Abstract
Three fibrillar collagen mRNAs, α1(I), α2(I), and α1(III), are coordinately upregulated in the activated hepatic stellate cell (hsc) in liver fibrosis. These three mRNAs contain sequences surrounding the start codon that can be folded into a stem-loop structure. We investigated the role of this stem-loop structure in expression of collagen α1(I) reporter mRNAs in hsc’s and fibroblasts. The stem-loop dramatically decreases accumulation of mRNAs in quiescent hsc’s and to a lesser extent in activated hsc’s and fibroblasts. The stem-loop decreases mRNA stability in fibroblasts. In activated hsc’s and fibroblasts, a protein complex binds to the stem-loop, and this binding requires the presence of a 7mG cap on the RNA. Placing the 3′ untranslated region (UTR) of collagen α1(I) mRNA in a reporter mRNA containing this stem-loop further increases the steady-state level in activated hsc’s. This 3′ UTR binds αCP, a protein implicated in increasing stability of collagen α1(I) mRNA in activated hsc’s (B. Stefanovic, C. Hellerbrand, M. Holcik, M. Briendl, S. A. Liebhaber, and D. A. Brenner, Mol. Cell. Biol. 17:5201–5209, 1997). A set of protein complexes assembles on the 7mG capped stem-loop RNA, and a 120-kDa protein is specifically cross-linked to this structure. Thus, collagen α1(I) mRNA is regulated by a complex interaction between the 5′ stem-loop and the 3′ UTR, which may optimize collagen production in activated hsc’s.
In liver fibrosis, activated hepatic stellate cells (hsc’s) are responsible for excessive production of extracellular matrix proteins (18, 41), including fibrillar collagens type I and III, fibronectin, proteoglycans, and collagen type IV (40, 47). In the normal liver, hsc’s are in a quiescent state and function to store vitamin A and to regulate the contractility of sinusoids (16, 29). When quiescent hsc’s are isolated from the liver and cultured on plastic, they spontaneously differentiate into activated hsc’s. This is accompanied by the same changes seen in liver fibrosis in vivo, including loss of retinoid stores, expression of markers of differentiation, and upregulation of extracellular matrix proteins, including fibrillar collagens (17, 19).
Collagen type I is a heterotrimeric molecule consisting of two α1(I) chains and one α2(I) chain (51). The steady-state level of collagen α1(I) mRNA increases 60- to 70-fold in activated hsc’s compared to quiescent hsc’s. The transcriptional rate of this gene increases about threefold, but the half-life of the mRNA increases from 1.5 to about 24 h (49). The stability of most mRNAs is regulated by sequences at their 3′ untranslated regions (3′ UTRs) and by protein factors that interact with these sequences (11, 45). We have identified a protein complex which binds the C-rich sequence in the collagen α1(I) mRNA 3′ UTR located 24 nucleotides (nt) downstream of the stop codon (49). This complex contains αCP, a poly(C) RNA binding protein, as a subunit, and its binding can be demonstrated only in extracts of activated hsc’s and not in extracts of quiescent hsc’s (49). Therefore, it is likely that the complex is involved in stabilization of collagen α1(I) mRNA in activated hsc’s. Interestingly, αCP binds a similar C-rich sequence to stabilize the α-globin mRNA (34, 52), another long-lived mRNA (53). The C-rich sequences in the 3′ UTRs of 15-lipoxygenase and tyrosine hydroxylase mRNAs can also bind αCP-containing complexes, but the functional role of this binding is unknown (32).
Although a role for 3′ UTRs in regulating mRNA stability is well established, a role for 5′ UTRs or for mRNA translation is not clear. Some mRNAs, such as c-myc mRNA (31) and histone mRNAs, require ongoing translation for degradation, while others are stabilized by transversing ribosomes (5). Furthermore, mRNAs containing premature stop codons are subjected to rapid degradation (1, 24, 27).
5′ UTRs can regulate the rate of translation initiation and thus indirectly the stability of mRNA. In general, mRNAs with long and highly structured 5′ UTRs are inefficiently translated (22, 30, 54). Stable stem-loops (SLs) (ΔG > 50 kcal/mol) (35) or SLs that bind RNA binding proteins (33) can block translation initiation if placed adjacent to the cap (33, 43). This block is by steric hindrance, since RNA binding proteins normally not involved in translation show this effect (33, 43). Binding of iron-regulatory proteins to the iron-responsive element (IRE) in the 5′ UTR of ferritin and erythroid 5-aminolevulinic acid synthase mRNAs regulates their translation in response to the iron stores in the cell (23). This regulation can be exerted only if the IRE is placed adjacent to the RNA cap.
In the 5′ UTRs of three collagen mRNAs, α1(I), α2(I), and α1(III), there is an SL structure encompassing the translation initiation codon (3, 4, 10, 36). This structure is located about 75 nt from the cap and has a stability of ΔG = 25 to 30 kcal/mol in different collagen mRNAs. These features make it unlikely that the structure can regulate translation as described for the IRE. However, the structure is well conserved in evolution, differing by 2 nt in Xenopus and human collagen mRNAs. A slightly different SL structure is also found around the translation start codon of the sea urchin collagen gene (14). A previous report did not demonstrate a regulatory role for this structure in mRNA steady-state levels or translation based on transfections of hybrid collagen-human growth hormone genes into fibroblasts and measurement of human growth hormone protein in cell medium (6).
We decided to investigate the role of the collagen α1(I) 5′ SL in gene expression in hsc’s and fibroblasts. We delivered a series of reporter genes containing the 5′ SL into quiescent and activated hsc’s by adenoviral vectors and measured their mRNA and protein levels intracellularly. We found that the SL dramatically decreases mRNA steady-state levels of reporter genes in quiescent hsc’s and to a lesser extent in activated hsc’s. The negative effect in activated hsc’s is partly reversed by placing the α1(I) 3′ UTR in these reporter genes. The inhibitory effect is, at least in part, due to a decreased stability of the reporter mRNA having the SL. We also provide evidence that some of these effects are mediated by a novel RNA binding activity that is distinct from αCP binding to the α1(I) 3′ UTR.
MATERIALS AND METHODS
Isolation and culture of hsc’s.
hsc’s were isolated from male Sprague-Dawley rats by in situ perfusion of the liver with pronase and collagenase followed by centrifugation of the cell suspension over a Stractan gradient as described elsewhere (17). The resulting hsc’s were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. The cells were then either used on day 2 (quiescent) or cultured for 14 days (activated).
Plasmid constructs.
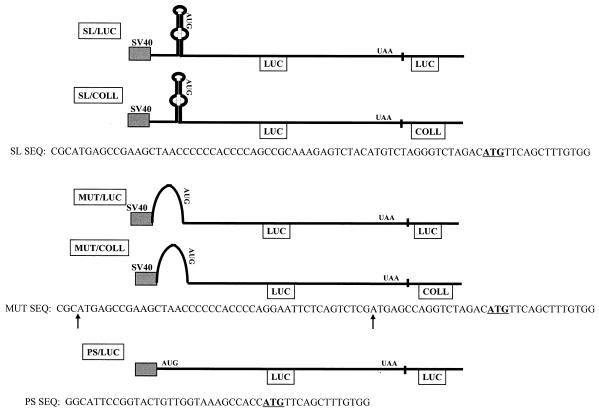
The pGL3 vector (Promega) was used to introduce a double-stranded oligonucleotide containing the mouse collagen α1(I) SL sequence (SL/LUC), a mutated form of the SL (MUT/LUC), a mutation of two short open reading frames in the 5′ UTR together with the SL mutation (GUG/MUT/LUC), or a control sequence having an optimal translation initiation site (PS/LUC) in frame with the luciferase coding region. In all of these constructs, the cloning was into HindIII-NcoI sites, so that the original luciferase initiator AUG codon was replaced with the first six codons of mouse collagen α1(I) mRNA to accommodate these changes. The sequences of the relevant part of the constructs are shown in Fig. 2. This hybrid protein had the same luciferase activity as the wild-type luciferase protein when tested in transient-transfection experiments. In two of the constructs, SL/LUC and MUT/LUC, the 3′ UTR of the luciferase gene was replaced by 316 nt of the mouse collagen α1(I) gene (COLL) 3′ UTR, containing the first polyadenylation signal, creating constructs SL/COLL and MUT/COLL. In addition, a 24-nt substitution in the αCP binding site was introduced by site-directed mutagenesis (37) into SL/COLL (SL/COLL-CP). To construct adenoviral vectors carrying these constructs, ClaI-SalI fragments from the above plasmids were recloned into the pACCMV.PLPASR(+) adenoviral shuttle vector (a kind gift of Frank L. Graham).
FIG. 2.
Constructs used in gene transfer experiments. All constructs are driven by the SV40 promoter. The sequence of the relevant region of the 5′ UTR is shown under each construct, extending from the fusion to the SV40 promoter to the start of the luciferase open reading frame. Two A-to-G changes which abolish two short open reading frames in construct GUG/MUT/LUC are indicated by arrows under the MUT sequence. The transcription start site is about 50 nt upstream of the sequence shown. The translation initiation codon is underlined. Different 3′ UTRs are also indicated. The constructs were named according to the sequence at the 5′ UTR and the sequence at the 3′ UTR.
To make riboprobes for RNase protection assays of reporter gene mRNAs, a 500-nt HindIII-AflIII fragment from the pGL3 vector was subcloned into HindIII-SmaI sites of pGEM3 vector (Promega). This plasmid was linearized with BstBI and transcribed with T7 polymerase to make a 380-nt antisense riboprobe which protects 330 nt of luciferase mRNA. To make riboprobes to measure glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs, pTRI-GAPDH-rat (Ambion) was linearized with StyI and transcribed with a T7 polymerase.
A 58-nt double-stranded oligonucleotide with the sequence of the mouse collagen α1(I) 5′ stem-loop (Fig. 1) was ligated in both orientations into the XmaI site of pGEM3. RNA and riboprobes used in binding experiments were transcribed in either the sense or the antisense orientation from these plasmids.
FIG. 1.
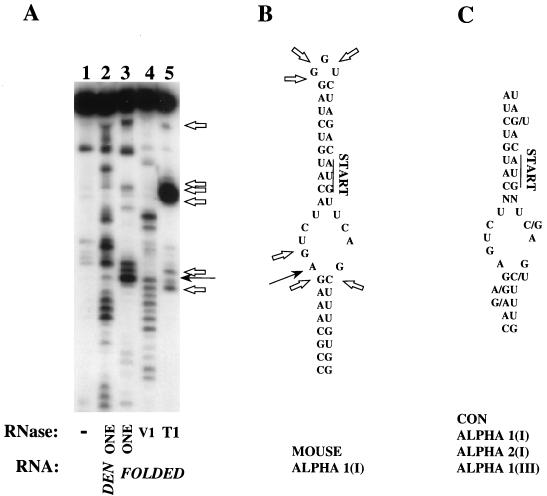
Collagen α1(I) 5′ SL structure. (A) Enzymatic probing of the synthetic 5′ SL RNA. 5′-end-labeled RNA with the sequence of the mouse collagen α1(I) SL was subjected to digestion with RNase ONE under denaturing conditions (lane 2), or in its folded conformation (lane 3), or with RNase V1 (lane 4) or RNase T1 (lane 5) in its folded conformation. Lane 1 is the input undigested RNA. Cleavage after G nucleotides is indicated by open arrows. A nucleotide attacked by all three enzymes is marked by a filled arrow. (B) Sequence of the mouse collagen α1(I) SL; nt 75 to 123 from the 5′ end are shown. The translation initiation codon is indicated by “START,” and an unusually stacked nucleotide is indicated by a black arrow. Cleavage sites of RNase T1 are indicated by open arrows. (C) Consensus (CON) sequence of the collagen 5′ SL derived from collagen α1(I) mRNA, α2(I) mRNA, and α1(III) mRNA of Xenopus (accession no. M63596), chicken (accession no. M17608, M17607, and K01481), mouse (accession no. M17491, X58251, and X70369), and human (accession no. M20789, Y00724, and M26939) cells. The translation initiation codon is indicated as for panel B.
Construction of viral vectors.
Reporter gene constructs in the pACCMV.PLPSR(+) vector were cotransfected together with ClaI-cut adenovirus DNA into L293 cells to allow homologous recombination as described elsewhere (21). Isolated plaques were picked and tested for luciferase expression after infection of Rat 1 cells. The expressing plaques were amplified and retested for expression. Viral titers were measured as described elsewhere (21), and viruses were stored in aliquots at −80°C for not more than 2 months. Two independent amplifications were performed, and their products were used in repeated experiments to allow for small fluctuations in virus viability seen upon prolonged storage. All viruses gave similar titers of 2 × 1010 to 4 × 1010 PFU/ml.
Infection of hsc’s and RNA and protein analysis.
Day 2 hsc’s or culture-activated hsc’s (about 2 × 106 cells) were supplemented with 2% serum, and adenoviral vectors were added at a multiplicity of infection (MOI) of 1,000. In control experiments, this MOI resulted in almost 100% of hsc’s expressing the transduced gene (28). The cells were incubated for 24 h, scraped from the plate, and resuspended in 1 ml of phosphate-buffered saline. A 100-μl aliquot was removed for estimation of luciferase activity. Total RNA was extracted from the rest of the cells by standard protocols (9) and used in the RNase protection assay. Luciferase activity was measured according to the manufacturer’s protocol (Analytical Luminescence Laboratories, Ann Arbor, Mich.) and expressed as relative luciferase units per micrograms of total protein. RNase protection analysis was performed as previously described (49), with incubation of the luciferase mRNA riboprobe together with the internal standard GAPDH riboprobe. The expression of luciferase mRNA was quantitated by phosphorimaging after normalization to GAPDH mRNA. Translation efficiency was calculated by dividing relative light units per microgram of protein by the mRNA levels. For the measurement of mRNA stability, Swiss 3T3 fibroblasts were infected with SL/LUC and MUT/LUC viruses as described above. After 24 h of incubation, the cells were divided into three plates, and after an additional 24 h, actinomycin D (ActD) (Sigma) was added at a concentration of 15 μg/ml. After addition of ActD, the cells were collected at 0, 5, and 10 h for the RNA analysis.
Analysis of RNA-protein binding.
The S130 postpolysomal supernatant of NIH 3T3 and Swiss 3T3 cells was made as described elsewhere (56). A cytoplasmic extract of hsc’s was made by incubating hsc’s in 10 mM Tris HCl (pH 7.6)–1 mM potassium acetate (KAc)–1.5 mM MgAc–2 mM dithiothreitol for 10 min on ice, followed by Dounce homogenization and 10 min of centrifugation at 14,000 × g. The supernatant was collected and immediately used in binding assays. Total protein was measured by the Bradford assay (7). Capped RNA probes used in binding experiments were made by in vitro transcription in the presence of a 10-fold excess of 7mGpppG cap analog (Pharmacia) with 30 μCi of [α-32P]GTP (48) and were gel purified. Cellular proteins (45 μg) were incubated with 40,000 cpm of the probe (4 ng) in 25 μl of 12 mM HEPES (pH 7.9)–15 mM KCl–0.25 mM EDTA–5 mM MgCl2–10% glycerol–200 ng of tRNA per μl–1 mM dithiothreitol buffer for 30 min on ice. The complexes were resolved on a 6% native acrylamide gel. The samples were UV cross-linked by irradiation for 20 min at 254 nm at a distance of 5 cm (UVGL-25 lamp; UVP), digested with 4 μg of RNase A and 1,280 U of RNase T1 (Gibco) for 1 h at 37°C, and analyzed on a sodium dodecyl sulfate–8% polyacrylamide gel.
Enzymatic mapping of the RNA structure.
5′-end-labeled SL RNA was made by in vitro transcription in the presence of 30 μCi of [γ-32P]GTP followed by gel purification. The 5′-end-labeled RNA was heated to 65°C and allowed to fold in 10 mM MgCl2–5 mM Tris HCl (pH 7.6)–25 mM KCl by slowly being cooled to ambient temperature (room temperature [RT]) for 20 min. Ten thousand counts per minute of the RNA in a total volume of 20 μl was digested with 0.25 U of RNase ONE (Promega) for 2.5 min at RT, 1.2 U of RNase T1 (Gibco) for 1 min at RT, or 0.2 U of RNase V1 (Pharmacia) for 10 min at RT. After the indicated times, the reactions were stopped by extraction with phenol-chloroform and the RNAs were precipitated with ethanol. As a control, the RNA was denatured by being heated to 55°C in 10 M urea–25 mM NaAc (pH 4.5) and digested at this temperature with 2.5 U of RNase ONE for 5 min. All samples were analyzed on a 12% denaturing acrylamide gel. Nucleotide assignments of the cleavage sites were made relative to the position of three G’s in the top loop as they were resolved in the RNase T1 lane.
In vitro translation.
A double-stranded oligonucleotide with the sequence of the T7 promoter was cloned into plasmids at a position 5′ to SL/LUC and MUT/LUC (see Fig. 2). After linearization of the templates with HpaI, the capped reporter mRNAs were synthesized in vitro with the Cap-Scribe kit (Boehringer). The integrity and concentration of mRNA were analyzed on agarose gels. mRNA (0.08 pM) was translated in a 25-μl reaction mixture with 50% rabbit reticulocyte lysate as prescribed by the manufacturer (Promega). After a 30-min incubation at 30°C, 5 μl of the reaction mixture was analyzed for luciferase activity.
Statistics.
Differences between experimental and control groups were analyzed by Student’s t test.
RESULTS
An SL structure at the 5′ end of collagen mRNAs.
Researchers have noted a potential folding of the 5′ end of three fibrillar collagen mRNAs into a structure consisting of a lower stem, a bulge, and an upper stem closed with a short loop (referred to as the 5′ SL structure in this paper) (55). This structure is found in collagen α1(I), α2(I), and collagen α1(III) mRNAs of all vertebrate species sequenced so far (3, 10, 15, 20, 36, 42, 44, 55). In addition, these mRNAs have two short open reading frames 5′ to the initiation codon. The sequence of the mouse collagen α1(I) 5′ SL is shown in Fig. 1B, and the consensus sequence of this structure is shown in Fig. 1C. In all cases, the translation initiation codon is located within the upper stem as indicated in Fig. 1B and C.
To determine if the SL of the mouse collagen α1(I) mRNA can fold into a higher-order structure, we probed a synthetic RNA containing this sequence (Fig. 1A). The 5′-end-labeled RNA was probed with nucleases that cut only single-stranded RNA (nucleases ONE and T1) and a nuclease which cuts only double-stranded RNA (nuclease V1) (13, 50). The enzyme concentrations were empirically determined to produce on average a single hit per molecule. First, the fully extended, denatured RNA was subjected to digestion with nuclease ONE, which shows no nucleotide preference for cutting. As expected, denatured RNA was digested throughout its length (lane 2, Fig. 1A). When this RNA was first allowed to fold, some regions of the molecule showed resistance to cutting with nuclease ONE (lane 3). These same regions were, however, susceptible to the attack of nuclease V1 (lane 4), suggesting a double-stranded conformation. Nuclease T1 cuts 3′ to G nucleotides in single-stranded regions, and this enzyme produced a cut at the G in the 3′ side of the bulge, strong cuts in the stretch of three G’s at the top of the loop, and cuts at two nonadjacent G’s at the 5′ side of the bulge (lane 5, open arrows). These same regions were also digested by nuclease ONE, suggesting that they are in single-stranded form (compare lanes 3 and 5). Based on the position of G nucleotides and the length of the fragments, double-stranded regions can be assigned to the two stems and single-stranded regions map to the bulge and the top loop. Thus, this SL RNA folds in vitro into a secondary structure similar to that shown in Fig. 1B but with one notable exception. There is a site which can be equally well cleaved by single-stranded RNA-specific and double-stranded RNA-specific nucleases (indicated by solid arrows in Fig. 1A and B). This cleavage maps to the first A nucleotide at the 5′ side of the bulge, indicating an unusual stacking of this nucleotide.
5′ SL structure negatively regulates expression in quiescent and activated hsc’s.
When freshly isolated hsc’s are cultured on uncoated plastic for 14 days, they spontaneously become activated and differentiate into myofibroblasts (17, 19). The hsc’s cultured for 2 days after isolation still show a quiescent phenotype (12, 19). To analyze the role of the α1(I) 5′ SL structure in gene expression in hsc’s, we used recombinant adenoviral vectors to efficiently deliver the reporter genes. The reporter genes were driven by the simian virus 40 (SV40) promoter and contained the same open reading frame, encoding the luciferase protein (Fig. 2). Constructs differ at their 5′ end: (i) the intact mouse collagen α1(I) SL with its two short open reading frames (SL/LUC), (ii) a mutated SL (MUT/LUC), or (iii) a mutated SL plus two mutated AUG codons of the preceding short open reading frames (GUG/MUT/LUC). Some constructs (SL/COLL and MUT/COLL) contain the mouse α1(I) 3′ UTR, including the first polyadenylation signal, instead of the 3′ UTR supplied by the LUC vector. The SL/COLL-CP construct contains a 24-nt mutation in the collagen α1(I) 3′ UTR that abolishes binding of the αCP complex (49).
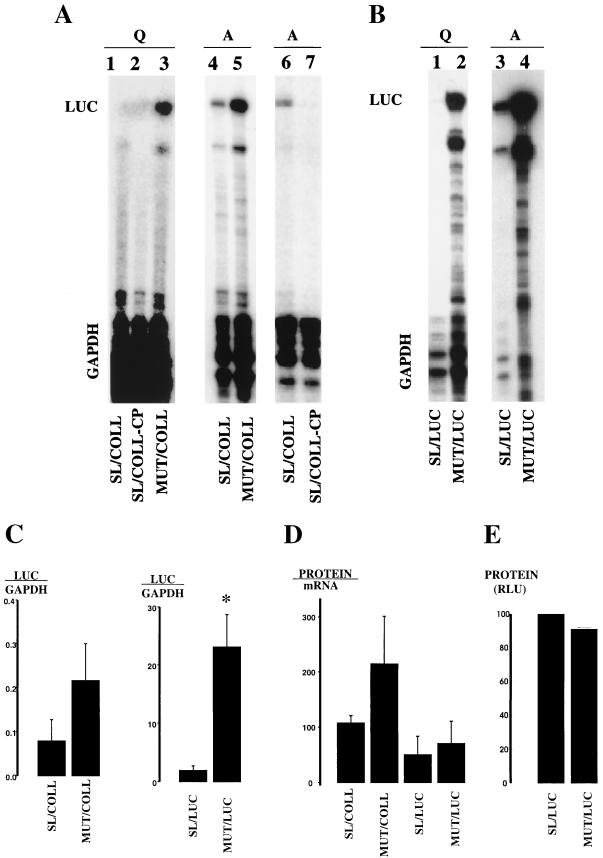
Figure 3 shows a representative RNase protection assay after gene delivery to quiescent or activated hsc’s. The results with constructs containing collagen 3′ UTR are shown in Fig. 3A. In quiescent hsc’s, two reporter mRNAs with the α1(I) 5′ SL accumulated at a very low, almost undetectable level (lanes 1 and 2). The mRNA in which this SL was disrupted accumulated at a much higher level (lane 3). In activated hsc’s, the inhibitory effect of the 5′ SL was less pronounced, and the mRNA with this SL is clearly detectable (compare lanes 4 and 5 to lanes 1 and 3). The negative effect of the 5′ SL on mRNA accumulation was seen regardless of the nature of the sequence at the 3′ UTR (Fig. 3B, constructs containing luciferase 3′ UTR). Again, the 5′ SL prevented accumulation of the mRNA in quiescent hsc’s (lane 1) and was less inhibitory in activated hsc’s (lane 3), while mRNAs without the SL were highly expressed in both cell types (lanes 2 and 4). However, constructs with LUC 3′ UTR are expressed at a higher level than are constructs with the COLL 3′ UTR, presumably due to a more efficient utilization of the poly(A) signal supplied by the optimized LUC vector. Consequently, the ratio of reporter mRNA signal to internal control GAPDH signal was higher for the LUC 3′ UTR constructs. These experiments were performed twice with two different virus preparations with the same results.
FIG. 3.
mRNA and protein levels of reporter genes in hsc’s. Expression of reporter mRNAs in hsc’s analyzed by RNase protection assay is shown. (A) Infection of hsc’s with constructs containing collagen 3′ UTR. Adenoviruses expressing the indicated constructs were added at an MOI of 1,000 to hsc’s cultured for 2 days (Q; lanes 1 to 3) and for 14 days (A; lanes 4 to 7). The assay was performed 24 h after infection with luciferase-specific and GAPDH-specific riboprobes. The bands protected by reporter mRNA are marked LUC, and bands protected by endogenous GAPDH mRNA, as an internal control, are marked GAPDH. (B) An analysis identical to that for panel A was performed with constructs containing luciferase 3′ UTR and expressed in day 2 hsc’s (Q; lanes 1 and 2) or activated hsc’s after 14 days in culture (A; lanes 3 and 4). (C) Quantitation of expression of reporter mRNAs in activated hsc’s. In three independent experiments, the intensity of LUC and GAPDH bands was measured by phosphorimaging, and the ratio is shown in arbitrary units. Only constructs with identical 3′ UTRs are compared to each other. ∗, P < 0.05. (D) Translation of reporter mRNAs in activated hsc’s. Luciferase activity was measured in each sample and normalized to total protein. The mRNA level was measured as the ratio of LUC to GAPDH by phosphorimaging. The protein level was divided by the mRNA level and is shown in arbitrary units for two independent experiments. (E) In vitro translation. Reporter mRNA (0.08 pM) was translated in rabbit reticulocyte lysate. Luciferase activity was measured after 30 min of incubation. The activity of SL/LUC mRNA was set at 100%. Results of two independent experiments are shown.
We also measured the luciferase protein translated from our reporter mRNAs in the cultured cells. In general, the protein level paralleled the mRNA level, suggesting that the SL did not have a major effect on translation (data not shown).
Short upstream open reading frames negatively regulate translation in some mRNAs (33). To address the role of the short open reading frames in the α1(I) mRNA, we mutated the two AUGs of the short open reading frames preceding the initiation codon in the context of the MUT/LUC reporter, creating the construct GUG/MUT/LUC. This construct was expressed similarly to the MUT/LUC construct, suggesting no additional effects of the short reading frames on mRNA steady-state level or translation (data not shown).
The negative effect of the 5′ SL is attenuated and modulated by the collagen α1(I) 3′ UTR in activated hsc’s.
The α1(I) 5′ SL inhibited accumulation of the mRNA containing the luciferase 3′ UTR about 12-fold in activated hsc’s (Fig. 3B, lanes 3 and 4). When the α1(I) 3′ UTR was added to reporter genes, the inhibitory effect of the 5′ SL on mRNA level was reduced to only threefold in activated hsc’s (Fig. 3A, lanes 4 and 5). The results of three independent experiments in activated hsc’s are graphed in Fig. 3C.
In extracts from activated but not quiescent hsc’s, the 3′ UTR of collagen α1(I) mRNA binds a protein complex containing αCP (49). To assess the role of αCP binding in activated hsc’s, we made a mutation in the αCP binding site within SL/COLL to create SL/COLL-CP. Mutating the αCP binding site dramatically decreased the steady-state level of reporter mRNA in activated hsc’s (Fig. 3A, lanes 6 and 7). When L293 cells were infected, the SL/COLL-CP virus produced luciferase protein to 52% of the level of that for the SL/COLL virus, excluding the possibility that this virus is nonfunctional. Thus, binding of αCP complex seems to be required for a high level of expression of this reporter mRNA in activated hsc’s.
To estimate the translational efficiency of the mRNAs in activated hsc’s, we measured luciferase activity in all samples and divided the protein level by the mRNA level (Fig. 3D). As in quiescent hsc’s, no major effect of the SL on translation was observed. The mRNAs with the α1(I) 3′ UTR are translated slightly better per molecule, but this is not significant. Similarly, we have seen no effect of the SL in in vitro translation experiments with these same mRNAs. Synthetic capped mRNAs with the sequence of SL/LUC and MUT/LUC shown in Fig. 2 were synthesized and translated in vitro in rabbit reticulocyte lysate. The results of two independent experiments, each performed in duplicate, are shown in Fig. 3E.
We conclude that the α1(I) 5′ SL strongly inhibits mRNA accumulation in quiescent hsc’s, while this inhibition is less pronounced in activated hsc’s. In activated hsc’s, binding of αCP complex to the 3′ UTR further attenuates the negative effect of the 5′ SL.
Collagen α1(I) 5′ SL destabilizes its mRNA.
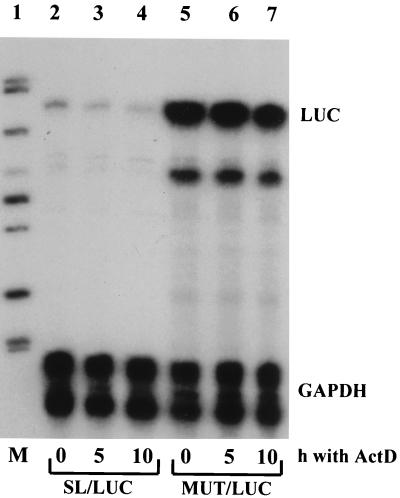
To provide insight into a mechanism by which the 5′ SL decreases the steady-state levels of mRNAs, we measured the stability of the reporter mRNAs, SL/LUC and MUT/LUC, after expression of these reporters in Swiss 3T3 fibroblasts (Fig. 4). Similar to hsc’s, SL/LUC mRNA was expressed at a lower level than was MUT/LUC mRNA (Fig. 4, lanes 2 and 5). After inhibition of transcription with ActD, the steady-state level of WT/LUC mRNA decreased with a half-life of about 5 h (lanes 2 to 4). In contrast, the steady-state level of MUT/LUC mRNA shows only a 15% decrease after 10 h of incubation with ActD (lanes 5 to 7), indicating a more stable mRNA. This result suggests that the SL destabilizes the mRNA and that this destabilization is, at least in part, responsible for the lesser accumulation of SL/LUC mRNA. The GAPDH mRNA used as an internal control in this experiment is a stable mRNA and shows no decay within 10 h, as reported previously (49).
FIG. 4.
Collagen α1(I) 5′ SL destabilizes its mRNA. SL/LUC mRNA (lanes 2 to 4) and MUT/LUC mRNA (lanes 5 to 7) were expressed in Swiss 3T3 fibroblasts by virus-mediated gene transfer. ActD at 15 μg/ml was added, and RNA was extracted at indicated time points (hours). The level of reporter mRNAs (LUC) and internal standard GAPDH mRNA (GAPDH) was estimated by RNase protection assay. Quantitation was performed by phosphorimaging and normalization to GAPDH. Lane 1 contains size markers.
The 5′ SL structure binds protein factors in activated hsc’s and in fibroblasts in a cap-dependent manner.
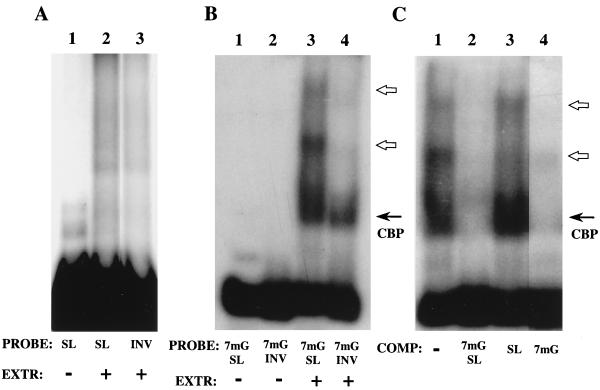
To identify protein factors that may interact with the 5′ SL and modulate its inhibitory activity, we performed RNA mobility shift experiments. First, we used extracts from collagen-producing NIH 3T3 and Swiss 3T3 fibroblasts to optimize the binding assay. S130 postpolysomal supernatant was prepared from these cells and incubated with radiolabeled 5′ SL RNA and inverted (INV) RNA. The INV RNA is antisense-transcribed sequence of the 5′ SL which served as a control in the binding experiments. The results with NIH 3T3 extracts are shown in Fig. 5; the extracts of Swiss 3T3 fibroblasts showed similar results. In the initial experiments with uncapped 5′ SL RNA and INV RNA, we could see no complex formation in a gel mobility assay (Fig. 5A). Then we capped these probes with 7mG and repeated the assay with the same S130 extract. This time, we detected a cap binding complex with both probes, as expected (Fig. 5B, solid arrow). However, additional complexes were seen only with the 5′ SL probe (Fig. 5B, lane 3, open arrows). This indicates that there are protein factors in fibroblasts, not associated with polysomes, which can bind the capped α1(I) 5′ SL RNA. To corroborate the importance of 7mG cap for binding, we performed competition experiments in which binding to capped α1(I) 5′ SL RNA was challenged with excess capped or uncapped 5′ SL RNAs as well as with an excess of 7mGpppG cap analog (Fig. 5C). An excess of capped 5′ SL RNA diminished formation of both the cap binding complex and the 5′ SL binding complexes (lane 2). An excess of this same uncapped RNA has a much weaker inhibitory effect on the 5′ SL-specific complexes (compare lanes 2 and 3). An excess of the cap analog completely abolished the formation of all complexes (lane 4). From these experiments, we conclude that the presence of the 7mG cap on the α1(I) SL RNA is required for efficient binding and competition for the protein factors in vitro.
FIG. 5.
Collagen α1(I) SL binding activity in NIH 3T3 cells. (A) S130 postpolysomal supernatant was incubated with radiolabeled probes, uncapped 5′ SL RNA (lane 2) or uncapped inverted SL RNA (lane 3), and the protein-RNA complexes were resolved on a native polyacrylamide gel. Lane 1 is the input 5′ SL RNA. (B) The same extract was incubated with capped probes, 7mG 5′ SL RNA (lane 3) or 7mG inverted SL RNA (lane 4). Lanes 1 and 2 are probes alone. A cap-dependent complex is shown as CBP, and the α1(I) 5′ SL-specific complexes are indicated by open arrows. (C) Competition of binding to the collagen α1(I) 5′ SL. The capped 5′ SL probe was incubated with NIH 3T3 postpolysomal supernatant in the absence of competitor (lane 1) or in the presence of a 100-fold molar excess of a specific competitor (capped 5′ SL RNA; lane 2), a 100-fold molar excess of uncapped 5′ SL RNA (lane 3), or 50 μM 7mGpppG cap analog (lane 4). Complexes were resolved on a 6% polyacrylamide native gel. Migrations of the cap binding complex and 5′ SL-specific complexes are indicated as described above.
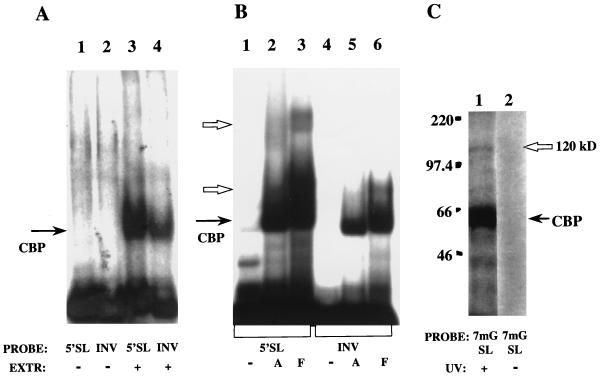
We next used extracts from day 2 hsc’s and activated hsc’s. Because of the limited supply of primary cells, these extracts were made as whole cytoplasmic extracts and not as S130 supernatants. We used capped 5′ SL RNA and INV RNA as probes. In quiescent hsc’s, no specific binding to the 5′ SL RNA was demonstrated, although formation of the cap binding complex is seen with both probes (Fig. 6A, lanes 3 and 4). Several extracts were prepared from different quiescent hsc isolates, but we were unable to see any 5′ SL-specific complexes. In contrast, in activated hsc’s a binding activity specific for the 5′ SL was obtained (Fig. 6B, compare lane 2 to lane 5). Extracts from fibroblasts and activated hsc’s form complexes with the 5′ SL RNA with similar but not identical mobilities. (A whole cytoplasmic extract was used in this experiment, and the relative mobilities of the complexes are slightly different from those in Fig. 5, for which S130 supernatants were used.) As a control, only a cap binding complex was assembled in both extracts with an INV RNA probe (lanes 5 and 6).
FIG. 6.
Collagen α1(I) SL binding proteins in hsc’s. (A) Gel mobility shift assay with cytoplasmic extract of day 2 hsc’s (lanes 3 and 4). 7mG capped 5′ SL and INV SL probes were used as described for Fig. 5. Lanes 1 and 2 are the input probes. Migration of the cap-dependent complex is marked as CBP. (B) Gel mobility shift analysis with cytoplasmic extract from activated hsc’s (lanes 2 and 5) and NIH 3T3 fibroblasts (lanes 3 and 6). Probes are as described for panel A with 5′ SL RNA (lane 1) and INV RNA (lane 4). The mobility of the cap binding complex is shown as CBP, and complexes specific for the 5′ SL are indicated by open arrows. (C) UV cross-linking of proteins to collagen 5′ SL RNA in activated hsc’s. A 7mG capped 5′ SL probe was incubated in a cytoplasmic extract of activated hsc’s and UV irradiated (lane 1), or UV irradiation was omitted (lane 2). After digestion with nucleases, cross-links were resolved on a sodium dodecyl sulfate–8% polyacrylamide gel. The cap-dependent cross-link is shown as CBP, and a specific 120-kDa protein is indicated by an open arrow. The migration of size markers (Amersham) is indicated on the left in kilodaltons.
A UV cross-linking experiment with extracts of activated hsc’s is shown in Fig. 6C. A 120-kDa protein (open arrow, lane 1) is covalently linked to the 5′ SL RNA. A very strong cap-dependent cross-link is also seen (arrow, lane 1). As a control, no protein-RNA adducts are formed without UV light (lane 2).
DISCUSSION
We have investigated the role of the conserved SL structure found at the 5′ ends of collagen α1(I) mRNA, α2(I) mRNA, and α1(III) mRNA. In liver fibrosis, these three collagen mRNAs are coordinately upregulated, resulting in increased production of fibrillar collagens (2, 38, 46). Since activated hsc’s are responsible for excessive collagen production in liver fibrosis (39), we used primary cultures of rat hsc’s. The inability to efficiently introduce genes into quiescent hsc’s has previously limited studies of gene expression in this cell type. However, delivery of genes by using adenoviral vectors is a receptor-mediated process, and infection with equal MOIs of a given population of cells results in uniform and reproducible gene transfer. Using this technology, we were able to successfully study gene expression in hsc’s with a quiescent phenotype only 2 days after isolation.
This report reveals some novel aspects of posttranscriptional regulation of collagen α1(I) gene expression in hsc’s. First, SL structure negatively regulates expression of reporter mRNAs in hsc’s, and this effect is more pronounced in quiescent hsc’s, suggesting that the SL may target mRNA for degradation (Fig. 3). The analysis of the stability of mRNAs showed destabilization of the mRNA containing the SL compared to mRNA with the mutated SL (Fig. 4). Second, mRNAs with the α1(I) SL have a higher steady-state level in activated hsc’s than in quiescent hsc’s, although SL mRNAs still do not accumulate to as high a level as do mRNAs without the SL. In activated hsc’s, we could demonstrate binding of protein factors to the 5′ SL (Fig. 6), which may inhibit mRNA degradation. Third, the α1(I) 3′ UTR containing the αCP binding site is required for high-level expression of mRNAs containing the destabilizing 5′ SL (Fig. 3A). Fourth, translation of mRNAs having the SL is as efficient as translation of mRNAs without the SL in hsc’s (Fig. 3D) and in vitro (Fig. 3E). Although the SL may target the mRNA for degradation, the surviving mRNA seems to be efficiently translated. This also suggests that the α1(I) collagen 5′ SL is not sufficiently stable to prevent translation once ribosome scanning has started (35).
Our study suggests that the SL primarily targets mRNAs for turnover in hsc’s, but how this targeting works is not yet clear. Enzymatic probing has revealed a higher-order structure for this SL with an unusually stacked A nucleotide at the 5′ side of the bulge (Fig. 1). Such sequences are often sites for protein binding, and interestingly, even sea urchin collagen mRNA has a similar A nucleotide in the context of two stems (14), suggesting evolutionary conservation of this fold. In quiescent hsc’s, we could not detect any protein binding to the SL in vitro (Fig. 6). If there is a protein, its binding may only be transient or it may bind only in vivo and require intact subcellular structures. In activated hsc’s, there is a protein factor(s) that binds in vitro to the collagen 5′ SL in a cap-dependent manner (Fig. 6). The complex is found in NIH 3T3 and Swiss 3T3 fibroblasts in the postpolysomal cytoplasmic fraction (Fig. 5). We do not know if this complex directly interacts with 7mG cap or with the cap binding protein, eIF4E (25, 26). Supershift experiments with anti-eIF4E antibody (a kind gift from C. H. Hagedorn) showed no change in gel mobility of the complex (data not shown). However, an excess of cap analog completely prevents formation of this complex in vitro (Fig. 5C). This complex may increase the steady-state level of the mRNAs by diverting them from the degradative pathway. A recent study demonstrated that the stability of the interleukin-2 mRNA is also regulated by a cis element in its 5′ UTR, but the trans-acting factors were not investigated (8). In addition, in the case of collagen α1(I) mRNA, a protein complex containing αCP binds to the 3′ UTR, and this binding can be seen only in extracts from activated hsc’s (49). We have postulated that this binding stabilizes collagen α1(I) mRNA. When we mutated the αCP binding site, the mRNA steady-state level decreased about 10-fold (Fig. 3A). In activated hsc’s, there may be an additional interaction between the 3′ UTR-αCP complex and the 5′ SL-cap complex, and this interaction may further stabilize the mRNA (Fig. 3C).
Expression of SL/COLL reporter mRNA resembles expression of endogenous collagen α1(I) mRNA in hsc’s; it is low in quiescent hsc’s and elevated in activated hsc’s (Fig. 3). Two findings point to the posttranscriptional regulation of this reporter mRNA in hsc’s, as has been described for the endogenous gene (49). The SV40 promoter seems to be equally active in both cell types, based on similar levels of expression of MUT/COLL and MUT/LUC in quiescent and activated hsc’s (Fig. 3). However, the SL clearly has a potential to decrease the stability of the mRNA (Fig. 4) and may modulate its steady-state level by interacting with protein factors. We cannot completely exclude the possibility that the SL negatively affects transcription, but since our constructs are driven by a heterologous promoter, this seems unlikely. Therefore, the regulation of SL/COLL mRNA in hsc’s has been achieved by a posttranscriptional mechanism and mediated by two sequences: 5′ SL and 3′ UTR. A previous report (6) failed to demonstrate a role for this SL in the regulation of reporter genes transiently or stably transfected into NIH 3T3 cells. These two studies differ in several aspects. The maximal inhibitory effect of the 5′ SL is demonstrated in undifferentiated primary cell cultures (quiescent hsc’s) in our study, compared to terminally differentiated immortal fibroblasts. Our study also uses a more sensitive luciferase assay to monitor the intracellular accumulation of reporter gene product. Furthermore, the earlier study used constructs that did not contain the collagen 3′ UTR, which modulates the expression of the collagen α1(I) gene. Using our constructs in transient transfections into NIH 3T3 cells, we demonstrated only about a threefold inhibition of the luciferase protein expression by the SL (data not shown). Posttranscriptional regulation of collagen α1(I) mRNA in hsc’s involves both αCP binding to the 3′ UTR and binding of a protein factor(s) to the 5′ SL. Binding to the α1(I) SL requires the presence of the 7mG cap structure. The role of the cap binding protein, eIF4E, or other translation initiation factors in assembly of this binding activity requires further investigation. Elucidating the interaction between the binding activities for the 5′ UTR and 3′ UTR of collagen α1(I) mRNA and cloning of the 5′ SL binding factors will greatly contribute to our understanding of collagen gene regulation.
REFERENCES
- 1.Applequist S E, Selg M, Raman C, Jack H-M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aycock R S, Seyer J M. Collagens of normal and cirrhotic human liver. Connect Tissue Res. 1989;23:19–31. doi: 10.3109/03008208909103901. [DOI] [PubMed] [Google Scholar]
- 3.Benson-Chanda V, Su M W, Weil D, Chu M L, Ramirez F. Cloning and analysis of the 5′ portion of the human type-III procollagen gene (COL3A1) Gene. 1989;78:255–265. doi: 10.1016/0378-1119(89)90228-x. [DOI] [PubMed] [Google Scholar]
- 4.Bernard M P, Myers J C, Chu M-L, Ramirez F, Eikenberry E F, Prockop D J. Structure of a cDNA for the Pro alpha 2 chain of human type I procollagen. Comparison with chick cDNA for Pro alpha 2(I) identifies structurally conserved features of the protein and the gene. Biochemistry. 1993;22:1139–1145. doi: 10.1021/bi00274a023. [DOI] [PubMed] [Google Scholar]
- 5.Blume J E, Shapiro D J. Ribosome loading, but not protein synthesis, is required for estrogen stabilization of Xenopus laevis vitellogenin mRNA. Nucleic Acids Res. 1989;17:9003–9014. doi: 10.1093/nar/17.22.9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bornstein P, McKay J, Devarayalu S, Cook S C. A highly conserved, 5′ untranslated, inverted repeat sequence is ineffective in translational control of the alpha 1(I) collagen gene. Nucleic Acids Res. 1988;16:9721–9736. doi: 10.1093/nar/16.20.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Chen C Y, Del Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.D’Alessio M, Bernard M, Pretorius P J, De Wet W, Ramirez F. Complete nucleotide sequence of the region encompassing the first twenty-five exons of the human pro alpha 1(I) collagen gene (COL1A1) Gene. 1988;67:105–115. doi: 10.1016/0378-1119(88)90013-3. [DOI] [PubMed] [Google Scholar]
- 11.Decker C J, Parker R. Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr Opin Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 12.DeLeeuw A M, McCarthy S P, Geerts A, Knook D L. Purified rat liver fat storing cells divide in culture and contain collagen. Hepatology. 1984;4:392–403. doi: 10.1002/hep.1840040307. [DOI] [PubMed] [Google Scholar]
- 13.Derrick W, Horowitz J M. Probing structural differences between native and in vitro transcribed Escherichia coli valine transfer RNA: evidence for stable base modification-dependent conformers. Nucleic Acids Res. 1993;21:4948–4953. doi: 10.1093/nar/21.21.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esposito J Y, Boute N, Deleage G, Garrone R. Characterization of two genes coding for a similar four-cysteine motif of the amino-terminal propeptide of a sea urchin fibrillar collagen. Eur J Biochem. 1995;234:59–65. doi: 10.1111/j.1432-1033.1995.059_c.x. [DOI] [PubMed] [Google Scholar]
- 15.Finer M H, Boedtker H, Doty P. Construction and characterization of cDNA clones encoding the 5′ end of the chicken pro alpha-1(I) collagen mRNA. Gene. 1987;56:71–78. doi: 10.1016/0378-1119(87)90159-4. [DOI] [PubMed] [Google Scholar]
- 16.Friedman S L. Hepatic stellate cells. Prog Liver Dis. 1996;14:101–130. [PubMed] [Google Scholar]
- 17.Friedman S L, Rockey D C, McGuire R F, Maher J J, Boyles J K, Yamasaki G. Isolated hepatic lipocytes and Kupffer cells from normal human liver: morphological and functional characteristics in primary culture. Hepatology. 1992;15:234–243. doi: 10.1002/hep.1840150211. [DOI] [PubMed] [Google Scholar]
- 18.Friedman S L, Roll F J, Boyles J, Bissell D M. Hepatic lipocytes: the principal collagen-producing cells of normal rat liver. Proc Natl Acad Sci USA. 1985;82:8681–8685. doi: 10.1073/pnas.82.24.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geerts A, Vrijsen R, Rauterberg J, Burt A, Schellinck P, Wisse E. In vitro differentiation of fat-storing cells parallels marked increases of collagen synthesis and secretion. J Hepatol. 1989;9:59–68. doi: 10.1016/0168-8278(89)90076-7. [DOI] [PubMed] [Google Scholar]
- 20.Glumoff V, Maekelae J K, Vuorio E. Cloning of cDNA for rat pro alpha 1(III) collagen mRNA. Different expression patterns of type I and type III collagen and fibronectin genes in experimental granulation tissue. Biochim Biophys Acta. 1994;1217:41–48. [PubMed] [Google Scholar]
- 21.Graham F L, Prevec L. Manipulation of adenovirus vectors. In: Murray E J, editor. Methods in molecular biology: gene transfer and expression protocols. Clifton, N.J: The Humana Press Inc.; 1991. pp. 109–128. [DOI] [PubMed] [Google Scholar]
- 22.Gray N K, Hentze M W. Regulation of protein synthesis by mRNA structure. Mol Biotechnol. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- 23.Gray N K, Quick S, Goossen B, Constable A, Hirling H, Kuhn L C, Hentze M W. Recombinant iron-regulatory factor functions as an iron-responsive-element-binding protein, a translational repressor and an aconitase. Eur J Biochem. 1993;218:657–667. doi: 10.1111/j.1432-1033.1993.tb18420.x. [DOI] [PubMed] [Google Scholar]
- 24.Hagan K W, Ruiz-Echevarria M J, Quan Y, Peltz S W. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol Cell Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagedorn C H, Spivak-Kroizman T, Friedland D E, Goss D J, Xie Y. Expression of functional eIF-4Ehuman: purification, detailed characterization, and its use in isolating eIF-4E binding proteins. Protein Expr Purif. 1997;9:53–60. doi: 10.1006/prep.1996.0661. [DOI] [PubMed] [Google Scholar]
- 26.Haghighat A, Sonenberg N. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J Biol Chem. 1997;272:21677–21680. doi: 10.1074/jbc.272.35.21677. [DOI] [PubMed] [Google Scholar]
- 27.He F, Brown A H, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hellerbrand C, Jobin C, Iimuro Y, Licato L L, Sartor R B, Brenner D A. Inhibition of NF kappa B in activated rat hepatic stellate cells by proteasome inhibitors and an I kappa B super-repressor. Hepatology. 1998;27:1285–1295. doi: 10.1002/hep.510270514. [DOI] [PubMed] [Google Scholar]
- 29.Hendriks H F J, Verhoofstad W A M M, Brouwer A, DeLeeuw A M, Knook D L. Perisinoidal fat-storing cells are the main vitamin A storage sites in rat liver. Exp Cell Res. 1985;160:138–149. doi: 10.1016/0014-4827(85)90243-5. [DOI] [PubMed] [Google Scholar]
- 30.Hensold J O, Stratton C A, Barth D. The conserved 5′-untranslated leader of Spi-1 (PU.1) mRNA is highly structured and potently inhibits translation in vitro but not in vivo. Nucleic Acids Res. 1995;25:2869–2876. doi: 10.1093/nar/25.14.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrick D J, Ross J. The half-life of c-myc mRNA in growing and serum-stimulated cells: influence of the coding and 3′ untranslated regions and role of ribosome translocation. Mol Cell Biol. 1994;14:2119–2128. doi: 10.1128/mcb.14.3.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holcik M, Liebhaber S A. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc Natl Acad Sci USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson R J, Wickens M. Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr Opin Genet Dev. 1997;7:233–241. doi: 10.1016/s0959-437x(97)80133-5. [DOI] [PubMed] [Google Scholar]
- 34.Kiledjian M, Wang X, Liebhaber S A. Identification of two KH domain proteins in the α-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 36.Kuivaniemei H, Tromp G, Chu M L, Prockop D J. Structure of a full-length cDNA clone for the prepro alpha 2(I) chain of human type I procollagen. Comparison with the chicken gene confirms unusual patterns of gene conservation. Biochem J. 1988;252:633–640. doi: 10.1042/bj2520633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maher J J, McGuire R F. Extracellular matrix gene expression increases preferentially in rat lipocytes and sinusoidal endothelial cells during hepatic fibrosis in vivo. J Clin Investig. 1990;86:1641–1648. doi: 10.1172/JCI114886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mak K M, Leo M A, Lieber C S. Alcoholic liver injury in baboon: transformation of lipocytes to transitional cells. Gastroenterology. 1984;87:188–200. [PubMed] [Google Scholar]
- 40.Milani S, Herbst H, Schuppan D, Hahn E G, Stein H. In situ hybridization for procollagen type I, III and IV mRNA in normal and fibrotic rat liver; evidence for predominant expression in nonparenchymal liver cells. Hepatology. 1989;10:84–92. doi: 10.1002/hep.1840100117. [DOI] [PubMed] [Google Scholar]
- 41.Milani S, Herbst H, Schuppan D, Kim K Y, Riecken E O, Stein H. Procollagen expression by nonparenchymal rat liver cells in experimental biliary fibrosis. Gastroenterology. 1990;98:175–184. doi: 10.1016/0016-5085(90)91307-r. [DOI] [PubMed] [Google Scholar]
- 42.Monson J M, Friedman J, McCarthy B J. DNA sequence analysis of a mouse pro-α1(I) procollagen gene: evidence for a mouse B1 element within the gene. Mol Cell Biol. 1982;2:1362–1371. doi: 10.1128/mcb.2.11.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muckenthaler M, Gunkel N, Stripecke R, Hentze M W. Regulated poly(A) tail shortening in somatic cells mediated by cap-proximal translational repressor proteins and ribosome association. RNA. 1997;3:983–995. [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips C L, Morgan A L, Lever L W, Wenstrup R J. Sequence analysis of a full-length cDNA for the murine pro alpha 2(I) collagen chain: comparison of the derived primary structure with human pro alpha 2(I) collagen. Genomics. 1992;13:1345–1346. doi: 10.1016/0888-7543(92)90065-z. [DOI] [PubMed] [Google Scholar]
- 45.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitt-Graff A, Chakroun G, Gabbiani G. Modulation of perisinusoidal cell cytoskeletal features during experimental hepatic fibrosis. Virchows Arch B Cell Pathol. 1993;422:99–107. doi: 10.1007/BF01607161. [DOI] [PubMed] [Google Scholar]
- 47.Schupan D. Structure of the extracellular matrix in normal and fibrotic liver: collagens and glycoproteins. Semin Liver Dis. 1990;10:1–10. doi: 10.1055/s-2008-1040452. [DOI] [PubMed] [Google Scholar]
- 48.Stefanovic B, Hackl W, Luhrmann R, Schumperli D. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stefanovic B, Hellerbrand C, Holcik M, Briendl M, Liebhaber S A, Brenner D A. Posttranscriptional regulation of collagen α1(I) mRNA in hepatic stellate cells. Mol Cell Biol. 1997;17:5201–5209. doi: 10.1128/mcb.17.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tranguch A, Kindelberger D, Rohlman C, Lee J-Y, Engelke D. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry. 1994;33:1778–1787. doi: 10.1021/bi00173a022. [DOI] [PubMed] [Google Scholar]
- 51.Vuorio E, de Crombrugghe B. The family of collagen genes. Annu Rev Biochem. 1990;59:837–872. doi: 10.1146/annurev.bi.59.070190.004201. [DOI] [PubMed] [Google Scholar]
- 52.Wang X, Kiledjian M, Weiss I M, Liebhaber S A. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability. Mol Cell Biol. 1995;15:1769–1777. doi: 10.1128/mcb.15.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss I M, Liebhaber S A. Erythroid cell-specific determinants of α-globin mRNA stability. Mol Cell Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood M, VanDongen H M A, VanDongen P E. The 5′-untranslated region of the N-methyl-D-aspartate receptor NR2A subunit controls efficiency of translation. J Biol Chem. 1996;271:8115–8120. doi: 10.1074/jbc.271.14.8115. [DOI] [PubMed] [Google Scholar]
- 55.Yamada Y, Mudryj M, de Crombrugghe B. A uniquely conserved regulatory signal is found around the translation initiation site in three different collagen genes. J Biol Chem. 1983;258:14914–14919. [PubMed] [Google Scholar]
- 56.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]