of Health Research Grant CA096991 (to X.A.Z.).
Abbreviations
- EM
Electron microscopy
- ERM
Ezrin/radixin/moesin
- Memtubs
Membrane tubular structures
- MVB
Multi-vesicular body
- TEM
Tetraspanin-enriched microdomain
References
- 1.Horejsi V, Vlcek C. Novel structurally distinct family of leucocyte surface glycoproteins including CD9, CD37, CD53 and CD63. FEBS Lett. 1991;288:1–4. doi: 10.1016/0014-5793(91)80988-F. [DOI] [PubMed] [Google Scholar]
- 2.Wright MD, Tomlinson MG. The ins and outs of the transmembrane 4 superfamily. Immunol Today. 1994;15:588–594. doi: 10.1016/0167-5699(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 3.Maecker HT, Todd SC, Levy S. The tetraspanin superfamily: molecular facilitators. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 4.Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 6.Richardson MM, Jennings LK, Zhang XA. Tetraspanins and tumor progression. Clin Exp Metastasis. 2011;28:261–270. doi: 10.1007/s10585-010-9365-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F, Kotha JP, Jennings LK, Zhang XA. Tetraspanins and vascular function. Cardiovasc Res. 2009;83:7–15. doi: 10.1093/cvr/cvp080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shum WW, Da Silva N, McKee M, Smith PJ, Brown D, Breton S. Transepithelial projections from basal cells are luminal sensors in pseudostratified epithelia. Cell. 2008;135:1108–1117. doi: 10.1016/j.cell.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, Reinecker HC. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 10.Bari R, Guo Q, Xia B, Zhang YH, Giesert EE, Levy S, Zheng JJ, Zhang XA. Tetraspanins regulate the protrusive activities of cell membrane. Biochem Biophys Res Commun. 2011;415:619–626. doi: 10.1016/j.bbrc.2011.10.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes H–H, Bukoreshtliev NV, Barroso JFV. Tunneling nanotubes: a new route for the exchange of components between animal cells. FEBS Lett. 2007;581:2194–2201. doi: 10.1016/j.febslet.2007.03.071. [DOI] [PubMed] [Google Scholar]
- 12.Sherer NM, Mothes W. Cytonemes and tunneling nanotubules in cell–cell communication and viral pathogenesis. Trends Cell Biol. 2008;18:414–420. doi: 10.1016/j.tcb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol. 2006;18:18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- 15.Monsky WL, Chen WT. Proteases of cell adhesion proteins in cancer. Semin Cancer Biol. 1993;4:251–258. [PubMed] [Google Scholar]
- 16.Gimona M, Buccione R, Courtneidge SA, Linder S. Assembly and biological role of podosomes and invadopodia. Curr Opin Cell Biol. 2008;20:235–241. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 17.McConnell RE, Higginbotham JN, Shifrin DA, Jr, Tabb DL, Coffey RJ, Tyska MJ. The enterocyte microvillus is a vesicle-generating organelle. J Cell Biol. 2009;185:1285–1298. doi: 10.1083/jcb.200902147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. The gamete fusion process is defective in eggs of CD9-deficient mice. Nat Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- 19.Runge KE, Evans JE, He Z-Y, Gupta S, McDonald KL, Stahlberg H, Primakoff P, Myles DG. Oocyte CD9 is enriched on the microvillar membrane and required for normal microvillar shape and distribution. Dev Biol. 2007;304:317–325. doi: 10.1016/j.ydbio.2006.12.041. [DOI] [PubMed] [Google Scholar]
- 20.Miyado K, Yoshida K, Yamagata K, Sakakibara K, Okabe M, Wang X, Miyamoto K, Akutsu H, Kondo T, Takahashi Y, Ban T, Ito C, Toshimori K, Nakamura A, Ito M, Miyado M, Mekada E, Umezawa A. The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc Natl Acad Sci USA. 2008;105:12921–12926. doi: 10.1073/pnas.0710608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Israels SJ, McMillan-Ward EM. Platelet tetraspanin complexes and their association with lipid rafts. Thromb Haemost. 2007;98:1081–1087. [PubMed] [Google Scholar]
- 22.Brisson C, Azorsa DO, Jennings LK, Moog S, Cazenave JP, Lanza F. Co-localization of CD9 and GPIIb-IIIa (alpha IIb beta 3 integrin) on activated platelet pseudopods and alpha-granule membranes. Histochem J. 1997;29:153–165. doi: 10.1023/A:1026437522882. [DOI] [PubMed] [Google Scholar]
- 23.Pan Y, Brown C, Wang X, Geisert EE. The developmental regulation of CD81 in the rat retina. Mol Vis. 2007;13:181–189. [PMC free article] [PubMed] [Google Scholar]
- 24.Zukauskas A, Merley A, Li D, Ang LH, Sciuto TE, Salman S, Dvorak AM, Dvorak HF, Jaminet SC. TM4SF1: a tetraspanin-like protein necessary for nanopodia formation and endothelial cell migration. Angiogenesis. 2011;14:345–354. doi: 10.1007/s10456-011-9218-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasioukhin V, Bauer C, Yin M, Fuchs E. Directed actin polymerization is the driving force for epithelial cell–cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/S0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 26.Jacinto A, Wood W, Balayo T, Turmaine M, Martinez-Arias A, Martin P. Dynamic actin-based epithelial adhesion and cell matching during Drosophila dorsal closure. Curr Biol. 2000;10:1420–1426. doi: 10.1016/S0960-9822(00)00796-X. [DOI] [PubMed] [Google Scholar]
- 27.Raich WB, Agbunag C, Hardin J. Rapid epithelial-sheet sealing in the Caenorhabditis elegans embryo requires cadherin-dependent filopodial priming. Curr Biol. 1999;9:1139–1146. doi: 10.1016/S0960-9822(00)80015-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhang F, Michaelson JE, Moshiach S, Sachs N, Zhao W, Sun Y, Sonnenberg A, Lahti JM, Huang H, Zhang XA. Tetraspanin CD151 maintains vascular stability by balancing the forces of cell adhesion and cytoskeletal tension. Blood. 2011;118:4274–4284. doi: 10.1182/blood-2011-03-339531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singethan K, Müller N, Schubert S, Lüttge D, Krementsov DN, Khurana SR, Krohne G, Schneider-Schaulies S, Thali M, Schneider-Schaulies J. CD9 clustering and formation of microvilli zippers between contacting cells regulates virus-induced cell fusion. Traffic. 2008;9:924–935. doi: 10.1111/j.1600-0854.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-España A, Chung PJ, Sarkar IN, Stiner E, Sun TT, Desalle R. Appearance of new tetraspanin genes during vertebrate evolution. Genomics. 2008;91:326–334. doi: 10.1016/j.ygeno.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 31.Huang S, Yuan S, Dong M, Su J, Yu C, Shen Y, Xie X, Yu Y, Yu X, Chen S, Zhang S, Pontarotti P, Xu A. The phylogenetic analysis of tetraspanins projects the evolution of cell–cell interactions from unicellular to multicellular organisms. Genomics. 2005;86:674–684. doi: 10.1016/j.ygeno.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 32.Kreidberg JA, Donovan MJ, Goldstein SL, Rennke H, Shepherd K, Jones RC, Jaenisch R. Alpha 3 beta 1 integrin has a crucial role in kidney and lung organogenesis. Development. 1996;122:3537–3547. doi: 10.1242/dev.122.11.3537. [DOI] [PubMed] [Google Scholar]
- 33.Sachs N, Kreft M, van den Bergh Weerman MA, Beynon AJ, Peters TA, Weening J, Sonnenberg A. Kidney failure in mice lacking the tetraspanin CD151. J Cell Biol. 2006;175:33–39. doi: 10.1083/jcb.200603073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baleato RM, Guthrie PL, Gubler MC, Ashman LK, Roselli S. Deletion of CD151 results in a strain-dependent glomerular disease due to severe alterations of the glomerular basement membrane. Am J Pathol. 2008;173:927–937. doi: 10.2353/ajpath.2008.071149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clergeot PH, Gourgues M, Cots J, Laurans F, Latorse MP, Pepin R, Tharreau D, Notteghem JL, Lebrun MH. PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea . Proc Natl Acad Sci USA. 2001;98:6963–6968. doi: 10.1073/pnas.111132998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–201277. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 37.Xu C, Zhang YH, Thangavel M, Richardson MM, Liu L, Zhou B, Zheng Y, Ostrom RS, Zhang XA. CD82 endocytosis and cholesterol-dependent reorganization of tetraspanin webs and lipid rafts. FASEB J. 2009;23:3273–3288. doi: 10.1096/fj.08-123414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quast T, Eppler F, Semmling V, Schild C, Homsi Y, Levy S, Lang T, Kurts C, Kolanus W. CD81 is essential for the formation of membrane protrusions and regulates Rac1-activation in adhesion-dependent immune cell migration. Blood. 2011;118:1818–1827. doi: 10.1182/blood-2010-12-326595. [DOI] [PubMed] [Google Scholar]
- 39.Wang HX, Kolesnikova TV, Denison C, Gygi SP, Hemler ME. The C-terminal tail of tetraspanin protein CD9 contributes to its function and molecular organization. J Cell Sci. 2011;124:2702–2710. doi: 10.1242/jcs.085449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shigeta M, Sanzen N, Ozawa M, Gu J, Hasegawa H, Sekiguchi K. CD151 regulates epithelial cell–cell adhesion through PKC- and Cdc42-dependent actin cytoskeletal reorganization. J Cell Biol. 2003;163:165–176. doi: 10.1083/jcb.200301075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bari R, Zhang YH, Zhang F, Wang NX, Stipp CS, Zheng JJ, Zhang XA. The transmembrane domain interactions are needed for kai1/cd82-mediated suppression of cancer invasion and metastasis. Am J Pathol. 2009;174:647–660. doi: 10.2353/ajpath.2009.080685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zyłkiewicz E, Nowakowska J, Maleszewski M. Decrease in CD9 content and reorganization of microvilli may contribute to the oolemma block to sperm penetration during fertilization of mouse oocyte. Zygote. 2010;18:195–201. doi: 10.1017/S0967199409990189. [DOI] [PubMed] [Google Scholar]
- 43.Jégou A, Ziyyat A, Barraud-Lange V, Perez E, Wolf JP, Pincet F, Gourier C. CD9 tetraspanin generates fusion competent sites on the egg membrane for mammalian fertilization. Proc Natl Acad Sci USA. 2011;108:10946–10951. doi: 10.1073/pnas.1017400108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kopczynski CC, Davis GW, Goodman CS. A neural tetraspanin, encoded by late bloomer, that facilitates synapse formation. Science. 1996;271:1867–1870. doi: 10.1126/science.271.5257.1867. [DOI] [PubMed] [Google Scholar]
- 45.Berditchevski F, Odintsova E. Tetraspanins as regulators of protein trafficking. Traffic. 2007;8:89–96. doi: 10.1111/j.1600-0854.2006.00515.x. [DOI] [PubMed] [Google Scholar]
- 46.Liu L, He B, Liu WM, Zhou D, Cox JV, Zhang XA. Tetraspanin CD151 promotes cell migration through regulating integrin trafficking. J Biol Chem. 2007;282:31631–31642. doi: 10.1074/jbc.M701165200. [DOI] [PubMed] [Google Scholar]
- 47.Volkman LE. Baculovirus infectivity and the actin cytoskeleton. Curr Drug Targets. 2007;8:1075–1083. doi: 10.2174/138945007782151379. [DOI] [PubMed] [Google Scholar]
- 48.Gamliel H, Polliack A. Virus–cell interactions as seen by scanning electron microscopy. Isr J Med Sci. 1979;15:647–652. [PubMed] [Google Scholar]
- 49.Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J Cell Biol. 2006;173:795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espenel C, Margeat E, Dosset P, Arduise C, Le Grimellec C, Royer CA, Boucheix C, Rubinstein E, Milhiet P-E. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. J Cell Biol. 2008;182:765–776. doi: 10.1083/jcb.200803010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arikawa K, Molday LL, Molday RS, Williams DS. Localization of peripherin/rds in the disk membranes of cone and rod photoreceptors: relationship to disk membrane morphogenesis and retinal degeneration. J Cell Biol. 1992;116:659–667. doi: 10.1083/jcb.116.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kedzierski W, Moghrabi WN, Allen AC, Jablonski-Stiemke MM, Azarian SM, Bok D, Travis GH. Three homologs of rds/peripherin in Xenopus laevis photoreceptors that exhibit covalent and non-covalent interactions. J Cell Sci. 1996;109:2551–2560. doi: 10.1242/jcs.109.10.2551. [DOI] [PubMed] [Google Scholar]
- 53.Conley SM, Stuck MW, Naash MI (2011) Structural and functional relationships between photoreceptor tetraspanins and other superfamily members. Cell Mol Life Sci Epub ahead of print [DOI] [PMC free article] [PubMed]
- 54.Wrigley JD, Ahmed T, Nevett CL, Findlay JB. Peripherin/rds influences membrane vesicle morphology. Implications for retinopathies. J Biol Chem. 2000;275:13191–13194. doi: 10.1074/jbc.C900853199. [DOI] [PubMed] [Google Scholar]
- 55.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 56.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 57.Niggli V, Rossy J. Ezrin/radixin/moesin: versatile controllers of signaling molecules and of the cortical cytoskeleton. Int J Biochem Cell Biol. 2008;40:344–349. doi: 10.1016/j.biocel.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 58.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 59.Sala-Valdes M, Ursa A, Charrin S, Rubinstein E, Hemler ME, Sanchez-Madrid F, Yanez-Mo M. EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ERM proteins. J Biol Chem. 2006;281:19665–19675. doi: 10.1074/jbc.M602116200. [DOI] [PubMed] [Google Scholar]
- 60.Coffey GP, Rajapaksa R, Liu R, Sharpe O, Kuo CC, Krauss SW, Sagi Y, Davis RE, Staudt LM, Sharman JP, Robinson WH, Levy S. Engagement of CD81 induces ezrin tyrosine phosphorylation and its cellular redistribution with filamentous actin. J Cell Sci. 2009;122:3137–3144. doi: 10.1242/jcs.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]