Summary
Although radiotherapy (RT) has achieved great success in the treatment of non-small cell lung cancer (NSCLC), local relapses still occur and abscopal effects are rarely seen even when it is combined with immune checkpoint blockers (ICBs). Here, we characterize the dynamic changes of tumor-infiltrating immune cells after RT in a therapy-resistant murine tumor model using single-cell transcriptomes and T cell receptor sequencing. At the early stage, the innate and adaptive immune systems are activated. At the late stage, however, the tumor immune microenvironment (TIME) shifts into immunosuppressive properties. Our study reveals that inhibition of CD39 combined with RT preferentially decreases the percentage of exhausted CD8+ T cells. Moreover, we find that the combination of V-domain immunoglobulin suppressor of T cell activation (VISTA) blockade and RT synergistically reduces immunosuppressive myeloid cells. Clinically, high VISTA expression is associated with poor prognosis in patients with NSCLC. Altogether, our data provide deep insight into acquired resistance to RT from an immune perspective and present rational combination strategies.
Keywords: NSCLC, radiotherapy, immunotherapy, scRNA-seq, CD8 T cells, TANs, tumor immune microenvironment
Graphical abstract

Highlights
-
•
scRNA-seq analyses reveal dynamic changes in tumor immune microenvironment after RT
-
•
The immune system is initially activated but shifts to a suppressive state over time
-
•
CD39i and RT combination reduces CD8+ T cell exhaustion and tumor growth
-
•
Anti-VISTA inhibits RT-induced aggregation of TANs and improves RT efficacy
Zhang et al. demonstrate how the tumor immune microenvironment dynamically changes after radiotherapy by using scRNA-seq and scTCR-seq. Based on that, CD39 inhibition and anti-VISTA blockade can enhance radiotherapy efficacy by activating CD8+ T cells and reducing the infiltration of TANs.
Introduction
Non-small cell lung cancer (NSCLC) remains the leading cause of cancer-related mortality despite considerable advances in surgery, radiotherapy, and targeted therapies.1 Immune checkpoint blockers (ICBs) have dramatically revolutionized cancer treatment paradigms, yet only a minority of patients with specific cancers can derive a durable clinical benefit.2,3 Although clinical trials have demonstrated that ICBs such as PD-1/PD-L1 antibodies show promising therapeutic effects against NSCLC, the response rate of unselected patients is still approximately 20%.4 Compared with immune-inflamed (“hot”) tumors, immune-desert (“cold”) tumors exhibit intrinsic resistance to ICBs by multiple mechanisms, such as low mutation load and rare infiltrating immune effector cells.5 Therefore, potential and effective combination strategies are required for further exploration to increase response rates.
Preclinical and clinical trials have demonstrated that radiotherapy has vast potential to improve the efficacy of immunotherapy.6,7 Therefore, combining radiotherapy and immunotherapy has generated much interest in the past few years, and many clinical trials are underway.8 Nevertheless, despite the high response rate, there remains a substantial fraction of patients with NSCLC that show refractory or acquired resistance to radiotherapy plus ICB therapy.9,10
It is critical to understand the immune mechanisms driving intrinsic or acquired resistance to optimize existing combination therapy. Moreover, previous studies revealed that radiotherapy may have both immunostimulatory (e.g., in situ vaccination and recruitment of T cells) and immunosuppressive effects (e.g., hypoxia and expansion of regulatory T cells).11,12,13 Unfortunately, owing to the heterogeneity and complexity of the tumor microenvironment (TME), the comprehensive and dynamic immune microenvironment changes after radiotherapy have not yet been fully characterized.
Here, we take advantage of the Lewis lung cancer model, which is poorly immunogenic, aggressive, and insensitive to various treatments, including radiotherapy and immunotherapy, to elucidate the mechanisms driving therapeutic resistance.14,15 By utilizing high-dimensional single-cell RNA sequencing (scRNA-seq) and paired T cell receptor sequencing (scTCR-seq), we thoroughly depicted the immune microenvironment changes and mechanisms after radiotherapy at early and late time points. Most importantly, we focused on CD8+ T cell reinvigoration and immunosuppressive tumor-associated neutrophil (TAN) inhibition via radiotherapy combined with CD39 or VISTA blockade to enhance efficacy.
Results
CD8+ TILs show increased infiltration but shift toward an exhausted state at the late stage after RT
To elucidate the dynamic tumor immune microenvironment (TIME) changes after radiotherapy (RT; 20 Gy), we performed droplet-based 5′ scRNA-seq and paired scTCR-seq on CD45+ immune cells in the ovalbumin-expressing Lewis lung carcinoma (LLC-OVA) model in three groups: a control group (10 days after inoculation), an RT-04 group (day 14), and an RT-10 group (day 20) (Figure 1A). Mice received RT (20 Gy) at day 10 after the tumor challenge. In total, 31,880 cells passed quality control. They were further identified as immune cell types, including monocytes/macrophages, neutrophils, dendritic cells (DCs), T cells, natural killer cells (NKs), B cells, plasma cells, and a small portion of non-immune cell types, including osteoclasts and fibroblasts (Figure 1B). The non-immune cells were excluded from subsequent analysis. The constitution of tumor-infiltrating immune cell types is shown in Figure 1C. The percentages of lymphocytes and neutrophils appreciably increased, while the percentages of monocytes and macrophages decreased 10 days after RT.
Figure 1.
CD8+ TILs show an exhausted state and clonal expansion at the late point after RT
(A) Schematic illustration of the study design.
(B) Uniform manifold approximation and projection (UMAP) plot of all cells passed quality control from all samples, colored by cell identities.
(C) Stacked bar plot showing the proportion of major immune cell types originating from different samples.
(D) t-Distributed stochastic neighbor embedding (t-SNE) plot of lymphocytes from all samples, colored by identified cell clusters.
(E) t-SNE showing the expression of selected genes in each cell cluster.
(F) Stacked bar plot showing the proportion of lymphocyte clusters originating from different samples.
(G) Violin plot of G2/M.Score and S.Score in each lymphocyte cluster.
(H) t-SNE showing the TCR distribution in lymphocyte clusters colored according to detected TCR (left) and clonal TCR (right). Clonal TCR indicates that the same TCR was detected in at least two cells.
(I) Bar plot showing the number of cells with clonal TCR in lymphocyte clusters.
(J) Volcano plot showing differentially expressed genes between clonal CD8-Tex and non-clonal CD8-Tex cells. The most significant genes are labeled in the graph.
See also Figure S1.
Reclustering of T cells and innate lymphoid cells revealed 14 clusters based on marker genes expression and TCR characteristics, including four subsets of CD4+ T cells (Th1, Th2, Th17, and Foxp3+ regulatory T cells [Tregs]); five subsets of CD8+ T cells (CD8-cytotoxic T lymphocyte [CTL], CD8-Gzmk, CD8-Ifit, CD8-central memory T cell [Tcm], and CD8-exhausted T cell [Tex]); two subsets of NKs (NK01-Gzmc, NK02-Klra8); two subsets of NKTs (NKT-αβ, NKT-γδ); and a subset of type 3 innate lymphoid cells (ILC3-NCR+) (Figures 1D, 1E, and S1A–S1D). Interestingly, similar to T cells, NKT cells could be generally divided into two groups based on differentially expressed TCR chains (Figures S1E and S1F). Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) revealed that NKT-γδ was related to cytokine secretion and T cell activation, while NKT-αβ was more related to granzyme-mediated apoptosis and cytolysis (Figure S1G).
The cytotoxic CD8-CTL subset showed the highest proliferation ability among the T cell subsets (Figure 1G). Moreover, the CD8-CTL subset increased at 4 days after RT, while the terminal exhausted CD8-Tex subset was significantly decreased (Figure 1F), suggesting the activation of antitumor immunity at an early time point. In addition, the proportions and proliferation of NKs and NKTs were also increased 4 days after RT (Figures 1F and S1H), suggesting activation of the innate immune systems.
RT can induce tumor neoantigens and greater diversity of the TCR repertoire.16,17 Consistently, the TCR clone diversity was increased at 4 days after RT (Figures 1H and 1I). However, we did not find an apparent clonal expansion of CTLs 4 days after RT. In fact, clonal expansion in almost all T cell subsets decreased shortly after RT (Figures 1H and 1I). Moreover, there was no TCR clonal expansion on NKTs, indicating that their TCR repertoires were highly diverse (Figure 1I). Taken together, these results indicated that RT might damage tumor-infiltrating T cells, which limited clonal expansion shortly after RT. However, other changes induced by RT, such as necrosis of tumor cells and release of tumor neoantigens, could recruit T cells, NKs, and NKTs from peripheral blood, which enhanced the antitumor property at that time point.
However, clonal expansion of terminal exhausted CD8-Tex and pre-exhausted CD8-Gzmk subsets was obviously increased at 10 days after RT (Figures 1H, 1I, and S1I). Moreover, compared with non-clonal CD8-Tex cells, the clonal cells highly expressed exhaustion genes, such as Pdcd1, Lag3, and Tigit, and expressed lower levels of genes related to immune memory and cytotoxicity, such as Tcf1, Sell, and Gzma (Figure 1J). Interestingly, clonal CD8-Tex cells highly expressed Cxcr6 (Figure 1J). Recently, CXCR6+ CD8+ tumor-infiltrating lymphocytes (TILs) were reported to have tumor antigen specificity and could maximize antitumor activity before progressing to irreversible dysfunction.18,19 In addition, the percentage of the cytotoxic CD8-CTL subset decreased 10 days after RT. These data implied that even though the proportion of CD8+ T cells was increased in total immune cells, they showed a trend toward exhaustion and dysfunction over time after RT.
Combining CD39 inhibition with RT suppresses tumor growth and reduces exhausted CD8+ TILs
Our scRNA-seq data analysis revealed that exhausted CD8+ TIL clusters clonally expanded after RT at the late time point, which was associated with RT and immunotherapy resistance.16,20 CD39 (Entpd1) was identified as a critical exhaustion marker21 and was overexpressed in exhausted CD8+ T cell subsets in pan-cancer and NSCLC scRNA-seq data22 (Figures 2A and S2A). Moreover, the expression of CD39 in CD8+ TILs was dramatically increased after RT (Figure 2B). Therefore, we investigated the combined effect of RT and CD39 inhibition. As described in Figure 2C, mice bearing LLC-OVA were treated with RT and/or POM-1, a small-molecule inhibitor of CD39. Notably, compared with isotype and monotherapies, combination therapy elicited significant tumor growth inhibition (Figures 2D and S2B). Additionally, we evaluated the antitumor activity of combination therapy in other refractory murine tumor models that were poorly immunogenic, highly aggressive, and resistant to both RT and ICBs, including B16F10 of melanoma and LLC models. Conversely, we also studied the immunogenic MC38 of a colon carcinoma model that was more responsive to ICBs and RT to reexamine the therapeutic efficacy. Similarly, combination therapy with RT and CD39i significantly improved antitumor activity and extended overall survival in B16F10, LLC, and MC38 models (Figures 2D and S2C–S2I). Besides, we observed a significantly higher complete response (CR) rate of 25% in mice treated with the combination of RT and CD39i in the MC38 model (Figure S2I). Furthermore, subcutaneous tumor rechallenge was performed on day 100, and the MC38 cells were eradicated in all mice of CR (Figures S2J and S2K), suggesting that combination therapy established durable antitumor immune memory.
Figure 2.
RT and CD39 inhibition combination therapy synergistically suppresses tumor growth and reduces exhausted CD8+ TILs
(A) Violin plot and boxplot showing the expression of CD39 (Entpd1) across CD8+ TILs metaclusters in pan-cancer. Tex, exhausted T cells.
(B) Representative histograms (left) and tabulated percentages of CD39+ cells among CD8+ TILs (n = 5 per group).
(C) A schematic summary of the treatment regimen.
(D) Tumor growth of LLC-OVA-bearing mice treated with the indicated treatments (left, n = 10 per group); tumor growth of B16F10-bearing and MC38-bearing mice treated with the indicated treatments (right, n = 8 per group).
(E) Percentage of intratumoral CD8+ T cells among CD45+ cells (n = 5 per group).
(F) Representative flow plot (left) and tabulated percentages (right) of effector memory phenotype (TEM; CD62Llow CD44high) and naive phenotype (TN; CD62Lhigh CD44low) cells among CD8+ TILs (n = 5 per group).
(G) Representative flow plot (left) and tabulated percentages (right) of IFNγ+ TNFα+, IFNγ+, and TNFα+ cells among CD8+ TILs (n = 5 per group).
(H) Percentage of GraB+ cells among CD8+ TILs (n = 5 per group).
(I) Representative histograms and tabulated percentages of TIM-3+, PD-1+, CD39+, and TIGIT+ cells among CD8+ TILs (n = 5 per group).
(J) Representative flow plot (left) and tabulated percentages of TOX+ cells among CD8+ TILs (n = 5 per group).
(K) Percentage of TCF-1+ PD-1+ cells among CD8+ TILs (n = 5 per group).
(L) Tumor growth of LLC-OVA-bearing mice treated with the indicated treatments (n = 10 per group). Statistical significance was calculated among four groups on day 28 (RT, CD39i +RT, αPD1+RT, CD39i+αPD1+RT).
(M) Survival curves of LLC-OVA-bearing mice treated with the indicated treatments (n = 10 per group).
Flow cytometry analysis was done 10 days after RT. Error bars depict means ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant, as determined by unpaired two-tailed Student’s t test (B), by one-way ANOVA with Tukey’s correction (E–K), by two-way ANOVA with Tukey’s correction (D and L), or by Kaplan-Meier method with log rank test (M). Data shown are representative of 2–3 independent experiments.
See also Figures S2 and S3.
To assess whether combination therapy could reinvigorate exhausted CD8+ T cells, we investigated the infiltration, proliferation, phenotype, function, and exhaustion state of CD8+ TILs at the late time point (10 days post-RT). Among all the groups, combination therapy exhibited the highest percentage of infiltrating CD8+ T cells (Figure 2E), although no significant difference was observed in proliferation (Ki67+; Figure S2L). In addition, compared with RT alone, the combination therapy further increased the proportion of the effector memory phenotype (TEM; CD62Llow CD44high) and decreased the proportion of the naive phenotype (TN; CD62Lhigh CD44low). In contrast, similar proportions of the central memory phenotype (TCM; CD62Lhigh CD44high) CD8+ TILs were observed among all the groups (Figures 2F and S2M). These results indicated that RT combined with CD39i contributed to enhanced infiltration of CD8+ TILs, especially effector-like CD8+ TILs.
To further evaluate the function of CD8+ T cells, we examined cytokine and cytotoxic granule secretion. Among all groups, CD8+ TILs from the combinational group comprised the highest frequencies of CD8+ TILs producing GzmB, tumor necrosis factor α (TNF-α), and interferon γ (IFN-γ) and co-producing TNF-α and IFN-γ (Figures 2G and 2H), particularly TNF-α. These results indicated that RT combined with CD39 inhibition increased the proportion of polyfunctional CD8+ TILs and mobilized effective antitumor immune responses.
Consistent with previous studies and our scRNA-seq results, although RT could induce cytokine production in CD8+ TILs to some degree, RT inevitably skewed CD8+ TILs toward an exhausted phenotype and further abrogated the therapeutic efficacy.15,23 To determine whether combination therapy could reinvigorate exhausted CD8+ TILs, we evaluated the dysfunction markers PD1, TIM3, CD39, TIGIT, and transcription factors, including TCF1 and TOX. Combination therapy significantly decreased the percentage of inhibitory receptor expression that was elevated after RT at the late time point (Figure 2I). TOX has been reported to transcriptionally regulate and maintain terminally exhausted CD8+ TILs.24 We further found a lower percentage of TOX+ CD8+ TILs in the combination group than in the RT group (Figure 2J). In contrast, Tcf1+ PD-1+ CD8+ TILs, defined as the stem-like phenotype, could differentiate into effector-like CD8+ TILs25 and showed the highest percentage in the combination group (Figure 2K). Considering that Tcf1+ PD-1+ CD8+ TIL accumulation was essential for ICB response,26 we proposed that additional PD-1 blockade would enhance the combination therapy. As expected, triple therapy further suppressed tumor growth (Figures 2L and S3A) and enhanced survival (Figure 2M). These results indicated that RT combined with CD39 inhibition decreased the percentage of exhausted CD8+ TILs and reprogrammed CD8+ TILs to acquire stem-like properties.
Combining CD39 inhibition with RT promotes TAM phenotypic changes and DC maturation
RT can release abundant proinflammatory extracellular ATP (eATP) by causing cell damage, and CD39 inhibitors can prevent eATP from further conversion into immunosuppressive adenosine (ADO).27 Furthermore, high levels of proinflammatory eATP in the TME could exert immunostimulatory effects by multiple mechanisms, including enhancing the proinflammatory phenotype of macrophages and tumor antigen presentation.27,28,29 Therefore, we examined changes in TAMs and observed that combination therapy not only dramatically reduced TAM infiltration but also skewed TAMs toward a proinflammatory M1-like phenotype (Figure S3B). Moreover, the combination therapy achieved the highest percentage of mature DCs, although the percentage of total DCs showed no difference among the groups (Figures S3C and S3D). Furthermore, there was no significant proportion change in monocytic myeloid-derived suppressor cells (M-MDSCs) and TANs in the combination group (Figure S3E), but the combination group exhibited the lowest expression level of CD39 (Figure S3F), suggesting less immunosuppressive activity. The massive infiltration of immunosuppressive myeloid cells and rapidly proliferating tumor cells might limit the efficacy of combination therapy. Considering that both RT and chemotherapy can cause cell death and can release large amounts of eATP, we next tried combination strategies of RT, CD39i, and carboplatin. Triple therapy significantly reduced tumor growth rates and prolonged survival (Figures S3G–S3I). These findings suggested that CD39 inhibition plus RT promoted M1-like TAM infiltration and DC maturation.
The function of DCs was partially impaired after RT
To gain further insights into the changes in DCs after RT, we characterized five DC subsets with marker genes: Itgae+ cDC1, Mki67+ cDC1-cycling, Itgam+ cDC2, Ccr7+ mature DC (mDC), and Siglech+ plasmacytoid DC (pDC) (Figures 3A and 3B). There was no remarkable change in the percentage of DCs in total immune cells after RT (Figure 1C). cDC1 was reported to prime CD8+ T cells by antigen cross-presentation, and cDC2 was able to activate CD4+ T cells.30,31 The percentage of proliferating cDC1 subsets was decreased at 10 days after RT, while the percentage of mature DC (mDC) was increased (Figure 3C).
Figure 3.
DCs consist of diverse subpopulations, and their function was partially impaired after RT
(A) t-SNE plot showing all DCs from all samples, colored by identified cell clusters.
(B) Violin plot showing the expression of marker genes in DC subsets.
(C) Stacked bar plot showing the proportion of DC clusters originating from different samples.
(D) Stratification of DC transcriptomes using scores for DC clusters. Single cells were colored by DC cluster annotation (left) or derivation of mDCs (medium). Stacked bar plot showing the proportion of derivation of mDC originating from different samples (right).
(E) Volcano plot showing differentially expressed genes between cDC1-derived mDCs and cDC2-derived mDCs. The most significant genes are labeled in the plot. p values were adjusted with the Benjamini-Hochberg method.
However, a high single dose of radiation might result in insufficient DC recruitment and activation.12 Moreover, compared with immature and semi-mature DCs, only fully mature DCs were immunogenic.32 Since mDCs could derive from both cDC1s and cDC2s, we next evaluated the derivation of mDCs. Compared with cDC2s, the cDC1-derived mature DC gradually decreased after RT, suggesting restriction of cDC1 maturation (Figure 3D). Moreover, consistent with a recent study in human and mouse NSCLC,33 cDC2-derived mDCs expressed higher Il1b and Cd274 levels, whereas cDC1-derived mDCs expressed higher Il12b (Figure 3E), suggesting that blocking cDC1 maturation may exacerbate the dysfunction of CD8+ T cells.33,34 Altogether, we observed that the function of DCs was partially impaired after RT, especially at the late time point.
Monocytes-macrophages show enhanced differentiation to the immunosuppressive direction at the late time point after RT
The immunosuppressive myeloid cells have been implicated in inducing CD8+ TIL exhaustion and supporting tumor progression. We next interrogated the heterogeneous phenotypes of myeloid compartments and evaluated their functional properties after RT. Apart from two cell-cycle-driven clusters (Figures S4A and S4B), we identified five monocyte and four macrophage subsets and annotated them according to the feature genes (Figure 4A). The Mo1 and Mo4 monocyte clusters were significantly enriched in peripheral blood mononuclear cells compared to tumors35 (Figure 4B). This indicated that Mo1 and Mo4 might be peripheral blood-derived subsets, while other monocyte and macrophage subpopulations were educated locally in tumors.36 Since then, we set Mo1 as the beginning of the differentiation and finished the trajectory with Monocle3. As shown in Figure 4C, three different differentiation directions were found. There was no obvious change in the proportion of Direction 1, indicating Direction 1 was almost not influenced by RT (Figure 4D). Feature genes of macrophages were found at the terminus of this direction (Figures 4E and S4C). Moreover, GO analysis showed that the terminal subset of direction 1 (M05_Mono-lift3) was related to active innate immune and IFN-related responses (Figure 4F).
Figure 4.
RT changes the monocyte/macrophage differentiation states linked to clinical outcomes
(A) UMAP plot showing monocytes and macrophages from all samples. Cells are colored by identified cell clusters.
(B) Boxplot showing the proportion of selected subsets in peripheral blood mononuclear cells per sample. p values were calculated with two-tailed Student’s t test.
(C) Trajectory analysis for the monocyte and macrophage clusters. Each dot represented an individual cell colored by clusters (top) or pseudotime (bottom).
(D) Stacked bar plot showing the proportion of monocyte and macrophage clusters from different samples.
(E) Heatmap showing the dynamic gene expression changes along the pseudotime of direction 1.
(F) Gene Ontology enrichment analysis of DEGs for Mo5 cluster. p values were adjusted with the Benjamini-Hochberg method.
(G) Differential enriched pathways in directions 2 (MΦ1 and MΦ2) and 3 (MΦ3 and MΦ4). Pathway activity was measured per cell by GSVA. The difference between two groups was measured by the Limma package. p values were adjusted with the Benjamini-Hochberg method.
(H) Heatmap showing the expression of selected genes in monocyte and macrophage clusters.
(I) Heatmap showing the dynamic expression of transcription factors along the pseudotime.
(J) UMAP plot showing myeloid cells from normal lung tissues and NSCLC tissues, colored by identified cell cluster.
(K) Stacked bar plot showing the proportion of myeloid cell clusters originating from different tissues.
(L) Heatmap showing the difference in pathway activities scored by GSVA per cell in different macrophage clusters. The t values from a lineal model are shown.
(M) The Kaplan-Meier overall survival curves of TCGA LUAD that were grouped by the gene signature of selected macrophage subsets.
See also Figure S4.
Unlike direction 1, direction 2 decreased progressively, whereas direction 3 increased after RT (Figure 4D). Gene set variation analysis (GSVA) revealed that antigen processing and presentation, positive regulation of T cell activation pathways, and phagocytosis were enriched in direction 2 (MΦ1 and MΦ2), corresponding to the “phagocytosis TAM” found in human cancer37,38 (Figure 4G). In contrast, neutrophil migration, response to hypoxia, and angiogenesis in direction 3 (MΦ3 and MΦ4) were similar to “angiogenesis TAM” in humans37,38 (Figure 5G). Consistently, MΦ1 and MΦ2 highly expressed major histocompatibility complex (MHC) class II molecules genes, while MΦ3 and MΦ4 highly expressed immunosuppressive and tissue repair genes, such as Arg1 and Spp1 (Figure 4H). Compared with MΦ1 and MΦ2, MΦ3 and MΦ4 expressed low levels of Sirpa, Cd276, Csf1r, and Trem2, which are current therapeutic targets on TAMs (Figure 4H), pending new strategies targeting immunosuppressive TAMs induced by RT. To interrogate the mechanism involved in the differentiation, we performed SCENIC analysis to evaluate the activity of transcription factors per cell. We found that HIF-1α was persistently activated when Mo2 tended to differentiate into direction 3 TAMs (Figure 4I). Conversely, HIF-1α regulon was detected only at a very low level in direction 2 TAMs (Figure 4I). This result suggested that the RT-induced differentiation of TAMs may be driven by HIF-1α.
Figure 5.
TANs gradually aggregate after RT
(A) UMAP plot of all neutrophils colored by cluster.
(B) Expression levels of immature markers (left) and mature markers (right) in neutrophils.
(C) Bubble heatmap showing selected genes in neutrophil clusters.
(D) Heatmap showing the difference in pathway activities scored by GSVA per cell in different neutrophil clusters. The t values from a lineal model are shown.
(E) Sankey diagrams showing the changes in the percentage of neutrophil clusters in CD45+ cells.
(F) Stacked bar plot showing the proportion of neutrophil clusters originating from different samples.
(G) Bubble heatmap showing interactions between chemokine receptors (left) and ligands (right) in all cell types.
(H) Bubble heatmap showing the expression of chemokine-coding genes in human lung NSCLC samples from public scRNA-seq datasets.
(I) The gene expression levels of Cxcl2 and Cxcl6 in colorectal cancer tissue pre- and post-RT. ∗p < 0.05; ∗∗p < 0.01, two-tailed Student’s t test.
(J) Heatmap showing gene expression in different clusters. Mega, megakaryocytes; Mono, monocytes.
To investigate the clinical value of direction 2 and 3 TAMs, we analyzed a single-cell dataset of human NSCLC.39 An appreciable difference was observed between myeloid cells from healthy and tumor tissues (Figures 4J and S4D–S4F). Tissue-resident macrophages (TRMs), also known as alveolar macrophages, were the major compartment of healthy tissue but were significantly decreased in NSCLC (Figure 4K). In contrast, several tumor-specific clusters, including Macro-HK2, Macro-HSP, and Macro-MT, were enriched in tumor tissues (Figure 4K). Via GSVA, we found a higher level of hypoxia and angiogenesis activity in Macro-HK2, Macro-HSP, and Macro-MT, suggesting that these clusters might serve as direction 3-like populations in humans (Figure 4L). Additionally, higher levels of signatures of these clusters were related to poor prognosis in the TCGA lung adenocarcinoma (LUAD) dataset, although Macro-MT did not show statistical significance (Figures 4M and S4G). Furthermore, the Macro-FOLR2 subset was also enriched in the TME and was functionally related to MHC class II antigen presentation (Figures 4K and 4L), suggesting this subset might serve as direction 2-like populations in humans. Moreover, the high signatures of the Macro-FOLR2 subset predicted favorable overall survival in LUAD (Figure S4G), resembling recently reported FOLR2+ macrophages associated with CD8+ T cell infiltration.40 Taken together, these results suggested that hypoxia-related direction 3 TAMs could be a potential target for patients with NSCLC, especially after RT.
Neutrophils gradually aggregate after RT
In addition to monocytes and macrophages, neutrophils were a major and important component of myoid cells in tumors. Notably, our results showed that neutrophils gradually aggregated after RT (Figure 1C). To further dissect the clusters and functions of neutrophils, we distinguished five subsets based on marker genes: Neutro-Hilpda, Neutro-Rsad2, Neutro-Lrg1, Neutro-ribo, and Neutro-Ly6g (Figures 5A and 5B). Some researchers introduced the N1 and N2 nomenclature to define TANs with antitumor and protumor functions, respectively.41 However, we found that both N1 (Cxcl10, Tnf, Ccl2, and Ccl3) and N2 (Arg2, Entpd1, Trem1, and Cd274) neutrophil-related genes42,43 were expressed in all subsets (Figure 5C). These results indicated that the definitions of N1 and N2 did not show clear dichotomies, suggesting the subpopulations may represent differential activation states (Figure 5D). In the process of accumulation of neutrophils after RT, all the subpopulations increased synchronously (Figure 5E). Only the Rsad2+ cluster showed short enrichment 4 days after RT, which may be related to the response to IFN (Figures 5D–5F).
To investigate what recruited neutrophils into the tumor region after RT, we performed ligand-receptor (L-R) analysis with cellphoneDB. Notably, among all the cytokine and chemokine receptors, only Ccr1 and Cxcr2 were expressed on neutrophils. Cross-talk between neutrophils and monocytes/macrophages via Cxcl1/Cxcl2/Cxcl3-Cxcr2 and Ccl3/Ccl7/Ccl8-Ccr1 may account for the recruitment of neutrophils (Figure 5G). Additionally, neutrophils themselves expressed the highest levels of Cxcl2 and Cxcl3, indicating that neutrophils had an aggregation tendency. Notably, direction 3 TAMs secreted more neutrophil-recruiting chemokines than direction 2 TAMs. Since direction 3 differentiation of macrophages was enhanced after RT, it could also explain why neutrophils accumulated in tumor regions after RT.
In human NSCLC samples,39,44 neutrophil-recruiting chemokines (Cxcl1, Cxcl2, Cxcl5, Cxcl6, Cxcl8) were also commonly expressed by cancer cells (Figure 5H). In addition, Cxcl2 and Cxcl6 were significantly upregulated after RT in human microsatellite-stable (MSS) colorectal cancer45 (Figure 5I), while data for NSCLC were still unavailable. Recent work demonstrated that neutrophils could promote resistance to RT.46,47 It was also reported that a higher level of Cxcl8 in peripheral blood could lead to resistance to chemotherapy or ICBs.48 Consistently, we analyzed single cells from ICB responders and non-responders with bladder cancer,48 finding that not only Cxcl8 but also Cxcl1, Cxcl2, and Cxcl3 were highly expressed among the non-responders, indicating that the accumulation of neutrophils may be related to resistance to ICB therapy (Figure 5J). Collectively, these results suggested that immunosuppressive neutrophils were recruited by chemokines and aggregated at the late time point after RT, which may be related to resistance to RT and immunotherapy and could be a potential target.
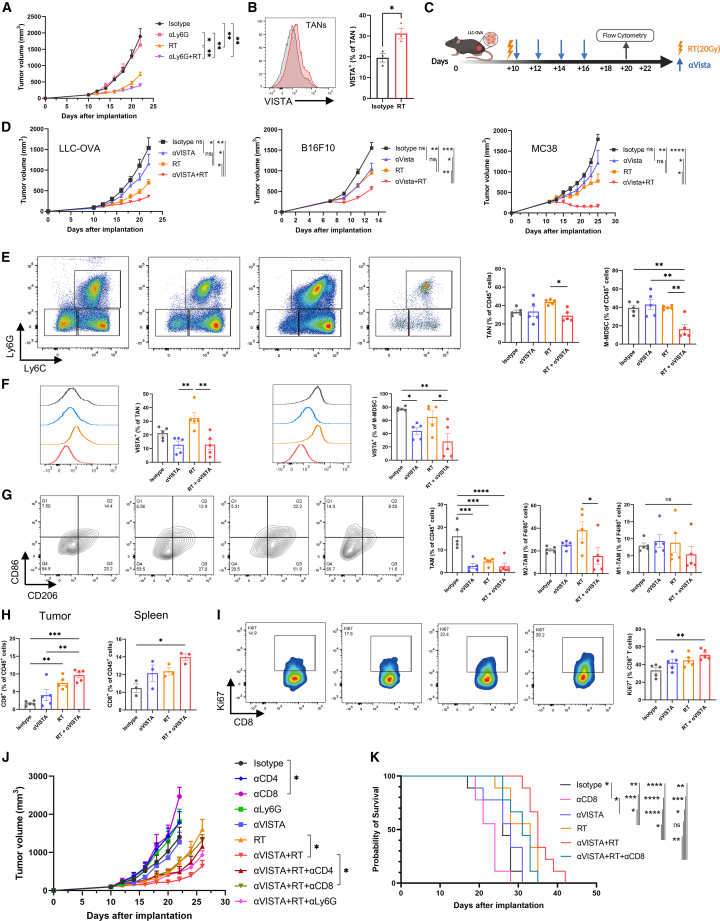
Combining anti-VISTA with RT synergistically reduces tumor growth and immunosuppressive myeloid element infiltration
Considering that neutrophils exhibited more immunosuppressive function in our data, we hypothesized that neutrophil accumulation impaired RT efficacy. We next depleted neutrophils by utilizing an anti-Ly6G antibody. Compared with RT alone, RT combined with anti-Ly6G further reduced the tumor volume (Figures 6A and S5A). However, considering that a neutralizing neutrophil therapeutic strategy might be inappropriate for clinical application due to its potential side effects, we next analyzed and explored myeloid targets in our data.
Figure 6.
RT and anti-VISTA combination therapy synergistically reduces tumor growth and immunosuppressive myeloid element infiltration
(A) Tumor growth of LLC-OVA-bearing mice treated with the indicated treatments (isotype, αLy6G, RT, αLy6G+RT, n = 8 per group).
(B) Representative histograms (left) and tabulated percentages of VISTA+ cells among intratumoral TANs (n = 3 per group).
(C) A schematic summary of the treatment regimen.
(D) Tumor growth of LLC-OVA-bearing mice treated with the indicated treatments (isotype, αVISTA, RT, αVISTA+RT, n = 10 per group, left); tumor growth of B16F10-bearing mice and MC38-bearing mice treated with the indicated treatments (right, n = 8 per group).
(E) Representative flow plot (left) and tabulated percentages (right) of intratumoral TANs (CD11b+Ly6G+Ly6Cint) and M-MDSCs (CD11b+Ly6G−Ly6Chigh) (n = 5 per group).
(F) Representative histograms and tabulated percentages of VISTA+ cells among intratumoral TANs and M-MDSCs (n = 5 per group).
(G) Representative flow plot (left) and tabulated percentages of M1-like (CD86+) and M2-like (CD206+) cells among intratumoral TAMs (CD11b+ F4/80+) (n = 5 per group).
(H) Percentages of CD8+ T cells in the tumor (left, n = 5 per group) and spleen (right, n = 3 per group).
(I) Representative flow plot (left) and tabulated percentage (right) of Ki67+ cells among CD8+ TILs (n = 5 per group).
(J) Tumor growth of LLC-OVA-bearing mice treated with the indicated treatments (n = 9 per group). Statistical significance was calculated among the first five groups on day 22 (isotype, αCD4, αCD8, αLy6G, αVISTA) and among the last five groups on day 26 (RT, αVISTA+RT, αVISTA+RT+αCD4/αCD8/αLy6G).
(K) Survival curves of LLC-OVA-bearing mice treated with the indicated treatments (n = 9 per group).
Flow cytometry analysis was done 10 days after RT. Error bars depict means ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant, as determined by unpaired two-tailed Student’s t test (B), by one-way ANOVA with Tukey’s correction (E–I), by two-way ANOVA with Tukey’s correction (A, D, and J), or by Kaplan-Meier method with log rank test (K). Data shown are representative of 2–3 independent experiments.
See also Figures S5 and S6.
VISTA has been reported to be predominantly expressed on myeloid compartments and implicated in patients with ICB resistance.49,50 In our study, VISTA was highly expressed in myeloid-derived cells at RNA and protein levels (Figures 6B, S5B, and S5C). Moreover, VISTA significantly increased in TANs 10 days after RT but not in M-MDSCs and TAMs (Figures 6B and S5C). To examine the antitumor effect of RT and VISTA blockade combination therapy, we treated LLC-OVA tumors with immunoglobulin G (IgG) isotype control (200 μg/dose, intraperitoneally [i.p.]), anti-VISTA (200 μg/dose, i.p.), RT (20 Gy), or anti-VISTA plus RT (Figure 6C). As a result, combination therapy synergistically inhibited tumor progression relative to isotype control and monotherapies (Figures 6D and S5D). The synergistic antitumor efficacy of anti-VISTA and RT was further tested in other mouse models, including B16F10, LLC, and MC38 models. Consistently, combination therapy group showed significantly lower growth kinetics and increased survival (Figures 6D and S5E–S5K). Furthermore, the combination therapy achieved a 37.5% CR rate and triggered sustained antitumor immune memory against rechallenged tumor cells in cured mice in the MC38 model (Figures S5L and S5M). Together, these four distinct murine models confirmed that VISTA blockade combined with RT elicited a robust systemic antitumor response and immune memory.
To evaluate whether additional anti-VISTA could inhibit the accumulation of TANs caused by RT, we analyzed the tumor myeloid cell infiltration at 10 days post-RT. In line with scRNA-seq data, the TAN percentage increased after RT alone but remarkably decreased in combination therapy (Figure 6E). Moreover, compared with isotype and monotherapies, combination therapy also included notable diminishment of M-MDSCs (Figure 6E). Furthermore, compared with RT, combination therapy significantly decreased VISTA expression in TANs and M-MDSCs (Figure 6F).
Considering that the spleen was the major peripheral reservoir for MDSCs,51 we analyzed splenic MDSCs. Interestingly, we observed splenomegaly in the isotype group and that spleen size markedly decreased after combination therapy (Figure S6A), which was consistent with the corresponding reduction of tumor burden. Moreover, combination therapy induced a significant decrease in TANs (Figure S6B). Additionally, the M-MDSCs also showed the most significant reduction in the combination group (Figure S6B). Accordantly, prior studies have reported that the decreasing splenic volume and splenic MDSCs were associated with desirable therapy responses.52,53 These data indicated that combination therapy reduced the accumulation of both peripheral and tumor-infiltrating TANs and M-MDSCs.
Due to the TME, especially hypoxia, being able to affect MDSCs’ differentiation to TAMs and DCs,54 we next analyzed the changes in TAMs and DCs. Compared with isotype control, either monotherapies or combination therapy decreased the total percentage of TAMs (Figure 6G). Furthermore, combination therapy remarkably reduced the percentage of M2-like TAMs (Figure 6G), despite maintaining a similar proportion of M1-like TAMs (Figure 6G). Moreover, there were no specific changes in proportions of total DCs (Figure S6C), but the percentage of mature DCs was increased in either monotherapy or combination therapy (Figure S6C). Collectively, these data indicated that combination therapy synergistically suppressed immunosuppressive myeloid element infiltration.
Combining anti-VISTA with RT synergistically promotes the activation and expansion of CD8+ TILs
As CD8+ T cells were critical for antitumor immunity, we next investigated the potential mechanisms of combination therapy by evaluating CD8+ T cells. Combination therapy established the highest expansion of CD8+ T cells in both tumor milieus and spleen (Figure 6H) and the highest proliferation (Ki67+) of CD8+ TILs (Figure 6I). Conversely, there was no specific change in either splenic and intratumoral CD4+ T cells or Tregs among the groups (Figures S6D and S6E). We further performed the depletion study via anti-CD4, anti-CD8, or anti-Ly6G neutralizing antibodies. The synergistic antitumor efficacy was partially abrogated by depletion of CD8+ T cells but not of CD4+ T cells or Ly6G+ neutrophils (Figures 6J, 6K, S6F, and S6G). These results suggested that combination therapy exerted antitumor effects in a CD8-dependent manner and that it increased intratumoral and peripheral CD8+ T cell expansion.
We further investigated the function and status of CD8+ TILs. Of note, the CD8+ TILs produced more TNF-α and IFN-γ and co-produced TNF-α+ IFN-γ+ in the combination group, particularly more IFN-γ (Figure 7A). Additionally, we measured the expression of the co-inhibitory receptors PD1, CD39, TIM3, and TIGIT in CD8+ TILs. Interestingly, compared with RT alone, the combination therapy significantly decreased the expression of CD39, TIM3, and TIGIT and the terminally exhausted subset (PD-1+TIM3+) in CD8+ TILs (Figures 7B and 7C). These findings revealed that the combination of anti-VISTA and RT further rescued antitumor functions of CD8+ TILs and reduced the dysfunctional phenotype.
Figure 7.
RT and anti-VISTA combination therapy promotes the activation and expansion of CD8+ TILs
(A) Representative flow plot (left) and tabulated percentages (right) of IFNγ+ TNFα+, IFNγ+, and TNFα+ cells among CD8+ TILs (n = 5 per group).
(B) Representative flow plot (left) and tabulated percentage (right) of TIM-3+ PD-1+ cells among CD8+ TILs (n = 5 per group).
(C) Representative histograms and tabulated percentages of PD-1+, TIM-3+, CD39+, and TIGIT+ cells among CD8+ TILs (n = 5 per group).
(D) Representative histograms of proliferation and tabulated percentages of proliferation index of CFSE-labeled CD8+ T cells after 72 h co-culture with TANs isolated from LLC-OVA tumors (n = 4 per group).
(E) A schematic summary of the treatment regimen.
(F) Tumor growth of LLC-OVA-bearing mice treated with the indicated treatments (n = 10 per group). Statistical significance was calculated among four groups on day 28 (RT, αVISTA+RT, αPD1+RT, αVISTA+αPD1+RT).
(G) Survival curves of LLC-OVA-bearing mice treated with the indicated treatments (n = 10 per group).
(H) Data are representative of VISTA-stained tumor or adjacent non-tumor tissues (scale bar, 100 μm) in LUSC (left) or LUAD (right) tissue microarrays.
(I) Quantitative analysis of VISTA expression in tumor or adjacent non-tumor tissues in LUSC (right, n = 90) or LUAD (left, n = 85) tissue microarrays.
(J) Survival curves of overall survival of patients with LUSC or LUAD stratified by the VISTA expression level.
Flow cytometry analysis was done 10 days after RT. Error bars depict means ± SEM. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, not significant, as determined by one-way ANOVA with Tukey’s correction (A–D), by two-way ANOVA with Tukey’s correction (F), by paired two-tailed Student’s t test (I), or by Kaplan-Meier method with log rank test (G and J). Data shown are representative of 2–3 independent experiments.
To investigate whether radiation-induced TANs directly inhibit CD8+ T cell function, we isolated TANs from tumors and co-cultured them with activated CD8+ T cells in vitro. Compared with the control group, CD8+ T cells co-cultured with radiation-induced TANs showed lower levels of proliferation capacity (CFSE) and activation marker CD69 (Figures 7D, S6H, and S6I). Notably, by adding anti-VISTA antibody, the decline in proliferation capacity and activation in CD8+ T cells was reversed compared with that in the RT group. These data suggested that radiation-induced TANs utilized VISTA to inhibit the proliferation and activation of CD8+ T cells.
Of note, we observed that PD-1 expression in CD8+ TILs did not show rapid downregulation in combination therapy (Figure 7C). Given these findings and that dual blockade of PD-1 and VISTA could exert synergistic antitumor effects,55 we hypothesized that triple therapy may further restrain tumor progression (Figure 7E). Indeed, triple-combination therapy was significantly superior to all other treatments (Figures 7F, 7G, and S6J). These data demonstrated that dual targeting of PD-1 and VISTA in combination with RT significantly limited tumor growth and elicited durable antitumor responses.
Overexpression of VISTA in tumors is associated with poor prognosis in patients with NSCLC
VISTA expression in the TME was correlated with opposite prognoses in different cancer types, suggesting that antagonistic VISTA therapy may specifically benefit particular patient subsets.56 Additionally, limited studies investigated VISTA expression level and prognostic value in NSCLC, and their conclusions were controversial.57 To interrogate the clinical relevance of VISTA in human NSCLC, we evaluated VISTA expression in LUAD (n = 180) and lung squamous cell carcinoma (LUSC) (n = 180) tissue microarrays by immunohistochemistry (IHC). The results showed that the VISTA protein level was significantly higher in tumor stroma than in paired peri-tumor tissues (Figures 7H and 7I). In addition, the patients with high VISTA protein expression had worse overall survival than patients with low VISTA expression in both LUAD and LUSC (Figure 7J). These data confirmed that VISTA could be a prognostic biomarker and a potential therapeutic target for NSCLC.
Discussion
Increasing evidence has suggested that RT has the potential to be a game changer in enhancing the depth and efficacy of responses to ICBs by modulating the immune microenvironment. However, RT has complex effects on the immune system that could augment immune activation or suppression. Therefore, a thorough understanding of the immunological effects of RT could aid the design of appropriate combination therapy of RT and immunotherapy in preclinical studies and further inform the clinical regimen.
Here, we systematically elaborated immune cell responses after RT at early and late time points by utilizing comprehensive scRNA-seq and scTCR-seq. Together, these data demonstrated that immune system activation occurred at the early time point, including increased proliferation and activation of NK, NKT, and cytotoxic CD8-CTL subsets. Similarly, previous studies demonstrated that radiation could contribute to in situ vaccination effects by inducing immunogenic cell death and MHC class I expression, which further prominently increased T cell priming and infiltration.58,59,60 However, at the late time point, the immune microenvironment exhibited a trend toward immunosuppression and RT resistance. Our data showed clonal expansion and an increased proportion of the exhausted CD8-Tex subset. Since tumor-reactive T cells were reported to exhibit exhaustion phenotypes due to persistent antigen stimulation,61,62 we speculated that RT-induced sustained neoantigen exposure and presentation caused CD8 exhaustion and clonal expansion. Meanwhile, clusters with unexpanded TCRs were like to be generated recently or to be bystanders. Moreover, our data demonstrated that RT enhanced the differentiation direction of proangiogenic macrophage populations. Further analysis showed that HIF-1α was persistently activated in the proangiogenic differentiation, suggesting that RT-induced hypoxia could enhance this differentiation. Nevertheless, transcriptome data on the HIF-1α activity were circumstantial evidence. Future studies are required to validate this speculation. Similarly, a recent study suggested that radiation-increased lactate induced HIF-1α signaling in MDSCs and contributed to the radioresistance.63 In addition, our analysis showed that RT dramatically induced immunosuppressive neutrophil accumulation. Recent studies also showed that neutrophils recruited to the TME after RT could facilitate tumor radioresistance or even enhanced metastatic propensity.47,64 Moreover, elevated circulating neutrophils were associated with adverse prognosis and poor local control in patients treated with radiation-based therapy.47,65
Based on the data analysis, we sought to explore the potential immunotherapy targets explored in clinical trials to improve and prolong the therapeutic effects of RT in two ways: (1) reducing the exhausted CD8+ T cell by a CD39 inhibitor and (2) inhibiting the accumulation of immunosuppressive myeloid cells, especially remarkably enriched TANs, by VISTA blockade.
CD39 has been reported to be a marker of exhausted CD8+ TILs and to be associated with poor prognosis and immunotherapy failure.21,66 Indeed, we found that CD39 expression was significantly elevated after RT in CD8+ TILs, and combination therapy (CD39 inhibitor and RT) exhibited remarkable tumor suppression. Additionally, we demonstrated that additional CD39 inhibition reduced the exhausted phenotype of CD8+ TILs after RT and synergistically increased the infiltration, proliferation, and cytokine production of CD8+ TILs. Interestingly, several studies have shown that CD39 expression distinguished tumor neoantigen-specific CD8+ TILs from bystander CD8+ TILs and portended survival and ICB responses.67,68,69 These conflicting findings suggested that CD39 was upregulated by antigen-driven, stimulated CD8+ T cells and that it acquired an exhausted phenotype to prevent an excessive immune response under normal conditions, which was detrimental to the tumor situation. Based on our results, we tended to speculate that suppressing reactive CD8+ TIL exhaustion by inhibiting CD39 improved the therapeutic efficacy.
Unlike CD73, inhibition of CD39 could not only reduce ADO accumulation but could also maintain proinflammatory eATP levels.70 Consistently, we also found that the combination therapy increased M1-like proinflammatory TAM infiltration and DC maturation. However, a recent study showed that CD39 deficiency adversely affected RT efficacy and exacerbated radiation-induced normal lung toxicity.71 In contrast to specific inhibition of CD39 activities, genetic defects of CD39 may lead to dysregulation of physiologic functions, including angiogenesis and inflammatory responses.72 Of note, despite POM1 is a potent inhibitor of CD39, it might be limited by off-target actions. Further studies are needed to verify the feasibility and efficacy of CD39 inhibition or blockade in combination with RT in clinical trials.
Considering that neutrophils are a substantial immune cell population in the lung cancer model43 and are even further abundantly increased after RT, we hypothesized the neutrophils were involved in CD8+ TIL dysfunction and that they impaired the efficacy of RT. To test this hypothesis, we first depleted neutrophils in combination with RT and found that this synergistically dampened tumor growth, suggesting that neutrophils played an immunosuppressive role to a greater extent in RT resistance. To explore better clinically applicable therapy, our next analysis identified that VISTA was expressed in TANs and significantly increased after RT. Furthermore, a combination of anti-VISTA and RT synergistically suppressed tumor growth and significantly decreased the aggregation of TANs that were elevated after RT, M-MDSCs and M2-like TAMs. Strikingly, the combination group also enhanced CD8+ TIL infiltration and activation. An early study showed that hypoxia upregulated VISTA expression via HIF-1α and that hypoxia-induced VISTA contributed to MDSC-mediated T cell suppression.73 Moreover, VISTA was identified as an acidic pH-selective ligand for the co-inhibitory receptor P-selectin glycoprotein ligand-1 (PSGL-1), and its immunoinhibitory activity was enhanced at the acidic pH of the TME.74 These results suggested that RT-induced hypoxia and acidic pH might potentially improve VISTA expression and function to cause an immunosuppressive milieu, which was turned into a synergistically therapeutic opportunity.
Limitations of the study
First, the mouse subcutaneous tumor model did not accurately mimic the TIME of human NSCLC. Nevertheless, obtaining NSCLC specimens before and after RT was relatively difficult. Second, by sorting immune cells for scRNA-seq and scTCR-seq, we could enrich low-abundance immune cell subpopulations and guarantee ideal sequencing depth. However, other compartments and states of the TME, such as metabolic status, hypoxia, tumor cells, fibroblasts, endothelial cells, and their cross-talk, were not explored in this study. Third, certain conclusions were made based on transcriptome-level analysis. Thus, further detailed and rigorous experimental validation is required, such as the differentiation of monocytes and macrophages and the regulation of transcription factors after RT. Fourth, our study is primarily a preclinical exploration. Further studies will be warranted to examine the possibility of clinical translation. Despite these limitations, our study illuminates dynamic TIME changes after RT and provides a rational basis for optimizing combinations of RT and ICBs, which have the potential to translate into clinical practice.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD45 (clone 30-F11) | Biolegend | Cat# 103132; RRID: AB_893340 |
| Anti-mouse CD8a (clone 53-6.7) | Biolegend | Cat# 100730; RRID: AB_493703 |
| Anti-mouse CD8a (clone 53-6.7) | Biolegend | Cat# 100752; RRID: AB_2563057 |
| Anti-mouse CD4 (clone GK1.5) | Biolegend | Cat# 100406; RRID: AB_312691 |
| Anti-mouse PD-1 (clone 29F.1A12) | Biolegend | Cat# 135220; RRID: AB_2562616 |
| Anti-mouse Tim-3 (clone RMT3-23) | Biolegend | Cat# 119738; RRID: AB_2810368 |
| Anti-mouse CD39 (clone Duha59) | Biolegend | Cat# 143806; RRID: AB_2563394 |
| Anti-mouse CD39 (clone Duha59) | Biolegend | Cat# 143804; RRID: AB_11218603 |
| Anti-mouse TIGIT (clone 1G9) | Biolegend | Cat# 142111; RRID: AB_2687311 |
| Anti-human/mouse Granzyme B (clone QA16A02) | Biolegend | Cat# 372204; RRID: AB_2687028 |
| Anti-mouse IFN-γ (clone XMG1.2) | Biolegend | Cat# 505808; RRID: AB_315402 |
| Anti-mouse TNF-α (clone MP6-XT22) | Biolegend | Cat# 506328; RRID: AB_2562902 |
| Anti-mouse/human CD44 (clone IM7) | Biolegend | Cat# 103008; RRID: AB_312959 |
| Anti-mouse CD62L (clone MEL-14) | Biolegend | Cat# 104412; RRID: AB_313099 |
| Anti-mouse/human CD11b (clone M1/70) | Biolegend | Cat# 101206; RRID: AB_312789 |
| Anti-mouse Ly-6G (clone 1A8) | Biolegend | Cat# 127652; RRID: AB_2616733 |
| Anti-mouse Ly-6C (clone HK1.4) | Biolegend | Cat# 128033; RRID: AB_2562351 |
| Anti-mouse F4/80 (clone BM8) | Biolegend | Cat# 123122; RRID: AB_893480 |
| Anti-mouse CD206 (clone C068C2) | Biolegend | Cat# 141717; RRID: AB_2562232 |
| Anti-mouse CD86 (clone GL-1) | Biolegend | Cat# 105037; RRID: AB_11204429 |
| Anti-mouse I-A/I-E (clone M5/114.15.2) | Biolegend | Cat# 107622; RRID: AB_493727 |
| Anti-mouse CD11c (clone N418) | Biolegend | Cat# 117318; RRID: AB_493568 |
| Anti-mouse VISTA (clone MIH63) | Biolegend | Cat# 150204; RRID: AB_2566412 |
| Anti-mouse/human Ki-67 (clone 11F6) | Biolegend | Cat# 151206; RRID: AB_2566801 |
| Anti-mouse CD69 (clone H1.2F3) | Biolegend | Cat# 104532; RRID: AB_2562327 |
| Rat IgG2b, κ Isotype Ctrl (clone H1.2F3) | Biolegend | Cat# 400626; RRID: AB_389343 |
| Rat IgG1, κ Isotype Ctrl (clone RTK2071) | Biolegend | Cat# 400407; RRID: AB_326513 |
| Rat IgG1, κ Isotype Ctrl (clone RTK2071) | Biolegend | Cat# 400429; RRID: AB_10900998 |
| Mouse IgG1, κ Isotype Ctrl (clone MOPC-21) | Biolegend | Cat# 400141; RRID: AB_2941933 |
| Rat IgG2a, κ Isotype Ctrl (clone RTK2758) | Biolegend | Cat# 400508; RRID: AB_326530 |
| Rat IgG2a, κ Isotype Ctrl (clone RTK2758) | Biolegend | Cat# 400535; RRID: AB_10933427 |
| Rat IgG2a, κ Isotype Ctrl (clone RTK2758) | Biolegend | Cat# 400539; RRID: AB_11126979 |
| Anti-mouse CD25 (clone PC61.5) | eBioscience | Cat# 12-0251-82; RRID: AB_465607 |
| Anti-mouse/Rat Foxp3 (clone FJK-16s) | eBioscience | Cat# 17-5773-82; RRID: AB_469457 |
| Rat IgG2a kappa Isotype Control (clone eBR2a) | eBioscience | Cat# 17-4321-81; RRID: AB_470181 |
| Anti-mouse TCF-7/TCF-1 (clone S33-966) | BD Biosciences | Cat# 564217; RRID: AB_2687845 |
| Mouse IgG1, κ Isotype Control (clone MOPC-21) | BD Biosciences | Cat# 554680; RRID: AB_395506 |
| Anti-human/mouse TOX (clone REA473) | Miltenyi Biotec | Cat# 130-118-335; RRID: AB_2751522 |
| Human IgG1 (clone REA293) | Miltenyi Biotec | Cat# 130-120-709; RRID: AB_2661686 |
| Anti-mouse CD16/32 (clone S17011E) | Biolegend | Cat# 156604; RRID: AB_2783138 |
| InVivoMAb anti-mouse VISTA (clone 13F3) | BioXcell | Cat# BE0310; RRID: AB_2736990 |
| InVivoMAb polyclonal Armenian hamster IgG | BioXcell | Cat# BE0091; RRID: AB_1107773 |
| InVivoMAb anti-mouse PD-1 (CD279) (clone 29F.1A12) | BioXcell | Cat# BE0273; RRID: AB_2687796 |
| InVivoMAb rat IgG2a isotype control (clone 2A3) | BioXcell | Cat# BE0089; RRID: AB_1107769 |
| InVivoMAb anti-mouse CD8α (clone YTS 169.4) | BioXcell | Cat# BE0117; RRID: AB_10950145 |
| InVivoMAb rat IgG2b isotype control (clone LTF-2) | BioXcell | Cat# BE0090; RRID: AB_1107780 |
| InVivoMAb anti-mouse CD4 (clone GK1.5) | BioXcell | Cat# BE0003-1; RRID: AB_1107636 |
| InVivoMAb anti-mouse Ly6G (clone 1A8) | BioXcell | Cat# BE0075-1; RRID: AB_1107721 |
| Recombinant Anti-VISTA antibody (clone EPR21050) | Abcam | Cat# ab238886; RRID: AB_2940797 |
| Purified anti-mouse CD3ε Antibody | Biolegend | Cat# 100359; RRID: AB_2616673 |
| Purified anti-mouse CD28 Antibody | Biolegend | Cat# 102121; RRID: AB_2810330 |
| Recombinant Murine IL-2 | PEPROTech | Cat# 212-12 |
| Biological samples | ||
| Human tissue microarrays of lung squamous cell carcinoma | Shanghai Outdo Biotech |
Cat# HLugS180Su02 |
| Human tissue microarrays of lung adenocarcinoma | Shanghai Outdo Biotech |
Cat# HLugA180Su04 |
| Chemicals, peptides, and recombinant proteins | ||
| DNase I | Roche | Cat# 10104159001 |
| Collagenase Type 4 | Rockland | Cat# MB-121-0100 |
| ACK Lysing Buffer | Thermo Fisher | Cat# A1049201 |
| Zombie UV™ Fixable Viability Kit | Biolegend | Cat# 423108 |
| Foxp3 / Transcription Factor Staining Buffer Set | eBioscience | Cat# 00-5523-00 |
| CFSE | eBioscience | Cat# 65-0850-84 |
| POM 1 | Santa Cruz Biotechnology | Cat# sc-203205 |
| Carboplatin | MedChemExpress | Cat# HY-17393 |
| Leukocyte Activation Cocktail | BD Biosciences | Cat# 550583; RRID: AB_2868893 |
| Critical commercial assays | ||
| Tumor Dissociation Kit, mouse | Miltenyi Biotec | Cat# 130-096-730 |
| Dead Cell Removal Kit | Miltenyi Biotec | Cat# 130-090-101 |
| CD45 (TIL) MicroBeads, mouse | Miltenyi Biotec | Cat# 130-110-618 |
| MojoSort™ Mouse CD8a Selection Kit | Biolegend | Cat# 480136 |
| MojoSort™ Mouse Ly-6G Selection Kit | Biolegend | Cat# 480124 |
| Chromium Single Cell 5’ Library Kit | 10X Genomics | PN-1000002 |
| Chromium Single Cell 5’ Gel Bead Kit | 10X Genomics | PN-1000003 |
| Chromium Single Cell 3’/5’ Library Construction Kit | 10X Genomics | PN-1000020 |
| Chromium Single Cell A Chip Kits | 10X Genomics | PN-1000009 |
| Chromium i7 Multiplex Kit | 10X Genomics | PN-120262 |
| Dynabeads MyOne™ SILANE | 10X Genomics | PN-2000048 |
| Deposited data | ||
| ScRNASeq data | This paper | GSA: CRA011692 |
| ScRNA-seq data of lung adenocarcinoma | Kim et al.39 | GEO: GSE131907 |
| ScRNA-seq data of advanced NSCLC | Wu et al.44 | GEO: GSE148071 |
| RNA-seq data of colon cancer treated with radiotherapy | Parikh et al.45 | GEO: GSE179351 |
| ScRNA-seq data of patients treated with atezolizumab | Yuen et al.48 | GEO: GSE145281 |
| TCGA LUAD bulk RNA-seq | The Cancer Genome Atlas (TCGA) | https://xenabrowser.net/datapages/ |
| Pan-cancer and NSCLC scRNA-seq data | Zheng et al.22 | http://cancer-pku.cn:3838/PanC_T/ |
| Mouse Cell Atlas (MCA) | Han et al.35 | http://bis.zju.edu.cn/MCA/ |
| Experimental models: Cell lines | ||
| MC38 | BMCR/NICR | Cat# 1101MOU-PUMC000523 |
| LLC | ATCC | Cat# CRL-1642 |
| B16F10 | ATCC | Cat# CRL-6475 |
| LLC-OVA | Maintained in our Lab | |
| Experimental models: Organisms/strains | ||
| C57/BL6J mice | SPF (Beijing) Biotechnology | N/A |
| Software and algorithms | ||
| FlowJo v10.6.2 | Tree Star | https://www.flowjo.com/ |
| GraphPad Prism v8.2.0 | GraphPad | https://www.graphpad.com/ |
| Cell Ranger v3.1.0 | 10X Genomics | http://10xgenomics.com/ |
| R v3.6.1 | R Core Team. | https://cran.r-project.org/ |
| Seurat v3.0.1 | Stuart et al.75 | https://satijalab.org/seurat/ |
| Harmony v0.1.0 | Korsunsky et al.76 | https://github.com/immunogenomics/harmony |
| clusterProfiler v3.10.1 | Yu et al.77 | http://bioconductor.org/packages/clusterProfiler/ |
| GSVA v1.42.0 | Hanzelmann et al.78 | https://github.com/rcastelo/GSVA |
| survminer v0.4.9 | survminer R package: Survival Data Analysis and Visualization. | https://cran.r-project.org/web/packages/survival/ |
| CellPhoneDB v 2.0.6 | Efremova et al.79 | https://github.com/Teichlab/cellphonedb |
| monocle3 v1.0.0 | Cao et al.80 | https://github.com/cole-trapnell-lab/monocle3/releases |
| pySCENIC v0.11.0 | Aibar et al.81 | https://github.com/aertslab/pySCENIC |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zhen Tao (ztao@tmu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell line
The mouse lung carcinoma line LLC, Ovalbumin-expressing LLC (LLC-OVA), melanoma cell line B16F10 and colon carcinoma MC38 were regularly tested for mycoplasma contamination. LLC, LLC-OVA and B16F10 cells were cultured at 37°C and 5% CO2 in DMEM medium (Gibco) with 10% fetal bovine serum (Sigma-Aldrich), 100 U/ml penicillin and 100 ug/ml streptomycin (Gibco). MC38 cells were cultured at 37°C and 5% CO2 in 1640 medium (Gibco) with 10% fetal bovine serum (Sigma-Aldrich), 100 U/ml penicillin and 100 ug/ml streptomycin (Gibco).
In vivo models
Six- to eight-week-old female C57BL/6J mice were purchased from SPF (Beijing) Biotechnology Co., Ltd. All mice were maintained under specific pathogen-free conditions. Animal studies were conducted under the approval of the Animal Ethical and Welfare Committee of Tianjin Medical University Cancer Institute and Hospital. Approximately 2 ×106 LLC-OVA cells,1 ×106 LLC cells, MC38 cells and B16F10 cells were inoculated subcutaneously into the right dorsal flank of mice. The mice were randomized to experimental groups when tumors grew to 80 to 130 mm3 in LLC-OVA and LLC models and 250-300 mm3 in MC38 and B16F10 models. Tumor volume was calculated as (length × width × width)/2.
Method details
Immunotherapies
For immune-checkpoint blockade therapy, anti-mouse VISTA (BioXCell; 13F3; BE0310) 200μg/dose and anti-mouse PD-1 (BioXCell; 29F.1A12; BE0273) 200μg/dose were intraperitoneally (i.p.) injected every 2 days (days 10,12,14 and 16). Polyclonal Armenian hamster IgG (BioXCell; BE0091) 200μg/dose and rat IgG2a isotype control (BioXCell; 2A3; BE0089) 200μg/dose was i.p. injected to the control (isotype) group every 2 days. For CD39 inhibition therapy, POM-1 (polyoxometalate-1) 5mg/kg/day (Santa Cruz Biotechnology; sc-203205), a CD39 inhibitor, was i.p. injected on a daily basis (days 10 to 20). The same volume of PBS was i.p. injected as an isotype control. For chemotherapy, carboplatin (MedChemExpress; HY-17393) 50mg/kg was i.p. injected once on day10. The same volume of PBS was i.p. injected as an isotype control.
For CD8+, CD4+ and Ly6G+ cell depletion, anti-mouse CD8α (BioXCell; YTS 169.4; BE0117), anti-mouse CD4 (BioXCell; GK1.5; BE0003-1) or anti-mouse Ly6G (BioXCell; 1A8; BE0075-1) monoclonal antibodies 200μg/dose were i.p. injected every 3 days (days 10,13,16 and 19). Rat IgG2b isotype control (BioXCell; LTF-2; BE0090) or rat IgG2a isotype control 200μg/dose was i.p. injected every 3 days. Depletion efficacy was evaluated by flow cytometry analysis of spleen samples (Figure S7A). For radiotherapy, a single fraction of 20 Gy was provided on day 10 and before other therapy initiation.
Radiation therapy
Radiotherapy was performed using a Varian 600CD (Varian Medical Systems, Palo Alto, CA) linear accelerator with 6 MV X-ray photon beams. Five separated radiation fields with a size of 2 cm × 2 cm each on the isocenter plane (source to the isocenter plane is 100 cm) were simultaneously formed by utilizing the jaw collimator and the multileaf collimator (MLC). The gantry and collimation angles were 0 degrees to guarantee the beam incident vertically downward. The tumor-bearing mice were anesthetized and placed in customized controlling devices, and they were placed on a 10cm thick solid water phantom with a source-to-tumor distance of 100 cm. To ensure an adequate dose, the tumor of each mouse was placed within 1 cm × 1 cm of the center of each radiation field while the rest of the body was placed away from the radiation fields as far as possible, and a 1.5 cm thick phantom for dose build-up was placed above the mice. Single fraction radiotherapy of 20 Gy dose treatment was applied with a 5 Gy/min absorbed dose rate. All experiments were carried out under the supervision of medical physicists and engineers. A schematic summary of the device and regimen was shown in Figure S7B.
Sample dissociation
Subcutaneous tumors or spleens were collected on the days indicated in the figures. Freshly isolated samples were dissected away from any surrounding fat, fibrous, necrotic areas and minced into small pieces in RPMI-1640 medium (Gibco) containing 10% fetal bovine serum. For scRNA-seq experiments, tumor samples were dissociated with mouse tumor dissociation kit (Miltenyi Biotec) according to manufacturer’s instructions and performed as follows: gentleMACS Program 37C_m_TDK_1 using gentleMACS Octo Dissociator with Heaters (Miltenyi Biotec). For flow cytometry analysis, tumor samples were digested with 0.1 mg/ml DNase I (Roche) and 0.5 mg/ml collagenase Type 4 (Rockland) for 30 min at 37°C. After the above two methods of dissociation, single-cell suspensions were filtered successively through 70-μm and 40-μm cell strainers (BD Falcon), followed by gentle mechanical dissociation of the remaining tissue left on the strainer with a plunger to increase cell yield. Filtered cells were incubated with ACK lysis buffer (Thermo Fisher) to lyse red blood cells. For scRNA-seq experiments, live cells were purified by Dead Cell Removal Kit (Miltenyi Biotec). Then, tumor-infiltrating CD45+ leukocytes were isolated by mouse CD45 (TIL) MicroBeads (Miltenyi Biotec).
Single-cell library preparation and sequencing
To obtain a sufficient number of CD45+ leukocytes and biological replicates of scRNA-seq experiments, three samples from the same group at each time point were pooled before magnetic beads sorting. Cellular virality was assessed by trypan blue staining and ensured to be higher than 90%. The concentration of single-cell suspension was adjusted to nearly 1000 live cells/μl. The single cells were loaded onto the 10 Genomics Chromium Controller and encapsulated using the Chromium Single Cell 5′ Reagent Kit and Gel Bead Kit (10× Genomics). 5′ gene expression libraries and TCR libraries were constructed following the manufacturer’s recommendations. All libraries were sequenced on the Illumina Nova-seq 6000 platform.
ScRNA-seq data processing and downstream analysis
The Cell Ranger toolkit (10x Genomics, v3.1.0) was utilized to align reads to the mm10 (v3.0.0) reference genome and generate the matrix of genes versus cells. Further quality control of each cell was applied as follows: (1) The number of unique molecular identifier (UMI) counts > 1000; (2) The number of detected genes > 300; (3) The proportion of mitochondrial gene expression <10%. A total of 31880 cells were included in further downstream analysis.
Unsupervised clustering and markers identification
Further analysis was performed with Seurat (v3.0.1)75 following recommended pipeline of Seurat (https://satijalab.org/seurat/). The top 3000 highly variable genes were identified for principal component analysis (PCA). Considering batch effect between different libraries, Harmony(v0.1.0)76 was used to remove the batch effect. Then the top 50 PCs were selected for further dimension reduction. Cell clusters were called by “FindClusters” function with resolution set as default. Clusters were manually annotated using differentially expressed genes from the “FindAllMarkers” function with selecting method “vst.” Cell cycle scores were assessed using “CellCycleScoring” function in Seurat.
Differentially expressed gene and pathway analysis
Differentially expressed genes were identified using “FindAllMarkers” or “FindMarkers” function in Seurat with FDR < 0.05 & average log2 fold change > 1. GO enrichment analysis was performed by clusterProfiler (v 3.10.1).77 For gene set enrichment analysis, gene set variation analysis (GSVA) was applied by using GSVA (v1.42.0)78 package. Differences between different cell groups were calculated with a linear model offered from the Limma package. Entpd1 expression in pan-cancer and NSCLC scRNA-seq data was analyzed through the online data browser (http://cancer-pku.cn:3838/PanC_T/).22
TCR analysis
The TCR-seq data were processed using Cell Ranger (v 3.1.0) against the mouse vdj reference genome supplied by 10× Genomics. TCR clonotypes were defined as TCRs sharing the same CDR3 sequences. Cells with the same TCR sequence (threshold value >1) were defined as clonal cells.
TCGA data and survival analysis
The Cancer Genome Atlas (TCGA) LUAD gene expression and survival data were downloaded from UCSC Xena (https://xenabrowser.net/datapages/). The samples were grouped into high and low expression subgroups by the mean expression values. Kaplan–Meier survival curves were plotted using the function “ggsurvplot” of R package survminer (v 0.4.9), and log-rank p values were calculated in R.
Cell-cell interaction analysis
The ligand-receptor pairs analysis between each cell type was assessed by CellPhoneDB (v 2.0.6).79 Only genes 1). coding cytokines, chemokines and their receptors 2). detected in over 10% cells of at least one cell cluster were used in this analysis.
Cell developmental trajectory inference
When investigating the origin of Mo1 and Mo4 monocyte clusters, peripheral blood mononuclear cells (PBMC) data from Mouse Cell Atlas (MCA) (http://bis.zju.edu.cn/MCA/) was utilized. The cell developmental trajectories of monocytes and macrophages were inferred using Monocle3 (v 1.0.0).80 Top 50 PCs were used and all other parameters were set as default. To measure the difference between differentiation directions based on transcription factors, SCENIC (v 0.11.0)81 was performed on all single cells.
Similarity analysis of clusters from different datasets
To compare the similarities of myeloid cell clusters from normal and tumor tissue datasets, we trained a logistic regression model using elastic net regularization, as previous research described.38
Generation of dendritic-cell subtype scores
As reported by a previous study,33 the cDC1 and cDC2 signature genes were defined as those with an absolute log2FC > 0.5 between the average expression of the cDC1 and cDC2 clusters. mDC genes were defined as those with the log2FC difference between mDC, and both DC1 and DC2 were more than 0.5.
Flow cytometry
Live/dead cell discrimination was performed using the Zombie UV™ Fixable Viability Kit (Biolegend). Single-cell suspensions were incubated with Mouse TrueStain FcX (Biolegend) to block the Fc receptor’s nonspecific binding and subsequently stained with antibodies using standard protocols. For intracellular cytokine labeling, cells were stimulated with Leukocyte Activation Cocktail with GolgiPlug (BD Biosciences) for 4 hours at 37°C with 5% CO2 in RPMI medium. Intracellular staining was performed with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s instructions.
Samples were analyzed using the anti-mouse specific antibodies obtained from the following companies: BioLegend: anti-CD45 (30-F11), anti-CD8a (53-6.7), anti-CD4 (GK1.5), anti-PD-1 (29F.1A12), anti-TIM-3 (RMT3-23), anti-CD39 (Duha59), anti-TIGIT (1G9), anti-Granzyme B (QA16A02), anti-IFN-γ (XMG1.2), anti-TNF-α (MP6-XT22), anti-CD44 (IM7), anti-CD62L (MEL-14), anti-CD11b (M1/70), anti-Ly6G (1A8), anti-Ly6C (HK1.4), anti-F4/80 (BM8), anti-CD206 (C068C2), anti-CD86 (GL-1), anti-I-A/I-E (M5/114.15.2), anti-CD11c (N418), anti-VISTA (MIH63), anti-Ki67 (11F6), anti-CD69 (H1.2F3); eBioscience: anti-CD25 (PC61.5), anti-FOXP3 (FJK-16s); BD Bioscience: anti-TCF-7/TCF-1 (S33-966); Miltenyi Biotec: anti-TOX (REA473).
All depicted flow cytometry plots were pregated on good-events (by FlowAI plugin), non-debris (by FSC and SSC), non-doublets, and viable (Zombie dye-) cells. Doublets were excluded by comparing SSC and FSC width and height to area. Representative flow cytometry gating was shown in Figure S7C. The respective gate was set based on fluorescence minus one (FMO). Samples were acquired on LSRFortessa (BD Biosciences) and analyzed using FlowJo (TreeStar).
In vitro T cell suppression assay
Control and RT group mice were injected LLC-OVA cells ten days apart to be consistent with the sequencing design and equalize tumor size on the day for co-culture experiments. Tumor tissues were processed as described in the “sample dissociation” section using mouse tumor dissociation kit (Miltenyi Biotec). TANs were purified using MojoSort Mouse Ly6G Selection Kit (BioLegend; 480124) and used in the suppression assay. The 96-well U-bottom plates were coated with 5ug/ml CD3 (Biolegend; 100340) at 4°C overnight. For the unstimulated control wells, add 50μl of sterile PBS. Mouse CD8 + T cells were isolated from spleen using MojoSort™ Mouse CD8a Selection Kit (BioLegend; 480136), labelled with 5μM CFSE (eBioscience; 65-0850-84) and cocultured with TANs (1:1 ratio) in complete RPMI1640 supplemented with 10% heat inactivated FBS, 1ug/ml CD28 (Biolegend; 102116) and 100U/ml IL2 (PeproTech; 212-12). To block VISTA in this assay, 10μg/ml anti-mouse VISTA (BioXCell; 13F3; BE0310) or polyclonal Armenian hamster IgG (BioXCell; BE0091) were added to the culture. After 72 hours, the proliferation of CFSE-labelled CD8 T cells was analysed by LSRFortessa (BD Biosciences) and modelled using FlowJo (Tree Star).
Tissue microarray and immunohistochemistry
Human tissue microarrays of lung squamous cell carcinoma (HLugS180Su02) and lung adenocarcinoma (HLugA180Su04) were purchased from Shanghai Outdo Biotech Company, Shanghai, China. Antibodies against VISTA (Abcam, EPR21050, 1: 1000 dilution) were used for immunohistochemistry staining. Two experienced pathologists reviewed histologic slides in a double-blinded manner. The immunostaining intensity was scored as 0 (negative), 1 (weak), 2 (moderate), and 3 (strong). The percentage of positive cells was documented as 0-100%. For semi-quantitative analysis, H-score (range, 0-300) was calculated by the following algorithm: 1 × [percentage of cells with 1+ staining] + 2 × [percentage of cells with 2+ staining] + 3 × [percentage of cells with 3+ staining].82 For survival analysis, the patients were equally divided into three subgroups according to H-score values in the tumor stroma area. Patients with the same values were in the same subgroup. The clinical and histopathological characteristics were described in Tables S1 and S2.
Quantification and statistical analysis
The mice were randomly grouped based on tumor volumes before treatments. There was no difference in tumor volume among groups after randomization. The sample sizes were typically n=5 or 10 or indicated in the corresponding figure legends and sufficient to determine statistical significance among groups. All experiments were conducted at least two independent times. Survival functions were performed by the Kaplan–Meier method with log-rank test to compare survival between groups. One-way or two-way analysis of variance (ANOVA) with Tukey’s correction was used for multiple group comparisons. Unpaired or paired two-tailed Student’s t-test was used for two different group comparisons. All analyses were performed by R (version 3.6.1) and GraphPad Prism (version 8.2.0). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001.
Acknowledgments
We are grateful to Guangzhou Genedenovo Biotechnology Co., Ltd. (Guangzhou, China) for assisting in scRNA-seq and scTCR-seq. We also appreciate Xiaoqi Wu (Genergy Biotechnology Shanghai Co., Ltd.), Dr. Jianming Zeng (University of Macau), and all the members of his bioinformatics team, biotrainee, for generously sharing their experience and codes. This work was supported by grants as listed here: The National Natural Science Foundation of China (grant no. 82073356); the Beijing-Tianjin-Hebei Basic Research Cooperation Special Project (grant no. 22JCZXJC00180); the Science & Technology Development Fund of Tianjin Education Commission for Higher Education (grant no. 2021ZD034); and the Bethune· Tumor Radiotherapy Translational Medicine Research Fund (grant no. FLZH202108).
Author contributions
Conceptualization, Z.T.; data curation, Y.Z. and J.H.; formal analysis, Y.Z. and J.H.; funding acquisition, Z.T. and K.C.; investigation, Y.Z., J.H., K.J., L.S., G.H., D.C., Y.D., and S.J.; methodology, Y.Z., K.J., Y.D., and S.J.; project administration, Z.T.; resources, Z.T., Y.D., S.J., J.W., and L.S.; software, Y.Z., J.H., Y.D., and S.J.; supervision, Z.T. and K.C.; visualization, Y.Z. and J.H.; writing – original draft, Y.Z. and J.H.; writing – review & editing, Z.T. and K.C.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: August 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101151.
Contributor Information
Ke Chen, Email: shenke@hust.edu.cn.
Zhen Tao, Email: ztao@tmu.edu.cn.
Supplemental information
Data and code availability
-
•
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA011692) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Kalbasi A., Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Syn N.L., Teng M.W.L., Mok T.S.K., Soo R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017;18:e731–e741. doi: 10.1016/S1470-2045(17)30607-1. [DOI] [PubMed] [Google Scholar]
- 4.Huang M.-Y., Jiang X.-M., Wang B.-L., Sun Y., Lu J.-J. Combination therapy with PD-1/PD-L1 blockade in non-small cell lung cancer: strategies and mechanisms. Pharmacol. Ther. 2021;219:107694. doi: 10.1016/j.pharmthera.2020.107694. [DOI] [PubMed] [Google Scholar]
- 5.Galon J., Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein M.B., Krishnan S., Hodge J.W., Chang J.Y. Immunotherapy and stereotactic ablative radiotherapy (ISABR): a curative approach? Nat. Rev. Clin. Oncol. 2016;13:516–524. doi: 10.1038/nrclinonc.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonia S.J., Villegas A., Daniel D., Vicente D., Murakami S., Hui R., Kurata T., Chiappori A., Lee K.H., de Wit M., et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med. 2018;379:2342–2350. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y., Gao M., Huang Z., Yu J., Meng X. SBRT combined with PD-1/PD-L1 inhibitors in NSCLC treatment: a focus on the mechanisms, advances, and future challenges. J. Hematol. Oncol. 2020;13:105. doi: 10.1186/s13045-020-00940-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altorki N.K., McGraw T.E., Borczuk A.C., Saxena A., Port J.L., Stiles B.M., Lee B.E., Sanfilippo N.J., Scheff R.J., Pua B.B., et al. Neoadjuvant durvalumab with or without stereotactic body radiotherapy in patients with early-stage non-small-cell lung cancer: a single-centre, randomised phase 2 trial. Lancet Oncol. 2021;22:824–835. doi: 10.1016/S1470-2045(21)00149-2. [DOI] [PubMed] [Google Scholar]
- 10.Theelen W.S.M.E., Peulen H.M.U., Lalezari F., van der Noort V., de Vries J.F., Aerts J.G.J.V., Dumoulin D.W., Bahce I., Niemeijer A.-L.N., de Langen A.J., et al. Effect of Pembrolizumab After Stereotactic Body Radiotherapy vs Pembrolizumab Alone on Tumor Response in Patients With Advanced Non-Small Cell Lung Cancer: Results of the PEMBRO-RT Phase 2 Randomized Clinical Trial. JAMA Oncol. 2019;5:1276–1282. doi: 10.1001/jamaoncol.2019.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng L., Liang H., Xu M., Yang X., Burnette B., Arina A., Li X.-D., Mauceri H., Beckett M., Darga T., et al. STING-Dependent Cytosolic DNA Sensing Promotes Radiation-Induced Type I Interferon-Dependent Antitumor Immunity in Immunogenic Tumors. Immunity. 2014;41:843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanpouille-Box C., Alard A., Aryankalayil M.J., Sarfraz Y., Diamond J.M., Schneider R.J., Inghirami G., Coleman C.N., Formenti S.C., Demaria S. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 2017;8:15618. doi: 10.1038/ncomms15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muroyama Y., Nirschl T.R., Kochel C.M., Lopez-Bujanda Z., Theodros D., Mao W., Carrera-Haro M.A., Ghasemzadeh A., Marciscano A.E., Velarde E., et al. Stereotactic Radiotherapy Increases Functionally Suppressive Regulatory T Cells in the Tumor Microenvironment. Cancer Immunol. Res. 2017;5:992–1004. doi: 10.1158/2326-6066.CIR-17-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao Z., McCall N.S., Wiedemann N., Vuagniaux G., Yuan Z., Lu B. SMAC Mimetic Debio 1143 and Ablative Radiation Therapy Synergize to Enhance Antitumor Immunity against Lung Cancer. Clin. Cancer Res. 2019;25:1113–1124. doi: 10.1158/1078-0432.CCR-17-3852. [DOI] [PubMed] [Google Scholar]
- 15.Lan Y., Moustafa M., Knoll M., Xu C., Furkel J., Lazorchak A., Yeung T.-L., Hasheminasab S.-M., Jenkins M.H., Meister S., et al. Simultaneous targeting of TGF-β/PD-L1 synergizes with radiotherapy by reprogramming the tumor microenvironment to overcome immune evasion. Cancer Cell. 2021;39:1388–1403.e10. doi: 10.1016/j.ccell.2021.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Twyman-Saint Victor C., Rech A.J., Maity A., Rengan R., Pauken K.E., Stelekati E., Benci J.L., Xu B., Dada H., Odorizzi P.M., et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520:373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou P., Chen D., Zhu B., Chen W., Xie Q., Wang Y., Tan Q., Yuan B., Zuo X., Huang C., et al. Stereotactic Body Radiotherapy Is Effective in Modifying the Tumor Genome and Tumor Immune Microenvironment in Non-Small Cell Lung Cancer or Lung Metastatic Carcinoma. Front. Immunol. 2020;11:594212. doi: 10.3389/fimmu.2020.594212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Pilato M., Kfuri-Rubens R., Pruessmann J.N., Ozga A.J., Messemaker M., Cadilha B.L., Sivakumar R., Cianciaruso C., Warner R.D., Marangoni F., et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell. 2021;184:4512–4530.e22. doi: 10.1016/j.cell.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B., Wang Y., Sun X., Deng G., Huang W., Wu X., Gu Y., Tian Z., Fan Z., Xu Q., et al. CXCR6 is required for antitumor efficacy of intratumoral CD8 T cell. J. Immunother. Cancer. 2021;9:e003100. doi: 10.1136/jitc-2021-003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yost K.E., Satpathy A.T., Wells D.K., Qi Y., Wang C., Kageyama R., McNamara K.L., Granja J.M., Sarin K.Y., Brown R.A., et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 2019;25:1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Canale F.P., Ramello M.C., Núñez N., Araujo Furlan C.L., Bossio S.N., Gorosito Serrán M., Tosello Boari J., Del Castillo A., Ledesma M., Sedlik C., et al. CD39 Expression Defines Cell Exhaustion in Tumor-Infiltrating CD8 T Cells. Cancer Res. 2018;78:115–128. doi: 10.1158/0008-5472.CAN-16-2684. [DOI] [PubMed] [Google Scholar]
- 22.Zheng L., Qin S., Si W., Wang A., Xing B., Gao R., Ren X., Wang L., Wu X., Zhang J., et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374:abe6474. doi: 10.1126/science.abe6474. [DOI] [PubMed] [Google Scholar]
- 23.Vendetti F.P., Karukonda P., Clump D.A., Teo T., Lalonde R., Nugent K., Ballew M., Kiesel B.F., Beumer J.H., Sarkar S.N., et al. ATR kinase inhibitor AZD6738 potentiates CD8+ T cell-dependent antitumor activity following radiation. J. Clin. Invest. 2018;128:3926–3940. doi: 10.1172/JCI96519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan O., Giles J.R., McDonald S., Manne S., Ngiow S.F., Patel K.P., Werner M.T., Huang A.C., Alexander K.A., Wu J.E., et al. TOX transcriptionally and epigenetically programs CD8 T cell exhaustion. Nature. 2019;571:211–218. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eberhardt C.S., Kissick H.T., Patel M.R., Cardenas M.A., Prokhnevska N., Obeng R.C., Nasti T.H., Griffith C.C., Im S.J., Wang X., et al. Functional HPV-specific PD-1 stem-like CD8 T cells in head and neck cancer. Nature. 2021;597:279–284. doi: 10.1038/s41586-021-03862-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui I., Schaeuble K., Chennupati V., Fuertes Marraco S.A., Calderon-Copete S., Pais Ferreira D., Carmona S.J., Scarpellino L., Gfeller D., Pradervand S., et al. Intratumoral Tcf1PD-1CD8 T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity. 2019;50:195–211.e10. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 27.Moesta A.K., Li X.-Y., Smyth M.J. Targeting CD39 in cancer. Nat. Rev. Immunol. 2020;20:739–755. doi: 10.1038/s41577-020-0376-4. [DOI] [PubMed] [Google Scholar]
- 28.Li X.-Y., Moesta A.K., Xiao C., Nakamura K., Casey M., Zhang H., Madore J., Lepletier A., Aguilera A.R., Sundarrajan A., et al. Targeting CD39 in Cancer Reveals an Extracellular ATP- and Inflammasome-Driven Tumor Immunity. Cancer Discov. 2019;9:1754–1773. doi: 10.1158/2159-8290.CD-19-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L., Hou Y., Deng C., Tao Z., Chen Z., Hu J., Chen K. Single cell sequencing reveals that CD39 inhibition mediates changes to the tumor microenvironment. Nat. Commun. 2022;13:6740. doi: 10.1038/s41467-022-34495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown C.C., Gudjonson H., Pritykin Y., Deep D., Lavallée V.P., Mendoza A., Fromme R., Mazutis L., Ariyan C., Leslie C., et al. Transcriptional Basis of Mouse and Human Dendritic Cell Heterogeneity. Cell. 2019;179:846–863.e24. doi: 10.1016/j.cell.2019.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson D.A., Murphy K.M., Briseño C.G. Development, Diversity, and Function of Dendritic Cells in Mouse and Human. Cold Spring Harb. Perspect. Biol. 2018;10:a028613. doi: 10.1101/cshperspect.a028613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz M.B., Schuler G. Immature, semi-mature and fully mature dendritic cells: which signals induce tolerance or immunity? Trends Immunol. 2002;23:445–449. doi: 10.1016/s1471-4906(02)02281-0. [DOI] [PubMed] [Google Scholar]
- 33.Maier B., Leader A.M., Chen S.T., Tung N., Chang C., LeBerichel J., Chudnovskiy A., Maskey S., Walker L., Finnigan J.P., et al. A conserved dendritic-cell regulatory program limits antitumour immunity. Nature. 2020;580:257–262. doi: 10.1038/s41586-020-2134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garris C.S., Arlauckas S.P., Kohler R.H., Trefny M.P., Garren S., Piot C., Engblom C., Pfirschke C., Siwicki M., Gungabeesoon J., et al. Successful Anti-PD-1 Cancer Immunotherapy Requires T Cell-Dendritic Cell Crosstalk Involving the Cytokines IFN-γ and IL-12. Immunity. 2018;49:1148–1161.e7. doi: 10.1016/j.immuni.2018.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F., et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Franklin R.A., Liao W., Sarkar A., Kim M.V., Bivona M.R., Liu K., Pamer E.G., Li M.O. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng S., Li Z., Gao R., Xing B., Gao Y., Yang Y., Qin S., Zhang L., Ouyang H., Du P., et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809.e23. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O'Brien S.A., He Y., Wang L., Zhang Q., Kim A., et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell. 2020;181:442–459.e29. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 39.Kim N., Kim H.K., Lee K., Hong Y., Cho J.H., Choi J.W., Lee J.-I., Suh Y.-L., Ku B.M., Eum H.H., et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nalio Ramos R., Missolo-Koussou Y., Gerber-Ferder Y., Bromley C.P., Bugatti M., Núñez N.G., Tosello Boari J., Richer W., Menger L., Denizeau J., et al. Tissue-resident FOLR2 macrophages associate with CD8 T cell infiltration in human breast cancer. Cell. 2022;185:1189–1207.e25. doi: 10.1016/j.cell.2022.02.021. [DOI] [PubMed] [Google Scholar]
- 41.Fridlender Z.G., Sun J., Kim S., Kapoor V., Cheng G., Ling L., Worthen G.S., Albelda S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: "N1" versus "N2" TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaillon S., Ponzetta A., Di Mitri D., Santoni A., Bonecchi R., Mantovani A. Neutrophil diversity and plasticity in tumour progression and therapy. Nat. Rev. Cancer. 2020;20:485–503. doi: 10.1038/s41568-020-0281-y. [DOI] [PubMed] [Google Scholar]
- 43.Tang K.H., Li S., Khodadadi-Jamayran A., Jen J., Han H., Guidry K., Chen T., Hao Y., Fedele C., Zebala J.A., et al. Combined Inhibition of SHP2 and CXCR1/2 Promotes Antitumor T-cell Response in NSCLC. Cancer Discov. 2022;12:47–61. doi: 10.1158/2159-8290.CD-21-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu F., Fan J., He Y., Xiong A., Yu J., Li Y., Zhang Y., Zhao W., Zhou F., Li W., et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 2021;12:2540. doi: 10.1038/s41467-021-22801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parikh A.R., Szabolcs A., Allen J.N., Clark J.W., Wo J.Y., Raabe M., Thel H., Hoyos D., Mehta A., Arshad S., et al. Radiation therapy enhances immunotherapy response in microsatellite stable colorectal and pancreatic adenocarcinoma in a phase II trial. Nat. Cancer. 2021;2:1124–1135. doi: 10.1038/s43018-021-00269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeon H.-Y., Ham S.W., Kim J.-K., Jin X., Lee S.Y., Shin Y.J., Choi C.-Y., Sa J.K., Kim S.H., Chun T., et al. Ly6G+ inflammatory cells enable the conversion of cancer cells to cancer stem cells in an irradiated glioblastoma model. Cell Death Differ. 2019;26:2139–2156. doi: 10.1038/s41418-019-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wisdom A.J., Hong C.S., Lin A.J., Xiang Y., Cooper D.E., Zhang J., Xu E.S., Kuo H.-C., Mowery Y.M., Carpenter D.J., et al. Neutrophils promote tumor resistance to radiation therapy. Proc. Natl. Acad. Sci. USA. 2019;116:18584–18589. doi: 10.1073/pnas.1901562116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuen K.C., Liu L.-F., Gupta V., Madireddi S., Keerthivasan S., Li C., Rishipathak D., Williams P., Kadel E.E., Koeppen H., et al. High systemic and tumor-associated IL-8 correlates with reduced clinical benefit of PD-L1 blockade. Nat. Med. 2020;26:693–698. doi: 10.1038/s41591-020-0860-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blando J., Sharma A., Higa M.G., Zhao H., Vence L., Yadav S.S., Kim J., Sepulveda A.M., Sharp M., Maitra A., et al. Comparison of immune infiltrates in melanoma and pancreatic cancer highlights VISTA as a potential target in pancreatic cancer. Proc. Natl. Acad. Sci. USA. 2019;116:1692–1697. doi: 10.1073/pnas.1811067116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J., Ward J.F., Pettaway C.A., Shi L.Z., Subudhi S.K., Vence L.M., Zhao H., Chen J., Chen H., Efstathiou E., et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat. Med. 2017;23:551–555. doi: 10.1038/nm.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steenbrugge J., De Jaeghere E.A., Meyer E., Denys H., De Wever O. Splenic Hematopoietic and Stromal Cells in Cancer Progression. Cancer Res. 2021;81:27–34. doi: 10.1158/0008-5472.CAN-20-2339. [DOI] [PubMed] [Google Scholar]
- 52.Hou Y., Liang H.L., Yu X., Liu Z., Cao X., Rao E., Huang X., Wang L., Li L., Bugno J., et al. Radiotherapy and immunotherapy converge on elimination of tumor-promoting erythroid progenitor cells through adaptive immunity. Sci. Transl. Med. 2021;13:eabb0130. doi: 10.1126/scitranslmed.abb0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tavukcuoglu E., Horzum U., Yanik H., Uner A., Yoyen-Ermis D., Nural S.K., Aydin B., Sokmensuer C., Karakoc D., Yilmaz K.B., et al. Human splenic polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC) are strategically located immune regulatory cells in cancer. Eur. J. Immunol. 2020;50:2067–2074. doi: 10.1002/eji.202048666. [DOI] [PubMed] [Google Scholar]
- 54.Marvel D., Gabrilovich D.I. Myeloid-derived suppressor cells in the tumor microenvironment: expect the unexpected. J. Clin. Invest. 2015;125:3356–3364. doi: 10.1172/JCI80005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J., Yuan Y., Chen W., Putra J., Suriawinata A.A., Schenk A.D., Miller H.E., Guleria I., Barth R.J., Huang Y.H., Wang L. Immune-checkpoint proteins VISTA and PD-1 nonredundantly regulate murine T-cell responses. Proc. Natl. Acad. Sci. USA. 2015;112:6682–6687. doi: 10.1073/pnas.1420370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hosseinkhani N., Derakhshani A., Shadbad M.A., Argentiero A., Racanelli V., Kazemi T., Mokhtarzadeh A., Brunetti O., Silvestris N., Baradaran B. The Role of V-Domain Ig Suppressor of T Cell Activation (VISTA) in Cancer Therapy: Lessons Learned and the Road Ahead. Front. Immunol. 2021;12:676181. doi: 10.3389/fimmu.2021.676181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan L., Tatineni J., Mahoney K.M., Freeman G.J. VISTA: A Mediator of Quiescence and a Promising Target in Cancer Immunotherapy. Trends Immunol. 2021;42:209–227. doi: 10.1016/j.it.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morris Z.S., Guy E.I., Francis D.M., Gressett M.M., Werner L.R., Carmichael L.L., Yang R.K., Armstrong E.A., Huang S., Navid F., et al. In Situ Tumor Vaccination by Combining Local Radiation and Tumor-Specific Antibody or Immunocytokine Treatments. Cancer Res. 2016;76:3929–3941. doi: 10.1158/0008-5472.CAN-15-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burnette B.C., Liang H., Lee Y., Chlewicki L., Khodarev N.N., Weichselbaum R.R., Fu Y.-X., Auh S.L. The efficacy of radiotherapy relies upon induction of type i interferon-dependent innate and adaptive immunity. Cancer Res. 2011;71:2488–2496. doi: 10.1158/0008-5472.CAN-10-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee Y., Auh S.L., Wang Y., Burnette B., Wang Y., Meng Y., Beckett M., Sharma R., Chin R., Tu T., et al. Therapeutic effects of ablative radiation on local tumor require CD8+ T cells: changing strategies for cancer treatment. Blood. 2009;114:589–595. doi: 10.1182/blood-2009-02-206870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caushi J.X., Zhang J., Ji Z., Vaghasia A., Zhang B., Hsiue E.H.-C., Mog B.J., Hou W., Justesen S., Blosser R., et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature. 2021;596:126–132. doi: 10.1038/s41586-021-03752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oliveira G., Stromhaug K., Klaeger S., Kula T., Frederick D.T., Le P.M., Forman J., Huang T., Li S., Zhang W., et al. Phenotype, specificity and avidity of antitumour CD8 T cells in melanoma. Nature. 2021;596:119–125. doi: 10.1038/s41586-021-03704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deng H., Kan A., Lyu N., He M., Huang X., Qiao S., Li S., Lu W., Xie Q., Chen H., et al. Tumor-derived lactate inhibit the efficacy of lenvatinib through regulating PD-L1 expression on neutrophil in hepatocellular carcinoma. J. Immunother. Cancer. 2021;9:e002305. doi: 10.1136/jitc-2020-002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nolan E., Bridgeman V.L., Ombrato L., Karoutas A., Rabas N., Sewnath C.A.N., Vasquez M., Rodrigues F.S., Horswell S., Faull P., et al. Radiation exposure elicits a neutrophil-driven response in healthy lung tissue that enhances metastatic colonization. Nat. Cancer. 2022;3:173–187. doi: 10.1038/s43018-022-00336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shinde-Jadhav S., Mansure J.J., Rayes R.F., Marcq G., Ayoub M., Skowronski R., Kool R., Bourdeau F., Brimo F., Spicer J., Kassouf W. Role of neutrophil extracellular traps in radiation resistance of invasive bladder cancer. Nat. Commun. 2021;12:2776. doi: 10.1038/s41467-021-23086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sade-Feldman M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell. 2018;175:998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Duhen T., Duhen R., Montler R., Moses J., Moudgil T., de Miranda N.F., Goodall C.P., Blair T.C., Fox B.A., McDermott J.E., et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 2018;9:2724. doi: 10.1038/s41467-018-05072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simoni Y., Becht E., Fehlings M., Loh C.Y., Koo S.-L., Teng K.W.W., Yeong J.P.S., Nahar R., Zhang T., Kared H., et al. Bystander CD8 T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–579. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 69.Chow A., Uddin F.Z., Liu M., Dobrin A., Nabet B.Y., Mangarin L., Lavin Y., Rizvi H., Tischfield S.E., Quintanal-Villalonga A., et al. The ectonucleotidase CD39 identifies tumor-reactive CD8+ T cells predictive of immune checkpoint blockade efficacy in human lung cancer. Immunity. 2023;56:93–106.e6. doi: 10.1016/j.immuni.2022.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Timperi E., Barnaba V. CD39 Regulation and Functions in T Cells. Int. J. Mol. Sci. 2021;22:8068. doi: 10.3390/ijms22158068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meyer A.V., Klein D., de Leve S., Szymonowicz K., Stuschke M., Robson S.C., Jendrossek V., Wirsdörfer F. Host CD39 Deficiency Affects Radiation-Induced Tumor Growth Delay and Aggravates Radiation-Induced Normal Tissue Toxicity. Front. Oncol. 2020;10:554883. doi: 10.3389/fonc.2020.554883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedman D.J., Künzli B.M., A-Rahim Y.I., Sevigny J., Berberat P.O., Enjyoji K., Csizmadia E., Friess H., Robson S.C. From the Cover: CD39 deletion exacerbates experimental murine colitis and human polymorphisms increase susceptibility to inflammatory bowel disease. Proc. Natl. Acad. Sci. USA. 2009;106:16788–16793. doi: 10.1073/pnas.0902869106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deng J., Li J., Sarde A., Lines J.L., Lee Y.-C., Qian D.C., Pechenick D.A., Manivanh R., Le Mercier I., Lowrey C.H., et al. Hypoxia-Induced VISTA Promotes the Suppressive Function of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Cancer Immunol. Res. 2019;7:1079–1090. doi: 10.1158/2326-6066.CIR-18-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnston R.J., Su L.J., Pinckney J., Critton D., Boyer E., Krishnakumar A., Corbett M., Rankin A.L., Dibella R., Campbell L., et al. VISTA is an acidic pH-selective ligand for PSGL-1. Nature. 2019;574:565–570. doi: 10.1038/s41586-019-1674-5. [DOI] [PubMed] [Google Scholar]
- 75.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.-R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 80.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.-C., Geurts P., Aerts J., et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Howitt B.E., Sun H.H., Roemer M.G.M., Kelley A., Chapuy B., Aviki E., Pak C., Connelly C., Gjini E., Shi Y., et al. Genetic Basis for PD-L1 Expression in Squamous Cell Carcinomas of the Cervix and Vulva. JAMA Oncol. 2016;2:518–522. doi: 10.1001/jamaoncol.2015.6326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA011692) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. This paper analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.