Abstract
Eukaryotic translation initiation factor 4E (eIF4E) is the mRNA 5′ cap binding protein, which plays an important role in the control of translation. The activity of eIF4E is regulated by a family of repressor proteins, the 4E-binding proteins (4E-BPs), whose binding to eIF4E is determined by their phosphorylation state. When hyperphosphorylated, 4E-BPs do not bind to eIF4E. Phosphorylation of the 4E-BPs is effected by the phosphatidylinositol (PI) 3-kinase signal transduction pathway and is inhibited by rapamycin through its binding to FRAP/mTOR (FK506 binding protein–rapamycin-associated protein or mammalian target of rapamycin). Phosphorylation of 4E-BPs can also be induced by protein synthesis inhibitors. These observations led to the proposal that FRAP/mTOR functions as a “sensor” of the translational apparatus (E. J. Brown and S. L. Schreiber, Cell 86:517–520, 1996). To test this model, we have employed the tetracycline-inducible system to increase eIF4E expression. Removal of tetracycline induced eIF4E expression up to fivefold over endogenous levels. Strikingly, upon induction of eIF4E, 4E-BP1 became dephosphorylated and the extent of dephosphorylation was proportional to the expression level of eIF4E. Dephosphorylation of p70S6k also occurred upon eIF4E induction. In contrast, the phosphorylation of Akt, an upstream effector of both p70S6k and 4E-BP phosphorylation, was not affected by eIF4E induction. We conclude that eIF4E engenders a negative feedback loop that targets a component of the PI 3-kinase signalling pathway which lies downstream of PI 3-kinase.
A key step in ribosome binding to the eukaryotic mRNA is the recognition of the cap structure (m7GpppX, where X is any nucleotide) by the cap binding protein complex, eukaryotic translation initiation factor 4F (eIF4F). eIF4F is composed of three subunits: eIF4E, a 24-kDa polypeptide which interacts directly with the cap structure; eIF4A, a 50-kDa polypeptide that exhibits RNA helicase activity; and eIF4G, which serves as a scaffolding protein for the binding of eIF4E, eIF4A, and eIF3, an initiation factor associated with the 40S ribosomal subunit (reviewed in references 62 and 66). eIF4F is an important target for translational control by extracellular stimuli, stress, and viral infections (30).
eIF4F function is regulated through multiple mechanisms, including transcriptional activation of eIF4E (39), changes in eIF4E phosphorylation state (reviewed in reference 67), and cleavage of eIF4G upon infection with some viruses, causing the shutoff of host protein synthesis (reviewed in references 7 and 23). In addition, members of a family of proteins, termed eIF4E-binding proteins (4E-BPs) (53), also called PHAS, for phosphorylated heat- and acid-stable protein (47), bind to eIF4E and prevent its interaction with eIF4G and thus the formation of the eIF4F complex (32, 49). Three mammalian members of the 4E-BP family have been cloned (53, 56). One functional analog of the 4E-BPs, p20, has been identified in Saccharomyces cerevisiae, although there is no sequence homology between the yeast protein and the mammalian 4E-BPs, other than the eIF4E binding site (3). The interaction of mammalian 4E-BPs with eIF4E is modulated by the extent of 4E-BP phosphorylation. The 4E-BPs strongly interact with eIF4E when in their hypophosphorylated state and dissociate from eIF4E upon hyperphosphorylation (47, 53).
Phosphorylation of 4E-BP is induced by growth factors, hormones, mitogens, G protein-coupled receptor agonists, and cytokines (for reviews see references 30, 45, and 67). A large body of evidence documents the involvement of the phosphatidylinositol (PI) 3-kinase signalling pathway in the phosphorylation of 4E-BP1 and a serine/threonine kinase, p70S6k, which phosphorylates ribosomal S6 protein (37). For example, the phosphorylation of both 4E-BP1 and p70S6k is strongly diminished by wortmannin, an inhibitor of PI 3-kinase (14, 17, 72). In addition, a dominant-negative mutant bearing a mutation of the 85-kDa regulatory subunit of the PI 3-kinase inhibits the phosphorylation and activity of p70S6k (14), and expression of an activated 110-kDa catalytic subunit of PI 3-kinase stimulates the phosphorylation of 4E-BP1 (30). Signalling from PI 3-kinase to 4E-BP1 and p70S6k is mediated at least in part through Akt (30, 40, 71).
Another kinase that modulates the activity of p70S6k and 4E-BP1 is FRAP/mTOR (FK506 binding protein [FKBP]–rapamycin-associated protein or mammalian target of rapamycin) (10, 12, 13, 15, 30, 34). FRAP/mTOR belongs to a growing family of kinases (PIK, PI 3-kinase related) which have homology in their catalytic domains with PI 3-kinase (reviewed in reference 36). The immunosuppressant rapamycin, in a complex with the immunophilin FKBP12, binds to a specific site in FRAP/mTOR and inhibits its activity (70). FRAP/mTOR phosphorylates 4E-BP1 and p70S6k in vitro (12, 13, 15). Furthermore, the signalling pathway which leads to S6 and 4E-BP1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70S6k (73). This explains the findings that treatment of cells with rapamycin decreases phosphorylation of 4E-BP1 and p70S6k and that addition of the FKBP12-rapamycin complex prevents phosphorylation of 4E-BP1 by FRAP/mTOR in an immune complex kinase assay (13).
In addition to extracellular stimuli, the rapamycin-sensitive phosphorylation of p70S6k and 4E-BP1 can be induced by inhibitors of protein synthesis, such as cycloheximide and anisomycin (51, 72). These and other observations led Brown and Schreiber to propose that FRAP/mTOR may be a component of a “sensor” apparatus for the activity of the translational machinery (11). Thus, there might exist a cellular feedback mechanism to regulate translational homeostasis. Were such a model correct, it might be expected that overexpression of eIF4E, which results in translation upregulation, would downregulate the FRAP/mTOR pathway. To address this idea, we established an eIF4E tetracycline-regulatable expression system to modulate cellular eIF4E levels.
MATERIALS AND METHODS
Vectors.
For construction of pRep9-CMVt-eIF4E, the coding sequence of eIF4E was cloned downstream of the tetracycline-responsive promoter CMVt, which consists of a minimal cytomegalovirus early promoter sequence fused to seven copies of a tet operator sequence. Murine eIF4E cDNA was excised from pBluescript KS-4E by digestion with HindIII/BamHI and ligated directionally into pRep9-CMVt (5).
Cell lines.
An NIH 3T3-derived cell line, S2-6, expressing Tetr-VP16 transactivator (tTA) (65), was transfected with 10 μg of pRep9-CMVt-eIF4E by calcium phosphate precipitation. G418-resistant colonies were isolated after 14 days of selection in medium containing 0.7 mg of G418/ml. Cells were maintained in medium containing 1 μg of tetracycline hydrochloride (Sigma) per ml. To induce expression of eIF4E, cells were rinsed three times in 1× phosphate-buffered saline (PBS) and grown in the absence of tetracycline for the times indicated in the figure legends.
Western blotting.
Cells were rinsed twice with ice-cold PBS and incubated with the appropriate lysis buffer. For analysis of eIF4E and 4E-BP1, the lysis buffer contained 10 mM Tris-HCl, pH 7.5; 1% Triton X-100; 50 mM KCl; 1 mM dithiothreitol (DTT); 2 mM MgCl2; 0.2 mM phenylmethylsulfonyl fluoride (PMSF). For analysis of p70S6k and Akt, the lysis buffer contained 50 mM sodium phosphate, pH 7.2; 2 mM EGTA; 25 mM NaF; 25 mM β-glycerophosphate; 0.5% Triton X-100; 100 μM Na3VO4; 1 mM PMSF; 1 μg of leupeptin/ml; 1 μg of pepstatin/ml; 1 mM benzamidine; 2 mM DTT. Lysates were centrifuged at 14,000 rpm for 10 min to remove cellular debris, and total protein concentration was determined in duplicate by using the bicinchoninic acid protein assay (Pierce) and quantitated against a standard curve of bovine serum albumin protein concentrations. For each experiment, an equal amount of total protein (25 to 50 μg) was electrophoresed on a sodium dodecyl sulfate (SDS)–8 to 15% polyacrylamide gel and electroblotted onto nitrocellulose membranes (Millipore) in 25 mM Tris-HCl, pH 7.5; 190 mM glycine; and 20% (vol/vol) methanol, at 75 V for 2 h. Filters were then blocked in Tris-buffered saline containing 0.2% Tween 20 (TBST) and 5% (wt/vol) dry milk at 25°C for 1 h. Membranes were incubated overnight at 4°C with one of the following antibodies at the indicated dilution: mouse monoclonal antiactin at 1:500 (ICN Biomedicals), rabbit polyclonal anti-eIF4E at 1:1,000 (26), rabbit polyclonal anti-4E-BP1 at 1:1,000 (31), rabbit polyclonal anti-p70S6k at 1:300 (Santa Cruz Biotechnology), rabbit polyclonal anti-phosphospecific p70S6k (Thr421/Ser424) at 1:1,000 (New England Biolabs), rabbit polyclonal anti-phosphospecific Akt (Ser473) at 1:1,000 (New England Biolabs), and goat polyclonal anti-Akt (C-20) at 1:1,000 (Santa Cruz Biotechnology). After they were washed with TBST, the membranes were incubated as follows: for rabbit polyclonal anti-4E-BP1, anti-p70S6k, anti-phosphospecific p70S6k, and anti-phosphospecific Akt antibodies, membranes were incubated with donkey anti-rabbit horseradish peroxidase-conjugated immunoglobulin G (IgG) (Amersham); and for goat anti-Akt (C-20), membranes were incubated with rabbit anti-goat horseradish peroxidase-conjugated IgG at 1:5,000 (Jackson Laboratories) for 30 min. Membranes were then washed with TBST three times, and the signals were detected by using an enhanced chemiluminescence (ECL) kit (Amersham) after exposure to an X-ray film (Du Pont). Membranes probed with the monoclonal antiactin antibody were incubated for 1 h with 0.1 μCi of 125I-labelled rabbit anti-mouse IgG (NEN) per ml, and for polyclonal anti-eIF4E, membranes were incubated for 1 h with 0.1 μCi of 125I-labelled protein A (Amersham) per ml. The membranes were washed three times in TBST and exposed to an X-ray film. Signals were quantified by using the Fuji Bas 2000 phosphorimager.
Far-Western analysis.
Heart muscle kinase (HMK)-eIF4E fusion protein was purified as described previously (53). HMK-eIF4E (3 μg) was incubated with HMK (20 U) (Sigma) in the presence of [γ-32P]-ATP (50 μCi) for 1 h at 37°C and purified by using a Sephadex G-50 column (Pharmacia Biotech). Total cellular protein (50 μg) was subjected to electrophoresis on an SDS–15% polyacrylamide gel and electroblotted onto a 0.2-μm-pore-size nitrocellulose membrane as described above for Western blotting. The membrane was blocked in 5% milk in HBB (25 mM HEPES-KOH, pH 7.7; 25 mM NaCl; 5 mM MgCl2; 0.1 mM EDTA; 1 mM DTT; 0.05% Nonidet P-40) for 2 h and then incubated overnight at 4°C in hybridization buffer (20 mM HEPES-KOH, pH 7.7; 75 mM KCl; 2.5 mM MgCl2; 0.1 mM EDTA; 1 mM DTT; 0.05% Nonidet P-40; 1% milk) containing 32P-labelled HMK-eIF4E (250,000 cpm/ml [9]). The membrane was washed four times with hybridization buffer and exposed to an X-ray film (Du Pont).
ODC assay.
Cells were rinsed twice in ice-cold PBS, scraped in lysis buffer (0.25 M Tris-HCl, pH 7.4; 1 mM EDTA; 1 mM DTT), and subjected to two freeze-thaw cycles. The lysate was centrifuged at 14,000 rpm for 10 min to remove cellular debris. Protein content of the lysate was determined in duplicate by using the bicinchoninic acid protein assay (Pierce) and quantitated against a standard curve of bovine serum albumin protein concentrations. Equal amounts of total protein (50 μg) were incubated with 2.5 μCi of 14C-labelled ornithine (Amersham) and 50 μM pyridoxal 5-phosphate for 1 h at 37°C. Incubations were performed in 96-well microtiter plates. Liberated 14CO2 was trapped in a covering 3MM paper saturated with a solution of barium hydroxide. The 3MM paper was rinsed with acetone, dried, and exposed to an X-ray film. Spots were cut from the paper, and the radioactivity was determined by using a beta-scintillation counter.
p70 S6 kinase assay.
Cell extract (20 μg) was diluted with extraction buffer (50 mM Tris-HCl, pH 8.0; 120 mM NaCl; 20 mM NaF; 1 mM benzamidine; 1 mM EDTA; 6 mM EGTA, 1% [vol/vol] Nonidet P-40; 0.1 mM PMSF) and incubated with rabbit polyclonal anti-p70S6k antibody (Santa Cruz Biotechnology) on ice for 2 h. A 37.5% suspension of protein A-Sepharose (Repligen) resin was added and incubated end over end at 4°C for 1 h. The resin was washed twice with extraction buffer and once with dilution buffer (50 mM morpholinepropanesulfonic acid [MOPS], pH 7.2; 5 mM MgCl2; 10 mM NaF; 30 mM β-glycerophosphate). p70S6k activity was assayed with 40S ribosomal subunits in a mixture (10 μl) containing 50 mM MOPS, pH 7.2; 1 mM DTT; 5 mM MgCl2; 5 mM p-nitrophenyl phosphate; 100 μM ATP; 0.5 μM protein kinase inhibitor (Sigma); 6 μCi of [32P]-ATP; 20 μg of 40S ribosomal subunits (44). The 40S ribosomes were a generous gift from G. Thomas. After incubation at 37°C for 30 min, reactions were terminated by addition of Laemmli buffer. Samples were resolved by SDS–12% polyacrylamide gel electrophoresis (PAGE), and S6 phosphorylation was visualized by autoradiography.
RESULTS
4E-BP1 dephosphorylation in eIF4E-transformed cells.
In an initial attempt to determine the effect of eIF4E overexpression on 4E-BP1 phosphorylation, we used an NIH 3T3-transformed cell line overexpressing eIF4E (46). We also used NIH 3T3 cell lines transformed with a mutant of another initiation factor, eIF2α (Ser51Ala, which cannot be phosphorylated [22]), as well as a mutant of its kinase (a dead-kinase mutant), the interferon-inducible PKR (protein kinase, double-stranded RNA dependent [42]). Overexpression of eIF2αala and a dominant-negative mutant of PKR, but not wild-type eIF2α (eIF2αwt), results in malignant transformation of NIH 3T3 cells (22, 42).
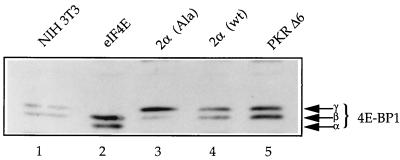
Murine 4E-BP1 can be resolved by SDS-PAGE into three bands as detected by Western blotting: the slowest-migrating, hyperphosphorylated isoform (γ) does not interact with eIF4E. The middle form (β) is less phosphorylated and binds to eIF4E with moderate affinity, while the fastest-migrating form (α) is hypophosphorylated and interacts strongly with eIF4E (8, 31, 48). In parental NIH 3T3 cells, 4E-BP1 was detected exclusively in its phosphorylated (γ and β) isoforms, while the α isoform was absent (Fig. 1, lane 1). However, in contrast to the parental cells, only the two faster-migrating isoforms (β and α) were detected (lane 2) in NIH 3T3 cells overexpressing eIF4E (46). This effect is similar to that seen in the presence of rapamycin (8). Overexpression of eIF2αala (lane 3) or eIF2αwt (lane 4) did not affect the state of 4E-BP1 phosphorylation in NIH 3T3 cells, as 4E-BP1 was present in the γ and β isoforms. The status of 4E-BP1 phosphorylation in NIH 3T3 cells expressing a dominant-negative mutant of PKR was also unchanged relative to that in parental NIH 3T3 cells (compare lanes 1 and 5). In addition, in NIH 3T3 cells which are transformed by overexpressing eIF4G (27), 4E-BP1 is not dephosphorylated (data not shown). Therefore, it appears that 4E-BP1 phosphorylation is altered in cells transformed with eIF4E but not in cells transformed by other initiation factors (this is further addressed in the Discussion section).
FIG. 1.
4E-BP1 phosphorylation state in transformed NIH 3T3 cells. Immunoblot analysis of total cell extract (50 μg) obtained from parental NIH 3T3 cells or NIH 3T3 cells overexpressing eIF4E (eIF4E), eIF2αala mutant [2α (Ala)] and wild type [2α (wt)], and mutant PKRΔ6. The blot was probed with a rabbit polyclonal anti-4E-BP1 antibody, as described in Materials and Methods. The three isoforms of 4E-BP1 are indicated.
Tetracycline-inducible eIF4E-expressing cell lines.
To demonstrate that the dephosphorylation of 4E-BP1 is a direct effect of eIF4E overexpression rather than the result of a secondary effect, such as an autocrine feedback loop or some aberration in the cell lines tested, we generated eIF4E inducible cell lines. We chose to use the tetracycline-inducible system rather than transient transfections, as the effects can be measured in shorter periods, thus minimizing possible autocrine effects. We utilized a tetracycline-inducible system in which both the eIF4E and the tTA genes are under the control of a tetracycline-inducible promoter, which is activated upon the removal of tetracycline (65).
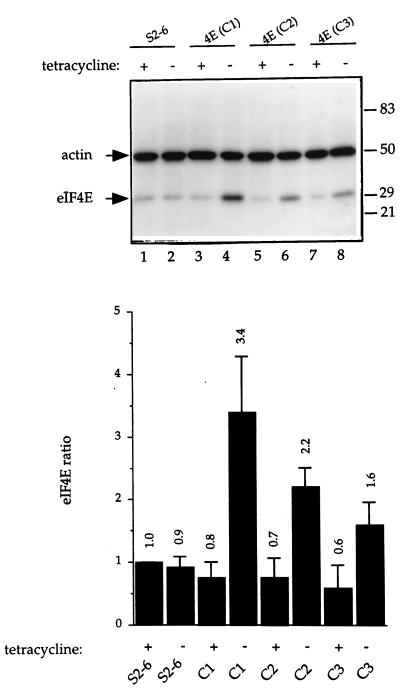
An NIH 3T3-derived stable cell line containing the autoregulatory tTA expression plasmid, S2-6 (65), was transfected with a CMV-eIF4E plasmid. G418-resistant cells were isolated and expanded, and the amount of eIF4E protein expressed was determined by Western blotting. Cell extract from each clone was assayed for eIF4E expression in cells grown in the presence (uninduced state) or absence (eIF4E-induced state) of tetracycline for 24 h. eIF4E expression was compared to that of control S2-6 cells cultured in the presence of tetracycline, and eIF4E protein levels were quantified and normalized against actin. The average (for three experiments) maximal increases in the amount of eIF4E after induction were 3.4-fold in clone 1 (Fig. 2 presents a representative experiment; compare lanes 4 and 1), 2.2-fold in clone 2 (compare lanes 6 and 1), and 1.6-fold in clone 3 (compare lanes 8 and 1) as compared to the amount in S2-6 cells. The amount of eIF4E in the stably transfected clones in the uninduced state was somewhat less (0.6- to 0.8-fold) than that found in the S2-6 cells (compare lane 1 to lanes 3, 5, and 7).
FIG. 2.
Expression of eIF4E in stably transfected tetracycline-inducible NIH 3T3 cells. (Upper panel) Immunoblot analysis was performed on total protein (25 μg) from cells cultured for 24 h in the presence or absence of tetracycline. S2-6 cells are the parental cells, which express tTA upon induction. C1, C2, and C3 are stably transfected cell lines expressing eIF4E. The blot was probed first with a rabbit polyclonal anti-eIF4E antibody, followed by a mouse monoclonal anti-actin antibody. After probing with 125I-labelled anti-rabbit and anti-mouse secondary antibodies, the amount of eIF4E expressed was quantitated with a Bas 2000 phosphorimager. Relative molecular masses for protein standards are indicated in kilodaltons to the right. (Lower panel) Quantitation of the results shown in the upper panel. eIF4E expression was normalized against actin levels and expressed as the ratio relative to the amount of eIF4E in S2-6 cells in the presence of tetracycline, which was set at 1. Values are the averages of three experiments, and the standard errors are shown.
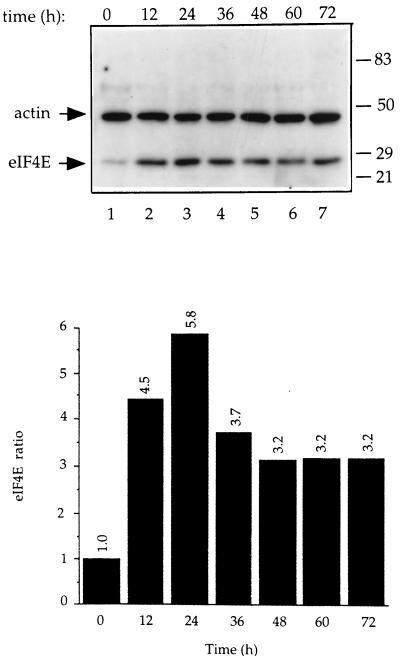
The expression kinetics of eIF4E were analyzed by Western blotting for clone 1, the clone expressing the highest level of eIF4E. Cells were cultured in the absence of tetracycline for 72 h, and the levels of eIF4E at several time points were compared to that in the cells in the uninduced state (tetracycline present, time zero) (Fig. 3, lane 1). eIF4E expression reached a maximal level (5.8-fold induction) at 24 h after removal of tetracycline (compare lanes 3 and 1), then declined to a 3.2-fold increase at 48 h, and remained stable over the remainder of the time course (lanes 5 to 7).
FIG. 3.
Time course of induction of eIF4E. (Upper panel) Total cell extract (25 μg) was prepared from C1 cells cultured in the absence of tetracycline for the indicated times. Extracts were immunoblotted and probed with a rabbit polyclonal anti-eIF4E antibody. The blot was subsequently probed with a mouse monoclonal anti-actin antibody to normalize for protein loading. After probing with 125I-labelled anti-rabbit and anti-mouse secondary antibodies, the amount of eIF4E expressed was quantitated with a Bas 2000 phosphorimager. (Lower panel) Quantitative analysis of the results presented in the upper panel. eIF4E expression levels were normalized against actin levels and are expressed as the ratios relative to the amount of eIF4E in C1 cells at time 0, which was set at 1. Results are representative of two experiments for which the values differed by less than 14%.
eIF4E expression increases ODC activity.
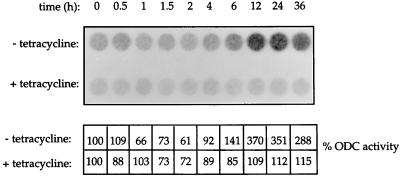
It was important to determine whether the overexpressed eIF4E is functional. We therefore examined ornithine decarboxylase (ODC) activity, which was previously shown to correlate with the amount of the protein (55, 59, 69). ODC is a downstream target of eIF4E, as its amount is increased in cells constitutively overexpressing eIF4E (61, 64). Within the first 4 h of eIF4E induction in clone 1 cells, ODC activity did not change significantly as compared to the activity measured in extracts prepared from cells cultured in the presence of tetracycline (Fig. 4, lower panel). However, ODC activity increased as early as 6 h post-induction (Fig. 4, upper panel; the activity was 141% of that at time zero), concomitant with the time line required for a detectable increase in the level of eIF4E. Maximal enzymatic activities were reached at 12 and 24 h (the activities were 370 and 350% as compared to that at time zero). Note that ODC activity starts to decline after 12 h, which could be due to the synthesis of polyamines that induce the degradation of ODC (50) or the downregulation of translation by 4E-BPs (see below). These results indicate that the induced eIF4E protein is active and bolster the original conclusion (61, 64) that the increase in ODC translation observed in cells constitutively overexpressing eIF4E is a direct result of eIF4E overexpression.
FIG. 4.
ODC activity is stimulated by eIF4E overexpression. C1 cells were maintained in the absence or presence of tetracycline for the indicated times. Total cell extract (50 μg) was assayed for ODC activity as described in Materials and Methods. ODC activity is expressed as the percentage of the value obtained at time 0. ODC activity at time 0 was 3.1 (without tetracycline) and 2.7 (in the presence of tetracycline) nmol of CO2/h/mg of protein. Results are representative of two experiments for which the activities differed by less than 26%.
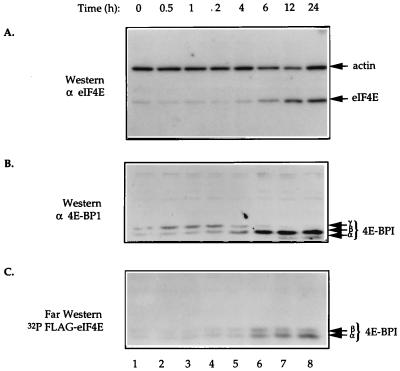
4E-BP1 is dephosphorylated upon eIF4E induction.
The inducible eIF4E-expressing cell lines were used to correlate 4E-BP1 phosphorylation status with eIF4E levels. A Western blot analysis of eIF4E induction for clone 1 cells was performed after removal of tetracycline. Overexpression of eIF4E was detected as early as 6 h postinduction (Fig. 5A, lane 6), and maximal eIF4E expression was detected after 24 h (lane 8). Western blot analysis of 4E-BP1 revealed a gradual dephosphorylation that coincided with the induction of eIF4E. Under uninduced conditions, only the slower-migrating phosphorylated isoforms (γ and β) of 4E-BP1 were detected (Fig. 5B, lane 1). However, 4E-BP1 was found only in its faster-migrating isoforms (β and α) at 12 and 24 h postinduction of eIF4E (lanes 7 and 8). Dephosphorylation of 4E-BP1 was detected as early as 4 and 6 h after induction of eIF4E (lanes 5 and 6), when eIF4E levels were also increased. At this time, the amount of the hyperphosphorylated (γ) isoform of 4E-BP1 began to decrease, while the β isoform began to predominate. It appears that the amount of 4E-BP1 is increased following eIF4E induction; however, because we are not completely sure whether our anti-4E-BP1 antibody has equal affinity for the various phosphorylation forms of 4E-BP1, we cannot conclude with certainty that there is increased synthesis or stability of 4E-BP1 upon induction of eIF4E.
FIG. 5.
eIF4E induction results in dephosphorylation of 4E-BP1. Cell extracts were prepared from C1 cells in the absence of tetracycline for the indicated times. (A) Total cell extract (25 μg) was resolved on an SDS–12.5% polyacrylamide gel, electroblotted onto a 0.45-μm-pore-size nitrocellulose membrane, and probed with rabbit polyclonal anti-eIF4E and mouse monoclonal anti-actin antibodies. (B) Total cell extract (50 μg) was resolved on an SDS–15% polyacrylamide gel, electroblotted onto a 0.2-μm-pore-size nitrocellulose membrane, and probed with a rabbit polyclonal anti-4E-BP1 antibody. (C) Far-Western analysis of cell extract (50 μg) by using 32P-labelled HMK-eIF4E as a probe was performed as described in Materials and Methods. In panels B and C, the different phosphorylation isoforms of 4E-BP1 are indicated with arrows.
To measure the binding capacity of 4E-BP1 to eIF4E over the time course of eIF4E induction, a far-Western analysis was performed, using a 32P-labelled recombinant eIF4E protein (53). Since phosphorylation of 4E-BP1 prevents its binding to eIF4E, only the α and β isoforms are detected in this assay (8, 31, 47, 53). The results of the far-Western analysis are consistent with those of the Western blot analysis of 4E-BP1 (compare Fig. 5C with Fig. 5B). The interaction of eIF4E with 4E-BP1 was first detected at 6 h postinduction, when a significant shift in 4E-BP1 phosphorylation occurred (Fig. 5C, lane 6), and this interaction was further strengthened as 4E-BP1 shifted predominantly to its β and α isoforms at 12 and 24 h postinduction (lanes 7 and 8). Taken together, these results show that upon induction of eIF4E, an increase in the amount of the hypophosphorylated (active) isoform of 4E-BP1 occurs, leading to increased interaction of 4E-BP1 with eIF4E.
The degree of 4E-BP1 dephosphorylation is directly proportional to eIF4E levels.
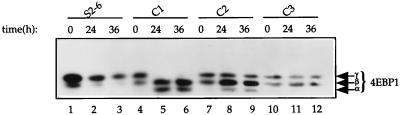
To further establish the correlation between eIF4E induction and 4E-BP1 dephosphorylation, we analyzed the phosphorylation of 4E-BP1 in eIF4E-expressing stably transfected inducible cells, which express different amounts of eIF4E upon induction. Cells were cultured in the absence of tetracycline for up to 36 h. Western blot analysis shows that in the uninduced state (0 h), 4E-BP1 was present in the phosphorylated state (γ and β isoforms) both in the parental S2-6 cells (Fig. 6, lane 1) and in the eIF4E stable transfectants (lanes 4, 7, and 10). Removal of tetracycline from S2-6 cells appears to cause a shift of the 4E-BP1 to the γ isoform (compare lane 1 to lanes 2 and 3; because the levels of total 4E-BP1 protein in lanes 2 and 3 were low in this experiment, the shift is not conclusive, but the shift was reproducible in other experiments). This shift is most probably due to activation of Akt as described below (see Fig. 8). Upon induction of eIF4E in the stable transfectants, 4E-BP1 was dephosphorylated to varying degrees. For clone 1, which exhibits the greatest overexpression of eIF4E, the dephosphorylation of 4E-BP1 was such that only the two faster-migrating isoforms (β and α) were detected after 24 h (compare lane 4 to lanes 5 and 6). For clone 2, which overexpresses eIF4E to a lesser degree than clone 1 (2.2-fold in contrast to 3.4-fold; see Fig. 2), 4E-BP1 dephosphorylation, upon eIF4E induction, was less dramatic than that for clone 1 (compare lane 7 to lanes 8 and 9). For clone 3, the cell line which overexpresses the least amount of eIF4E (1.6-fold) (Fig. 2), phosphorylation of 4E-BP1 was altered to the smallest extent upon eIF4E induction (compare lane 10 to lanes 11 and 12). In conclusion, the degree of 4E-BP1 dephosphorylation is directly proportional to the level of eIF4E expression, in that higher eIF4E expression is correlated with greater dephosphorylation of 4E-BP1. Taken together, these results suggest that an upstream element in a signal transduction pathway that regulates 4E-BP1 phosphorylation is affected by eIF4E overexpression.
FIG. 6.
The extent of 4E-BP1 dephosphorylation is proportional to the level of eIF4E expression. Parental cells (S2-6) and eIF4E-expressing cell clones C1, C2, and C3 were cultured in the absence of tetracycline for the indicated times. Total cell extract (50 μg) was immunoblotted and probed with a rabbit polyclonal anti-4E-BP1 antibody. Arrows indicate the different phosphorylation isoforms of 4E-BP1.
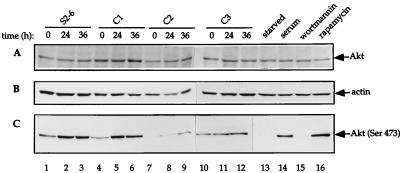
FIG. 8.
Akt phosphorylation is not inhibited by eIF4E overexpression. Tetracycline-inducible cells (S2-6, C1, C2, and C3) were incubated in the absence of tetracycline for the indicated times (lanes 1 through 12); NIH 3T3 cells were grown in Dulbecco’s minimal essential medium containing 0.5% fetal bovine serum (FBS) for 16 h (lanes 13 through 16), after which cells were either mock treated (lane 13), incubated for 30 min in the presence of 100 nM wortmannin (lane 15) or 20 ng of rapamycin/ml (lane 16) prior to stimulation with 10% FBS for 1 h, or stimulated with 10% FBS alone (lane 14). Cells were then harvested, and total cell extract (50 μg) was subjected to electrophoresis on an SDS–8% polyacrylamide gel, electroblotted, and probed by using the following: (A) goat polyclonal anti-Akt antibody (C-20), which recognizes Akt irrespective of its phosphorylation state; (B) mouse monoclonal anti-actin antibody; (C) rabbit polyclonal anti-phosphospecific (Ser473) Akt antibody. The result shown is a representative of two independent experiments.
eIF4E overexpression influences p70S6k phosphorylation.
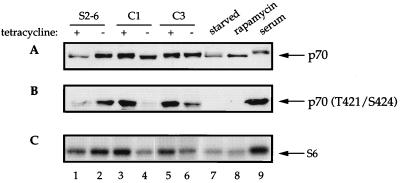
Since the signalling pathway acting upon 4E-BP1 and p70S6k bifurcates downstream of a common effector in the PI 3-kinase-FRAP/mTOR signalling pathway (73), we reasoned that the direct target of the negative feedback loop engendered by overexpression of eIF4E could be an upstream component of the shared pathway. Hence, eIF4E overexpression should affect both 4E-BP1 and p70S6k phosphorylation. Consequently, we examined the state of p70S6k phosphorylation upon induction of eIF4E. The activity of p70S6k is regulated by phosphorylation (e.g., see reference 4; see also reviews in references 37, 57, and 58), which influences the migration of the protein on SDS-PAGE (18). Decreased migration of p70S6k was observed upon addition of 10% serum to serum-starved NIH 3T3 cells (Fig. 7A; compare lane 9 to lane 7). A small shift to a slower-migrating form occurs upon removal of tetracycline in the parental cell line (compare lane 1 to lane 2). When eIF4E was induced, p70S6k migrated more rapidly, indicating hypophosphorylation of the protein. The shift was most evident in clone 1 cells, where the level of eIF4E is the highest (compare lane 4 to lane 3). In comparison to clone 1, a much less pronounced shift was noted for clone 3 (compare lane 6 to lane 5). These data show that the extent of this shift was largely proportional to the amount of eIF4E synthesized in the two clones tested.
FIG. 7.
Effect of eIF4E induction on p70S6k phosphorylation. eIF4E-expressing cells were maintained in the presence or absence of tetracycline for 36 h (lanes 1 through 6); NIH 3T3 cells were grown in Dulbecco’s minimal essential medium containing 0.5% fetal bovine serum (FBS) for 16 h (lanes 7 to 9), after which cells were either mock treated (lane 7), incubated for 30 min in the presence of 20 ng/ml of rapamycin prior to stimulation with 10% FBS for 1 h (lane 8), or stimulated with 10% FBS alone (lane 9). Total cell extract (50 μg) was electrophoresed on two separate SDS–8% polyacrylamide gels and electroblotted onto a 0.45-μm-pore-size nitrocellulose membrane. One membrane was probed with a rabbit polyclonal anti-p70S6k antibody which recognizes p70S6k irrespective of its phosphorylation state (A), while the other was probed with a rabbit polyclonal anti-phosphopeptide antibody which is specific for phospho-Thr421 and phospho-Ser424 in p70S6k (B). (C) Total cell extract (20 μg) was immunoprecipitated with rabbit polyclonal anti-p70S6k antibody. The immunoprecipitate was assayed for p70S6k activity by using 40S ribosomal subunits as a substrate as described in Materials and Methods. The figure is a representative of three independent experiments.
To substantiate these conclusions, Western blot analysis using a phosphospecific p70S6k antibody which recognizes phospho-Thr421 and phospho-Ser424 was performed (35, 75). These sites are contained in the autoinhibitory domain of the kinase and become phosphorylated upon activation of the PI 3-kinase pathway (58). To demonstrate that the phosphospecific antibody binds only to the phosphorylated form of p70S6k, serum-deprived NIH 3T3 cells were stimulated with serum to induce phosphorylation. The antibody failed to interact with p70S6k from serum-deprived or rapamycin-treated cells, but it bound to p70S6k from serum-fed cells (Fig. 7B, compare lanes 7 and 8 to lane 9). The phosphorylation of p70S6k at Thr421 and Ser424 is consistent with the shifts in mobility described above. Thr421 and Ser424 phosphorylation was increased in the parental S2-6 cells upon removal of tetracycline (compare lane 1 to lane 2), an effect similar to the observed effect on 4E-BP1 phosphorylation, which was probably due to phosphorylation of Akt (see Fig. 8). In the eIF4E-overexpressing cells, p70S6k phosphorylation at Thr421 and Ser424 decreased, and this reduction was proportional to the amount of eIF4E expressed upon induction in the two cell clones tested. The decrease in phosphorylation was most evident in clone 1 cells, where the level of eIF4E is the highest (compare lane 4 to lane 3). In comparison to clone 1, a lesser decrease in phosphorylation of p70S6k at Thr421 and Ser424 was noted for clone 3 (compare lane 6 to lane 5).
Finally, it was imperative to show directly that eIF4E overexpression affects p70S6k activity. An immune-complex kinase assay was performed using extracts from eIF4E-expressing cells as the source of the kinase and 40S ribosomal subunits as the source of the substrate. S6 phosphorylation increased (1.4-fold) upon the removal of tetracycline in the parental S2-6 cells, consistent with the increase in phosphorylation at Thr421 and Ser424 (Fig. 7C; compare lane 1 to lane 2). In contrast, induction of eIF4E resulted in decrease in S6 phosphorylation in eIF4E-transfected cells (compare lanes 3 and 4 and lanes 5 and 6). The decrease in S6 phosphorylation was proportional to the level of eIF4E in the two cell clones tested. The decrease in p70S6k activity was most noticeable in clone 1 cells, where S6 phosphorylation decreased 62% (compare lane 4 to lane 3). For clone 3, a lesser decrease in p70S6k activity was noted, as S6 phosphorylation decreased only by 35% (compare lane 6 to lane 5).
eIF4E does not inhibit Akt phosphorylation.
The protein Akt is a serine/threonine kinase (6), which is activated by PI 3-kinase (20, 25). Phosphorylation of Ser473 and Thr308 activates Akt (2, 68). Akt acts upstream in the 4E-BP1 signalling pathway and stimulates the phosphorylation of 4E-BP1 (30, 40, 71). In addition, Akt stimulates the phosphorylation of p70S6k (14, 40, 41). The mechanism by which Akt stimulates the phosphorylation of 4E-BP1 and p70S6k is not known, although it was recently reported that Akt activates FRAP/mTOR (63). To determine whether the target of the negative feedback loop induced by eIF4E overexpression lies upstream or downstream of Akt, we examined whether Akt phosphorylation is influenced by eIF4E. The levels of Akt protein did not change upon eIF4E induction or upon serum withdrawal in the different cell lines (Fig. 8A; an anti-actin antibody was used to normalize for loading variability [Fig. 8B]). We monitored the phosphorylation of Akt at Ser473 during a 36-h time course following tetracycline removal by using a phosphospecific antibody against Ser473. The antibody did not detect Akt in serum-deprived cells, as expected (Fig. 8C, lane 13). Upon stimulation of NIH 3T3 cells with serum, Akt became phosphorylated and was detected by the antibody (lane 14). In addition, wortmannin treatment blocked the phosphorylation of Akt (lane 15). Since rapamycin exerts its effect downstream of Akt, phosphorylation of Akt in rapamycin-treated cells, as expected, was not altered (lane 16). Western blot analysis performed by using the phosphospecific antibody showed that the phosphorylation of Akt at Ser473 was elevated upon induction in the eIF4E-transfected cells rather than decreased, as would be expected were eIF4E to inhibit Akt phosphorylation (compare lanes 4 to 6, 7 to 9, and 10 to 12). Note, however, that an increase of Akt phosphorylation was observed in the parental cells upon removal of tetracycline (compare lanes 1 through 3). This increase can explain the increase in phosphorylation of 4E-BP1 and p70S6k upon removal of tetracycline in the parental cells (see Fig. 5, 6, and 7). Although an explanation for the increase is not immediately clear, it is possible that the tTA causes Akt phosphorylation and activation. Our results show that eIF4E does not downregulate Akt phosphorylation. Furthermore, a decrease in the phosphorylation of Akt was not observed in a cell line stably overexpressing eIF4E (data not shown). This excludes the possibility that an upstream component signalling to Akt is affected by eIF4E. However, an intriguing possibility not addressed here is that eIF4E inhibits Akt activity through direct binding or indirectly by another mechanism. In conclusion, the target of the negative feedback loop established by eIF4E overexpression must lie at or downstream of Akt.
DISCUSSION
Translation plays an important role in control of cell growth (19). Rates of protein synthesis are also coupled to cell cycle progression, since inhibition of translation leads to arrest of cell growth, generally at the G1 phase (52). Because the control of the cell cycle is of paramount importance to cell growth and to the organism’s survival, there exist many checkpoints to ensure its integrity and appropriate functioning. It is conceivable that checkpoint mechanisms may function to monitor the activity of the translational machinery and to compensate for changes in the levels or activities of translation factors. One such checkpoint mechanism has been proposed by Brown and Schreiber (11) to explain the increase in the phosphorylation of p70S6k and/or 4E-BP1 observed in cells treated with protein synthesis inhibitors, such as anisomycin and cycloheximide (51, 72). Based on these observations it was postulated that FRAP/mTOR, an upstream regulator of the phosphorylation of p70S6k and 4E-BP1 (10, 13, 15, 30), is a sensor of translational activity in the cell and a target of a feedback loop. This feedback loop would then act to maintain translational homeostasis in the cell in order to prevent uncontrolled growth and proliferation, or cell death.
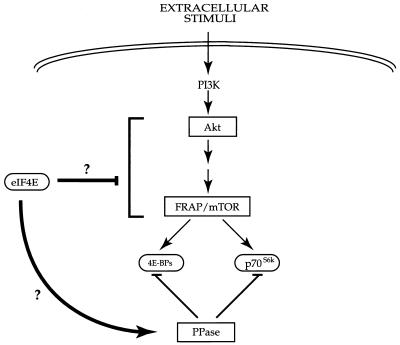
Here we report evidence for the existence of a potential translational homeostasis mechanism regulated by a negative feedback mechanism in response to an increase in the concentration of an endogenous translation factor, eIF4E. A model (depicted in Fig. 9) summarizes the PI 3-kinase signal transduction pathway which results in the phosphorylation of 4E-BPs and p70S6k. Both in a cell line stably overexpressing eIF4E and in cell lines in which eIF4E overexpression is inducible, dephosphorylation of 4E-BP1 correlates well with eIF4E protein upregulation. We have also noted dephosphorylation of 4E-BP2 upon overexpression of eIF4E (39a). Significantly, upregulation of eIF4E also results in the dephosphorylation of p70S6k. Akt, which is an upstream effector of 4E-BP1 (30, 40, 71) and might lie upstream of FRAP/mTOR (63; reviewed in reference 67) in this signalling pathway, is not dephosphorylated when eIF4E is overexpressed, but it could potentially be inhibited by direct interaction with eIF4E or by another mechanism, indicating that the negative feedback target lies at or downstream of Akt.
FIG. 9.
Proposed model for translational homeostasis in response to changes in eIF4E concentration. eIF4E incorporation into an eIF4F complex is regulated by the PI 3-kinase pathway, through Akt and FRAP/mTOR, which leads to the phosphorylation of 4E-BPs and p70S6k. Forced expression of eIF4E engenders a negative feedback loop by downregulation of a component of the Akt-FRAP/mTOR pathway or by activating a phosphatase (PPase) which dephosphorylates 4E-BP1 and p70S6k. This results in the dephosphorylation of p70S6k and 4E-BP1.
We do not know whether the negative feedback effect of eIF4E is by a direct or indirect stimulation of translation of a negative regulator. Since the phosphorylation of both 4E-BP1 and p70S6k is reported to be either directly or indirectly affected by FRAP/mTOR (10, 13, 15), the latter appears to be one intriguing candidate for the target of eIF4E. However, it is equally possible that eIF4E enhances the activity of 4E-BP1 and p70S6k-directed phosphatase(s) (Fig. 9). To directly address the question of whether FRAP/mTOR activity is modulated by eIF4E overexpression, we have attempted to determine whether in vitro FRAP/mTOR autokinase activity (10) or FRAP/mTOR-associated 4E-BP1 kinase activity (13) was enhanced upon upregulation of eIF4E expression. However, we failed to observe any significant changes in these activities following eIF4E upregulation (1, 28a). This result is not entirely surprising in view of the findings of Avruch and collaborators that FRAP/mTOR activity, in response to amino acid depletion followed by readdition or insulin stimulation, is not changed when assayed in an in vitro kinase assay (34, 35). Hara et al. also noted that FRAP/mTOR from rapamycin-treated cells, which is completely inactive in vivo, showed no decrease in autokinase and 4E-BP1 kinase activity in vitro, and they thus concluded that inhibition of FRAP/mTOR is apparently readily reversible during cell extract preparation (35). However, it should be noted that in other experiments FRAP/mTOR activity on 4E-BP1 in an in vitro kinase assay was shown to be stimulated by insulin by 1.8-fold (63). Also, we did not detect any changes in the level of mTOR upon eIF4E induction (39a).
The finding that eIF4E overexpression causes dephosphorylation of p70S6k at Thr421 and Ser424 is of interest. These Ser/Thr residues, which are two of the four initially identified sites of mitogen-induced phosphorylation, are followed by proline residues and are located in the carboxy terminus of p70S6k (24, 70). The carboxy terminus is thought to act as a pseudosubstrate autoinhibitory domain through interaction with its amino terminus. Phosphorylation of Ser411, Ser418, Ser424, and Thr421 has been postulated to disrupt the interaction between the amino and carboxy termini and allow the phosphorylation of Thr229 and Thr389, resulting in the activation of p70S6k (21). There are conflicting reports as to the rapamycin sensitivity of the phosphorylation of Thr421 and Ser424. In metabolic labelling experiments, followed by tryptic map analyses, it was shown that phosphorylation at these residues is rapamycin insensitive (33, 54). However, more recent experiments using a phosphospecific antibody against phosphorylated Thr421 and Ser424 peptides demonstrated that the phosphorylation of these sites in response to insulin is rapamycin sensitive (35, 75).
The expression of a dominant-negative PKR, a mutant of eIF2α, and even eIF4G did not cause the dephosphorylation of 4E-BP1. Expression of either of these proteins in NIH 3T3 cells results in cellular transformation (22, 27, 42), presumably because of an increase in translation of key growth-promoting proteins. Thus, the effects reported here cannot be elicited by all translation factors. It is also noteworthy that several colon tumor cell lines and tumor tissues which overexpress eIF4E also show an increase in the hypophosphorylated isoforms of 4E-BP1 (39a).
Some translational inhibitors (such as anisomycin and cycloheximide) stimulate, in addition to the FRAP/mTOR pathway, the p38 mitogen-activated protein kinase and JNK/SAPK pathways (16, 38, 43). Although anisomycin and cycloheximide exert this effect at low concentrations which do not inhibit general translation (16), it is possible that translation of sensitive mRNAs is inhibited. A downstream target of the p38 kinase is the serine/threonine kinase, Mnk1 (28, 60), which phosphorylates eIF4E on the physiologically relevant Ser 209 (74) and is thought to be required for optimal eIF4E activity (29). Thus, a translational feedback mechanism might be achieved by the integration of signals from several signal transduction pathways, via the activity of different effector proteins.
ACKNOWLEDGMENTS
We thank the anonymous reviewers for their insightful comments on the manuscript. We thank Robert D. Polakiewicz and Michael J. Comb for phosphospecific antibodies against p70S6k and Akt, George Thomas for providing 40S ribosomes, and Kazuei Igarashi for providing NIH 3T3 cells overexpressing eIF4G. We thank Christine DeMaria and Andrew Craig for valuable comments on the manuscript. We also thank Colin Lister and Chantal Binda for technical assistance and Avak Kahvejian for assistance in preparation of the manuscript.
This research was supported by a grant from the National Institute of Canada and the Howard Hughes Medical Institute International Scholar Program to N.S. N.S. is a Medical Research Council of Canada Distinguished Scientist and a Howard Hughes International Scholar. K.K. is a recipient of a predoctoral studentship from the Medical Research Council of Canada. S.P. is a recipient of a postdoctoral fellowship from the Association pour la Recherche sur le Cancer. A.-C.G. is a recipient of a 1967 Centennial Studentship from the National Science and Engineering Research Council of Canada.
REFERENCES
- 1.Abraham, R. T. Personal communication.
- 2.Alessi D R, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 3.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballou L M, Siegmann M, Thomas G. S6 kinase in quiescent Swiss mouse 3T3 cells is activated by phosphorylation in response to serum treatment. Proc Natl Acad Sci USA. 1988;85:7154–7158. doi: 10.1073/pnas.85.19.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauparlant P, Lin R, Hiscott J. The role of the C-terminal domain of IκBα in protein degradation and stabilization. J Biol Chem. 1996;271:10690–10696. doi: 10.1074/jbc.271.18.10690. [DOI] [PubMed] [Google Scholar]
- 6.Bellacosa A, Testa J R, Staal S P, Tsichlis P N. A retroviral oncogene, Akt, encoding a serine-threonine kinase containing an SH2-like region. Science. 1991;254:274–277. doi: 10.1126/science.254.5029.274. [DOI] [PubMed] [Google Scholar]
- 7.Belsham G J, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beretta L, Gingras A C, Svitkin Y V, Hall M N, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 9.Blanar M A, Rutter W J. Interaction cloning: identification of a helix-loop-helix zipper protein that interacts with c-Fos. Science. 1992;256:1014–1018. doi: 10.1126/science.1589769. [DOI] [PubMed] [Google Scholar]
- 10.Brown E J, Beal P A, Keith C T, Chen J, Shin T B, Schreiber S L. Control of p70 S6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 11.Brown E J, Schreiber S L. A signalling pathway to translational control. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 12.Brunn G J, Fadden P, Haystead T A J, Lawrence J., Jr The mammalian target of rapamycin phosphorylates sites having a (Ser/Thr)-Pro motif and is activated by antibodies to a region near its COOH terminus. J Biol Chem. 1997;272:32547–32550. doi: 10.1074/jbc.272.51.32547. [DOI] [PubMed] [Google Scholar]
- 13.Brunn G J, Hudson C C, Sekulic A, Williams J M, Hosoi H, Houghton P J, Lawrence J, Jr, Abraham R T. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 14.Burgering B M, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 15.Burnett P E, Barrow R K, Cohen N A, Snyder S H, Sabatini D M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cano E, Hazzalin C A, Mahadevan L C. Anisomycin-activated protein kinases p45 and p55 but not mitogen-activated protein kinases ERK-1 and -2 are implicated in the induction of c-fos and c-jun. Mol Cell Biol. 1994;14:7352–7362. doi: 10.1128/mcb.14.11.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou M M, Blenis J. The 70 kDa S6 kinase: regulation of a kinase with multiple roles in mitogenic signalling. Curr Opin Cell Biol. 1995;7:806–814. doi: 10.1016/0955-0674(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 18.Chung J, Kuo C V, Crabtree G R, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of signalling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 19.Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 20.Datta K, Bellacosa A, Chan T O, Tsichlis P N. Akt is a direct target of the phosphatidylinositol 3-kinase. Activation by growth factors, v-src and v-Ha-ras, in Sf9 and mammalian cells. J Biol Chem. 1996;271:30835–30839. doi: 10.1074/jbc.271.48.30835. [DOI] [PubMed] [Google Scholar]
- 21.Dennis P B, Pullen N, Pearson R B, Kozma S C, Thomas G. Phosphorylation sites in the autoinhibitory domain participate in p70s6k activation loop phosphorylation. J Biol Chem. 1998;273:14845–14852. doi: 10.1074/jbc.273.24.14845. [DOI] [PubMed] [Google Scholar]
- 22.Donzé O, Jagus R, Koromilas A E, Hershey J W, Sonenberg N. Abrogation of translation initiation factor eIF-2α phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14:3828–3834. doi: 10.1002/j.1460-2075.1995.tb00052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrenfled E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–573. [Google Scholar]
- 24.Ferrari S, Bannwarth W, Morley S J, Totty N F, Thomas G. Activation of p70s6k is associated with phosphorylation of four clustered sites displaying Ser/Thr-Pro motifs. Proc Natl Acad Sci USA. 1992;89:7282–7286. doi: 10.1073/pnas.89.15.7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke T F, Kaplan D R, Cantley L C, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 26.Frederickson R M, Montine K S, Sonenberg N. Phosphorylation of eukaryotic translation initiation factor 4E is increased in Src-transformed cell lines. Mol Cell Biol. 1991;11:2896–2900. doi: 10.1128/mcb.11.5.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuchi-Shimogori T, Ishii I, Kashiwagi K, Mashiba H, Ekimoto H, Igarashi K. Malignant transformation by overproduction of translation initiation factor eIF4G. Cancer Res. 1997;57:5041–5044. [PubMed] [Google Scholar]
- 28.Fukunaga R, Hunter T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 1997;16:1921–1933. doi: 10.1093/emboj/16.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Gingras, A.-C. Unpublished observations.
- 29.Gingras A-C, Kennedy S G, O’Leary M A, Sonenberg N, Hay N. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt (PKB) signalling pathway. Genes Dev. 1998;12:502–513. doi: 10.1101/gad.12.4.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gingras, A.-C., B. Raught, and N. Sonenberg. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulation of translation. Annu. Rev. Biochem., in press. [DOI] [PubMed]
- 31.Gingras A-C, Svitkin Y V, Belsham G J, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han J-W, Pearson R B, Dennis P B, Thomas G. Rapamycin, wortmannin, and methylxanthine SQ20006 inactivate p70s6k by inducing dephosphorylation of the same subset of sites. J Biol Chem. 1995;270:21396–21403. doi: 10.1074/jbc.270.36.21396. [DOI] [PubMed] [Google Scholar]
- 34.Hara K, Yonezawa K, Kozlowski M T, Sugimoto T, Andrabi K, Weng Q P, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 35.Hara K, Yonezawa K, Weng Q P, Kozlowski M T, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 36.Hoekstra M F. Responses to DNA damage and regulation of cell cycle checkpoints by the ATM protein kinase family. Curr Opin Genet Dev. 1997;7:170–175. doi: 10.1016/s0959-437x(97)80125-6. [DOI] [PubMed] [Google Scholar]
- 37.Jefferies H B J, Thomas G. Ribosomal protein S6 phosphorylation and signal transduction. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 389–409. [Google Scholar]
- 38.Jiang Y, Chen C, Li Z, Guo W, Gegner J A, Lin S, Han J. Characterization of the structure and function of a new mitogen-activated protein kinase (p38β) J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 39.Jones R M, Branda J, Johnston K A, Polymenis M, Gadd M, Rustgi A, Callanan L, Schmidt E V. An essential E box in the promoter of the gene encoding the mRNA cap-binding protein (eukaryotic initiation factor 4E) is a target for activation by c-myc. Mol Cell Biol. 1996;16:4754–4764. doi: 10.1128/mcb.16.9.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Khaleghpour, K. Unpublished observations.
- 40.Kohn A D, Barthel A, Kovacina K S, Boge A, Wallach B, Summers S A, Birnbaum M J, Scott P H, Lawrence J C, Roth R A. Construction and characterization of a conditionally active version of the serine/threonine kinase Akt. J Biol Chem. 1998;273:11937–11943. doi: 10.1074/jbc.273.19.11937. [DOI] [PubMed] [Google Scholar]
- 41.Kohn A D, Takeuchi F, Roth R A. Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J Biol Chem. 1996;271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 42.Koromilas A E, Roy S, Barber G N, Katze M G, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 43.Kyriakis J M, Avruch J. pp54 microtubule-associated protein 2 kinase. A novel serine/threonine protein kinase regulated by phosphorylation and stimulated by poly-l-lysine. J Biol Chem. 1990;265:17355–17363. [PubMed] [Google Scholar]
- 44.Lane H A, Thomas G. Purification and properties of mitogen-activated S6 kinase from rat liver and 3T3 cells. Methods Enzymol. 1991;200:268–291. doi: 10.1016/0076-6879(91)00146-n. [DOI] [PubMed] [Google Scholar]
- 45.Lawrence J, Jr, Abraham R T. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem Sci. 1997;22:345–349. doi: 10.1016/s0968-0004(97)01101-8. [DOI] [PubMed] [Google Scholar]
- 46.Lazaris-Karatzas A, Montine K S, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 47.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J C. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 48.Lin T A, Kong X, Saltiel A R, Blackshear P J, Lawrence J C. Control of PHAS-I by insulin in 3T3-L1 adipocytes—synthesis, degradation, and phosphorylation by a rapamycin-sensitive and mitogen-activated protein kinase-independent pathway. J Biol Chem. 1995;270:18531–18538. doi: 10.1074/jbc.270.31.18531. [DOI] [PubMed] [Google Scholar]
- 49.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsufuji S, Matsufuji T, Miyazaki Y, Murakami Y, Atkins J F, Gesteland R F, Hayashi S. Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell. 1995;80:51–60. doi: 10.1016/0092-8674(95)90450-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen F C, Ostergaard L, Nielsen J, Christiansen J. Growth-dependent translation of IGF-II mRNA by a rapamycin-sensitive pathway. Nature. 1995;377:358–362. doi: 10.1038/377358a0. [DOI] [PubMed] [Google Scholar]
- 52.Norbury C, Nurse P. Animal cell cycles and their control. Annu Rev Biochem. 1992;61:441–470. doi: 10.1146/annurev.bi.61.070192.002301. [DOI] [PubMed] [Google Scholar]
- 53.Pause A, Belsham G J, Gingras A C, Donzé O, Lin T A, Lawrence J, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 54.Pearson R B, Dennis P B, Han J W, Williamson N A, Kozma S C, Wettenhall R E, Thomas G. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 1995;14:5279–5287. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pegg A E. Recent advances in the biochemistry of polyamines in eukaryotes. Biochem J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poulin F, Gingras A-C, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding family. J Biol Chem. 1998;273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 57.Proud C G. p70 S6 kinase: an enigma with variations. Trends Biochem Sci. 1996;21:181–185. [PubMed] [Google Scholar]
- 58.Pullen N, Thomas G. The modular phosphorylation and activation of p70s6k. FEBS Lett. 1997;410:78–82. doi: 10.1016/s0014-5793(97)00323-2. [DOI] [PubMed] [Google Scholar]
- 59.Pyronnet S, Gingras A C, Bouisson M, Kowalski-Chauvel A, Seva C, Vaysse N, Sonenberg N, Pradayrol L. Gastrin induces phosphorylation of eIF4E binding protein 1 and translation initiation of ornithine decarboxylase mRNA. Oncogene. 1998;16:2219–2227. doi: 10.1038/sj.onc.1201748. [DOI] [PubMed] [Google Scholar]
- 60.Pyronnet S, Imataka H, Gingras A C, Fukunaga R, Hunter T, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J. 1999;18:270–279. doi: 10.1093/emboj/18.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc Natl Acad Sci USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 63.Scott P H, Brunn G J, Kohn A D, Roth R A, Lawrence J C. Evidence of insulin-stimulated phosphorylation and activation of the mammalian target of rapamycin mediated by a protein kinase B signalling pathway. Proc Natl Acad Sci USA. 1998;98:7772–7777. doi: 10.1073/pnas.95.13.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shantz L M, Pegg A E. Overproduction of ornithine decarboxylase caused by relief of translational repression is associated with neoplastic transformation. Cancer Res. 1994;54:2313–2316. [PubMed] [Google Scholar]
- 65.Shockett P, Difilippantonio M, Hellman N, Schatz D G. A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice. Proc Natl Acad Sci USA. 1995;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sonenberg N. mRNA 5′ cap-binding protein eIF4E and control of cell growth. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 245–269. [Google Scholar]
- 67.Sonenberg N, Gingras A C. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr Opin Cell Biol. 1998;10:268–275. doi: 10.1016/s0955-0674(98)80150-6. [DOI] [PubMed] [Google Scholar]
- 68.Stokoe D, Stephens L R, Copeland T, Gaffney P R, Reese C B, Painter G F, Holmes A B, McCormick F, Hawkins P T. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 69.Tabor C W, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 70.Thomas G, Hall M N. TOR signalling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 71.Ueki K, Yamamoto-Honda R, Kaburagi Y, Yamauchi T, Tobe K, Burgering B M, Coffer P J, Komuro I, Akanuma Y, Yazaki Y, Kadowaki T. Potential role of protein kinase B in insulin-induced glucose transport, glycogen synthesis, and protein synthesis. J Biol Chem. 1998;273:5315–5322. doi: 10.1074/jbc.273.9.5315. [DOI] [PubMed] [Google Scholar]
- 72.von Manteuffel S R, Gingras A C, Ming X F, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.von Manteuffel S R, Dennis P B, Pullen N, Gingras A-C, Sonenberg N, Thomas G. The insulin-induced signalling pathway leading to S6 and initiation factor 4E binding protein 1 phosphorylation bifurcates at a rapamycin-sensitive point immediately upstream of p70s6k. Mol Cell Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waskiewicz A J, Flynn A, Proud C G, Cooper J A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16:1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weng Q P, Kozlowski M, Belham C, Zhang A, Comb M J, Avruch J. Regulation of the p70 S6 kinase by phosphorylation in vivo. J Biol Chem. 1998;273:16621–16629. doi: 10.1074/jbc.273.26.16621. [DOI] [PubMed] [Google Scholar]