Abstract
Heme plays key regulatory roles in numerous molecular and cellular processes for systems that sense or use oxygen. In the yeast Saccharomyces cerevisiae, oxygen sensing and heme signaling are mediated by heme activator protein 1 (Hap1). Hap1 contains seven heme-responsive motifs (HRMs): six are clustered in the heme domain, and a seventh is near the activation domain. To determine the functional role of HRMs and to define which parts of Hap1 mediate heme regulation, we carried out a systematic analysis of Hap1 mutants with various regions deleted or mutated. Strikingly, the data show that HRM1 to -6, located in the previously designated Hap1 heme domain, have little impact on heme regulation. All seven HRMs are dispensable for Hap1 repression in the absence of heme, but HRM7 is required for Hap1 activation by heme. More importantly, we show that a novel class of repression modules—RPM1, encompassing residues 245 to 278; RPM2, encompassing residues 1061 to 1185; and RPM3, encompassing residues 203 to 244—is critical for Hap1 repression in the absence of heme. Biochemical analysis indicates that RPMs mediate Hap1 repression, at least partly, by the formation of a previously identified higher-order complex termed the high-molecular-weight complex (HMC), while HRMs mediate heme activation by permitting heme binding and the disassembly of the HMC. These findings provide significant new insights into the molecular interactions critical for Hap1 repression in the absence of heme and Hap1 activation by heme.
Heme plays a central role in oxygen sensing and utilization in all living organisms. It not only serves as a prosthetic group in numerous enzymes and proteins that use, transport, and store oxygen, but it also directly regulates various molecular and cellular processes for systems that use or sense oxygen (18). For example, heme stimulates erythroid cell differentiation, hepatic cell differentiation, and nerve cell differentiation (24, 25). Heme promotes the transcription of genes encoding globin chains in erythroid cells and genes encoding cytochromes in hepatic cells (2, 5, 23), controls protein synthesis through the heme-regulated inhibitor (HRI) kinase in reticulocytes (3, 4), and regulates the assembly and degradation of many protein or enzyme complexes, such as hemoglobin, heme lyase, and δ-aminolevulinate (δALA) synthase (δALAS) (13, 14, 18, 26). How does heme regulate these diverse processes? Recent evidence suggests that heme regulates different processes through a short sequence motif, the heme-responsive motif (HRM) (14, 33). HRMs are found in heme-regulated proteins of diverse functions, such as the Saccharomyces cerevisiae heme-responsive transcriptional activator Hap1 (33), NF-E2 (16), HRI kinase (3, 4), the leader sequence of δALAS (14), heme lyase (26) and heme oxygenase (17).
Hap1 is a transcriptional activator whose activity is directly and stringently controlled by heme. It activates transcription of genes encoding functions required for respiration and for controlling oxidative damage in the yeast S. cerevisiae (36). As the heme concentration rises, Hap1 activity increases accordingly and reaches its limit at micromolar heme concentrations (31). Hap1 contains five known functional domains (20, 30, 33, 34): the Zn2-Cys6 cluster, the dimerization domain, the heme domain, the HRM7 domain, and the activation domain. Recent experiments show that three of these domains—the dimerization domain, the heme domain, and the HRM7 domain—are all important for heme regulation (35). Two of these heme regulatory domains contain HRMs: six are located in the heme domain, and one is in the HRM7 domain (33). Deletion of the heme domain renders Hap1 constitutively active and unresponsive to heme (6, 32, 35). Moreover, a synthetic peptide containing one HRM was shown to bind directly to heme in vitro (33). These results suggest that HRMs in Hap1 and other hemoproteins could bind to heme and mediate heme regulation. However, recent experiments from various laboratories have generated conflicting results on the functional role of HRMs. For example, while the HRMs in heme lyase are critical for heme binding and enzymatic activity (26), the HRMs in heme oxygenase and NF-E2 are not important for protein functions (16, 17). In Hap1, the role of HRMs is inferred from the existence of HRM1 to HRM6 in the heme domain, but there is no direct functional evidence to demonstrate the role of HRMs in vivo. Thus, the role of HRMs in heme regulation must be rigorously ascertained. In addition, the three heme regulatory domains span a large internal Hap1 region from amino acid position 117 to 1308, and one of these domains, the dimerization domain, does not contain any HRM (35). Therefore, very likely, unidentified modules in these domains also play important roles in heme regulation. Furthermore, previous evidence suggests that heme regulation of Hap1 involves the disassembly of a higher-order complex, termed the high-molecular-weight complex (HMC) (6, 32, 35). When the cellular heme concentration falls, Hap1 is bound by proteins, including Hsp90 and Ydj1, and forms an HMC (35). Heme disrupts the HMC and allows Hap1 to bind to DNA with high affinity (6, 32, 35). Two lines of evidence strongly support the idea that HMC formation is linked to Hap1 repression in the absence of heme. First, the HMC binds to DNA with very low affinity. As the heme concentration increases in vitro, the HMC is gradually transformed into a smaller, dimeric Hap1 complex with high DNA-binding affinity (6, 32). This transformation correlates with the gradual increase of Hap1 activity in response to higher heme concentrations in vivo (31, 32, 35). Second, previously characterized Hap1 mutants defective in HMC formation are also derepressed and gain a high level of activity in the absence of heme (35). Until now, the Hap1 elements that are critical for HMC formation and Hap1 repression have been largely unknown.
In this report, we describe a systematic functional analysis aimed at defining Hap1 elements critical for heme regulation. We have also begun to dissect the complex molecular interactions responsible for Hap1 repression in the absence of heme and for subsequent Hap1 activation by heme. We found that two classes of elements are important for heme regulation. One class is composed of three newly identified repression modules that mediate Hap1 repression in the absence of heme. Another class is composed of the previously identified HRMs, in particular HRM7, which mediate heme activation. Analysis of the HMCs formed by Hap1 mutants suggests that the repression modules mediate Hap1 repression, at least in part, by the formation of the HMC. These results provide significant new insights into mechanisms of heme regulation and transcriptional activation.
MATERIALS AND METHODS
Yeast strains and reporters.
The yeast strains used in this study were MHY200 (MATa ura3-52 leu2-3,112 his4-519 ade1-100 hem1-Δ100 hap1::LEU2) (10), L51 (MATa ura3-52 leu2-3,112 his4-519 ade1-100 hap1::LEU2 trp1::HisG) (31), and JEL1 (MATα leu2 trp1 ura3-52 nprbl-1122 pep4-3 ΔHis3::pGAL10-GAL4). The UAS1/CYC1-lacZ reporter plasmid has been described previously (27).
Construction of deletion mutants in the Hap1 heme domain.
To construct Hap1 heme domain deletion mutants, DNA fragments containing the coding sequences for amino acid residues in the corresponding regions of both the DNA-binding domain and the heme domain in HH4-HH (Fig. 1) were generated by PCR amplification, cleaved with the BamHI restriction enzyme, and inserted into the BamHI site of the Hap1 expression plasmid SD5-HAP1 (27). The expression plasmid for Δheme was generated as previously described (35). Correct clones were identified by restriction digestion and confirmed by sequencing.
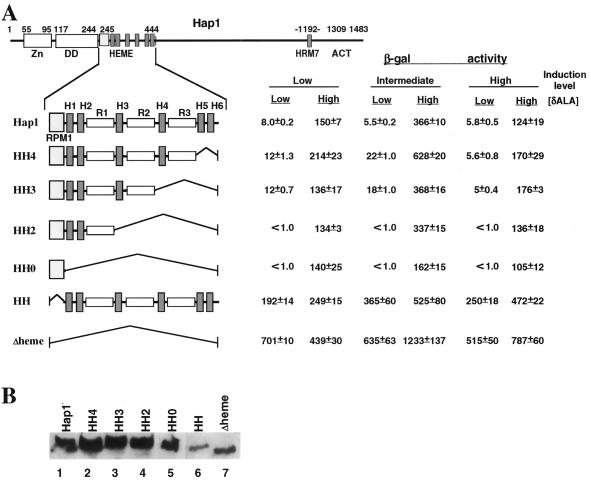
FIG. 1.
Deletion analysis of functional modules in the heme domain critical for heme regulation. (A) Structures of Hap1 derivatives with deletions in the heme domain and their transcriptional activity in the presence and absence of heme. Shown are the zinc cluster (residues 55 to 95), the dimerization domain (DD) (residues 117 to 244), the previously designated heme domain (residues 245 to 444), HRM7 (near residue 1192), and the activation domain (ACT) (residues 1309 to 1483). In the heme domain, the lightly shaded box represents RPM1 (residues 245 to 278); the heavily shaded boxes represent the six heme-responsive motifs; the open boxes represent three 17-amino-acid repeats; and the angled lines represent the deleted regions. Residues 387 to 444, 345 to 444, 321 to 444, 278 to 444, 245 to 278, and 245 to 444 are deleted in HH4, HH3, HH2, HH0, HH, and Δheme, respectively. MHY200 cells were cotransformed with an expression vector producing wild-type Hap1 or mutants and a UAS1-TATA-lacZ reporter. Cells were induced with 0.25% galactose–1.75% glucose (low expression level), 1% galactose–1% glucose (intermediate expression level), and 2% galactose (high expression level), respectively. β-Galactosidase (β-gal) activities (mean ± standard error) were detected in heme-deficient cells (low [δALA]) and heme-sufficient cells (high [δALA]). (B) Western blot showing the expression levels of wild-type Hap1 and deletion mutants in yeast cells. Yeast extracts (150 μg) containing wild-type Hap1 (lane 1), HH4 (lane 2), HH3 (lane 3), HH2 (lane 4), HH0 (lane 5), HH (lane 6), and Δheme (lane 7) were analyzed on an SDS–7% polyacrylamide gel, transferred, and detected by using an antibody against GST-Hap1. Extracts were prepared from cells induced with 2% galactose.
Generation of deletion mutants in the HRM7 domain and the dimerization domain.
To construct the expression plasmids for the Hap1 mutants with residues 447 to 1309 or 746 to 1309 deleted (H7-d1 and H7-d2, Fig. 2), the plasmid pHAP1(Δ447-1309) or pHAP1(Δ746-1309) (20) was cleaved with BstEII and KpnI. The fragment containing HAP1 sequences was then inserted into the BstEII-KpnI site of SD5-HAP1.
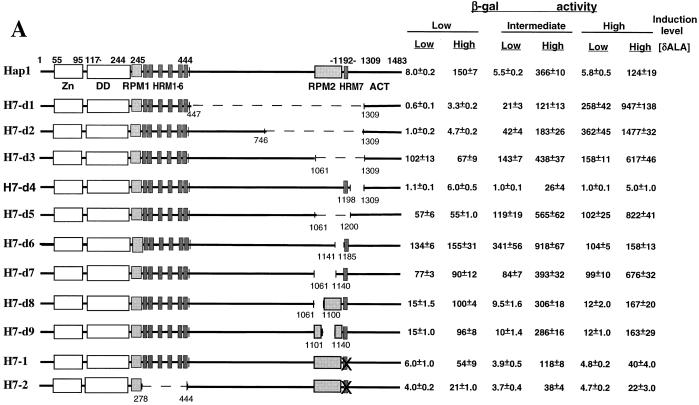
FIG. 2.
Deletion and mutational analyses of Hap1 modules in the HRM7 domain. (A) Domain structures of wild-type Hap1 and mutants and their activity in the presence and absence of heme. Shown here are the zinc cluster (Zn), the dimerization domain (DD), RPM1, HRM1 to -6, RPM2 (residues 1161 to 1185), HRM7, and the activation domain (ACT). Deleted regions are marked by dashed lines. In H7-1 and H7-2, the Cys residue in the HRM7 motif is mutated to Ala. Cells were induced with 0.25% galactose–1.75% glucose (low expression level), 1% galactose–1% glucose (intermediate expression level), and 2% galactose (high expression level), respectively. β-Galactosidase (β-gal) activities were detected in heme-deficient cells (low [δALA]) and heme-sufficient cells (high [δALA]). (B) Western blots showing the expression levels of wild-type Hap1 and mutants in yeast cells. Extracts (100 μg) containing wild-type Hap1 (lanes 1 and 8), H7-d3 (lane 2), H7-d5 (lane 3), H7-d7 (lane 4), H7-d6 (lane 5), H7-d8 (lane 6), and H7-d9 (lane 7); 50 μg of extracts containing H7-d1 (lane 9), H7-d2 (lane 10), H7-d4 (lane 11), and H7-1 (lane 12); and 20 μg of H7-2 (lane 13) were analyzed on SDS–7% polyacrylamide gels, transferred, and detected by using an antibody against GST-Hap1. The minor bands below the main band were due to Hap1 degradation. Extracts were prepared from cells induced with 2% galactose.
To construct the other deletion mutants and H7-1 shown in Fig. 2, the BglII-KpnI fragment encoding Hap1 residues 746 to 1309 from the HAP1 expression vector (SD5-HAP1) (27) was inserted into a vector derived from the Stratagene Bluescript II KS(+) vector (a BglII site was inserted into the NotI site). Single-stranded uracil DNA was then generated in the dut ung strain CJ 236 (Bio-Rad). Oligonucleotides encoding mutated amino acid residues, with 16 bases complementary to the region on either side of the deleted or mutated sequence, were annealed to the single-stranded uracil DNA. Double-stranded DNA was synthesized and transformed into the wild-type MV 1190 strain, and plasmid DNA was isolated from the transformants. Deletion mutants were identified by restriction digestion. A HaeII site was created at the mutated region in H7-1 and used to identify the mutant. BglII-KpnI fragments from confirmed mutants were inserted into SD5-HAP1 to reconstruct the expression plasmids for the mutants. Mutant H7-2 was generated by inserting the BglII-KpnI fragment released from H7-1 into the HH0 plasmid (see Fig. 1) cleaved with BglII and KpnI. Mutants were further confirmed by DNA sequencing. Mutants with deletions in the dimerization domain (see Fig. 3) were generated in the same manner except that a KS(+) vector containing the BamHI fragment of SD5-HAP1 (27) was used in the mutagenesis. Oligonucleotide sequences are available upon request.
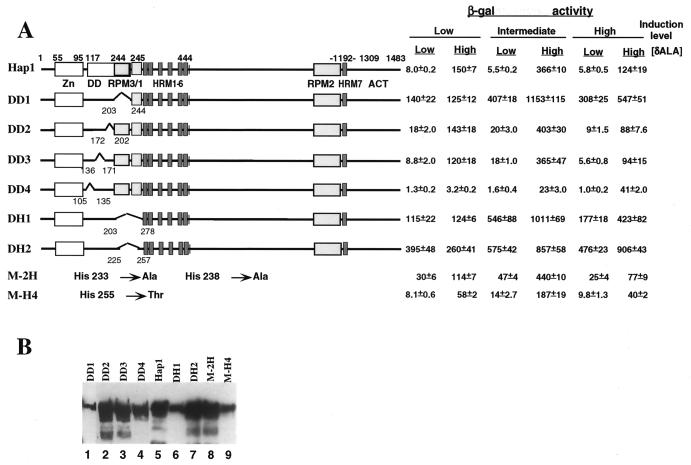
FIG. 3.
Deletion and mutational analyses of the Hap1 dimerization domain. (A) Hap1 mutants with deletions in the dimerization domain and their activity in the presence and absence of heme. Shown here are the zinc cluster (Zn), the dimerization domain (DD), RPM3 and RPM1 (RPM3/1), RPM2, HRM1 to -6, HRM7, and the activation domain (ACT). In DD1 to DD4, DH1, and DH2, residues 203 to 244, 172 to 202, 136 to 171, 105 to 135, 203 to 278, and 225 to 257 are deleted. MHY200 cells were cotransformed with an expression vector producing wild-type Hap1 or mutants and a UAS1-TATA-lacZ reporter. Cells were induced with 0.25% galactose–1.75% glucose (low expression level), 1% galactose–1% glucose (intermediate expression level), and 2% galactose (high expression level), respectively. β-Galactosidase (β-gal) activities (mean ± standard error) of wild-type Hap1 and mutants were detected in heme-deficient cells (low [δALA]) and heme-sufficient cells (high [δALA]). The mutations in the mutants M-2H and M-H4 are shown. The primary amino acid sequence of Hap1 residues 201 to 280 is SSSLSISNKY DNDELDLTKD FDLLHIKSNG TIHLGATHWL SIMKGDPYLK LLWGHIFAMR EKLNEWYYQK NSYSKLKSSK (boldface type indicates the four His residues). (B) Western blot showing the expression levels of wild-type Hap1 and mutants in yeast cells. Extracts (100 μg) containing DD2 (lane 2) and DD3 (lane 3) and 200 μg of extracts containing wild-type Hap1 (lane 5), DD1 (lane 1), DD4 (lane 4), DH1 (lane 6), DH2 (lane 7), M-2H (lane 8), and M-H4 (lane 9) were analyzed on an SDS–7% polyacrylamide gel, transferred, and detected by using an antibody against GST-Hap1. Extracts were prepared from cells induced with 2% galactose. The lower bands below the major band were due to degraded Hap1 fragments.
Preparation of yeast extracts and DNA mobility shift assays.
Extracts were prepared according to previously established protocols (32, 35). Briefly, yeast L51 or JEL1 cells bearing expression plasmids were grown to an optical density of 0.5 and induced with 2% galactose for specified times (see the figure legends). Cells were harvested and resuspended in 3 packed cell volumes of buffer (20 mM Tris, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM dithiothreitol, 0.3 M NaCl, 1 mM phenylmethylsulfonyl fluoride, pepstatin [1 μg/ml], leupeptin [1 μg/ml]). Cells were then permeabilized by agitation with 4 packed cell volumes of glass beads, and extracts were collected as described previously (21). This method consistently generated extracts with protein concentrations of approximately 10 mg/ml.
DNA-binding reactions were carried out in a 20-μl volume with 5% glycerol, 4 mM Tris (pH 8), 40 mM NaCl, 4 mM MgCl2, 10 mM dithiothreitol 3 μg of salmon sperm DNA, 10 μM ZnOAc2, and 300 μg of bovine serum albumin per ml as described previously (32). Approximately 0.01 pmol of labeled UAS1/CYC1 or UAS/CYC7 and 20 μg of protein extracts were used in each reaction. The reaction mixtures were incubated at room temperature for 1 h and then loaded onto 3.5% polyacrylamide gels in 1/3× Tris-borate-EDTA (1/3TBE) for electrophoresis at 4°C. The intensity of bands representing the HMCs and dimeric complexes was quantified by using the PhosphoImage system (Molecular Dynamics).
Western blotting.
Whole-cell extracts for Western blotting were first separated on sodium dodecyl sulfate (SDS)–7% polyacrylamide gels and then transferred to polyvinylidene difluoride or nitrocellulose membranes. Hap1 was visualized by using a purified antibody against glutathione S-transferase–Hap1 (GST-Hap1) (residues 1 to 171) and a chemiluminescence Western blotting kit (Boehringer Mannheim), as described previously (35).
β-Galactosidase assays.
To determine β-galactosidase levels from reporter genes in cells containing wild-type Hap1 and mutants in the presence and absence of heme, yeast high-copy-number 2μm replicating plasmids expressing wild-type Hap1 and mutants from the GAL1-10 promoter were transformed into the strain MHY200 (10) bearing the UAS1/CYC1-lacZ reporter. Cells were grown in synthetic complete medium containing 2% raffinose with limiting amounts of heme precursor δALA (2 μg/ml) or high amounts of δALA (250 μg/ml) to an optical density of approximately 0.5. Cells were then induced partially with 0.25% galactose–1.75% glucose or 1% galactose–1% glucose or completely with 2% galactose for 7 h and harvested for determination of β-galactosidase activity as described previously (32).
RESULTS
Deletion analysis shows that residues 245 to 278, not the HRMs in the heme domain, are responsible for mediating heme regulation.
Hap1 contains seven HRMs: six clustered in the heme domain and one near the activation domain. Interestingly, three copies of a 17-amino-acid repeat are found in the heme domain between HRM2 and -3, HRM3 and -4, and HRM4 and -5 (Fig. 1). We postulated that, in vivo, the HRMs are responsible for heme binding, whereas the 17-amino-acid repeats provide interaction surfaces for other components of the HMC to bind and repress Hap1 in the absence of heme. We thought that all these HRMs and the 17-amino-acid repeats would collectively contribute to a high degree of heme regulation. If this model is correct, the extent of Hap1 heme responsiveness would gradually diminish with the progressive deletion of the HRMs and 17-amino-acid repeats. We therefore examined the effect of deletion of the HRMs and 17-amino-acid repeats on Hap1 heme responsiveness. However, in vivo analysis of Hap1 mutants defective in heme regulation is difficult because of their toxicity under heme-deficient conditions (20, 33). None of the previously generated Hap1 deletion mutants was examined under heme-deficient conditions (20, 33), so the extent of heme responsiveness of these mutants could not be determined. To overcome this problem, we expressed wild-type and Hap1 mutants from the inducible GAL1-10 promoter, which is not heme regulated (9). However, the inducible GAL1-10 promoter is a strong promoter. The full induction of this promoter can lead to the overexpression of the Hap1 proteins, which may minimize the effects of mutations on heme regulation. We therefore examined the Hap1 proteins at a low expression level, an intermediate expression level, and a high expression level (27). The low expression level was achieved by adding 0.25% galactose–1.75% glucose to the culture grown on noninducing, nonrepressing raffinose medium. Under this condition, the activity of the GAL1-10 promoter-lacZ reporter is indistinguishable from the basal level (8, 12). Intermediate and high expression levels were achieved by adding 1% galactose–1% glucose and 2% galactose to the cultures, respectively (27). Under the condition for the intermediate expression level, the activity of the GAL1-10 promoter is about 5% of that under the high (full) induction condition.
As expected, at all expression levels, Hap1 activity was low under heme-deficient conditions (as described in the legend to Fig. 1A). In heme-sufficient cells, Hap1 activity was highest at the intermediate expression level, very likely because this Hap1 level was high enough to achieve maximum transcriptional activity but not enough to cause squelching (7), which might occur when Hap1 was expressed at a high level. In fact, in heme-sufficient cells, most mutants showed the highest activity when they were expressed at an intermediate level (Fig. 1 and later figures). Nevertheless, overexpressing wild-type Hap1 or mutants did not significantly change the extent of heme responsiveness (Fig. 1A and later figures). Strikingly, progressive deletion and even complete deletion of the HRMs and 17-amino-acid repeats did not abolish Hap1 repression in the absence of heme (low [δALA]) (HH4 to HH0 in Fig. 1A), or Hap1 activation by heme (high [δALA]). HH2 and HH0 were even more heme responsive: they were much more repressed than wild-type Hap1 but were activated by heme to nearly the same level as wild-type Hap1. These results suggest that HRM1 to -6 and 17-amino-acid repeats in the heme domain are not responsible for heme regulation.
However, deletion of residues 245 to 278 (HH in Fig. 1A) outside HRM1 to -6 and 17-amino-acid repeats had a drastic effect on heme regulation: at all expression levels, HH activity in the absence of heme was at least as high as that of wild-type Hap1 in the presence of heme (compare HH and Hap1 in Fig. 1A), suggesting that the mutant was completely derepressed in the absence of heme. Deletion of the whole heme domain (Δheme) also caused the complete derepression of Hap1. It is not clear why, at a low expression level, the activity of Δheme was higher in the absence of heme than in the presence of heme. In fact, all derepressed deletion mutants (Fig. 1; also see Fig. 2 and 3) were barely stimulated by heme at the low expression level. To rule out the possibility that different protein levels, caused by variations in the stability of mutants, contributed to the derepression of HH and Δheme and to varying transcriptional activities of other mutants, we carried out Western blotting analysis. The amounts of wild-type Hap1 or mutants were detectable by Western blotting only at the high expression level, but we expected the relative amounts to remain approximately the same at all expression levels. As shown in Fig. 1B, fusions from HH4 to HH0 were expressed at a level identical to wild-type Hap1, suggesting that the gradual decrease in the activity of these mutants was not attributable to the difference in protein levels. Furthermore, the constitutive mutants HH and Δheme were produced at a lower level than that of wild-type Hap1 or other mutants, suggesting that the high transcriptional activity of HH and Δheme was not due to a high protein level. These results demonstrate that residues 245 to 278, not the HRMs or 17-amino-acid repeats, are responsible for Hap1 repression in the absence of heme. The region encompassing residues 245 to 278 is named repression module 1 (RPM1) (Fig. 1A) hereafter in this work.
Functional analysis of the HRM7 domain shows that the module encompassing residues 1061 to 1185, not HRM7, is critical for Hap1 repression in the absence of heme.
Previous studies identified an HRM (HRM7) containing amino acid residue 1192. Mutations of amino acid residues near HRM7 affect Hap1 heme responsiveness (10, 33), suggesting that the domain encompassing HRM7 is important for heme regulation of Hap1. However, this domain contains a large region with very little informative sequence or few structural characteristics, except for HRM7. We therefore determined which parts of this region contribute to heme regulation of Hap1 by systematically testing various deletions and mutations in this domain for their effects on Hap1 activity (Fig. 2). As shown in Fig. 2A, mutants of the HRM7 domain were analyzed under low, intermediate, and high expression levels (Fig. 2A). At the low expression level, H7-d1, with residues 447 to 1309 deleted, and H7-d2, with residues 746 to 1309 deleted, were much less active than wild-type Hap1 or mutants with smaller deletions, probably because these large deletions significantly perturbed the protein structure (note that these proteins were stably expressed as shown by Western analysis [Fig. 2B]). At the intermediate expression level, these mutants showed a moderate level of activity both in the absence and presence of heme, and heme stimulated their activity about four- to sixfold (Fig. 2A). At the high expression level, these two mutants were derepressed in the absence of heme. They showed an activity in the absence of heme higher than that of wild-type Hap1 in the presence of heme (Fig. 2A). Heme hyperstimulated the mutants’ activity three- to fivefold. To pinpoint the residues critical for heme regulation, we deleted a smaller region, residues 1061 to 1309 (H7-d3). At all expression levels, H7-d3 was derepressed in the absence of heme. At the intermediate or high expression level, H7-d3 was further stimulated about fourfold by heme. The results show that the residues critical for Hap1 repression in the HRM7 domain are located within the region encompassing residues 1061 to 1309.
To narrow down the critical residues, mutants with smaller deletions were generated. Deletion of residues 1061 to 1200 (H7-d5) caused Hap1 derepression in the absence of heme, while deletion of residues 1198 to 1309 (H7-d4) abolished Hap1 activity even in the presence of heme (Fig. 2A). The region encompassing residues 1061 to 1200 was further divided into two segments and deleted separately. Deletion of residues 1061 to 1140 (H7-d7) (Fig. 2A) caused Hap1 derepression in the absence of heme. At intermediate and high expression levels, the mutant was hyperinduced by heme by fourfold. Interestingly, deletion of residues 1141 to 1185 (H7-d6) (Fig. 2A), immediately adjacent to the HRM7 motif, also caused the full derepression of Hap1. These results show that residues 1061 to 1140 and residues 1141 to 1185 are both critical for Hap1 repression in the absence of heme. We therefore defined the region encompassing residues 1061 to 1185 as RPM2. Residues 1061 to 1140 in RPM2 were further divided into two parts, residues 1061 to 1100 (H7-d8) (Fig. 2A) and residues 1101 to 1140 (H7-d9) (Fig. 2A), and each part was deleted separately. Notably, these deletions did not cause Hap1 to be derepressed, suggesting that these parts provide redundant functions in Hap1 repression. Western blotting analysis showed that all mutants were produced at levels similar to the wild-type level (Fig. 2B). The small variation of protein levels among mutants did not correlate with the variation in the mutant activity (Fig. 2). For example, the constitutive mutants H7-d1, H7-d2, H7-d3, H7-d5, and H7-d7 were expressed at a slightly lower level than the totally inactive mutant H7-d4 and the repressed mutants H7-d8 and H7-d9. Therefore, heme responsiveness of these mutants was not dictated by their expression levels. These experiments show that not the HRM7 motif but rather RPM2, which contains residues 1061 to 1185, is critical for Hap1 repression in the absence of heme. This module appears to be composed of three parts: residues 1061 to 1100, residues 1101 to 1140, and residues 1141 to 1185. Residues 1141 to 1185 can cooperate with either residues 1061 to 1100 (in H7-d9) or residues 1101 to 1140 (in H7-d8) to repress Hap1 in the absence of heme.
Evidently, HRM7 is not responsible for Hap1 repression. To further investigate the role of the HRM7 motif in heme regulation, we generated a mutant containing a Cys-to-Ala change at position 1193. This Cys residue within the HRM7 motif is critical for heme binding in vitro (33). As shown in Fig. 2A, mutating HRM7 (mutant H7-1) did not affect Hap1 repression in the absence of heme but reduced its activity in the presence of heme, at all expression levels. The reduction in heme activation was further reinforced in the double mutant H7-2 (Fig. 2A), with HRM1 to -6 deleted and HRM7 mutated. Not surprisingly, H7-2 showed an even lower activity than H7-1 in the presence of heme but remained repressed in the absence of heme. The low activity of H7-1 and H7-2 was not attributable to low protein expression levels, because Western blotting analysis showed that these mutants were produced at an even higher level than that of wild-type Hap1 (Fig. 2B). These results suggest that the HRM7 motif has no role in repressing Hap1 in the absence of heme but plays a role in heme activation of Hap1 or transcriptional activation by Hap1 (see also below).
A module encompassing residues 203 to 244 in the dimerization domain is critical for Hap1 repression in the absence of heme.
Previous experiments suggest that the Hap1 dimerization domain is also important for heme regulation (35). However, it is not clear from these experiments whether the entire dimerization domain or just a segment of the domain is important for heme regulation. We therefore divided the dimerization domain into four segments and determined whether deletion of each segment affects Hap1 heme responsiveness. These segments contain residues 203 to 244, 172 to 202, 136 to 171, and 105 to 135, respectively. As shown in Fig. 3A, deletion of residues 203 to 244 (mutant DD1) caused the complete derepression of Hap1. At all expression levels, the activity of DD1 in the absence of heme was nearly as high as or higher than that of wild-type Hap1 in the presence of heme, even though the protein level of DD1 was much lower than that of wild-type Hap1 (Fig. 3). Deletion of residues 172 to 202 (DD2) or 136 to 171 (DD3) did not considerably affect Hap1 repression or activation by heme. Deletion of residues 105 to 135 (DD4) caused Hap1 activity to be lower, both in the presence and absence of heme. Because heme significantly stimulated the activity of DD4 at the intermediate (14-fold) or high (41-fold) expression level, the low activity in heme-sufficient cells was very likely caused by the low transcriptional activity of the mutant, not by deficiency in heme activation. Again, Western blotting analysis showed that all deletion mutants except for the derepressed mutant DD1 were produced at a level similar to the wild-type Hap1 level (Fig. 3B). These results show that the segment containing residues 203 to 244, termed RPM3 hereafter in this work, is critical for Hap1 repression.
RPM3, which contains residues 203 to 244, is contiguous to RPM1, which contains residues 245 to 278 (Fig. 3A). These two modules may closely cooperate to mediate heme regulation. Inspection of the sequences in these regions revealed four His residues: His 225, His 233, His 238, and His 255 (as described in the legend to Fig. 3). Because His residues are often involved in heme binding in hemoglobin and certain cytochromes, we speculated that these four residues might contribute to heme binding and, hence, heme regulation. If so, then deletion of the region containing these four His residues would abolish heme regulation. As shown in Fig. 3A, deletion of residues 225 to 257 (DH2), like deletion of residues 203 to 278 containing both RPM1 and RPM2 (DH1), caused Hap1 derepression at all expression levels, although the protein levels of these mutants appeared to be lower than that of wild-type Hap1 (Fig. 3B). DH2 had a slightly higher activity, both in the presence and absence of heme, than did DD1 and DH1, probably because its protein level was slightly higher (Fig. 3B). These results show that the region containing the four His residues is indeed critical for Hap1 repression in the absence of heme. To explore further the possibility that the four His residues are critical for heme regulation, we generated two mutants: mutant M-2H with His 233 and His 238 changed to Ala and mutant M-H4 with His 255 changed to Thr. Mutant M-2H was partially derepressed in the absence of heme and further stimulated four- to eightfold by heme, at all expression levels (Fig. 3A). Mutation of the His 255 to Thr in mutant M-H4 had no effect on Hap1 repression in the absence of heme, but its activity was lower in the presence of heme, probably because its protein level was lower (Fig. 3B). These results suggest that His 233 and His 238 are important but are not the only residues responsible for heme regulation.
Some derepressed mutants are defective in forming HMC in the absence of heme.
Our in vivo functional analysis of Hap1 mutants identified three new repression modules, RPM1 to -3, that are important for Hap1 repression in the absence of heme. Previous experiments suggest that a higher-order complex, termed HMC, plays an important role in Hap1 repression (6, 32, 35). If RPMs mediate Hap1 repression by permitting HMC formation, then derepressed mutants would be defective in HMC formation. Therefore, we determined whether the mutants described in Fig. 1 to 3 can form the HMC. Previously, it was shown that in the absence of heme, Hap1 forms the HMC even when its expression level is low (but high enough for detecting the HMC) (6, 32). As the Hap1 concentration increases, the amount of the HMC gradually increases and reaches the highest level when non-Hap1 proteins in the HMC become limiting. Excess Hap1 forms the dimeric complex even in the absence of heme (6, 32). Under the conditions for the low and intermediate expression levels used for β-galactosidase assays, the amounts of wild-type or mutant Hap1 proteins were not sufficient for the detection of the HMC or the dimeric complex (27). We therefore expressed various amounts of wild-type and mutant Hap1 proteins by varying the induction times, as previously described (Fig. 4A) (6, 32). As shown in previous studies (6, 32) and in Fig. 4A, when the Hap1 amount was low, the HMC, not the dimeric complex, was formed in the absence of heme (Fig. 4A, lane 1 [only three different amounts are shown in Fig. 4A for brevity]). Heme disrupted the HMC and permitted Hap1 to bind to DNA with much higher affinity (lane 2). As the Hap1 amount increased, more HMC formed (lanes 3 and 5). The dimeric complex also formed in the absence of heme at the highest Hap1 level (lane 5).
FIG. 4.
The effect of protein expression levels on the formation of the HMC. (A) Formation of the HMC and the dimeric complex by increasing amounts of wild-type Hap1. Yeast cells bearing the expression plasmid for wild-type Hap1 under the control of the GAL1-10 promoter grown in 2% raffinose were induced with 2% galactose for 1 h (lanes 1 and 2), 4 h (lanes 3 and 4), and 8 h (lanes 5 and 6). Extracts were prepared from these cells and analyzed in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of heme by DNA mobility shift assays on 3.5% polyacrylamide gels. (B) Formation of the HMC and dimeric complex by increasing amounts of H7-d8. Yeast cells bearing the expression plasmid for H7-d8 under the control of the GAL1-10 promoter grown in 2% raffinose were induced with 2% galactose for 1 h (lanes 1 and 2), 2 h (lanes 3 and 4), and 4 h (lanes 5 and 6). Extracts were prepared from these cells and analyzed in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of heme by DNA mobility shift assays. Note that H7-d8, like wild-type Hap1, also formed the dimeric complex in the absence of heme when extracts were prepared from cells induced with galactose for longer periods (Fig. 5B). (C) Formation of the HMC and dimeric complex by increasing amounts of Δheme. Yeast cells bearing the expression plasmid for Δheme under the control of the GAL1-10 promoter grown in 2% raffinose were induced with 2% galactose for 1 h (lanes 1 and 2), 2 h (lanes 3 and 4), and 4 h (lanes 5 and 6). Extracts were prepared from these cells and analyzed in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of heme by DNA mobility shift assays. (D) Formation of the HMC and dimeric complex by increasing amounts of H7-d6. Yeast cells bearing the expression plasmid for H7-d6 under the control of the GAL1-10 promoter grown in 2% raffinose were induced with 2% galactose for 1 h (lanes 1 and 2), 2 h (lanes 3 and 4), and 4 h (lanes 5 and 6). The band below the dimeric complex was very likely due to degradation of the mutant protein. Extracts were prepared from these cells and analyzed in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of heme by DNA mobility shift assays.
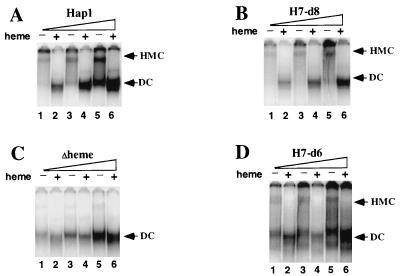
If HMC formation is required for Hap1 repression in the absence of heme, then repressed mutants would form the HMC at the same level as wild-type Hap1, whereas derepressed mutants would have reduced levels of HMC. Indeed, as shown in Fig. 4, the repressed mutant H7-d8, like wild-type Hap1, formed more HMC as its amount increased (Fig. 4B) (note that H7-d8 was not overexpressed enough to form the dimeric complex in the absence of heme; after a longer induction period, as shown in Fig. 5B, H7-d8 did form the dimeric complex). However, unlike wild-type Hap1 and H7-d8, the Δheme mutant, which was derepressed approximately 100-fold relative to wild type Hap1 (Fig. 1A), formed very little HMC (about 2% of that formed by wild-type Hap1) (compare lanes 1, 3, and 5 in Fig. 4C to those in Fig. 4A). Instead, even in the absence of heme, a significant amount of the dimeric complex was formed at all expression levels. Similarly, another derepressed mutant, H7-d6, which was derepressed approximately 20- to 50-fold relative to wild-type Hap1 (Fig. 2A), formed significantly less HMC (about 10% that of wild-type formed by Hap1) at all expression levels (compare Fig. 4D to 4A). H7-d6 also formed the dimeric complex in the absence of heme, even when its amount was low (Fig. 4D, lanes 1 and 2). These titration experiments show that in the absence of heme, wild-type Hap1 and the repressed H7-d8 mutant preferentially formed the HMC whereas the derepressed Δheme and H7-d6 mutants preferentially formed the dimeric complex.
FIG. 5.
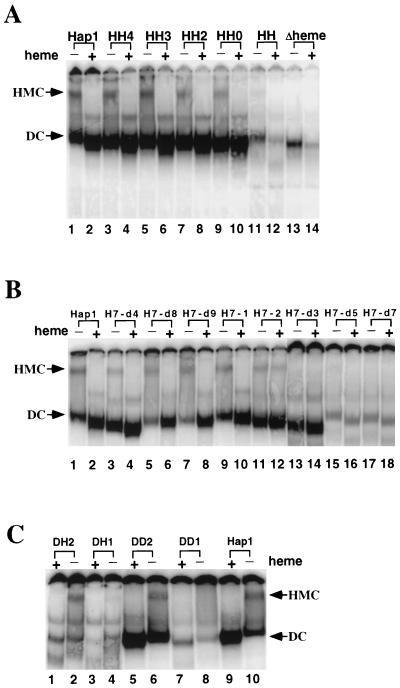
Analysis of DNA-binding complexes formed by wild-type Hap1 and mutants. (A) DNA-binding complexes formed by deletion mutants in the heme domain. Extracts containing wild-type Hap1 (lanes 1 and 2), HH4 (lanes 3 and 4), HH3 (lanes 5 and 6), HH2 (lanes 7 and 8), HH0 (lanes 9 and 10), HH (lanes 11 and 12), and Δheme (lanes 13 and 14) were incubated with radiolabeled DNA in the absence (lanes 1, 3, 5, 7, 9, 11, and 13) or presence (lanes 2, 4, 6, 8, 10, 12, and 14) of heme. The reaction mixtures were analyzed on a 3.5% polyacrylamide gel. (B) DNA-binding complexes formed by deletion mutants in the HRM7 domain. Extracts containing Hap1 (lanes 1 and 2), H7-d4 (lanes 3 and 4), H7-d8 (lanes 5 and 6), H7-d9 (lanes 7 and 8), H7-1 (lanes 9 and 10), H7-2 (lanes 11 and 12), H7-d3 (lanes 13 and 14), H7-d5 (lanes 15 and 16), and H7-d7 (lanes 17 and 18) were incubated with radiolabeled DNA in the absence (lanes 1, 3, 5, 7, 9, 11, 13, 15, and 17) or presence (lanes 2, 4, 6, 8, 10, 12, 14, 16, and 18) of heme. H7-d1 and H7-d2 are not shown here because no distinctive bands were observed when they were subjected to DNA mobility shift assays. (C) DNA-binding complexes formed by deletion mutants in the dimerization domain. Extracts containing DH2 (lanes 1 and 2), DH1 (lanes 3 and 4), DD2 (lanes 5 and 6), DD1 (lanes 7 and 8), and wild-type Hap1 (lanes 9 and 10) were incubated with radiolabeled DNA in the presence (lanes 1, 3, 5, 7, and 9) or absence (lanes 2, 4, 6, 8, and 10) of heme. DD3 and DD4 behaved the same as DD2 and are not shown here. The positions of the HMC and dimeric complex (DC) are marked. All extracts used here were prepared from JEL1 cells bearing expression plasmids induced with 2% galactose for 8 h.
These and previous experiments (6, 32) show that overexpressing Hap1 does not interfere with HMC formation but optimizes the sensitivity of detection of the HMC. Therefore, to determine whether other mutants can form the HMC, we analyzed extracts prepared from cells expressing high levels of mutants. Although this approach cannot detect subtle differences in the HMC, it provides the optimum conditions for determining whether a mutant can form the HMC. If a mutant cannot form the HMC when overexpressed, it is unlikely to form the HMC when expressed at lower levels. As shown in Fig. 5, several derepressed mutants—HH and Δheme (Fig. 5A); H7-d3, H7-d5, and H7-d7 (Fig. 5B); and DD1 and DH1 (Fig. 5C)—did not form a discernible amount of the HMC under the test condition (the signal at the position of the HMC band for the mutants was less than 5% of that for wild-type Hap1). Instead, they formed the dimeric complex and bound strongly to DNA in the absence of heme. Heme did not stimulate DNA binding by these mutants; it actually weakened DNA binding by HH and Δheme (Fig. 5A). DNA binding by DD1 was stimulated slightly by heme, but no discernible HMC band was detected (Fig. 5C, lane 8). (H7-d1 and H7-d2 are not shown here because no distinctive bands were observed when extracts containing these mutants were analyzed, whether or not heme was present.) In sum, eight mutants, seven shown in Fig. 5 and H7-d6 shown in Fig. 4D, appeared to preferentially form the dimeric complex rather than the HMC. This suggests that they are defective or partially defective in HMC formation, although it is still possible that they can form the HMC if their protein levels in extracts are made as high as that of wild-type Hap1 (the protein levels of derepressed mutants in extracts were usually lower than that of wild-type Hap1 [Fig. 1B, 2B, and 3B]). One derepressed mutant (DH2) (Fig. 5C) formed a stable HMC. All repressed mutants formed the HMC (see mutants HH4 to HH0 in Fig. 5A; mutants H7-d4, H7-d8, H7-d9, H7-1, and H7-2 in Fig. 5B; and mutant DD2 in Fig. 5C; other mutants of the dimerization domain behaved identically to DD2 [not shown]). The intensity of HMCs formed by these mutants was about the same (70 to 120%) as that formed by wild-type Hap1. These results suggest that repressed mutants always retain the ability to form the HMC, while many but not all derepressed mutants are defective in forming the HMC.
A higher heme concentration is required to disrupt the HMC when the HRM7 motif is mutated.
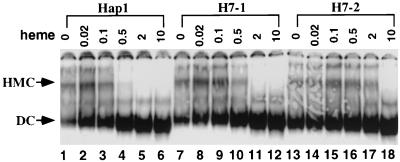
Mutants H7-1 and H7-2 had a much lower activity than wild-type Hap1 even in heme-sufficient cells, but these mutants were expressed at an even higher level than wild-type Hap1 (Fig. 2B). Thus, the reduced activity most likely results from hyperrepression caused by an HMC insensitive to heme or the loss of transcription-activating activity of the activation domain. Since the activation domain usually acts independently of other domains and since mutating amino acid residues to Ala often does not cause drastic changes in protein conformation (29), it is unlikely that the mutations in H7-1 and H7-2 weakened the activation domain. However, HRMs can bind to heme directly in vitro (33), and Fig. 5B reveals that the HMC formed by H7-2 was not totally disrupted by heme (lane 12). Therefore, it is conceivable that H7-1 and H7-2 might be defective in heme binding, thereby forming HMCs less sensitive to heme and, therefore, hyperrepressed. If this line of reasoning is correct, then H7-1, which was more repressed than wild-type Hap1 (Fig. 2), would form an HMC that is more resistant to heme than the HMC formed by wild-type Hap1. Likewise, H7-2, which was more repressed than H7-1, would form an HMC that is even more resistant to heme than the HMC formed by H7-1. This is indeed the case as shown in Fig. 6. The HMC formed by wild-type Hap1 required only 0.1 to 0.5 ng of heme per μl for dissociation (Fig. 6, lanes 1 to 6), and H7-1 required 0.5 to 2 ng/μl (Fig. 6, lanes 7 to 12), while H7-2 required 2 to 10 ng/μl for dissociation (Fig. 6, lanes 13 to 18). These results suggest that H7-1 forms a more stable HMC than wild-type Hap1 and H7-2 forms an even more stable HMC than H7-1. We also attempted to determine whether the activity of H7-1 and H7-2 can be enhanced by the addition of high levels of the heme precursor, deuteroporphyrin IX, or δALA. However, we did not observe any significant increase under high concentrations of deuteroporphyrin IX or δALA, very likely because heme concentration in vivo cannot reach the same level as in vitro due to the characteristics of enzymes involved in heme synthesis. Nonetheless, the data on H7-1 and H7-2 shown in Fig. 2 and 6, together with the previous finding that a peptide containing an HRM can bind heme directly, strongly support the idea that HRMs can bind to heme and mediate heme activation of Hap1 in vivo.
FIG. 6.
Effect of various heme concentrations on the HMCs formed by wild-type Hap1, H7-1, and H7-2. Extracts containing wild-type Hap1 (lanes 1 to 6), H7-1 (lanes 7 to 12), and H7-2 (lanes 13 to 18) were incubated with radiolabeled DNA under the following heme concentrations (in nanograms per microliter): 0 (lanes 1, 7, and 13), 0.02 (lanes 2, 8, and 14), 0.1 (lanes 3, 9, and 15), 0.5 (lanes 4, 10, and 16), 2 (lanes 5, 11, and 17) and 10 (lanes 6, 12, and 18). The reaction mixtures were analyzed on a 3.5% polyacrylamide gel. The positions of the HMC and dimeric complex (DC) are marked.
DISCUSSION
This report provides significant new insights into the mechanism by which Hap1 is repressed in the absence of heme and is subsequently activated by heme. Three themes emerge from the analyses of various Hap1 mutants. First, three novel repression modules, RPM1 to -3, located in the previously designated dimerization domain, the heme domain, and the HRM7 domain, are responsible for Hap1 repression in the absence of heme. Second, the repression modules RPM1 to -3 mediate Hap1 repression, at least in part, through the formation of the HMC. Third, HRMs, in particular HRM7, mediate heme activation of Hap1, evidently by permitting heme binding and the disassembly of the HMC. These themes are further discussed below.
Novel modules critical for Hap1 repression in the absence of heme.
Three novel repression modules, RPM1 to -3, not the previously identified HRMs, are critical for Hap1 repression in the absence of heme. The first module, RPM1 (Fig. 1), lies within residues 245 to 278 in the previously designated heme domain. The second module, RPM2 (Fig. 2), lies within residues 1061 to 1185 near the HRM7 motif. The third module, RPM3 (Fig. 3A), lies within the previously designated dimerization domain and encompasses residues 203 to 244. RPM1 to -3 act synergistically to repress Hap1 activity in the absence of heme because deletion of any one of these modules caused Hap1 derepression in the absence of heme (see Fig. 1 to 3). Intriguingly, the data show that the distinctive HRM1 to -6 and the three 17-amino-acid repeats in the previously designated heme domain have no effect on Hap1 repression in the absence of heme, although HRM1 to -6 appear to play a minor role in heme activation (see below).
Possible mechanisms by which RPMs mediate Hap1 repression.
Previous experiments linked HMC formation to Hap1 repression (35). The data here show that RPM1 to -3 are essential for Hap1 repression. We imagine two possible mechanisms by which RPMs can confer Hap1 repression. One possibility is that RPMs interact with Hap1 DNA-binding and/or activation domains, thereby masking and repressing the DNA-binding and/or activation domains by intramolecular interactions. Deletion of any RPM would therefore disrupt the interactions and result in unmasking of these domains, leading to Hap1 derepression. Alternatively, RPM1 to -3 may be involved in HMC formation and thus repress Hap1. Deletion of any RPM would then disrupt the HMC, causing Hap1 derepression. The data suggest that RPMs are at least partially involved in forming the HMC (see below), but do not rule out the possibility that intramolecular interactions are important for Hap1 repression. These two possibilities are not mutually exclusive. Perhaps both intramolecular interactions and HMC formation play important roles in Hap1 repression.
Previous reports (6, 32) and data shown in Fig. 4 show that in the absence of heme, as the amount of wild-type Hap1 and H7-d8 increased, the HMC was formed first, followed by the dimeric complex, which formed when the HMC became saturated. Unlike wild-type Hap1 and H7-d8, 8 of 11 derepressed mutants—including HH and Δheme (Fig. 5A); H7-d3, H7-d5, and H7-d7 (Fig. 5B); H7-d6 (Fig. 4D); and DH1 and DD1 (Fig. 5C)—preferentially formed the dimeric complex, whether or not the HMC was observed. Therefore, we conclude that the derepressed mutants are defective in forming the HMC. One argument against this idea is that the protein levels of many derepressed mutants may be lower than those of wild-type Hap1 and repressed mutants. Thus, the potential HMCs formed by derepressed mutants could be more difficult to detect. However, in the absence of heme, the derepressed mutants formed a significant amount of dimeric complex (Fig. 4C and D and 5). For wild-type Hap1 (6, 32) and the repressed mutant H7-d8 (Fig. 4A and B and 5C), this larger amount of dimeric complex was formed only when they were overexpressed and when the HMC became saturated. Although these derepressed mutants may still form the HMC if the protein levels were higher, they appeared to preferentially form the dimeric complex and were at least partially defective in forming the HMC. Another argument is that the derepressed mutants may form defective HMCs that cannot bind to DNA, making their detection difficult. Our data cannot totally exclude this possibility, but we do not think it is likely for the following reasons. First, many derepressed mutants that showed reduced dimer binding also had a lower protein level (HH and Δheme [Fig. 1B and 5A], H7-d5 and H7-d7 [Fig. 2B and 5B], and DH1 and DD1 [Fig. 3B and 5C]). Therefore, the reduced dimer binding was very likely due to reduced protein levels, not due to reduced affinity. Second, previous and recent data show that Hap1 mutations similarly affect HMC binding and Hap1 dimer binding and that the HMC and Hap1 dimer recognize various DNA sites in the same manner (11, 32). It is therefore unlikely that mutations in the derepressed mutants selectively affected HMC binding but not dimer binding. On the basis of these arguments, the conclusion that these derepressed mutants are at least partially defective in forming the HMC appears to be plausible. This deficiency may be in part responsible for the loss of repression of these mutants. It is not clear why DH2 (Fig. 5C) is derepressed even though it forms the HMC. Perhaps the HMC formed by DH2 adopts an altered conformation so that Hap1 can activate transcription even when associated with other proteins. This occurs in the Gal4-Gal80 regulatory system in which activated Gal4 is still associated with its repressor Gal80 (1, 15, 19, 22). Alternatively, perhaps the DNA-binding and activation domains of DH2 are totally unmasked even though the HMC is intact.
Regardless of whether RPMs act by permitting HMC formation, the data clearly show that the three RPMs act cooperatively in repressing Hap1. Interestingly, protein structure analysis indicates that residues 248 to 267 in RPM1 have a high potential of forming an α-helix. The helical-wheel representation of this module indicates that most of the hydrophobic residues are clustered on one face of the helix. This hydrophobic surface may interact with other proteins in the HMC or the Hap1 DNA-binding and activation domains, thereby repressing Hap1. In addition, residues 1089 to 1111 and residues 1110 to 1134 in RPM2 have a strong tendency to form an α-helix. These structural modules may provide important intramolecular or intermolecular interactions critical for repressing Hap1.
Role of HRMs in heme regulation.
Our data show that HRMs have no impact on Hap1 repression in the absence of heme (Fig. 1 and 2). Rather, the HRMs, in particular HRM7, are important for mediating heme activation. HRM7 appeared to play a predominant role in mediating heme activation. HRM7 alone (HH0 in Fig. 1A), but not HRM1 to -6 (H7-1 in Fig. 2A), was sufficient for mediating efficient activation of Hap1 by heme. HRM1 to -6 appeared to play an auxiliary role in mediating heme activation, which was revealed only when HRM7 was mutated in H7-2 (compare HH0 with wild-type Hap1 in Fig. 1A and H7-2 with H7-1 in Fig. 2A). The analysis of HMCs formed by H7-1 and H7-2 (Fig. 6) further suggests that HRMs mediate heme activation by allowing heme binding and subsequent disassembly of the HMC. H7-1 lacks a functional HRM7, so it would bind to heme less efficiently than wild-type Hap1. Consequently, the mutant HMC required a higher heme concentration to be disrupted (Fig. 6), leading to hyperrepression in vivo (Fig. 2A). H7-2 lacks both HRM7 and the six HRMs in the heme domain, so it would bind to heme even less efficiently than H7-1. As a result, the mutant HMC required an even higher heme concentration to be disrupted (Fig. 6) and was even more repressed than H7-1 (Fig. 2A). HRMs do not appear to be the only element responsible for heme binding and activation, because the mutant H7-2 was still stimulated by heme (Fig. 2A). Perhaps other residues in Hap1 also bind to heme and potentiate Hap1 activation, or perhaps this effect is mediated through heme binding to other proteins in the HMC. Nevertheless, the data shown here strongly suggest that HRMs, in particular HRM7, play an important role in heme binding and in mediating heme activation of Hap1. Further, the data show that not all HRMs are important for heme action although they all have the capability to bind to heme in vitro (33). In the case of Hap1, it is very surprising that deletion of all six HRMs in the heme domain did not diminish Hap1 heme responsiveness (Fig. 1). This surprising finding shows that the functional importance of HRMs in other hemoproteins cannot be inferred from their mere presence but must be determined by functional analysis. This explains why mutating the HRMs in heme lyase and ALAS affects their functions while mutating the HRMs in heme oxygenase and NF-E2 has no effect on the protein functions (14, 16, 17, 26).
New model for heme regulation of Hap1.
Our studies provide a new model for heme regulation of Hap1. The data show that two distinct classes of elements—the repression modules RPM1 to -3 and HRMs—are responsible for heme regulation of Hap1. The functions of both RPMs and HRMs are linked to the HMC. RPMs appear to permit the formation of the HMC, thereby allowing Hap1 to be repressed in the absence of heme. Therefore, deletion of the RPMs can cause disruption or partial disruption of the HMC, leading to Hap1 derepression (Fig. 1 to 5). HRMs appear to permit heme binding and the disassembly of the HMC, thereby leading to Hap1 activation by heme. Therefore, mutating HRMs causes the formation of mutant HMCs insensitive to heme and hyperrepression of the mutants (see H7-1 and H7-2 in Fig. 2 and 6).
These studies on Hap1 heme regulation and transcriptional activation may help reveal how the activities of other heme regulatory proteins are regulated. For example, modules in addition to HRMs may play important roles in mediating heme action in other hemoproteins including HRI kinase, heme oxygenase, and NF-E2 (3, 14, 16, 17, 26). In particular, HRI kinase binds to Hsp90 and Hsp70, and this interaction appears to be important for the regulation of HRI kinase activity (3, 28). Perhaps similar modules in Hap1 and HRI kinase interact with these molecular chaperones and mediate heme regulation. The analyses here allow us to make two general inferences on the precise and sophisticated mechanisms by which the activity of many regulators can be controlled. First, multiple modules in distant regions of one protein can act together to confer tight regulation. All three RPMs of Hap1 together mediate a high level of regulation, whereas each alone can mediate only a few fold of regulation (Fig. 1 to 3). Multiple modules may also be involved in the regulation of numerous other transcription factors, even though these modules may not be important for DNA binding or transcription-activating activity. Second, regulation of a transcription factor can be partitioned into repression and activation. Distinct classes of modules may be required for each function. This may be a general mechanism by which the activity of a regulator can be controlled by different signals in a precise and stringent manner.
ACKNOWLEDGMENTS
We thank W. Jelinek, H. Salomon, J. Qin, D. Conrad, J. Borowiec, and H. Lee for critical reviewing and proofreading of the manuscript.
This work was supported by grants from the National Institutes of Health (GM53453) and the National Science Foundation (MCB-96174720) to L.Z.
REFERENCES
- 1.Blank T E, Woods M P, Lebo C M, Xin P, Hopper J E. Novel Gal3 proteins showing altered Gal80p binding cause constitutive transcription of Gal4p-activated genes in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2566–2575. doi: 10.1128/mcb.17.5.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charnay P, Maniatis T. Transcriptional regulation of globin gene expression in the human erythroid cell line K562. Science. 1983;220:1281–1283. doi: 10.1126/science.6574602. [DOI] [PubMed] [Google Scholar]
- 3.Chen J J, London I M. Regulation of protein synthesis by heme-regulated eIF-2 alpha kinase. Trends Biochem Sci. 1995;20:105–108. doi: 10.1016/s0968-0004(00)88975-6. [DOI] [PubMed] [Google Scholar]
- 4.Chen J J, Throop M S, Gehrke L, Kuo I, Pal J K, Brodsky M, London I M. Cloning of the cDNA of the heme-regulated eukaryotic initiation factor 2 alpha (eIF-2 alpha) kinase of rabbit reticulocytes: homology to yeast GCN2 protein kinase and human double-stranded-RNA-dependent eIF-2 alpha kinase. Proc Natl Acad Sci USA. 1991;88:7729–7733. doi: 10.1073/pnas.88.17.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean A, Ley T J, Humphries R K, Fordis M, Schechter A N. Inducible transcription of five globin genes in K562 human leukemia cells. Proc Natl Acad Sci USA. 1983;80:5515–5519. doi: 10.1073/pnas.80.18.5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fytlovich S, Gervais M, Agrimonti C, Guiard B. Evidence for an interaction between the CYP1 (HAP1) activator and a cellular factor during heme-dependent transcriptional regulation in the yeast Saccharomyces cerevisiae. EMBO J. 1993;12:1209–1218. doi: 10.1002/j.1460-2075.1993.tb05762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 8.Guarente L, Lalonde B, Gifford P, Alani E. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell. 1984;36:503–511. doi: 10.1016/0092-8674(84)90243-5. [DOI] [PubMed] [Google Scholar]
- 9.Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- 10.Haldi M L, Guarente L. Multiple domains mediate heme control of the yeast activator HAP1. Mol Gen Genet. 1995;248:229–235. doi: 10.1007/BF02190805. [DOI] [PubMed] [Google Scholar]
- 11.Hon, T., A. Hach, D. Tamalis, Y.-H. Zhu, and L. Zhang. The yeast heme responsive transcriptional activator HAP1 is a preexisting dimer in the absence of heme. Submitted for publication. [DOI] [PubMed]
- 12.Johnston M, Flick J S, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3834–3841. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komar A A, Kommer A, Krasheninnikov I A, Spirin A S. Cotranslational folding of globin. J Biol Chem. 1997;272:10646–10651. doi: 10.1074/jbc.272.16.10646. [DOI] [PubMed] [Google Scholar]
- 14.Lathrop J T, Timko M P. Regulation by heme of mitochondrial protein transport through a conserved amino acid motif. Science. 1993;259:522–525. doi: 10.1126/science.8424176. [DOI] [PubMed] [Google Scholar]
- 15.Leuther K K, Johnston S A. Nondissociation of GAL4 and GAL80 in vivo after galactose induction. Science. 1992;256:1333–1335. doi: 10.1126/science.1598579. [DOI] [PubMed] [Google Scholar]
- 16.Liu D. Ph.D. thesis. State University of New York, Health Science Center at Brooklyn; 1998. [Google Scholar]
- 17.McCoubrey W K, Jr, Huang T J, Maines M D. Heme oxygenase-2 is a hemoprotein and binds heme through heme regulatory motifs that are not involved in heme catalysis. J Biol Chem. 1997;272:12568–12574. doi: 10.1074/jbc.272.19.12568. [DOI] [PubMed] [Google Scholar]
- 18.Padmanaban G, Venkateswar V, Rangarajan P N. Haem as a multifunctional regulator. Trends Biochem Sci. 1989;14:492–496. doi: 10.1016/0968-0004(89)90182-5. [DOI] [PubMed] [Google Scholar]
- 19.Parthun M R, Jaehning J A. A transcriptionally active form of GAL4 is phosphorylated and associated with GAL80. Mol Cell Biol. 1992;12:4981–4987. doi: 10.1128/mcb.12.11.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeifer K, Kim K S, Kogan S, Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989;56:291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- 21.Pfeifer K, Prezant T, Guarente L. Yeast HAP1 activator binds to two upstream activation sites of different sequence. Cell. 1987;49:19–27. doi: 10.1016/0092-8674(87)90751-3. [DOI] [PubMed] [Google Scholar]
- 22.Platt A, Reece R J. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rangarajan P N, Padmanaban G. Regulation of cytochrome P-450b/e gene expression by a heme- and phenobarbitone-modulated transcription factor. Proc Natl Acad Sci USA. 1989;86:3963–3967. doi: 10.1073/pnas.86.11.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sassa S. Novel effects of heme and heme-related compounds in biological systems. Curr Med Chem. 1996;3:273–290. [Google Scholar]
- 25.Sassa S, Nagai T. The role of heme in gene expression. Int J Hematol. 1996;63:167–178. doi: 10.1016/0925-5710(96)00449-5. [DOI] [PubMed] [Google Scholar]
- 26.Steiner H, Kispal G, Zollner A, Haid A, Neupert W, Lill R. Heme binding to a conserved Cys-Pro-Val motif is crucial for the catalytic function of mitochondrial heme lyases. J Biol Chem. 1996;271:32605–32611. doi: 10.1074/jbc.271.51.32605. [DOI] [PubMed] [Google Scholar]
- 27.Turcotte B, Guarente L. HAP1 positive control mutants specific for one of two binding sites. Genes Dev. 1992;6:2001–2009. doi: 10.1101/gad.6.10.2001. [DOI] [PubMed] [Google Scholar]
- 28.Uma S, Hartson S D, Chen J J, Matts R L. Hsp90 is obligatory for the heme-regulated eIF-2α kinase to acquire and maintain an activable conformation. J Biol Chem. 1997;272:11648–11656. doi: 10.1074/jbc.272.17.11648. [DOI] [PubMed] [Google Scholar]
- 29.Wells J A. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Bermingham M O, Turcotte B, Guarente L. Antibody-promoted dimerization bypasses the regulation of DNA binding by the heme domain of the yeast transcriptional activator HAP1. Proc Natl Acad Sci USA. 1993;90:2851–2855. doi: 10.1073/pnas.90.7.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Guarente L. Evidence that TUP1/SSN6 has a positive effect on the activity of the yeast activator HAP1. Genetics. 1994;136:813–817. doi: 10.1093/genetics/136.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Guarente L. HAP1 is nuclear but is bound to a cellular factor in the absence of heme. J Biol Chem. 1994;269:14643–14647. [PubMed] [Google Scholar]
- 33.Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14:313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L, Guarente L. The yeast activator HAP1—a GAL4 family member—binds DNA in a directly repeated orientation. Genes Dev. 1994;8:2110–2119. doi: 10.1101/gad.8.17.2110. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, Hach A, Wang C. Molecular mechanism governing heme signaling in yeast: a higher-order complex mediates heme regulation of the transcriptional activator HAP1. Mol Cell Biol. 1998;18:3819–3828. doi: 10.1128/mcb.18.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zitomer R S, Lowry C V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]