Summary
Effective triage of high-risk human papillomavirus (hrHPV)+ women is warranted to avoid unnecessary referral and overtreatment. Molecular triage tests have recently begun to impact cervical intraepithelial neoplasia grade 3 (CIN3) or cervical cancer (CC), termed CIN3+, detection. We find that zinc finger protein 671 methylation (ZNF671m) test has superior performance for CIN3+ detection in all single molecular triage tests, including HPV16/18 genotyping, paired box gene 1 methylation (PAX1m), and ZNF671m, in the training set. Using ZNF671m test instead of Thinprep cytologic test (TCT) as a single triage strategy or as a combined triage strategy with HPV16/18 genotyping has achieved comparable sensitivity but higher specificity for CIN3+ detection among 391 hrHPV+ women in the validation set. Little attention has been paid to the women with hrHPV− status but detected CIN3+. We find that the CIN3+ risk after a negative result could be reduced further by triage using ZNF671m in hrHPV− patients.
Keywords: DNA methylation, ZNF671, hrHPV, ASCUS, CIN3+ detection, Thinprep cytologic test, cervical cancer
Graphical abstract
Highlights
-
•
ZNF671 methylation test shows superior performance for CIN3+ detection
-
•
ZNF671 methylation test can be a promising alternative to cytology triage
-
•
Triage by ZNF671 methylation test reduces the CIN3+ risk in hrHPV− women
-
•
ZNF671 methylation test is more accessible in areas with poor medical resources
Zhu et al. evaluate the performance of ZNF671 methylation test for CIN3+ detection. ZNF671 methylation test can be a promising alternative to cytology triage. In addition, triage by ZNF671 methylation test reduces the CIN3+ risk further in hrHPV− women.
Introduction
Cervical cancer (CC) is the fourth most common cancer in women worldwide,1 with approximately 604,127 new cases of CC and 341,831 related deaths in 2020.1 More than 80% of CC-related deaths occur in developing countries.2,3,4 In China, 47,739 deaths and 106,430 new cases of CC have been recorded, accounting for 18.7% of diagnoses and 15.3% of deaths globally.5 CC screening programs can lower the incidence of CC6; however, the incidence rates are still high in low-/middle-income countries.7 It is challenging to solve female health and decrease the death rate of CC.
HPV infection is a major contributor of CC, and persistent HPV infection,8,9,10 especially high-risk HPV (hrHPV) infection, leads to the progression of CC.11,12,13 hrHPV testing is a highly sensitive screening method for detecting clinically relevant lesions in CC.14 A known disadvantage of hrHPV testing is the suboptimal specificity resulting in the overtreatment of women with a transient hrHPV infection since most women infected with hrHPV will clear such an infection without development.15,16 To overcome the disadvantage of hrHPV testing, Thinprep cytologic test (TCT [ASCUS+]) or HPV16/18 genotyping are recommended as additional triage tests to identify women who require further investigation. However, the effectiveness of these triage strategies is still suboptimal and unstable due to the subjective interpretation and suboptimal sensitivity of TCT(ASCUS+) and the low sensitivity of HPV16/18 genotyping.17,18
Molecular triage tests are more objective and more acceptable for early cervical screening, as large numbers of PCR laboratories established throughout the epidemic of COVID-19.19 Increasing evidence showed that DNA methylation testing could be a promising cervical screening method due to its objective outcome and good performance for cervical precancer and cancer detection. Gene DNA methylation testing always has higher specificity than hrHPV testing or has higher sensitivity than TCT. Many genes, such as SOX1,20 PAX1,21,22 zinc finger protein 582 (ZNF582),23 ASTN1/DLX1/ITGA4/RXFP3/SOX17/ZNF671,24,25,26 JAM3,27 EPB41L3,28 CADM1/MAL/miR124-2,29,30 FAM19A4,31,32 and POU4f3,33 have been reported as promising markers for CIN2+/CIN3+ detection. We analyzed all the reported genes and selected some good-performance genes to be validated in our samples of Chinese women (data not shown). Our results showed that ZNF671 had a great performance in detecting CIN3+.
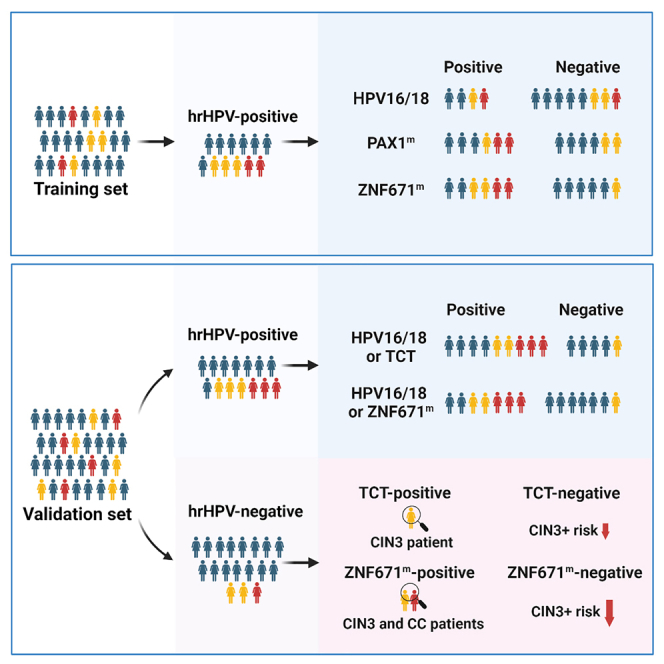
PAX1, a methylated gene that has been reported as a promising maker for management of HPV+ women in the Chinese population, was also included in our current study.21,22 HPV16/18 genotyping, ZNF671 methylation (ZNF671m), and PAX1 methylation (PAX1m) in 281 cervical scrapings were performed to evaluate and compare the performance of different molecular triage strategies for CIN3+ detection in hrHPV+ women. Moreover, validation of the superior molecular triage strategy was performed in an independent cohort of 391 hrHPV+ cases, and the performance of the superior triage test was compared with TCT in the validation set. Last, we evaluated the performance of ZNF671m and PAX1m or TCT for CIN3+ in 179 or 242 hrHPV− women.
Results
Patient clinicopathological characteristics
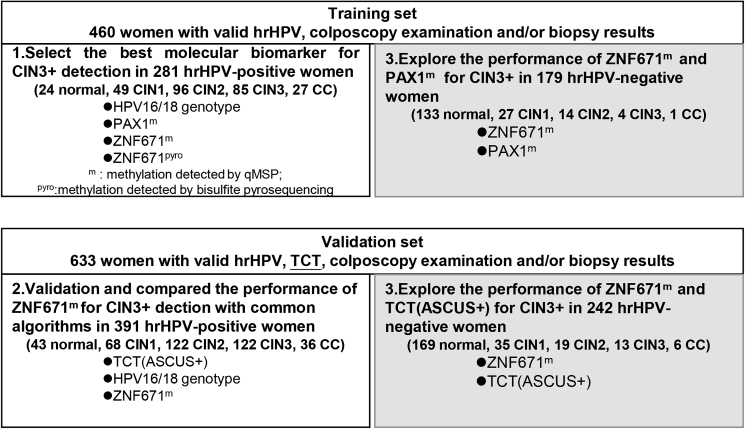
Cohort demographic characteristics and clinical information are summarized in Table 1. In the training set, colposcopy and biopsy results showed 157 (34.13%) women with normal uterine cervix, 76 cases (16.52%) of CIN1, 110 (23.91%) cases of CIN2, 89 (19.35%) cases of CIN3, and 28 (6.09%) cases of CC. The training set included 281 (61.09%) hrHPV+ and 179 (38.91%) hrHPV− women. In the validation set, colposcopy and biopsy results showed 212 (33.49%) women with normal uterine cervix, 103 cases (16.27%) of CIN1, 141 (22.27%) cases of CIN2, 135 (21.33%) cases of CIN3, and 42 (6.64%) cases of CC. The validation set included 391 (61.77%) hrHPV+ and 242 (38.23%) hrHPV− women. The study protocol is shown in Figure 1.
Table 1.
Population and test characteristics by histologic category
| Characteristic | Cutoff | Histological results |
Total | ||||
|---|---|---|---|---|---|---|---|
| Normal | CIN1 | CIN2 | CIN3 | CC | |||
| Training set | |||||||
| N (%) | – | 157 (34.13) | 76 (16.52) | 110 (23.91) | 89 (19.35) | 28 (6.09) | 460 (100.00) |
| Age (years) | – | 44.45 ± 9.43 | 47.68 ± 9.06 | 43.45 ± 10.57 | 45.20 ± 8.80 | 50.64 ± 8.00 | 45.27 ± 9.62 |
| HPV16/18 (%) | – | 3 (1.91) | 9 (11.84) | 32 (29.09) | 36 (40.45) | 21 (75.00) | 101 (21.96) |
| hrHPV (%) | – | 24 (15.29) | 49 (64.47) | 96 (87.27) | 85 (95.51) | 27 (96.43) | 281 (61.09) |
| ZNF671m | ΔCp ≤ 11.20 | 10 (6.37) | 9 (11.84) | 22 (20.00) | 64 (71.91) | 28 (100.00) | 133 (28.91) |
| ZNF671pyro | m% ≥ 4.25 | 14 (8.92) | 12 (15.79) | 17 (15.45) | 45 (50.56) | 26 (92.86) | 114 (24.78) |
| PAX1m | ΔCp ≤ 11.84 | 42 (26.75) | 15 (19.74) | 34 (30.91) | 59 (66.29) | 27 (96.43) | 177 (38.48) |
| Validation set | |||||||
| N (%) | – | 212 (33.49) | 103 (16.27) | 141 (22.27) | 135 (21.33) | 42 (6.64) | 633 (100.00) |
| Age (years) | – | 43.94 ± 11.03 | 42.02 ± 10.45 | 40.17 ± 10.92 | 44.91 ± 10.85 | 54.02 ± 7.75 | 43.66 ± 11.14 |
| HPV16/18 (%) | – | 9 (4.25) | 20 (19.42) | 42 (29.79) | 64 (47.41) | 29 (69.05) | 164 (25.91) |
| hrHPV (%) | 43 (20.28) | 68 (66.02) | 122 (86.52) | 122 (90.37) | 36 (85.71) | 391 (61.77) | |
| ZNF671m | ΔCp≤11.20 | 35 (16.51) | 26 (25.24) | 45 (31.91) | 99 (73.33) | 39 (92.86) | 244 (38.55) |
| TCT | NILM | 187 (88.21) | 35 (33.98) | 37 (26.24) | 25 (18.52) | 4 (9.52) | 288 (45.50) |
| ASCUS | 9 (4.25) | 32 (31.07) | 38 (26.95) | 24 (17.78) | 6 (14.29) | 109 (17.22) | |
| LSIL | 4 (1.89) | 30 (29.13) | 32 (22.70) | 19 (14.07) | 0 (0.00) | 85 (13.43) | |
| ASC-H/AGC/HSIL+ | 12 (5.66) | 6 (5.83) | 34 (24.11) | 67 (49.63) | 32 (76.19) | 151 (23.85) | |
m, methylation detected by qMSP; Cp, crossing point; Pyro, methylation detected by bisulfite pyrosequencing.
Figure 1.
Study protocol
m, methylation detected by qMSP; pyro, methylation detected by bisulfite pyrosequencing.
Performance of molecular triage testing for CIN3+ in 281 hrHPV+ women (training set)
We analyzed and compared the area under the curve (AUC) of HPV16/18, hrHPV, ZNF671m, ZNF671Pyro, and PAX1m in cervical scrapings by receiver operating characteristic (ROC) curves for distinguishing different diagnosis groups (CIN1−/CIN2+, CIN2−/CIN3+). As shown in Figure S1, the performance of ZNF671m was better than ZNF671Pyro in our study, and we could only use ZNF671m in our follow analysis.
As shown in Table 2, HPV16/18 genotyping exhibited a sensitivity of 50.89% for CIN3+ with a specificity of 73.69%. While the PAX1m test and the ZNF671m test achieved significantly higher sensitivity (relative sensitivity of PAX1m test: 1.46, 95% confidence interval [CI]: 1.19–1.78; relative sensitivity of ZNF671m test: 1.56, 95% CI: 1.29–1.89) with comparable specificity (relative specificity of PAX1m test: 0.90, 95% CI: 0.78–1.04; relative specificity of ZNF671m test: 1.08, 95% CI: 0.96–1.22) compared with HPV16/18 genotyping.
Table 2.
Performance of molecular triage for the detection of CIN3+ among 281 hrHPV+ women (training set)
| Triage algorithms | Colposcopy referral rates(%) (n/N) | Sensitivity (%) (n/N) 95% CI | Specificity (%) (n/N) 95% CI | PPV (%) (n/N) 95% CI | NPV (%) (n/N) 95% CI | NNC | Compared with HPV16/18 |
Compared with HPV16/18 or PAX1m |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Relative sensitivity (95% CI) | Relative specificity (95% CI) | Relative sensitivity (95% CI) | Relative specificity (95% CI) | |||||||
| HPV16/18 | 35.94 (101/281) | 50.89 (57/112) 41.33–60.40 | 73.96 (125/169) 66.56–80.26 | 56.44 (57/101) 46.22–66.16 | 69.44 (125/180) 62.08–75.96 | 1.77 | 1 | 1 | – | – |
| PAX1m | 49.47 (139/281) | 74.11 (83/112) 64.82–81.71 | 66.86 (113/169) 59.16–73.79 | 59.71 (83/139) 51.04–67.84 | 79.58 (113/142) 71.83–85.69 | 1.67 | 1.46 (1.19–1.78) | 0.90 (0.78–1.04) | – | – |
| ZNF671m | 43.77 (123/281) | 79.46 (89/112) 70.58–86.28 | 79.88 (135/169) 72.88–85.48 | 72.36 (89/123) 63.44–79.85 | 85.44 (135/158) 78.75–90.36 | 1.38 | 1.56 (1.29–1.89) | 1.08 (0.96–1.22) | – | – |
| HPV16/18 or PAX1m | 65.12 (183/281) | 84.82 (95/112) 76.53–90.66 | 47.93 (81/169) 40.24–55.71 | 51.91 (95/183) 44.44–59.30 | 82.65 (81/98) 73.39–89.29 | 1.93 | 1.67 (1.41–1.96) | 0.65 (0.57–0.74) | 1 | 1 |
| HPV16/18 or ZNF671m | 59.79 (168/281) | 86.61 (97/112) 78.56–92.06 | 57.99 (98/169) 50.15–65.45 | 57.74 (97/168) 49.88–65.24 | 86.73 (98/113) 78.74–92.13 |
1.73 | 1.70 (1.44–2.01) | 0.78 (0.72–0.86) | 1.02 (0.94–1.11) | 1.21 (1.06–1.38) |
| ZNF671m or PAX1m | 58.36 (164/281) | 87.50 (98/112) 79.59–92.75 | 60.95 (103/169) 53.13–68.26 | 59.76 (98/164) 51.80–67.24 | 88.03 (103/117) 80.42–93.06 | 1.67 | 1.72 (1.42–2.09) | 0.82 (0.70–0.97) | 1.03 (0.95–1.12) | 1.27 (1.11–1.46) |
| HPV16/18 or ZNF671m or PAX1m | 71.89 (202/281) | 93.75 (105/112) 87.09–97.23 | 42.60 (72/169) 35.11–50.44 | 51.98 (105/202) 44.87–59.01 | 91.14 (72/79) 82.04–96.06 | 1.92 | 1.84 (1.55–2.20) | 0.58 (0.50–0.67) | 1.11 (1.04–1.18) | 0.89 (0.82–0.96) |
m, methylation; PPV, positive predictive values; NPV, positive predictive values; NNC, the number of colposcopies needed for detection per CIN3+ case.
When combing two or three tests to triage women with positive hrHPV, HPV16/18 genotyping with PAX1m or ZNF671m demonstrated an increase in sensitivity by 67% or 70% but a decrease in specificity by 35% and 22%. Combining the three tests to triage hrHPV+ women could further increase sensitivity by 84% but decreased specificity by 42%. Compared with HPV16/18 or PAX1m triage test, HPV16/18 or ZNF671m triage test had higher specificity (relative specificity: 1.21, 95% CI: 1.06–1.38).
NNC (the number of colposcopies needed for detection per CIN3+ case) reflects the clinical efficiency of molecular triage tests. Triage strategies involving HPV16/18 or ZNF671m or PAX1m had the highest referral rate for colposcopy (71.89%) and required 1.92 colposcopies for detection per CIN3+ case (Table 2). HPV16/18 genotyping had the lowest colposcopy referral rate (35.94%) but missed 1/2 CIN3+ cases. ZNF671m alone had the lowest NNC (1.38) for CIN3+ with a slightly lower colposcopy referral rate (43.77%). The performance for CIN2+ detection is shown in Table S1.
Validation and comparison of the performance of ZNF671m testing with common triage algorithms for CIN3+ in 391 hrHPV+ women (validation set)
As shown in Table 3, the sensitivity of ZNF671m test for detecting CIN3+ was 78.48% (95% CI: 71.10%–84.45%), which was not significantly lower than that of TCT(ASCUS+), with a relative sensitivity of 0.92 (95% CI: 0.84–1). However, the specificity of ZNF671m test (67.38%, 95% CI: 60.90%–73.28%) was significantly higher than that of TCT(ASCUS+) (33.05%, 95% CI: 27.12%–39.54%), with relative specificity at 2.04 (95% CI: 1.68–2.48). The sensitivity of HPV16/18 (58.86%, 95% CI: 50.75%–66.53%) genotyping was lower than that of TCT(ASCUS+), but the specificity was higher (69.53%, 95% CI: 63.12%–75.28%). Regarding colposcopy efficiency, ZNF671m test required an NNC of 1.61 colposcopies to detect a single CIN3+ case, which is less than that required for TCT(ASCUS+) (NNC = 2.16) and for HPV16/18 (NNC = 1.76).
Table 3.
Performance of triage for the detection of CIN3+ among 391 hrHPV+ women (validation set)
| Triage algorithms | Colposcopy referral rates (%) (n/N) | Sensitivity (%) (n/N) 95% CI | Specificity (%) (n/N) 95% CI | PPV (%) (n/N) 95% CI | NPV (%) (n/N) 95% CI | NNC | Compared with TCT(ASCUS+) |
Compared with HPV16/18 or TCT(ASCUS+) |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Relative Sensitivity (95%CI) | Relative specificity (95%CI) | Relative Sensitivity (95%CI) | Relative specificity (95%CI) | |||||||
| TCT(ASCUS+) | 74.42 (291/391) | 85.44 (135/158) 78.75–90.36 | 33.05 (77/233) 27.12–39.54 | 46.39 (135/291) 40.58–52.30 | 77.00 (77/100) 67.31–84.58 | 2.16 | 1 | 1 | – | – |
| HPV16/18 | 41.94 (164/391) | 58.86 (93/158) 50.75–66.53 | 69.53 (162/233) 63.12–75.28 | 56.71 (93/164) 48.75–64.34 | 71.37 (162/227) 64.94–77.06 | 1.76 | 0.69 (0.59–0.80) | 2.10 (1.71–2.59) | – | – |
| ZNF671m | 51.15 (200/391) | 78.48 (124/158) 71.10–84.45 | 67.38 (157/233) 60.90–73.28 | 62.00 (124/200) 54.85–68.68 | 82.20 (157/191) 75.87–87.20 | 1.61 | 0.92 (0.84–1.00) | 2.04 (1.68–2.48) | – | – |
| HPV16/18 or TCT(ASCUS+) | 85.17 (333/391) | 94.94 (150/158) 89.93–97.63 | 21.46 (50/233) 16.48–27.40 | 45.05 (150/333) 39.64–50.57 | 86.21 (50/58) 74.07–93.44 | 2.22 | 1.11 (1.05–1.17) | 0.65 (0.55–0.77) | 1 | 1 |
| HPV16/18 or ZNF671m | 67.26 (263/391) | 90.51 (143/158) 84.56–94.41 | 48.50 (113/233) 41.95–55.10 | 54.37 (143/263) 48.14–60.47 | 88.28 (113/128) 81.11–93.07 | 1.84 | 1.06 (0.98–1.15) | 1.47 (1.17–1.83) | 0.95 (0.90–1.01) | 2.26 (1.78–2.87) |
| ZNF671m or TCT(ASCUS+) | 82.10 (321/391) | 93.04 (147/158) 87.58–96.30 | 25.32 (59/233) 19.97–31.50 | 45.79 (147/321) 40.27–51.42 | 84.29 (59/70) 73.20–91.52 | 2.18 | 1.09 (1.04–1.14) | 0.77 (0.68–0.87) | 0.98 (0.93–1.03) | 1.18 (0.97–1.44) |
| HPV16/18 or ZNF671m or TCT(ASCUS+) | 89.26 (349/391) | 98.10 (155/158) 94.12–99.51 | 16.74 (39/233) 12.31–22.30 | 44.41 (155/349) 39.15–49.80 | 92.86 (39/42) 79.45–98.14 | 2.25 | 1.15 (1.08–1.23) | 0.51 (0.41–0.63) | 1.03 (1.00–1.06) | 0.78 (0.67–0.90) |
m, methylation; PPV, positive predictive values; NPV, positive predictive values; NNC, the number of colposcopies needed for detection per CIN3+ case.
Compared with the combined test, all combined tests were shown to have a significantly higher sensitivity than that of TCT(ASCUS+), except HPV16/18 or ZNF671m test. Meanwhile, only HPV16/18 or ZNF671m test (48.50%, 95% CI: 41.95%–55.10%) had a significantly higher specificity than that of TCT(ASCUS+) (33.05%, 95%CI: 27.12%–39.54%), with a relative specificity at 1.47 (95% CI: 1.17–1.83). Compared with HPV16/18 or TCT(ASCUS+), the common triage in hrHPV+ women,HPV16/18 or ZNF671m test increases the specificity by 126% with comparable sensitivity. The NNC of HPV16/18 or ZNF671m test (NNC = 1.84) was less than that of HPV16/18 or TCT(ASCUS+) (NNC = 2.22), with a lower colposcopy referral rate (67.26% vs. 85.17%). The performance for CIN2+ detection is shown in Table S2.
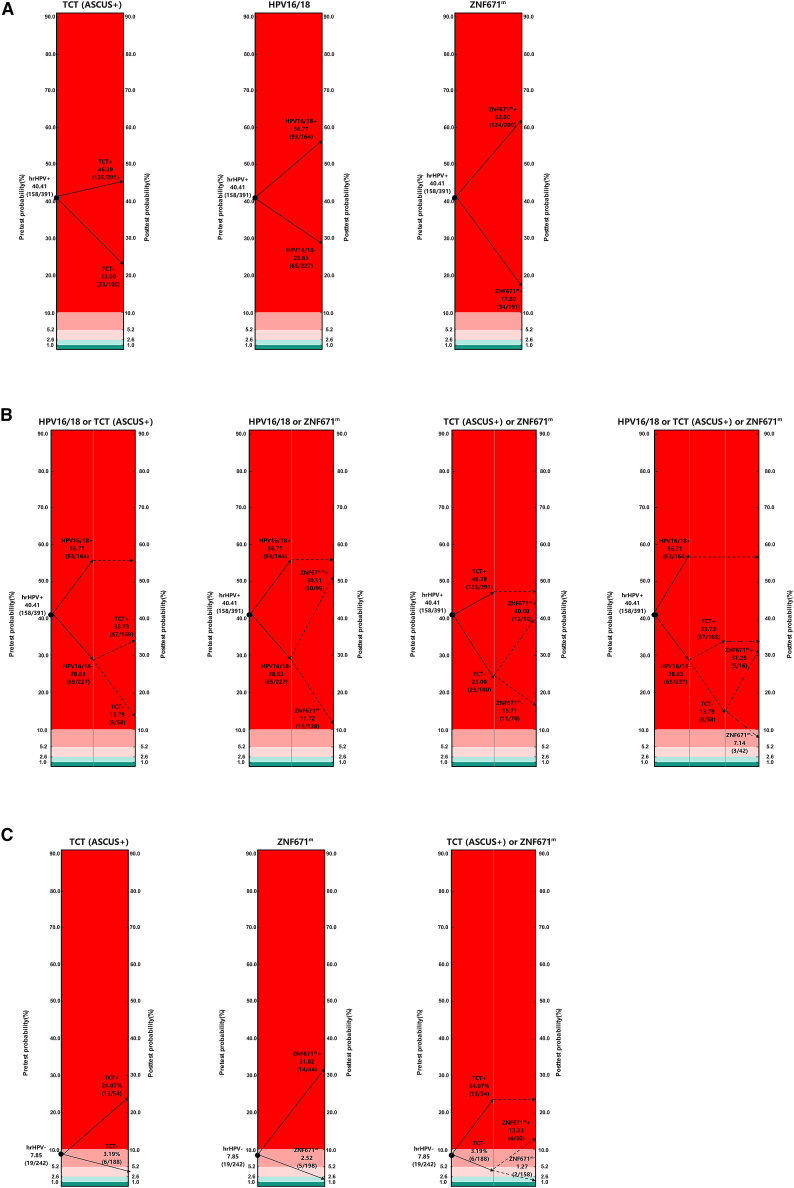
Pre- and post-test CIN3+ risk after applying different triage strategies in hrHPV+ women
Figures 2A and 2B display the pre- and post-test CIN3+ risk after applying different triage strategies in hrHPV+ women. Before triaging, the CIN3+ risk was 40.41% among 391 hrHPV+ women. Our results showed that the CIN3+ risks of all single triage strategies were decreased but still high (TCT−: 23%; HPV16/18−: 28.63%); only the risk of CIN3+ by ZNF671m was lower than 20%, despite the fact that the pro-test risks did not fall into the safe or medium-risk zones in women who were negative by any of the single triage methods. After the second-step triage test using TCT(ASCUS+) or ZNF671m in non-16/18+ women, the risk of CIN3+ could be reduced to 13.79% or 11.72%, respectively, and was close to the boundary value of risk (10%). The pro-test risks further be reduced by third-step triage test. The CIN3+ risk was 7.14% among the three-test-negative population. However, none of the triages mentioned above had a sufficient risk reaching 1% or 2.6%.
Figure 2.
Pre- and post-test CIN3+ risk after applying different triage strategies in hrHPV+ women or in hrHPV− women
(A) Pre- and post-test CIN3+ risk by single triage algorithms in hrHPV+ women (n = 391).
(B) Pre- and post-test CIN3+ risk by combined triage algorithms in hrHPV+ women (n = 391).
(C) Pre- and post-test CIN3+ risk after triage in hrHPV− women (n = 242).
CIN3+ risk: red area, >10%; flesh pink area, between 10% and 5.2%; pink area, between 5.2% and 2.6%; light green area, between 2.6% and 1%; dark green area, <1%. Benchmarks are defined at risk levels of 1% (safe) and 10% (high risk) (applied in Europe) and 2.6% (safe) and 5.2% (high-risk) (applied in the United States) for CIN3+. High-risk zone: red area; medium risk: flesh pink area and pink area; safe zone: light green area and dark green area.
Pre- and post-test CIN3+ risk after applying different triage strategies in hrHPV− women
In the past few years, CIN3+ risk was always calculated in the hrHPV+ population. The CIN3+ risk in the hrHPV− population was unclear. In our current study, the CIN3+ risk was 7.85% (>2.6% or >1%) among 242 hrHPV− women. It suggested that it might be necessary to triage hrHPV− women by second-step test. We looked back and analyzed the performance of ZNF671m, PAX1m, or TCT(ASCUS+) for CIN3+ detection. As shown in Table 4, ZNF671m alone could be an effective test to detect the missed cases by hrHPV screening. Figure 2C displays the CIN3+ risk for different triage algorithms in hrHPV− women. Results indicated that women with hrHPV− status who were given a second-step triage test with ZNF671m test had 2.52% chance of detecting CIN3+, falling into the light green zone.
Table 4.
Performance of triage for the detection of CIN3+ among hrHPV− women
| Dataset | Triage algorithms | Colposcopy referral rates(%) (n/N) | Sensitivity (%) (n/N) 95% CI | Specificity (%) (n/N) 95%CI | PPV (%) (n/N) 95% CI | NPV (%) (n/N) 95% CI | Missed CIN3+ cases | Relative sensitivity (95% CI) | Relative specificity (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| 179 hrHPV− women in training set | PAX1m | 21.23 (38/179) | 60.00 (3/5) 17.04–92.74 | 79.89 (139/174) 73.00–85.42 | 7.89 (3/38) 2.06–22.48 | 98.58 (139/141) 94.45–99.75 | 2 (2 CIN3) | 1 | 1 |
| ZNF671m | 5.59 (10/179) | 60.00 (3/5) 17.04–92.74 | 95.98 (167/174) 91.56–98.26 | 30.00 (3/10) 8.09–64.63 | 98.82 (167/169) 95.32–99.79 | 2 (2 CIN3) | 1 | 1.20 (1.11–1.30) | |
| ZNF671m or PAX1m | 22.91 (41/179) | 60.00 (3/5) 17.04–92.74 | 78.16 (136/174) 71.14–83.91 | 7.32 (3/41) 1.91–21.01 | 98.55 (136/138) 94.33–99.75 | 2 (2 CIN3) | 1 | 0.98 (0.95–1.00) | |
| 242 hrHPV− women in validation set | TCT(ASCUS+) | 22.31 (54/242) | 68.42 (13/19) 43.50–86.44 | 81.61 (182/223) 75.77–86.35 | 24.07 (13/54) 13.92–37.94 | 96.81 (182/188) 92.86–98.70 | 6 (4 CIN3 and 2 CC) | 1 | 1 |
| ZNF671m | 18.18 (44/242) | 73.68 (14/19) 48.58–89.88 | 86.55 (193/223) 81.19–90.60 | 31.82 (14/44) 19.07–47.71 | 97.47 (193/198) 93.88–99.07 | 5 (4 CIN3 and 1 CC) | 1.08 (0.73–1.58) | 1.06 (0.98–1.15) | |
| ZNF671m or TCT(ASCUS+) | 34.71 (84/242) | 89.47 (17/19) 65.46–98.16 | 69.96 (156/223) 63.41–75.80 | 20.24 (17/84) 12.56–30.70 | 98.73 (156/158) 95.03–99.78 | 2 (2 CIN3) | 1.31 (1.00–1.70) | 0.86 (0.81–0.91) |
m, methylation; PPV, positive predictive values; NPV, positive predictive values; NNC, the number of colposcopies needed for detection per CIN3+ case.
The cost of different algorithms for detection of CIN3+ in validation set
ZNF671m test showed better performance for CIN3+ detection than TCT(ASCUS+). However, whether the ZNF671m test had a cost advantage is not clear. The cost of ZNF671m test ($11.62), calculated according to the data in our laboratory, is near the cost of TCT ($10.23).34 Due to the sensitivity of HPV16/18 or ZNF671m being comparable with the sensitivity of HPV16/18 or TCT(ASCUS+) in hrHPV+ women (Table 3), we calculated the triage cost needed for detection per CIN3+ case. The results showed that the triage cost needed for detection per CIN3+ case of HPV16/18 or ZNF671m ($88) was slightly lower than that of HPV16/18 or TCT(ASCUS+) ($95) in hrHPV+ women (Table 5). The results showed that the cost needed for detection per CIN3+ case of HPV16/18 or ZNF671m ($160) was also slightly lower than that of HPV16/18 or TCT(ASCUS+) ($164) when using them as a combination test in all women in the validation set (Table S3). Taken together, the results suggested that HPV16/18 or ZNF671m could slightly reduce the cost and ensure the performance for CIN3+ detection (Tables 3, 4, 5, and S3).
Table 5.
The triage cost for detection of CIN3+ among 391 hrHPV+ women (validation set)
| Triage algorithms | Total triage costs ($) | The triage cost needed to detect per CIN3+ cases ($) |
|---|---|---|
| TCT(ASCUS+) | 12,925.94 | 95.75 |
| HPV16/18 | 5031.30 | 54.10 |
| ZNF671m | 10,679.42 | 86.12 |
| HPV16/18 or TCT(ASCUS+) | 14,214.44 | 94.76 |
| HPV16/18 or ZNF671m | 12,612.18 | 88.20 |
| ZNF671m or TCT(ASCUS+) | 18,389.99 | 125.10 |
| HPV16/18 or ZNF671m or TCT(ASCUS+) | 19,248.99 | 124.19 |
The total triage costs including triage test (excluded HPV16/18 test) cost, referral service cost, colposcopy cost, and pathological examination cost. The cost data (per person) of TCT ($10.23), referral service ($1.55), colposcopy ($9.30), and pathological examination ($24.79) were collected from the published literature.34 The cost of ZNF671m test was calculated according the data in our laboratory ($11.62).
Discussion
Effective triage and management of hrHPV+ women is warranted to avoid unnecessary referral and overtreatment. In the current study, we demonstrated that ZNF671m test alone or combined with HPV16/18 genotyping had better performance for CIN3+ detection than TCT(ASCUS+) to triage gynecological hrHPV+ outpatients. Moreover, we explored the triage possibilities for CIN3+ of these triage algorithms among the hrHPV− population in gynecological clinic. The CIN3+ risk after a negative result could be reduced further by triage using ZNF671m in hrHPV− outpatients.
The incidence of cervical lesions has declined significantly since the introduction of hrHPV testing as a primary screening strategy.35,36 A known disadvantage of hrHPV testing is the suboptimal specificity resulting in severe overtreatment of women with a transient hrHPV infection.15,16 In our current study, hrHPV testing showed a good sensitivity (91.84%) but low specificity (49.69%) for CIN3+ detection, as reported in other previous study.14 This leads to potential overtreatment of a large number of women who may have regression lesions.37,38 Unnecessary damage, especially among young women of reproductive age, can be avoided by providing accurate stratified classification strategies to identify CIN3+. Women who need further treatment would benefit from TCT(ASCUS+) or from HPV16/18 genotyping as an additional triage test.39,40
In the present study, we found that ZNF671m test had higher specificity than TCT (ASUCS+), at a relative specificity 2.04, and had comparable sensitivity with TCT (ASUCS+). The colposcopy referral rate of ZNF671m test was 23% lower than that of TCT (ASUCS+). Compared with HPV16/18 genotyping, ZNF671m test had a significantly higher sensitivity (78.48% vs. 58.86%) and comparable specificity (67.38% vs. 69.53%). In addition, the NNC of ZNF671m test was the lowest one in all single triage tests. Currently, HPV16/18 or TCT(ASCUS+) was the most common triage in hrHPV+ women. Our results showed that compared with HPV16/18 or TCT(ASCUS+), HPV16/18 or ZNF671m test increases the specificity by 126% with comparable sensitivity. The NNC of HPV16/18 or ZNF671m test (NNC = 1.84) was less than that of HPV16/18 or TCT(ASCUS+) (NNC = 2.22), with a lower colposcopy referral rate (67.26% vs. 85.17%) (Table 3). Moreover, the cost of triage needed for detection per CIN3+ case of HPV16/18 or ZNF671m ($88) was slightly lower than that of HPV16/18 or TCT(ASCUS+) ($95) among 391 hrHPV+ women (Table 5). All results together show that ZNF671m could be a promising alternative to cytology triage in the hrHPV+ population.
In addition to the above advantages, using ZNF671m test instead of TCT(ASCUS+) as a triage strategy for CIN3+ detection was more objective and was no longer limited and affected by the skill of doctor. ZNF671m test could reach more hospitals, as large numbers of PCR laboratories were established throughout the epidemic of COVID-19.19 In addition, the patients are more likely to prefer ZNF671m test because there is no additional cervical scraping cell collection required when triage is done by ZNF671m test. ZNF671m test, as a molecular test, has possibilities of self-sampling and a central laboratory operating system and thus may be a more reasonable choice to develop higher-quality triage methods. It can greatly increase the efficiency and amount of tests and can significantly reduce the cost, making inexpensive testing more accessible and easier to perform in areas with poor medical resources.
Moreover, little attention has been paid to a small portion of women with hrHPV− status but detected CIN3+. One explanation is that the women screened in gynecology clinics may not to be new to screening and may have a history of hrHPV infection and antiviral therapy and, therefore, have negative hrHPV results at this screening. Another explanation is that a small percentage of CIN and CC resulted from other rare and poorly characterized viral types.41 As shown in our results (Table 4), ZNF671m alone could be an effective test to detect the missed cases by hrHPV screening. In addition, the CIN3+ risk after a negative result could be reduced further by triage using ZNF671m test in hrHPV− patients. ZNF671m test could reduce some missed cases caused by the above reasons and did not increase the cost compared with TCT(ASCUS+) (Table S3).
Although there have been some studies on the performance of a six-gene methylation panel (ASTN1/ITGA4/RXFP3/SOX17/DLX1/ZNF671) for CIN2+/CIN3+ detection.24,26 The score of ZNF671 was 0.5, which was higher than other genes (0.2 or 0.1). It showed that ZNF671 played a major role in calculating the methylation score (>0.5 termed as methylation panel positive).25,42 The results were consistent with our results. The six-gene methylation panel may more effective but less cost effective for CIN3+ than ZNF671m test only in hrHPV+ women. In addition, the performance of six-gene methylation in hrHPV− women is still unclear.
When using hrHPV testing as primary screening, triaging hrHPV+ women by HPV16/18 or ZNF671m test could greatly reduce referral rates to colposcopy and, even more importantly, improve positive predictive values and prevent overtreatment. In addition, triaging hrHPV− women by ZNF671m could reduce the missed CIN3+ cases as much as possible. Our study suggests that ZNF671m could be a promising alternative to cytology triage, especially in areas with poor medical resources. However, prospective population-based studies are necessary for further implementation of this screening protocol.
Limitations of the study
In this study, we only evaluated the performance of different tests for CIN3+ detection in gynecology clinic-screened women. In addition, only the samples with exact colposcopy and/or histology results were included in our study. The patients who were not referred to colposcopy were not included. So, the CIN3+ risk calculated in our study was higher than the actual risk after application triage strategies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| cervical scrapings | Xiangya Hospital of Central South University, the Second Xiangya Hospital of Central South University, the Third Xiangya Hospital of Central South University, the First Affiliated Hospital of Shantou University Medical College, Loudi Central Hospital and Zhengzhou Central Hospital Affiliated to Zhengzhou University | This study |
| Critical commercial assays | ||
| Human Papillomavirus DNA (Genotype) Diagnostic Kit | SANSURE | S3027E |
| EZ DNA Methylation-Direct ™ Kit | ZYMO RESEARCH | D5021 |
| HiPure Universal DNA Kit | Magen Biotech | D3018-02 |
| Uterine cervix cancer gene methylating Diagnostic Kit(PCR-Fluorescence Probing) | HOOMYA | HOOMYA |
| Oligonucleotides | ||
| ZNF671-F: GGTGGAGGTGTTGGGAAA | This paper | N/A |
| ZNF671-R: 5′- CTAAAACACAAAAACTA CAAACACTTTAC-3′ |
This paper | N/A |
| ZNF671-S: 5′- GTTTGATGTTTTGT AGGGA-3′ |
This paper | N/A |
| ZNF671-QF:5′- GTCGTTTTCGGTA GTTGTTCGC -3′ |
This paper | N/A |
| ZNF671-QR: 5′- AAAAACGCAACAC CCACCC -3′ |
This paper | N/A |
| ZNF671-P: 5′-FAM- TTTACGGTTTTA TGGCGGA-MGB-3′ |
This paper | N/A |
| β-actin-QF: 5′-AGGAGGTTTGGATTTT TAATTGTGTATAG-3′ |
This paper | N/A |
| β-actin-QR:5′-AACTCACCACTACAAAA ATCAAACCA -3′ |
This paper | N/A |
| β-actin-P:5′-VIC-TGGTAGAGAGGAAG GTGG-MGB-3′ |
This paper | N/A |
| Software and algorithms | ||
| SPSS software | SPSS Inc | version 22.0 |
| R | R-project | version 3.6.3 |
Resource availability
Lead contact
Further information and requests will be fulfilled by the Lead Contact, Qing Li (liqing9251026@csu.edu.cn).
Materials availability
No new materials or reagents were generated in this study.
Experimental model and subject details
Specimens and clinical data
Between Jan 2020 and July 2022, cervical scrapings were collected from gynecologic outpatients older than 18 years of age with exact colposcopy and/or histology results from Xiangya Hospital of Central South University, the Second Xiangya Hospital of Central South University, the Third Xiangya Hospital of Central South University, the First Affiliated Hospital of Shantou University Medical College, Loudi Central Hospital and Zhengzhou Central Hospital Affiliated to Zhengzhou University. This study was undertaken in accordance with the Declaration of Helsinki, and the protocol was approved by the local ethical committees where applicable (no.2019103 and no.202101007). All patients gave written informed consent before participation in this study. A history of cervical cancer or existence of other cancer, cervical surgery, anti-HPV vaccinations, or pregnancy were excluded from this study.
The training set included women who had a normal uterine cervix (n = 157), CIN1 (n=76), CIN2 (n=110), CIN3 (n=89) and CC (n=28) diagnosed according to the histologic reports. The final diagnosis was made by tissue-proven pathology in the CIN1+ test result group. When the biopsy results revealed CIN3+, the patients underwent cervical conization or major surgery. The validation set was composed of 633 cases (212 normal, 103 CIN1, 141 CIN2, 135 CIN3 and 42 CC). Clinical, histological and pathological characteristics of all patients were showed in Table 1. Data on hrHPV status, TCT and local pathology diagnoses of patients were obtained from the parent institutes. Methylation detection tests for PAX1 and ZNF671 and hrHPV genotyping were carried out by using residual cervical cells from cytological tests. The laboratory tests were performed blinded to all other triage results and were detailed below.
Method details
DNA preparation
The residual cervical scrapings cells were stored in preservation solution (JIANG SU JIANYOU MEDICAL TECHNOLOGY, Jiangsu, China) at −20°C. The residual cervical cells were centrifuged and washed with phosphate-buffered saline. Genomic DNA (gDNA) was extracted from the cells using the HiPure Universal DNA Kit (Magen Biotech, Guangzhou, China). The concentrations of gDNA in each sample were measured using a BioSpec-nano spectrophotometer (Shimadzu Corporation, Tokyo, Japan).
HrHPV DNA test
DNA were prepared as described above. HrHPV DNA detection were performed by High-risk Human Papillomavirus DNA (Genotype) Diagnostic Kit (SANSURE, Changsha, China). This kit is an in vitro nucleic acid amplification test for the detection of hrHPV (type16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66 and 68). The test results can be used for identification of HPV genotypes. In our current study, we followed the WHO guideline for screening and treatment of cervical pre-cancer lesions for cervical cancer prevention, second edition and only regarded 14 genotypes as hrHPV, included type16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68.39
Pyrosequencing
After determination of the amount of gDNA, up to 500 ng of gDNA was subjected to bisulfite conversion using EZ DNA Methylation-Direct ™ Kit (ZYMO RESEARCH, USA). Pyrosequencing was performed to evaluate 2 CpG sites of ZNF671 promoter region (NC_000019.10: 57727411-57727425, ACGGTTTTATGGCGG) on bisulfite-treated DNA in 460 samples from training set. PCR amplification and pyrosequencing primers for target sites were designed using PyroMark Assay Design 2.0. Bisulfite-treated DNA was then amplified using TaKaRa Ex Taq (TaKaRa, Beijing, China). The primers were listed as follows: ZNF671-F:5′- GGTGGAGGTGTTGGGAAA -3′; ZNF671-R: 5′- CTAAAACACAAAAACTACAAACACTTTAC-3′; ZNF671-S: 5′- GTTTGATGTTTTGTAGGGA-3′.The reaction mixture including 13.5 μl of nuclease-free water, 2 μL of 10 × Ex Taq Buffer, 2 μL of dNTP, 0.4 μL former primer and reverse primer, 0.2 μL of Ex Taq HS and 1.5 uL of bisulfite-treated DNA. The PCR product act as a template in Pyrosequencing reactions, using the PyroMark Q24 instrument (Qiagen, MD) according to the manufacturer’s recommended protocol.43 Raw data were analyzed using the Pyromark Q24 analysis software (Qiagen, MD). The average percentage of methylation across 2 CpG sites were obtained.
DNA quantitative methylation-specific PCR
After bisulfite treatment, quantitative methylation-specific PCR (qMSP) was performed on the ABI 7500 Real-Time PCR System (Life Tech, USA) to measure the methylation status of the PAX1 and ZNF671 genes. The methylation status of the PAX1 were detected by Uterine cervix cancer gene methylating Diagnostic Kit (PCR-Fluorescence Probing) (HOOMYA, Changsha, China). The methylation status of ZNF671 genes were detected by qPCR kits developed by our team, with β-actin as an internal reference gene. The primers were listed as follows: ZNF671-QF:5′- GTCGTTTTCGGTAGTTGTTCGC -3′; ZNF671-QR: 5′- AAAAACGCAACACCCACCC -3′; ZNF671-P: 5′-FAM- TTTACGGTTTTATGGCGGA-MGB-3′. β-actin-QF: 5′-AGGAGGTTTGGATTTTTAATTGTGTATAG-3′; β-actin-QR: 5′-AACTCACCACTACAAAAATCAAACCA -3′; β-actin-P: 5′-VIC-TGGTAGAGAGGAAGGTGG-MGB-3′. And the total volume of qMSP reaction mixture was 20 μL, including 18.5 μL of PCR mixture and 1.5 μL of bisulfite template DNA. The mixture included 0.4 μM β-actin-QF, 0.4 μM β-actin-QR, 0.4 μM β-actin-P, 0.45 μM ZNF671-QF, 0.45 μM ZNF671-QR, 0.45 μM ZNF671-P, PCR buffer (1×), 0.25 mM dNTP and 1U/reaction Ex Taq HS. The following qMSP cycle was performed: pre-incubation, 5 min at 95°C; amplification, 50 cycles of 15 s at 95°C, 30 s at 60°C. The Fluorescence data were collected during the annealing or extension step. The crossing point (Cp) value for β-actin (Cp ≤ 35) was set as the validity indicator for the testing. The DNA methylation was calculated as follow: ΔCp = Cptest gene − Cpβ-actin.
Quantification and statistical analysis
SPSS software (version 22.0, Chicago, IL, USA) and R (version 3.6.3) were used for all statistical analyses. Characteristics of patients in Table 1 are presented as means ± SD. Receiver operating characteristic (ROC) curves were generated to confirmed the accuracy of different detection methods. The area under the curve (AUC) for each method to determine different diagnosis groups, including CIN2+/CIN1- and CIN3+/CIN2-, was calculated. The optimal cutoff value (ΔCp) of PAX1m or ΔCp of ZNF671m or average percentage of methylation of ZNF671pyro in training set was generated using the Youden index from 460 subjects. The optimal cutoff value (ΔCp) of ZNF671m in validation set was generated using the Youden index from 1093 subjects.
Sensitivity, specificity, positive predictive values (PPV), negative predictive values (NPV) and with a 95% confidence interval (CI) were calculated. Relative sensitivity, relative specificity and 95% CI were calculated according to the formula reported in previous study.44 If the 95% CI did not contain the value 1, the difference was considered significant between given triage strategies and the reference triage. To evaluate the efficiency of different triage algorithms for detecting CIN2+ or CIN3+ cases, we measured the colposcopy referral rates and number of colposcopies needed to detect per CIN3+ cases (NNC).45 Pre- and post-test CIN3+ risk after application different triage strategies were calculated and visualized as Figure 2.46
Acknowledgments
We sincerely thank all participants in the study. This research was supported by the National Natural Science Foundation(NNSF) of China (81973401). The graphical abstract was created with BioRender.com.
Author contributions
P.Z., J.X., H.Z., and Q.L. conceived and designed the study. P.Z., R.L., Y.H., Y.A., L.Y., and Xiong Li acquired data and performed the data analysis. Y.C. and S.L. provided technical support. J.X., D.Y., Xiang Li, L.L., J.H., B.W., Q.N., S.W., L.D., and P.C. obtained informed consent and acquired patient samples and clinical information. P.Z. wrote the first draft of the manuscript. Q.L. supervised the study. All authors read and approved the final manuscript.
Declaration of interests
Q.L., H.Z., P.Z., Y.C., Y.A., and L.Y. are inventors on a patent related to the detection of ZNF671 gene methylation (publication number: CN112941189A, publication date: June 11, 2021).
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 8, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101143.
Supplemental information
Data and code availability
The data reported in this study are available from the lead contact upon request. This paper does not report original code. No custom computer code was used for data collection, which was performed using open-source software. All analyses used previously published software or methods. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Forouzanfar M.H., Foreman K.J., Delossantos A.M., Lozano R., Lopez A.D., Murray C.J.L., Naghavi M. Breast and cervical cancer in 187 countries between 1980 and 2010: a systematic analysis. Lancet. 2011;378:1461–1484. doi: 10.1016/s0140-6736(11)61351-2. [DOI] [PubMed] [Google Scholar]
- 3.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA. Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA. Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 5.Bruni L., Albero G., Serrano B., Mena M., Gómez D., Munoz J., Bosch F., de Sanjosé S. 2019. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in Mali. Summary Report. [Google Scholar]
- 6.WHO . 2nd edn. World Health Organization; 2014. Comprehensive Cervical Cancer Control: A Guide to Essential Practice. [PubMed] [Google Scholar]
- 7.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 8.Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Muñoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999;189:12–19. doi: 10.1002/(sici)1096-9896(199909)189:1<12::Aid-path431>3.0.Co;2-f. [DOI] [PubMed] [Google Scholar]
- 9.Kulasingam S.L., Hughes J.P., Kiviat N.B., Mao C., Weiss N.S., Kuypers J.M., Koutsky L.A. Evaluation of human papillomavirus testing in primary screening for cervical abnormalities: comparison of sensitivity, specificity, and frequency of referral. JAMA. 2002;288:1749–1757. doi: 10.1001/jama.288.14.1749. [DOI] [PubMed] [Google Scholar]
- 10.Kim J.J. Practice-based evidence for primary HPV testing in the United States. J. Natl. Cancer Inst. 2014;106:dju213. doi: 10.1093/jnci/dju213. [DOI] [PubMed] [Google Scholar]
- 11.Ho G.Y., Bierman R., Beardsley L., Chang C.J., Burk R.D. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 1998;338:423–428. doi: 10.1056/nejm199802123380703. [DOI] [PubMed] [Google Scholar]
- 12.Muñoz N., Bosch F.X., de Sanjosé S., Herrero R., Castellsagué X., Shah K.V., Snijders P.J.F., Meijer C.J.L.M., International Agency for Research on Cancer Multicenter Cervical Cancer Study Group Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 13.de Villiers E.M. Cross-roads in the classification of papillomaviruses. Virology. 2013;445:2–10. doi: 10.1016/j.virol.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Goodman A. HPV testing as a screen for cervical cancer. Bmj. 2015;350:h2372. doi: 10.1136/bmj.h2372. [DOI] [PubMed] [Google Scholar]
- 15.de Sanjosé S., Brotons M., Pavón M.A. The natural history of human papillomavirus infection. Best Pract. Res. Clin. Obstet. Gynaecol. 2018;47:2–13. doi: 10.1016/j.bpobgyn.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Wentzensen N., Schiffman M., Palmer T., Arbyn M. Triage of HPV positive women in cervical cancer screening. J. Clin. Virol. 2016;76(Suppl 1) doi: 10.1016/j.jcv.2015.11.015. S49-s55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Q.J., Hu S.Y., Guo H.Q., Zhang W.H., Zhang X., Chen W., Cao J., Jiang Y., Zhao F.H., Qiao Y.L. Liquid-based cytology and human papillomavirus testing: a pooled analysis using the data from 13 population-based cervical cancer screening studies from China. Gynecol. Oncol. 2014;133:172–179. doi: 10.1016/j.ygyno.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Swid M.A., Monaco S.E. Should screening for cervical cancer go to primary human papillomavirus testing and eliminate cytology? Mod. Pathol. 2022;35:858–864. doi: 10.1038/s41379-022-01052-4. [DOI] [PubMed] [Google Scholar]
- 19.Serrano B., Ibáñez R., Robles C., Peremiquel-Trillas P., de Sanjosé S., Bruni L. Worldwide use of HPV self-sampling for cervical cancer screening. Prev. Med. 2022;154:106900. doi: 10.1016/j.ypmed.2021.106900. [DOI] [PubMed] [Google Scholar]
- 20.Rogeri C.D., Silveira H.C.S., Causin R.L., Villa L.L., Stein M.D., de Carvalho A.C., Arantes L.M.R.B., Scapulatempo-Neto C., Possati-Resende J.C., Antoniazzi M., et al. Methylation of the hsa-miR-124, SOX1, TERT, and LMX1A genes as biomarkers for precursor lesions in cervical cancer. Gynecol. Oncol. 2018;150:545–551. doi: 10.1016/j.ygyno.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Kan Y.Y., Liou Y.L., Wang H.J., Chen C.Y., Sung L.C., Chang C.F., Liao C.I. PAX1 methylation as a potential biomarker for cervical cancer screening. Int. J. Gynecol. Cancer. 2014;24:928–934. doi: 10.1097/igc.0000000000000155. [DOI] [PubMed] [Google Scholar]
- 22.Liou Y.L., Zhang T.L., Yan T., Yeh C.T., Kang Y.N., Cao L., Wu N., Chang C.F., Wang H.J., Yen C., et al. Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clin. Epigenetics. 2016;8:66. doi: 10.1186/s13148-016-0232-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liou Y.L., Zhang Y., Liu Y., Cao L., Qin C.Z., Zhang T.L., Chang C.F., Wang H.J., Lin S.Y., Chu T.Y., et al. Comparison of HPV genotyping and methylated ZNF582 as triage for women with equivocal liquid-based cytology results. Clin. Epigenetics. 2015;7:50. doi: 10.1186/s13148-015-0084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitz M., Eichelkraut K., Schmidt D., Zeiser I., Hilal Z., Tettenborn Z., Hansel A., Ikenberg H. Performance of a DNA methylation marker panel using liquid-based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer. 2018;18:1197. doi: 10.1186/s12885-018-5125-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansel A., Steinbach D., Greinke C., Schmitz M., Eiselt J., Scheungraber C., Gajda M., Hoyer H., Runnebaum I.B., Dürst M. A promising DNA methylation signature for the triage of high-risk human papillomavirus DNA-positive women. PLoS One. 2014;9:e91905. doi: 10.1371/journal.pone.0091905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz M., Wunsch K., Hoyer H., Scheungraber C., Runnebaum I.B., Hansel A., Dürst M. Performance of a methylation specific real-time PCR assay as a triage test for HPV-positive women. Clin. Epigenetics. 2017;9:118. doi: 10.1186/s13148-017-0419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin A., Zhang Q., Kong X., Jia L., Yang Z., Meng L., Li L., Wang X., Qiao Y., Lu N., et al. JAM3 methylation status as a biomarker for diagnosis of preneoplastic and neoplastic lesions of the cervix. Oncotarget. 2015;6:44373–44387. doi: 10.18632/oncotarget.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brentnall A.R., Vasiljevic N., Scibior-Bentkowska D., Cadman L., Austin J., Cuzick J., Lorincz A.T. HPV33 DNA methylation measurement improves cervical pre-cancer risk estimation of an HPV16, HPV18, HPV31 and ∖textit{EPB41L3} methylation classifier. Cancer Biomark. 2015;15:669–675. doi: 10.3233/cbm-150507. [DOI] [PubMed] [Google Scholar]
- 29.Kelly H., Benavente Y., Pavon M.A., De Sanjose S., Mayaud P., Lorincz A.T. Performance of DNA methylation assays for detection of high-grade cervical intraepithelial neoplasia (CIN2+): a systematic review and meta-analysis. Br. J. Cancer. 2019;121:954–965. doi: 10.1038/s41416-019-0593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Strooper L.M.A., van Zummeren M., Steenbergen R.D.M., Bleeker M.C.G., Hesselink A.T., Wisman G.B.A., Snijders P.J.F., Heideman D.A.M., Meijer C.J.L.M. CADM1, MAL and miR124-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J. Clin. Pathol. 2014;67:1067–1071. doi: 10.1136/jclinpath-2014-202616. [DOI] [PubMed] [Google Scholar]
- 31.De Strooper L.M.A., Verhoef V.M.J., Berkhof J., Hesselink A.T., de Bruin H.M.E., van Kemenade F.J., Bosgraaf R.P., Bekkers R.L.M., Massuger L.F.A.G., Melchers W.J.G., et al. Validation of the FAM19A4/mir124-2 DNA methylation test for both lavage- and brush-based self-samples to detect cervical (pre)cancer in HPV-positive women. Gynecol. Oncol. 2016;141:341–347. doi: 10.1016/j.ygyno.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bu Q., Wang S., Ma J., Zhou X., Hu G., Deng H., Sun X., Hong X., Wu H., Zhang L., Luo X. The clinical significance of FAM19A4 methylation in high-risk HPV-positive cervical samples for the detection of cervical (pre)cancer in Chinese women. BMC Cancer. 2018;18:1182. doi: 10.1186/s12885-018-4877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pun P.B., Liao Y.P., Su P.H., Wang H.C., Chen Y.C., Hsu Y.W., Huang R.L., Chang C.C., Lai H.C. Triage of high-risk human papillomavirus-positive women by methylated POU4F3. Clin. Epigenetics. 2015;7:85. doi: 10.1186/s13148-015-0122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia C., Xu X., Zhao X., Hu S., Qiao Y., Zhang Y., Hutubessy R., Basu P., Broutet N., Jit M., Zhao F. Effectiveness and cost-effectiveness of eliminating cervical cancer through a tailored optimal pathway: a modeling study. BMC Med. 2021;19:62. doi: 10.1186/s12916-021-01930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halford J.A., Batty T., Boost T., Duhig J., Hall J., Lee C., Walker K. Comparison of the sensitivity of conventional cytology and the ThinPrep Imaging System for 1,083 biopsy confirmed high-grade squamous lesions. Diagn. Cytopathol. 2010;38:318–326. doi: 10.1002/dc.21199. [DOI] [PubMed] [Google Scholar]
- 36.Özcan Z., Kimiloğlu E., Iğdem A.A., Erdoğan N. Comparison of the Diagnostic Utility of Manual Screening and the ThinPrep Imaging System in Liquid-Based Cervical Cytology. Turk Patoloji Derg. 2020;36:135–141. doi: 10.5146/tjpath.2019.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moscicki A.-B., Ma Y., Wibbelsman C., Darragh T.M., Powers A., Farhat S., Shiboski S. Rate of and Risks for Regression of Cervical intraepithelial Neoplasia 2 in Adolescents and Young Women. Obstet. Gynecol. 2010;116:1373–1380. doi: 10.1097/AOG.0b013e3181fe777f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ovestad I.T., Gudlaugsson E., Skaland I., Malpica A., Munk A.C., Janssen E.A.M., Baak J.P. The impact of epithelial biomarkers, local immune response and human papillomavirus genotype in the regression of cervical intraepithelial neoplasia grades 2-3. J. Clin. Pathol. 2011;64:303–307. doi: 10.1136/jcp.2010.083626. [DOI] [PubMed] [Google Scholar]
- 39.WHO Guideline for Screening and Treatment of Cervical Pre-cancer Lesions for Cervical Cancer Prevention . Vol. 7. World Health Organization; 2021. p. 6. [PubMed] [Google Scholar]
- 40.Melnikow J., Henderson J.T., Burda B.U., Senger C.A., Durbin S., Weyrich M.S. Screening for Cervical Cancer With High-Risk Human Papillomavirus Testing: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA. 2018;320:687–705. doi: 10.1001/jama.2018.10400. [DOI] [PubMed] [Google Scholar]
- 41.Chrysostomou A.C., Stylianou D.C., Constantinidou A., Kostrikis L.G. Cervical Cancer Screening Programs in Europe: The Transition Towards HPV Vaccination and Population-Based HPV Testing. Viruses. 2018;10:729. doi: 10.3390/v10120729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L., Zhao X., Hu S., Chen S., Zhao S., Dong L., Carvalho A.L., Muwonge R., Zhao F., Basu P. Triage performance and predictive value of the human gene methylation panel among women positive on self-collected HPV test: Results from a prospective cohort study. Int. J. Cancer. 2022;151:878–887. doi: 10.1002/ijc.34041. [DOI] [PubMed] [Google Scholar]
- 43.Iwagami S., Baba Y., Watanabe M., Shigaki H., Miyake K., Ishimoto T., Iwatsuki M., Sakamaki K., Ohashi Y., Baba H. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann. Surg. 2013;257:449–455. doi: 10.1097/SLA.0b013e31826d8602. [DOI] [PubMed] [Google Scholar]
- 44.Cheng H., Macaluso M. Comparison of the accuracy of two tests with a confirmatory procedure limited to positive results. Epidemiology. 1997;8:104–106. doi: 10.1097/00001648-199701000-00017. [DOI] [PubMed] [Google Scholar]
- 45.Rezhake R., Chen F., Hu S.Y., Zhao X.L., Zhang X., Cao J., Qiao Y.L., Zhao F.H., Arbyn M. Triage options to manage high-risk human papillomavirus-positive women: A population-based cross-sectional study from rural China. Int. J. Cancer. 2020;147:2053–2064. doi: 10.1002/ijc.33001. [DOI] [PubMed] [Google Scholar]
- 46.Dick S., Vink F.J., Heideman D.A.M., Lissenberg-Witte B.I., Meijer C.J.L.M., Berkhof J. Risk-stratification of HPV-positive women with low-grade cytology by FAM19A4/miR124-2 methylation and HPV genotyping. Br. J. Cancer. 2022;126:259–264. doi: 10.1038/s41416-021-01614-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this study are available from the lead contact upon request. This paper does not report original code. No custom computer code was used for data collection, which was performed using open-source software. All analyses used previously published software or methods. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.